Abstract
Introduction
Breast cancer is one of the most common cancers in the world. Long noncoding RNA 00504 (LINC00504) was reported to be a functional gene in some tumours but not breast. Accordingly, the purpose of this article is to study the function of LINC00504 in breast cancer.
Methods
qPCR assay was used to detect the expression of LINC00504 in tissue and cell lines. The online database and chromatin immunoprecipitation assay (ChIP) were employed to confirm the transcription factor of LINC00504. Cell function assays including cell proliferation, migration and invasion were designed to detect the function of LINC00504 in vitro and in vivo. Luciferase reporter assay and RNA immunoprecipitation (RIP) assay were used to confirm the relationship between LINC00504 and miR‐140-5p. And Western blot assay was employed for testing the key protein.
Results
We found that LINC00504 is upregulated in breast cancer. In addition, we found that the transcription factor regulatory factor X5 (RFX5) can strongly bind to the LINC00504 promoter region and subsequently increase its transcriptional activity. We also found that the manipulation of RFX5 expression can significantly affect LINC00504 expression, which suggested that RFX5 can transcriptionally activate LINC00504 in breast cancer (BC). Knockdown of LINC00504 inhibits cell proliferation, migration and invasion in vitro and in vivo. We further found that LINCOO504 inhibits miR‐140-5p, which decreases the levels of VEGFA. The further results showed that miR-140-5p was one of the target gene of LINC00504. The WB assay demonstrated that the E-cadherin was increased and Vimentin was decreased when knocking down of LINC00504 and they can be rescued while adding the inhibitors of miR-140-5p.
Discussion
Our results demonstrated the mechanism by which the LINC00504–miR‐140-5p–VEGFA axis participates in breast cancer cell proliferation and invasion and may lead to new lncRNA-based diagnostic or therapeutic strategies for breast cancer.
Keywords: breast cancer, LINC00504, proliferation, invasion, miR-140-5p
Introduction
Breast cancer is the most common type of gynaecological tumour in China.1 Despite efforts focused on diagnostic techniques and patient management, there has been little progress in improving the overall survival of breast carcinoma patients. In addition, it has been reported that there is a clearly increasing trend in the incidence and mortality rates of breast cancer.2,3 The development of suitable therapies to increase patient survival rates has been limited because the pathophysiological mechanisms that contribute to breast carcinoma are largely unknown.4 Therefore, revealing the molecular mechanisms underlying the development and progression of breast carcinoma is necessary for developing effective therapies.
Long noncoding RNAs (lncRNAs) are a new category of noncoding RNAs that contain over 200 nucleotides and lack protein coding ability.5–7 Increasing evidence has shown that lncRNAs play important roles in various human cancers, including breast cancer,8 liver cancer,9 and gastric cancer.10 LncRNAs have been associated every stage of cell life, including cell proliferation, differentiation, apoptosis, and motility. For instance, knockdown of the long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer.11 Therefore, identifying the mechanism by which LncRNAs are regulated is essential for tumour diagnosis and therapy. A long noncoding RNA, called LINC00504, is a newly identified lncRNA. Feng J et al first reported that the noncoding RNA LINC00504 interacts with c-Myc to regulate tumour metabolism in colon cancer.12 However, the expression of LINC00504 and its biological effects in breast cancer have not been reported.
In this study, we demonstrated that LINC00504 is upregulated in human breast cancer tissues and cell lines. In addition, we revealed that LINC00504 performs its oncogenic function by regulating the miR-140-5p–VEGFR pathway during breast cancer development. These results might provide new insight into the treatment of breast cancer.
Materials and Methods
Tissue Specimens
Twenty-eight breast cancer (BC) tissue specimens and matched adjacent normal tissues were collected from 6 stage 1–2 patients and 22 stage 3–4 patients at Guangdong Provincial People’s Hospital. All the histological diagnoses of the BC and normal tissues were independently reviewed and confirmed by two pathologists. All the tissues were immediately frozen in liquid nitrogen and stored at −80°C before use. Before collection, written informed consent was obtained from each patient, and the study protocol was approved by the Ethics Committee of Guangdong Provincial People’s Hospital Institutional Review Board (Certificate Number: NO. GDREC2018218H(R1)). And this protocol also was conducted in accordance with the Declaration of Helsinki.
Cell Culture
The human normal breast cell line MCF-10A was purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). The human breast cancer cell lines BT549, T47D, MCF-7, SKBR3 and MDA-MB-231 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All the cells, except the MCF10A cells, were cultured with 10% foetal bovine serum (FBS; Gibco-BRL/Life Technologies, Paisley, UK) in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; HyClone Laboratories, Inc., Logan, UT, USA) supplemented with antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin; Gibco-BRL, Grand Island, NY, USA) at 37°C in 5% CO2 in a humidified incubator. The MCF10A cells were maintained in complete media [DMEM/F12 (50:50 mix) supplemented with 5% horse serum, 10 mg/mL 10 mM HEPES, insulin, 20 ng/mL epidermal growth factor, 0.5 mg/mL hydrocortisone and 100 ng/mL cholera toxin].
QRT-PCR Analysis
Total RNA was extracted from the tissues and cell lines with TRIzol reagent (15596026, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. First-stand cDNA was generated using the PrimeScript RT Reagent Kit (RR037A, TaKaRa, Dalian, China). SYBR Premix Ex Taq (RR420A, TaKaRa, Dalian, China) was used to detect LINC00504 and miR-140-5p expression. PCR was carried out at least in triplicate, and the results were analysed on an ABI 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA). The expression of LINC00504 and miR-140-5p was normalized to the expression of β-actin and U6, and the relative expression levels were calculated using the 2−ΔΔCT method. The gene-specific primers for LINC00504, miR-140-5p and their endogenous controls were designed and synthesized, as previously described.13,14 The primers for RFX5 were RFX5-F: 5`-GATGAGCCTGATGCTAAGAGC-3` and RFX5-R: 5`-CCCTCTACTTTGTTCTGCACG-3`.
Chromatin Immunoprecipitation Assay (ChIP)
The ChIP assay was performed to show that RFX5 directly interacted with the RFX5 binding sites in the LINC00504 promoter, and the EZ-Zyme Chromatin Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was used according to the manufacturer`s instructions. Briefly, MDA-MB-231 and SKBR3 cells were crosslinked using formaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 10 min. Subsequently, chromatin was fragmented into 100–300-bp pieces by using the sonication method, and 1% of the supernatant was preserved at −80°C as an input control. The remaining DNA fragments were immunoprecipitated with RFX5-specific antibodies (PA1-32133, Thermo Fisher Scientific) or IgG (2729S, CST) as the control. Finally, after purification, the DNA fragments were subjected to ChIP-PCR analysis with the following primers covering the LINC00504 promoter sequences: ChIP-PCR-F: 5`-AACATTCTCTCCACATGGGC-3`, and ChIP-PCR-R: 5`-TCAGTTCAGGCACGTAATGC-3`.
Plasmid, Transfection and Lentivirus Transduction
The RFX5 ORF region was amplified from MDA-MB-231 cDNA with the following primers: RFX5-ORF-F: 5ʹ-ATGGCAGAAGATGAGCCTGATG-3ʹ and RFX5-ORF-R: 5ʹ-TTATGGGGGTGTTGCTTTTGGG-3ʹ. The RFX5-ORF was subcloned into the pcDNA 3.1 vector, and all of the recombinant plasmids were verified by sequencing. Oligonucleotide transfection was performed with Lipofectamine 2000 reagent (11668019, Invitrogen, Carlsbad, CA, USA). The modified miR-140-5p mimics (5ʹ-CAGUGGUUUUACCCUAUGGUAG-3ʹ), microRNA negative control (NC) (5ʹ-UUGUACUACACAAAAGUACUG-3ʹ), miRNA inhibitors (miR-140-5p (in), 5ʹ-CUACCAUAGGGUAAAACCACUG-3ʹ), miRNA inhibitor NC (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) and si-RFX5 (#1: 5ʹ-CCTGCAAGATGTACAGAAA-3ʹ, #2: 5ʹ-CCTTATCCCAGGAGCATAA-3ʹ) were synthesized by Genepharma (Shanghai, China). The final transfection concentration of each mimic was 10 nM. The DNA encoding short hairpin RNA (shRNA) specifically targeting LINC00504 (sh-LINC00504 #1 and #2) was designed by Takara (Dalian, China) and cloned into the lentiviral vector packaging system, as previously described.12
Cell Cycle Analysis
Sh-LINC00504 and its control (sh-NC) were transfected into SKBR3 and MDA-BM-231 cells for 48 h. Then, the cells were lysed in precooled ethanol (75%) and incubated at 4°C for 4 h. Next, cold PBS was used to wash the cells, followed by staining with BD PharmingenTM PI/RNase for 30 mins at room temperature. Finally, the cells in different cell cycle phases (G0/G1, S, and G2) were analysed by flow cytometry. A total of 10,000 cells were measured for each sample.
Cell Proliferation Analysis
Cell proliferation was detected using the Cell Counting Kit-8 (CCK-8, CK04; Dojindo, Tokyo, Japan) assay. A total of 1000 cells transfected with shRNA (shLINC00504#1 and #2) or control were seeded in 96-well plates and incubated at 37°C with 5% CO2. At 0, 24, 48, 72 and 96 h, CCK‐8 assay solution (10 μL) was added to each well. The absorbance at 450 nm was measured with an enzyme immunoassay analyser (Thermo Fisher Scientific, Shanghai, China).
Colony Formation Assays
For colony formation assays, the cells were seeded into 12-well plates (5x102 cells/well) and cultured for 7–10 days. Subsequently, the colonies were fixed with 4% paraformaldehyde (PFA, P0099, Beyotime Institute of Biotechnology, Shanghai, China) for 10 min at room temperature, stained with 0.1% crystal violet (332488, Sigma-Aldrich; Merck KGaA) at room temperature for 10 min, and washed with water 3 times, and then, the number of colonies (>50 cells) was manually counted.
Wound Healing Assay
After transfection, the cells (1x10^5 cells/well) were seeded into 6-well plates, and before starting the assay, cells were starved in foetal bovine serum (FBS)-free culture medium overnight. Then, a wound was made using a 200-µL pipette tip. Next, the cells were incubated with 2% FBS medium. The wound was imaged at 0 h and 36 h.
Transwell Invasion Assay
A Transwell chamber (8‐μm pore size, Corning, Cambridge, MA, USA) was used to perform the cell invasion assays. Cells transfected with sh-LINC00504, sh-control or cotransfected sh-LINC00504 and miR-140-5p were resuspended in 200 μL of serum‐free medium and adjusted to a density of 1 ×10^6 cells/mL. Then, the cells (2x10^5/well) were cultured in the upper chamber with Matrigel (BD Biosciences), and complete medium containing 20% FBS was added to the lower chamber. After incubation for 36 h at 37°C, the cells adhering to the lower surface of the Transwell membrane were fixed in 20% methanol and stained with 0.1% crystal violet. Then, cell counting was performed under an inverted phase‐contrast microscope (Olympus) according to observation of the cells in 5 random fields. In addition, the number of invaded cells was analysed.
Immunofluorescence
After transfection, SKBR3 and MDA-MB-231 cells were cultured on glass slides and fixed with 4% paraformaldehyde for 15 min. After being washed with phosphate-buffered saline (PBS) three times, the cells were incubated with blocking buffer (PBS solution containing 3% foetal bovine serum (FBS), 1% goat serum, and 0.1% Triton X-100) for 2 h at room temperature. Then, the cells were incubated with the primary antibodies anti-VIM (ab8978, Abcam) and anti-CDH1 (ab76055, Abcam) diluted in PBS at 4°C overnight. After another round of washing with PBS, the slides were incubated with anti-mouse IgG(H+L), F(ab`)2 Fragment (Alexa Fluor® 488 Conjugate) (#4408, CST) for 1 h in the dark. Then, the slides that underwent the third round of washing were mounted with Pro-Long® Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole (DAPI: Molecular Probes, Eugene, OR, USA). The slides were observed by an LSM 800 confocal microscope (Carl Zeiss AG, Oberkochen, Germany).
Xenograft Mouse Model
A lentivirus-based system was used to specifically downregulate LINC00504; briefly, recombinant lentiviral plasmids carrying sh-LINC00504 (Plko.1 lentiviral vector) were successfully established and used to transfect SKBR3 cells. Then, SKBR3 cells (1 × 107) were subcutaneously (SC) injected into 5-week-old athymic nude mice (Bikai, Shanghai, China). The tumour size was measured every 6 days, and the tumour volume was calculated as 0.5 × L× W2, with L representing the length and w representing the width. The mice were euthanized 30 days after injection, and the tumours were weighed after sampling. All the animal related procedures were approved by the Animal Care and Use Committee of Guangdong Provincial People’s Hospital Research Ethics Committee. And this protocol also was conducted in accordance with the Rules of 3Rs, which refers to replacing and reducing animals in experiments, and refining procedures to make them less harmful.
Luciferase Reporter Assay
Microcode bioinformatics tools (https://cm.jefferson.edu/rna22/Interactive/ and http://www.mircode.org/) were used to identify the potential binding sequences of miR-140-5p and LINC00504. SKBR3 and MDA-MB-231 cells were seeded in 24-well plates. The cells were cotransfected with miR-140-5p or microRNA NC and luciferase reporter constructs containing WT-LIC00504 or Mut-LINC00504, as previously described.13 The luciferase activity was detected using a Dual Luciferase Reporter Gene Assay Kit (RG027, Beyotime Institute of Biotechnology, Shanghai, China), according to the manufacturer’s protocol. Mutations in the putative binding sites of LINC00504 were made using a Quick-change site-directed mutagenesis kit (200518, Agilent Technologies, Santa Clara, USA). The primers were as follows: LINC00504, Forward primer: 5ʹ-GTGACTCGAGCTTGCCTCTGCCATGT-3ʹ, Reverse primer: 5ʹ-GTGAGCGGCCGCTTTCAGAGTGAAACAATACTT-3ʹ; Mut-LINC00504, Forward primer: 5`-GGATTACAGGCGTGAGGGTGAC-3`; and Reverse primer: 5`-GTCACCCTCACGCCTGTAATCC-3`.
RNA Immunoprecipitation (RIP) Assay
RIP was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation kit (17–700, Millipore, Bedford, MA), according to the manufacturer’s instructions. Briefly, cells were transfected with miR-140-5p mimics, microRNA NC mimics, and LINC00504 and then resuspended and lysed in lysis buffer. Simultaneously, protein A/G magnetic beads were vortexed and resuspended in 500 µL RIP wash buffer. Next, cell lysates were centrifuged at 4°C for 10 min, and the supernatant was added to the resuspended magnetic beads and incubated with anti-Ago2 (ab186733, Abcam) or anti-IgG (ab133470, Abcam) antibodies. The samples were mixed on a vertical mixer at 4°C for more than 3 h. The samples were briefly centrifuged to shake the sample to the bottom of the tube, and the precipitate was further digested with Proteinase K (Applied Biosystems) to extract the RNA. Finally, qRT-PCR assays were used to determine the relative enrichment of LINC00504 mRNA.
Western Blot
Cells were lysed with lysis buffer (RIPA) containing protease inhibitors. The total protein concentration was determined by the bicinchoninic acid (BCA) assay (P0012, Beyotime Institute of Biotechnology, Shanghai, China). The total protein (20 µg) was separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride (1620177, PVDF, Bio-Rad, Hercules, USA) membranes. After blocking with 5% skim milk for 60 min, the membranes were incubated with primary antibodies against VEGFA (ab1316, Abcam), E-cadherin (ab76055, Abcam), vimentin (ab8978, Abcam), and GAPDH (ab8245, Abcam) overnight followed by incubation with Alexa Fluor Plus 800 fluorescence-conjugated secondary antibodies (A32730, Thermo Fisher, USA) (1:10,000) for 30 min. The bands were detected by an infrared laser imaging system (Odyssey; Li-Cor, Lincoln, NE, USA).
Statistical Analysis
The statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL, USA). The data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Differences between two groups or more than two groups were evaluated by Student’s t-test or one‐way analysis of variance (ANOVA), respectively. Spearman rank-correlation was performed to calculate the correlation coefficient between the LINC00504 and miR-140-5p expression levels. **p < 0.01, *p < 0.05 was considered to indicate a statistically significant difference.
Results
LINC00504 is Upregulated in Breast Cancer Tissues and Cells
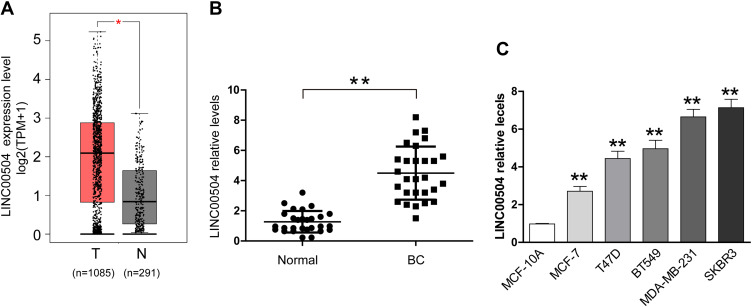
Bioinformatics analysis was used to identify the expression of LINC00504 in breast cancer patients and healthy subjects in The Cancer Genome Atlas (TCGA) database. We found a significant increase in the level of LINC00504 in breast cancer tissues compared to that in normal tissues (Figure 1A). Furthermore, real-time qPCR was used to investigate the expression of LINC00504 in breast cancer tissues and adjacent normal tissues. The results showed that LINC00504 was expressed at higher levels in tumour tissues than in adjacent normal tissues, which also confirmed the bioinformatics results (Figure 1B). We also measured LINC00504 expression in five human breast cancer cell lines (BT549, T47D, MCF-7, SKBR3 and MDA-MB-231) and in the normal breast cell line MCF-10A. LINC00504 expression was significantly higher in the five breast cancer cell lines than in the normal breast cell line (Figure 1C). We also analyse the relationship of LINC00504 and Ki67 (a key protein of proliferation) and the result showed that they were positively correlated (Supplementary Figure 1A). These results demonstrated that LINC00504 was upregulated in breast cancer. We then selected two cell lines with higher expression (MDA-MB-231 and SKBR3) for subsequent experiments.
Figure 1.
LINC00504 was upregulated in breast cancer tissues and cells. (A) The expression of LINC00504 in breast cancer patients and healthy subjects in The Cancer Genome Atlas (TCGA) database was assessed. (B) qRT-PCR assay was used to assess the expression of LINC00504 in 28 pairs of breast cancer tissues and adjacent noncancerous tissues. (C) Breast cancer cell lines (BT549, T47D, MCF-7, SKBR3 and MDA-MB-231) and the normal breast cell line MCF-10A. The data represent the mean ± standard deviation (SD). *P<0.05, **P<0.01.
The Transcription of LINC00504 Was Critically Activated by the Transcription Factor (TF) RFX5
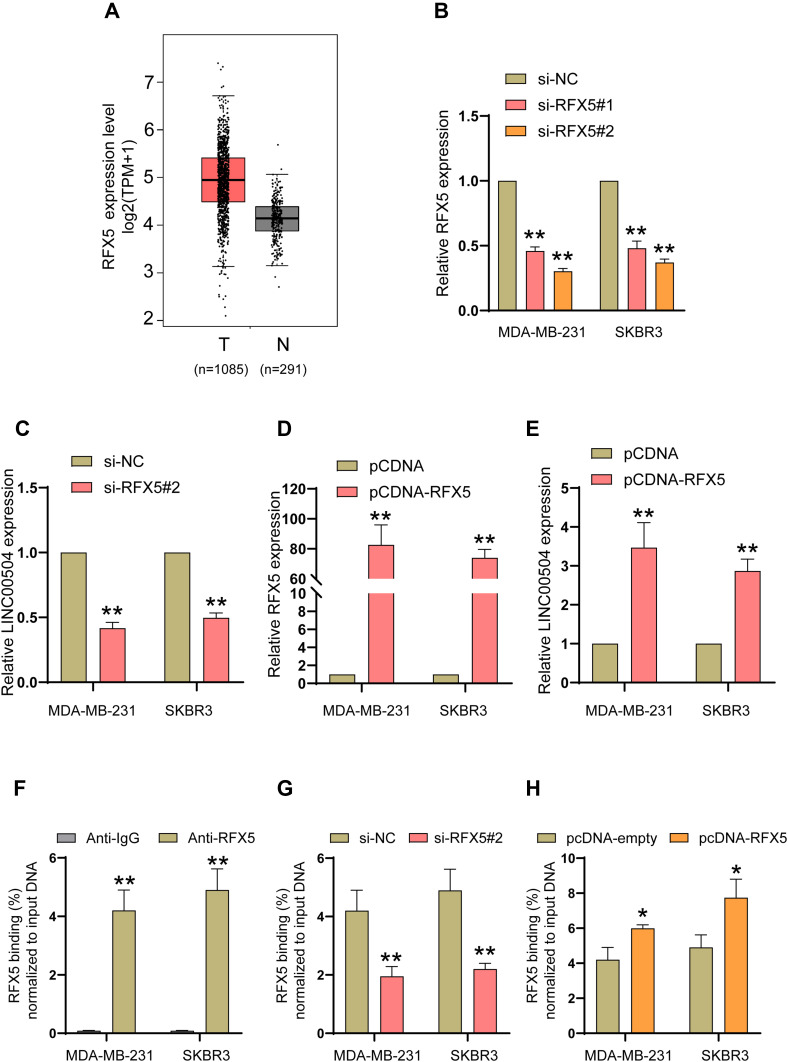
What causes the abnormal expression of LINC00504? Here, we attempted to determine the regulators involved in LINC00504 overexpression to reveal the upstream mechanism of LINC00504 in breast cancer. We first explored publicly available data from the UCSC (http://genome.ucsc.edu/) and JARSPAR (http://jaspar.genereg.net/) online databases to analyse the upstream sequence (2Kb) of LINC00504. And some important transcription factors such as EZH1, ZFHX3, RFX5 and EP300 can act as the candidate, combined with their expressions in BC, RFX5, which is upregulated in breast cancer (Figure 2A), caught our attention. To verify the upstream regulatory mechanism by which RFX5 regulates LINC00504, we first analyse the relationship of them in the online database, and we found that they were positively correlated (Supplementary Figure 1B), and then we used siRNAs against RFX5 to knockdown RFX5 in MDA-MB-231 and SKBR3 cells (Figure 2B), and qRT-PCR assays were performed to determine the LINC00504 level. The results showed that silencing RFX5 effectively impaired the expression level of LINC00504 (Figure 2C). In contrast, when we used pcDNA-RFX5 to overexpress RFX5 in MDA-MB-231 and SKBR3 cells (Figure 2D), the opposite effect on the LINC00504 levels was observed (Figure 2E). Furthermore, we conducted ChIP assays and showed that RFX5 directly interacted with the RFX5 binding sites within the LINC00504 promoter in MDA-MB-231 and SKBR3 cells (Figure 2F). Subsequently, we found that RFX5 enrichment on the LINC00504 promoter was significantly decreased or increased when RFX5 expression was decreased or increased, respectively (Figure 2G and H). Taken together, these results suggest that RFX5 directly activates the transcription of LINC00504 in BC.
Figure 2.
The key transcription factor RFX5 critically activated the transcription of LINC00504. (A) The expression of RFX5 in breast cancer patients and healthy subjects in The Cancer Genome Atlas (TCGA) database was assessed. (B and D) The expression level of RFX5 was detected in MDA-MB-231 and SKBR3 cells after transfection with RFX5 siRNAs or pcDNA-RFX5. (C and E) The effect of the altered expression level of RFX5 on the regulation of LINC00504. (F) ChIP-qPCR assays showing RFX5 binding to the LINC00504 promoter in BC cells. (G and H) ChIP-qPCR assays revealed the enrichment of RFX5 on the LINC00504 promoter in BC cells after transfection with RFX5 siRNA or pcDNA-RFX5. *P<0.05/**P<0.01.
Knockdown of LINC00504 Suppressed Breast Cancer Cell Proliferation, Migration and Invasion in vitro and in vivo
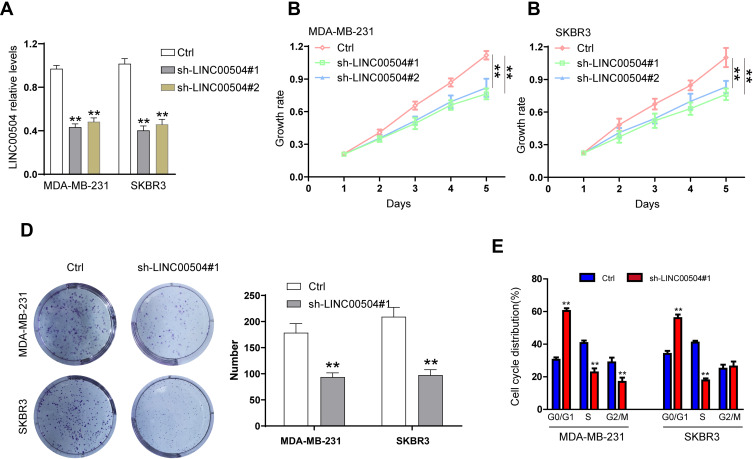
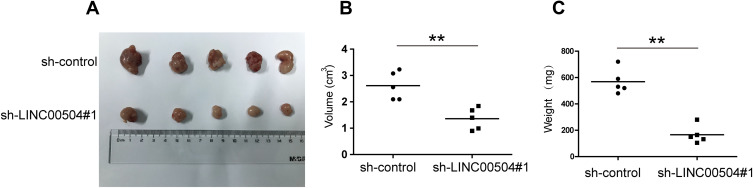
The physiological role of LINC00504 was explored using SKBR3 and MDA-MB-231 cells that were transfected with sh-control and sh-LINC00504 (#1 and #2). The significant decrease in LINC00504 expression in these sh-LINC00504-transfected cells was confirmed by qRT‐PCR (Figure 3A). Then, we performed CCK-8 assays to analyse the effect of the knockdown of LINC00504 on the proliferation of BC cells. We found that knockdown of LINC00504 inhibited the proliferation of the two cell lines compared to their corresponding controls (Figure 3B and C). Similarly, LINC00504 knockdown dramatically decreased the colony formation of SKBR3 and MDA-MB-231 cells (Figure 3D). Cell cycle analysis was used to explore the cell distribution at different cell phases. The results showed that knockdown of LINC00504 arrested SKBR3 and MDA-MB-231 cells at G0/G1 phase (Figure 3E). In xenograft mouse models, we found that LINC00504 knockdown significantly inhibited tumour growth (Figure 4A–C).
Figure 3.
The effect of LINC00504 knockdown on the proliferation and cell cycle distribution of breast cancer cells in vitro. (A) SKBR3 and MDA-MB-231 cells were transfected with sh-control or sh-LINC00504 (#1 and #2), and LINC00504 expression was detected by qRT-PCR after transfection of the two cell lines. (B–D) Transfected cell proliferation was detected by CCK-8 and colony formation assays. (E) Flow cytometry was used to analyse the distribution of the transfected cells in different cell cycle phases (G0/G1, S, and G2). **P<0.01.
Figure 4.
The effect of LINC00504 knockdown on proliferation in vivo. (A) Representative photos of xenografts. (B) The volumes in the subcutaneous xenografts were measured and calculated once a week for 5 weeks. (C) The tumour weight was measured at the end of the experiments. The data are presented as the mean ± SD, **p<0.01.
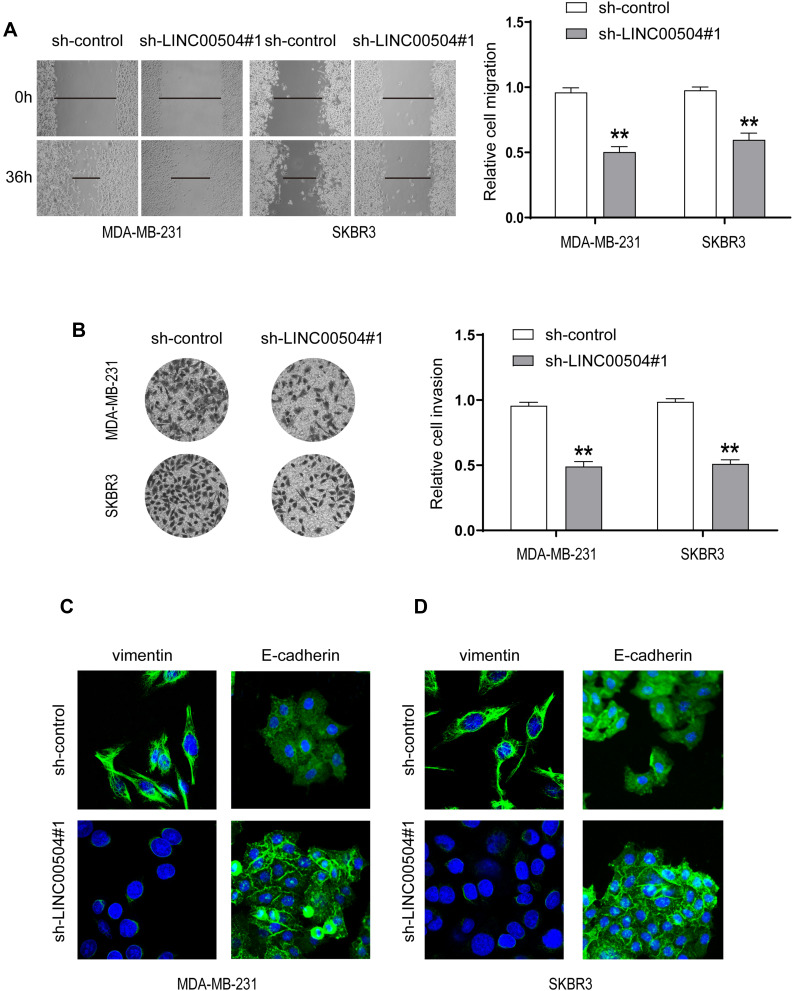
Next, we examined whether LINC00504 can influence breast cancer cell migration and invasion. The results of wound healing and Transwell assays showed that LINC00504 knockdown suppressed breast cancer cell migration and invasion (Figure 5A and B). Immunofluorescence was used to detect the changes in Vimentin and E-cadherin, which are characteristic proteins of epithelial–mesenchymal transition (EMT).15,16 The results demonstrated that knockdown of LINC00504 inhibited the expression of Vimentin and enhanced the expression of E-cadherin (Figure 5C and D), which also proved that LINC00504 boosts breast cancer cell migration and invasion. Collectively, LINC00504 knockdown impaired further progression of breast cancer cells.
Figure 5.
The effect of LINC00504 knockdown on the migration, invasion and EMT of breast cancer cells. (A and B) The migration and invasion capacities of transfected MDA-MB-231 and SKBR3 cells were assessed by wound healing (magnification, 100×) and Transwell assays (magnification, 200×). (C and D) Immunofluorescence was used to detect the changes in VIM and E-cadherin in the transfected cells (magnification for VIM, 630×, magnification for E-cadherin, 400×). **P<0.01.
MIR-140-5p Was a Target of LINC00504 in Breast Cancer
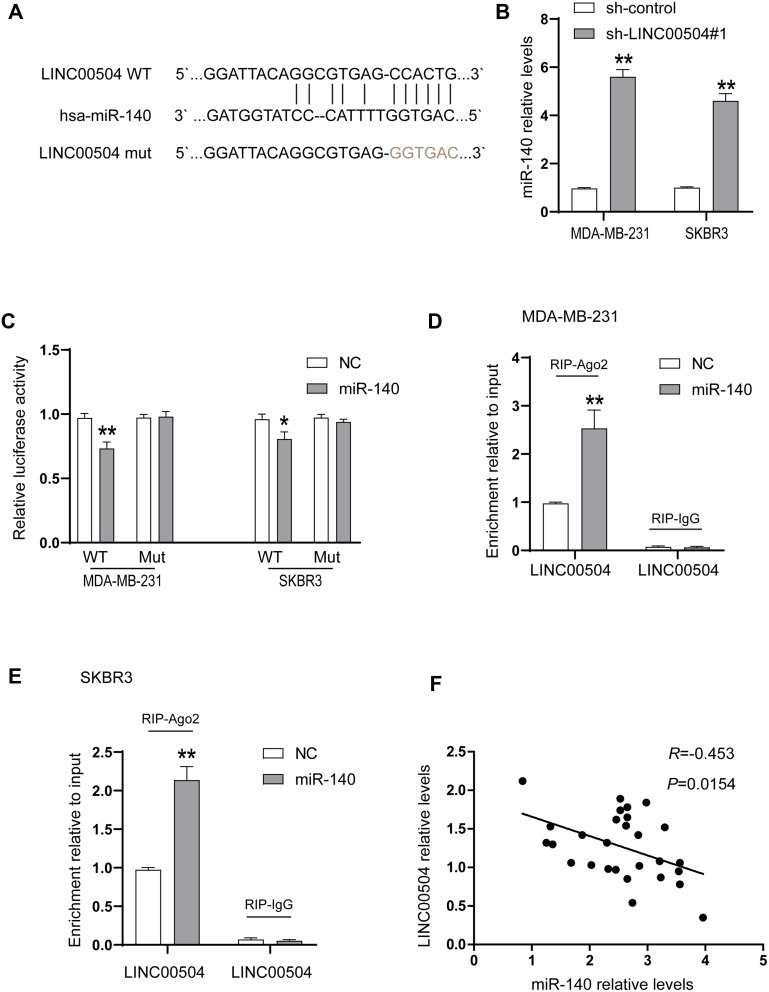
It has been reported that lncRNAs can act as Competing Endogenous RNAs (ceRNAs) to exert their regulatory functions. To further determine the underlying mechanism responsible for the function of LINC00504 in breast cancer, we examined a set of miRNAs that were predicted to interact with LINC00504 using the prediction software miRcode and RNA22. MIR-140-5p was found to be a promising target of LINC00504, and the predicted binding site of miR-140-5p in the LINC00504 sequence is shown in Figure 6A. After transfection with sh-LINC00504, qRT-PCR analysis revealed that miR-140-5p expression was significantly increased after LINC00504 was knocked down (Figure 6B). To further verify the interaction between miR-140-5p and LINC00504, luciferase reporter vectors were constructed that contained a wild‐type (wt) or mutated (mut) miR-140-5p‐binding site in LINC00504. The luciferase reporter assay results showed that miR-140-5p suppressed the luciferase activity of the LINC00504‐wt reporter vector but barely influenced that of the LINC00504‐mut reporter vector (Figure 6C). Furthermore, a RIP assay was used to examine the potentially endogenous interaction between LINC00504 and miR-140-5p. The data showed that LINC00504 was substantially enriched by miR-140-5p overexpression with anti-Ago2 in MDA-MB-231 and SKBR3 cells (Figure 6D and E). Using Spearman correlation analysis, we found that the levels of LINC00504 were reciprocally correlated with those of miR-140-5p in breast cancer tissue samples (Figure 6F). We also detected the expression level of miR-140-5p in BC cells after transfection with RFX5 siRNA, the results showed that the expression of miR-140-5p were upregulated in both MDA-MB-231 and SKBR3 cells (Supplementary Figure 1C). These data indicated that miR-140-5p is a direct target of LINC00504 in breast cancer.
Figure 6.
LINC00504 was associated with miR-140-5p in SKBR3 and MDA-MB-231 cells. (A) The potential binding sites between LINC00504 and miR-140-5p and the mutant in the seed region. (B) The expression of miR-140-5p was detected in sh-control- or sh-LINC00504-transfected cells. (C) Luciferase reporter assays were performed by transfecting LINC00504-WT or LINC00504-Mut constructs into SKBR3 and MDA-MB-231 cells with miR-NC mimics or miR-140-5p. (D and E) SKBR3 and MDA-MB-231 cells were transfected with miR-NC mimics or miR-140-5p mimics, followed by the assessment of LINC00504 mRNA enrichment with anti-Ago2 by qRT-PCR. Anti IgG served as the control. (F) Relationship between the levels of LINC00504 and miR-140-5p in breast cancer tissues. *P<0.05/**P<0.01.
LINC00504 Promoted the Progression of Breast Cancer by the miR-140-5p-VEGFA Axis
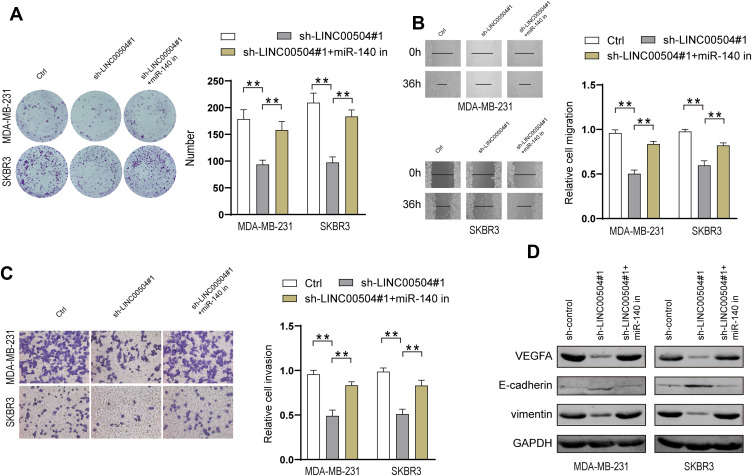
Next, we explored the effect of miR-140-5p on breast cancer progression, which was driven by LINC00504. We knocked down LINC00504 and inhibited miR-140-5p in SKBR3 and MDA-MB-231 cells. Colony formation assay, migration and invasion assays showed that LINC00504 knockdown significantly inhibited cell proliferation, migration and invasion, while miR-140-5p inhibition abrogated these effects (Figure 7A–C); these results demonstrated that miR-140-5p plays a key role in the LINC00504-related oncogenic effects on breast cancer cells. It has been reported that microRNA-140-5p inhibits invasion and angiogenesis by targeting VEGFA in breast cancer.17 Thus, we hypothesized that VEGFA is involved in the LINC00504/miR-140-5p‐dependent malignant progression of breast cancer cells. We simultaneously knocked down LINC00504 and inhibited miR-140-5p in SKBR3 and MDA-MB-231 cells. The expression of VEGFA, (E-Cadherin) and VIM was detected by Western blotting. The results showed that VEGFA and VIM were downregulated, while the expression of CDH1 was increased when LINC00504 was knocked down. However, the opposite expression of these proteins was observed when LINC00504 was knocked down and miR-140-5p was simultaneously inhibited (Figure 7D). These results suggested that LINC00504 induced tumour development via inhibition of miR-140-5p and by targeting VEGFA.
Figure 7.
miR-140-5p-VEGFA axis was regulated by LINC00504. (A) SKBR3 and MDA-MB-231 cells were transfected with sh-control, sh-LINC00504 or sh-LINC00504+miR-140-5p inhibitor, followed by colony formation assays to assess cell proliferation, (B) wound healing assays to assess cell migration (magnification, 100×) and (C) Transwell assays to assess invasion (magnification, 200×). (D) Western blot analysis of the VEGFA, CDH1, and VIM levels in MDA-MB-231 and SKBR3 cells. **P<0.01.
Discussion
Breast cancer is the most common and lethal malignant tumour in females worldwide.18 The incidence of breast cancer has increased rapidly in recent years.19 Early breast cancer often does not have typical symptoms and signs, and it is not easily detected. Breast cancer has often progressed to the middle and late stages at diagnosis.20 However, the underlying mechanism that regulates breast cancer development remains largely unknown. It is crucial to develop novel molecular biomarkers for the diagnosis and prognosis of breast cancer. Here, we found that LINC00504 was significantly upregulated in breast cancer. In addition, LINC00504 was critical for the proliferation, migration and invasion of breast cancer cells, which indicated that LINC00504 may be a new biomarker for breast cancer.
Regulatory factor X-5 (RFX5) belongs to the RFX family and is a transcription factor that encodes a DNA binding protein.21,22 In this study, our ChIP-qPCR assay data showed that RFX5 can directly bind to the LINC00504 promoter region. Moreover, we found that RFX5 enrichment on the LINC00504 promoter was significantly decreased or increased when RFX5 expression was decreased or increased, respectively. Notably, we provided the first evidence of the upstream mechanism of LINC00504 in BC and elucidated that the key transcription factor RFX5 plays a key role in the activation of LINC00504 transcription. LINC00504 may have more than one transcription factor, but RFX5 is the key transcription factor.
Emerging evidence has shown that dysregulation of lncRNAs is involved in tumorigenesis and progression of breast cancer,23–25 suggesting the possibility of lncRNAs serving as novel targets for breast cancer diagnosis and therapy. LINC00504 is a newly found lncRNA that is highly expressed in colon cancer. Feng J et al first reported that LINC00504 interacts with c‐Myc to regulate tumour metabolism in colon cancer.12 However, the detailed function and underlying mechanism of LINC00504 in breast cancer remain unclear. In this study, we showed that LINC00504 was highly expressed in breast cancer tissues and cell lines. By using in vitro and in vivo assays, we showed that LINC00504 knockdown remarkably inhibited cell proliferation. Similarly, a clear weakening trend of cell migration and invasion was observed with LINC00504 depletion in breast cancer. Moreover, we first demonstrated that LINC00504 knockdown resulted in decreased EMT in breast cancer cells. All these results suggested that LINC00504 might contribute to the metastasis of breast cancer.
Previous evidence has shown that lncRNAs can serve as competitive endogenous RNAs (ceRNAs) to sponge miRNAs.26–28 To further explore the underlying molecular mechanism by which LINC00504 regulates breast cancer, we predicted and found that miR-140-5p was a promising candidate. The gene encoding miRNA-140-5p is located on chromosome 16 and has been proven to function in several cancer cells.29–31 For instance, Yunfeng et al demonstrated that miR-140-5p could suppress the tumour growth and metastasis of non-small cell lung cancer by targeting IGF1R.32 In hypopharyngeal squamous cell carcinoma, miRNA-140-5p suppresses tumour cell migration and invasion by targeting the ADAM10-mediated Notch1 signalling pathway.33 In addition, miRNA-140-5p inhibits invasion and angiogenesis by targeting VEGF-A in breast cancer.34 The above studies indicate that miRNA-140-5p may be a tumour suppressor. To further investigate the correlation between LINC00504 and miRNA-140-5p in breast cancer tumorigenesis, we performed luciferase reporter assays. The results showed that LINC00504 directly combined with miRNA-140-5p in breast cancer cells. We also found that LINC00504 significantly inhibited miRNA-140-5p expression, enhanced VEGFA and VIM expression and reduced CDH1 levels. Taken together, our results indicated that the oncogene LINC00504 promoted breast cancer progression by negatively regulating miRNA-140-5p, a tumour suppressor, by targeting VEGFA. LINC00504/miRNA-140-5p may act as a novel therapeutic target for the treatment of breast cancer.
Funding Statement
This study was supported by the Guangzhou Science and Technology Program (grant No. 201803010094), the Natural Science Foundation of Guangdong (grant No. 2020A151501158), and the Medical Scientific Research Foundation of Guangdong Province of China (grant No. A2019477).
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study protocol was approved by the Ethics Committee of Guangdong Provincial People’s Hospital Institutional Review Board (Certificate Number: NO.GDREC2018218H(R1)), and all patients signed informed consent forms. And this protocol was also conducted in accordance with the Declaration of Helsinki.
The animal study protocol was approved by GuangdongProvincial People’s Hospital Research Ethics Community (Certificate Number: No. GDREC2018218A). And this protocol also was conducted in accordance with the Rules of 3Rs, which refers to replacing and reducing animals in experiments, and refining procedures to make them less harmful.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors reported no conflicts of interest for this work.
References
- 1.Li T, Mello-Thoms C, Brennan PC. Descriptive epidemiology of breast cancer in China: incidence, mortality, survival and prevalence. Breast Cancer Res Treat. 2016;159(3):395–406. doi: 10.1007/s10549-016-3947-0 [DOI] [PubMed] [Google Scholar]
- 2.Libson S, Lippman M. A review of clinical aspects of breast cancer. Int Rev Psychiatry. 2014;26(1):4–15. doi: 10.3109/09540261.2013.852971 [DOI] [PubMed] [Google Scholar]
- 3.Zuo TT, Zheng RS, Zeng HM, Zhang SW, Chen WQ. Female breast cancer incidence and mortality in China, 2013. Thoracic Cancer. 2017;8(3):214–218. doi: 10.1111/1759-7714.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadoo KA, Traina TA, King TA. Advances in molecular and clinical subtyping of breast cancer and their implications for therapy. Surg Oncol Clin N Am. 2013;22(4):823–840. doi: 10.1016/j.soc.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 5.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. [DOI] [PubMed] [Google Scholar]
- 6.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Gao S, Li H, Lv M, Lu C. Long noncoding RNAs (lncRNAs) in triple negative breast cancer. J Cell Physiol. 2017;232(12):3226–3233. doi: 10.1002/jcp.25830 [DOI] [PubMed] [Google Scholar]
- 9.Birgani MT, Hajjari M, Shahrisa A, et al. Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol Oncol Res. 2018;24(2):329–337. doi: 10.1007/s12253-017-0241-3 [DOI] [PubMed] [Google Scholar]
- 10.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. doi: 10.18632/oncotarget.6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res. 2016;23:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Ma J, Liu S, Wang J, Chen Y. A noncoding RNA LINC00504 interacts with c-Myc to regulate tumor metabolism in colon cancer. J Cell Biochem. 2019;120(9):14725–14734. doi: 10.1002/jcb.28733 [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, He X, Chen Y, Cao D. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol Cell Biochem. 2020;464(1–2):39–50. doi: 10.1007/s11010-019-03647-z [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123–1133. doi: 10.1159/000430237 [DOI] [PubMed] [Google Scholar]
- 15.Wu S, Du Y, Beckford J, Alachkar H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J Transl Med. 2018;16(1):170. doi: 10.1186/s12967-018-1539-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma F, Li W, Liu C, et al. MiR-23a promotes TGF-β1-induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/β-catenin signaling. Oncotarget. 2017;8(41):69538–69550. doi: 10.18632/oncotarget.18422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Qin T, Li J, et al. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24(9):386–392. doi: 10.1038/cgt.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia. 2017;59(5):368–379. doi: 10.1016/j.rx.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 19.Paluch-Shimon S, Warner E. Breast cancer in young women: challenges, progress, and barriers. Curr Opin Support Palliat Care. 2015;9(3):268–270. doi: 10.1097/SPC.0000000000000152 [DOI] [PubMed] [Google Scholar]
- 20.Jin L, Han B, Siegel E, Cui Y, Giuliano A, Cui X. Breast cancer lung metastasis: molecular biology and therapeutic implications. Cancer Biol Ther. 2018;19(10):858–868. doi: 10.1080/15384047.2018.1456599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24(5):803–807. doi: 10.1093/nar/24.5.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aftab S, Semenec L, Chu J, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8(1):226. doi: 10.1186/1471-2148-8-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. doi: 10.1016/j.bbrc.2017.02.094 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Zhou J, Wang Z, Wang P, Li S. Upregulation of SOX2 activated LncRNA PVT1 expression promotes breast cancer cell growth and invasion. Biochem Biophys Res Commun. 2017;493(1):429–436. doi: 10.1016/j.bbrc.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 25.Tracy KM, Tye CE, Ghule PN, et al. Mitotically-associated lncRNA (MANCR) affects genomic stability and cell division in aggressive breast cancer. Mol Cancer Res. 2018;16(4):587–598. doi: 10.1158/1541-7786.MCR-17-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1887–1899. doi: 10.1016/j.bbamcr.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 27.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–157. doi: 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130(18):1965–1975. doi: 10.1182/blood-2017-06-788695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, He F, OuYang S, et al. miR-140-5p could suppress tumor proliferation and progression by targeting TGFBRI/SMAD2/3 and IGF-1R/AKT signaling pathways in Wilms’ tumor. BMC Cancer. 2019;19:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flamini V, Dudley E, Jiang WG, Cui Y. Distinct mechanisms by which two forms of miR-140 suppress the malignant properties of lung cancer cells. Oncotarget. 2018;9(92):36474–36491. doi: 10.18632/oncotarget.26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfson B, Eades G, Zhou Q. Roles of microRNA-140 in stem cell-associated early stage breast cancer. World J Stem Cells. 2014;6(5):591–597. doi: 10.4252/wjsc.v6.i5.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yunfeng Y, Shen Y, Xue L, Fan H, Filleur S. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8(9):e73604. doi: 10.1371/journal.pone.0073604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing P, Sa N, Liu X, Liu X, Xu W. MicroR-140-5p suppresses tumor cell migration and invasion by targeting ADAM10-mediated Notch1 signaling pathway in hypopharyngeal squamous cell carcinoma. Exp Mol Pathol. 2016;100(1):132–138. doi: 10.1016/j.yexmp.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 34.Varghese E, Liskova A, Kubatka P, Mathews Samuel S, Büsselberg D. Anti-angiogenic effects of phytochemicals on miRNA regulating breast cancer progression. Biomolecules. 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]