Abstract
The cytokinin (CK) phytohormones have long been known to activate cell proliferation in plants. However, how CKs regulate cell division and cell expansion remains unclear. Here, we reveal that a basic helix–loop–helix transcription factor, CYTOKININ-RESPONSIVE GROWTH REGULATOR (CKG), mediates CK-dependent regulation of cell expansion and cell cycle progression in Arabidopsis thaliana. The overexpression of CKG increased cell size in a ploidy-independent manner and promoted entry into the S phase of the cell cycle, especially at the seedling stage. Furthermore, CKG enhanced organ growth in a pleiotropic fashion, from embryogenesis to reproductive stages, particularly of cotyledons. In contrast, ckg loss-of-function mutants exhibited smaller cotyledons. CKG mainly regulates the expression of genes involved in the regulation of the cell cycle including WEE1. We propose that CKG provides a regulatory module that connects cell cycle progression and organ growth to CK responses.
Cytokinin-responsive transcription factor controls cell cycle and cell expansion, leading to organ growth in Arabidopsis.
Introduction
Cytokinins (CKs) are a class of phytohormones that regulate numerous growth and development processes such as cell proliferation and fate determination, vasculature development, sink/source relationship, leaf senescence, stress, and defense responses, as well as responses to environmental cues (Roitsch and Ehneß, 2000; Mähönen et al., 2006; Hejatko et al., 2009; Choi et al., 2010; Nishiyama et al., 2011; Raines et al., 2016; Arnaud et al., 2017; Poitout et al., 2018; reviewed in Hwang et al., 2012; Kieber and Schaller, 2018; Cortleven et al., 2019; Wybouw and De Rybel, 2019).
CKs are perceived by a series of two-component histidine kinase receptors located at the plasma membrane or the endoplasmic reticulum (ER): ARABIDOPSIS HISTIDINE KINASE (AHK) AHK2, AHK3, and AHK4, also known as CYTOKININ RESPONSE1 (CRE1) and WOODENLEG (WOL). Activated receptors then phosphorylate ARABIDOPSIS PHOSPHOTRANSFER PROTEINs, which act as shuttles between the plasma membrane or the ER and the nucleus to phosphorylate type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs; reviewed in Hwang et al., 2012; Kieber and Schaller, 2014). Type-B ARRs target various CK-responsive genes, including the negative CK signaling regulators type-A ARRs in a feedback loop (Hwang and Sheen, 2001; Zubo et al., 2017). In addition, CYTOKININ RESPONSE FACTORs, which belong to the ETHYLENE RESPONSE FACTOR/APETALA2 (AP2)-type transcription factors, are involved in CK signaling (Rashotte et al., 2006; Kim, 2016).
CKs also play a critical role in the regulation of stem cell niches in primary meristems. For example, the shoot apical meristem (SAM) of the ahk2 ahk3 ahk4 triple mutant is much smaller than that of the wild-type (Higuchi et al., 2004; Nishimura et al., 2004). Conversely, the overexpression of a constitutively active form of the type-A ARR7 severely affects SAM formation and subsequent shoot proliferation (Leibfried et al., 2005; Bartrina et al., 2011). Likewise, loss of the CYTOKININ OXIDASE-DEHYDROGENASE (CKX) CK-degrading enzymes increases SAM size in Arabidopsis thaliana (Bartrina et al., 2011), whereas a mutation of the CK biosynthetic gene LONELY GUY4 (LOG4) reduces SAM size and perturbs meristem functions in rice (Oryza sativa; Kurakawa et al., 2007).
In contrast to the positive role of CKs in SAM cell proliferation, the size of the root apical meristem (RAM) increases in the ipt3 ipt5 ipt7 triple mutant for the CK biosynthesis genes ISOPENTENYL TRANSFERASEs (IPTs), as well as in the ahk3 single mutant and a double mutant of type-B ARR1 and ARR12, due to a reduced differentiation rate of meristematic cells (Dello Ioio et al., 2007). In addition, CKs promote the transition from cell division to cell expansion in the RAM by initiating an early onset of endoreduplication via ARR2 (Takahashi et al., 2013). CKs also induce the expression of the cell cycle regulator CYCLIN D3 (CYCD3), which controls the transition from the G1 to the S phase of the cell cycle in the SAM. In addition, the CK biosynthesis inhibitor lovastatin blocks the G2/M phase transition in tobacco (Nicotiana tabacum) cell cultures (Redig et al., 1996; Riou-Khamlichi et al., 1999; Dewitte et al., 2007). It is clear that CK-mediated plant growth and development is tightly associated with the cell-cycle progression machinery, but how they are connected is not fully understood. The opposite mode of action exhibited by CKs between the SAM and RAM suggests that CKs regulate the cell cycle in a cell-type specific manner through different downstream targets or via additional but yet-unknown specific modulators of CK signaling.
In this study, we reveal that CYTOKININ-RESPONSIVE GROWTH REGULATOR (CKG), previously named basic helix–loop–helix (bHLH) transcription factor bHLH137 (Toledo-Ortiz et al., 2003), directly targets cell-cycle regulators and mediates organ growth in a CK-dependent manner in Arabidopsis. We propose that the CK–CKG module provides an important regulatory node for the cell cycle during plant development.
Results
CKG is a novel regulator of CK responses
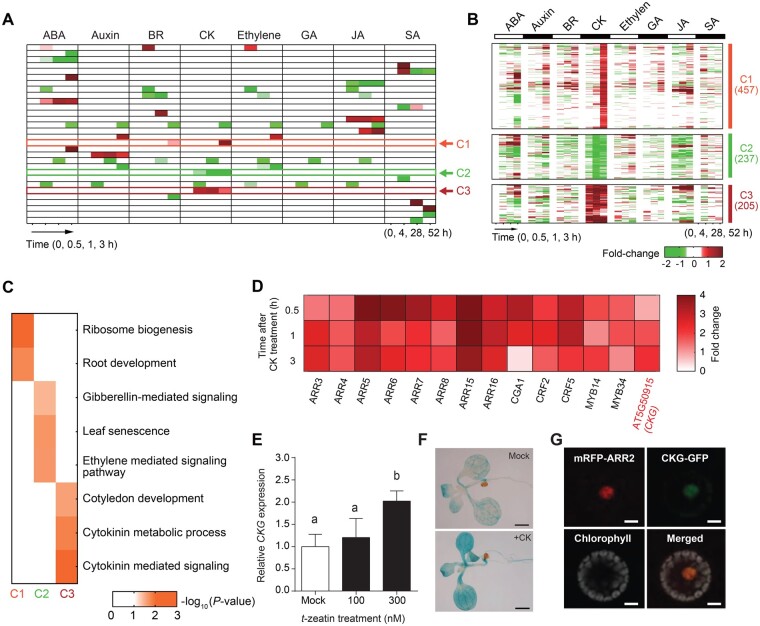
To identify novel transcription factors that specifically respond to CKs, we first analyzed transcriptome data from a time-course data set that tested the response of Arabidopsis seedlings to the phytohormones abscisic acid, auxin, brassinosteroids, CKs, ethylene, gibberellins, jasmonic acid, and salicylic acid (GSE39384, E-TABM-51), using the systematic analytical tool iNID (Choi et al., 2014). Using non-negative matrix factorization clustering, we classified phytohormone responses into 30 differential expression patterns (DEPs) that described either phytohormone-specific regulation or complex regulation of multiple phytohormones on gene expression (Figure 1A). Among them, three DEPs (arrows in Figure 1A) were associated with CK-specific regulation and designated as clusters C1, C2, and C3 (Figure 1B), containing 457, 237, and 205 genes, respectively. Arabidopsis seedlings were treated with CK for 30 min, 1 h, and 3 h in the original data set, and we observed that each cluster followed a distinct CK response profile. The C1 cluster included genes that were upregulated in response to CKs only at the 3-h time point. In contrast, the C3 cluster consisted of genes induced as early as 30 min after CK treatment and remained highly expressed thereafter. Genes that belonged to the C2 cluster exhibited the opposite expression pattern from those in C3.
Figure 1.
Transcriptome-wide discovery of potential CK signaling regulators. A, Differential gene expression patterns in hormone responses. Red and green colors indicate up- and downregulated gene expression, respectively, in time points under different hormone-treated conditions. ABA, abscisic acid; BR, brassinosteroid; GA, gibberellin; JA, jasmonic acid; SA, salicylic acid. B, The three clusters of gene expression patterns associated with CK response. Red and green indicate up- and downregulation in time points, respectively, under CK-treated conditions. Color bar, the gradient of log2(fold changes). C, Gene ontology biological processes represented by the genes in each cluster. Color bar, the gradient of −log10(P-values) from Fisher’s exact test. D, Fourteen transcription factors in cluster 3 show the significant expression changes after CK treatment (P < 0.001). Color bar, the gradient of log2(fold changes). E, CKG expression was induced by CK in a dose-dependent manner. Seven-day-old seedlings were treated with or without CK for 30 min. The level of CKG expression in the absence of CK was set to 1, and its relative value is presented. Error bars, sd (n = 3). F, CK induced the expression of pCKG:GUS in seedlings. CK treatment, 300-nM t-zeatin for 1 h. Scale bar, 1 mm. G, CKG localizes in the nucleus. 35S:CKG-GFP was co-transfected with 35S:mRFP-ARR2 as a nucleus marker. Scale bar, 10 μm. Different letters indicate significant differences at P < 0.05 based on Tukey’s honestly significant difference (HSD) test.
To determine whether these three clusters were associated with biological processes related to CKs, we investigated potential enrichment in gene ontology (GO) biological processes represented by their constituent genes (Figure 1C;P < 0.05, Supplemental Figure S1). The C3 cluster included genes involved in “cytokinin metabolic process” and “cytokinin-mediated signaling”. The C1 cluster was associated with “ribosome biogenesis” and “root development”, while genes from the C2 cluster returned the GO biological processes “gibberellin-mediated signaling”, “leaf senescence”, and “ethylene-mediated signaling pathway”. These results suggest that the C3 cluster is tightly connected to CKs, while the other two clusters are not directly linked to CK-mediated responses.
Therefore, we further characterized the genes belonging to cluster C3 to identify new regulators of CK responses: we selected a set of transcription factor-encoding genes that (1) were co-expressed with primary CK-responsive ARRs (ARR3/4/5/6/7/8/15/16) and (2) that showed significant expression changes after CK treatment (P < 0.001, Figure 1D). Among them, CYTOKININ-RESPONSIVE GATA FACTOR1 (CGA1) and cytokinin response factor2/5 have been previously characterized in the context of CK responses (Rashotte et al., 2006; Kollmer et al., 2011), while MYB34 regulates indolic glucosinolate biosynthesis (Celenza et al., 2005; Frerigmann and Gigolashvili, 2014). MYB14 also functions in cold tolerance by regulating the expression of C-REPEAT/DRE BINDING FACTOR (CBF) genes (Chen et al., 2013). However, the function of AT5G50915, hereafter named CKG, has not been characterized, and is the focus of this work.
To examine the potential role of CKG as a regulator or mediator of CK responses, we first confirmed the induction of CKG expression by CK treatment. In agreement with the transcriptome results, we observed that CKs did upregulate the expression of CKG in 7-d-old seedlings in a dose-dependent manner (Figure 1E). We also generated a promoter reporter line by placing the β-glucuronidase gene GUS under the control of the CKG promoter (pCKG:GUS). We detected strong GUS signal in cotyledons, root, leaves, ovules, vasculature, embryos, inflorescence, and flowers (Supplemental Figure S2A). GUS signal was further induced by CK treatment (Figure 1F). CKG is predicted to encode a member of the bHLH family of transcription factors (Toledo-Ortiz et al., 2003). To determine the subcellular localization of CKG, we fused the CKG coding sequence to the green fluorescent protein (GFP), and transfected Arabidopsis protoplasts with the resulting 35S:CKG–GFP construct together with the nuclear marker gene mRFP-ARR2 (encoding a fusion between ARR2, a transcription regulator of CK signaling, and the red fluorescent protein; Hwang and Sheen, 2001). The CKG–GFP fusion clearly co-localized with mRFP-ARR2 in the nucleus (Figure 1G). We further examined CKG expression in the CK signaling mutant arr2 and a line overexpressing ARR2, as well as the CK degradation mutant CKX2, which degrades endogenous CK, overexpressing line (Werner et al., 2003). CKG was highly expressed regardless of the CK treatment in the type-B ARR2-overexpressing line compared to the wild-type Col-0, but the CK-responsive induction of CKG expression was hampered in the arr2 mutant and the CKX2-overexpressing line (Supplemental Figure S2B). We then turned to a protoplast reporter assay using pARR6:LUC as a readout for CK signaling (Hwang and Sheen, 2001). Overexpression of CKG increased transcriptional activation of the ARR6 promoter, and this effect was further enhanced by treatment with CK (Supplemental Figure S2C). These results indicate that CKG may function as a mediator of a canonical CK signaling cascade.
CKG enhances the growth of various organs
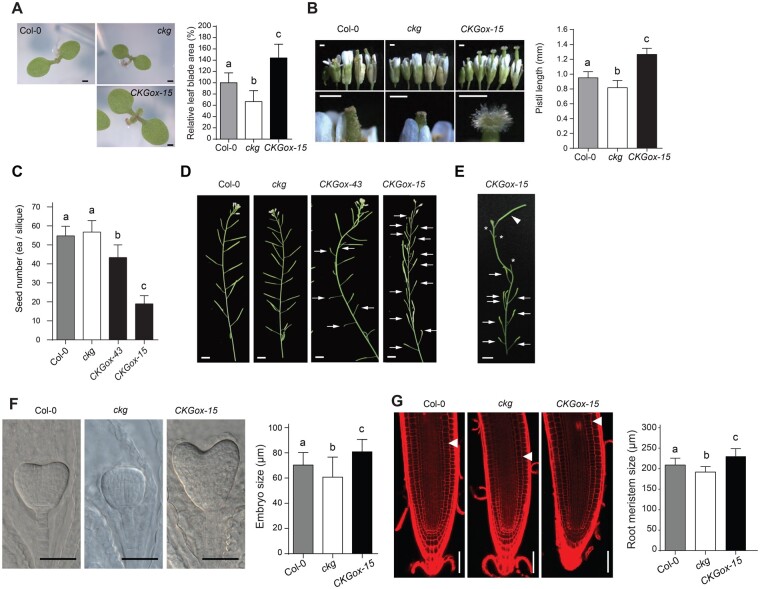
To further unravel the biological function of CKG, we generated lines overexpressing CKG and selected two individual lines with high CKG expression (Supplemental Figure S3C). We also identified a loss-of-function allele of CKG: the T-DNA insertion line SALK_141414C named ckg, which has severely reduced CKG transcript levels and is therefore considered a null allele (Supplemental Figure S3, A and C). Seedlings of the CKGox-15 line, with the highest CKG expression level, showed larger cotyledons compared to Col-0, while the ckg mutant had smaller cotyledons (Figure 2A). Indeed, cotyledon size of 7-d-old seedlings increased by over 40% in CKGox-15 seedlings, but decreased by 30% in the ckg mutant, relative to Col-0.
Figure 2.
CKG promotes the growth of various organs at different developmental stages. A, (Left) representative cotyledons of Col-0, ckg-knockout, and CKG-overexpressing lines. Scale bar, 500 µm. (Right) Cotyledons of 7-d-old CKGox-15 seedlings are larger than those of Col-0 by 40%, but ckg cotyledons are smaller than Col-0 by over 30%. Error bars, sd (n ≥ 35). B, (Left) representative flowers of Col-0, ckg, and CKGox-15 lines at serial developmental stages. In the CKGox-15 line, pistils elongated fast, and stigma development was enhanced. Scale bars, 500 µm. (Right) pistils of the CKGox-15 line were longer than those of Col-0, but the ckg mutant had shorter pistils at 2 d after pollination. Error bars, sd (n = 6). C, CKG affects the number of seeds. CKGox lines exhibited a low self-fertilization rate. Error bars, sd (n = 30). D, Alteration in fertilization by CKG expression levels affects silique development. CKGox lines had a portion of unfertilized siliques (arrows), and few siliques in the CKGox-15 line were normally developed by natural fertilization. Scale bar, 5 mm. E, The normal development of siliques in the CKGox-15 line after artificial pollination. Arrowheads indicate a pollinated silique, asterisks indicate stumps after cutting of peduncles, and arrows indicate unfertilized siliques. Scale bar, 5 mm. F, (Left) Representative embryos at the heart stage of Col-0, ckg, and CKGox-15 lines. Scale bar, 50 µm. (Right) Embryos of the CKGox-15 line at 4 d after pollination were larger than those of Col-0, but ckg embryos were smaller than Col-0 embryos. Error bars, sd (n = 20). G, (Left) Representative root tips of Col-0, ckg and CKGox-15 lines. White arrowheads indicate the transition zone (TZ). Scale bar, 50 µm. Right; the sizes of root meristems in ckg and CKGox-15 lines were significantly reduced and increased compared to Col-0, respectively. Error bars, sd (n = 12). Different letters indicate significant differences at P < 0.05 based on Tukey’s honestly significant difference (HSD) test
Since we had observed high pCKG:GUS signal in flowers (Supplemental Figure S2A), we paid close attention to any phenotype in reproductive structures and measured pistil length in all genotypes. Arabidopsis undergoes self-pollination (autogamy) by promoting the growth of the pistil through a circle of stamens shortly after flower buds open. Pistils from CKGox-15 flowers outgrew the stamen at an earlier stage and were longer than Col-0, whereas ckg flowers had slightly shorter pistils (Figure 2B). The outgrowth of pistils in CKG-overexpressing lines resulted in individual siliques with fewer seeds (Figure 2C). In addition, the CKGox-15 line formed many aborted siliques that contained few seeds (Figure 2D). However, the CKGox-15 line was fully fertile, as hand pollination restored normal seed set (Figure 2E).
CKG is expressed in the egg cell and central cells (Johnston et al., 2007), and in the embryo (Supplemental Figure S2A), which suggests that CKG functions in early ovule and embryo development. Developing seeds of the CKGox-15 line exhibited larger embryos at 4 d after pollination compared to Col-0 and the ckg mutant (Figure 2F). We also observed that most radicles of CKGox-15 embryos were bent and skewed within the seed, which is likely due to enhanced outgrowth of the cotyledons and embryonic root within a limited space. A fraction of ckg mutant embryos showed a defective and abnormal development not seen in Col-0 or the CKGox-15 line (Supplemental Figure S4). In addition to alterations in reproductive structures, ectopic expression of CKG in the overexpression lines increased the size of root meristems compared to Col-0, while the ckg mutant showed smaller root meristems (Figure 2G), which indicates that CKG may drive early cell elongation or differentiation in the root meristem. We also generated a CKG deletion line with clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), named CKG-CAS9 (Supplemental Figure S3, A and B). CKG-CAS9 plants showed a similar phenotype as the ckg mutant—short pistil length, and smaller cotyledons, embryo, and root meristem compared to Col-0 (Supplemental Figure S5).
The visible phenotypes associated with variation in CKG levels therefore point to cell and organ growth as a CK response, leading us to hypothesize that cell cycle progression and cell expansion might be directly under the control of CKG.
CKG regulates cell cycle and macromolecule synthesis genes
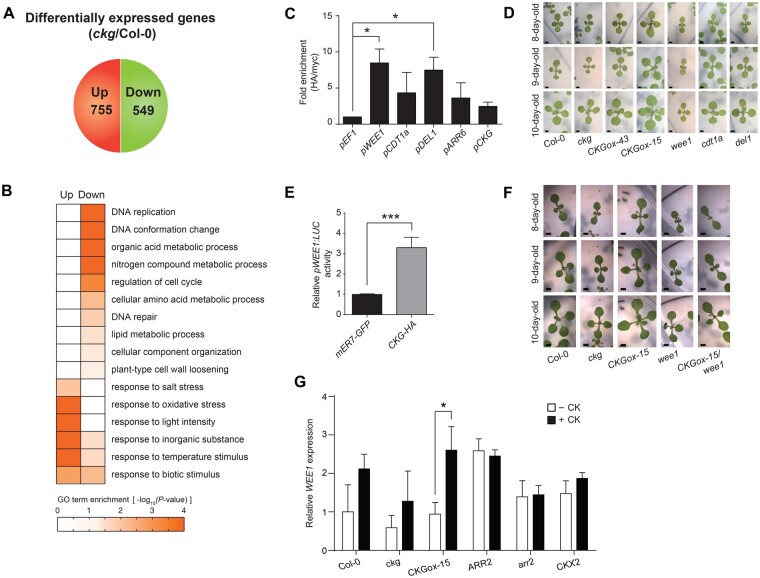
To test whether genes related to cell cycle progression and cell expansion were regulated by CKG, we used microarrays to profile transcript levels in the cotyledons of 7-d-old Col-0 and ckg seedlings. We identified differentially expressed genes (DEGs) in ckg relative to Col-0, based on an absolute fold-change of 1.5 or more and with an associated P< 0.05. Among the DEGs, 755 genes and 549 genes were up- and downregulated, respectively (Figure 3A;Supplemental Figure S6 and Supplemental Data Set S1).
Figure 3.
The downstream targets of CKG are mainly involved in the cell cycle and macromolecule synthesis. A, The number of up- and downregulated genes in the ckg mutant compared to Col-0. Among the genes with P < 0.05, those with log2(fold changes) > 0.58 and less than −0.58 were classified as the up- and downregulated genes, respectively. B, GO terms represented by the up- and downregulated genes in the ckg mutant. The downregulated genes were involved in cell cycle, DNA replication, and macromolecule metabolic processes, whereas the upregulated genes were mainly related to response to biotic and abiotic stresses. C, CKG directly binds to the promoters of core cell cycle regulators. Protoplasts were transfected with 35S:CKG-HA, and ChIP assays of the designated promoters were performed with anti-HA antibody and anti-myc antibody as background. The EF1 promoter was used as a negative control. Fold enrichment was measured by calculating the ratios between normalized results from anti-HA antibody and anti-myc antibody. Error bars, sd (n = 3). D, ckg and wee1 mutants showed similar phenotypes in the cotyledon and leaf growth. CKGox lines grew faster than Col-0, but the ckg and wee1 mutants were smaller than Col-0. The other mutants did not show a growth difference compared to Col-0. Scale bar, 1 mm. E, CKG increases pWEE1:LUC reporter activity. Protoplasts were transfected with pWEE1:LUC and 35S:CKG-HA. mER7-GFP was used as a control and normalized to 1. Error bars, sd (n = 4). F, CKGox-15/wee1 and wee1 plants showed smaller leaves compared to Col-0. Scale bar, 1 mm. G, WEE1 expression is induced by CKG in a CK-dependent manner. Seven-day-old seedlings of Col-0, ckg, CKGox-15, ARR2-overexpressing, arr2, and CKX2-overexpressing lines were treated with or without CK. The expression of WEE1 was highly upregulated in the CKGox-15 line after CK treatment, but not in the arr2 and CKX2-overexpressing lines. The level of WEE1 expression in Col-0 without CK was set to 1, and its relative value was presented. Error bars, sd (n = 3). CK treatment, 300-nM t-zeatin for 3 h. Asterisks indicate significant differences based on the P-value of two-tailed Student’s t test (*< 0.05; ** < 0.01; ***< 0.001)
To investigate the biological processes related to these DEGs, we determined the GO terms that were enriched in the DEGs that were up- and downregulated using BiNGO (Maere et al., 2005; Figure 3B;Supplemental Data Set S2). Interestingly, downregulated DEGs in the ckg mutant showed strong enrichment with cell cycle- and organ growth-related biological processes (“DNA replication”, “DNA conformation change”, “regulation of cell cycle”, and “nitrogen/amino acid metabolic process”). Upregulated DEGs in the ckg mutant included many genes associated with various stress responses (“response to salt/oxidative/light/temperature/biotic stress”). These results indicated that CKG likely regulates organ growth via the modulation of transcript levels for genes involved in cell cycle progression and macromolecule synthesis.
We then tested whether CKG directly controlled the expression of the core cell cycle regulators (Menges et al., 2005; Gutierrez, 2009) that were downregulated in the ckg mutant by chromatin immunoprecipitation (ChIP) analysis using an anti-HA antibody on chromatin extracted from 35S:CKG-HA transfected protoplasts. Since bHLH transcription factors are known to bind to the E-box motif (CANNTG; Heim et al., 2003), we looked for the E-box in the promoters of candidate genes and designed the relevant primer sets for ChIP analysis (Supplemental Figure S7 and Supplemental Table S1). Indeed, we observed direct binding of CKG to the promoters of WEE1, DP-E2F-like 1 (DEL1), and the Arabidopsis homologue of yeast CDC10 Target1a (CDT1a; Figure 3C). In addition, CKG binding was highly enriched at the ARR6 promoter regions. CKG binding was also highly enriched at the CKG promoter regions, suggesting that CKG reinforces its activity through a positive feedback loop to allow sufficient gene regulation. To test the stability of CKG protein, we transfected protoplasts with 35S:CKG-HA and treated with/without CK (t-zeatin) and MG132, a 26S proteasome inhibitor with cycloheximide, a protein synthesis inhibitor. The protein level of CKG started to decrease in 10 min, and MG132 treatment delayed CKG degradation. CK did not affect the stability of CKG protein (Supplemental Figure S8).
To elucidate the genetic relationship between CKG and individual target genes during vegetative growth, we first monitored growth daily for the ckg mutant, CKG overexpressing lines, as well as selected mutants in target genes. wee1 mutants in particular exhibited a retarded growth phenotype similar to that of the ckg mutant (Figure 3D). WEE1 is well conserved in eukaryotes and functions as a kinase controlling cell cycle progression during the S phase, especially under DNA replication stress (O’Connell et al., 1997; Kellogg, 2003). However, the other selected mutants did not show any visible phenotypes, which might be due to functional redundancy with related genes. Our data therefore suggest that CKG directly controlled WEE1 expression to regulate cell growth downstream of CK signaling.
To test this hypothesis, we examined the induction of WEE1 transcription by CKG using a protoplast reporter assay. Co-transfection of 35S:CKG-HA induced pWEE1:LUC promoter activity, confirming our assumptions (Figure 3E). As an independent genetic test, we crossed the CKG overexpressing line with the wee1 mutant to determine the genetic hierarchy between CKG and WEE1. CKGox-15/wee1 plants had the same retarded growth phenotype as the wee1 mutant, placing WEE1 downstream of CKG (Figure 3F). We also measured WEE1 expression levels in the CKGox-15 line, the ckg mutant, and CK-related transgenic lines with or without CK treatment. WEE1 expression was induced by CK treatment in the wild-type (Figure 3G) and was further enhanced in the CKGox-15 line but disrupted in the ckg mutant. Although WEE1 was highly expressed in the ARR2 overexpressing line regardless of CK treatment, CK-dependent induction of WEE1 was abolished in the arr2 mutant and the CKX2 overexpressing line. Collectively, these data suggest that CKG is a direct transcriptional regulator of WEE1 during CK signaling.
CKG regulates cell growth and CK-mediated cell cycle transition independently of ploidy
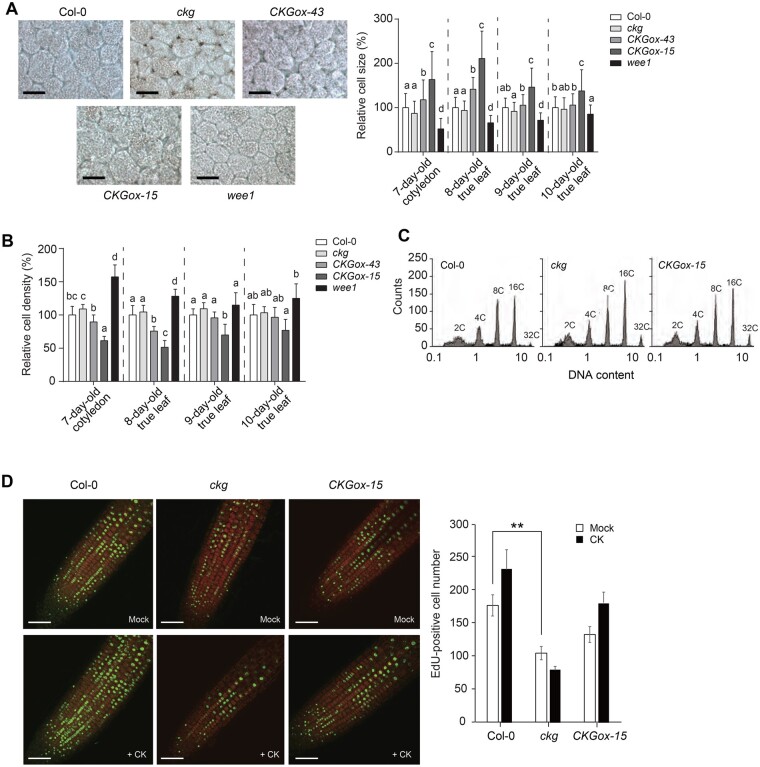
To further unravel the function of CKG and WEE1 during cell growth, we measured the sizes of palisade cells in 7-d-old cotyledons and 8- to 10-d-old true leaves of the ckg mutant, the CKGox lines and the wee1 mutant. The CKGox lines displayed larger cells, whereas the ckg mutant exhibited smaller cells relative to Col-0 (Figure 4A). The sizes of palisade cells in the wee1 mutant were smaller than those of both Col-0 and the ckg mutant. These results suggest that CKG and WEE1 affect cell expansion in a similar fashion. While CKG had a profound effect on leaf growth at early growth stages, mature leaves from the CKGox lines and the ckg mutant were much closer in size and shape to those of Col-0 (Supplemental Figure S9). In addition, the CKGox-15 line had an early flowering phenotype, which may be due to a shortened vegetative phase by a developmental and growth promotion during early growth stages (Supplemental Figure S10). In agreement with cell size, the cotyledons and leaves of the CKGox lines, especially CKGox-15, had a lower cell density compared to Col-0, while the ckg and wee1 mutants exhibited higher cell densities (CKGox-15: 39% decrease over Col-0; ckg: 9% increase over Col-0; wee1: 57% increase over Col-0; Figure 4B 7-d-old cotyledon).
Figure 4.
CKG functions as a growth regulator of cotyledons and leaves. A, (Left) Representative palisade cells of 7-d-old cotyledons of Col-0, ckg, CKGox, and wee1 lines. Scale bar, 50 µm. (Right) Relative cell size of cotyledons and leaves of Col-0, ckg, CKGox, and wee1 lines. The ckg and wee1 mutations resulted in smaller cotyledon and leaf cells compared to Col-0, while CKGox lines exhibited larger cells. Error bars, sd (n 100). B, Relative cell densities of cotyledons and leaves at different growth stages. Due to the changes in cell size, cell densities of CKGox lines were low, but those of ckg and wee1 mutants were relatively high throughout all growth stages. Error bars, sd (n4). C, The ploidy of Col-0, CKGox-15, and ckg plants was similar. Flow cytometric analysis was performed with 7-d-old cotyledons. D, S phase entries of mitotic cells in Col-0, ckg, and CKGox-15 lines. S phase entries were visualized by EdU-staining of 6-d-old root tips. The seedlings were grown in the presence or absence of 100-nM t-zeatin, and then treated with EdU for 2 h. Scale bar, 50 µm. Error bars, sd (n8). Different letters indicate significant differences at P < 0.05 based on Tukey’s honestly significant difference (HSD) test. Asterisks indicate significant differences based on the P-value of two-tailed Student’s t test (*P< 0.05; **P < 0.01; ***P < 0.001)
Since cell size is largely related to ploidy levels, we measured DNA content contained in the nuclei of 7-d-old cotyledons for all genotypes. However, we failed to observe notable changes in ploidy for the CKGox-15 line or the ckg mutant when compared to Col-0 (Figure 4C). Similarly, the wee1 mutation did not affect ploidy levels, as expected (De Schutter et al., 2007). Therefore, CKG and WEE1 regulate cell expansion in a ploidy-independent manner. Finally, we tested whether CKG was involved in cell cycle transitions under the control of CKs by monitoring 5-ethynyl-2′-deoxyuridine (EdU) incorporation into the nuclei of 6-d-old seedlings that are undergoing DNA replication and thus have entered the S phase (Kotogány et al., 2010). The number of EdU-positive cells was significantly lower in the ckg mutant relative to Col-0, but remained unchanged in the CKGox-15 line (Figure 4D). The number of EdU-positive cells rose in response to CK treatment in Col-0, but not in the ckg mutant (Figure 4D). Our data therefore support the hypothesis that CKG mediates the G1/S transition during cell cycle progression as well as cell cycle speed during root growth in response to CKs.
Discussion
Identification of a novel transcription factor involved in CK-mediated regulation of cell size and cell cycle progression
We exploited a microarray data set cataloging plant responses to various phytohormones to identify transcriptional regulators specific for CKs. Our analysis revealed multiple candidate genes, several of which had been previously described as being involved in CK signaling, thus validating our approach. We named another CK-specific gene CKG (for CYTOKININ-RESPONSIVE GROWTH REGULATOR) and focused on its characterization. CKG transcript levels were induced by CK treatment, and CKG overexpression led to the activation of transcription from the pARR6:LUC reporter, positioning CKG downstream of CK perception and identifying it as a positive factor along the CK signaling cascade. A phenotypic analysis of CKG-overexpressing lines alongside a loss-of-function mutant demonstrated that CKG increases cell size, plant growth rates and entry into the S phase of the cell cycle. In sum, CKG is a key regulator participating in cell growth and cell cycle regulation in response to CK.
CK responses in cell growth regulation are complicated because of their opposite actions in the SAM and the RAM. To explain these diametrically opposed features of CK signaling, many reports have proposed the presence of cell- and tissue-specific regulators of CK signaling (reviewed in Takahashi et al., 2013; Zhang et al., 2013; Schaller et al., 2014). Moreover, CKs are connected to auxin signaling, as ARR1 and ARR12 upregulate SHORT HYPOCOTYL2 (SHY2) expression, which itself minimizes auxin transport by repressing the expression of the auxin transporters PIN-FORMED (PIN) and related genes. This modulation of auxin transport controls root meristem size by affecting the balance between the rates of cell proliferation and differentiation (Ioio et al., 2008; Moubayidin et al., 2010). Moreover, CK and auxin interact for the regulation of SAM activity via MONOPTEROS (MP, also known as AUXIN RESPONSE FACTOR5 or ARF5), which represses ARR7 and ARR15 expression in the central zone (Zhao et al., 2010). However, the roles of CKs in root cell proliferation cannot be easily summarized by a linear signaling pathway, as CKs affect each growth stage and tissue differently. Other phytohormones such as jasmonic acid, abscisic acid, strigolactones, gibberellins, ethylene, and brassinosteroids are also involved in the regulation of the cell cycle, but their signal integration is largely unknown (Del Pozo et al., 2005; reviewed in Vanstraelen and Benková, 2012; Takatsuka and Umeda, 2014). In addition, CK levels can affect the size of the root meristem. The transcription factor PHABULOSA (PHB) induces CK biosynthesis in the distal parts of the root meristem, which results in the activation of ARR1 in the elongation/differentiation zone, where ARR1 then represses both PHB and the microRNA MIR165A, a negative regulator of PHB, thereby forming an incoherent regulatory loop that controls meristem size (Dello Ioio et al., 2012). Another level of regulation comes from the oscillation of endogenous CK contents as a function of the cell cycle, during which CKs mainly accumulate during the S and M stages (Redig et al., 1996). Hartig and Beck (2005) also presented evidence for the accumulation of CKs at stage transitions such as the G1/S and the M/G1 transitions. Hartig and Beck proposed that the transient peak in CK levels and low CK content at the end of each phase were equally important for cell cycle stage transitions. Overall, the regulation of the cell cycle by CKs should be considered holistically by integrating CK-responsive factors with the target tissue or cell type, cell cycle stage and signaling from other phytohormones.
Menges et al. (2005) profiled the transcriptome of a synchronized MM2d Arabidopsis cell line over the course of the cell cycle. They reported that CKG expression was constant over all cell cycle stages, although CKG target genes (WEE1, CDT1a, and DEL1) were induced at specific stages, which suggests that the regulation of CKG expression and its targets is distinct and reflects the stage of the cell cycle. In addition, several bHLH proteins form homo- or heterodimers in mammals, in the fruit fly (Drosophila melanogaster; Evan and Littlewood, 1998), and in plants (Halliday et al., 1999; Lingam et al., 2011; Liu et al., 2013). The potential for combinatorial dimerization may result in the observed distinct transcriptional profiles (Heim et al., 2003). Likewise, CKG may interact with other factors that are themselves cell cycle stage-specific, a notion that is supported by the fact that CKG protein has a short half-life (Supplemental Figure S4). Therefore, CKG may coordinate cell cycle-related genes together with other transcription regulators that respond to various signals, including CKs, to promote entry into the S phase of the cell cycle.
CKG regulates cell size
CKs induce CKG expression, which in turn regulates the expression of cell cycle genes and various macromolecular synthesis genes. CKG physically binds to promoters of cell cycle-promoting factors such as WEE1, CDT1a, and DEL1, consistent with a role for CKG in cell cycle regulation in response to CK signaling. Although we failed to observe any significant differences in leaf size in cdt1a or del1 mutants relative to Col-0 (Figure 3D), we hypothesize that redundancy may partially mask their contribution to cell cycle progression and cell division. Indeed, DEL1 has two closely related genes in the Arabidopsis genome, while yeast CDT1 has two Arabidopsis homologs. In contrast, the wee1 mutant exhibited smaller cell size in cotyledons when compared to the wild-type Col-0 and also showed delayed growth. The WEE1 kinase is a cell cycle checkpoint that measures DNA damage and cell size at the G2/M transition (O'Connell et al., 1997; Kellogg, 2003), so we had expected to see cell division and endoreduplication phenotypes in the wee1 mutant. Transgenic lines overexpressing WEE1 showed no significant differences in cell size, although limited overexpression of WEE1 using a heat shock-inducible system resulted in cell enlargement (De Schutter et al., 2007), which resolved the function of WEE1 in the promotion of cell expansion. However, ploidy levels remained normal in various tissues from wee1 plants (De Schutter et al., 2007), which fits well with the absence of ploidy changes in CKG gain- and loss-of-function lines and indicates that CKG and WEE1 may not affect endoreduplication or DNA volume and thus increase cell size independently of ploidy levels. WEE1 expression is also induced by DNA damage by the two upstream kinases ATAXIA-TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR). There is little evidence to suggest that WEE1 may participate in cell size increase. Therefore, even though we can firmly link CKG and WEE1 in the control of cell size, we cannot exclude the existence of an additional player that regulates cell size and that would act downstream of CKG but upstream of WEE1. We do note that the ploidy-independent cell size increase observed here may be partially explained by enhanced macromolecule synthesis, as pointed out by our transcriptome analysis.
Although ploidy level is a major factor controlling cell size, other ploidy-independent factors control cell size as well, for instance, the level of cellular vacuolization or the induction of genes involved in macromolecule biosynthesis (reviewed in Sugimoto-Shirasu and Roberts, 2003). It is worth noting that the expression of genes related to macromolecule biosynthesis is reduced in the ckg mutant, which suggests that changes in cell size reflect macromolecule biosynthesis potential. Like other cellular events, cell growth needs to be accompanied by the production of macromolecules such as nucleic acids, proteins, carbohydrates, and lipids to sustain growth. In particular, macromolecules such as cellulose and phospholipids are essential structural components for cell wall expansion and the addition of new membrane at the plasma membrane, endomembrane systems, and organelles.
The effect of ploidy on cell size can be easily explained by the resulting increase in gene dosage, but cannot explain how CKG affects cellular growth. Instead, CKG may specifically induce the expression of genes related to cell growth, resulting in larger cells without resorting to ploidy changes. The finding that overexpression of CKG later during plant development is not accompanied by changes in overall plant size also suggests that CKG-mediated effects on cell growth are somewhat restricted and do not go over a pre-determined growth promotion threshold.
In this work, we propose that CKG is a key transcription factor controlling cell cycle regulators to increase organ size and growth rates in a ploidy-independent manner. CKG is a mediator of a canonical CK signaling cascade, mainly downstream of a type-B ARR2, connecting to WEE1 in the control of cell size and cycle (Supplemental Figure S11). It is of note that CK, however, still induces CKG expression in the arr2 mutant (Figure S2B), suggesting that other type-B ARRs may play a role in CK-mediated CKG expression. However, the exact nature of the regulatory mechanisms behind CKG-mediated cell and organ growth remain to be fully investigated. For example, how does CKG regulate the progression through cell cycle stages and what are its co-regulators? What other factors does CKG interact with to modulate gene expression? It will be informative to study how CKG is connected to other known cell growth regulators that are under the control of CKs. In addition, CKG affects cell size regulation at an early growth and development stage, which might constitute a useful trait that can be harnessed to promote growth, early flowering, and higher productivity of leaf vegetables.
Methods
Plant materials and growth conditions
Arabidopsis thaliana accession Col-0 was used as the wild-type control and as the genetic background of the transgenic lines. The plants were grown in a controlled growth chamber (23°C, 16-h light/8-h dark). T-DNA insertion lines of ckg (SALK_141414C), wee1 (SALK_039890C), cdt1a (GABI_025G08), del1 (SALK_105648), and arr2 (SALK_016143) were provided by the Arabidopsis Biological Resource Center. The ARR2 and CKX2 overexpression lines were previously described (Choi et al., 2010). Plants for protoplast isolation were grown in short-day conditions (23°C, 10-h light/14-h dark). To generate transgenic plants overexpressing CKG-HA, the CKG coding sequence was cloned into the pCB302ES vector containing the CaMV 35S promoter and a HA epitope tag. The expression level of CKG was verified by reverse transcription quantitative polymerase chain reaction (RT-qPCR) using a CKG-specific primer set and also by western blot with a horseradish peroxidase-conjugated high-affinity anti-HA antibody (1:2,000, Anti-HA-Peroxidase, High Affinity, Roche, catalogue no. 12013819001). To generate pCKG:GUS transgenic plants, a 2-kb sequence of the CKG promoter was cloned into pCAMBIA1303. The primers used for cloning are listed in Supplemental Table S2.
RT-qPCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s manual. One microgram of total RNA was used to synthesize cDNA using oligo(-dT) primers and ImProm-II reverse transcriptase (Promega). RT-qPCR was performed with gene-specific primers (CKG, WEE1) with the Light Cycler 2.0 (Roche) and the SYBR Premix Ex Taq system (Takara). EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (EIF4A1) was used as an internal control. The primers used for RT-qPCR are listed in Supplemental Table S2.
Plasmid constructs and protoplast transient expression assay
The full-length cDNA of CKG was cloned into plant expression vectors containing the CaMV 35S promoter and an HA epitope tag. For the reporter assay, 2 × 104 protoplasts were transfected with 20 μg of total plasmid DNA composed of different combinations of the reporters (pARR6:LUC, pWEE1:LUC), effector (35S:CKG-HA), and an internal control (mER7-GFP). For the pARR6:LUC reporter assay, transfected protoplasts were incubated for 3 h and then treated with 100-nM t-zeatin for 3 h at room temperature. To confirm the subcellular localization of CKG, a CKG–GFP fusion construct was co-transfected with the mRFP-ARR2 construct in protoplasts and observed with a confocal microscope (LSM800, Carl Zeiss). pARR6:LUC, mRFP-ARR2, and mER7-GFP constructs were previously described (Hwang and Sheen, 2001; Ryu et al., 2007; Choi et al., 2010). The primers used for cloning are listed in Supplemental Table S2.
CKG deletion by CRISPR-Cas9
Protospacer oligos targeting CKG were designed from http://crispr.hzau.edu.cn/CRISPR2/ (Liu et al., 2017) and cloned as described by (Schiml et al., 2016). Briefly, two protospacer oligos targeting CKG were inserted into each BbsI-digested pEn_C1.1. The sgRNA 1 containing first protospacer oligos from pEn_C1.1 was inserted into pDe-CAS9 by Bsu36I and MluI digestion and ligation. The sgRNA 2 containing second protospacer oligos from pEn_C1.1 was inserted into pDe-CAS9 containing sgRNA 1 by Gateway Cloning LR reaction, generating pDe-CAS containing both sgRNA 1 and 2. This construct was transformed in Col-0. The region surrounding the protospacer oligo target site was amplified from genomic DNA and inserted into pGEM-T Easy Vector. Targeted gene deletions were detected by sequencing of the pGEM-T Easy Vector containing amplified PCR products. The primers used for protospacer oligos and confirmation of CKG deletion are listed in Supplemental Table S2.
GUS staining
For determination of CKG expression, Arabidopsis samples were stained with GUS-staining buffer (100-mM Tris–HCl (pH 7.0), 2-mM ferricyanide, and 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronidase)) for 3 h. For the CKG induction experiment by CK treatment, the seedlings were transferred to 1/2 B5 medium with or without 300-nM t-zeatin and incubated for 3 h, and then observed with a stereomicroscope and a microscope (Axioplan2, Carl Zeiss).
Phenotypic analysis
For analysis of cotyledons and rosette leaves, seedlings were grown on 1/2 B5 medium (Duchefa) containing 1% sucrose (w/v; Duchefa) and 0.8% type-M agar (w/v; Sigma) at 23°C with a 16-h light/8-h dark photoperiod. As previously described with minor modifications (Jun et al., 2013), cotyledons and leaves were fixed with fixation solution (2.5% acetic acid (v/v), 2.5% formaldehyde (v/v), and 45% ethanol (v/v) in water) under a vacuum, and the size of palisade cells in the region between the mid-vein and leaf margin in the center of the leaf were measured using a microscope (Axioplan2, Carl Zeiss) and ImageJ software (NIH Image). Pistil lengths were measured at 2 d after pollination using a microscope (Axioplan2, Carl Zeiss). For analysis of embryos, developing seeds at 4 d after pollination were dissected under a stereomicroscope, and embryo sizes were measured using a microscope (Axioskop2, Carl Zeiss). To analyze root meristem size, sample preparations were performed as previously described (Malamy and Benfey, 1997), and measured using a confocal microscope (LSM800, Carl Zeiss).
Microarray
For microarray gene expression analysis, two independent biological replicates for Col-0 and three independent biological replicates for ckg cotyledons were analyzed with the Arabidopsis V4 Oligo Microarray (Agilent) according to the standard Agilent protocols. The total RNA integrity was measured by using the Bioanalyzer 2100 (Agilent). Fluorescent signals (probe intensities) were scanned using the Agilent microarray scanner. Log2-intensities of whole samples were normalized using quantile normalization (Bolstad et al., 2003). To identify DEGs between two conditions, P-values were computed as previously described (Chae et al., 2013): (1) we computed t-statistics from the two-tailed t test for individual genes with assumption of unequal variance. Log2(median differences) were computed for individual genes; (2) empirical null distribution of the t-statistics and log2(median difference) was derived from random permutations of whole samples; (3) P-values of t test and log2(median difference) were computed based on corresponding empirical null distribution ; and (4) two P-values were integrated by Liptak–Stouffer’s Z method (Hwang et al., 2005). DEGs were selected if they showed an over 1.5-fold change and P < 0.05. Microarray data were deposited in NCBI Gene Expression Omnibus (accession ID: GSE131029).
Functional enrichment analysis
BiNGO was used to identify GO biological processes significantly represented in DEGs (Maere et al., 2005). We used Benjamini and Hochberg false discovery rate corrected P-value provided from BiNGO to compute the enrichment P-value.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described with minor modifications (Lee et al., 2007; Cho et al., 2014). Protoplasts were transfected with 20 μg of 35S:CKG-HA constructs. After 6 h, protoplasts were crosslinked with 1% formaldehyde (v/v) for 10 min and quenched with 125-mM glycine. Harvested protoplasts were ground and resuspended with nuclei isolation buffer (0.25-M sucrose, 15-mM 1,4-piperazinediethanesulfonic acid (PIPES) (pH 6.8), 5-mM MgCl2, 60-mM KCl, 15-mM NaCl, 1-mM CaCl2, 1% Triton X-100 (v/v), and 1-mM phenylmethylsulphonyl fluoride). The nuclear pellets were resuspended in nuclei lysis buffer (50-mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) (pH 7.0), 150-mM NaCl, 1-mM ethylenediaminetetraacetic acid (EDTA), 1% sodium dodecyl sulfate (SDS) (w/v), 1% Triton X-100 (v/v), 1-mM phenylmethylsulphonyl fluoride, and 1X Protease inhibitor cocktail for plant cell and tissue extracts (Sigma)). The chromatins were isolated and sheared by sonication to produce 0.5–1-kb DNA fragments. Chromatin fractions were diluted tenfold with nuclei lysis buffer and pre-cleared with protein G agarose/salmon sperm DNA (Millipore) for 2 h at 4°C. The protein–DNA complexes were immunoprecipitated with anti-HA antibody (1:1,000, anti-HA tag antibody – ChIP Grade, Abcam, catalogue no. ab9110) and anti-myc antibody (1:1,000, Myc-Tag (9B11) Mouse mAb, Cell Signaling Technology, catalogue no. 2276) for 9 h at 4°C and further incubated with protein G agarose/salmon sperm DNA for 2 h at 4°C. After washing with low salt buffer (150-mM NaCl, 20-mM Tris–HCl (pH 8.0), 0.2% SDS (w/v), 0.5% Triton X-100 (v/v), and 2-mM EDTA) for 5 min at 4°C and high-salt buffer (500-mM NaCl, 20-mM Tris–HCl (pH 8.0), 0.2% SDS (w/v), 0.5% Triton X-100 (v/v), and 2-mM EDTA) for 5 min at 4°C, the immunocomplexes were eluted twice by elution buffer (0.5% SDS (w/v) and 0.1-M NaHCO3) and then reverse crosslinked with 200-mM NaCl for 6 h at 65°C. After removing proteins with proteinase K, DNAs were purified by phenol–chloroform extraction and recovered by ethanol precipitation. Precipitated DNAs were resuspended with TE buffer (10-mM Tris–HCl (pH 8.0) and 1-mM EDTA) and used as qPCR templates with ChIP primer sets (Supplemental Table S2).
Ploidy analysis
Ploidy levels were measured using the ploidy analyzer PA-I (Partec) as described previously (Sugimoto-Shirasu et al., 2002). At least 7,000 nuclei isolated from cotyledons of 7-d-old seedlings were used for each ploidy measurement.
EdU staining
Six-day-old seedlings were treated with 1-µM EdU (Invitrogen Click-iT EdU Imaging Kit) for 2 h, transferred to fixative (4% formaldehyde (v/v)), permeabilized with 0.1% Triton X-100 (v/v) for 30 min and washed with 3% bovine serum albumin (BSA) (w/v) in phosphate-buffered saline (PBS). The seedlings were incubated in Click-iT reaction cocktail for 30 min in the dark, then washed with 3% BSA (w/v) in PBS three times before observation by confocal microscope (LSM510, Carl Zeiss).
Statistical analysis
Quantitative data were subjected to two-tailed Student’s t tests or analysis of variance (ANOVA) test. For Student’s t tests, asterisks indicate the P-values (*< 0.05; **< 0.01; ***< 0.001). For ANOVA test, different letters indicate significant differences at P < 0.05 based on Tukey’s honestly significant difference test.
Accession numbers
The sequence data from this article can be found in The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: ARR2 (AT4G16110), ARR6 (AT5G62920), CDT1a (AT2G31270), CKG (AT5G50915), CKX2 (AT2G19500), DEL1 (AT3G48160), EF1 (AT5G60390), EIF4A1 (AT3G13920), and WEE1 (AT1G02970).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression patterns of representative genes in C1 and C2 cluster (related to Figure 1C).
Supplemental Figure S2. CK-dependent expression of CKG and its expression pattern in tissues.
Supplemental Figure S3. T-DNA insertion site and CRISPR-Cas9-meditated deletion site in CKG, and CKG expression levels in ckg and CKGox transgenic lines.
Supplemental Figure S4. Abnormal embryo growth in CKGox-15 and ckg plants.
Supplemental Figure S5. The CKG-CAS9 deletion line shows similar phenotypes to the ckg mutant.
Supplemental Figure S6. PCA plot of Col-0 and ckg microarray samples.
Supplemental Figure S7. Promoter regions amplified for the ChIP assay in Figure 3C.
Supplemental Figure S8. CK does not affect the stability of CKG proteins.
Supplemental Figure S9. The growth phenotypes of wee1, ckg, and CKGox lines.
Supplemental Figure S10. The early bolting phenotypes of CKGox-15 plants.
Supplemental Figure S11. Model of CK-mediated CKG action for the cell cycle.
Supplemental Data Set S1. List of DEGs related to Figure 3A.
Supplemental Data Set S2. Gene ontology of up- and downregulated genes in the ckg mutant.
Supplemental Table S1. CKG target gene candidates.
Supplemental Table S2. Primer list.
Supplementary Material
Acknowledgments
We are grateful to Professor Gyung-Tae Kim (Dong-A University, Korea) for the advice. We thank Hoyoung Nam for sample preparation.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2020R1A2C3012750).
Conflict of interest statement. Authors declare that they have no conflict of interest.
J.P., S.L, and I.H. designed this study. S.L., D.C., and D.H. analyzed and processed the microarray data. J.P. and S.L. performed transgenic, cell biological, and other functional analyses. G.P. performed phenotypic analysis of embryos and floral organs. M.U. performed ploidy analysis. H.C. performed the EdU staining assay. J.P., S.L., and I.H. wrote the manuscript. Y.C., M.U., and I.H. supervised the work.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ildoo Hwang (ihwang@postech.ac.kr).
References
- Arnaud D, Lee S, Takebayashi Y, Choi D, Choi J, Sakakibara H, Hwang I (2017) Cytokinin-mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in Arabidopsis. Plant Cell 29: 543–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu M-H, Lee B, Hwang D, Park KS (2013) A systems approach for decoding mitochondrial retrograde signaling pathways. Science Signal 6: rs4. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Rep 31: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Ryu H., Rho S., Hill K., Smith S., Audenaert D., Park J., Han S., Beeckman T., Bennett M.J.. , et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16: 66–76 [DOI] [PubMed] [Google Scholar]
- Choi D, Choi J, Kang B, Lee S, Cho YH, Hwang I, Hwang D (2014) iNID: an analytical framework for identifying network models for interplays among developmental signaling in Arabidopsis. Mol Plant 7: 792–813 [DOI] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek K-H, Hwang I (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19: 284–295 [DOI] [PubMed] [Google Scholar]
- Cortleven A, Leuendorf JE, Frank M, Pezzetta D, Bolt S, Schmülling T (2019) Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ 42: 998–1018 [DOI] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D. , et al. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo JC, Lopez-Matas MA, Ramirez-Parra E, Gutierrez C (2005) Hormonal control of the plant cell cycle. Physiol Plant 123: 173–183 [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK. , et al. (2012). A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol 22: 1699–1704 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V. , et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G, Littlewood T (1998) A matter of life and cell death. Science 281: 1317–1322 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T (2014) MYB34, MYB51 and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7: 814–828 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (2009) The Arabidopsis cell division cycle. Arabidopsis Book 7:e0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH (1999) poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci 96: 5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig K, Beck E (2005) Endogenous cytokinin oscillations control cell cycle progression of tobacco BY-2 Cells. Plant Biol 7: 33–40 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hejatko J, Ryu H, Kim GT, Dobesova R, Choi S, Choi SM, Soucek P, Horak J, Pekarova B, Palme K. , et al. (2009). The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21:2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S. , et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Rust A, Ramsey S, Smith J, Leslie D, Weston A, de Atauri P, Aitchison J, Hood L, Siegel A. , et al. (2005). A data integration methodology for systems biology. Proc Natl Acad Sci USA 102: 17296–17301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Ioio RD, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Johnston A, Meier P, Gheyselinck J, Wuest S, Federer M, Schlagenhauf E, Becker J, Grossniklaus U (2007) Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol 8: R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Okushima Y, Nam J, Umeda M, Kim G-T (2013) Kip-related protein 3 is required for control of endoreduplication in the shoot apical meristem and leaves of Arabidopsis. Mol Cells 35: 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR (2003) Wee1-dependent mechanisms required for coordination of cell growth and cell division. J Cell Sci 116: 4883–4890 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. Arabidopsis Book 12:e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development 145: dev149344. [DOI] [PubMed] [Google Scholar]
- Kim J (2016) CYTOKININ RESPONSE FACTORs gating environmental signals and hormones. Trends Plant Sci 21: 993–996 [DOI] [PubMed] [Google Scholar]
- Kollmer I, Werner T, Schmulling T (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Kotogány E, Dudits D, Horváth GV, Ayaydin, F (2010) A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen L-L (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10: 530–532 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. Plos Genet 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T. , et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MJ, Raleigh JM, Verkade HM, Nurse P (1997) Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J 16: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout A, Crabos A, Petřík I, Novák O, Krouk G, Lacombe B, Ruffel S (2018) Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-Zeatin in shoots. Plant Cell 30: 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines T, Shanks C, Cheng C-Y, McPherson D, Argueso CT, Kim HJ, Franco-Zorrilla JM, López-Vidriero I, Solano R, Vaňková R. , et al. (2016). The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J 85: 134–147 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redig P, Shaul O, Inzé D, Van Montagu M, Van Onckelen H (1996) Levels of endogenous cytokinins, indole-3-acetic acid and abscisic acid during the cell cycle of synchronized tobacco BY-2 cells. FEBS Lett 391: 175–180 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Roitsch T, Ehneß R (2000) Regulation of source/sink relations by cytokinins. Plant Growth Regul 32: 359–367 [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Street IH, Kieber JJ (2014) Cytokinin and the cell cycle. Curr Opin Plant Biol 21: 7–15 [DOI] [PubMed] [Google Scholar]
- Schiml S, Fauser F, Puchta H (2016) CRISPR/Cas-mediated site-specific mutagenesis in Arabidopsis thaliana using Cas9 nucleases and paired nickases. Methods Mol Biol 1469: 111–122 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC (2002) DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol 12: 1782–1786 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, Kim Y, Katagiri Y, Okushima Y, Matsunaga S, Hwang I, Umeda M (2013) Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol 23: 1812–1817 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M (2014) Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65: 2633–2643 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28: 463–487 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw B, De Rybel B (2019) Cytokinin - a developing story. Trends Plant Sci 24: 177–185 [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Swarup R, Bennett M, Schaller GE, Kieber JJ (2013) Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol 23: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang W, Hill K, Raines T, Solano R. , et al. (2017) Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci USA 114: E5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.