An Arabidopsis calmodulin-binding protein is a critical transcriptional activator in immune responses; loss of the protein activates an effector-triggered immunity pathway and induces autoimmunity.
Abstract
Plants use a dual defense system to cope with microbial pathogens. The first involves pathogen-associated molecular pattern-triggered immunity which is conferred by membrane receptors, and the second involves effector-triggered immunity (ETI), which is conferred by disease-resistance proteins (nucleotide-binding leucine-rich repeat-containing proteins; NLRs). Calmodulin-Binding Protein 60 (CBP60) family transcription factors are crucial for pathogen defense: CBP60g and Systemic Acquired Resistance Deficient 1 (SARD1) positively regulate immunity, whereas CBP60a negatively regulates immunity. The roles of other Arabidopsis (Arabidopsis thaliana) CBP60s remain unclear. We report that CBP60b positively regulates immunity and is redundant with—yet distinct from—CBP60g and SARD1. By combining chromatin immunoprecipitation-PCRs and luciferase reporter assays, we demonstrate that CBP60b is a transcriptional activator of immunity genes. Surprisingly, CBP60b loss-of-function results in autoimmunity, exhibiting a phenotype similar to that of CBP60b gain-of-function. Mutations at the ENHANCED DISEASE SUSCEPTIBILITY 1-PHYTOALEXIN DEFICIENT 4-dependent ETI pathway fully suppressed the defects of CBP60b loss-of-function but not those of CBP60b gain-of-function, suggesting that CBP60b is monitored by NLRs. Functional loss of SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1, an R-gene, partially rescued the phenotype of cbp60b, further supporting that CBP60b is a protein targeted by pathogen effectors, that is, a guardee. Unlike CBP60g and SARD1, CBP60b is constitutively and highly expressed in unchallenged plants. Transcriptional and genetic studies further suggest that CBP60b plays a role redundant with CBP60g and SARD1 in pathogen-induced defense, whereas CBP60b has a distinct role in basal defense, partially via direct regulation of CBP60g and SARD1.
Introduction
Plants are constantly challenged by pathogenic microbes during development. To survive these biotic stresses, plants have evolved two strategies to surveil and respond to pathogens, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI; Jones and Dangl, 2006; Dodds and Rathjen, 2010; Peng et al., 2018). Plants recognize specific molecular patterns on the surface of pathogens, that is, PAMPs, by plasma membrane (PM)-localized pattern recognition receptors (PRRs; Zipfel, 2008). PRRs recognize and bind to specific PAMPs, leading to PTI. To counteract PTI, pathogens inject effector proteins directly to plant cells, avoiding the recognition by PRRs. These effectors target PRRs or their downstream signaling components to increase pathogen fitness. In the arms-race, plants have evolved disease-resistance (R) genes, which encode nucleotide-binding, leucine-rich repeat (NB-LRR) containing proteins (NLRs; Jones and Dangl, 2006; Cui et al., 2015). Upon recognition of their cognate effector proteins, NLRs induce ETI, which often involves hypersensitive response and culminates in cell death of affected tissues (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Cui et al., 2015; Zhou and Zhang, 2020).
NLRs are classified into three categories based on their N-terminal domains. Toll interleukin-1 receptor (TIR)-NB-LRR (TNL) contains a TIR domain; coiled-coil (CC)-NB-LRR (CNLs) contains a CC domain; resistance to powdery mildew 8 (RPW8)-NB-LRR (RNL) contains an RPW8-like (CCR) domain (Li et al., 2015; Jubic et al., 2019; Zhou and Zhang, 2020). Most classic TNLs and CNLs can directly or indirectly recognize effectors and are thus called sensor NLRs (Jubic et al., 2019; Zhou and Zhang, 2020). RNLs are helper NLRs (hNLRs) that participate in effector recognition by working together with TNLs or CNLs in the initiation of ETI (Castel et al., 2019; Jubic et al., 2019; Wu et al., 2019; Zhou and Zhang, 2020). NLR gain-of-function results in autoimmunity, reflected by a small and dwarf status, precocious cell death, a high level of salicylic acid (SA), constitutive expression of PATHOGENESIS-RELATED (PR) genes, and enhanced pathogen resistance (Zhang et al., 2003; Bi et al., 2011; Rodriguez et al., 2016; van Wersch et al., 2016; Chakraborty et al., 2018).
Three lipase-like proteins, ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), SENESCENCE-ASSOCIATED GENE 101 (SAG101), and PHYTOALEXIN DEFICIENT 4 (PAD4), form either an EDS1-PAD4 or EDS1-SAG101 complex to mediate TNL-induced ETI (Feys et al., 2005; Wiermer et al., 2005; Cheng et al., 2011; Zhu et al., 2011; Wagner et al., 2013; Xu et al., 2015), in addition to a role in PTI and systemic acquired resistance (SAR; Zhou and Zhang, 2020). TNL signaling transmitted through the EDS1-PAD4 complex may involve three hNLRs, that is ACTIVE DISEASE RESISTANCE 1 (ADR1), ADR1-like 1 (ADR1-L1), and ADR1-L2 (Dong et al., 2016; Castel et al., 2019; Lapin et al., 2019; Wu et al., 2019). Alternatively, N REQUIRED GENE 1a (NRG1a) and NRG1b serve as the hNLRs for EDS1-SAG101-mediated TNL signaling (Qi et al., 2018; Castel et al., 2019; Lapin et al., 2019; Wu et al., 2019). ETI induced by most CNLs requires NON-RACE-SPECIFIC DISEASE RESISTANCE 1 (NDR1; Jones and Dangl, 2006; Dodds and Rathjen, 2010; Knepper et al., 2011; Cui et al., 2015). Mutations of the EDS1-PAD4 or EDS1-SAG101 pathway suppress autoimmunity caused by gain-of-function of the cognate NLRs (Cheng et al., 2011; Xu et al., 2015).
Not only do NLRs recognize pathogen effectors to induce ETI, they also sense and respond to targets of pathogen effectors, a phenomenon described as the “guard hypothesis” (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Kourelis and van der Hoorn, 2018). According to the guard hypothesis, pathogen effectors target plant immune proteins, or “guardees,” to facilitate infection. Plants counteract this strategy by using specific NLRs to monitor these immune proteins, whose defects activate NLRs to induce ETI (Cui et al., 2015; Rodriguez et al., 2016). A well-studied example is the mitogen-activated protein (MAP) kinase cascade MEKK1-MKK1/MKK2-MPK4 in PTI (Rasmussen et al., 2012). Mutations of the MAPK pathway components cause ectopic activation of NLRs, leading to autoimmunity. Autoimmunity of mekk1, mkk1;mkk2, or mpk4 is suppressed by functional loss of PAD4 or its guards NLRs, SUMM2 (SUPPRESSOR OF mkk1;mkk2), and SMN1 (MEKK1N OVEREXPRESSION-INDUCED DWARF 1; Zhang et al., 2012; Takagi et al., 2019).
Calmodulin-binding protein 60 (CBP60)-like proteins are atypical transcription factors in plants (Ding and Redkar, 2018). There are eight members encoded in the Arabidopsis (Arabidopsis thaliana) genome, CBP60a-g, and SAR Deficient 1 (SARD1; Ding and Redkar, 2018). CBP60g and SARD1 mediate immune gene expression during pathogen invasion and are critical for disease resistance (Wang et al., 2009, 2011; Zhang et al., 2010b; Truman and Glazebrook, 2012; Sun et al., 2015; Qin et al., 2018). They directly bind to the promoters of SA INDUCTION DEFICIIENT2 (SID2)/ISOCHORISMATE SYNTHASE1 to promote SA biosynthesis and immune gene expression (Zhang et al., 2010b). They also bind to and activate the expression of other immune genes in the ETI, PTI, and SAR pathways (Sun et al., 2015, 2018). Functional loss of CBP60g and SARD1 rendered plants more sensitive to pathogens (Zhang et al., 2010b; Wang et al., 2011). Another member of this family, CBP60a, was shown to negatively regulate immunity (Truman et al., 2013), although the mechanism of its action is unclear.
By combining chromatin immunoprecipitation (ChIP)-PCRs and luciferase (LUC) reporter assays, we report that CBP60b positively regulates immunity through direct binding and activation of immune genes. CBP60b gain-of-function resulted in dwarfism, enhanced PR expression, and hyposensitivity to Pto DC3000, consistent with its being a positive regulator of immunity. However, CBP60b loss-of-function also caused autoimmunity. Mutations at the EDS1-PAD4-dependent ETI pathway fully suppressed the defects of CBP60b loss-of-function but not those of CBP60b gain-of-function, suggesting that autoimmunity of cbp60b was due to ectopic activation of EDS1-PAD4-initiated NLR signaling. Indeed, functional loss of SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 (SNC1), an NLR-coding gene, partially rescued the phenotype of cbp60b. Unlike CBP60g and SARD1, CBP60b is expressed constitutively and highly in unchallenged plants. Based on transcriptional and genetic studies, we further show that CBP60b plays a redundant role with CBP60g and SARD1 in pathogen-induced defense, whereas CBP60b has a distinct role in basal defense, partially through direct regulation of CBP60g and SARD1.
Results
CBP60b is constitutively expressed and induced by pathogen challenge
To determine the function of the other five Arabidopsis CBP60s that belong to the same subfamily (Ding and Redkar, 2018), namely CBP60b, CBP60c, CBP60d, CBP60e, and CBP60f, we first examined their expression patterns by generating genomic:GUS reporter lines. By histochemical GUS staining, we determined that CBP60b was highly expressed in various tissues and developmental stages, including seedlings (Supplemental Figure S1, A), primary and lateral roots (Supplemental Figure S1, B and C), shoot apical meristem (SAM; Supplemental Figure S1, D), leaves including trichrome and guard cells (Supplemental Figure S1, E, F and H), and inflorescences (Supplemental Figure S1, G). By contrast, the other members of this subfamily, that is CBP60c (Supplemental Figure S2, A–D), CBP60d (Supplemental Figure S2, E–H), CBP60e (Supplemental Figure S2, I–L), and CBP60f (Supplemental Figure S2, M–P) were expressed at lower levels, often only in root tips. These results suggested that CBP60b is a dominant member of the CBP60 subfamily in plant growth.
CBP60b loss-of-function compromises plant growth
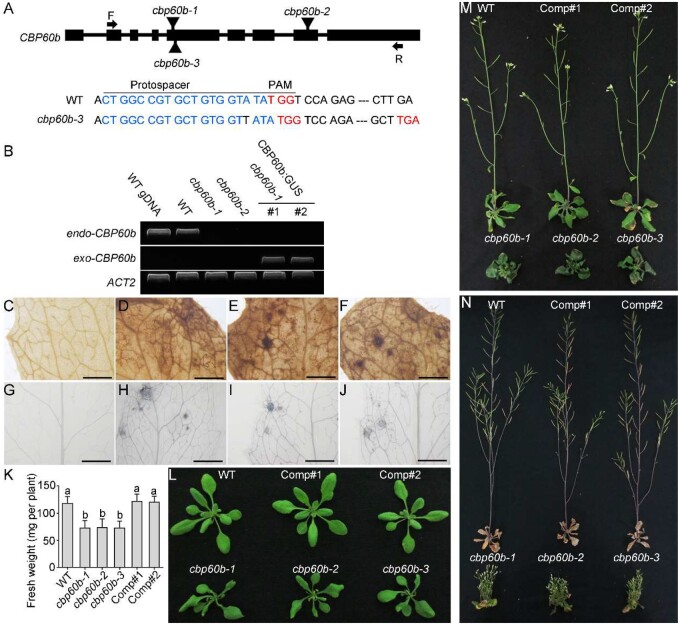
To determine the function of CBP60b, we took a reverse genetic approach by characterizing its mutants. Two T-DNA insertion lines were obtained from stock centers, cbp60b-1 (SAIL_40_E09) and cbp60b-2 (GK-521D01), both of which do not express the full-length transcript of CBP60b (Figure 1, A and B). We also generated a third allele, cbp60b-3, by CRISPR/Cas9-based genomic editing (Figure 1, A). The cbp60b plants were all smaller than those of wild-type (WT) at the same developmental stages (Figure 1, K–N). By examination of leaves at 3 weeks after germination (WAG) with 3,3′-diaminobenzidine (DAB) staining or Trypan blue staining, we determined that the cbp60b mutant leaves contained a high level of hydrogen peroxide (Figure 1, D–F) and showed enhanced cell death (Figure 1, H–J) compared with that of WT (Figure 1, C and G). The cbp60b mutants failed to transition from vegetative to reproductive growth, unlike WT (Figure 1, M). At late stages, WT plants had almost finished reproductive growth such that they set seeds and rosette leaves senesced (Figure 1, N). However, the cbp60b mutants were dwarfed and bushy (Figure 1, N). Introducing the genomic:GUS translational fusion construct CBP60b:GUS rescued the defects of cbp60b-1 (Figure 1, B and K–N), indicating that the phenotype was indeed caused by functional loss of CBP60b. Because all three mutants were comparable in phenotypes, the following experiments were performed mostly with cbp60b-1 unless noted otherwise.
Figure 1.
Functional loss of CBP60b compromised plant growth. A, Schematic illustration of CBP60b genomic locus. Boxes represent exons. T-DNA insertions (cbp60b-1 and cbp60b-2) and CRISPR/Cas9-generated cbp60b-3 are labeled on the genomic locus. A base pair insertion at the protospacer region caused a pre-stop codon of CBP60b in cbp60b-3. PAM sequences and stop codons are indicated in red; protospacer sequences are indicated in blue. B, Transcript analysis showing endogenous and exogenous CBP60b in various genetic backgrounds. C–F, Representative DAB staining for hydrogen peroxide in WT (C), cbp60b-1 (D), cbp60b-2 (E), or cbp60b-3 (F). G–J, Representative Trypan blue staining for the level of cell death in WT (G), cbp60b-1 (H), cbp60b-2 (I), or cbp60b-3 (J). K, Fresh weight of the indicated genotypes. Values are means ± standard deviation (sd, n > 15). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). L–N, Representative plant growth at three WAG (L), five WAG (M), or seven WAG (N). Comp indicates CBP60b:GUS;cbp60b-1 lines. Bars = 1 mm. See also Supplemental Figures S1, S2.
CBP60b loss-of-function results in autoimmunity
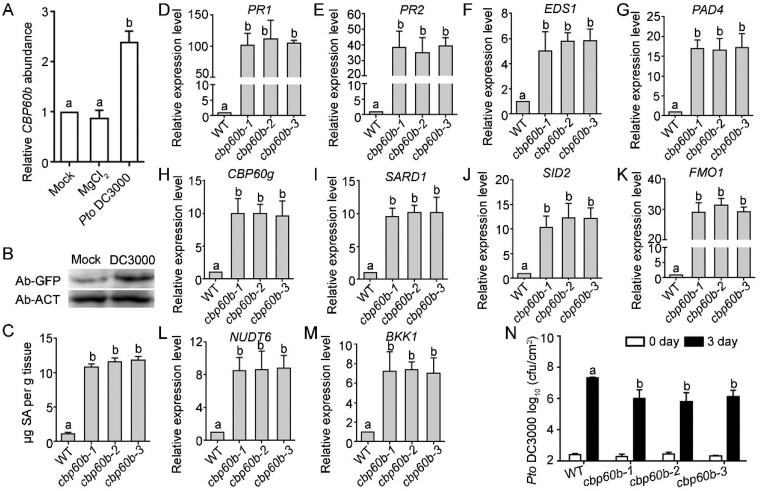
Phenotypes of the cbp60b mutants, such as small size, high H2O2, and increased cell death, resembled that of lesion mimic mutants (Bruggeman et al., 2015) and mutants of autoimmunity (van Wersch et al., 2016; Chakraborty et al., 2018). Since CBP60g and SARD1 are critical players in immunity (Sun et al., 2015), we investigated the possibility that functional loss of CBP60b resulted in autoimmunity. First, we examined whether CBP60b was transcriptionally regulated by pathogens, which is quite common for regulatory genes in immunity (Zhou and Zhang, 2020). Indeed, inoculation of Pseudomonas syringae pv. tomato (Pto) DC3000 significantly induced the expression of CBP60b when compared with that by mock or MgCl2 treatment (Figure 2, A). In comparison, the transcription of all four other CBP60s in the same subfamily was not substantially upregulated in response to DC3000 (Supplemental Figure S3). We confirmed the induction of CBP60b by Pto DC3000 on protein level by western blot assays (Figure 2, B). Autoimmunity is often associated with elevated SA levels (van Wersch et al., 2016). Next, by examining total SA in leaves of three WAG plants, we determined that SA levels were significantly elevated in the cbp60b mutants, when compared with that in WT (Figure 2, C). Third, elevated immunity involves enhanced expression of immune genes that are otherwise at basal levels (van Wersch et al., 2016). We examined the expression of immune response genes in WT versus in the cbp60b mutants by reverse transcription-quantitative PCRs (RT-qPCRs). PR1 and PR2 were highly expressed in the non-challenged cbp60b mutants, significantly different from that in WT (Figure 2, D–E). By RT-qPCRs, we determined that EDS1 (Figure 2, F), PAD4 (Figure 2, G), SID2 (Figure 2, J), and FMO1 (Figure 2, K) were all significantly upregulated in the cbp60b mutants, suggesting the activation of ETI pathway, of SA biosynthesis, and of the SAR pathway.
Figure 2.
Immune response is constitutively activated in CBP60b loss-of-function. A and B, Transcript analysis of CBP60b by RT-qPCRs (A) or western blot analysis showing protein abundance of CBP60b-GFP (in plants transformed with CBP60b genomic:GFP translational fusion construct (CBP60b:GFP)) at 24 h after infiltration (B). For A, values are means ± se (n = 3). Each biological replicates include leaves from five plants. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). Anti-ACTIN was used as the internal loading control in the Western blot assays. C, Total SAs in the indicated genotypes. Values are means ± sd (n = 4). Each biological replicate includes the fifth to sixth leaves from >10 plants. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). D–M, Relative transcript abundance of immune genes, including PR1 (D), PR2 (E), EDS1 (F), PAD4 (G), CBP60g (H), SARD1 (I), SID2 (J), FMO1 (K), NUDT6 (L), and BKK1 (M). Values are means ± se (n = 3). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). N, Sensitivity of the indicated genotypes to Pto DC3000. Plants at five WAG were infiltrated with Pto DC3000 (OD600 = 0.0001). Bacterial growth was determined 2 h post-inoculation (0 day) or 3 d post-inoculation (3 d). cfu, colony-forming unit. Values are means ± sd (n = 4). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). Experiment was repeated three times with similar results. See also Supplemental Figures S1–S3.
Because EDS1, PAD4, SID2, and FMO1 are target genes of CBP60g and SARD1 (Sun et al., 2015), we examined whether CBP60g and SARD1 were upregulated in the cbp60b mutants. It was indeed the case (Figure 2, H–I). Consistently, other downstream genes of CBP60g and SARD1, such as NUDT6 and BKK1, encoding either a negative or positive regulator in immunity (Sun et al., 2015), were significantly upregulated in the cbp60b mutants (Figure 2, L and M). Finally, we examined the sensitivity of the cbp60b mutants to Pto DC3000. Consistent with the elevated SA and immune gene expression levels, the cbp60b mutants contained significantly fewer bacteria colonies than WT at 3 d post-inoculation (Figure 2, N). These results indicated that functional loss of CBP60b caused autoimmunity.
CBP60b is a transcriptional activator of immune genes
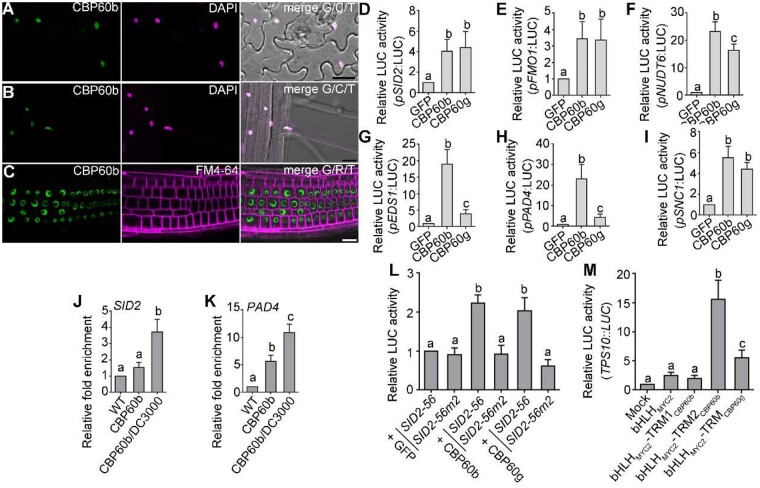
That functional loss of CBP60b resulted in autoimmunity suggested that CBP60b is a negative regulator of immunity. Since all members of the CBP60 family contain a conserved DNA-binding domain (DBD; Supplemental Figure S4, D), we investigated whether CBP60b also regulates transcription. By examining the subcellular localization of CBP60b-GFP in the CBP60b:GFP transgenic plants, we determined that CBP60b was targeted to the nucleus (Figure 3, A–C). Based on the analysis of DBD in SARD1 (Zhang et al., 2010b), we identified the DBD of CBP60b and generated a CBP60bΔ206-210-GFP construct in which five highly conserved amino acids (AAs) within the DBD were deleted to abolish its DNA binding (Supplemental Figure S4, D). The five amino acid deletion did not affect the nuclear targeting or stability of CBP60b based on fluorescence imaging (Supplemental Figure S5, B and C). However, introducing CBP60bΔ206-210-GFP into cbp60b could not rescue its growth defects (Supplemental Figure S5, A, D, and E), indicating that CBP60b functions through transcriptional regulation.
Figure 3.
CBP60b positively regulates immune gene expression. A–C, Representative CLSM of leaf pavement cells (A) from the CBP60b:GFP;cbp60b plants at three WAG, CLSM of root elongation zone (B) or root meristematic zone (C) from the CBP60b:GFP;cbp60b seedlings at five DAG. DAPI staining (magenta) was used to show the nuclei in (A and B). FM4-64 staining was used to show the PM in (C). A–C, bars = 20 μm. D–I, Quantitative luminescence comparison showing the LUC activity of pSID2:LUC (D), pFMO1:LUC (E), pNUDT6:LUC (F), pEDS1:LUC (G), pPAD4:LUC (H), and pSNC1:LUC (I). The promoter:LUC constructs were co-transfected with 35S:GFP (GFP), 35S:CBP60b-GFP (CBP60b), or 35S:CBP60g-HA (CBP60g). Values are means ± se (n = 4). J and K, ChIP-PCRs of CBP60b on SID2 (J) and PAD4 (K). ChIP signal was quantified as the percentage of total input DNA by qPCR, and normalized to corresponding fragment in WT (set as 1). Plants used are WT or cbp60b-1 complemented with the CBP60b genomic:GFP fusion construct (CBP60b:GFP;cbp60b, labeled on figure as CBP60b). For Pto DC3000 treatment, CBP60b:GFP;cbp60b plants at 5 WAG were infiltrated with Pto DC3000 (OD600 = 0.0001) 24 h before collecting and cross-linking with 1% formaldehyde. Values are means ± se (n = 3). L, Quantitative luminescence comparison. SID2-56:LUC or SID2-56m:LUC containing two mutations of gaaatt to gaaggg at the consensus motifs of pSID2 (SID2-56m2) was co-expressed with 35S:GFP, 35S:CBP60b-GFP, or 35S:CBP60g-HA. M, Quantitative luminescence comparison. TPS10::LUC was transfected with 35S:GFP (mock), 35S:bHLHMYC2-GFP, 35S:bHLHMYC2-TRM1CBP60b-GFP, 35S:bHLHMYC2-TRM2CBP60b-GFP, or 35S:bHLHMYC2-TRMCBP60g-GFP. LUC reporter activity was determined 16 h post-transfection. Values are means ± se (n = 4). From (D) to (M), different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). See also Supplemental Figures S4, S5.
Because CBP60b shares the conserved DBD with CBP60g and SARD1 (Supplemental Figure S4, D), we tested the possibility that CBP60b is able to bind to the same target genes as CBP60g and SARD1. The promoters of SID2 and PAD4 were demonstrated to be targets of CBP60g and SARD1 (Sun et al., 2015). Indeed, CBP60b-GFP was significantly enriched at the same sites of pSID2 and pPAD4 (Figure 3, J and K) as those targeted by CBP60g and SARD1 (Sun et al., 2015), based on ChIP-PCRs using the complemented plants expressing CBP60b-GFP in cbp60b.
CBP60b targeted the same downstream genes as CBP60g and SARD1 while immune response genes were ectopically expressed in CBP60b loss-of-function plants (Figure 2), suggesting CBP60b is a transcriptional repressor in immune gene expression. To test this hypothesis, we generated constructs expressing the LUC reporter gene under the control of the promoter of SID2 (Figure 3, D), FMO1 (Figure 3, E), NUDT6 (Figure 3, F), EDS1 (Figure 3, G), PAD4 (Figure 3, H), or SNC1 (Figure 3, I), all of which have been demonstrated to be targeted by CBP60g and SARD1 (Sun et al., 2015) or enriched with the G(A/T)AATT(T/G) motifs (Truman and Glazebrook, 2012; Sun et al., 2015). These reporter constructs were transformed into Arabidopsis protoplasts together with 35S:GFP, 35S:CBP60b-GFP, or 35S:CBP60g-HA (Qin et al., 2018) to examine the LUC reporter expression levels. For all six reporter constructs, the expression of CBP60g-HA resulted in a significantly higher LUC activity than that of GFP alone (Figure 3, D–I), consistent with the regulation of these genes by CBP60g (Sun et al., 2015; Qin et al., 2018). Surprisingly, CBP60b activates the expression of the same target genes as CBP60g (Figure 3, D–I). By contrast, co-expressing CBP60bΔ206-210-GFP did not induce LUC activities of the target promoters despite that the protein was substantially accumulated in the nucleus (Supplemental Figure S5), supporting that CBP60b is a transcriptional activator.
To provide further evidence that CBP60b targeted the promoters by recognizing the consensus motif of the CBP60 family (Truman and Glazebrook, 2012; Sun et al., 2015), we performed the LUC activity assays using a 56-bp sequence of pSID2 (SID2-56) and SID2-56m2, in which two consensus motifs were mutated (Sun et al., 2015). Indeed, CBP60b or CBP60g induced a significantly higher LUC activity in SID2-56:LUC than in SID2-56m2:LUC (Figure 3, L), supporting the hypothesis that CBP60b recognizes the consensus motif G(A/T)AATT(T/G). The LUC assays suggested that CBP60b is a transcriptional activator. To provide further evidence, we selected two transcriptional regulatory motifs (TRMs) of CBP60b to test in LUC activation assays, chosen according to a similar study on CBP60g (Qin et al., 2018). We generated constructs expressing TRM1CBP60b and TRM2CBP60b (Supplemental Figure S4, E) fused with the basic helix–loop–helix (bHLH) domain of MYC2 (bHLHMYC2) and GFP. The TERPENE SYNTHASE gene 10 (TPS10):LUC reporter, containing the promoter sequence of TPS10, was co-transformed with GFP, bHLHMYC2-GFP, bHLHMYC2-TRM1CBP60b-GFP, bHLHMYC2-TRM2CBP60b-GFP, or bHLHMYC2-TRMCBP60g-GFP. Co-expression with bHLHMYC2-TRM2CBP60b-GFP strongly activated the TPS10:LUC reporter whereas bHLHMYC2-GFP or bHLHMYC2-TRM1CBP60b-GFP did not (Figure 3, M). These results supported CBP60b being a transcriptional activator rather than repressor.
Overexpressing CBP60b results in constitutive immune responses
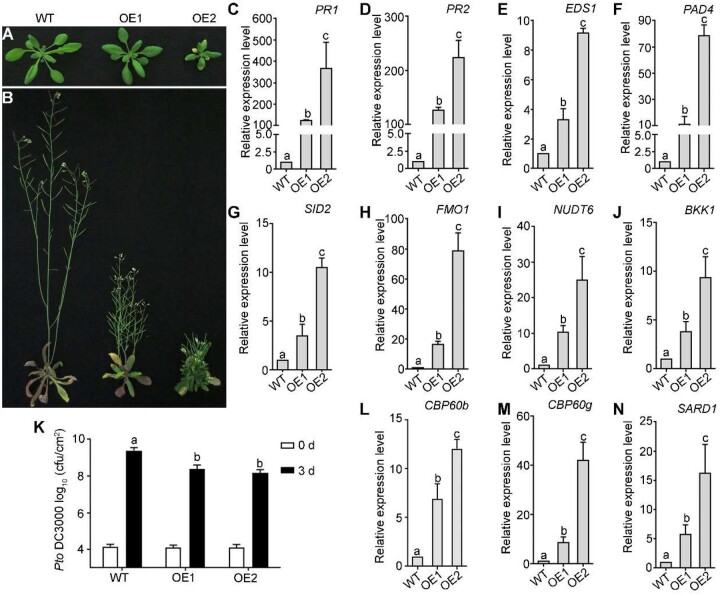
To provide further evidence supporting a positive role of CBP60b in immunity, we generated transgenic plants overexpressing CBP60b. Interestingly, overexpressing CBP60b-GFP (CBP60b-OE; Figure 4, L) resulted in growth morphologies similar to that of the cbp60b mutants, such that plants had smaller rosette leaves and were dwarf (Figure 4, A and B). CBP60b-OE lines exhibited enhanced expression of immune genes including PR1, PR2, EDS1, PAD4, CBP60g, SARD1, SID2, FMO1, NUDT6, and BKK1 (Figure 4, C–I, M, and N), supporting its role in transcriptional activation of immune response genes. At the same time, the OE plants were hyposensitive, compared with WT, to Pto DC3000 (Figure 4, K). These results supported CBP60b being a positive regulator in immune responses.
Figure 4.
Overexpressing CBP60b results in constitutive immune responses. A and B, Representative growth of WT and two independent lines overexpressing CBP60b-GFP (OE1 and OE2) at three WAG (A) or five WAG (B) under LD conditions. C–J, Relative transcript abundance of immune genes including PR1 (C), PR2 (D), EDS1 (E), PAD4 (F), SID2 (G), FMO1 (H), NUDT6 (I), and BKK1 (J). K, Sensitivity of the indicated genotypes to Pto DC3000. Plants at five WAG were infiltrated with Pto DC3000 (OD600 = 0.0001). Bacterial growth was determined 2 h post-inoculation (0 day) or 3 d post-inoculation (3 d). L, Transcript abundance of CBP60b in WT and two OE lines by RT-qPCRs. M and N, Relative transcript abundance of CBP60g (M) and SARD1 (N). Values are means ± se (n = 3) for (C)–(N). Experiment was repeated three times with similar results. From (C) to (N), different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). See also Supplemental Figures S4, S5.
Mutations at the EDS1-PAD4 pathway suppress the autoimmunity of CBP60b loss- but not gain-of-function
That CBP60b is a transcriptional activator for immune genes (Figures 3, 4) did not reconcile with the facts that immune genes were highly upregulated in the cbp60b mutants (Figure 2) and cbp60b mutants demonstrated autoimmunity (Figure 1). We thus considered the possibility that CBP60b is a guardee monitored by NLR proteins. According to the guard hypothesis, functionally impaired guardees activate cognate NLR proteins to induce ETI, leading to autoimmunity (Jones and Dangl, 2006). To test whether defects of the cbp60b mutants were due to guard-activation-induced ETI, we introduced eds1-1 (Parker et al., 1996) or pad4-1 (Dayadevi et al., 1999) into cbp60b, producing double mutants. Indeed, mutations at EDS1 or PAD4 completely suppressed the defective growth of cbp60b (Figure 5, A and B), strongly supporting the idea that autoimmunity of cbp60b had resulted from ectopic activation of the EDS1-PAD4 pathway. To provide further support, we mutated EDS1 or PAD4 in cbp60b by CRISPR/Cas9 (Supplemental Figure S6). Consistently, mutations at EDS1 or PAD4 fully suppressed the defective growth of cbp60b (Figure 5, A and B), the ectopic expression of SID2 and PR genes in cbp60b (Figure 5, I–K), and the hyposensitivity of cbp60b toward Pto DC3000 (Figure 5, G). By contrast, mutations at EDS1 and PAD4 did not affect the phenotype of CBP60b gain-of-function (Supplemental Figure S7), suggesting that autoimmunity observed in CBP60b gain-of-function was different from the autoimmunity in CBP60b loss-of-function. In addition, mutations at SAG101 did not affect the phenotype of cbp60b (Supplemental Figures S6, GS8, A). These results suggested that CBP60b loss-of-function resulted in autoimmunity via EDS1-PAD4-mediated TNL activation.
Figure 5.
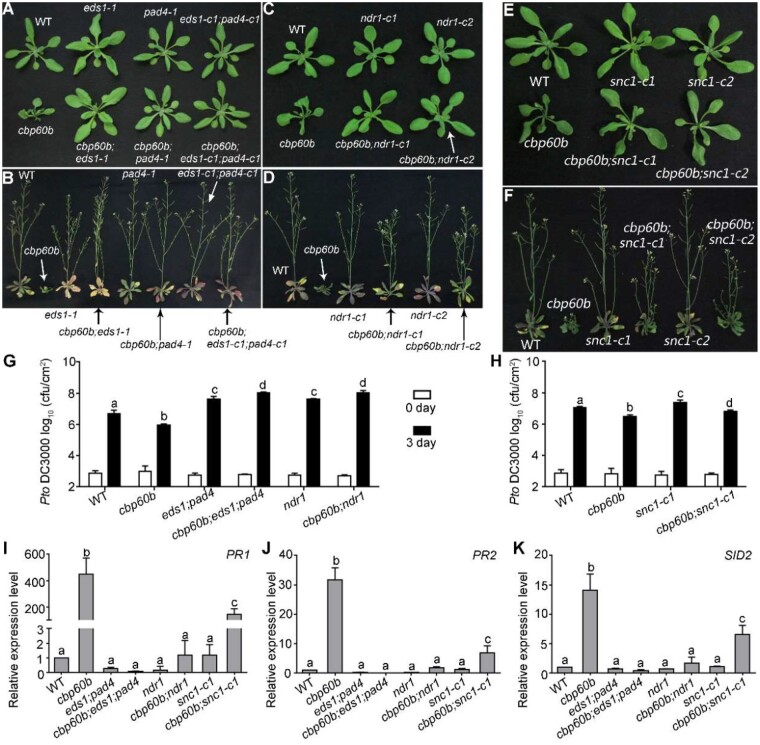
The autoimmunity of cbp60b was fully suppressed by mutations at EDS1-PAD4 and partially suppressed by mutations at NDR1 or SNC1. A and B, Representative growth of WT, cbp60b, eds1-1, pad4-1, eds1-c1;pad4-c1, cbp60b;pad4-1, cbp60b;eds1-1, and cbp60b;eds1-c1;pad4-c1 at three WAG (A) or five WAG (B) under LD conditions. C and D, Representative growth of WT, cbp60b, ndr1-c1, ndr1-c2, cbp60b;ndr1-c1, and cbp60b;ndr1-c2 at three WAG (C) or five WAG (D) under LD conditions. Arrows in (A–D) were used to point at the plant with specified genotype. E and F, Representative growth of WT, cbp60b-1, snc1-c1, snc1-c2, cbp60b-1;snc1-c1, and cbp60b-1;snc1-c2 at three WAG (E) or five WAG (F) under LD conditions. G and H, Sensitivity of the indicated genotypes to Pto DC3000. Plants at five WAG were infiltrated with Pto DC3000 (OD600 = 0.0001). Bacterial growth was determined 2 h post-inoculation (0 day) or 3 d post-inoculation (3 d). Values are means ± sd (n = 4). Experiment was repeated three times with similar results. I–K, Relative transcript abundance of PR1 (I), PR2 (J), and SID2 (K). Values are means ± se (n = 4). RNAs were extracted from leaves of three WAG plants under LD conditions. Results are means ± se (n = 3). Different letters in (G–K) indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). See also Supplemental Figures S6–S10.
To provide further evidence supporting the involvement of the EDS1-PAD4 pathway in the monitoring of CBP60b and to provide more details about the pathway involved, we mutated different hNLR genes in cbp60b (Supplemental Figure S6). Functional loss of the three ADR1 family members, but not the two NRG1s, suppressed the autoimmunity phenotype of cbp60b (Supplemental Figure S8), confirming that autoimmunity of cbp60b depends on EDS1-PAD4-mediated TNL activation. On the other hand, mutations of the CNL pathway, that is functional loss of NDR1 (Supplemental Figure S6), partially rescued the defective growth of cbp60b such that rosettes of the cbp60b;ndr1 double mutant were comparable to those of WT during vegetative growth, but the cbp60b;ndr1 double mutant was semi-dwarf during reproductive growth (Figure 5, C and D). Consistent with the recovered morphology, functional loss of NDR1 significantly suppressed the ectopic expression of PR genes (Figure 5, I and K>) and hyposensitivity to Pto DC3000 (Figure 5, G) in cbp60b. These results suggested that the CNL pathway is also involved in the monitoring of CBP60b. These results supported the idea that CBP60b loss-of-function resulted in autoimmunity through EDS1-PAD4- and NDR1-mediated TNL and CNL activation.
Functional loss of SNC1 partially rescued the autoimmunity of cbp60b
One of the R genes, SNC1, genetically interacts with CBP60g and SARD1 such that introducing snc1, a SNC1 gain-of-function mutant, into cbp60g;sard1 resulted in growth defects reminiscent of autoimmunity (Sun et al., 2018). Introducing snc1 into cbp60b resulted in a significant increase of PR genes and further reduction of plant growth (Supplemental Figure S9), suggesting a synergistic interaction between CBP60b and SNC1. Thus, we generated SNC1 loss-of-function mutations in cbp60b by CRISPR/Cas9 (Supplemental Figure S9). Indeed, the cbp60b;snc1-c1 and cbp60b;snc1-c2 double mutants grew substantially better than the cbp60b single mutant (Figure 5, E and F). SNC1 loss-of-function also significantly reduced the elevated expression of SID2 and PR genes in cbp60b (Figure 5, I–K). Consistent with the reduced PR genes, the cbp60b;snc1-c1 double mutants were hypersensitive to Pto DC3000 compared with cbp60b (Figure 5, H). These results suggested that SNC1 is one of the R genes monitoring the CBP60b pathway or that SNC1 might be involved in the amplification of NLR signaling in cbp60b.
CBP60b is redundant with, yet distinct from, CBP60g and SARD1 in immunity
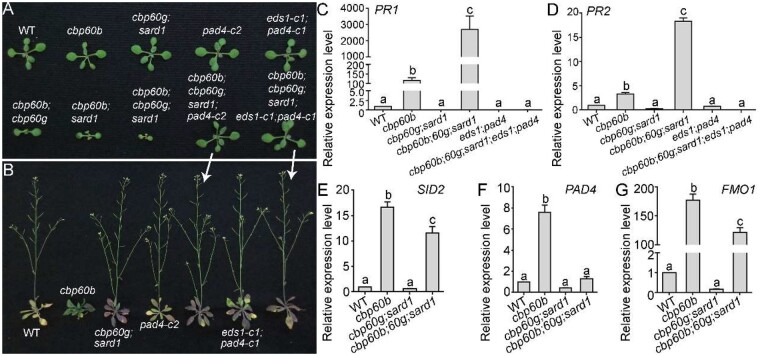
CBP60b targets the same immune genes, while positively regulating their expression, as do CBP60g and SARD1, suggesting functional redundancy among the three CBP60 members. To examine their genetic relationships, we generated a hierarchy of mutants of CBP60b, CBP60g, and SARD1. Introducing either cbp60g or sard1 (Zhang et al., 2010b) into cbp60b caused a more severe phenotype while the cbp60b;cbp60g;sard1 triple mutants were extremely weak and died after two WAG (Figure 6, A and B). The expression of PR genes was much higher in the triple mutant than in cbp60b (Figure 6, C and D), suggesting an enhanced autoimmunity in the cbp60b;cbp60g;sard1 triple mutant. The enhanced expression of SID2, FMO1, or PAD4 in cbp60b was abolished or significantly reduced in the cbp60b;cbp60g;sard1 triple mutant (Figure 6, E–G), suggesting that upregulated expression of immune genes in cbp60b partially depended on the upregulation of CBP60g and SARD1 (Figure 2, H and I).
Figure 6.
CBP60b is redundant with CBP60g and SARD1 in immunity. A and B, Representative WT, cbp60b, cbp60g;sard1, pad4-c2, eds1-c1;pad4-c1, cbp60b;cbp60g, cbp60b;sard1, cbp60b;cbp60g;sard1, cbp60b;cbp60g;sard1;pad4-c2, and cbp60b;cbp60g;sard1;eds1-c1;pad4-c1 plants at two WAG (A) or five WAG (B) under LD conditions. Note that the cbp60b;cbp60g;sard1 triple mutant plants die around two WAG and thus a representative plant is not included in the image taken at five WAG. C–G, Relative transcript abundance of PR1 (C), PR2 (D), SID2 (E), PAD4 (F), or FMO1 (G) in the indicated genotypes. Values are means ± se (n = 4). RNAs were extracted from leaves of two WAG plants under LD conditions. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
To provide further evidence supporting that the severe phenotype of the cbp60b;cbp60g;sard1 triple mutant was due to enhanced autoimmunity, as would be expected if the three CBP60s are all monitored by EDS1-PAD4-dependent NLR signaling, we mutated EDS1 or PAD4 in the cbp60b;cbp60g;sard1 triple mutant. Indeed, mutations of EDS1 or PAD4 completely rescued the growth morphology and lethality of the cbp60b;cbp60g;sard1 triple mutant (Figure 6, A and B) as well as the ectopic expression of PR genes (Figure 6, C and D), supporting the redundancy among CBP60g, SARD1, and CBP60b being guardees of EDS1-PAD4 mediated NLRs.
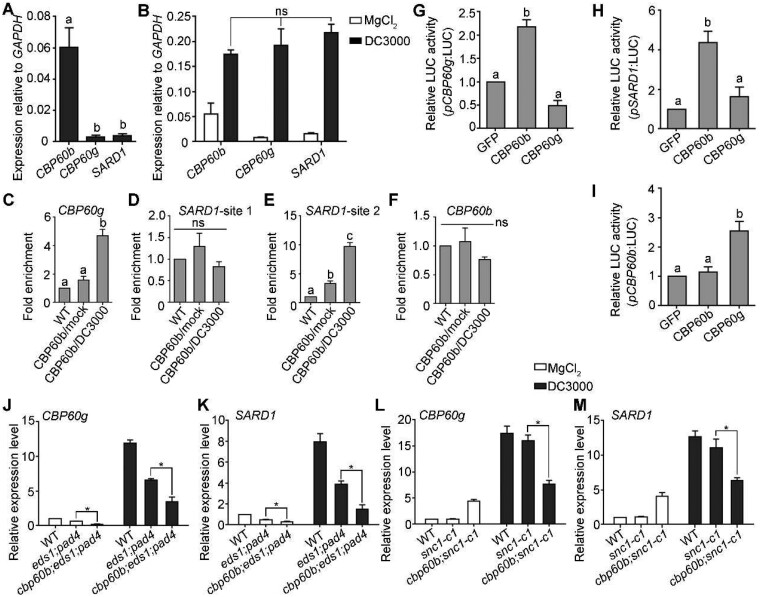
The redundancy between CBP60b and CBP60g or SARD1 predicts that cbp60g;sard1 and cbp60b would exhibit similar levels of autoimmunity, which is not the case (Zhang et al., 2010b; Wang et al., 2011). By examining the expression of these CBP60 members, we determined that the expression of CBP60b was much higher than that of CBP60g or SARD1 under non-challenged conditions (Figure 7, A), suggesting that CBP60b is the dominant CBP60 in non-challenged plants. That explains the lack of autoimmunity in the cbp60g;sard1 double mutant (Zhang et al., 2010a; Wang et al., 2011). Pto DC3000 induces expression of the three genes at comparable levels (Figure 7, B), suggesting comparable contributions of the three CBP60 members upon pathogen invasion.
Figure 7.
CBP60b plays a key role in immunity partially through direct regulation of CBP60g and SARD1. A, Expression of CBP60b, CBP60g, or SARD1 relative to GAPDH. RNAs were extracted from five WAG under SD conditions. B, Expression of CBP60b, CBP60g, or SARD1 relative to GAPDH. Plants at five WAG under SD conditions were infiltrated with 10 mM MgCl2 (mock) or DC3000 (OD600 = 0.0001) and RNAs were extracted from infiltrated leaves 24 h post-inoculation. C–F, ChIP-PCRs of CBP60b on CBP60g (C), SARD1 (D–E), and CBP60b (F). ChIP signal was quantified as the percentage of total input DNA by qPCR, and normalized to corresponding fragment in WT (set as 1). Plants used are cbp60b-1 complemented with the CBP60b genomic:GFP fusion construct (CBP60b:GFP;cbp60b). For Pto DC3000 treatment, CBP60b:GFP;cbp60b plants at five WAG were infiltrated with Pto DC3000 (OD600 = 0.0001) 24 h before collecting and cross-linking with 1% formaldehyde. ns, not significant. G–I, Quantitative luminescence showing the LUC activity of pCBP60g:LUC (G), pSARD1:LUC (H), or pCBP60b:LUC (I). The promoter:LUC constructs were co-transfected with 35S:GFP, 35S:CBP60b-GFP, or 35S:CBP60g-HA. From (A) to (I), values for are means ± se (n = 3). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). J–M, Relative transcript abundance of CBP60g (J, L) and SARD1 (K, M) upon mock treatment (MgCl2) or DC3000 treatment in designated genotypes. Plants at five WAG were infiltrated with MgCl2 or Pto DC3000 (OD600 = 0.0001). RNAs were extracted from infiltrated leaves at 24 h post-treatment. Results are means ± se (n = 3). Asterisks indicate significant difference (t test, P < 0.05).
Because both CBP60g and SARD1 were upregulated in CBP60b-OE lines (Figure 4) and the consensus motif targeted by CBP60b (this study), CBP60g, and SARD1 (Sun et al., 2015) was enriched in their own promoters (Supplemental Figure S4, B and C), we tested whether CBP60b activates the expression of CBP60g and SARD1. By ChIP-PCRs, we determined that CBP60b targeted the promoters of CBP60g and SARD1 at regions containing the consensus motif G(A/T)AATT(T/G) (Figure 7, C and D) but not its own promoter (Figure 7, F) despite the presence of the same motif (Supplemental Figure S4, A). Next, we generated constructs expressing the LUC reporter gene under the control of the promoter of CBP60g, SARD1, or CBP60b. These reporter constructs were co-transformed with 35S:GFP, 35S:CBP60b-GFP, or 35S:CBP60g-HA. Based on LUC reporter expression levels, CBP60b activated the expression of CBP60g and SARD1, but not of itself (Figure 7, G–I).
The EDS1-PAD4 pathway induces the upregulation of CBP60g and SARD1 upon pathogen invasion (Wang et al., 2011). To determine whether the upregulation of CBP60g and SARD1 in unchallenged cbp60b (Figure 2) was solely due to ectopic activation of the EDS1-PAD4-mediated NLR activation, we examined the expression of CBP60g and SARD1 in various genetic materials. Under normal growth conditions, the expression of CBP60g and SARD1 was significantly lower in cbp60b;eds1;pad4 than that in cbp60b (Figure 7, J–K), suggesting that the upregulation of CBP60g and SARD1 in the cbp60b mutant was indeed due to ectopic activation of EDS1-PAD4-mediated NLR activation. Importantly, CBP60g and SARD1 were both significantly lower in cbp60b;eds1;pad4 than that in eds1;pad4 (Figure 7, J and K), indicating an EDS1-PAD4-independent activation of CBP60g and SARD1 by CBP60b. Upon Pto DC3000 treatment, there was a significant reduction of CBP60g and SARD1 in cbp60b;eds1;pad4 compared with that in eds1;pad4 (Figure 7, J and K). In addition, both CBP60g and SARD1 expressions showed a significant reduction in the cbp60b;snc1-c1 double mutant compared with that in snc1-c1 (Figure 7, L and M). These results suggested that although the enhanced expression of CBP60g and SARD1 in cbp60b was induced by the activation of EDS1-PAD4-mediated NLR activation, CBP60b activates the expression of CBP60g and SARD1 under both non-challenged and challenged conditions in an EDS1-PAD4-independent way.
Discussion
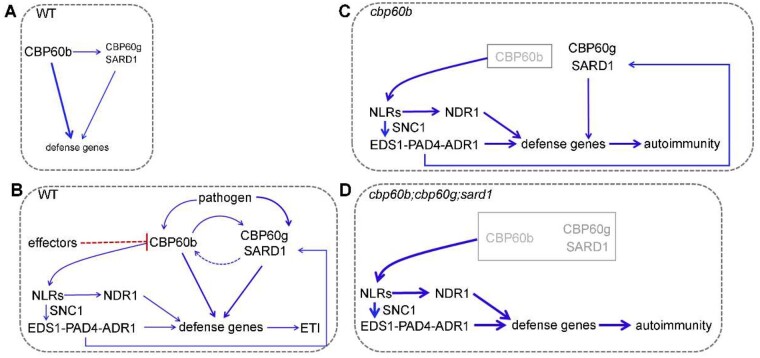
In this study, we report a central role of CBP60b in immunity (Figure 8). Under non-challenged conditions, CBP60b mediates the basal expression of immune genes including CBP60g and SARD1 so that plants maintain basal immunity without compromising resources for growth (Figure 8, A). Upon pathogen invasion, the three CBP60 genes, CBP60b, CBP60g, and SARD1, are induced and activated (Figure 8, B). CBP60b positively regulates immunity by directly and indirectly activating the expression of immune genes (Figure 8, B). Because CBP60b and its signaling pathways play such an important role in immunity, specific NLR proteins are used to monitor CBP60b and/or its signaling pathway to avoid immune deficiency caused by pathogen effectors whose identities remain to be determined (Figure 8, C). Functional loss of CBP60b or all three CBP60s induces autoimmunity by ectopic activity of NLRs, which was suppressed by functional loss of the EDS1-PAD4-ADR1s pathway, the NDR1 pathway, or the TNL-class R gene SNC1 (Figure 8, C and D). Although a previous study reported that the cbp60b-1 mutant was comparable with WT to P. syringae pv maculicola (Pma) strain ES4326 (Truman et al., 2013), the discrepancy is likely due to using Pto DC3000, which is more virulent than Pma ES4326 (Sreekanta et al., 2015), or different plant growth conditions. It is fairly common that lesion mimic (autoimmune) phenotypes vary a lot across the laboratories or plant materials used. We examined pathogen resistance using cbp60b leaves for which strong morphological defects appeared.
Figure 8.
CBP60b positively regulates immunity: a working model. Under normal growth conditions (A), CBP60b is the dominant CBP60 maintaining basal immunity. Pathogen invasion (B) induces transcriptional upregulation of all three CBP60bs while unknown pathogen effectors attack CBP60 (red dashed line). Pathogen attack induces activation of NLRs, which through the EDS1-PAD4-ADR1 and NDR1 pathway initiates ETI. In cbp60b (C) or cbp60b;cbp60g;sard1 (D), NLRs are strongly activated and plants exhibit autoimmunity. CBP60b regulates immunity partially through direct activation of CBP60g and SARD1. Thickness of the arrows indicates induction strength; blue arrows indicate positive action.
CBP60b plays a central role in the transcriptional regulation of plant immunity. First, by ChIP-PCRs and LUC assays, we showed that CBP60b directly bound to the promoters of immune genes, and its binding ability was substantially enhanced after pathogen invasion. We showed that CBP60b directly bound to the promoters of CBP60g and SARD1 (Figure 7), and activated their expression, in addition to genes involved in SA, SAR, ETI, and PTI pathways (Figure 3). These results supported a key role of CBP60b in the transcriptional regulation of immunity. Second, overexpressing CBP60b promoted the expression of immune genes and caused hyposensitivity to Pto DC3000 (Figure 4), similar to that caused by SARD1 overexpression (Zhang et al., 2010a). Third, genetic and phenotypic analysis of the cbp60b;cbp60g;sard1 triple mutant suggested a redundancy between CBP60b and CBP60g or SARD1 (Figure 6), supporting a positive regulatory role of CBP60b in immunity. Finally, the expression of CBP60g or SARD1 was significantly lower in the cbp60b;eds1;pad4 triple mutant than in the eds1;pad4 double mutants (Figure 7), supporting a CBP60b-dependent and EDS1-PAD4-independent transcriptional activation of CBP60g and SARD1.
Some key immune components, such as MEKK1-MKK1/MKK2-MPK4, SAUL1 (Senescence-associated E3 Ubiquitin Ligase 1), and Exo70B1 (a subunit of the exocyst complex), are guarded/monitored by NLR proteins (Zhao et al., 2015; Liang et al., 2019). Mutating the guardees led to autoimmunity, which was suppressed by introducing eds1 or pad4 or mutants of their cognate NLRs (Zhao et al., 2015; Liang et al., 2019). We propose that CBP60b may be a guardee targeted by effectors and monitored by NLRs. Several lines of evidence support this idea. First, CBP60b loss-of-function results in autoimmunity (Figure 2). Second, functional loss of the EDS1-PAD4-ADR1s or NDR1 pathways is fully suppressed, whereas that of the SNC1 partially suppressed, the autoimmunity of cbp60b (Figure 5). Third, mutations of all three CBP60s caused a strong autoimmunity that was fully suppressed by eds1 or pad4 (Figure 6). On the other hand, the downstream signaling components of CBP60b could also be guarded. The cbp60g;sard1 double mutant grew comparably to WT ( Zhang et al., 2010b; Wang et al., 2011) while the double mutation caused a severe autoimmunity phenotype when introduced into cbp60b (Figure 6), suggesting that their downstream components were monitored by NLRs. In addition, loss of SNC1 function only partially suppressed the autoimmunity of cbp60b (Figure 5), indicating that more TNL- and CNL-class R proteins are involved. It is interesting that the mutation of the DBD of CBP60b did not rescue the function of the protein as a negative regulator of immunity (Supplemental Figure S5). There are two possible explanations. First, the DBD-defective CBP60b may be able to activate R proteins. Alternatively, because the DBD-defective CBP60b fails to activate downstream genes (Supplemental Figure S5), whereas downstream components of CBP60b signaling could also be targeted by R proteins, the defective CBP60b, therefore, did not rescue its negative function in immunity. Identifying more guard NLRs by EMS mutagenesis or by overexpressing dominant-negative NLRs (Lolle et al., 2017) in the future may give better insights into the regulation of CBP60b and its signaling in immunity.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) Columbia-0 was used as the WT. Plant materials including sard1 (SALK_138476), cbp60g (SALK_023199), eds1-1, pad4-1, and snc1 were described previously (Parker et al., 1996; Dayadevi et al., 1999; Zhang et al., 2003, 2010a). The cbp60b-1 (SAIL_40_E09) and cbp60b-2 (GK-521D01-020205) mutants were obtained from Arabidopsis Biological Resource Center. The following mutants, including cbp60b-3, eds1-c1, pad4-c1, pad4-c2, ndr1-c1, ndr1-c2, sag101-c1, sag101-c2, adr1-c1, adr1-c2, adr1-L1-c1, adr1-L2-c1, adr1-L2-c2, nrg1a-c1, nrg1a-c2, nrg1b-c1, nrg1b-c2, snc1-c1, and snc1-c2, were generated using CRISPR-Cas9 as described (Supplemental Figure S6). Plants were grown in a growth chamber at 22°C under long day (LD) conditions (16-h light/8-h dark) or short day (SD) conditions (12-h light/12-h dark). Arabidopsis plants were transformed and selected as described (Zhou et al., 2013).
RT-PCRs and RT-qPCRs
RNAs were extracted from the fifth to sixth leaves of five WAG plants under SD conditions unless noted otherwise. For MgCl2 or Pto DC3000 treatment, plants at five WAG were infiltrated with 10 mM MgCl2 or Pto DC3000 (OD600 = 0.0001) and RNAs were extracted at 24 h after infiltration. Total RNAs were isolated using Ultrapure RNA Kit (Cwbiotech, Beijing, China) according to the instruction of manufacturers. Reverse transcription was performed with ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). RT-qPCRs were performed with the ABI QuantStudio 6 Flex using SYBR Green real-time PCR master mix (Toyobo). GAPDH was used as a quantitative control for RT-qPCRs. All experiments were repeated in three to four biological replicates. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). All primers are listed in Supplemental Table S1.
Plasmid construction
The genomic-GFP or GUS translational fusion constructs (CBP60b:GFP, CBP60b:GUS, CBP60c:GUS, CBP60d:GUS, CBP60e:GUS, and CBP60f:GUS) were generated by double digestion and ligation as follows: PCR fragments were amplified with the primer pair ZP5055/ZP5053 or ZP5055/ZP5054 for CBP60b (3531 bp), ZP7134/ZP7135 for CBP60c (3645 bp), ZP7128/ZP7129 for CBP60d (4362 bp), ZP7132/ZP7133 for CBP60e (3595 bp), and ZP7130/ZP7131 for CBP60f (3332 bp). The PCR fragments were inserted into the destination vector GW:GFP pre-digested with BamHI/ApaI or GW:GUS pre-digested with BamHI/XbaI using pEASY-Uni Seamless Cloning and Assembly Kit (Transgen Biotech, Beijing, China).
For the expression construct pCBP60b:GW-GFP, the sequence of CBP60b promoter (844 bp) was amplified with the primer pair ZP6570/ZP6571. PCR fragment was digested with SacI/SpeI and inserted into 35S:GW-GFP pre-digested with SacI/SpeI. The Gateway system (Invitrogen, Carlsbad, CA, USA) was used to generate pCBP60b:CBP60b-GFP and pCBP60b:CBP60bΔ206-210-GFP. The pENTR/D/TOPO vector (Invitrogen) was used to generate entry vectors. Primers for generating entry vectors were as follows: ZP7136/ZP7137 for CBP60b, ZP6576/ZP6577 for CBP60bΔ206-210. The entry vectors were used in LR reactions with the destination vector pCBP60b:GW-GFP to generate pCBP60b:CBP60b-GFP and pCBP60b:CBP60bΔ206-210-GFP.
For constructs used in dual LUC reporter assays in Arabidopsis protoplasts, 35S:CBP60b-GFP and 35S:CBP60bΔ206-210-GFP were generated by LR reactions. 35S:CBP60g-HA was described previously (Qin et al., 2018). A fragment encoding the bHLHMYC2 domain (446–525 AA) was amplified with the primer pair ZP9701/ZP9719 ligated into pENTR/D/TOPO (Invitrogen). PCR fragments encoding TRM1CBP60b, TRM2CBP60b, or TRM1CBP60g (211–440 AA) were generated by using the primer pairs ZP9703/ZP9704, ZP9703/ZP9808, and ZP9705/ZP9706, respectively. The PCR fragments were inserted into the entry vector containing coding sequences of bHLHMYC2 to generate entry vectors containing bHLHMYC2-TRM1CBP60b, bHLHMYC2-TRM2CBP60b, or bHLHMYC2-TRM1CBP60g using pEASY-Uni Seamless Cloning and Assembly Kit. The corresponding entry vectors were used in LR reactions with the destination vector 35S:GW-GFP to generate 35S:bHLHMYC2-GFP, 35S:bHLHMYC2-TRM1CBP60b-GFP, 35S:bHLHMYC2-TRM2CBP60b-GFP, or 35S:bHLHMYC2-TRM1CBP60g-GFP. Promoter fragments used in LUC reporter assays were generated by corresponding primer pairs (Supplemental Table S1). PCR fragments of these promoters were inserted into pGreen-0800 vector pre-digested with KpnI/SalI (Hellens et al., 2005) using pEASY-Uni Seamless Cloning and Assembly Kit to generate LUC reporter constructs. The 56 bp sequence of pSID2 (SID2-56) and that containing two mutated consensus motifs (SID2-56m2) were synthesized in vitro and inserted into pGreen-0800 as described (Hellens et al., 2005).
CRISPR/Cas9 constructs were generated with gene-specific sequences amplified with corresponding primers (Supplemental Table S1) and with pCBC-DT1T2 as the template (Xing et al., 2014). PCR fragments were inserted into pHEE401E using restriction–ligation reactions as described (Wang et al., 2015). Sequencing with target-specific primers (Supplemental Table S1) was performed to verify genomic editing of the targets. For the generation of mutants by the CRISPR-Cas9 genome editing, gene-specific Cas9 constructs were first introduced into the heterozygous cbp60b-1 plants (cbp60b-1/+). T1 plants with the cbp60b-1/+ background were selected. From progenies of these T1 plants, WT or the homozygous cbp60b-1 plants with the same mutation/transgene were used for genetic comparation. In the following generations, the Cas9 vector was cleared out from the background.
All PCR amplifications used Phusion hot start high-fidelity DNA polymerase at the annealing temperature and extension times recommended by the manufacturer. The Bioneer PCR purification kit and Bioneer Spin miniprep kit were used for PCR product recovery and plasmid DNA extraction, respectively. All constructs were sequenced and analyzed using Vector NTI. Primers are listed in Supplemental Table S1.
Pathogen inoculation and bacterial growth assays
Pto DC3000 strains were cultured at room temperature in King’s B medium (protease peptone, 10 mg/mL; glycerol, 15 mg/mL; K2HPO4, 1.5 mg/mL; MgSO4, 5 mM, pH 7.0) supplemented with 12.5 mg/mL rifampicin as described (Wang et al., 2009). Plants at four WAG were used for inoculation. Pto DC3000 suspensions in 10 mM MgCl2 (OD600 = 0.0001) were infiltrated into the abaxial sides of mature leaves using a needleless 1-mL syringe. Determination of bacterial titers was as described (Feys et al., 2005).
Dual LUC reporter assays in Arabidopsis protoplasts
Arabidopsis protoplasts were co-transformed with various combinations of effectors and reporters using the PEG method as described (Yoo et al., 2007). LUC activity was tested with a Double-LUC Reporter Assay Kit (TransGen Biotech) using the Dual-Light Chemiluminescent Reporter Gene Assay System (Berthold, Baden-Wuerttemberg, Germany). The ratio of LUC driven by the promoters to Renilla LUC driven by the 35S promoter was calculated to determine the transcriptional activities, as described (Tian et al., 2020). All experiments were repeated in three to four biological replicates. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
ChIP
The EpiQuik Plant ChIP kit (Epigentek, Farmingdale, NY, USA; P-2014) was used to perform the ChIP assays. Approximately 1 g of leaf tissue was harvested from uninoculated or Pto DC3000 (OD600 = 0.0001)-inoculated CBP60b:GFP;cbp60b-1 plants at 24 h after infiltration. Chromatin was immunoprecipitated with an anti-GFP antibody (TransGen Biotech) 24 h post-inoculation. The primers used for qPCRs are listed in Supplemental Table S1. The primers for PAD4 and SID2 were as described (Sun et al., 2015). All experiments were repeated in three biological replicates. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
Chemical staining
Histochemical GUS staining was performed as described (Xing et al., 2014). For SAM staining, plants at five DAG were incubated for 6 h in the dark with 5-bromo-4-chloro-3-indolyl-d-GlcUA, followed by fixation, clearing, and paraffin embedding (Sigma–Aldrich, St. Louis, MO, USA). For FM4-64 staining of roots, roots of five DAG seeding were dipped in liquid MS media supplemented with 4 µM FM4-64 for 5 min, washed out twice with liquid MS medium without FM4-64, and examined under a confocal microscope. 4′,6-Diamidino-2-phenylindole (DAPI) staining of leaf pavement cells were performed as followed: leaf epidermal peels were incubated with 1 ng/mL DAPI for 10 min before imaging. For Trypan blue staining, leaves of two to three WAG were boiled in Trypan blue (Beyotime Biotech, Shanghai, China) for 2 min, destained in 4 g/mL chloral hydrate, and then placed on slides in 50% glycerol for visualization. For DAB staining, leaves were vacuum-infiltrated for 15 min with 1 mg/mL DAB solution, placed in the dark for 16 h, and then destained in 90% ethanol before imaging. For ROS and cell death staining, the fifth to sixth true leaves were collected from plants at three WAG under LD conditions.
Western blot assays
Western blot analysis was performed as described (Li et al., 2018). Anti-GFP antibody (TransGen Biotech, HT801-01, 1:2,000 dilution) was used to recognize CBP60b-GFP while anti-ACTIN (TransGen Biotech, HC201-01, 1:2,000 dilution) was used as the internal loading control.
Fluorescence imaging
Fluorescence imaging of CBP60b-GFP or its variants was performed with Zeiss LSM880 (Zeiss, Oberkochen, Germany) confocal laser-scanning microscope (CLSM) with a 488-nm argon laser/BP 505–550 filter as described (Li et al., 2018).
Quantification of SA levels
For SA quantification, leaves of five WAG plants under SD conditions were harvested and SA levels were analyzed using high-pressure liquid chromatography.
Accession numbers
All genes involved can be found in TAIR under the following accession numbers: AT5G57580 for CBP60b, AT2G18750 for CBP60c, AT4G25800 for CBP60d, AT2G24300 for CBP60e, AT4G31000 for CBP60f, AT5G26920 for CBP60g, AT1G73805 for SARD1, AT3G48090 for EDS1, AT3G52430 for PAD4, AT5G14930 for SAG101, AT3G20600 for NDR1, AT1G33560 for ADR1, AT4G33300 for ADR1-L1, AT5G04720 for ADR1-L2, AT5G66900 for NRG1a, AT5G66910 for NRG1b, AT1G74710 for SID2, AT1G19250 for FMO1, AT2G14610 for PR1, AT3G57260 for PR2, AT2G04450 for NUDT6, AT2G45760 for BAP2, AT2G13790 for BKK1/SERK4, AT1G32640 for MYC2, AT2G24210 for TPS10, and AT4G16890 for SNC1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. CBP60b is expressed in diverse tissues and developmental stages.
Supplemental Figure S2. Expression of the other CBP60b subfamily members by histochemical GUS staining of genomic:GUS reporter lines.
Supplemental Figure S3. Expression of the other CBP60b subfamily members is not responsive to Pto DC3000.
Supplemental Figure S4. Analysis of CBP60 promoters and their protein-coding domains.
Supplemental Figure S5. CBP60b is a transcriptional activator.
Supplemental Figure S6. Generation of mutants by CRISPR/Cas9.
Supplemental Figure S7. Mutations at EDS1 or PAD4 did not affect the phenotype of CBP60b gain-of-function.
Supplemental Figure S8. Autoimmunity of cbp60b depends on the EDS1-PAD4-ADR1s pathway, but not EDS1-SAG101-NRG1s pathway.
Supplemental Figure S9. SNC1 gain-of-function enhanced the autoimmunity of cbp60b.
Supplemental Table S1. Oligos used in this study.
Supplementary Material
Acknowledgments
The authors thank Profs. Jianmin Zhou for constructive comments, Jie Zhang for the CBP60g-HA construct and the cbp60g;sard1 double mutant, Gang Li for pGreenII 0800 construct, and Zhaohui Chu for eds1-1 and pad4-1. They also thank ABRC for other mutant seeds. The authors declare that there is no conflict of interests.
L.-S.L. initiated the project. L.-S.L. conducted all experiments with the technical assistance of J.Y., E.L., T.M., M.L., L.-M.G., and G.W. S.L. and Y.Z. supervised this project. S.L., Y.Z., and L.-S.L. wrote the article. S.L. secured the funding.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Sha Li (shali@sdau.edu.cn).
Funding
This research was supported by the Natural Science Foundation of China (31771558 and 31970332 to S.L.).
Conflict of interest statement. None declared.
References
- Bi D, Johnson KC, Zhu Z, Huang Y, Chen F, Zhang Y, Li X (2011) Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front Plant Sci 2:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG (2019) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222:966–980 [DOI] [PubMed] [Google Scholar]
- Chakraborty J, Ghosh P, Das S (2018) Autoimmunity in plants. Planta 248:751–767 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108:14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66:487–511 [DOI] [PubMed] [Google Scholar]
- Dayadevi J, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96:13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, Redkar A (2018) Pathogens suppress host transcription factors for rampant proliferation. Trends Plant Sci 23:950–953 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11:539–548 [DOI] [PubMed] [Google Scholar]
- Dong OX, Tong M, Bonardi V, El Kasmi F, Woloshen V, Wunsch LK, Dangl JL, Li X (2016) TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol 210:960–973 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17:2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50:82–94 [DOI] [PubMed] [Google Scholar]
- Knepper C, Savory EA, Day B (2011) The role of NDR1 in pathogen perception and plant defense signaling. Plant Signal Behav 6:1114–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019) A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31:2430–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Cui Y, Ge FR, Chai S, Zhang WT, Feng QN, Jiang L, Li S, Zhang Y (2018) AGC1.5 kinase phosphorylates RopGEFs to control pollen tube growth. Mol Plant 11:1198–1209 [DOI] [PubMed] [Google Scholar]
- Li X, Kapos P, Zhang YL (2015) NLRs in plants. Curr Opin Immunol 32:114–121 [DOI] [PubMed] [Google Scholar]
- Liang W, van Wersch S, Tong M, Li X (2019) TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol 221:2054–2066 [DOI] [PubMed] [Google Scholar]
- Lolle S, Greeff C, Petersen K, Roux M, Jensen MK, Bressendorff S, Rodriguez E, Somark K, Mundy J, Petersen M (2017) Matching NLR immune receptors to autoimmunity in camta3 mutants using antimorphic nlr alleles. Cell Host Microbe 21:518–529 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Andem F, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasíiíca specified by severa1 different RPP genes. Plant Cell 8:2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, Zhang Y (2018) Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact 31:403–409 [DOI] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115:E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wang K, Sun L, Xing H, Wang S, Li L, Chen S, Guo HS, Zhang J (2018) The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 7: e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J (2012) MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci 3:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, El Ghoul H, Mundy J, Petersen M (2016) Making sense of plant autoimmunity and ‘negative regulators’. FEBS J 283:1385–1391 [DOI] [PubMed] [Google Scholar]
- Sreekanta S, Bethke G, Hatsugai N, Tsuda K, Thao A, Wang L, Katagiri F, Glazebrook J (2015) The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol 207:78–90 [DOI] [PubMed] [Google Scholar]
- Sun T, Liang W, Zhang Y, Li X (2018) Negative regulation of resistance protein-mediated immunity by master transcription factors SARD1 and CBP60g. J Integr Plant Biol 60:1023–1027 [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y (2015) ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat Commun 6:10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Hamano K, Takagi H, Morimoto T, Akimitsu K, Terauchi R, Shirasu K, Ichimura K (2019) Disruption of the MAMP-induced MEKK1-MKK1/MKK2-MPK4 pathway activates the TNL immune receptor SMN1/RPS6. Plant Cell Physiol 60:778–787 [DOI] [PubMed] [Google Scholar]
- Tian T, Ma L, Liu Y, Xu D, Chen Q, Li G (2020) Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32:1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Glazebrook J ( 2012) Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol 12:1471–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Sreekanta S, Lu Y, Bethke G, Tsuda K, Katagiri F, Glazebrook J (2013) The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol 163:1741–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch R, Li X, Zhang YL (2016) Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front Plant Sci 7:1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14:619–630 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5:e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen le V, Katagiri F, Glazebrook J (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67:1029–1041 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8:383–389 [DOI] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X (2019) Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222:938–953 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:1471–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhu C, Cevik V, Johnson K, Liu Y, Sohn K, Jones JD, Holub EB, Li X (2015) Autoimmunity conferred by chs3-2D relies on CSA1, its adjacent TNL-encoding neighbour. Sci Rep 5:8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocol 2:1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15:2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010a) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107:18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Xu SH, Ding PT, Wang DM, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010b) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107:18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11:253–263 [DOI] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, Tang D (2015) A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet 11:e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhang YL (2020) Plant immunity: danger perception and signaling. Cell 181:978–989 [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y (2013) Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25:1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P (2011) SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog 7:e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20:10–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.