Abstract
Physical dormancy in seeds exists widely in seed plants and plays a vital role in maintaining natural seed banks. The outermost cuticle of the seed coat forms a water-impermeable layer, which is critical for establishing seed physical dormancy. We previously set up the legume plant Medicago truncatula as an excellent model for studying seed physical dormancy, and our studies revealed that a class II KNOTTED-like homeobox, KNOX4, is a transcription factor critical for controlling hardseededness. Here we report the function of a seed coat β-ketoacyl-CoA synthase, KCS12. The expression level of KCS12 is significantly downregulated in the knox4 mutant. The KCS12 gene is predominantly expressed in the seed coat, and seed development in the M. truncatula kcs12 mutant is altered. Further investigation demonstrated that kcs12 mutant seeds lost physical dormancy and were able to absorb water without scarification treatment. Chemical analysis revealed that concentrations of C24:0 lipid polyester monomers are significantly decreased in mutant seeds, indicating that KCS12 is an enzyme that controls the production of very long chain lipid species in the seed coat. A chromatin immunoprecipitation assay demonstrated that the expression of KCS12 in the seed coat is directly regulated by the KNOX4 transcription factor. These findings define a molecular mechanism by which KNOX4 and KCS12 control formation of the seed coat and seed physical dormancy.
Analysis of mutants in the model legume Medicago truncatula reveals a role for a seed coat fatty acid elongase complex subunit in seed physical dormancy.
Introduction
Seed physical dormancy, also named hardseededness, exists in at least 17 plant families, and is very common in legume species (Finch-Savage and Leubner-Metzger, 2006; Bolingue et al., 2010; Smýkal et al., 2014). Compared to physiological dormancy, which is controlled by endogenous hormones, physical dormancy is caused by a water-impermeable palisade cell layer covered with intact cuticles, which inhibits seed germination by blocking water and oxygen from the outside environment (Finkelstein et al., 2008; Graeber et al., 2012). Only after the cuticles are removed by scarification can the seeds absorb water and germinate. Physical dormancy, as an adaptive trait in seed plants, plays a crucial role in protecting seeds and maintaining seed banks in soil for various timespans, sometimes even for hundreds of years (Rolston, 1978).
In contrast to the abundant information on seed physiological dormancy from studies conducted in Arabidopsis thaliana, research on the molecular mechanisms underlying the formation of physical dormancy is very limited. One important reason is the lack of a suitable model plant system. In past years, Medicago truncatula has been used as a model legume to study compound leaf development and nodule development (Kang et al., 2016; Zhao et al., 2019; Zhou et al., 2019). We have noticed that M. truncatula seeds exhibit typical physical dormancy. Anatomical analysis in M. truncatula using light microscopy showed several structural layers in the seed coat region, from inside to outside: parenchyma cells, hourglass cells, and palisade cells covered with cuticular layer (Chai et al., 2016). The mature seeds of M. truncatula do not absorb water or germinate without scarification. With a population of 20,000 mutants tagged with a retrotransposon, M. truncatula has become an ideal model for studying physical dormancy. In a previous study, we identified a class II KNOTTED-like homeobox (KNOX4) mutant lacking physical dormancy, which exhibited dysfunctional palisade cuticles in the mutant seed (Chai et al., 2016). As a crucial transcription factor in physical dormancy, KNOX4 regulates expression of the cuticle pathway genes. In the knox4 mutant, the content of many lipid monomers including very long chain fatty acids (VLCFAs) significantly decreased, indicating enzymes related to biosynthesis of VLCFAs may be involved in the formation of physical dormancy.
VLCFAs, which contain more than 20 carbons (20C) and their derivatives, are usually present in most organisms. In plants, VLCFAs are generated by fatty acid elongase (FAE) complexes, which is formed from a β-ketoacyl-CoA synthase (KCS), a ketoacyl-CoA reductase (KCR), a hydroxyacyl-CoA dehydratase (HCD), and an enoyl-CoA reductase (ECR; Fehling and Mukherjee, 1991; Haslam and Kunst, 2013). All FAE complexes share the same KCR, HCD, and ECR enzymes, while different KCSs determine the substrate and product specificities. KCSs are also thought to be the rate-limiting enzymes in the entire process catalyzed by the FAE complex. There are 21 KCS genes in Arabidopsis genome and they each showed different catalytic activities. KCS1 affects wax formation by catalyzing the elongation of multiple fatty acids (Todd et al., 1999); KCS2 and KCS20 are functionally redundant in cuticular wax and root suberin biosynthesis (Lee et al., 2009); KCS6 is involved in stem and leaf wax biosynthesis as well as tryphine/pollen coat synthesis in the tapetum (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002); KCS9 is involved in the synthesis of cuticular waxes, suberin, sphingolipids, and phospholipids (Kim et al., 2013); KCS10 suppresses epidermal cell interaction in Arabidopsis during vegetative development (Yephremov et al., 1999; Pruitt et al., 2000); the mutant plants of KCS13 exhibit much higher stomatal density at elevated CO2 (Lake et al., 2002); KCS16 catalyzes the elongation of C34–C38 acyl-CoAs in Arabidopsis leaf trichrome (Hegebarth et al., 2017). Recently, a homolog of Arabidopsis FDH/KCS10 was found to be involved in lateral organ development and cuticular wax synthesis in M. truncatula (Yang et al., 2021). AD1, an epidermally expressed KCS gene, is involved in cuticular wax biosynthesis in maize (Liu et al., 2020). Although certain KCS enzymes have been associated with the elongation of fatty acyl-CoAs to C20 and longer, to date, no functional KCS gene involved in seed coat development has been reported. It is not clear whether any KCS gene is associated with seed physical dormancy, and the regulatory network of physical dormancy formation remains largely elusive.
In this study, we identified a seed coat-specific KCS gene involved in physical dormancy formation in M. truncatula and named it KCS12 according to phylogenetic analysis. KCS12 is predominantly expressed in M. truncatula seed coat. The loss of function of KCS12 caused the mutant seeds to lose their physical dormancy and absorb water easily without scarification. The C24 very long chain lipid monomer composition significantly decreased in the kcs12 mutant. The expression of KCS12 is under the control of KNOX4, a key regulator for physical dormancy formation. Our data imply that VLCFAs play an important role in the formation of physical dormancy by controlling composition integrity of the seed coat cuticle.
Results
Identification of the KCS gene that specifically expresses in the seed coat of M. truncatula
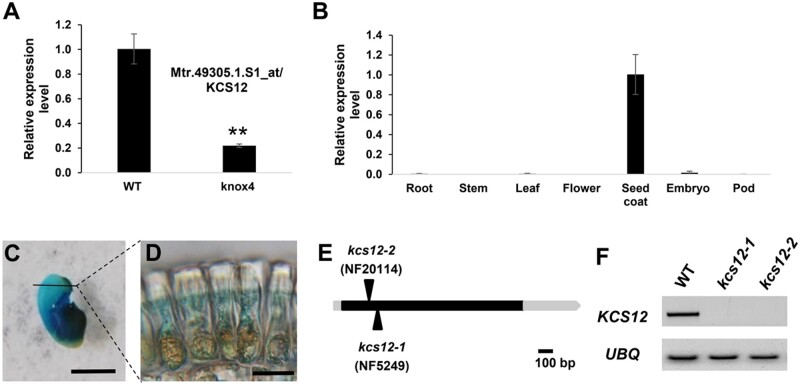
While analyzing the knox4 mutant in M. truncatula (Chai et al., 2016), we found that the expression level of one KCS (Mtr.49305.1.S1_at) gene was significantly decreased in the seed coat of knox4 (Figure 1A). We named this gene M. truncatula KCS12 with reference to its closest KCS ortholog in Arabidopsis (Supplemental Figure S1). Based on current sequencing information, there are 28 KCS members in the M. truncatula genome. According to the M. truncatula gene expression atlas (mtgea.noble.org/v2), KCS12 has a relatively high expression level in seed coat tissue compared to other KCS genes (Supplemental Figure S2). To confirm the expression pattern of the KCS12 gene, total RNAs were isolonated from root, stem, leaf, flower, pod, and seed coat. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis showed that KCS12 has the highest expression in the seed coat (Figure 1B). To detect detailed spatial expression of MtKCS12 in seed coat, we isolated the MtKCS12 promoter, fused it with the β-glucuronidase reporter gene and introduced the construct (MtKCS12 promoter::GUS) into wild-type M. truncatula. Strong GUS expression was observed in the seed coat (Figure 1C) and the palisade cells (Figure 1, C and D).
Figure 1.
Isolation and characterization of kcs12 mutant in M. truncatula. A, Relative expression levels of KCS12 in seed coat of wild-type and knox4 mutant, determined by RT-qPCR. B, RT-qPCR analysis of KCS12 expression in different plant organs. C, Expression analysis of KCS12 promoter::GUS construct in transgenic plant, bar = 0.2 cm. D, Magnification of cross section from the black line in (C) showing outmost epidermal palisade cells, bar = 10 µm. E, Schematic diagram of the gene structure of KCS12 and Tnt1 insertion sites. 5′- and 3′-untranslated regions and coding region of KCS12 gene are labelled with gray boxes and black box, respectively. Vertical arrows mark the locations of Tnt1 retrotransposon in the kcs12 mutant. F, RT-PCR analysis of KCS12 transcript levels in the wild-type and kcs12 mutants, UBQ was used as the loading control. Asterisks indicate significant differences between mutant and WT (**P < 0.01) in pairwise comparisons by a two-tailed Student’s t test.
Quantitative analysis of the kcs12 mutant seed developmental phenotype
To investigate the function of KCS12 in M. truncatula, two Tnt1 insertion lines (NF5249, kcs12-1; NF20114, kcs12-2) were identified from the M. truncatula mutant collection. Sequencing of the Tnt1 insertion products showed that both Tnt1 insertions were located in the exon of KCS12 (Figure 1E). To examine the expression levels of KCS12 in these two Tnt1 insertion lines, total RNA was isolated from the wild-type, kcs12-1 and kcs12-2 mutant; RT-PCR analysis confirmed that no KCS full transcript was produced in either mutant (Figure 1F).
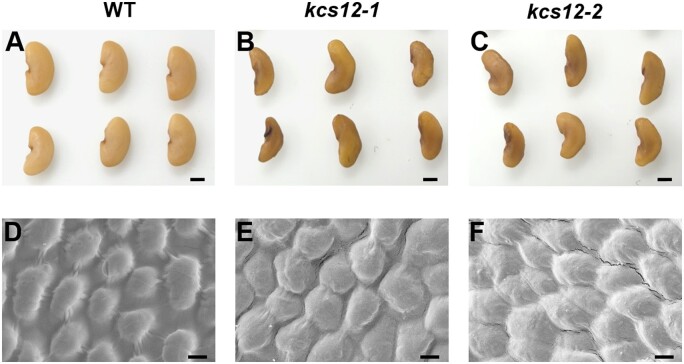
The kcs12-1 and kcs12-2 mutant plants did not exhibit any obvious abnormal growth phenotypes. Interestingly, we noticed that the kcs12 mutant mature seeds exhibited an irregular seed shape phenotype (Figure 2, A–C). Scanning electron microscopy (SEM) was employed to detect the changes in seed coat surface morphology between the kcs12 mutant and wild-type, and showed the surface of wild-type seeds was very smooth with regularly arranged dome-like epidermal cells (Figure 2D), while the mutant seed surface was slightly irregular with wrinkles (Figure 2, E and F).
Figure 2.
Seed phenotype of wild-type and kcs12 mutants in M. truncatula. A–C, Morphologies of mature seeds from wild-type and kcs12 mutants, bars = 1 mm. D–F, SEM observations of mature seed coats from wild-type and kcs12 mutant seeds. Wild-type seeds exhibited smooth, dome-like epidermal cells, while kcs12 mutants seeds displayed irregular epidermal cells with wrinkles, bars = 2 µm.
To quantitatively analyze the changes of seed size and shape, we took into account various parameters of seed: maximum length, waist width, perimeter, area, ratio of length and width (LWR), and seed shape compactness (4*π∗s/(p∗p); Supplemental Figure S3). Overall, for kcs12-1 and kcs12-2, the seed lengths of the two mutants were 98% and 94% of the wild-type seeds, respectively. The widths of the mutant seeds were 81% of the wild-type seeds. The LWRs of the mutant seeds were 118% and 113% of the wild-type seeds, respectively. The perimeters of the mutant seeds were 93% and 92% of the wild-type seeds, respectively. The areas of the mutant seeds were 80% and 79% of the wild-type seeds, respectively. The seed shape compactness measurements of the mutants were 92% and 93% of the wild-type seeds, respectively. Moreover, the seed-weights of the two kcs12 mutants were 64% and 57% of that of the wild-type, respectively (Supplemental Figure S4). These data indicate that the loss-of-function of KCS12 impacts seed morphology and weight.
Loss of seed physical dormancy in the kcs12 mutant
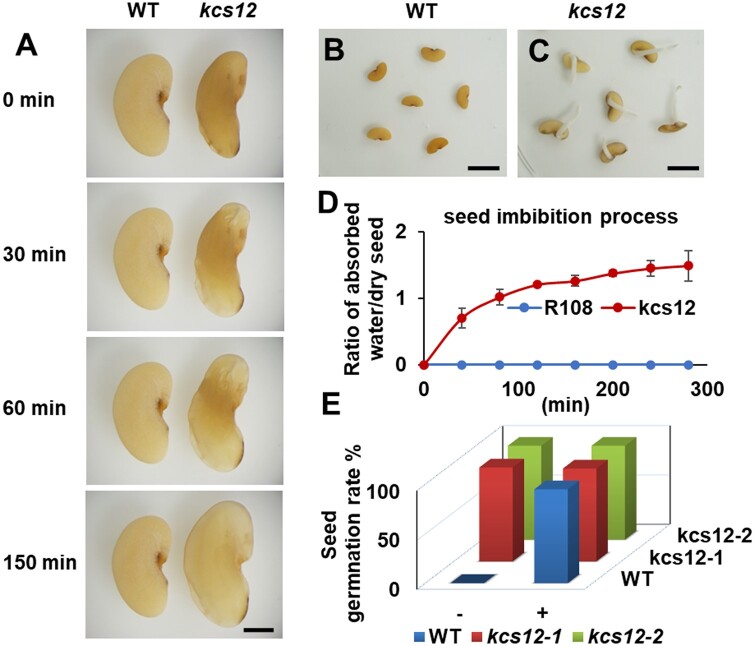
To investigate the impact of KCS12 deficiency on seed physical dormancy, we performed seed coat permeability analysis. Without any scarification, the kcs12 mutant seeds were able to imbibe water and germinate, while wild-type seeds did not imbibe water even after days in sterile water (Figure 3, A–C). The imbibition process of the seeds from wild-type and kcs12 mutant in sterile water without any scarification treatment is shown in Supplemental Movie S1. The imbibition process was also quantified in real-time by tracking the ratio of the weight of absorbed water to the weight of dry seed (Figure 3D). After scarification treatment, the wild-type seeds absorbed water and showed germination rates similar to the kcs12 mutant seeds with and without scarification (Figure 3E). These data indicate that the mutant seeds lost physical dormancy, while the seed germination vigor was not impacted.
Figure 3.
Analysis of seed physical dormancy of wild-type and kcs12 mutant in M. truncatula. A, Representative photographs of wild-type and kcs12 mutant seeds during the imbibition process in sterile water, bar = 1mm. B, C, Germination of wild-type and kcs12 mutant seeds without scarification, bars = 5 mm. D, Imbibition process of seeds from wild-type and kcs12 mutant in sterile water. Error bars represent sd from three biological replicates. E, Germination rates of wild-type and kcs12 mutant seeds, with (+) and without (−) scarification treatment, in sterile water for 2 d.
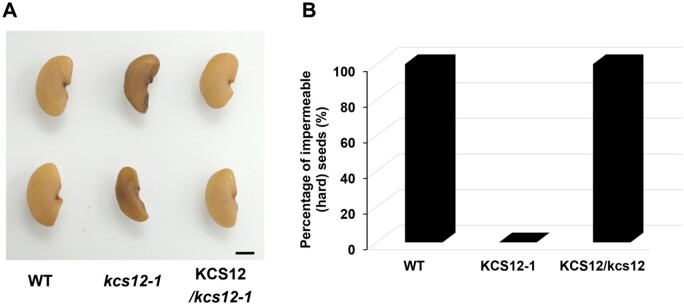
To further confirm the seed phenotype was caused by the loss-of-function of KCS12, we performed a complementation experiment. The KCS12 coding sequence driven by its native promoter was transformed into a homozygous kcs12-1 plant. Seeds harvested from the transgenic plants exhibited a similar phenotype compared to that of the wild-type seeds (Figure 4A). Seed permeability analysis was also performed with the wild-type, kcs12 mutant and complementation lines, and the results showed that the transferred KCS12 gene completely restored physical dormancy of the kcs12 mutant seeds (Figure 4B).
Figure 4.
Genetic complementation analysis of the kcs12 mutant. A, Mature seed morphologies of wild-type, kcs12 mutant, and a complementation line in the kcs12 mutant background, bar = 1 mm. B, Seed permeability analysis of wild-type, kcs12 mutant, and the complementation line.
KCS12 is involved in VLCFA synthesis in M. truncatula seed
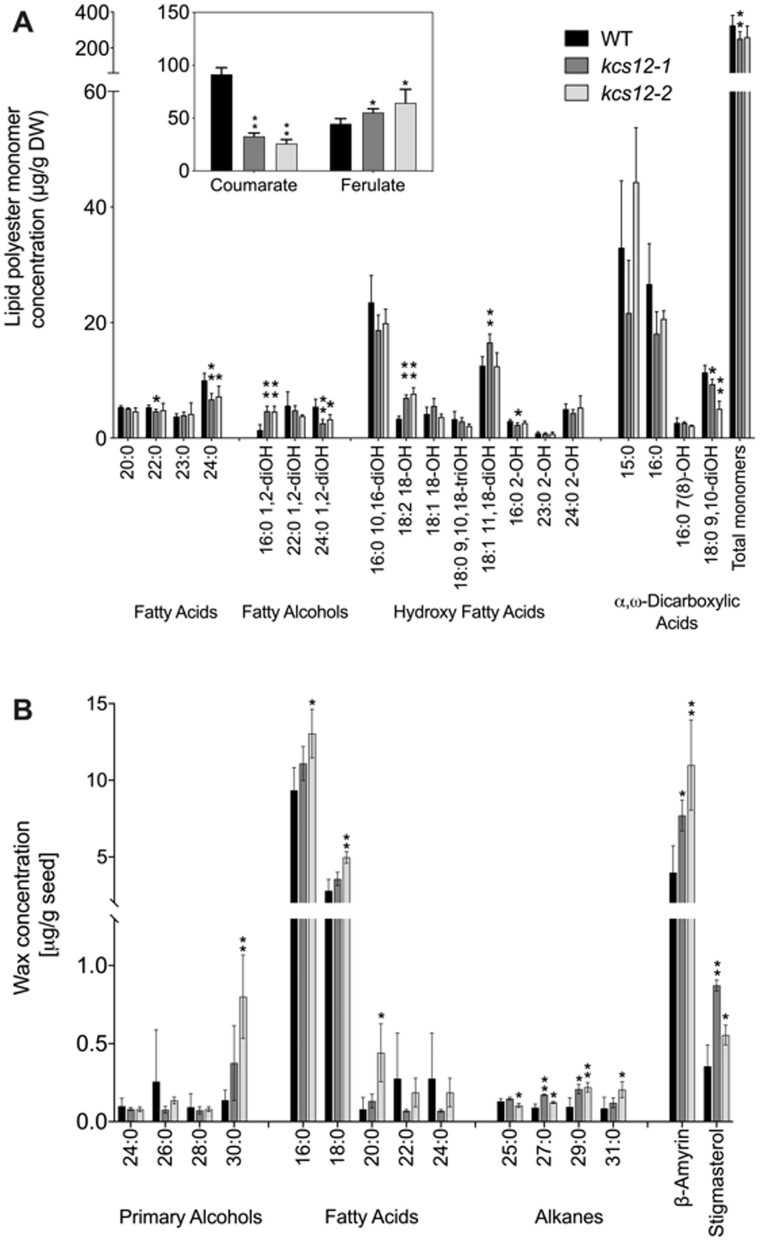
The palisade cuticle is an important part of the legume seed coat, imparting impermeability and contributing to seed physical dormancy (Smýkal et al., 2014). To analyze the structural change in palisade cells, cross sections of seed coat were prepared and stained with toluidine blue; a decrease of palisade cuticle was observed in the kcs12 mutant (Supplemental Figure S5). To quantitively evaluate the impact of loss of function of KCS12 on seed coat cuticle, we analyzed cuticle composition, including lipid polyester monomers and chloroform-soluble waxes, in mature wild-type and kcs12 mutant seeds. Because M. truncatula is a hard seeded species and methods to separate seed coats or seed coat-enriched fractions such as those developed for Arabidopsis (Perry and Wang, 2003) do not work for such seeds, we analyzed lipid polyesters in thoroughly solvent-extracted whole seed tissues. The total lipid polyester monomers of kcs12-1 mutant seeds showed a 22% reduction compared to wild-type seeds, but no significant difference was found between kcs12-2 total monomer loads and wild-type controls (Figure 5A). At the individual monomer level, a significant decrease was observed in the loads of 24:0 fatty acids and 24:0 1,2-diOH fatty alcohol in both kcs12-1 and kcs12-2 mutants. Conversely, a significant increase was observed in two long chain components, 16:0 1,2-diOH fatty alcohol and 18:2 18-OH hydroxy fatty acid, in both kcs12 mutants (Figure 5A). In addition, the concentration of a phenolic monomer changed in the mutants; whereas the amount of coumaric acid was reduced by ca. 70%, ferulic acid was moderately increased (Figure 5A, inset). Seed surface waxes (Figure 5B) were mostly composed of long chain (C16-C18) fatty acids, β-amyrin, and stigmasterol, with only small contributions of VLCFAs (20:0–24:0) and other compounds typically found in plant cuticular waxes (i.e. alcohols, alkanes). The significant increases in very long chain components of waxes, namely 30:0 fatty alcohol, 20:0 fatty acid, and 27:0–31:0 alkanes, observed in kcs12 mutants suggest that KCS12 may negatively impact the synthesis of these wax lipids. Further analysis of the fatty acids released by transesterification of M. truncatula seed extractable lipids, namely neutral (oil) and polar (membrane) lipids, showed differences in VLCFA content (Supplemental Figure S6). In fact, significant increases in the concentrations of 14:0, 18:0, 20:0, 22:0, and 24:0 fatty acid methyl esters were observed in both mutants. These results suggest that mutation of KCS12 has differential impacts on VLCFA components in seed coat extracellular lipid polyesters, while very long chain components of waxes, seed oils, or membrane components were either unchanged or increased in the mutants.
Figure 5.
Extracellular lipid composition of wild-type and kcs12 mutant seeds in M. truncatula. A, Identified lipid polyester monomers released by NaOMe-catalyzed polymerization of whole delipidated seed residues are shown. B, Seed wax components extracted by immersion in chloroform. Values in (A) and (B) are means from four replicates; error bars represent sd. Asterisks indicate significant differences between each mutant and WT (*P < 0.05; **P < 0.01) using two‐tailed unpaired Student’s t test.
The expression of KCS12 is controlled by transcription factor KNOX4
Both KNOX4 and KCS12 showed high expression levels in seed coat and both mutant seeds lost physical dormancy. As the expression level of KCS12 dramatically decreased in the knox4 mutant compared to that of wild-type (Figure 1A), we hypothesized that KNOX4 may regulate the KCS12 expression level as an important regulatory pathway to control seed physical dormancy. To investigate how KCS12 is regulated by KNOX4 transcription factor, we performed a chromatin immunoprecipitation (ChIP) assay with the Pro35S::GFP-KNOX transgenic plants in the knox4 mutant background (Figure 6, A and B). With seed coat tissues as material, the ChIP-PCR data demonstrated that KNOX4 protein could directly bind the promoter region of KCS12. Based on these molecular data, we proposed a model for seed coat development related to VLCFA biosynthesis in M. truncatula. KCS12 is a necessary enzyme for the reaction of VLCFA biosynthesis in seed coat. The expression of KCS12 in seed coat is determined by KNOX4 transcription factor (Supplemental Figure S7). The KNOX-KCS12 module determines biosynthesis of the VLCFAs in seed coat, and is critical for the seed physical dormancy formation.
Figure 6.
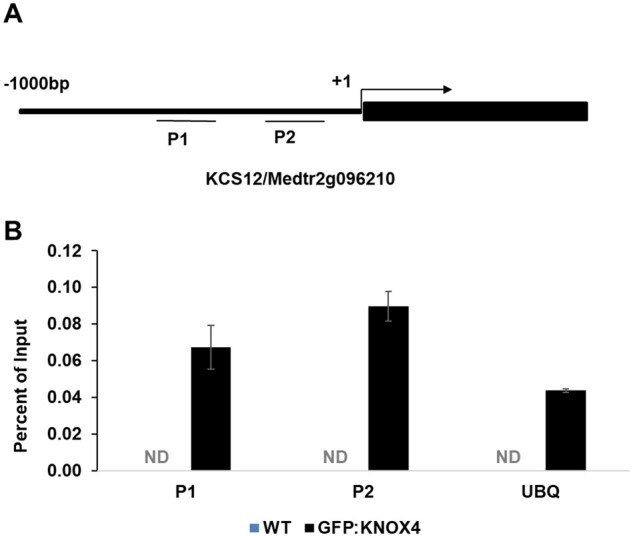
KCS12 is regulated by the KNOX4 transcription factor. A, PCR amplification regions of the KCS12 gene. P1 and P2 indicate two promoter regions. Coding regions are indicated by black box. B, ChIP analysis of the interaction between KNOX4 and the promoter of KCS12, performed with GFP antibody. P1 and P2 are promoter regions as indicated in (A). UBQ is used as control. ND, Not detectable. Error bars represent sd from three biological replicates.
Discussion
Physical dormancy is an interesting ecological phenomenon and important agronomic trait. During the domestication process of crops, physical dormancy has a tendency to be lost or partially lost. To date, information regarding molecular mechanisms controlling physical dormancy is very limited. By using M. truncatula as a model plant, here we discovered that the seed coat KCS12 is involved in seed physical dormancy by controlling the production of specific VLCFAs that are part of the seed coat lipid polyesters.
Involvement of KCS in seed coat permeability
Previous anatomical and biochemical studies showed the integrity of the outmost cuticle layer of seed coat cell wall has a crucial role in providing impermeability to the seed coat and thus in establishing physical dormancy (Ma et al., 2004; Chai et al., 2016). Our hypothesis is that genes involved in cuticle biosynthesis could have an indirect function in establishing physical dormancy. In plants, VLCFAs have been proven to be essential components for the formation of leaf cuticular waxes, sphingolipids, and membrane lipids. KCSs, the critical enzymes of VLCFA biosynthesis that determine chain-length and specificity, exhibit differential expression patterns in different organs or tissues at different development stages (Todd et al., 1999; Yephremov et al., 1999; Fiebig et al., 2000; Hooker et al., 2002; Shao et al., 2007; Kim et al., 2013; Hegebarth et al., 2017). Previous research implied that the role of KCSs was closely related to their expression patterns. For example, among 21 putative KCS genes in Arabidopsis, different KCS genes showed specific functions in the corresponding tissues such as rosette leaves, flowers, siliques, and trichromes. In this study, we identified KCS12 as the gene preferentially expressed in seed coat. Our study revealed that KCS12 gene played a special role in the biosynthesis of seed coat lipid polyester components (but not in the biosynthesis of their associated waxes) and the formation of seed physical dormancy.
KCS12 is involved in seed physical dormancy through the biosynthesis of C24 lipid polyester components
Cuticle components are ubiquitously present in many plant tissues exposed to air, and show a wide spectrum of chain length in their hydrocarbon chains, ranging from 16C to 38C. The plant cuticle is composed of a polyester of fatty acid derivatives, cutin, and solvent-extractable waxes. A related extracellular lipid polyester, suberin, is found in the cell walls of periderms and certain internal tissues such as root endodermis and seed coats. Acyl chains with 16C to 24C are typically found in suberin, whereas cutin mostly contains monomers with 16C and 18C. On the other hand, cuticular waxes co-deposited with cutin and suberin are composed of aliphatics—typically alkanes, fatty acids, aldehydes, and alcohols—with 22C to 38C (Joubès et al., 2008; Li-Beisson et al., 2013). In Arabidopsis, KCS is the component of the FAE multienzyme complex that confers substrate chain length specificity and tissue specificity (Joubès et al., 2008), producing VLCF acyl-CoAs ranging from 20C to 30C (Guo et al., 2016). According to our lipid polyester component analysis, 24:0 fatty acids and 24:0 1,2-dihydroxy fatty alcohol showed significant decreases in the kcs12 mutants compared to wild-type. The fact that increases in VLCFAs concentrations were found in waxes, polar, or neutral lipids, combined with the observation that normal loads of 2-hydroxy fatty acids were present, indicates that the KCS12 gene may be involved in the biosynthesis of C24 VLCFAs incorporated into seed coat polyesters. It is interesting that the M. truncatula seed lipid polyester monomers include very long chain (VLC) alcohols, and particularly 1,2-fattydiols. Fatty alcohols are usually found in suberin but not in cutin, and VLC aliphatics are also unusual in cutin. Because our biochemical experiments were performed in whole seeds, these monomers could be part of a rather unusual cutin polyester, or of suberized cell walls that might also be present in the seed coat. We also observed a >50% decrease in methyl coumarate. Coumarate is a product of the phenylpropanoid pathway, therefore, the large decrease in its concentration in both mutant alleles cannot be interpreted as a direct consequence of the lack of KCS12 activity. However, the small but significant deficiency in VLCFA content combined with the steep decrease in coumarate might have changed seed coat permeability (Shao et al., 2007), which is a putative reason for the change in physical dormancy in kcs12. One could speculate that this hydroxycinnamic acid is in part normally bound to the lipid polymer via aliphatics that are affected in the mutant. In this scenario, the loss of such monomers would result in a concomitant decrease in coumaric acid.
Evolution and conservation of the KCS gene family
The outermost cuticle layer of the land plant aerial surface is essential for plant survival in the terrestrial environment. The KCS gene family expansion process occurred during land plant evolution. In the green plant lineage, like green algae Chlamydomonas reinhardtii and Volvox carteri, a single-copy KCS gene was found. In contrast, duplication and expansion of KCS was observed in land plants (Guo et al., 2016). Selaginella moellendorffii contains 7 KCS genes, while Arabidopsis has 21 KCS genes and M. truncatula possesses 28 KCS genes. In general, most angiosperms keep an average of 20 KCS genes. The cuticle formation in seed coat is crucial in seed plants for the formation of physical dormancy to protect seeds for survival in unfavorable conditions. Since they encode a key enzyme in VLCFA biosynthesis, the KCS-like genes are involved in the evolutionary origins of seed plants. Acyl chains of various lengths are thought to be important for biophysical properties of the cuticle function. KCS12-like genes might have diverged from other KCS genes in seed plants, and these genes play a conserved function in the seed coat cuticle formation during the evolution of plants (Joubès et al., 2008).
Conclusion
In summary, we have identified a seed coat-specific KCS enzyme involved in the biosynthesis of ester-bonded 24:0 fatty acids and fatty diols, adding a missing piece to KCS gene family research. Our study indicates that such components may be critical for seed physical dormancy formation and may impact seed development. Furthermore, KNOX4-KCS12 regulatory pathway provides a regulatory mechanism for seed physical dormancy formation.
Materials and methods
Plant materials and growth conditions
Medicago truncatula ecotype R108 was used in all the experiments in this paper. The kcs12 mutant alleles, kcs12-1(NF5249) and kcs12-2 (NF20114), were identified from a tobacco (Nicotiana tabacum) Tnt1 retrotransposon-tagged mutant collections of M. truncatula. Wild-type and kcs12 mutant plants were grown at 22°C/20°C day/night temperature, 16-h/8-h day/night photoperiod and 70%–80% relative humidity in the greenhouse.
RNA extraction, RT-PCR, and RT-qPCR
Total RNA was extracted from various tissues, including root, stem, leaf, flower, and seed coat, from mature plants with the RNeasy Plant Mini Kit (Qiagen). Two micrograms of RNA were used for reverse transcription, according to the manufacturer’s instructions (SuperScriptIII, Invitrogen). Quantitative real-time PCR was used to determine the expression level of KCS12 in different tissues. The UBQ gene was used as reference gene. qPCR was performed with an ABI prism 7900 HT sequence detection system (Applied Biosystems). SYBR Green was used as the detection reagent. Data were analyzed using SDS 2.2.1 software (Applied Biosystems).
Microscopic observations of seed coat
The intact seeds were used for imbibition assays. Seed imbibition process was observed and recorded under a microscope. Wild-type seeds were scarified with sandpaper. Dry mature seeds were used for SEM observation, which was performed as described previously (Chai et al., 2016). To investigate seed coat structure, mature seeds from wild-type and kcs12 mutants were fixed with 2.5% (vol/vol) glutaraldehyde in sodium phosphate buffer (pH 7.0) in 4°C fridge overnight. They were then washed with 1-mL sodium phosphate buffer three times. After that, the samples were dehydrated in a series of ethanol dilutions and embedded in activated Technovit 7100 resin I. Serial 3-μm cross sections (approximately) were cut in the middle of the seed. Semithin sections were stained with 1% (w/v) toluidine blue O [with 1% (w/v) sodium borate], and pictures were taken with a Nikon Microphot-2 (Nikon Corporation).
GUS staining
To generate the KCS12::GUS construct, a 2.31-kb promoter region of KCS12 was amplified using primers KCS-PF/KCS-PR (Supplemental Table S1) and transferred into the pHGWFS7 vector using the Gateway LR reaction (Invitrogen). The final construct was introduced into the Agrobacterium tumefaciens EHA105 strain for stable transformation. The seeds from 20 d after pollination were collected for GUS staining. GUS activity was histochemically detected as previously described (Jefferson et al., 1987). Cross sectioning was performed to detect the GUS signal in seed coat.
Seed imbibition
About 100 mg mature seeds were soaked in sterile water at room temperature and the whole weight was measured every 30 min in three independent experiments. The absorbed water equals the whole weight of the imbibition seeds minus the original weight of dry seeds.
Seed size data analysis
Around 100 mature seeds were used for seed size data analysis. Mature seeds were photographed using a scanner, and then seed length, width, perimeter, and area were measured by computer software. For seed weight, 100 seeds from wild-type and from the kcs12 mutants were weighed.
Lipid analysis
To characterize lipid polyester monomers, dry seed samples from wild-type and kcs12 mutant were ground, and dry residues were depolymerized by methanolysis in the presence of sodium methoxide following published methods (Molina et al., 2006; Jenkin and Molina, 2015). The depolymerization products extracted by CH2Cl2 were derivatized by treatment with N,O-bis-trimethylsilyl-trifluoroacetamide (BSTFA) to form trimethylsilyl ethers, and the monomers were analyzed by gas chromatography–mass spectrometry (GC–MS) on a TRACE 1300 Thermo Scientific GC with a Thermo Scientific ISQ Single Quadrupole MS detector.
Seed surface waxes were extracted from 0.1 g seed samples with 1-min immersion in 5-mL chloroform with gentle mixing. Three internal quantification standards were added: 1 μg each of n-tetracosane (24:0 alkane), 1-pentadecanol (15:0-OH), and heptadecanoic acid (17:0). The chloroform extracts were then evaporated under a gentle stream of nitrogen and derivatized by dissolving the samples in 100 µL of BSTFA plus 100 µL of pyridine. The samples were incubated for 10 min at 110°C. The derivatized samples were evaporated under a nitrogen gas, resuspended in hexanes and analyzed by GC–MS on a TRACE 1300 Thermo Scientific GC with a Thermo Scientific ISQ Single Quadrupole MS detector. Splitless injection was used with a TraceGOLD-5MS GC capillary column (30 m × 0.25 mm i.d., and 0.10 µm film thickness; Thermo Fisher Scientific Inc, Mississauga, ON, Canada) and a helium flow set at 1.0 mL min−1. Temperature settings were as follows: inlet 310°C, detector 300°C, oven temperature set at 150°C for 3 min and then increased to 320°C at a rate of 5°C min−1, with a final hold at 320°C for 10 min. The mass spectrometer ran in scan mode over 60–650 amu (electron impact ionization), with peaks quantified on the basis of their total ion current. Wax components were identified by their relative retention times and by comparing their spectra to a mass spectral reference library (NIST 2011).
Total lipids were analyzed in whole seeds using the method described previously (Li et al., 2006).
ChIP assay
Immunoprecipitation was performed using 300 mg fresh seed coat from transgenic plants expressing Pro35S::GFP-KNOX in the knox4 mutant background (Chai et al., 2016). Briefly, fresh seed coat was harvested and fixed in 1% formaldehyde and washed three times with PBS buffer. Anti-GFP antibody (Roche) was used for the immunoprecipitation. The ChIP assay was performed following previous methods (Johnson et al., 2002; Zhou et al., 2015). The final precipitated DNA was dissolved in TE buffer (10-mM Tris, 1-mM EDTA at pH 8.0) and used for the quantitative PCR detection. The M. truncatula ubiquitin gene was used as the qPCR control (Supplemental Table S1).
Phylogenetic analysis
The amino acid sequences of KCS family members from Arabidopsis and M. truncatula were used for phylogenetic analysis. Alignments were performed with Clustal Mega with default parameters (www.ebi.ac.uk/Tools/msa/clustalo/). The MEG8 program was used to make the phylogenetic tree, and the parameter was neighbor joining with 1,000 bootstrap replications (www.megasoftware.net).
Accession numbers
The GenBank accession number for KCS is: AES67563.1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of 49 KCS protein sequences using neighbor-joining method in M. truncatula and Arabidopsis.
Supplemental Figure S2. Expression profiles of KCS family genes in M. truncatula in different plant organs.
Supplemental Figure S3. Schematic diagram of parameters of seed size data.
Supplemental Figure S4. Physical properties of mature seeds from wild-type and kcs12 mutants in M. truncatula.
Supplemental Figure S5. Transverse sections of seed coat at maturation stage in the wild-type and kcs12-1 mutant.
Supplemental Figure S6. Fatty acid methyl esters released from M. truncatula seed neutral (oil) and polar (membrane) lipids.
Supplemental Figure S7. A proposed model for seed coat development regulated by VLCFAs biosynthesis in M. truncatula.
Supplemental Table S1. Primers used in this study.
Supplemental Movie S1. Imbibition process of wild-type (left) and kcs12-1 (right) mutant seeds in sterile water.
Supplementary Material
Acknowledgments
We thank Dr Kirankumar Mysore for providing the Tnt1 mutants.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31972958), Noble Research Institute and the First Class Grassland Science Discipline Program of Shandong Province, China.
Conflict of interest statement. The authors declare no conflict of interest.
M.C. and Z.-Y.W. designed the research. M.C., I.C., A.S., I.M., S.W., Z.Z., W.L., J.D., H.X., J.Y., and J.S. performed the experiments. M.C., F.L., Q.J., and I.M. analyzed the data. M.C., I.M., and Z.-Y.W. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Zeng-Yu Wang (zywang@qau.edu.cn).
References
- Bolingue W, Vu BL, Leprince O, Buitink J (2010) Characterization of dormancy behaviour in seeds of the model legume Medicago truncatula. Seed Sci Res 20: 97–107 [Google Scholar]
- Chai M, Zhou C, Molina I, Fu C, Nakashima J, Li G, Zhang W, Park J, Tang Y, Jiang Q, Wang Z-Y (2016) A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc Natl Acad Sci USA 113: 6997–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling E, Mukherjee KD (1991) Acyl-CoA elongase from a higher plant (Lunaria annua): metabolic intermediates of very-long-chain acyl-CoA products and substrate specificity. Biochim Biophy Acta 1082: 239–246 [DOI] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35: 1769–1786 [DOI] [PubMed] [Google Scholar]
- Guo H-S, Zhang Y-M, Sun X-Q, Li M-M, Hang Y-Y, Xue J-Y (2016) Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol Genet Genom 291: 739–752 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Kunst L (2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci 210: 93–107 [DOI] [PubMed] [Google Scholar]
- Hegebarth D, Buschhaus C, Joubès J, Thoraval D, Bird D, Jetter R (2017) Arabidopsis ketoacyl‐CoA synthase 16 (KCS16) forms C36/C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ 40: 1761–1776 [DOI] [PubMed] [Google Scholar]
- Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkin S, Molina I (2015) Isolation and compositional analysis of plant cuticle lipid polyester monomers. J Vis Exp 105: 53386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547. [DOI] [PubMed] [Google Scholar]
- Kang Y, Li M, Senjuti S, Jerome V (2016) A snapshot of functional genetic studies in Medicago truncatula. Front Plant Sci 7: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung JH, Lee SB, Go YS, Kim HJ, Cahoon R, Cahoon EB, Markham JE, Suh MC (2013) Arabidopsis 3-ketoacyl-CoA synthase 9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol 162: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Woodward FI, Quick WP (2002) Long‐distance CO2 signalling in plants. J Exp Bot 53: 183–193 [DOI] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC (2009) Two Arabidopsis 3‐ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP (2013) Acyl-lipid metabolism. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu X, Bourgault R, Galli M, Strable J, Gallavotti A (2020) The FUSED LEAVES1-ADHERENT1 regulatory module is required for maize cuticle development and organ separation. New Phytol 229: 388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Ewa C, Tasneem M, Peterson CA, Mark G (2004) Cracks in the palisade cuticle of soybean seed coats correlate with their permeability to water. Ann Bot 94: 213–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Perry SE, Wang H (2003) Rapid isolation of Arabidopsis thaliana developing embryos. BioTechniques 35: 278–280, 282 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada J-P, Ploense SE, Grossniklaus U, Lolle SJ (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolston MP (1978) Water impermeable seed dormancy. Bot Rev 44: 365–396 [Google Scholar]
- Shao S, Meyer CJ, Ma F, Peterson CA, Bernards MA (2007) The outermost cuticle of soybean seeds: chemical composition and function during imbibition. J Exp Bot 58: 1071–1082 [DOI] [PubMed] [Google Scholar]
- Smýkal P, Vernoud V, Blair MW, Soukup A, Thompson RD (2014) The role of the testa during development and in establishment of dormancy of the legume seed. Front Plant Sci 5: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3‐ketoacyl‐CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Yang T, Li Y, Liu Y, He L, Chen J (2021) The 3-ketoacyl-CoA synthase WFL is involved in lateral organ development and cuticular wax synthesis in Medicago truncatula. Plant Mol Biol 105: 193–204 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu R, Xu Y, Wang M, Zhou C (2019) AGLF provides C-function in floral organ identity through transcriptional regulation of AGAMOUS in Medicago truncatula. Proc Natl Acad Sci USA 116: 5176–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Zhao Y, Wang H, Wang Z-Y (2019) Transforming compound leaf patterning by manipulating REVOLUTA in Medicago truncatula. Plant J 100: 562–571 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.