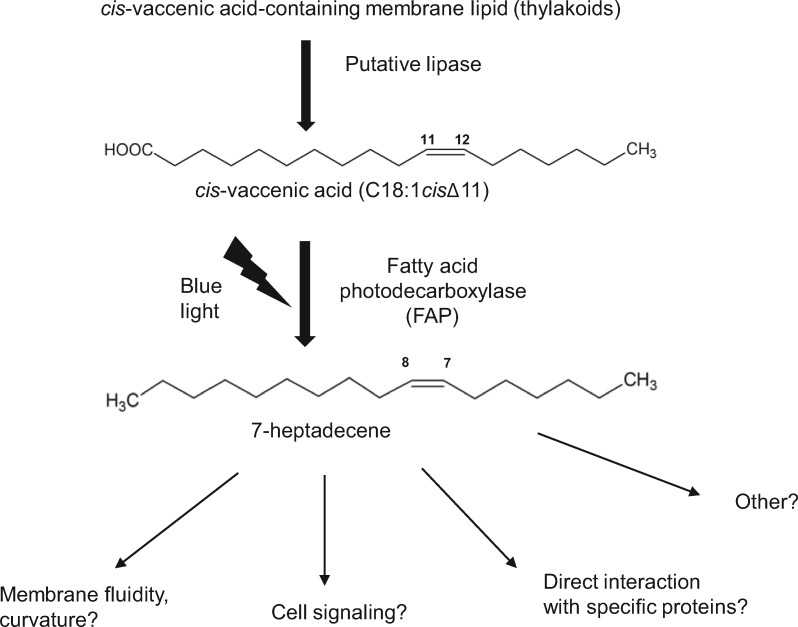
Most algal lineages have the ability to produce hydrocarbons in thylakoids of the chloroplast.
Abstract
Fatty acid photodecarboxylase (FAP) is one of the few enzymes that require light for their catalytic cycle (photoenzymes). FAP was first identified in the microalga Chlorella variabilis NC64A, and belongs to an algae-specific subgroup of the glucose–methanol–choline oxidoreductase family. While the FAP from C. variabilis and its Chlamydomonas reinhardtii homolog CrFAP have demonstrated in vitro activities, their activities and physiological functions have not been studied in vivo. Furthermore, the conservation of FAP activity beyond green microalgae remains hypothetical. Here, using a C. reinhardtii FAP knockout line (fap), we showed that CrFAP is responsible for the formation of 7-heptadecene, the only hydrocarbon of this alga. We further showed that CrFAP was predominantly membrane-associated and that >90% of 7-heptadecene was recovered in the thylakoid fraction. In the fap mutant, photosynthetic activity was not affected under standard growth conditions, but was reduced after cold acclimation when light intensity varied. A phylogenetic analysis that included sequences from Tara Ocean identified almost 200 putative FAPs and indicated that FAP was acquired early after primary endosymbiosis. Within Bikonta, FAP was retained in secondary photosynthetic endosymbiosis lineages but absent from those that lost the plastid. Characterization of recombinant FAPs from various algal genera (Nannochloropsis, Ectocarpus, Galdieria, Chondrus) provided experimental evidence that FAP photochemical activity was present in red and brown algae, and was not limited to unicellular species. These results thus indicate that FAP was conserved during the evolution of most algal lineages where photosynthesis was retained, and suggest that its function is linked to photosynthetic membranes.
Introduction
Most organisms have the ability to synthesize highly hydrophobic compounds made only of carbon and hydrogen called hydrocarbons (HCs). Many HCs are isoprenoids but others like n-alkanes and their unsaturated analogues (n-alkenes) derive from fatty acids (Herman and Zhang, 2016). In plants, C29–C35 n-alkanes are synthesized in the epidermis from very-long-chain fatty acids, and secreted onto the surface of aerial organs (Lee and Suh, 2013). Plant n-alkanes are important for adaptation to the terrestrial environment because they constitute a major part of the wax layer of the extracellular cuticle that prevents the loss of internal water. Occurrence of n-alka(e)nes has also been reported in microorganisms such as microalgae (Sorigué et al., 2016) and cyanobacteria (Schirmer et al., 2010). The magnitude and biogeochemical importance of HC production by marine cyanobacteria (and possibly microalgae) is an intriguing question (Lea-Smith et al., 2015; Valentine and Reddy, 2015). Microbial HCs are presumably mostly located in membranes, but their physiological function has not been studied much. While in cyanobacteria n-alka(e)nes have been proposed to play roles in cell growth, cell division, photosynthesis, and salt tolerance (Berla et al., 2015; Lea-Smith et al., 2016; Yamamori et al., 2018; Knoot and Pakrasi, 2019), in microalgae the biological function of n-alka(e)nes is still completely unknown. Besides the elucidation of their biological roles, n-alkanes and n-alkenes have also attracted attention because of their interest as fuels, cosmetics, lubricants, and as synthons in organic chemistry (Jetter and Kunst, 2008; Jiménez-Díaz et al., 2017). Bio-based alka(e)ne production would be highly desirable to replace part of petroleum-derived HCs, and is currently the focus of intense research efforts (Zhou et al., 2018; Liu and Li, 2020).
A number of n-alka(e)ne-forming enzymes have been identified and characterized in the last decade, and it is now clear that conversion of fatty acids to HCs occurs through a variety of reactions and proteins (Herman and Zhang, 2016). Besides, the biosynthetic enzymes involved in the same types of reactions are not conserved across phylogenetic groups. For instance, it has been shown in bacteria that synthesis of terminal olefins (1-alkenes) occurs through decarboxylation of a saturated long-chain fatty acid, and that this reaction is catalyzed by a cytochrome P450 in Jeotgalicoccus spp. (Rude et al., 2011) and by a nonheme iron oxidase in Pseudomonas (Rui et al., 2014). In the bacterium Micrococcus luteus, yet another pathway has been discovered, which consists of the head-to-head condensation of fatty acids to form very-long-chain n-alkenes with internal double bonds (Beller et al., 2010). Cyanobacterial n-alka(e)nes are produced by two distinct pathways (Schirmer et al., 2010; Mendez-Perez et al., 2011), which have been shown to be mutually exclusive (Coates et al., 2014). In insects, a cytochrome P450 oxidative decarbonylase acts on an aldehyde to produce cuticular alka(e)nes (Qiu et al., 2012). The plant pathway producing the very-long-chain n-alkanes of the cuticular waxes may also involve an aldehyde intermediate, and is known to require the ECERIFERUM (CER) proteins: CER1 and CER3 (Bernard et al., 2012), which are homologous to the oxygen-dependent membrane class of diiron fatty acid desaturases.
In microalgae, we have shown that C15–C17 n-alka(e)nes occur in Chlorella variabilis NC64A (named C. variabilis from here on), and that they are synthesized through decarboxylation of long-chain fatty acids (Sorigué et al., 2016). A C. variabilis protein with a fatty acid decarboxylase activity was then identified as a photoenzyme (Sorigué et al., 2017), a rare type of enzyme that requires photons at each catalytic cycle (Björn, 2015). The C. variabilis protein was thus named fatty acid photodecarboxylase (FAP, E.C. 4.1.1.106). It is one of the few photoenzymes discovered so far, the others being the reactional centers of the photosystems, DNA photolyases, and light-dependent protochlorophyllide oxidoreductase. FAP belongs to a family of flavoproteins (Sorigué et al., 2017), the glucose–methanol–choline (GMC) oxidoreductases, which includes a large variety of enzymes present in prokaryotic and eukaryotic organisms (Zámocký et al., 2004). FAP activity thus represents an additional type of chemistry in the GMC oxidoreductase family (Sorigué et al., 2017). Molecular phylogenetic analysis has shown that C. variabilis FAP and CrFAP belong to an algal branch of GMC oxidoreductases. However, whether FAP activity is conserved in other algal lineages beyond green algae and whether FAP is indeed responsible for n-alka(e)ne formation in vivo remain to be demonstrated. Besides, the subcellular location and role of FAP in algal cells have not yet been investigated.
In this work, we isolate and characterize in C. reinhardtii an insertional mutant deficient in FAP (fap mutant strain). We show that FAP is indeed responsible for the formation of 7-heptadecene, the only fatty acid-derived HC present in this alga. In addition, we provide evidence for a thylakoid localization of C. reinhardtii FAP and its alkene product. We also show that growth and photosynthesis are not affected in the knockout under laboratory conditions, but photosynthetic efficiency is impacted under cold conditions when light intensity varies. Finally, we build a large molecular phylogeny of GMC oxidoreductases based on TARA Ocean data and identify almost 200 uncharacterized putative FAP sequences across algal lineages. Experimental evidence demonstrates that FAP photochemical activity is conserved in red and brown algae and, since it is also present in macroalgae, is not limited to unicellular species.
Results
FAP is responsible for alkene synthesis in C. reinhardtii
Chlamydomonas reinhardtii (hereafter named C. reinhardtii) has been previously shown to produce 7-heptadecene (C17:1-alkene) from cis-vaccenic acid (Sorigué et al., 2016) and to have a FAP homolog that can also perform photodecarboxylation of fatty acids in vitro (Sorigué et al., 2017). Although it seemed likely that FAP proteins are indeed responsible for the synthesis of alka(e)nes produced by C. reinhardtii and C. variabilis, the possibility that the alkenes are formed in vivo by another enzyme could not be ruled out. In order to address this issue and investigate the biological role of FAP, a C. reinhardtii strain mutated for FAP was isolated from the Chlamydomonas Library Project (CLiP; Li et al., 2016). This strain showed undetectable levels of FAP protein (Figure 1) and was named fap-1. The only fatty acid-derived HC in C. reinhardtii, i.e. 7-heptadecene, could not be detected in fap-1 knockout or in the two other fap mutants isolated (Figure 1). After performing nuclear complementation of fap-1 using the genomic FAP gene expressed under the promotor PsaD, four independent transformants with different expression levels of C. reinhardtii FAP (CrFAP) were isolated (named Cp-1–4). In these complemented strains, production of 7-heptadecene was clearly related to FAP amount (Figure 1). These results thus demonstrate that CrFAP is responsible for alkene formation in vivo in C. reinhardtii. Cp-4 was one of the three complemented strains with 7-heptadecene levels similar to the wild-type (WT) strains used for physiological studies (see “Materials and methods” for the isolation of the WT, fap, and Cp strains).
Figure 1.
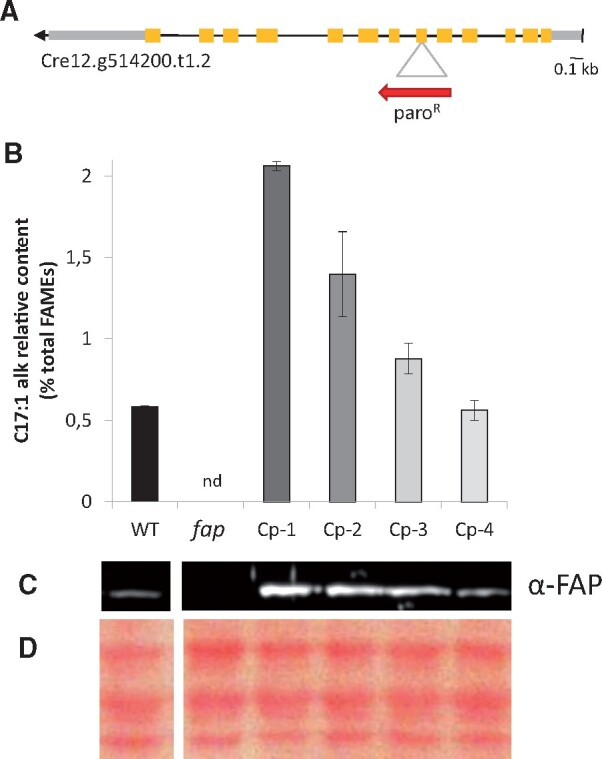
FAP level corresponds to the amount of 7-heptadecene in C. reinhardtii strains mutated in FAP. A, CrFAP gene structure and site of cassette insertion in fap-1 knockout strain. Yellow boxes: exons; gray boxes: untranslated regions; red arrow: antibiotic resistance cassette. B, Relative content of total fatty-acid-derived HCs measured on whole cells; the only fatty acid-derived HC of C. reinhardtii is 7-heptadecene (abbreviated as C17:1 alk). Values are mean ± sd of n = 3 independent experiments for each strain. nd: not detected. C, Immunoblot of total protein extract probed with anti-FAP antibody. D, Loading control of the immunoblot (Ponceau staining). B, C, and D, WT: wild type strains; fap: FAP knockout strains; Cp-1–4: strains complemented with FAP gene (fap-1 background). In C and D, each track corresponds to the genotype situated just above in (B).
FAP activity is conserved beyond green microalgae
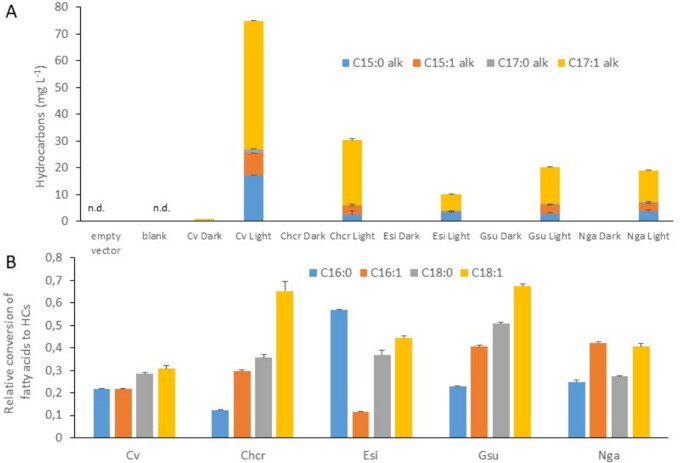
Molecular phylogeny of GMC oxidoreductases has previously shown that CrFAP and the FAP from C. variabilis (CvFAP) are present in an evolutionary branch containing only sequences from algae (Sorigué et al., 2017). The term “algae” is used here in the classical sense of photosynthetic organisms that have chlorophyll a as their primary photosynthetic pigment and lack a sterile covering of cells around the reproductive cells (Lee, 2008). To investigate whether FAP activity has been conserved in algal groups other than green algae, genes encoding putative FAPs from selected algal lineages were cloned and expressed in Escherichia coli, and the bacterial HC content was analyzed. Considering the basal position of red algae, we decided to explore FAP activity in Rhodophytes, selecting the microalga Galdieria sulphuraria and the macroalga Chondrus crispus. For algae deriving from secondary endosymbiosis, we also chose the microalga Nannochloropsis gaditana and the macroalga Ectocarpus silicosus. When cultivated under light, all E. coli strains expressing the various FAPs produced a range of n-alkanes and n-alkenes with different chain lengths (C15–C17) in various proportions (Figure 2A). The small amount of HCs detected in E. coli cells cultivated under dark was not observed in the nontransformed strain or culture medium, and is thus likely to be due to brief illumination required in the cell treatment process. Interestingly, among the FAPs investigated, we observed some profiles of conversion of E. coli fatty acids to HCs that are different from that of CvFAP (Figure 2B). Taken together, these results therefore show that FAP photoenzymatic activity (1) is present in red algae, (2) has been conserved in algae with secondary plastids, (3) is not limited to unicellular algae, and (4) is likely to be diverse with respect to fatty acid specificity.
Figure 2.
Hydrocarbons produced in E. coli strains expressing various microalgal FAPs. HC content was analyzed by GC–MS after transmethylation of whole cells. Data are means ± sd of n = 3 independent cultures. Cv, C. variabilis NC64A; Chcr, Chondrus crispus; Esi, Ectocarpus silicosus; Gsu, Galdieria sulphuraria; Nga, Nannochloropsis gaditana.; nd, not detected. A, Amount of hydrocarbon produced in E. coli strains cultivated under light or dark. Empty vector strain was cultivated under light. Blank corresponds to culture medium alone. B, Relative conversion of E. coli major fatty acids to corresponding HCs. Data represent the amount of a HC produced under light in a FAP-expressing strain divided by the amount of the fatty acid precursor and by the total amount of HCs.
Identification of a reservoir of putative FAPs
To provide a wider picture of the occurrence and evolution of putative FAP photoenzymes within algal groups and to increase the reservoir of FAPs for future biotechnological purposes, a large phylogenetic analysis of GMC oxidoreductases sequences was conducted. We used GMC oxidoreductases retrieved from public databases, from sequenced algal genomes (Blaby-Haas and Merchant, 2019) and from the Tara Ocean project (de Vargas et al., 2015). Tara data gave a unique opportunity to enlarge the FAP dataset with marine algal species that may not be easy to grow under laboratory conditions and whose genomes have not been sequenced. Protein sequences sharing between 50% and 33% of homology with the sequence of C. variabilis FAP were retrieved using Basic Local Alignment Search Tool (BLAST) searches in algal genomes and in the TARA dataset (Supplemental Tables S1 and S2). Over 500 putative GMC oxidoreductases were thus identified. Additional putative GMC oxidoreductases from various taxa of the three domains of life were selected from public databases in order to have a representative diversity of the whole GMC oxidoreductase superfamily.
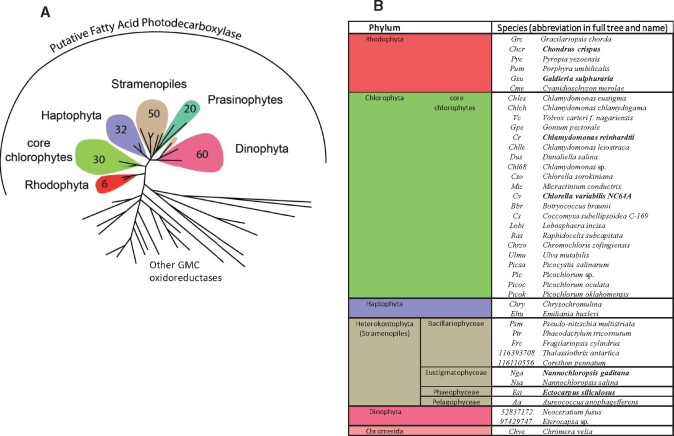
Molecular phylogenetic analysis identified almost 200 sequences of putative GMC oxidoreductases that grouped with the five FAPs that have demonstrated biochemical activity (CvFAP, CrFAP, and the four additional FAPs characterized in this study). These sequences constitute a reservoir of putative FAPs. Sequences in this FAP group all belonged to algal species (Figure 3; Supplemental Figure S1). Logo sequences built with the putative FAPs exhibited highly conserved patterns, some of which were absent in the other GMC oxidoreductases (Supplemental Figure S2). Interestingly, the residues C432 and R451 of CvFAP active site (Sorigué et al., 2017; Heyes et al., 2020) were strictly conserved, and this feature was specific to the FAP group. It is thus likely that all proteins of this group are genuine FAPs.
Figure 3.
Identification of a set of putative FAPs across algal groups. A, Simplified circular tree of GMC oxidoreductases showing the number of putative FAP sequences found in each group of algae. All 198 identified putative FAPs belong to algae and have been found in TARA data (161 FAPs) or in sequenced algal genomes (37 FAPs). The tree was built using maximum likelihood algorithm using GMC oxidoreductases from various kingdoms. Annotations are focused on the branch of putative FAPs; other branches are other GMC oxidoreductases. Branches have been collapsed; full tree is available in Supplemental Figure S1. B, Names of algae species with at least one putative FAP. For most algal groups, the number of species listed in B is lower than that indicated in A because many species from TARA data have no annotation down to the species level. When the biochemical activity of the FAP homolog is demonstrated (Sorigué et al., 2017 or this study), species names are indicated in bold.
Sequences from plants as well as other streptophytes (including charophytes) did not group with algal FAPs (Figure 3; Supplemental Figure S1). Absence of FAP in charophytes indicated early loss of FAP function in streptophytes. No putative FAP sequences could be found in cyanobacteria, although this group is highly represented in TARA data (de Vargas et al., 2015). Phylogeny within the FAP branch indicated that red algae (rhodophytes) sequences were the most basal. Interestingly, FAP sequences from secondary endosymbiosis-derived species appeared to be more closely related to FAPs of green algae (chlorophytes) than red algae.
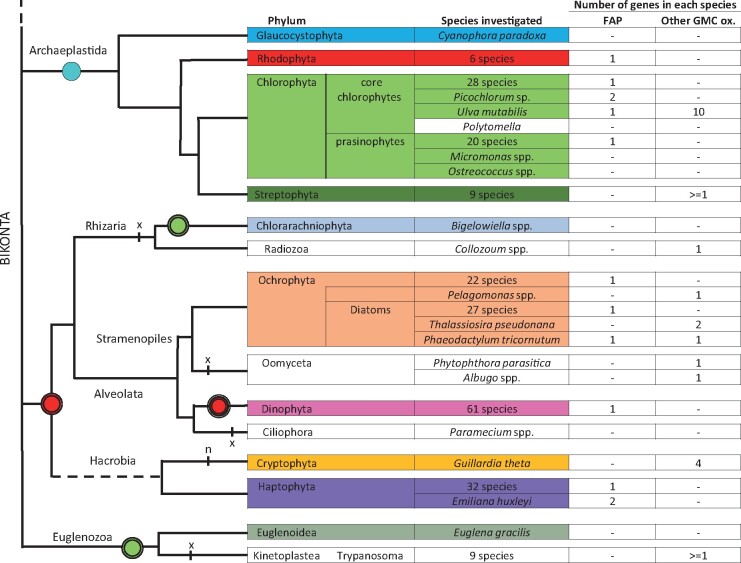
Overall, the putative FAPs that could be identified in algae were present in a variety of algal groups, including stramenopiles (heterokonts), haptophytes, and dinophytes. Most eukaryotic algae harbored one putative FAP and no other GMC oxidoreductase, but a few algae contained no FAP and/or several non-FAP GMC oxidoreductases (Figure 4). Indeed, no putative FAP could be found in the sequenced algal genomes of the glaucocystophyte Cyanophora paradoxa; Mamiellophyceae species: Ostreoccocus spp., Micromonas spp., and Bathycoccus spp.; and the diatom Thalassiosira pseudonana. Conversely, only a few algal sequences could be found in other branches of the GMC oxidoreductase family. Existence in the diatom Thalassiosira pseudonana of a GMC oxidoreductase grouping with bacterial choline dehydrogenase was supported by one sequence from the sequenced genome (Tps-GMC) and one sequence from Tara (48230190). A Tara sequence annotated as a Pelagomonas protein (5166790) also turned out not to be located on the FAP clade. The cryptophyte Guillardia theta contained three different GMC oxidoreductases in three different branches but none of them grouped with FAPs. Ulva mutabilis had 11 predicted GMCs, but only one was in the FAP clade. The 10 other members of this multigene family of Ulva appeared to form a group close to plant GMC oxidoreductases. Although exceptions similar to these probably exist in algal diversity, the general picture is that most algae have only one GMC oxidoreductase and that it groups with the characterized FAP clade in the phylogenetic tree.
Figure 4.
Overview of the number of FAP homologs and other GMC oxidoreductases identified in eukaryotic algae and other bikonts. In most groups, there is one FAP and no other GMC oxidoreductase. Remarkable species whose number of FAP or other GMC oxidoreductases depart from this rule are listed individually. Hyphens indicate that no protein could be identified by BLAST searches. A widely accepted phylogeny of the Bikonta is used. The dashed line connects the tree of the Bikonta to the rest of the phylogenetic tree of the Eukaryotes. Photosynthetic groups or species are colored. Rounds correspond to endosymbiosis: blue round with one black circle, primary endosymbiosis; green and red rounds with two black circles, secondary endosymbiosis; red round with three black circles is for tertiary endosymbiosis in some (not all) Dinophyta; red or green rounds indicate red or green plastid origin, respectively; n: nucleomorph; x: secondary plastid loss.
Chlamydomonas reinhardtii FAP binds membranes, and most of its products are found in the thylakoid fraction
CrFAP is predicted by PredAlgo (Tardif et al., 2012), a software dedicated to the analysis of subcellular targeting sequences in green algae, to be localized to the chloroplast. Consistently, CrFAP is found in a set of 996 proteins proposed to be chloroplastic in C. reinhardtii (Terashima et al., 2011). A broader study of putative FAP targeting peptides, also using PredAlgo, indicated that FAPs from various green and red algae were largely predicted to be chloroplastic (Supplemental Figure S3). In algae with secondary plastids (i.e. containing three or four membranes), the presence of a signal peptide was consistent with targeting to the ER or chloroplast ER (CER) membrane. Further analysis performed using ASAFind (Gruber et al., 2015), a prediction tool designed to recognize CER targeting motifs in signal peptides, indicated that such a motif was present in Ectocarpus silicosus and N. gaditana. Taken together, these results suggest that FAP homologs are very likely to be localized to chloroplasts in green algae, in red algae, and also in at least some of the algae that acquired plastids through secondary endosymbiosis.
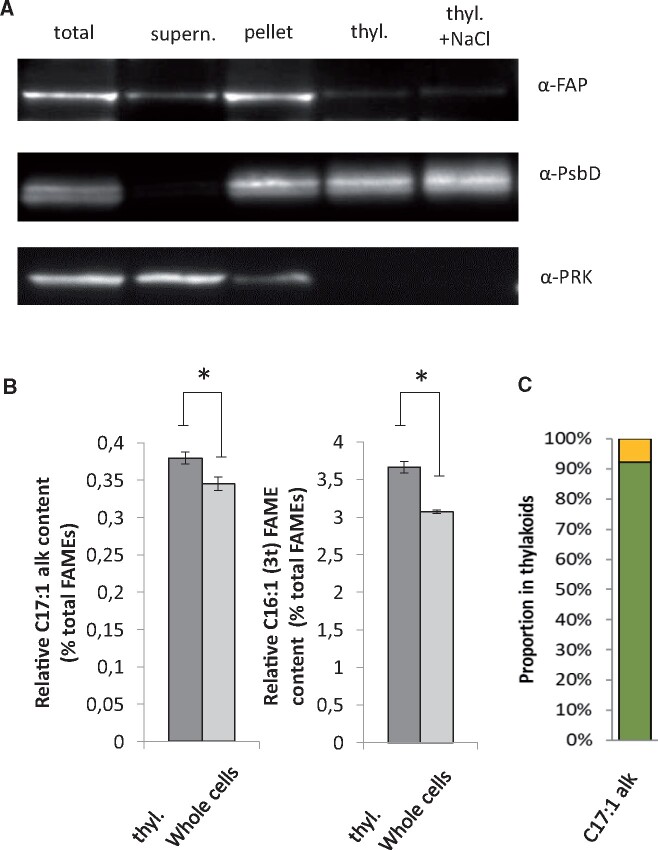
Since CrFAP is a soluble protein (Sorigué et al., 2017) and is predicted to be plastidial, we then sought to determine whether it is a stromal protein with no affinity for membranes or if it has some ability to bind thylakoid membranes. Subcellular fractionation of C. reinhardtii cells was therefore performed (Figure 5A). Most of the FAP protein was found in the total membrane fraction. Thylakoid-enriched membranes were then isolated from whole cells using a sucrose-gradient purification procedure (Figure 5A). Co-purification with thylakoids was performed using the D2 protein (PsbD), a thylakoid membrane protein from the photosystem II (PSII) core complex. The fact that the phosphoribulokinase, used as a control for stroma, could barely be detected in our thylakoid fraction, indicated the presence of little intact chloroplasts or cells. After the thylakoid purification procedure, the amount of CrFAP still bound to this fraction varied between purifications, but the protein was always detectable. A salt wash experiment confirmed that the CrFAP remaining in the thylakoid fraction was tightly bound to the thylakoids, and thus not likely to be a contaminating stromal protein. When analyzing the percentage of 7-heptadecene in total fatty acids in whole cells versus thylakoid-enriched membranes, a slight but significant enrichment in alkene was found in the thylakoid fraction (Figure 5B). Most importantly, by using the fatty acid C16:1(3t) as a marker of the thylakoid lipids, it could be estimated that the enrichment in 7-heptadecene corresponds, in fact, to the localization of >90% of this compound to the thylakoid fraction (Figure 5C). Taken together, these results indicated that CrFAP is a chloroplast-targeted soluble protein that has the ability to bind membranes (including the thylakoid fraction) and whose n-alka(e)nes products are mostly associated to the thylakoid fraction.
Figure 5.
FAP and its 7-heptadecene product are present in a thylakoid membrane-enriched fraction of C. reinhardtii. A, Western blot of protein extracts from whole WT cells before (total) and after centrifugation (supernatant and pellet), and from a thylakoid membrane-enriched fraction without (thyl.) or with (thyl. + NaCl) an additional wash with 0.5-M NaCl. Photosystem II D2 protein (PsbD) and phosphoribulokinase (PRK) are thylakoid and stromal proteins, respectively. Proteins were loaded on a constant chlorophyll basis. B, Relative content in 7-heptadecene and C16:1(3t) fatty acid in whole cells and in the thylakoid fraction (thyl.). The C16:1(3t) fatty acid is almost exclusively present in thylakoids and shown for comparison. Values are mean ± sd of n = 4 independent experiments. (*) denote P < 0.05 in two-sided Student’s t test. C, Estimation of the proportion of total 7-heptadecene present in thylakoids (in green) and elsewhere in the cell (in yellow). Proportion of 7-heptadecene in thylakoids was estimated using C16:1(3t) fatty acid as a thylakoid marker (see “Materials and methods” for calculation).
7-Heptadecene content varies with cell cycle in C. reinhardtii
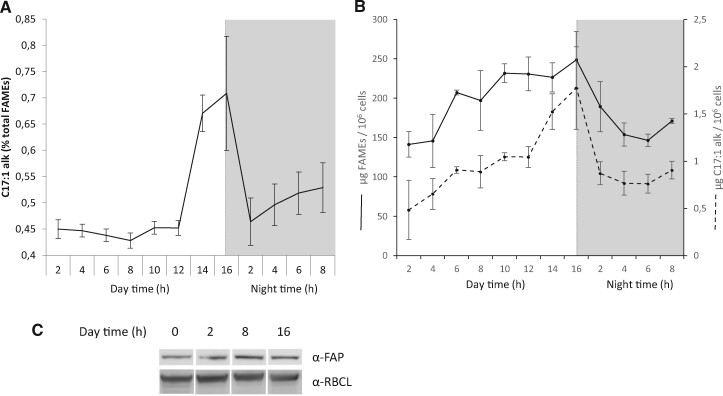
The lack of FAP and HCs in chloroplasts of C. reinhardtii did not result in any obvious differences in the overall organization of cells or chloroplasts as seen by transmission electron microscopy (TEM; Supplemental Figure S4). To try to gather clues on FAP function, publicly available FAP transcriptomic data were mined. Transcriptomic data (Zones et al., 2015) show that FAP has a similar expression pattern as those genes encoding proteins of the photosynthetic apparatus (Supplemental Figure S5). In order to determine whether FAP product varied with time, we monitored total fatty acids and 7-heptadecene content during a day–night cycles in synchronized C. reinhardtii cells. While a constant level of 7-heptadecene representing 0.45% of total fatty acids was found most of the time, a substantial peak (0.7%) was observed before the night (after 12 h of light), and a steep decrease in 7-heptadecene content relative to total fatty acids occurred at the beginning of the night (Figure 6A). This decrease in relative 7-heptadecene content was concomitant with cell division, as evidenced by the drop in total fatty acid content per cell that occurred at that time (Figure 6B). Interestingly, the peak in 7-heptadecene (Figure 6A) was not associated with an increase in total FAP amount between 8 h and 16 h (Figure 6C).
Figure 6.
Variation of 7-heptadecene compared to total fatty acids during C. reinhardtii cell cycle. A, 7-Heptadecene content of cells expressed as a percent of total FAMEs. Values are mean ± sd (n = 3 biological replicates). B, Total fatty acid and 7-heptadecene content per million cells during cell cycle. Total fatty acids were analyzed as FAMEs by GC–MS. A and B, Data are mean ± sd of n = 3 independent cultures. The orange dashed line indicates the end of day time and the beginning of night time (gray shading). C, Immunodetection of FAP during day time. The large Rubisco subunit (RBCL) was used as a loading control.
Fatty acid and membrane lipid compositions differ in the fap mutants
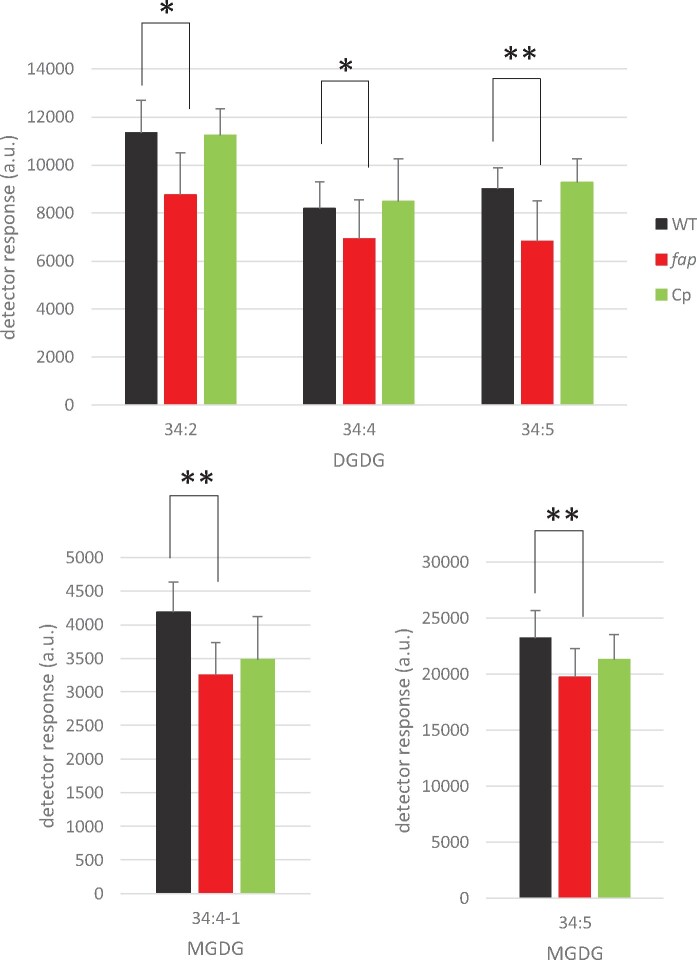
Since 7-heptadecene content varied during cell cycle and may thus play a role in cell division, growth of the WT and fap strains were analyzed. Growth at 25°C was compared in photoautotrophic conditions (mineral medium [MM]) and in mixotrophic conditions (Tris–acetate–phosphate medium [TAP]). No difference between WT and fap could be detected, neither in growth rates nor in cell volumes under these conditions (Supplemental Figure S6). In addition, no difference between WT and fap strains could be observed when cells were grown under various concentrations of sodium chloride (Supplemental Figure S7). Although the lack of HCs had no effect on growth, fatty acid profiling showed some differences in C16:1(9), C18:1(9), and C18:3(9,12,15) (Supplemental Figure S8). Synchronized cells also exhibited no growth differences between WT and fap strains, but exhibited differences in the dynamics of some fatty acid species (Supplemental Figure S9). Changes in fatty acid profiles prompted us to perform a lipidomic analysis by ultra-performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS). Interestingly, this analysis revealed that a limited set of lipid molecular species were significantly different between WT and fap and that they were all plastidial lipids belonging to the galactolipid classes digalactosyldiacylglycerol (DGDG) and monogalactosyldiacylglycerol (MGDG; Figure 7; Supplemental Figure S10). The decrease in the relative content of these galactolipid species appeared to be fully restored by complementation in the case of DGDG but not of MGDG (Figure 7). Taken together, these results indicate that the lack of 7-heptadecene in the fap mutant causes a change in thylakoid lipid composition, which is evidenced by the decrease in the relative content of at least three galactolipid species belonging to the DGDG class.
Figure 7.
Identification of lipid molecular species significantly different between C. reinhardtii WT and fap strains. Relative abundance of glycerolipids was measured by LC–MS/MS analysis of total lipid extracts of whole cells. Only glycerolipid molecular species showing significant differences between WT and fap strains are shown here (see Supplemental Figure S12 for complete results). WT, fap, and Cp: wild-type, FAP knockout and complementation strains, respectively. Lipid extracts from the three strains were loaded on a constant total fatty acid basis. Data are mean ± sd of n = 9 independent cultures (using three different strains for each of the three genotypes). MGDG34:4-1 is one of the two species of MGDG 34:1 species (which differ by C18:3 fatty acid isomers). Asterisks indicate significant differences according to a Mann–Whitney U-test at *P < 0.05 or **P < 0.01. Cells were grown in TAP medium, under 80 µmol photons m−2 s−1 in Erlenmeyers.
FAP is not strongly associated to photosynthetic complexes, and lack of HCs has no effect on their organization
In cyanobacteria, a role of HCs in photosynthesis has been suggested (Berla et al., 2015) but is controversial (Lea-Smith et al., 2016). In C. reinhardtii, there was no difference in the 77K chlorophyll fluorescence spectrum between WT, complementation and fap mutant strains, which indicated no major changes in antenna distribution around photosystems (Supplemental Figure S11A). No difference in photosynthetic efficiency could be detected among WT, complementation, and fap strains grown under standard laboratory conditions (Supplemental Figure S11B). Membrane inlet mass spectrometry (MIMS) experiments conducted to quantify O2 exchange showed no difference in respiration or photosynthesis rates between the two genotypes (Supplemental Figure S11C). Native electrophoresis of proteins from purified thylakoids and FAP immunodetection revealed that FAP could only be detected at an apparent molecular size of the monomeric FAP (Figure 8), indicating no strong association with proteins of photosynthetic complexes. Besides, no difference in organization of photosynthetic complexes between WT and fap could be seen on the native protein electrophoresis.
Figure 8.
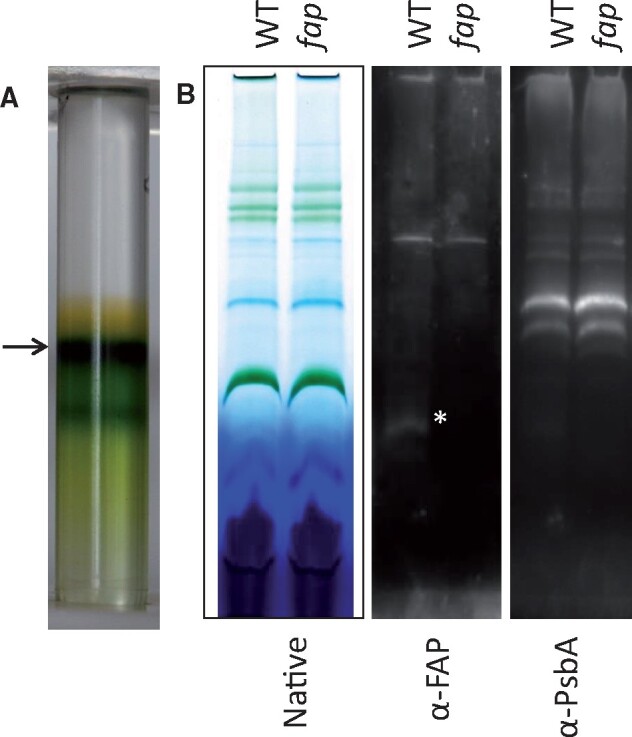
Thylakoid purification and immunoblot analysis. A, Purification of C. reinhardtii thylakoids using a sucrose density gradient. The thylakoid fraction collected is indicated by an arrow. B, Blue native polyacrylamide gel of solubilized proteins (0.5% digitonine, 0.5% α-DM) and corresponding immunodetection. WT: wild-type strain, fap: FAP knockout strain. PsbA: Photosystem II D1 protein. Asterisk indicates FAP band.
Photosynthesis is affected under light and cold stress in the fap mutants
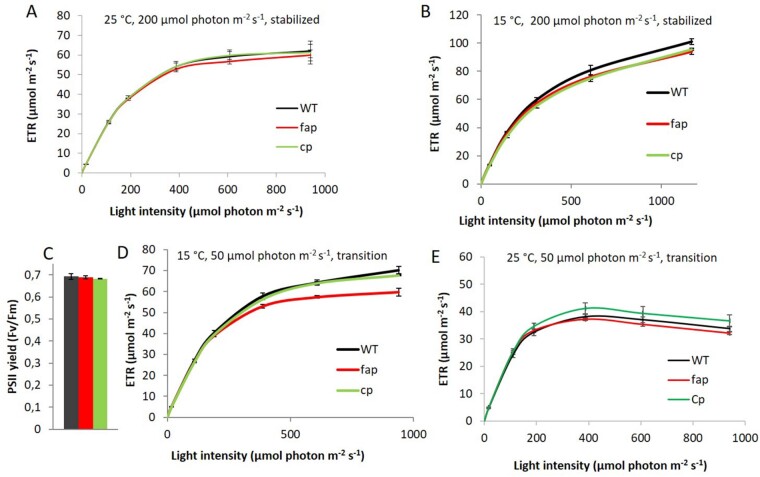
Lack of HCs in the fap strain did not cause changes in the photosynthesis activity under standard growth conditions. However, since significant modifications in the composition of membrane lipids could be detected, we explored harsher conditions to further challenge photosynthetic membranes. We chose to investigate chilling temperatures because cold is well-known to affect both membrane physical properties and photosynthesis (Los et al., 2013). Using a multicultivator in turbidostat mode, we first stabilized cultures at 25°C under medium light (200 µmol photons m−2 s−1), finding the apparent electron transfer rate (ETR) showed no difference under this condition (Figure 9A). When cooling down the culture to 15°C and after 3 d of acclimation, both ETR and 77 K chlorophyll fluorescence spectra still showed no differences (Figure 9B;Supplemental Figure S11A). However, after 1 d at a lower light intensity (50 µmol photons m−2s−1, 15°C), while the maximal PSII yield was equal for all the strains (Figure 9C), the ETR was lower for the mutant when measured at high light intensities (Figure 9D). Interestingly, longer acclimation to this condition (3 d) led to the disappearance of this phenotype. This transient phenotype of the fap KO was clearly linked to low temperature because shifting cultures at 25°C from medium light to low light did not result in any difference in maximal PSII yield or ETR among the strains (Figure 9E).
Figure 9.
Photosynthetic acclimation to cold conditions in C. reinhardtii WT and fap strains. Apparent ETR at various light intensities for cells grown in photoautotrophic conditions at 25°C (A) and 15°C after 3 d acclimation (B). PSII yield (C) and ETR (D) at 15°C after exposure to lower light for one day (50 µmol photon m−2 s−1). E, ETR at 25°C after transition to lower light for one day (50 µmol photon m−2 s−1). WT, fap, and Cp: wild-type, FAP knockout, and complementation strains, respectively. Data are mean ± sd of n = 3 independent cultures (corresponding to three different strains for each genotype).
Note: Figure Replacement Requested.
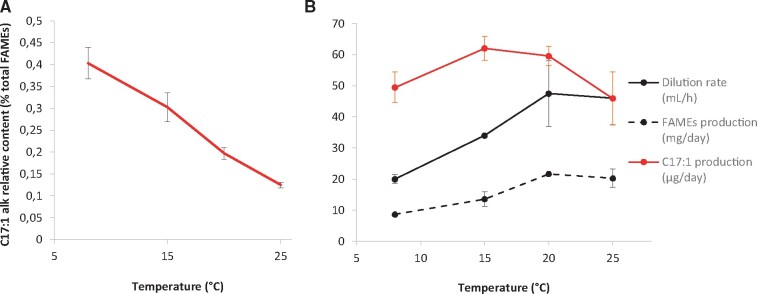
In order to provide support for a possible link between HCs and cold acclimation, 7-heptadecene content was quantified under various growth temperatures. Relative HC content in WT cells clearly increased under cold conditions (Figure 10). Enrichment of WT membranes in HC was due to a slower decrease in HC synthesis compared to fatty acid synthesis (Figure 10B). As expected, an increase in the relative content of polyunsaturated species occurred upon cold treatment in both WT and fap strains (Supplemental Figure S12), but no difference in the dynamics of fatty acid remodeling was observed between the two strains.
Figure 10.
Hydrocarbon production in C. reinhardtii WT cells cultivated at low temperatures. A, HC content in cells as percentage of total FAMEs. B, Dilution rate of culture medium and production of FAMEs and HCs. For A and B, Cells were grown in TAP medium in photobioreactors in turbidostat mode under 50 µmol photons m−2 s−1. HC and fatty acid content were analyzed by GC–MS after transmethylation. Data are mean ± sd of n = 3 independent cultures.
Discussion
Here, we report the isolation and characterization of an insertional C. reinhardtii mutant deficient in FAP, and we perform phylogenetic and functional analyses of algal homologs. We show that FAP and the vast majority of its 7-heptadecene product are associated to thylakoid membranes. It is also shown that a gene encoding for a putative FAP is present in most algal lineages and indeed encodes a functional FAP in some species of red algae, and of secondary algae, including some macroalgae. By studying a FAP knockout C. reinhardtii mutant, we provide evidence that lack of HCs is associated correlated with small changes in galactolipid composition, but has no impact on photosynthesis and growth in C. reinhardtii under standard culture conditions. However, in the absence of HCs generated by FAP, the photosynthetic activity is transiently affected during cold acclimation when light intensity varies. The possible significance of these results for algal physiology, as well as FAP function and evolution, is discussed below.
CrFAP localization and regulation in algal cells
Based on the characterization of a fap mutant, we first show that CrFAP is responsible for the synthesis of all fatty acid-derived HCs found in C. reinhardtii cells (Figure 1). Our result clearly demonstrates that the FAP activity measured in vitro for CrFAP (Sorigué et al., 2017) is not a promiscuous secondary activity and indeed corresponds to a genuine biological activity, namely the light-driven synthesis of 7-heptadecene from cis-vaccenic acid (Figure 11). Also, the results from the fap knockout line demonstrate that no other enzyme is able to synthesize 7-heptadecene in C. reinhardtii. Furthermore, the HC production was found to correspond to the quantity of CrFAP present in complementation lines. Thus, FAP is a limiting factor for 7-heptadecene production in vivo, but the lipase(s) that may act upstream of FAP to generate the free cis-vaccenic acid does not limit the pathway. This conclusion is consistent with a previous observation that 7-heptadecene is increased eight-fold in C. reinhardtii when CvFAP is overexpressed (Yunus et al., 2018).
Figure 11.
Proposed pathway for hydrocarbon formation from fatty acids in C. reinhardtii and putative roles. The only fatty acid-derived HC in C. reinhardtii (7-heptadecene) is generated from cis-vaccenic acid by FAP only when cells are exposed to blue light. The fatty acid precursor must be released from a thylakoid lipid by an unknown lipase. The 7-heptadecene FAP product may play several roles in thylakoid membranes depending on temperature and light conditions.
CrFAP is a soluble protein (Sorigué et al., 2017). Using subcellular fractionation of C. reinhardtii cells and anti-CrFAP antibodies, we show here that CrFAP is predominantly found in the total membrane fraction (Figure 5A). Based on the predicted plastid localization of CrFAP (Supplemental Figure S3) and the fact that the thylakoid fraction harbors >90% of the 7-heptadecene product (Figure 5C), we propose that in vivo CrFAP is a soluble chloroplastic protein that can bind to and unbind from thylakoid membranes. The capacity of CrFAP to bind to thylakoids is supported by the observation that after the thylakoid purification procedure, a fraction of CrFAP is still tightly bound to this fraction (Figure 5A). Plastidial localization may be a general feature of FAPs because presence of plastid transit peptides in FAPs seems to be a general rule in green and red algae (for primary plastids) and is also predicted for some secondary endosymbiosis-derived algae (Supplemental Figure S3).
Concerning the regulation of FAP activity in membranes, it is clear that the peak in 7-heptadecene during the cell cycle (Figure 6A) is not due to an increase in the total amount of FAP protein (Figure 6C), and another mode of regulation of FAP activity must exist. Availability of free fatty acid substrates provided by a lipase acting upstream of FAP is a possibility (Figure 11). The peak in CrFAP gene expression during the day (Supplemental Figure S5) may correspond to the necessity of a high turnover of the CrFAP protein due to photodamage to the protein. This would be consistent with the idea that C. variabilis FAP has a radical-based mechanism (Sorigué et al., 2017), which may be the cause of photoinactivation of the protein (Lakavath et al., 2020) and degradation under excessive light (Moulin et al., 2019).
Role of CrFAP products in membranes
In a work reporting cyanobacterial mutants devoid of fatty acid-derived HCs, it has been suggested that HCs are located in membranes and may play a role in cell division (Lea-Smith et al., 2016). In the proposed cyanobacterial model, integration of HCs into the lipid bilayer would be responsible for membrane flexibility and curvature. HCs may play a similar role in thylakoid membranes of green algae. In light of this connection, it is interesting to note that in C. reinhardtii, HC production follows lipid production during cell growth except just before mitosis (Figure 6, A and B). The ratio of HCs to fatty acid methyl esters (FAMEs) decreased at the beginning of night, when cells are dividing, indicating that the extra amount of HCs synthesized before the night must be somehow lost or metabolized during cell division. Simple mechanisms that could explain HC loss during cell division involve enrichment in HCs at breaking points of plastidial membranes before cell division, exclusion from these membranes during division, and/or loss to the gas phase of the culture due to HC volatility (Supplemental Figure 5B).
HCs might thus impact local flexibility of algal plastidial membranes and participate in lipid membrane remodeling during cell division. However, under standard culture conditions, the presence of HCs is apparently not critical for chloroplast structure (Supplemental Figure 4), cell size and cell division rate (Supplemental Figure 6). A role of cyanobacterial HCs in tolerance to salt stress has also been suggested (Yamamori et al., 2018). In C. reinhardtii, contrary to what has been shown in cyanobacteria, no difference could be detected in growth under increasing salt concentrations (Supplemental Figure 7). One could thus hypothesize that even if HCs are produced in chloroplasts and accumulate in thylakoids, their function might be different from that in cyanobacteria. It is also possible that laboratory culture conditions used for C. reinhardtii (this study) are far from natural growth conditions where HCs may be necessary. Alternatively, a compensation mechanism for HC loss may operate differently in C. reinhardtii and in cyanobacteria. In C. reinhardtii, part of this mechanism may involve changes in membrane lipid composition. Interestingly, lipidomic analysis under standard growth conditions unraveled specific changes in DGDG molecular species (Figure 7) but no other significant differences in other class of lipids (Supplemental Figure S8). Taken together, these results suggest that in C. reinhardtii, HCs play no crucial role in cell division and growth under standard conditions. Cells may adapt to a lack of HCs by some changes in the composition of membranes, which could specifically involve some DGDG galactolipids. Alternatively, or in addition to this proposed effect on properties of the membrane lipid phase, it cannot be ruled out that 7-heptadecene may act locally to disrupt or enhance some specific protein–protein interactions, or may play a yet to be defined role, such as acting as a signaling molecule or its precursor (Figure 11).
The fact that FAP gene expression follows that of photosynthesis genes in day–night cycles, the likely localization of FAP in plastids of green and red algae (as well as in some secondary algae), and the localization of part of FAP and almost all its alkene product in C. reinhardtii thylakoids, point toward a role of FAPs in the photosynthetic function of algal cells. This idea is strongly reinforced by the conservation of the FAP-encoding gene in many eukaryotic algae but not in nonphotosynthetic protists (Figures 3 and 4) and in Polytomella, an algae that has kept some of its plastidial function but lost photosynthesis (Smith and Lee, 2014). As standard culture conditions did not reveal any photosynthesis phenotype in C. reinhardtii fap mutant (Supplemental Figure S11), more challenging conditions involving colder temperatures and variations in light intensity were tested. These experiments have revealed a difference between WT and fap mutant in the photosynthesis activity during cold acclimation after light intensity varied from medium to low (Figure 9). This difference could be highlighted under the high-light ETR measurement conditions. Interestingly, colder temperatures were associated with membrane enrichment in HCs (Figure 10) and fatty acid profiles followed similar variations with temperature in both WT and fap strains (Supplemental Figure S12). Taken together, these observations indicate that adaptations in membrane lipid composition compensate partly for the loss of HCs in standard growth conditions, but not in harsher conditions, such as light intensity variations under cold temperatures.
At temperatures lower than 20°C, it is clear that cell division and FAME production slow down (Figure 10B). Interestingly, HC production does not follow the same pattern, and increases with decreasing temperatures (except at 8°C). Enrichment of membranes in HCs with decreasing temperatures is consistent with our proposal that HCs are lost during cell division (Supplemental Figure S5): lower temperature causes reduced division and would hence result in lower HC losses. Whether the sustained production of HC at low temperature is an adaptation mechanism to low temperature or not, remains to be elucidated.
Conservation of FAP in algae
According to molecular phylogeny (Figures 3 and 4), FAP proteins appear to be specific to algae and highly conserved in many algae species. A noticeable exception is the Mamiellophyceae class of the green algae. Algae are a common denomination that covers photosynthetic eukaryotes which mainly live in aquatic environments. This polyphyletic group includes organisms derived from a first endosymbiosis, as well as organisms derived from a secondary, or even tertiary, endosymbiosis. However, a functional FAP can be found in Chlorophyta (green algae), Rhodophyta (Chondrus and Galdieria), and Stramenopiles (in the Phaeophyceae Ectocarpus and the Eustigmatophyceae Nannochloropsis), as demonstrated by heterologous expression in E. coli of the corresponding identified FAPs (Figure 2). FAP activity was therefore conserved during secondary endosymbiotic event(s) that gave rise to the red algae lineage. Moreover, FAP activity is not specific to the unicellular state, as FAPs were also functional in the pluricellular algae (macroalgae) E. silicosus and C. crispus. Considering homology of sequences, FAP function is thus expected to be present in most algal phyla, including Haptophyta and Dinophyta. Importantly, some amino acid residues that are likely to be involved in fatty acid substrate stabilization or photocatalysis, such as CvFAP Arg451 or Cys432 (Sorigué et al., 2017; Heyes et al., 2020), are strictly conserved in the 198 putative FAPs (Supplemental Figure S2). This observation reinforces the idea that all the putative FAPs identified in this work have the ability to photo-produce HCs from fatty acids.
FAP neofunctionalization from GMC oxidoreductases may have occurred early during evolution of algae, almost concomitantly with the very first endosymbiosis shared by green and red algae. No GMC could be found in Glaucocystophyta (Figure 4), which may indicate that this neofunctionalization event has occurred after separation of this group from red and green algae. However, it should be noted that so far only one complete Glaucocystophyta genome is available. Absence of FAP in charophytes (Supplemental Figure 1) indicates early loss of FAP function in Streptophyta. Phylogenetic analysis indicates that FAPs from secondary endosymbiosis lineages are more closely related to core Chlorophyta than to Rhodophyta. FAP could thus be one of the genes that was inherited from green algae by horizontal gene transfer (Moustafa et al., 2009).
Concerning the conservation of FAP activity, it should be noted that some of the FAPs selected for heterologous expression had fatty acid preference profiles different from that of CvFAP (Figure 2B). For example, while CvFAP converted E. coli fatty acids into HCs with a slight preference for C18 over C16 fatty acids, Chondrus and Galdieria FAPs showed higher preference for C16 over C18, and saturated over unsaturated fatty acids. In contrast, Ectocarpus FAP had a strong preference for C16:0 over C16:1 fatty acid, and N. gaditana FAP preferred unsaturated over saturated chain for both C16 and C18 fatty acids. This indicates that the algal biodiversity is likely to contain FAPs with fatty acid specificities different from the FAPs of C. variabilis and C. reinhardtii. FAPs with different properties may be useful for a biobased production of specific HCs, for instance shorter chain semi-volatile HCs for liquid fuels (Moulin et al., 2019).
In conclusion, the results presented here show that FAP activity is conserved beyond green microalgae, and identify a big reservoir of FAPs that may be useful for biotechnological applications. It also provides some important clues for future studies aiming at unravelling the exact role of the FAP photoenzyme in eukaryotic algae.
Materials and methods
Strains and culture conditions
The fap mutant and its corresponding WT strain of C. reinhardtii were ordered from the CLiP library (Li et al., 2016). Upon receipt, strains were plated on TAP medium and streaked to allow formation of single colonies. For each strain, after 1-week growth in the dark, three single-clone-derived colonies were randomly chosen for characterization. WT strains are CC-4533 cw15 mt− for mating type minus and CC-5155 cw15 mt+ (Jonikas CMJ030 F5 backcross strain) [isolate E8] for mating type plus. Mutant LMJ.RY0402.226794 was used in this study, which is predicted to harbor a first insertional cassette in the coding sequence of Cre12.g514200 which encodes FAP. A second insertion in the line LMJ.RY0402.226794 was predicted in Cre14.g628702. To remove this side mutation, we backcrossed the mutant strain to CC-5155. Analysis of one full tetrad showed two progeny strains resistant to paromomycin and which were mutated in FAP gene. The region of Cre14.g628702 was amplified by polymerase chain reaction (PCR) and sequenced for the four progeny strains of the tetrad. No insertion was actually found, therefore a potential insertion at this locus was ruled out. Work on mutant strains was conducted on one isolated parental strain with the mutation from LMJ.RY0402.226794, and on two mutants from the backcross with CC5155 (full tetrad). These three strains are thereafter named fap-1, fap-2, and fap-3. WT strains were parental strain CC5155 (WT-1) and single colony-derived lines of background strain CC4533 (WT-2, WT-3). The average of the three WT strains and the average of the three knockout strains are labeled as WT and fap, respectively. For liquid culture experiments, cells were grown in 24 deep well plates of 25 mL culture, under 100 µmol photons m−2 s−1 with constant shaking at 25°C. Cells were grown in TAP or MM (Harris, 1989) for mixotrophic and autotrophic conditions, respectively. Cell growth was followed using a cell counter Multisizer (Beckman Coulter). For the day–night cycle experiment, cells were cultivated autotrophically in 1L-photobiorectors in turbidostat mode (OD880nm at 0.4) under 16 h of light (40 µmol photons m−2 s−1) and 8 h of dark at 25°C. For photosynthesis analysis, cells were grown autotrophically in 80 mL photobioreactors (multicultivator, Photon Systems Instruments) in turbidostat mode (OD680nm at 0.8). Conditions were 25°C, with medium light intensity (200 µmol photons m−2 s−1) or low light intensity (50 µmol photons m−2 s−1); or 15°C, with medium or low light intensity. All cultures were performed under ambient air.
Complementation of the fap mutants
The construct for complementation of the knockout strain for FAP gene was performed using pSL-Hyg vector containing an AphVII cassette conferring hygromycin resistance. This vector allowing nuclear transformation was kindly provided by Pr. Steven Ball (University of Lille, France). A WT copy of the FAP gene was obtained by PCR of WT genomic DNA. It was cloned into TOPO-XL vector. pSL-Hyg vector and FAP gene were digested with EcoRV and SpeI and ligated. Then, the vector was linearized with PvuI and was electroporated into the fap strains. Level of complementation was verified by immunoblot (to assess quantity of protein) and by transmethylation of whole cells (to assess quantity of HCs). The two fap mutants from a full tetrad were complemented. Three complemented strains with HC levels similar to WT were kept for physiological studies (e.g. Cp-4 shown in Figure 1).
SDS–PAGE and immunodetection
Cells (10–15 mL) were harvested by centrifugation at 3,000g for 2 min. Pellets were then frozen in liquid nitrogen and stored at -80°C until use. Pellets were resuspended in 400 mL 1% (w/v) sodium dodecyl sulfate (SDS) and then 1.6 mL acetone precooled to −20°C was added. After overnight incubation at -20°C, samples were centrifuged (20,800 g, 10 min, 4°C). Supernatant was removed and used for chlorophyll quantification using a UVmc spectrophotometer (SAFAS). Pellets were resuspended to 1-mg chlorophyll mL−1 in Lithium dodecyl sulfate buffer in the presence of NuPAGE reducing agent (ThermoFischer), and loaded on 10% (w/v) polyacrylamide gel electrophoresis (PAGE) Bis–Tris SDS gel. To load equal protein amounts for immunoblot analysis, protein contents were estimated by Coomassie Brilliant Blue staining of the gel using an Odyssey IR Imager (LICOR). After gel electrophoresis, proteins were transferred to nitrocellulose membranes for 75 min at 10 V using a semi-dry set up. Membranes were blocked in Tris-buffered saline Tween 20 buffer with milk 5% (w/v) overnight at 4°C and then incubated at room temperature in the presence of the following antibodies: anti-Cyt f, anti-AtpB, anti-PsaD, anti-PsbA, anti-LHCSR3 (Agrisera), or anti-FAP (see below). After 2-h incubation, primary antibody was removed by rinsing three times in TBST, and a peroxidase-coupled secondary antibody was added for at least 1 h. Luminescence was detected with a Gbox imaging system (Syngene).
Production of anti-CrFAP antibodies
A codon-optimized synthetic gene encoding C. reinhardtii FAP (Sorigué et al., 2017) was cloned into the pLIC7 expression vector, allowing the production of a recombinant FAP fused to TEV-cleavable His-tagged E. coli thioredoxin. Production was performed in the E. coli BL21 Star (DE3) strain initially grown at 37°C in TB medium. Induction was initiated at an OD600nm of 0.8 by adding 0.5-mM isopropyl b-d-thiogalactoside (IPTG), and cultures were then grown at 20°C. Following overnight incubation, cells were centrifuged and protein was purified as described previously (Sorigué et al., 2017). Purity of the purified protein was controlled on SDS–PAGE, and it was brought to a final concentration of 2 mg mL−1 using an Amicon-Ultra device (Millipore). Polyclonal antibodies against FAP were raised in rabbits (ProteoGenix, Schiltigheim, France).
Analysis of HCs and fatty acids
For quantification of HCs, approximately one hundred million cells were pelleted by centrifugation in glass tubes. Transmethylation was conducted by adding 2 mL of methanol containing 5% (v/v) sulfuric acid to the cell pellet. Internal standards (10 µg of hexadecane and 20 µg of triheptadecanoylglycerol) were added for quantification. Reaction was carried out for 90 min at 85°C in sealed glass tubes. After cooling down, 1 mL of 0.9% (w/v) NaCl and 500 µL of hexane were added to the samples to allow phase separation and recovery of FAMEs and HCs in the hexane phase. Samples were mixed and then centrifuged to allow phase separation. Two microliter of the hexane phase was injected in the apparatus of gas chromatography coupled to mass spectrometry and flame ionization detection (GC–MS/FID). Analyses were carried out on an Agilent 7890A gas chromatographer coupled to an Agilent 5975C mass spectrometer (simple quadrupole). A Zebron 7HG-G007-11 (Phenomenex) polar capillary column (length 30 m, internal diameter 0.25 mm, and film thickness 0.25 mm) was used. Helium carrier gas was at 1 mL min−1. Oven temperature was programmed with an initial 2-min hold time at 35°C, a first ramp from 35°C to 150°C at 15°C min−1, followed by a 1-min hold time at 170°C then a second ramp from 170°C to 240°C at 5°C min−1 and a final 2-min hold time at 240°C. The MS was run in full scan over 40 to 350 amu (electron impact ionization at 70 eV), and peaks of FAMEs and HCs were quantified based on the FID signal using the internal standards C17:0 FAME and hexadecane, respectively.
Chlorophyll fluorescence measurements and MIMS analysis
Chlorophyll fluorescence measurements were performed using a pulse amplitude-modulated fluorimeter (Dual-PAM 100) upon 15-min dark-adaptation under continuous stirring. Detection pulses (10 mmol photons m−2 s−1 blue light) were supplied at a 100-Hz frequency. Basal fluorescence (F0) was measured in the dark prior to the first saturating flash. Red saturating flashes (6,000 mmol photons m−2 s−1, 600 ms) were delivered to measure Fm (in the dark) and Fm′ (in the light). PSII maximum yields were calculated as (Fm – F0)/Fm, and PSII yield for each light intensity was measured with a saturating flash after 2–3 min of illumination and was calculated from (Fm′ – F)/Fm′. The apparent ETR was calculated as the product of light intensity and PSII yield. MIMS was used to measure gas exchange as described previously (Burlacot et al., 2018) .
Transmission electron microscopy
Cells were grown photoautotrophically in photobioreactors under 40 µmol photons m−2 s−1 in turbidostat (OD880nm at 0.4). The algal cells were collected by centrifugation (1,500g, 1 min) and were immediately fixed with 2.5% (v/v) glutaraldehyde in 0.1 M, pH 7.4 sodium cacodylate buffer for 2 d at 4°C. They were then washed by resuspending 5 min in the same buffer for three times. Samples were post-osmicated with 1% (w/v) osmium tetroxyde in cacodylate buffer for 1 h, dehydrated through a graded ethanol series, and finally embedded in monomeric resin Epon 812. All chemicals used for histological preparation were purchased from Electron Microscopy Sciences (Hatfield, USA). Ultra-thin sections for TEM (90 nm) were obtained by an ultramicrotome UCT (Leica Microsystems GmbH, Wetzlar, Germany) and mounted on copper grids and examined in a Tecnai G2 Biotwin Electron Microscope (ThermoFisher Scientific FEI, Eindhoven, the Netherlands) using an accelerating voltage of 100 kV and equipped with a CCD camera Megaview III (Olympus Soft imaging Solutions GmbH, Münster, Germany).
Isolation of thylakoids and native PAGE
Thylakoids were isolated according to a protocol described previously (Chua and Bennoun, 1975). All steps were performed on ice or at 4°C with as little light as possible. Briefly, cells were pelleted and resuspended in 8-mL 25-mM HEPES, 5-mM MgCl2, 0.3-M sucrose, pH 7.5 with a protease inhibitor cocktail for plant cell and tissue extracts (Sigma P9599). Cells were disrupted with French press at a pressure of 6,000 psi. Total membranes were collected by centrifugation (1,000g, 10 min) and washed first in 5-mM HEPES, 10-mM EDTA, 0.3-M sucrose, pH 7.5 and then in 5-mM HEPES, 10-mM EDTA, 1.8-M sucrose, pH 7.5. Sucrose gradient was 0.5-M sucrose (5 mL), 1.3-M sucrose (2 mL), and 1.8-M sucrose, initially containing thylakoids (5 mL). After ultracentrifugation (274,000g, 1h), thylakoids were collected at the interface between 0.5 and 1.3 M sucrose. They were washed with 5-mM HEPES, 10-mM EDTA, pH 7.5 (with or without 0.5-M NaCl), and resuspended at 1-mg mL−1 chlorophyll for subsequent SDS–PAGE analysis. For nondenaturing conditions, thylakoids were resuspended in NativePAGE sample buffer (Life technologies) at 1-mg mL−1 chlorophyll, then thylakoids were solubilized for 30 min on ice in the same volume of 1% (w/v) n-dodecyl-alpha-d-maltoside, 1% (w/v) digitonine (final concentrations: 0.5-mg mL−1 chlorophyll, 0.5% (w/v) n-dodecyl-alpha-d-maltoside, and 0.5% (w/v) digitonine). For each sample, 20 µL were then loaded with 2 µL of G-250 sample additive (Life technologies) on 4–16% (w/v) NativePAGE gels (Life technologies). Cathode running buffer (Life technologies) was supplemented with 0.02% (w/v) G-250 for two-thirds of the migration, and with 0.002% (w/v) G-250 for the remaining third. Annotation of observed bands was done according to a previous publication (Pagliano et al., 2012). For immunoblot analysis, native gel was incubated in Tris Glycine SDS buffer, 10% (v/v) ethanol for 15 min and transferred onto polyvinylidene fluoride membrane using XCell II Blot module (25v, 1 h). Immunodetection was performed as described above.
Based on C16:1(3t) FAME, we determined a factor of enrichment expected for a compound that would be located exclusively within thylakoids [ratio EF=C16:1 (3t) FAMEwhole cells/C16:1 (3t) FAMEthylakoids]. Considering the amount of C17:1 alkene found in thylakoids, we calculated the expected content in whole cells, which equals C17:1 alkthylakoids*ratio EF. This calculated value for thylakoids was divided by the value for whole cells that had been determined experimentally, which gives the proportion of C17:1 alkene that is present in thylakoids.
Lipidomic analysis by UPLC-MS/MS
Lipid molecular species analysis was done by UPLC-MS/MS. Lipids were first extracted with a modified hot isopropanol method. Briefly, C. reinhardtii cells were harvested by centrifugation at 1,000 g for 5 min in glass tubes. Pellets were immediately resuspended in 1 mL of hot isopropanol (85°C) containing 0.01% (w/v) butylated hydroxytoluene. Sealed tubes were heated at 85°C for 10 min to inactivate lipases. Internal standards were added. Lipids were then extracted in 3-mL methyl tert-butyl ether (MTBE) with a phase separation with 1 mL of water. Organic phase was collected and aqueous phase was washed with an additional milliliter of MTBE. Organic phases were evaporated under a gentle nitrogen stream and resuspended in 500 μL of a mixture of acetonitrile/isopropanol/ammonium acetate 10 mM (65:30:5, v/v/v). Lipid molecular species were analyzed on an ultimate RS 3000 UPLC system (ThermoFisher, Waltham, USA) connected to a quadrupole-time-of-flight 5,600 mass spectrometer (AB Sci ex, Framingham, MA, USA) equipped with a duo-spray ion source operating in positive mode. Lipid extracts were first separated on a Kinetex™ (Kinetex, Atlanta, USA) C182.1 × 150 mm 1.7 μm column (Phenomenex, Torrance, USA). Two solvent mixtures, acetonitrile–water (60:40, v/v) and isopropanol–acetonitrile (90:10, v/v), both containing 10-mM ammonium formate at pH 3.8, were used as eluents A and B, respectively. A 32-min-long binary gradient elution was performed; eluent B was increased from 27% to 97% in 20 min and the mixture was maintained for 5 min; eluent B was then decreased to 27% and the mixture maintained for another 7 min for column re-equilibration. The flowrate was 0.3 mL min−1 and the column oven temperature was maintained at 45°C. Lipid identification was based on retention time and on mass accuracy peaks from the MS survey scan compared with theoretical masses and fragment ions from MS/MS scan. Relative quantification was achieved with multiquant software (AB Sciex, USA) on the basis of intensity values of extracted masses of the different lipids previously identified. Detector response was normalized by the quantity of FAME previously measured by GC–MS for each sample.
Phylogenetic analysis and logo sequences
The CvFAP protein sequence was used as bait and blasted against different databases using tBLASTn or BLASTp (including NCBI, Phytozome, Fernbase, and Unigene TARA ocean databases). Sequences from the BLAST were pooled with reference sequences from a previous tree of the GMC oxidoreductase superfamily (Zámocký et al., 2004). Alignment of sequences was done with Muscle algorithm (Edgar, 2004) and viewed with Seaview software (Gouy et al., 2010). Selection of conserved sites was done with Gblock. A set of 226 conserved positions were used for tree construction using maximum likelihood algorithm (PhyML, with LG matrix) with 100 replicates for bootstrap analysis. Annotation of the tree was done using annotation data provided by TARA. FAP Logo sequence was based on 35 sequences including at least one sequence of each taxa from the phylogeny. GMC logo sequence was based on sequences of non-FAP GMC oxidoreductases. Alignment of sequences was done using Muscle algorithm and viewed with Seaview software. Construction of Logo sequences was done using WebLogo (https://weblogo.berkeley.edu/logo.cgi).
Heterologous expression of FAPs in E. coli
Coding sequences of putative FAPs from G. sulphuraria (Supplemental Figure S13) and N. gaditana (Supplemental Figure S14) were directly amplified from cDNAs obtained by reverse transcription of total RNAs. The FAP homologs from Galdieria, Chondrus, and Ectocarpus were codon-optimized and synthesized, then cloned into pLIC07 as described before for CvFAP and CrFAP (Sorigué et al., 2017). Potential transit peptides were removed for better expression in E. coli, and N-terminal sequences were as follows: GFDRSREFDYVIVGGG for Galdieria; SSEAATTYDYIIVGGG for Chondrus; LQSVSMKAPAAVASSTYDYIIVGGG for Nannochloropsis; SMSVAEEGHKFIIIGGG for Ectocarpus. Escherichia coli was cultivated in Terrific broth medium at 37°C until OD600nm reached 0.8. Expression was then induced with 0.5-mM IPTG and cultures transferred at 22°C under 100 µmol photons m−2 s−1.
77K fluorescence emission spectra
Low temperature spectra were measured on whole cells at 77K using a SAFAS Xenius optical fiber fluorescence spectrophotometer (Dang et al., 2014). About 200 µL of light-adapted cell suspension was frozen in a liquid nitrogen bath cryostat. The excitation wavelength used was 440 nm, and detection wavelength ranged from 600 to 800 nm with a 5 nm split. Fluorescence emission spectra were all normalized to the 686 nm signal.
Accession numbers
FAP is referenced under the enzyme classification number E.C. 4.1.1.106. Accession numbers of FAP genes expressed in this study are XP_005842992 (C. variabilis NC64A), XP_001703004 (C. reinhardtii), CBJ25560 (E. silicosus), and XP_005714951 (C. crispus).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of GMC oxidoreductase superfamily.
Supplemental Figure S2. Logo sequences for FAPs and other GMC oxidoreductases.
Supplemental Figure S3. FAP is predicted to be localized in plastids in many algae
Supplemental Figure S4. Ultrastructure of C. reinhardtii WT and fap strains.
Supplemental Figure S5. FAP gene expression during day–night cycles and hypothetical mechanism that may explain HC loss.
Supplemental Figure S6. Growth curves and cell volume of WT and fap strains.
Supplemental Figure S7. Growth of WT and fap strains using various concentrations of salt.
Supplemental Figure S8. Fatty acid profile in mixotrophic conditions.
Supplemental Figure S9. Variation in the proportion of each fatty acid in total fatty acids during cell cycle.
Supplemental Figure S10. Profiles of major glycerolipid species in WT, fap, and complementation strains.
Supplemental Figure S11. Photosynthetic activity in fap and WT strains.
Supplemental Figure S12. Fatty acid acclimation to cold conditions.
Supplemental Figure S13. Sequence of G. sulphuraria FAP deduced from the cDNA cloned.
Supplemental Figure S14. Sequence of N. gaditana FAP deduced from the cDNA cloned.
Supplemental Table S1. Multiple alignment of FAP sequences used for the phylogenetic analysis (text file).
Supplemental Table S2. List of GMC oxidoreductases used for the phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Dr Olivier Vallon for help with analysis of some sequenced algal genomes and helpful discussions. Thanks are due to Dr Quentin Carradec and Dr Patrick Wincker for providing access to TARA sequences and to Dr Florence Corellou, Dr Eric Maréchal, and Prof. Stefano Caffarri for useful discussions. Help of Dr Philippe Ortet and Emmanuelle Billon with analysis of TARA sequences is also acknowledged.
Funding
This project received funding from CEA (DRF Impulsion Invention E2FAP to F.B.) and from Agence Nationale de la Recherche (PHOTOALKANE, N° ANR-18-CE43-0008-01, to G.P.). This work was also supported by the HelioBiotec platform funded by the EU, the Région Sud, the French Ministry of Research, and the C commissariat à l'énergie atomique et aux énergies alternatives (CEA). S.L.Y.M. has received a PhD scholarship from Ecole Normale Supérieure Paris and the French Ministry of Education and Research.
Conflict of interest statement. None declared.
F.B. conceived the original research project; F.B., S.L.Y.M., and G.P. designed the experiments and analyzed the data; S.L.Y.M., A.B.-A., S.C., S.B., D.S., B.L., A.B., P.P.S. performed experiments; M.F. performed the transmission electronmicroscopic study; S.L.Y.M. performed phylogenetic analysis; F.B. and S.M. wrote the article with contributions from Y.L.-B. and G.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Fred Beisson (frederic.beisson@cea.fr).
References
- Beller HR, Goh E-B, Keasling JD (2010) Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol 76:1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berla BM Saha R Maranas CD Pakrasi HB (2015) Cyanobacterial alkanes modulate photosynthetic cyclic electron flow to assist growth under cold stress. Sci Rep 5: 14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure J-D, Haslam RP, Napier JA, Lessire R, Joubès J (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24:3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björn LO, editor (2015) Photoactive proteins. InPhotobiology, The Science of Light and Life, Ed 3. Springer, New York, pp 139–150 [Google Scholar]
- Blaby-Haas CE, Merchant SS (2019) Comparative and functional algal genomics. Annu Rev Plant Biol 70:605–638 [DOI] [PubMed] [Google Scholar]
- Burlacot A, Sawyer A, Cuiné S, Auroy-Tarrago P, Blangy S, Happe T, Peltier G (2018) Flavodiiron-mediated O2 photoreduction links H2 production with CO2 fixation during the anaerobic induction of photosynthesis. Plant Physiol 177:1639–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Bennoun P (1975) Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA 72: 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates RC, Podell S, Korobeynikov A, Lapidus A, Pevzner P, Sherman DH, Allen EE, Gerwick L, Gerwick WH(2014) Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS One 9:e85140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang K-V, Plet J, Tolleter D, Jokel M, Cuiné S, Carrier P, Auroy P, Richaud P, Johnson X, Alric J, et al. (2014) Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26:3036–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, et al. (2015) Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224 [DOI] [PubMed] [Google Scholar]
- Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T (2015) Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J 81:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- Herman NA, Zhang W (2016) Enzymes for fatty acid-based hydrocarbon biosynthesis. Curr Opin Chem Biol 35:22–28 [DOI] [PubMed] [Google Scholar]
- Heyes DJ, Lakavath B, Hardman SJO, Sakuma M, Hedison TM, Scrutton NS (2020) Photochemical mechanism of light-driven fatty acid photodecarboxylase. ACS Catal 10:6691–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R, Kunst L (2008) Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J 54:670–683 [DOI] [PubMed] [Google Scholar]
- Jiménez-Díaz L, Caballero A, Pérez-Hernández N, Segura A (2017) Microbial alkane production for jet fuel industry: motivation, state of the art and perspectives. Microb Biotechnol 10:103–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoot CJ, Pakrasi HB (2019) Diverse hydrocarbon biosynthetic enzymes can substitute for olefin synthase in the cyanobacterium Synechococcus sp. PCC 7002. Sci Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakavath B, Hedison TM, Heyes DJ, Shanmugam M, Sakuma M, Hoeven R, Tilakaratna V, Scrutton NS (2020) Radical-based photoinactivation of fatty acid photodecarboxylases. Anal Biochem 600:113749. [DOI] [PubMed] [Google Scholar]
- Lea-Smith DJ, Biller SJ, Davey MP, Cotton CAR, Perez Sepulveda BM, Turchyn AV, Scanlan DJ, Smith AG, Chisholm SW, Howe CJ (2015) Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc Natl Acad Sci USA 112:13591–13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea-Smith DJ, Ortiz-Suarez ML, Lenn T, Nürnberg DJ, Baers LL, Davey MP, Parolini L, Huber RG, Cotton CAR, Mastroianni G, et al. (2016) Hydrocarbons are essential for optimal cell size, division, and growth of cyanobacteria1[OPEN]. Plant Physiol 172:1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RE, editor (2008) Basic characteristics of the algae. InPhycology, Ed 4. Cambridge University Press, New York, p 3 [Google Scholar]
- Lee SB, Suh MC (2013) Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol Plant 6:246–249 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. (2016) An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28:367–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Li S (2020) Biosynthesis of fatty acid-derived hydrocarbons: perspectives on enzymology and enzyme engineering. Curr Opin Biotechnol 62:7–14 [DOI] [PubMed] [Google Scholar]
- Los DA, Mironov KS, Allakhverdiev SI (2013) Regulatory role of membrane fluidity in gene expression and physiological functions. Photosynth Res 116:489–509 [DOI] [PubMed] [Google Scholar]
- Mendez-Perez D, Begemann MB, Pfleger BF (2011) Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Environ Microbiol 77:4264–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin S, Légeret B, Blangy S, Sorigué D, Burlacot A, Auroy P, Li-Beisson Y, Peltier G, Beisson F (2019) Continuous photoproduction of hydrocarbon drop-in fuel by microbial cell factories. Sci Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726 [DOI] [PubMed] [Google Scholar]
- Pagliano C, Barera S, Chimirri F, Saracco G, Barber J (2012) Comparison of the α and β isomeric forms of the detergent n-dodecyl-D-maltoside for solubilizing photosynthetic complexes from pea thylakoid membranes. Biochim Biophys Acta 1817:1506–1515 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, Fricaux T, Taquet N, Blomquist GJ, Feyereisen R (2012) An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci USA 109:14858–14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A (2011) Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol 77:1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Z, Li X, Zhu X, Liu J, Domigan B, Barr I, Cate JHD, Zhang W (2014) Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc Natl Acad Sci USA 111:18237–18242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB (2010) Microbial biosynthesis of alkanes. Science 329:559–562 [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW (2014) A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella1[W][OPEN]. Plant Physiol 164:1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorigué D, Légeret B, Cuiné S, Blangy S, Moulin S, Billon E, Richaud P, Brugière S, Couté Y, Nurizzo D, et al. (2017) An algal photoenzyme converts fatty acids to hydrocarbons. Science 357:903–907 [DOI] [PubMed] [Google Scholar]
- Sorigué D, Légeret B, Cuiné S, Morales P, Mirabella B, Guédeney G, Li-Beisson Y, Jetter R, Peltier G, Beisson F (2016) Microalgae synthesize hydrocarbons from long-chain fatty acids via a light-dependent pathway. Plant Physiol 171:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29:3625–3639 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57:151–168 [DOI] [PubMed] [Google Scholar]
- Valentine DL, Reddy CM (2015) Latent hydrocarbons from cyanobacteria. Proc Natl Acad Sci USA 112:13434–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T, Kageyama H, Tanaka Y, Takabe T (2018) Requirement of alkanes for salt tolerance of Cyanobacteria: characterization of alkane synthesis genes from salt-sensitive Synechococcus elongatus PCC7942 and salt-tolerant Aphanothece halophytica. Lett Appl Microbiol 67:299–305 [DOI] [PubMed] [Google Scholar]
- Yunus IS, Wichmann J, Wördenweber R, Lauersen KJ, Kruse O, Jones PR (2018) Synthetic metabolic pathways for photobiological conversion of CO2 into hydrocarbon fuel. Metab Eng 49:201–211 [DOI] [PubMed] [Google Scholar]
- Zámocký M, Hallberg M, Ludwig R, Divne C, Haltrich D (2004) Ancestral gene fusion in cellobiose dehydrogenases reflects a specific evolution of GMC oxidoreductases in fungi. Gene 338:1–14 [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Kerkhoven EJ, Nielsen J (2018) Barriers and opportunities in bio-based production of hydrocarbons. Nature Energy 3:925–935 [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27:2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.