Abstract
Magnesium (Mg) and calcium (Ca) are essential mineral nutrients poorly supplied in many human food systems. In grazing livestock, Mg and Ca deficiencies are costly welfare issues. Here, we report a Brassica rapa loss-of-function schengen3 (sgn3) mutant, braA.sgn3.a-1, which accumulates twice as much Mg and a third more Ca in its leaves. We mapped braA.sgn3.a to a single recessive locus using a forward ionomic screen of chemically mutagenized lines with subsequent backcrossing and linked-read sequencing of second back-crossed, second filial generation (BC2F2) segregants. Confocal imaging revealed a disrupted root endodermal diffusion barrier, consistent with SGN3 encoding a receptor-like kinase required for normal formation of Casparian strips, as reported in thale cress (Arabidopsis thaliana). Analysis of the spatial distribution of elements showed elevated extracellular Mg concentrations in leaves of braA.sgn3.a-1, hypothesized to result from preferential export of excessive Mg from cells to ensure suitable cellular concentrations. This work confirms a conserved role of SGN3 in controlling nutrient homeostasis in B. rapa, and reveals mechanisms by which plants are able to deal with perturbed shoot element concentrations resulting from a “leaky” root endodermal barrier. Characterization of variation in leaf Mg and Ca accumulation across a mutagenized population of B. rapa shows promise for using such populations in breeding programs to increase edible concentrations of essential human and animal nutrients.
“Leaky” root Casparian strips lead to accumulation of two-fold greater magnesium and ∼35% greater calcium concentrations in leaves of a Brassica rapa mutant.
Introduction
More than 2 billion people worldwide suffer from dietary micronutrient deficiencies, in particular of iron, iodine, vitamin A, and zinc (Gödecke et al., 2018). This is linked to ∼3% of the total global burden of disease and represents more than 70 million disability adjusted life years lost annually (Stein, 2014). Less attention is focused on other essential mineral nutrients of potential dietary concern such as magnesium (Mg) and calcium (Ca; Kumssa, et al., 2015a, 2015b). However, food supply data suggest that around half the global human population is at risk of dietary Ca deficiency, rising to more than 95% in 16 countries in Africa (Joy et al., 2014; Kumssa et al., 2015a).
Magnesium deficiency risks are reportedly greater in high-income countries (HICs) than in low-income countries (LICs; Kumssa et al., 2015b). For example, dietary intake surveys report that ∼9% of the combined USA and UK adult populations (Broadley and White, 2010), and 27% and 50% of UK boys and girls, respectively, aged 11–18 years (Public Health England, 2018), consume Mg at amounts below the Lower Reference Nutrient Intake (LRNI). Low Mg intake in HICs may be due to a high incidence of the consumption of processed foods (Rosanoff and Kumssa, 2020). Low dietary supply of Ca, especially in many LICs, is due to limited consumption of animal source foods and leafy vegetables, and high consumption of low-Ca starchy staples (Kumssa et al., 2015a). Dietary deficiencies of Mg (hypomagnesemia) and Ca (hypocalcemia) are also a pressing welfare and economic issue in many livestock food systems, especially in temperate regions (Kumssa et al., 2019; Friend et al., 2020). These can result from insufficient dietary Mg and Ca as well as high levels of dietary potassium (K), which can interfere with the uptake of Mg and Ca in the gut. Deficiency of these nutrients in livestock can lead to conditions including milk fever, grass tetany, and staggers (Suttle, 2010; Schonewille, 2013; Friend et al., 2020), which can induce coma and death of affected animals if not treated.
Biofortification of crop plants with Mg and Ca has been proposed to improve dietary intakes in both human (White and Broadley, 2009) and livestock (Kumssa et al., 2019; Penrose et al., 2020) food systems. For example, varieties of pasture grasses containing 30%–50% greater leaf concentrations of Mg and 10%–18% more Ca than typical varieties were selectively bred in the 1970s and 1980s (Kumssa et al., 2019; Penrose et al., 2020). However, these have not yet been widely adopted in production systems. In Brassica crops, shoot Mg and Ca concentrations are highly heritable traits (Broadley et al., 2008). Associated genetic loci and candidate genes with allelic variation in thale cress (Arabidopsis thaliana) have previously been reported (Graham et al., 2014; Alcock et al., 2017). However, to date, direct evidence of a causal link between gene and function in Mg and Ca accumulation has not yet been reported for any crop species.
Here, we report the isolation of a mutant line of Brassica rapa subsp. trilocularis (Roxb.) that accumulates more than twice as much leaf Mg as wild-type (WT) plants on a concentration basis, as well as ∼35% more Ca. These traits are associated with reduced leaf K concentration, but without apparent deleterious effects on growth or development. We identified a single SNP within a putative ortholog of A. thaliana SCHENGEN3 (SGN3) as the causative mutation and demonstrate its effects on ionomic and physiological traits, including disrupted root endodermal extracellular diffusion barrier function.
Results
EMS mutagenesis to generate ionomic variation in B. rapa
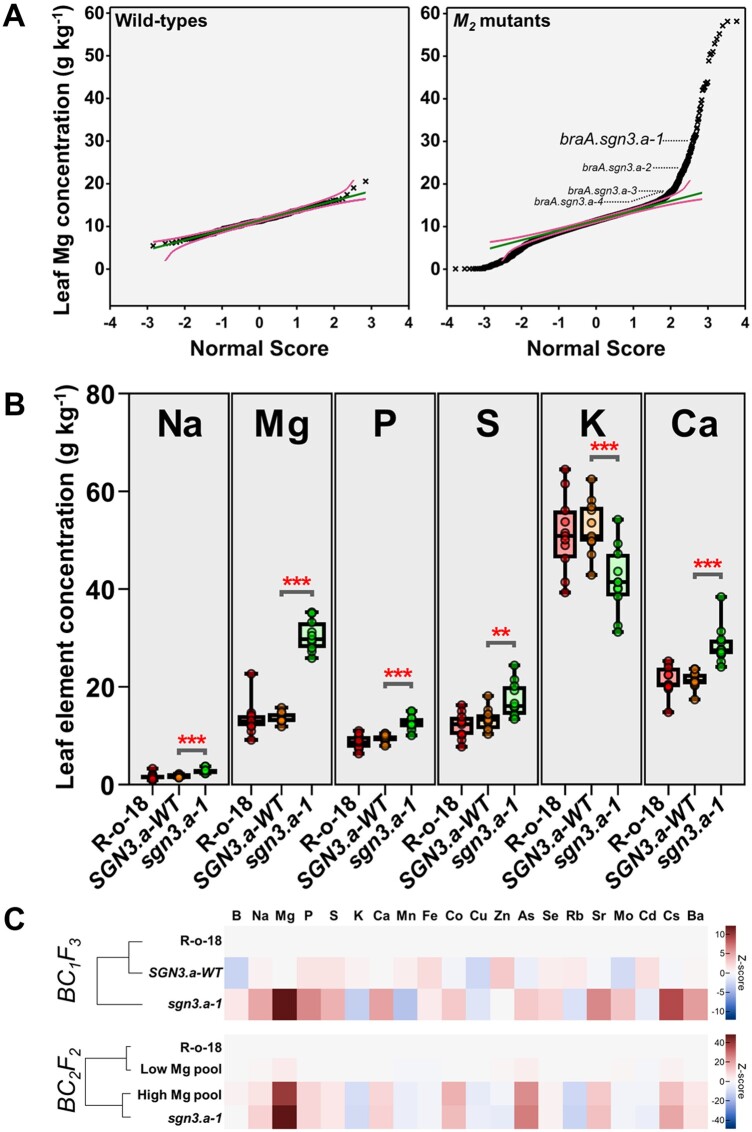
To identify B. rapa mutant lines with elevated concentrations of Mg and Ca, we characterized ionomic variation across a population of 3,464 B. rapa subsp. trilocularis var. R-o-18 M2 lines previously subjected to ethyl methane sulfonate (EMS) mutagenesis (Figure 1; Stephenson et al., 2010). Similar approaches have successfully isolated ionome mutants in closely related A. thaliana (Lahner et al., 2003; Huang and Salt, 2016). Extensive variation in leaf Mg concentration was observed across the mutant population, with maximum leaf concentrations more than twice as large in mutants than in wild-type plants, on a dry weight (DW) basis (Figure 2A;Supplemental Table S1). Similar levels of variation were also observed for leaf Ca concentration across the mutagenized population (Supplemental Table S1).
Figure 1.
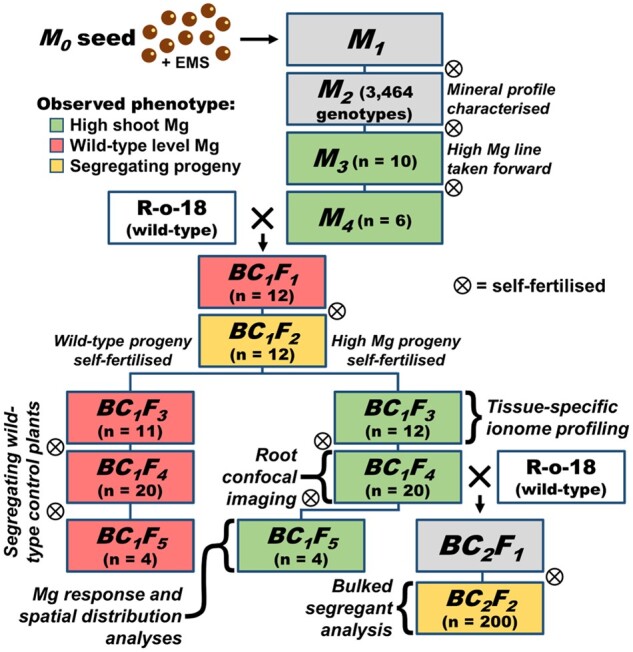
Descent of B. rapa braA.sgn3.a-1 from EMS application to M0 seed, through two backcrosses to wild-type (WT) R-o-18, leading to high and low Mg segregant groups and segregating F2 populations. Cell color represents Mg phenotype among sibling plants (where analyzed) where green = high Mg phenotype in all sibling plants, red = low Mg phenotype in all sibling plants, and yellow = segregating sibling plants. Specific analyses performed indicated next to relevant generations for reference. M = mutant generation. BC = back-cross. F = filial generation.
Figure 2.
Isolation and ionomic characterization of braA.sgn3.a-1. A, M2 generation fourth leaf magnesium (Mg) concentrations (g kg-DW−1; y-axis) in WT (n = 269) and EMS mutagenized plants (n = 7,611) as a function of expected values with a normal distribution (x-axis). Green lines in each panel show expected distributions based on WT samples with 95% simultaneous confidence bands shown as magenta lines. Outlier samples with Mg concentrations < LOD and > 5 standard deviations above global mean were removed. braA.sgn3.a-1 and candidate alternative alleles are indicated. B, Fourth leaf concentrations of elements present at greater than 0.1% DW (w/w) in BC1F3 generation braA.sgn3.a-1 (sgn3.a-1), WT segregants (SGN3.a-1) and un-mutagenized R-o-18 (all n = 11). Boxes represent interquartile range with the median shown. Whiskers show entire data range, with data points shown. Stars above boxes represent differences (two-tailed T tests) between braA.sgn3.a-1 and BraA.SGN3.a-WT at **P < 0.01, and ***P < 0.001. C, Heatmap showing full ionomic profiles (Z-scores) of BC1F3 and BC2F2 mutant and WT plants as measured by ICP-MS. Ionome profiles of braA.sgn3.a-1 and BraA.SGN3.a-WT relative to WT R-o-18 shown for BC1F3 (all n = 11). Ionome profiles of high (n = 56) and low (n = 144) Mg segregants relative to R-o-18 (n = 10) shown for BC2F2, with sgn3.a-1 control plant (n = 10) ionomic profiles included for reference.
We selected self-fertilized M3 seed from these lines for further study by identifying lines with leaf Mg concentrations over three standard deviations above the mutant population-wide mean (n = 83 genotypes; leaf Mg concentrations from 22.5 to 58.2 g kg-DW−1). Seven of these genotypes went on to set viable seed, one of which maintained the high Mg phenotype beyond the M3 generation (Supplemental Tables S2, S3). Male donor M4 plants of this mutant line were backcrossed to wild-type R-o-18 plants to reduce background mutation load and to generate segregating wild-type plants (Figure 1). All plants of the backcrossed, first filial generation (BC1F1) exhibited leaf Mg concentrations that were statistically indistinguishable from R-o-18 wild-type plants (t(22) = −0.73, P = 0.473), indicating a recessive mutation (Supplemental Table S4). This was further demonstrated in BC1F2; progeny derived from self-fertilizing four F1 parents, each derived from one of two independent backcross events, all presented segregating leaf Mg phenotypes, with 32.6% of all BC1F2 plants exhibiting the high leaf Mg trait (n = 46; Supplemental Table S5).
High Mg lines were maintained by single-seed descent through to BC1F5 (Figure 1). Figure 2B shows leaf concentrations of elements present at greater than 0.1% DW (w/w; Na, Mg, P, S, K, and Ca) in both high and low Mg BC1F3 segregants (typical of all generations). High and low Mg segregants are labeled sgn3.a-1 and SGN3.a-WT, respectively, corresponding to the causative mutation described in the next section. Plants of SGN3.a-WT have similar concentrations of each of Na, Mg, P, S, K, and Ca as wild-type R-o-18 plants, indicating loss of the mutation responsible for the high Mg trait. Concentrations of Na, Mg, P, Ca (all P < 0.001; DF = 20) and S (t(20) = 3.24, p = 0.004) were all greater in sgn3.a-1 than SGN3.a-WT. The greatest difference was observed for Mg, with a mean leaf concentration of 30.5 g kg-DW−1 in sgn3.a-1 compared with 13.6 g kg-DW−1 in SGN3.a-WT, corresponding to a 2.24-fold difference. The second largest increase was observed for Ca concentration, with a mean leaf concentration of 28.5 g kg-DW−1 in sgn3.a-1 compared with 21.4 g kg-DW−1 in SGN3.a-WT (33.2% increase). Conversely, leaf K concentration was 20.5% lower in sgn3.a-1 than SGN3.a-WT (P < 0.001), with mean concentrations of 41.8 and 52.6 g kg-DW−1, respectively (Figure 2B;Supplemental Table S6). Similar differences in element concentrations between sgn3.a-1 and SGN3.a-WT were observed in other leaf tissues, including leaves 1 and 8. Larger Mg concentrations were also observed in cauline leaves, flowers, mature pods, and root tissue of sgn3.a-1 than SGN3.a-WT (Supplemental Figure S1 and Supplemental Table S7). Complete ionomic profiles of BC1F3sgn3.a-1 and SGN3.a-WT are represented via Z-scores in Figure 2C, where more positive Z-scores indicate greater elemental concentrations compared to R-o-18, and more negative Z-scores indicate lower elemental concentrations compared to R-o-18. Seed yields of BC2F2 plants were statistically indistinguishable between high and low Mg segregants (t(65) = −0.87, P = 0.388; Supplemental Table S8).
Mutant ionomic perturbations are linked to the loss-of-function of SCHENGEN3, a receptor-like kinase responsible for Casparian strip formation
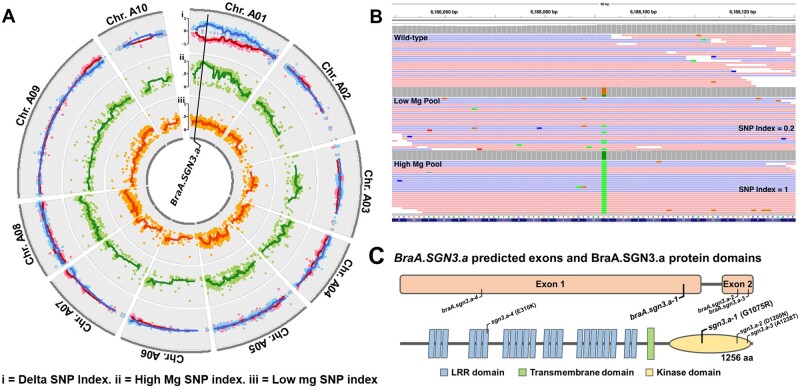
We identified the mutation controlling the high Mg mutant phenotype through bulked-segregant analysis. A population of 200 BC2F2 plants was characterized for ionomic perturbations (Supplemental Table S9). Linear discriminant analysis based on concentrations of elements present at greater than 0.1% DW (w/w; Na, Mg, P, S, K, and Ca) revealed two distinct segregation groups (segregation ratio 56:144; Supplemental Figure S2), with mean leaf Mg concentrations of 28.6 and 11.7 g kg-DW−1, respectively. These values closely corresponded to mean leaf Mg concentrations in BC1F4 high Mg reference (29.2 g kg-DW−1) and wild-type R-o-18 plants (11.0 g kg-DW−1). Bulked segregant analysis revealed a single, high confidence QTL associated with ionomic variation between the two segregant groups. This was identified using locus-specific SNP Indices for each pooled sample, i.e. the proportion of mapped reads that displayed the alternative SNP call at each locus in each pool (Takagi et al., 2013). The QTL was defined by a continuous region of Delta SNP Indices (difference between SNP Indices in each segregant group at a given locus) of greater than 0.6, from position 5,758,675 to 8,187,360 on chromosome A01 (Supplemental Table S10). This was linked to a region of elevated SNP indices in the high Mg segregant pool (Figure 3A, Track ii) and lower SNP indices in the low Mg segregant pool (Figure 3A, Track iii). Delta SNP indices in this region averaged 0.62 (Figure 3A, Track i), approximating the theoretically expected Delta SNP index of 0.67 at the causative locus. Thirty SNPs were identified within the candidate QTL, of which eight fell within coding regions. Four of these led to no amino acid change, and two had a high Mg SNP Index of less than one, so were disregarded. One of the remaining two candidate SNPs emerged as likely to be responsible for the observed ionomic perturbations, based on predicted gene function. This was found at position A01:6,186,092, with SNP indexes of 1.00 and 0.196 in high and low Mg segregant pools, respectively (Figure 3B;Supplemental Figure S3). This SNP corresponded to a mutation in the gene A01p012600.1_BraROA, a putative ortholog of A. thaliana SCHENGEN3 (At.SGN3; AT4G20140; CDS alignment E = 0) referred to here as BraA.SGN3.a.
Figure 3.
Genetic characterization of braA.sgn3.a-1. A, SNP Indices representing allelic depth of called, filtered SNPs in low (track iii; orange) and high (track ii; green) magnesium (Mg) DNA pools with delta SNP index shown as high minus low Mg (blue) and low minus high Mg (red) SNP indices (track i). Dots represent individual SNPs. Lines represent moving average SNP index around each SNP ± 9 SNP calls. Genomic location of BraA.SGN3.a indicated. B, Allelic frequency (SNP index) of high confidence SNP within BraA.SGN3.a in R-o-18 and high and low Mg DNA pools. Twenty-five randomly selected reads shown for each pool; for all reads see Supplemental Figure S3). C, BraA.SGN3.a predicted exons and BraA.SGN3.a protein domains (as predicted by ExPASy Prosite and TMHMM 2.0). Genomic location of braA.sgn3.a-1 mutation and candidate alternative alleles shown, with predicted amino acid change.
In A. thaliana, SGN3 encodes a leucine-rich repeat receptor-like kinase, localized to the endodermal plasma-membrane precisely around the Casparian strip (Pfister et al., 2014). At.SGN3 is an essential receptor in the SCHENGEN pathway, enabling the Casparian strip to fuse into a single contiguous band (Fujita et al., 2020). At.SGN3 is activated following binding of stele-expressed CIF (CASPARIAN STRIP INTEGRITY FACTOR) ligands (Doblas et al., 2017a; Okuda et al., 2020), after which SGN1, a kinase expressed exclusively at the outer plasma membrane of the endodermis (Alassimone et al., 2016), transduces the signal, leading to localized lignin accumulation (Fujita et al., 2020). Once the endodermal barrier is sealed, CIF ligands are contained within the stele, and thus CIF-activated SGN3 is unable to signal through SGN1 (Doblas et al., 2017a). The system represents a surveillance framework, enabling lignification of the Casparian strip until complete closure or repair, mediated through SGN3.
Mutant alleles of At.SGN3 possess discontinuous Casparian strips resulting in a “leaky” extracellular diffusion barrier. This causes bidirectional leaking of solutes across the endodermis to and from the xylem, leading to increased Mg and reduced K in leaves (Karley and White, 2009; Pfister et al., 2014). BraA.SGN3.a has high sequence similarity to At.SGN3 (85.1% shared CDS bases) and conserved predicted protein topology (Figure 3C), presumably as a result of recent species divergence (Wang et al., 2011). The candidate SNP identified here results in a glycine to arginine amino acid substitution within the kinase domain, toward the protein C-terminal end. This is in close proximity to a number of amino acid substitutions previously identified in A. thaliana that lead to loss-of-function of At.SGN3 (Pfister et al., 2014). This forms compelling evidence that the candidate SNP at position A01:6,186,092 is responsible for the ionomic perturbations observed in the mutant line described here, which we name braA.sgn3.a-1.
Additional braA.sgn3.a alleles share the high shoot Mg phenotype
Additional braA.sgn3.a alleles were sought by mining the RevGen UK exome capture database of EMS-induced mutations in B. rapa (Stephenson et al., 2010). Eighteen of the described mutant genotypes were also present in our forward ionomic screen database. Three genotypes emerged as additional candidate mutant alleles, since at least one of the screened plants of each genotype (presumed to be segregating) had leaf Mg concentrations >50% greater than wild-type concentrations. The leaf Mg concentration of each of these is indicated in Figure 2A, and the genomic loci indicated in Figure 3C. Two of the three additional candidate alleles, here named braA.sgn3.a-2 and braA.sgn3.a-3, fell within the protein kinase domain, close to the location of braA.sgn3.a-1. This provides additional evidence that a mutation in this region is responsible for the observed high Mg and Ca phenotypes reported here.
braA.sgn3.a-1 shares both ionomic and root anatomical perturbations with A. thaliana sgn3 including a disrupted endodermal barrier
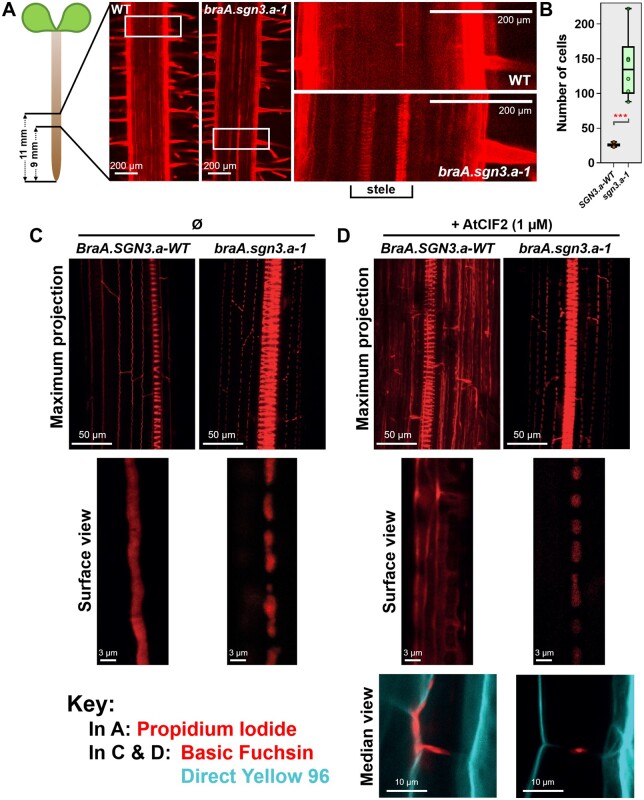
The shoot ionome of braA.sgn3.a-1 has striking similarities to At.sgn3 (Pfister et al., 2014). Notably, both mutant genotypes have greater shoot Mg concentrations than wild-type plants, and lower shoot K concentrations (Supplemental Figure S4). Root anatomical perturbations are also highly similar in sgn3 mutants of both species. In regions close to the root tip, all root cell files were similarly visible between braA.sgn3.a-1 and BraA.SGN3.a-WT seedlings following staining with the apoplastic tracer propidium iodide (PI; Supplemental Figure S5). In roots of braA.sgn3.a-1 however, PI was able to penetrate into the root stele in zones expected to have undergone endodermal differentiation at least to the stage of forming sealed Casparian strips (Doblas et al., 2017b). This was evidenced by the visibility of xylem vessels in such regions of PI stained roots (Figure 4A), suggesting that Casparian strips were not functioning as effective endodermal barriers. While PI can generally be expected to enter the root stele toward the developing root apex in wild-type plants (Alassimone et al., 2010), PI was detected much further from the root tip in braA.sgn3.a-1. In BraA.SGN3.a-WT, PI penetration to the stele was blocked after an average of 26 epidermal cells from the onset of elongation (n = 7), compared with an average of 139 epidermal cells in braA.sgn3.a-1 (n = 6; Figure 4B;Supplemental Table S11).
Figure 4.
Characterization of root endodermal barrier anatomy. A, Confocal images of 5-d-old roots of R-o-18 WT and braA.sgn3.a-1 mutant plants stained with PI (shown in red). Images are centered approximately 10 mm from root tip. Regions of 1.55 × 0.78 mm shown on left side. White boxes indicate position of enlarged regions shown to the right. Note visibility of xylem tissue in stele of braA.sgn3.a-1, indicating perturbed endodermal barrier. B, Number of epidermal cells from the onset of elongation until the endodermal cells blocked the PI penetration to the stele of roots of BraA.SGN3.a-WT (SGN3.a-WT) and braA.sgn3.a-1 (sgn3.a-1) mutant plants. Boxes represent interquartile range with the median shown. Whiskers show entire data range, with data points shown. Stars represent difference (two-tailed T test) between braA.sgn3.a-1 and BraA.SGN3.a-WT at P < 0.001. C, Confocal images of 4-d-old roots of BraA.SGN3.a-WT and braA.sgn3.a-1 stained with the lignin stain Basic Fuchsin (shown in red). Roots were imaged at 40 cells after the onset of elongation. Maximum projection of Z-stack of endodermal cells and enlarged surface view of the top of endodermal cells shown. Note non-contiguous lignin band in braA.sgn3.a-1, indicating a disrupted endodermal barrier. D, Confocal images of 4-d-old roots of BraA.SGN3.a-WT and braA.sgn3.a-1 as in (C) but in the presence of exogenous AtCIF2 peptide. Basic Fuchsin shown in red. Note ectopic lignin deposits in roots of BraA.SGN3.a-WT in the presence of AtCIF2, whereas braA.sgn3.a-1 fails to show a similar response. Endodermal cell median view shown below, with additional Direct Yellow 96 staining shown in cyan to visualize cellulose/cell wall.
We further confirmed the conserved role of B. rapa sgn3 in enabling the Casparian strip to fuse into a single contiguous band, through visualizing lignin deposition. In At.sgn3 mutants, lignin staining previously revealed discontinuities in endodermal lignification (Doblas et al., 2017a). Highly comparable results were observed here: in roots of BraA.SGN3a-WT, lignin staining with Basic Fuchsin showed continuous endodermal lignification, whereas roots of braA.sgn3.a-1 exhibited characteristically discontinuous lignification (Figure 4C). In addition, roots of braA.sgn3.a-1 did not respond to exogenous CIF2, a SGN3 ligand crucial for ensuring proper Casparian strip development. In wild-type A. thaliana, application of 1 µM exogenous CIF2 peptide leads to considerable ectopic lignin deposition, whilst no response was observed in At.sgn3 mutants (Doblas et al., 2017a). Application of 1 µM of exogenous AtCIF2 to BraA.SGN3.a-WT similarly led to widespread ectopic lignin deposition throughout the wild-type root endodermis (Figure 4D). The enlarged, median view indicates that this ectopic lignin was concentrated on the outer half of the endodermis. This presumably corresponds to the outer endodermal localization of SGN1, which transduces the CIF2 signal leading to localized lignin accumulation (Fujita et al., 2020). Exogenous CIF2 peptide application to braA.sgn3.a-1 induced no observable response (Figure 4D), further implying loss-of-function of SGN3 in this genotype, leading to a lack of transduction of the SGN3-dependent CIF2 signal.
braA.sgn3.a-1 preferentially accumulates multiple elements in leaf epidermal cells and in the apoplast
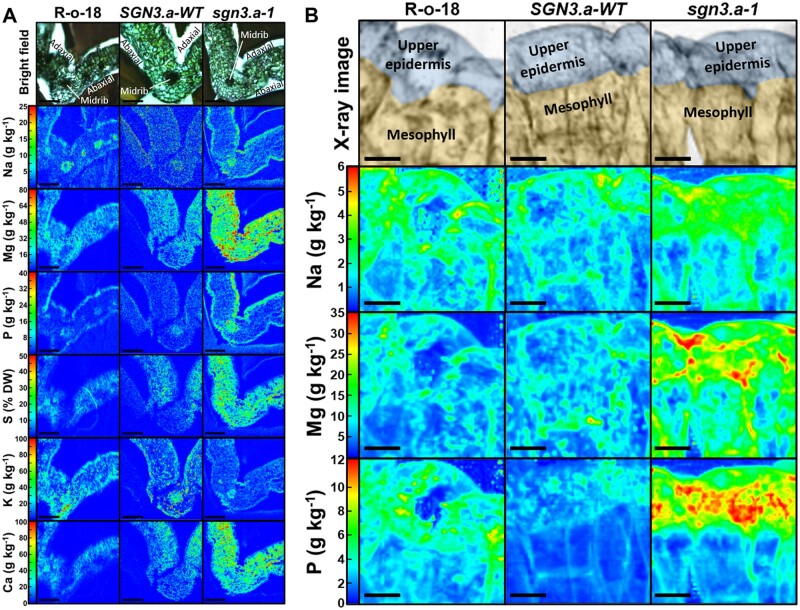
We mapped the spatial distribution of elements in leaf and seed tissue of braA.sgn3.a-1 and wild-type plants. Leaf Mg was present at larger concentrations in braA.sgn3.a-1 than wild-types in upper epidermal cells (t(5) = 4.95, P = 0.004) and across mesophyll tissue (t(5) = 8.08, P < 0.001; Figure 5A;Supplemental Figure S6). Concentrations of Ca (t(5) = 4.90, P = 0.004) and S (t(5) = 5.44, P = 0.003) were also elevated in mesophyll tissue of braA.sgn3.a-1. Concentrations of Na (t(5) = 3.41, P = 0.019) and P (t(5) = 3.42, P = 0.019) were elevated in upper leaf epidermal cells in braA.sgn3.a-1, with Na additionally elevated in lower epidermal cells (t(5) = 3.58, P = 0.016). Leaf K concentrations appeared to be lower across all leaf cell types in braA.sgn3.a-1 compared to wild-types, particularly in leaf epidermal cells (Figure 5A). However, this was accompanied by considerable variation between replicates (Supplemental Figure S6 and Supplemental Table S12). At an intracellular scale, large quantities of Na, Mg, and P appear to be stored either in upper epidermal cells or in the apoplast in braA.sgn3.a-1 (Figure 5B).
Figure 5.
Leaf spatial distribution of elements. A, Quantified spatial distributions of sodium (Na) magnesium (Mg), phosphorus (P), sulphur (S), potassium (K), and calcium (Ca) in representative first or second leaves of R-o-18, BraA.SGN3.a-WT (SGN3.a-WT) and braA.sgn3.a-1 (sgn3.a-1) as analyzed by micro-particle induced X-ray emission (µPIXE). Scale bar = 200 µm. B, Quantified spatial distributions of Na, Mg, and P in upper epidermal and mesophyll cells of R-o-18, SGN3.a-WT and sgn3.a-1 as analyzed by X-ray fluorescence. Scale bar = 20 µm. X-ray images in top panels digitally colored to show localization of upper epidermis and mesophyll.
There were no obvious differences in elemental localization between braA.sgn3.a-1 and wild-type seeds (Supplemental Figure S7). The spatial distribution of elements within wild-type seeds of B. rapa has not been reported previously. Notable observations are greater Ca and Mg concentrations in the seed coat compared with the rest of the seed, and greater concentrations of P and S across seeds compared with leaves. Elevated concentrations of P in seeds were previously observed in B. napus (Thomas et al., 2016). Elevated seed S was also observed in B. napus, though this was largely limited to vegetable and fodder varieties rather than in modern oilseed varieties (Thomas et al., 2016), where the latter have been subject to selection for lower seed concentrations of S-containing glucosinolates (Borpatragohain et al., 2016).
Magnesium concentrations in shoots of braA.sgn3.a-1 are not limited by the flux saturation observed in wild-type plants
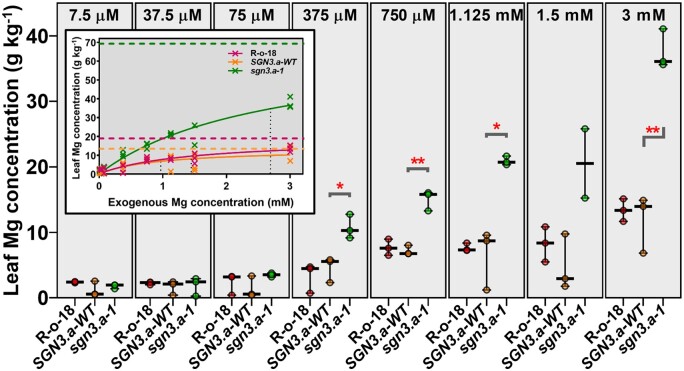
Plants of braA.sgn3.a-1, BraA.SGN3.a-WT, and wild-type R-o-18 were grown using the nutrient film technique (NFT) with solution Mg concentrations ranging from 0.0075 to 3 mM. A solution Mg concentration of 0.75 mM is routinely considered as optimum (Broadley et al., 2003; Neugebauer et al., 2018). Leaf Mg concentrations of all genotypes were positively correlated with solution Mg concentration (Figure 6;Supplemental Table S13; Simple Linear Regression analysis, all P < 0.001). However, the response of shoot Mg concentration to increasing solution Mg concentration was greater in braA.sgn3.a-1 (R2 = 93.0) than in BraA.SGN3.a-WT (R2 = 53.7) or wild-type R-o-18 (R2 = 83.2). Leaf Mg concentration was significantly greater in braA.sgn3.a-1 than BraA.SGN3.a-WT plants at solution Mg concentrations of 375 µM (t(4) = 4.01, P = 0.016), 750 µM (t(4) = 8.02, P < 0.001), 1.125 mM (t(4) = 5.38, P = 0.030), and 3 mM (t(4) = 8.31, P < 0.001), with differences becoming more marked as solution Mg concentration increased. The inset in Figure 6 shows the Michaelis–Menten relationships between shoot Mg concentration and solution Mg concentration for each genotype, used here to demonstrate the rate of Mg uptake based on exogenous concentration, and theoretical maximum that could be accumulated. At the solution Mg concentrations investigated here, both BraA.SGN3.a-WT and wild-type R-o-18 genotypes are close to approaching their Vmax, but braA.sgn3.a-1 is not. This indicates that, unlike BraA.SGN3.a-WT and wild-type R-o-18, Mg accumulation in the shoot of braA.sgn3.a-1 is not kinetically restricted at a solution Mg concentration of 3 mM, which suggests a greater apoplastic flux of Mg to the xylem of braA.sgn3.a-1. This further supports the hypothesis that the ionomic perturbations observed here are the result of a defective root endodermal barrier.
Figure 6.
Leaf Mg concentrations of BC1F5 plants grown under different exogenous Mg concentrations (indicated at top of each panel). Median value plotted in each panel where whiskers represent range with data points shown (n = 3 for each genotype for each treatment). braA.sgn3.a-1 abbreviated to sgn3.a-1, BraA.SGN3.a-WT abbreviated to SGN3.a-WT. Stars represent differences (two-tailed T tests) between braA.sgn3.a-1 and BraA.SGN3.a-WT at *P < 0.05 and **P < 0.01. Figure inset shows Michaelis–Menten kinetics curves for each genotype with Vmax and Km lines shown as dashed and dotted lines, respectively
Discussion
This work has led to the isolation of a mutant line of B. rapa, braA.sgn3.a-1, that accumulates twice as much leaf Mg and a third more Ca than wild-types, as a result of loss-of-function of SCHENGEN3 (SGN3). In A. thaliana, SGN3 encodes an essential receptor in the SCHENGEN pathway, responsible for fusing the ring-like structure surrounding root endodermal cells into a contiguous, nonpermeable band known as the Casparian strip (Fujita et al., 2020). The basis for elevated concentrations of multiple elements in leaves of braA.sgn3.a-1 is associated with disrupted formation of the root extracellular endodermal diffusion barrier, which results in a greater accumulation of elements from the external environment. We hypothesize that elevated concentrations of affected elements can be maintained due to relatively small concentration gradients between the xylem of braA.sgn3.a-1 and the external environment. The difference in accumulation between elements, such as that between the divalent cations Mg and Ca, could be linked to the magnitude of the concentration gradient between the apoplast and the endodermis. The impacts of the mutation in braA.sgn3.a-1 on shoot K concentrations may be linked to the high mobility of K within the plant (Karley and White, 2009). While there is also considerable recirculation of Mg, K is typically present at greater concentrations than other nutrients in the shoot, as well as in the xylem (Ahmad and Maathuis, 2014). An ineffective endodermal barrier could conceivably permit K to leak back into the apoplast, impacting maintenance of wild-type shoot K concentrations. At a physiological level, this work has also revealed insights into the effects of loss-of-function of sgn3 on the plant ionome, particularly in terms of inter- and intra-cellular distributions of elements. Increased concentrations of Na, Mg, and P in the upper epidermis or apoplast of leaf tissue of braA.sgn3.a-1 may be due to preferential export of excess quantities of these elements from mesophyll cells to maintain nontoxic levels, as observed across various hyperaccumulators (Leitenmaier and Küpper, 2013).
Brassica rapa includes the crop types Chinese cabbage, pak choi, turnip, and forage rape. Furthermore, B. rapa is a close relative of both B. juncea (vegetable and forage/fodder types) and B. napus (rapeseed/canola, swede/rutabaga, forage/fodder types; Wang et al., 2011). As allotetraploid species comprising a full set of diploid chromosomes from B. rapa, it is possible to introgress traits to both B. juncea and B. napus from B. rapa through conventional breeding (Wang et al., 2017). Brassica crop varieties with altered levels of the glucosinolate, glucoraphanin, have previously been developed and become commercially available (Faulkner et al., 1998; Traka et al., 2013). However, to our knowledge, a commercially viable Brassica cultivar with elevated mineral nutrient concentrations, or a crop cultivar of any species biofortified with Mg or Ca, has not previously been developed. Crossing braA.sgn3.a-1 with suitable cultivars could potentially generate new varieties with the capacity to boost human and animal dietary intakes of Mg and Ca. The reduced leaf K concentration trait in braA.sgn3.a-1 may be of additional interest in livestock forage breeding, due to the potential for K to interfere with Mg and Ca uptake in the gut (Pelletier et al., 2008). However, Casparian strip mutants are known to have multiple impacts on the leaf ionome, as well as water transport and transpiration traits (Huang and Salt, 2016; Wang et al., 2019). It is also not currently known how braA.sgn3.a-1 performs in low-K environments. Thus, further detailed characterization of braA.sgn3.a-1 is required before it is considered for introduction into commercial Brassica cultivars.
Conclusions
From a forward ionomic screen, this study has identified considerable variation in leaf Mg and Ca concentrations across an EMS-mutagenized population of B. rapa. Notably, the range of leaf concentrations of these elements is greater in the EMS population than that available in natural populations of B. napus (Thomas et al., 2016), B. oleracea (Broadley et al., 2008), or across many other species within the Brassicaceae family (Neugebauer et al., 2018). Isolation of braA.sgn3.a-1, which accumulates over twice as much Mg and a third more Ca than wild-types without any apparent impact on seed production, shows promise for using mutagenized germplasm in breeding programs to increase edible concentrations of essential human nutrients. Of significance to the usability of this germplasm for generating new cultivars is that it originates from a GMO-free background. However, further work is required in order to evaluate potential negative impacts of the induced mutations before it is introduced into Brassica breeding programs.
Materials and methods
Forward ionomic screen
Seeds of 3,464 M2 genotypes of the inbred line R-o-18 of yellow sarson (B. rapa subsp. trilocularis (Roxb.) Hanelt, 1986), previously subjected to 0.3% (v/v) EMS mutagenesis (Stephenson et al., 2010), were sown directly into fine-grade compost-based growing media (Levington Seed & Modular + Sand -F2S; Everris Ltd, Ipswich, UK) in 1 L pots and established in a single-skinned polytunnel (Visqueen Luminance Skin, Northern Polytunnels, Colne, UK) at the Sutton Bonington Campus of the University of Nottingham (52°49′58.9″ N, 1°14′59.2″ W). Pots were arranged in a randomized block design across three runs, with each mutant genotype represented once per run (n = 10,392) along with 100 R-o-18 wild-type plants per run (n = 300; Supplemental Table S14). Plants were watered twice daily by hand, and after 3 weeks were supplemented by Vitafeed® 2-1-4 nutrient solution (N-P-K: 16-8-32 + micronutrients; Vitax Ltd., Coalville, Leicestershire, UK) at a rate of 3 g L−1. Plants were covered with 380 × 900 mm microperforated pollination bags (Focus Packaging & Design Ltd, Brigg, UK) once inflorescences began to show to prevent cross-pollination. Seeds were harvested and stored at −10°C in a chilling cabinet. Generations M3–M4 and BC1F1–BC1F4 were grown in a glasshouse with supplementary artificial lighting (12 h light d−1; Philips Master GreenPower SON-T 400 W bulbs controlled by Grasslin Uni 45 timer) and were otherwise treated as above.
Three leaves were sampled from each M2 plant at the rosette stage (typically six to eight true-leaves showing). Leaf samples were dried in individual paper bags at 50°C for 48 h and then hand crushed. Leaf sub-samples of between 0.05 and 0.1 g DW (exact weights recorded) were digested using a microwave system as described previously (Thomas et al., 2016). External certified reference material (IAEA-359 Cabbage; LGC, Teddington, UK) was included in each digestion run to validate digestion efficiency.
Inductively coupled plasma mass spectroscopy (ICP-MS; Thermo Fisher Scientific iCAPQ, Thermo Fisher Scientific, Bremen, Germany) was used to analyze the concentrations of elements as described by Thomas et al. (2016). Of 10,692 potential samples, 2,713 were not analyzed due to insufficient sample being available. Element-specific limits of detection (LODs) were reported as three times the standard deviation (SD) of the operational blank concentrations, assuming a notional starting DW of 0.100 g. Elements with mean concentrations less than or close to the LOD were excluded from further analysis. Data were retained for 13 elements (Na, Mg, K, Ca, Mn, Fe, Cu, Zn, Rb, Sr, Mo, Cs, and Ba). Individual values below element-specific LODs elements were also removed from further analysis (3,996 out of 103,727 values), as well as values over five SDs above the arithmetic mean for each retained element (285 values).
Isolation and descent of braA.sgn3.a-1
M 2 genotypes with leaf Mg concentrations over three SDs above the population-wide mean Mg concentration were considered as candidates for further study. This amounted to 83 genotypes, of which 7 set seed. M3 and M4 plants of each of these lines were subsequently grown and leaf element concentrations quantified as above. Plants of a genotype that retained the high Mg trait to M4 were backcrossed to wild-type R-o-18 plants via emasculation of wild-type plants and hand-pollination with mutant pollen, to reduce background mutation load. Resulting plants were maintained by single-seed descent from generations BC1F1–BC1F5. In each generation, leaf element concentrations of all plants were quantified by ICP-MS to track the high Mg trait. The BC1F2 population was observed to be segregating for the high Mg trait, so individual high and low Mg lines were derived by self-fertilizing plants of contrasting phenotypes (Figure 1).
Tissue-specific ionome profiling
Additional ionome profiling took place on BC1F3 plants. Six plants of each of the high and low Mg segregants and wild-type R-o-18 genotypes of this generation were sampled at the rosette stage and roots were hand-washed in deionized H2O and dried in an incubator (60°C; 24 h). Root samples were grouped into three pools to attain adequate sample weights for ICP-MS analysis. An additional six plants of each genotype were grown to maturity and the first, eighth, and cauline leaves, immature pods/flowers, mature pods, stem, and mature seeds were sampled at appropriate points in the plants’ life cycle. Each sample was microwave digested and analyzed by ICP-MS as described above. Arabidopsis thaliana sgn3 mutant ionome data were extracted from the Ionomics Hub (https://www.ionomicshub.org/; Baxter et al., 2007).
Bulked-segregant analysis
Generating a segregating F2population
BC 1 F 4 mutant plants (Supplemental Table S15) were backcrossed to female wild-type R-o-18 (Figure 1). Plants of the resulting BC2F1 population were grown in glasshouse conditions as described above and self-fertilized by covering with microperforated pollination bags. BC2F2 seed from a single sibling plant was harvested for use in bulked-segregant analysis experiments. A total of 200 BC2F2, 10 wild-type R-o-18, and 10 BC1F4 high Mg reference seeds were sown directly into homogenized, high nutrient compost (Levington M3) in 2 L pots. These were distributed in the glasshouse in a randomized blocked design generated in the R package blocksdesign (version 3.8; Edmondson, 2019) with 10 blocks, each comprising 20 BC2F2 plants, 1 wild-type R-o-18 plant, and 1 BC1F4 high Mg reference plant (Supplemental Table S16). Twenty-one days after germination, third and fourth leaves were taken from each plant, dried for 5 d at 50°C, microwave-digested, and analyzed by ICP-MS as described above. Linear discriminant analysis of leaf concentrations of Na, Mg, P, S, K, and Ca, BC2F2 plants revealed two distinct segregation groups (segregation ratio 56:144) with ionomic profiles highly similar to either BC1F4 high Mg reference or wild-type R-o-18 plants (Supplemental Figure S2).
High molecular weight DNA extraction
First and second leaf tips from each segregant group and R-o-18 wild-type control plants were sampled for high molecular weight (HMW) DNA extraction. DNA was extracted using a custom CTAB extraction protocol, with HMW modifications. Wide-bore pipette tips were used throughout to prevent DNA shearing, and centrifuge speeds did not exceed 5,000g. Samples were each subjected to RNase A (Thermo Scientific EN0531) treatment, washed using equal volumes of chloroform/isoamyl alcohol (24:1), and finally resuspended in elution buffer (EB; 10-mM Tris–HCl, pH 8.5) in 1.5-mL Eppendorf Protein LowBind® tubes (Eppendorf 30108116).
DNA concentrations were quantified using a Qubit 4 Fluorometer (Invitrogen) and dsDNA BR (Broad Range) Assay Kit (ThermoFisher Q32850; Supplemental Table S17). The 40 highest yielding samples of each of the high and low Mg BC2F2 segregant groups were then pooled into separate, concentration-normalized samples. Remaining samples were retained as back-ups. Twenty R-o-18 wild-type control DNA samples were pooled into an additional concentration-normalized sample. Samples were concentrated using Agencourt AMPure XP (Beckman Coulter A63881) and size selected to remove fragments <15 kb. Sage Blue Pippin was used with the 0.75% High Pass Plus >15 kb Cassette and U1 marker (Sage; BPLUS10). Resulting products were concentrated again using AmpureXP before elution in 15 µL of EB.
Linked-read sequencing
10× Genomics Chromium Genome Library kit and Gel Bead Kit v2 (10× Genomics; 120258) were used with the Chromium Genome Chip Kit v2 (10× Genomics; 120257) the Chromium i7 Multiplex Kit (10× Genomics; 120262) and the Chromium Controller (10× Genomics; 110203) to generate linked-read libraries. The 10× Genomics Chromium Genome Library protocol was followed (https://www.10xgenomics.com/resources/support-documentation/?solution=genome&workflow-step=library-preparation) with an input of 0.8 ng of genomic DNA and unique i7 barcode pools used for each genotype. Sequencing was performed on an Illumina NextSeq500 using a High Output 300 Cycle (150 PE) kit with the following configuration: Read 1 – 150 bases; I7 – 8 bases; I5 – 0 bases; Read 2 – 150 bases. All sequence files are available in the European Nucleotide Archive (ENA) under the accession PRJEB40556.
Bioinformatics
Raw Illumina NextSeq500 reads from the four lanes run were demultiplexed using the longranger (version 2.2.2; 10× Genomics, Pleasanton, CA, USA) mkfastq pipeline, which is a wrapper around the bcl2fastq tool (version 2.19.0.316; Illumina, Inc., San Diego, CA, USA), separating out reads into the original libraries by presence of specific i7 barcodes. The longranger wgs pipeline was then used to call genotypes for the three libraries (R-o-18, Low Mg pool and High Mg pool) using GATK (version 4.0.3.0; Van der Auwera et al., 2013) against the reference B. rapa subsp. trilocularis R-o-18 (v2.2; Whole Genome Shotgun project deposited at DDBJ/ENA/GenBank under the accession JADBGQ000000000. The version described in this paper is JADBGQ010000000. The NCBI Assembly number is GCA_017639395.1). Fold coverage (read depth) was 84.6× for the high Mg DNA pool, 75.8× for the low Mg pool, and 88.1× for the R-o-18 wild-type pool. All 10× data handling was performed on a high-performance compute node with 2× Intel Xeon Gold 6138 20C 2.0GHz CPU and 192 Gb RAM.
To isolate SNPs presumed to have resulted from EMS mutagenesis, raw variant call files were filtered, using grep (GNU operating system), to only include G to A and C to T base changes that passed initial longranger/GATK filtering (PASS). High and low Mg pooled sample sequence data were then processed using the isec tool in bcftools (version 1.10, https://samtools.github.io/bcftools/) to identify sites present in both samples. Resulting SNP calls were stripped of SNPs also present in the R-o-18 DNA pool, using the isec tool in bcftools, and filtered to remove SNP calls with QUAL scores of less than 50, read depth of less than 20 and/or with the label “10X_RESCUED_MOLECULE”.
SNP index scores for each SNP in each pool were calculated using reference and alternate allelic depth scores following Takagi et al. (2013). Two-way delta SNP index scores were then calculated at each SNP locus, by subtracting the SNP index of the low Mg pool from that of the high Mg pool, and vice versa. The SNP index in the high Mg pool was expected to tend to 1 at the causative locus, meaning all reads at that locus, and thus all plants in the pool, share an alternative SNP call. In the low Mg pool, average SNP index was predicted to tend to 0.33, since approximately two-thirds of BC2F2 plants in the pool were expected to be heterozygous for the causative SNP. Delta SNP index of 0.67 was theoretically anticipated at the causative locus. A candidate QTL was identified by considering all SNPs for which the moving average (±9 SNPs) Delta SNP index was greater than 0.60. DNA sequences of exons containing SNPs were aligned to B. rapa version 1 coding sequences (Chiifu-401-42; Wang et al., 2011), to identify previously annotated gene identities. Coding sequences of genes containing SNPs within the candidate QTL were then aligned to the A. thaliana genome using BLAST functions in EnsemblPlants (Howe et al., 2020) to predict function.
Root confocal imaging
Seeds of BC1F4 high and low Mg segregant and R-o-18 lines were surface sterilized, sown onto plates containing 0.5 MS (Murashige and Skoog), pH 5.8 with 0.8%–1% agar (w/v), stratified at 4°C in the dark for 24–48 h and grown vertically in a growth chamber with 16-h photoperiod (150 μmol m−2 s−1) set to 22°C during the day and 19°C at night. After 5 d, whole seedlings were sampled and soaked in PI solution (10 µg·mL−1; Invitrogen) for 15 min in the dark to stain living cell walls. Roots were rinsed twice in deionized water, mounted on glass slides with water, and covered with a coverslip. Roots were imaged using a Leica SP8 (Leica Microsystems GmbH, Wetzlar, Germany) confocal microscope. A 514-nm laser (intensity 19.9%) was used to excite PI fluorescence, which was detected using a photomultiplier tube at between 592 and 726 nm. Gains were optimized and kept consistent throughout the experiment. Images were generated using Z-stack functions in LAS-X Leica software. Images are the average of four frames per pixel, with six Z-steps captured per region. Using a Leica CTR5000 fluorescence microscope, 20× magnification, we additionally quantified the number of epidermal cells from the onset of elongation until the endodermal cells blocked the PI penetration to the stele.
For CIF2 experiments, seedlings were grown in the presence or absence of 1-µM AtCIF2 peptide (DY(SO3H)GHSSPKPKLVRPPFKLIPN; Cambridge Peptides Ltd) in the growth media. Seedlings were sampled after 4 d, fixed with 4% paraformaldehyde for 120 min at 20°C, washed twice for 1 min with 1× phosphate-buffered saline, and transferred to ClearSee solution for 24 h as described previously (Ursache et al., 2017). Seedlings were then stained overnight in 0.2% Basic Fuchsin (Fluka, Analytical) in ClearSee for lignin visualization. Basic Fuchsin solution was removed and the seedlings were washed three times for 120 min in ClearSee with gentle shaking. Seedlings were then stained with 0.1% Direct Yellow 96 (Sigma) in ClearSee for 1 h to stain cell walls. The roots were washed three times for 120 min in ClearSee with gentle shaking. Roots were carefully placed on a microscope slide with ClearSee and covered with a coverslip. Roots were imaged at 40 cells after the onset of elongation using a Leica SP8 confocal microscope, 63× objective (NA = 1.2), by performing a Z-stack on the top endodermal cells. Excitation at 594 nm (intensity 3%) and an emission band-path of 600–650 nm were used for Basic Fuchsin, and excitation at 488 nm (intensity 1%) and an emission band-path of 500–540 nm was used for Direct yellow 96. Gains were optimized and kept consistent for all experiments.
Mapping the spatial distribution of elements
BC 1 F 5 seeds of high and low Mg segregant and R-o-18 lines were sown directly onto homogenized, high nutrient compost (Levington M3) in 750 mL pots and grown in the glasshouse. Plants were arranged in a one-way randomized design generated in GenStat (17th Edition, VSN International, Hemel Hempsted, UK). After 23 d, whole plants were sampled. Compost was washed off by hand and roots were wrapped in damp, 2-ply blue paper tissue. Whole plants were placed into individual, polythene bags in a protected box and sent via DHL Express 12:00 to the University of Ljubljana, Biotechnical Faculty (46°03′04.5″N 14°28′13.1″E). On arrival, sub-samples of first or second true-leaves (1–2 cm from tip) were prepared immediately for freezing as described previously (Vogel-Mikuš et al., 2014). Frozen samples were placed into a Leica CM3050 Cryostat microtome (Leica, Bensheim, Germany) with chamber and cutting head temperature set to −20°C and allowed to warm to this temperature. Sixty and twenty-five micrometer sections were then cut transversely from leaf samples. Sections were freeze-dried in a Christ LDC-1M Alpha 2-4 freezedryer (Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany) at −25°C and a pressure of 0.08 mbar for 3 d, then mounted onto custom aluminium holders between two layers of Pioloform foil (approx. 400 nm thickness; SPI supplies, West Chester, PA, USA).
Localization of elements within 60-μm leaf cross sections was assessed by micro-PIXE (Pongrac et al., 2013) at the nuclear microprobe in the tandetron accelerator laboratory at the Jožef Stefan Institute (Ljubljana, Slovenia; Simičič et al., 2002). The sample area ranged from 800 × 800 to 1200 × 1200 μm, at approximately 4 μm resolution. Na, Mg, P, S, K, and Ca were detected by a silicon drift detector (e2v SiriusSD®; High Wycombe, Buckinghamshire, UK). Proton dose was measured using a rotating, in-beam chopper to enable element quantification (Vogel-Mikuš et al., 2009). On–off axis scanning transmission ion microscopy was performed to determine beam exit energy from the sample (Pallon et al., 2004; Vavpetič et al., 2013). The micro-PIXE spectra were analyzed using GeoPIXE II (Ryan, 2000). Seeds were cut transversely by hand using platinum-coated stainless steel razor blades (Science Services, Germany), deposited on a double-sided carbon tape (SPI Supplies, USA) and covered with a layer of Pioloform foil. Sections were then analyzed as described for leaves.
Cellular level spatial distribution of elements within 25-μm cross-sections was determined using the TwinMic beamline at the Elettra Synchrotron (Trieste, Italy; Gianoncelli et al., 2016). Beam energy was set to 2,319 eV, and a max photon flux of 8 × 1010 photons s−1 mA−1. For each sample, an 80 × 80 μm region of the cross-section comprising upper epidermal and palisade mesophyll cells was analyzed in a vacuum (10−7 mbar) at a resolution of 1.2 μm. Elements mapped with low-energy X-ray fluorescence were O, F, Na, Mg, Al, Si, P, Fe, and Ni. Resulting 2-D X-ray count data were quantified through methods described previously (Kump and Vogel-Mikuš, 2017). Spatial distribution maps were generated in PyMCA (version 5.2.2; Solé et al., 2007).
Response to exogenous Mg concentrations
Plants of the BC1F5 generation were grown under eight different Mg treatments using the NFT (Broadley et al., 2003). Seeds were established in 25 × 25 × 40 mm rockwool plugs (Grodan, Roermond, The Netherlands) soaked with half strength Hoagland’s solution (Hoagland’s No. 2 Basal Salt Mixture; Sigma–Aldrich, Darmstadt, Germany) then transferred to eight, 4-m-long gullies, each with one of the following concentrations of MgCl2: 0.0075, 0.0375, 0.075, 0.375, 0.75, 1.125, 1.5, and 3 mM. Solution concentrations of other elements were as described by Broadley et al. (2003). Nutrient solutions were buffered to pH 6. The position of each of the eight Mg treatments was randomly distributed by gully (Supplemental Table S18). Within each gully, three plants per genotype were allocated based on a one-way, blocked design generated in GenStat (Supplemental Table S5). After 21 d of growth under NFT conditions, whole third and fourth leaves were sampled and pooled from each plant, dried, microwave digested, and analyzed by ICP-MS as described above.
Additional figure generation and statistical analysis
Figure 2A was generated using Probability Distribution functions in Genstat (20th edition). Figures 2B, 4B, 6 and Supplemental Figures S1, S2B, S4, S6, and S7B were generated in GraphPad Prism 8 (San Diego, CA, USA). Figure 2C was generated using heatmap.2 functions in R package gplots v3.1.0. Figure 3A was generated using R package OmicCircos (Hu and Yan, 2020). Supplemental Figure S2A was generated in Genstat using linear discriminant analysis functions. Tests of significant differences between genotypes were carried out using one-way ANOVA, two-sided T tests and F tests in Genstat, and two-sided Z tests calculated in Microsoft Excel. Simple Linear Regression functions in Genstat were used to detect trends in the response to exogenous Mg concentrations experiment.
Data availability
The genomic DNA data generated by 10× Genomics Chromium/Illumina sequencing in this work are accessible in the ENA (https://www.ebi.ac.uk/ena/browser/home) under Project identifier PRJEB40556. Arabidopsis thaliana ionomics data used for comparison in Supplemental Figure S4 are freely available at https://www.ionomicshub.org/. Raw data for all other figures are available in the supplemental tables.
Availability of biological materials
M 3 seed of all genotypes included in the initial ionomic screen is available from the RevGen service at the John Innes Centre, UK (http://revgenuk.jic.ac.uk/). Seed from all other generations is available on request to qualified researchers, without undue reservation.
Accession numbers
Sequence data for the main genes from this article can be found in the EMBL/TAIR data libraries under accession numbers Bra013446 (B. rapa SGN3) and AT4G20140 (Arabidopsis thaliana SGN3).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. BC1F3 tissue-specific element concentrations.
Supplemental Figure S2. BC2F2 segregant groupings.
Supplemental Figure S3. Reads aligned to BraA.SGN3.a showing locations of SNP calls.
Supplemental Figure S4. braA.sgn3.a-1 leaf ionome compared with A. thaliana sgn3-3
Supplemental Figure S5. braA.sgn3.a-1 and BraA.SGN3.a-WT root tips stained with PI.
Supplemental Figure S6. BC1F5 tissue-specific micro-PIXE element concentrations.
Supplemental Figure S7. Seed element spatial distribution.
Supplemental Table S1. M2 forward ionomic screen leaf Mg, K, and Ca concentration data following processing.
Supplemental Table S2. M3 leaf mineral concentration data following processing.
Supplemental Table S3. M4 leaf mineral concentration data following processing.
Supplemental Table S4. BC1F1 leaf mineral concentration data following processing.
Supplemental Table S5. BC1F2 leaf mineral concentration data following processing.
Supplemental Table S6. BC1F3 fourth leaf mineral concentration data following processing.
Supplemental Table S7. BC1F3 other tissue mineral concentration data following processing.
Supplemental Table S8. Seed yield of a subset of BC2F2 plants showing mutant and wild-type Mg concentrations.
Supplemental Table S9. BC2F2 leaf mineral concentration data following processing.
Supplemental Table S10. Variant call data representing G to A and C to T SNP calls present in both high and low Mg DNA pools.
Supplemental Table S11. Number of epidermal cells from the onset of elongation until the endodermal cells blocked PI penetration into the stele of roots of BraA.SGN3.a-WT and braA.sgn3.a-1 mutant plants.
Supplemental Table S12. Leaf cell specific concentrations of Na, Mg, P, S, K, and Ca in first or second leaves of R-o-18, BraA.SGN3.a-WT (SGN3.a-WT) and braA.sgn3.a-1 (sgn3.a-1).
Supplemental Table S13. Magnesium response experiment leaf mineral concentration data following processing.
Supplemental Table S14. Position of plants with polytunnel for all three runs in the initial M2 generation forward ionomic mineral screen.
Supplemental Table S15. BC1F4 leaf mineral concentration data following processing.
Supplemental Table S16. BC2F2 segregating population block design.
Supplemental Table S17. Qubit dsDNA concentrations of segregating BC2F2 population.
Supplemental Table S18. Mg response experiment one-way design in randomized blocks.
Supplementary Material
Acknowledgments
The authors thank Helen Bowen (University of Warwick) for support with glasshouse studies and Scott Young (University of Nottingham) for support and advice in undertaking element analyses. DNA sequencing was performed at Deep Seq, the University of Nottingham Next Generation Sequencing Facility. Bioinfomatics work was undertaken on Augustus, part of the High Performance Computing facilities at the University of Nottingham, UK. They acknowledge Elettra Sincrotrone Trieste for providing access to its synchrotron radiation facilities (project number 20175128) and they thank Alessandra Gianoncelli and Matteo Altissimo for assistance in using beamline “TwinMic”. Further they thank David Alcock for assistance with preparation of samples for export to Slovenia.
Funding
This work was supported by a series of projects and studentships:
The UK Biotechnology and Biological Sciences Research Council (BBSRC) and the UK Natural Environment Research Council (NERC), through a Sustainable Agriculture Research and Innovation Club (SARIC) project, Magnesium Network (MAG-NET): Integrating Soil-Crop-Animal Pathways to Improve Ruminant Health [grant number BB/N004302/1].
The Future Food Beacon of Excellence, University of Nottingham, UK (T.D.A., G.R.).
The BBSRC, through a studentship funded by the Renewable Industrial Products from Rapeseed (RIPR) Program (T.D.A.) [grant number BB/L002124/1].
The BBSRC, through a studentship (CLT) funded by a Crop Improvement Research Club (CIRC) project, Delivering Low-Cost, High-Throughput Root Phenotyping Screens for Arable Crops [grant number BB/J019631/1].
The BBSRC, through an Industry Partnering Award, Biofortifying Brassica with Calcium (Ca) and Magnesium (Mg) for Human Health [grant number BB-G013969-1].
The BBSRC, through a Targeted Priority Studentship in Crop Science (S.Ó.), Improving the Zinc (Zn) Composition of Brassica Crops [grant number BBSSE200613215].
P.J.W. was supported by the Rural and Environment Science and Analytical Services Division (RESAS) of the Scottish Government.
P.P., K.V.-M., and M.K. were supported by the Slovenian Research Agency through programs P1-0212 and P1-0112 and projects N7-0077, N1-0105, N1-0090, J7-9418, and J7-9398.
Conflict of interest statement. None declared.
T.D.A. performed the genetic screens, physiology experiments, high molecular weight DNA extractions and sequencing, X-ray fluorescence and confocal microscopy imaging, data analyses, and led on the writing of the manuscript. C.L.T. performed the genetic screens, physiology experiments, and data analyses. S.Ó. performed the genetic screens and data analyses. T.D.A., C.L.T., and S.Ó. maintained germplasm for all generations beyond M2. P.P. and M.K. performed and processed micro-PIXE spatial distribution mapping experiments. M.W. performed bioinformatics analysis of sequence data. C.M. performed the high molecular weight DNA sample preparation and linked-read sequencing. G.R. performed the staining experiments, CIF2 treatment, and confocal microscopy imaging. K.V.-M. advised on X-ray fluorescence experiments, and data analyses. R.H. advised on glasshouse experiments and maintained plant populations. L.W. undertook element analyses. P.S. and L.Ø. produced germplasm for initial forward genetic screen. J.A.I. and G.J.K. provided Brassica rapa var. R-o-18 reference genome data. J.P.H. advised on bioinformatics analysis. J.P.H., G.J.K., D.E.S., N.S.G., P.J.W., and M.R.B. contributed to project conception, management, funding, and data analyses. All authors have read, edited, and approved the final version of the manuscript. M.R.B. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Martin R. Broadley (martin.broadley@nottingham.ac.uk).
References
- Ahmad I, Maathuis FJ (2014) Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J Plant Physiol 171: 708–714 [DOI] [PubMed] [Google Scholar]
- Alassimone J, Fujita S, Doblas VG, van Dop M, Barberon M, Kalmbach L, Vermeer JEM, Rojas-Murcia N, Santuari L, Hardtke CS, et al. (2016) Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat Plants 2: 16113. [DOI] [PubMed] [Google Scholar]
- Alassimone J, Naseer S, Geldner N (2010) A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA 107: 5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock TD, Havlickova L, He Z, Bancroft I, White PJ, Broadley MR, Graham NS (2017) Identification of candidate genes for calcium and magnesium accumulation in Brassica napus L. by association genetics. Front Plant Sci 8: 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE (2007) Purdue ionomics information management system. An integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borpatragohain P, Rose TJ, King GJ (2016) Fire and brimstone: molecular interactions between sulfur and glucosinolate biosynthesis in model and crop Brassicaceae. Front Plant Sci 7: 1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, Bowen HC, Cotterill HL, Hammond JP, Meacham MC, Mead A, White PJ (2003) Variation in the shoot calcium content of angiosperms. J Exp Bot 54: 1431–1446 [DOI] [PubMed] [Google Scholar]
- Broadley MR, Hammond JP, King GJ, Astley D, Bowen HC, Meacham MC, Mead A, Pink DAC, Teakle GR, Hayden RM, et al. (2008) Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea. Plant Physiol 146: 1707–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, White PJ (2010) Eats roots and leaves. Can edible horticultural crops address dietary calcium, magnesium and potassium deficiencies? Proc Nutr Soc 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017a) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Geldner N, Barberon M (2017b) The endodermis, a tightly controlled barrier for nutrients. Curr Opin Plant Biol 39: 136–143 [DOI] [PubMed] [Google Scholar]
- Edmondson RN (2019) blocksdesign: nested and crossed block designs for factorial, fractional factorial and unstructured treatment sets. R package version 3.8. https://CRAN.R-project.org/package=blocksdesign (accessed 20 November 2020). [Google Scholar]
- Faulkner K, Mithen R, Williamson G (1998) Selective increase of the potential anticarcinogen 4-methylsulphinylbutyl glucosinolate in broccoli. Carcinogenesis 19: 605–609 [DOI] [PubMed] [Google Scholar]
- Friend MA, Bhanugopan MS, McGrath SR, Edwards JH, Hancock S, Loudon K, Miller D, McGilchrist P, Refshauge G, Robertson SM, et al. (2020) Do calcium and magnesium deficiencies in reproducing ewes contribute to high lamb mortality? Anim Prod Sci 60: 733–751 [Google Scholar]
- Fujita S, De Bellis D, Edel KH, Köster P, Andersen TG, Schmid‐Siegert E, Tendon VD, Pfister A, Marhavý P, Ursache R, et al. (2020). SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianoncelli A, Kourousias G, Merolle L, Altissimo M, Bianco A (2016) Current status of the TwinMic beamline at Elettra: a soft X-ray transmission and emission microscopy station. J Synchrotron Radiat 23: 1526–1537 [DOI] [PubMed] [Google Scholar]
- Gödecke T, Stein AJ, Qaim M (2018) The global burden of chronic and hidden hunger: trends and determinants. Glob Food Secur 17: 21–29 [Google Scholar]
- Graham NS, Hammond JP, Lysenko A, Mayes S, Lochlainn SO, Blasco B, Bowen HC, Rawlings CJ, Rios JJ, Welham S, et al. (2014) Genetical and comparative genomics of Brassica under altered Ca supply identifies Arabidopsis Ca-transporter orthologs. Plant Cell 26: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yan C (2020) OmicCircos: high-quality circular visualization of omics data. R package version 1.26.0. https://bioconductor.org/packages/OmicCircos/ (accessed 20 November 2020).
- Huang XY, Salt DE (2016) Plant ionomics: from elemental profiling to environmental adaptation. Mol Plant 9: 787–797 [DOI] [PubMed] [Google Scholar]
- Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L, et al. (2020) Ensembl Genomes 2020—enabling non-vertebrate genomic research. Nucleic Acids Res 48: D689–D695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy EJM, Ander EL, Young SD, Black CR, Watts MJ, Chilimba ADC, Chilima B, Siyame EWP, Kalimbira AA, Hurst R, et al. (2014) Dietary mineral supplies in Africa. Physiol Plant 151: 208–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karley AJ, White PJ (2009) Moving cationic minerals to edible tissues: potassium, magnesium, calcium. Curr Opin Plant Biol 12: 291–298 [DOI] [PubMed] [Google Scholar]
- Kump P, Vogel-Mikuš K (2017) Quantification of 2D elemental distribution maps of intermediate-thick biological sections by low energy synchrotron µ-X-ray fluorescence spectrometry. J Instrum 13: C05014 [Google Scholar]
- Kumssa DB, Penrose B, Bone PA, Lovatt JA, Broadley MR, Kendall N, Ander EL (2019) A reconnaissance survey of farmers’ awareness of hypomagnesaemic tetany in UK cattle and sheep farms. PLoS ONE 14: e0223868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumssa DB, Joy EJM, Ander EL, Watts MJ, Young SD, Walker S, Broadley MR (2015a) Dietary calcium and zinc deficiency risks are decreasing but remain prevalent. Scient Rep 5: 10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumssa DB, Joy EJM, Ander EL, Watts MJ, Young SD, Rosanoff A, White PJ, Walker S, Broadley MR (2015b) Global magnesium supply in the food chain. Crop Pasture Sci 66: 1278–1289 [Google Scholar]
- Kumssa DB, Lovatt JA, Graham NS, Palmer S, Hayden R, Wilson L, Young SD, Lark RM, Penrose B, Ander EL, et al. (2019) Magnesium biofortification of Italian ryegrass (Lolium multiflorum L.) via agronomy and breeding as a potential way to reduce grass tetany in grazing ruminants. Plant Soil 457: 25–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al. (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Leitenmaier B, Küpper H (2013) Compartmentation and complexation of metals in hyperaccumulator plants. Front Plant Sci 4: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer K, Broadley MR, El-Serehy HA, George TS, McNicol JW, Moraes MF, White PJ (2018) Variation in the angiosperm ionome. Physiol Plant 163: 306–322 [DOI] [PubMed] [Google Scholar]
- Okuda S, Fujita S, Moretti A, Hohmann U, Doblas VG, Ma Y, Pfister A, Brandt B, Geldner N, Hothorn M (2020) Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2. Proc Natl Acad Sci USA 117: 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallon J, Auzelyte V, Elfman M, Garmer M, Kristiansson P, Malmqvist K, Nilsson C, Shariff A, Wegdén M (2004) An off-axis STIM procedure for precise mass determination and imaging. Nucl Instrum Methods Phys Res B 219–220: 988–993 [Google Scholar]
- Pelletier S, Bélanger G, Tremblay GF, Chantigny MH, Allard G (2008) Dietary cation–anion difference and tetany index of timothy forage fertilized with liquid swine manure .Agron J 100: 213–220 [Google Scholar]
- Penrose B, Lovatt JA, Palmer S, Thomson R, Broadley MR (2020) Revisiting variation in leaf magnesium concentrations in forage grasses for improved animal health. Plant Soil 457: 43–55 [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, Vermeer JEM, Yamazaki M, Li G, Maurel C, Takano J, et al. (2014) A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3: e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongrac P, Vogel-Mikuš K, Regvar M, Kaligarič M, Vavpetič P, Kelemen M, Grlj N, Shelef O, Golan-Goldhirsh A, Rachmilevitch S, et al. (2013) On the distribution and evaluation of Na, Mg and Cl in leaves of selected halophytes. Nucl Instrum Methods Phys Res B 306: 144–149 [Google Scholar]
- Roberts C, Steer T, Maplethorpe N, Cox L, Meadows S, Nicholson S, Page P, Swan G; Public Health England (2018) National Diet and Nutrition Survey. Results from Years 7 and 8 (combined) of the Rolling Programme (2014/15 to 2015/16). PHE Publications, London, UK [Google Scholar]
- Rosanoff A, Kumssa DB (2020) Impact of rising body weight and cereal grain food processing on human magnesium nutrition. Plant Soil 457: 5–23 [Google Scholar]
- Ryan CG (2000) Quantitative trace element imaging using PIXE and the nuclear microprobe. Int J Imag Syst Technol 11: 219–230 [Google Scholar]
- Schonewille JT (2013) Magnesium in dairy cow nutrition: an overview. Plant Soil 368: 167–178 [Google Scholar]
- Simičič J, Pelicon P, Budnar M, Šmit Ž (2002) The performance of the Ljubljana ion microprobe. Nucl Instrum Methods Phys Res B 190: 283–286 [Google Scholar]
- Solé VA, Papillon E, Cotte M, Walter P, Susini J (2007) A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra. Spectrochim Acta Part B 62: 63–68 [Google Scholar]
- Stein AJ (2014) Rethinking the measurement of undernutrition in a broader health context: should we look at possible causes or actual effects? Glob Food Secur 3: 193–199 [Google Scholar]
- Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Østergaard L (2010) A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle NF (2010) Mineral Nutrition of Livestock, Ed 4. CABI, Oxfordshire, UK
- Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka S, Uemura A, Utsushi H, Tamiru M, Takuno S, et al. (2013) QTL‐seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74: 174. [DOI] [PubMed] [Google Scholar]
- Thomas CL, Alcock TD, Graham NS, Hayden R, Matterson S, Wilson L, Young SD, Dupuy LX, White PJ, Hammond JP, et al. (2016) Root morphology and seed and leaf ionomic traits in a Brassica napus L. diversity panel show wide phenotypic variation and are characteristic of crop habit. BMC Plant Biol 16: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka MH, Saha S, Huseby S, Kopriva S, Walley PG, Barker GC, Moore J, Mero G, den Bosch F, Constant H, et al. (2013) Genetic regulation of glucoraphanin accumulation in Beneforté broccoli. New Phytol 198: 1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N (2017) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J 93: 399–412 [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy‐Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013) From FastQ data to high‐confidence variant calls: the genome analysis toolkit best practices pipeline. Curr ProtocBioinform 43: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavpetič P, Pelicon P, Vogel-Mikuš K, Grlja N, Pongrac P, Jeromel L, Ogrinc N, Regvar M (2013) Micro-PIXE on thin plant tissue samples in frozen hydrated state: a novel addition to JSI nuclear microprobe. Nucl Instrum Methods Phys Res B 306: 140–143 [Google Scholar]
- Vogel-Mikuš K, Pelicon P, Vavpetič P, Kreft I, Regvar M (2009) Elemental analysis of edible grains by micro-PIXE: common buckwheat case study. Nucl Instrum Methods Phys Res B 267: 2884–2889 [Google Scholar]
- Vogel-Mikuš K, Pongrac P, Pelicon P (2014) Micro-PIXE elemental mapping for ionome studies of crop plants. Int J PIXE 24: 217–233 [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035. [DOI] [PubMed] [Google Scholar]
- Wang X, Long Y, Wang N, Zou J, Ding G, Broadley MR, White PJ, Yuan P, Zhang Q, Luo Z, et al. (2017) Breeding histories and selection criteria for oilseed rape in Europe and China identified by genome wide pedigree dissection. Scient Rep 7: 1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Calvo-Polanco M, Reyt G, Barberon M, Champeyroux C, Santoni V, Maurel C, Franke RB, Ljung K, Novak O, et al. (2019) Surveillance of cell wall diffusion barrier integrity modulates water and solute transport in plants. Scient Rep 9: 4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2009) Biofortification of crops with seven mineral elements often lacking in human diets—iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol 182: 49–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic DNA data generated by 10× Genomics Chromium/Illumina sequencing in this work are accessible in the ENA (https://www.ebi.ac.uk/ena/browser/home) under Project identifier PRJEB40556. Arabidopsis thaliana ionomics data used for comparison in Supplemental Figure S4 are freely available at https://www.ionomicshub.org/. Raw data for all other figures are available in the supplemental tables.