Abstract
Breast cancer is a vastly heterogeneous disease encompassing a panoply of special histological subtypes. Although rare breast tumors have largely not been investigated systematically in large scale genomics series, recent studies have shed light on the genetic underpinnings of special histologic subtypes of breast cancer. Genomic analyses of estrogen receptor-positive special histologic types of breast cancer have not resulted in the identification of novel pathognomonic genetic alterations in addition to the confirmation of the presence of CDH1 loss-of-function mutations in invasive lobular carcinomas. By contrast, the analyses of triple-negative breast cancers have demonstrated that low-grade triple-negative breast cancers categorically differ from the common forms of high-grade triple-negative disease biologically and phenotypically and are underpinned by specific fusion genes or hotspot mutations. A subset of low-grade triple-negative disease has been shown to harbor highly recurrent if not pathognomonic genetic alterations, such as ETV6-NTRK3 fusion gene in secretory carcinomas, the MYB-NFIB fusion gene, MYBL1 rearrangements or MYB gene amplification in adenoid cystic carcinomas, and HRAS Q61 hotspot mutations coupled with mutations in PI3K pathway genes in estrogen receptor-negative adenomyoepitheliomas. A subset of these pathognomonic genetic alterations (e.g., NTRK1/2/3 fusion genes) now constitute an FDA approved indication for the use of TRK inhibitors in the advanced/metastatic setting. These studies have also corroborated that salivary gland-like tumors of the breast, other than acinic cell carcinomas, harbor the repertoire of somatic genetic alterations detected in their salivary gland counterparts. Reassuringly, the systematic study of special histologic types of breast cancer utilizing state-of-the-art sequencing approaches, rather than rendering pathology obsolete, has actually strengthened the importance of breast cancer histologic typing and is providing additional ancillary markers for the diagnosis of these rare but fascinating entities.
Introduction
Breast cancer is a heterogeneous disease comprising various entities with distinctive phenotypes, biological and molecular features, clinical presentations and responses to therapy as well as clinical outcomes [1]. The World Health Organization classification recognizes common forms of the disease, the so-called invasive ductal carcinomas of no special type (IDC-NSTs), and over 20 special types of breast cancer, which display consistent histologic features and account for ~20% of all invasive breast cancers [1–3].
Despite the evidence that tumors from each of these special histologic types of the disease are more homogeneous at the molecular level than IDC-NSTs [1, 4], information about the subtyping of the disease has largely not been investigated in large scale genomics studies and taken into account for clinical management [5], principally due to the limited inter-observer agreement for the diagnosis of special histologic types of breast cancer and the relative rarity of each of the entities.
Recent studies, however, have begun to tackle the genomic underpinnings of special histologic types of breast cancer in a systematically manner [6–9], and the results of these analyses have been transformative. This new body of knowledge has revealed (i) that genotypic–phenotypic correlations do exist in breast cancer, (ii) that at the molecular level, special histologic types of breast cancer are more homogeneous than IDC-NSTs, (iii) novel cancer driver genes and hotspot mutations, and (iv) the histologic context of oncogenes mutated at high frequencies in other cancer types but rarely mutated in breast cancer.
Genotypic–phenotypic correlations in rare types of breast cancer
We and others have posited that special types of breast cancer would constitute extremes of phenotype as defined by histology, and, therefore, would be underpinned by either pathognomonic genetic alterations or specific constellations of genetic alterations [10]. Targeted sequencing, whole-exome sequencing, and RNA-sequencing analyses performed in the last decade have shed light into the genomic basis of special histologic types of breast cancer. Although these analyses have yielded limited novel findings in the context of estrogen receptor (ER)-positive special types of breast cancer, with the confirmation of CDH1 loss-of-function mutations being present in >80% of invasive lobular carcinomas [6, 7, 11] (Fig. 1), the use of these technologies in the study of rare forms of triple-negative breast cancers (TNBCs) has been transformative. In fact, studies from our group and others have indeed revealed specific genotypic–phenotypic associations and novel convergent phenotypes in rare forms of low-grade TNBCs, in particular in the context of salivary gland-like tumors of the breast.
Fig. 1. Genetic alterations in special histologic subtypes of estrogen receptor-positive breast cancer.
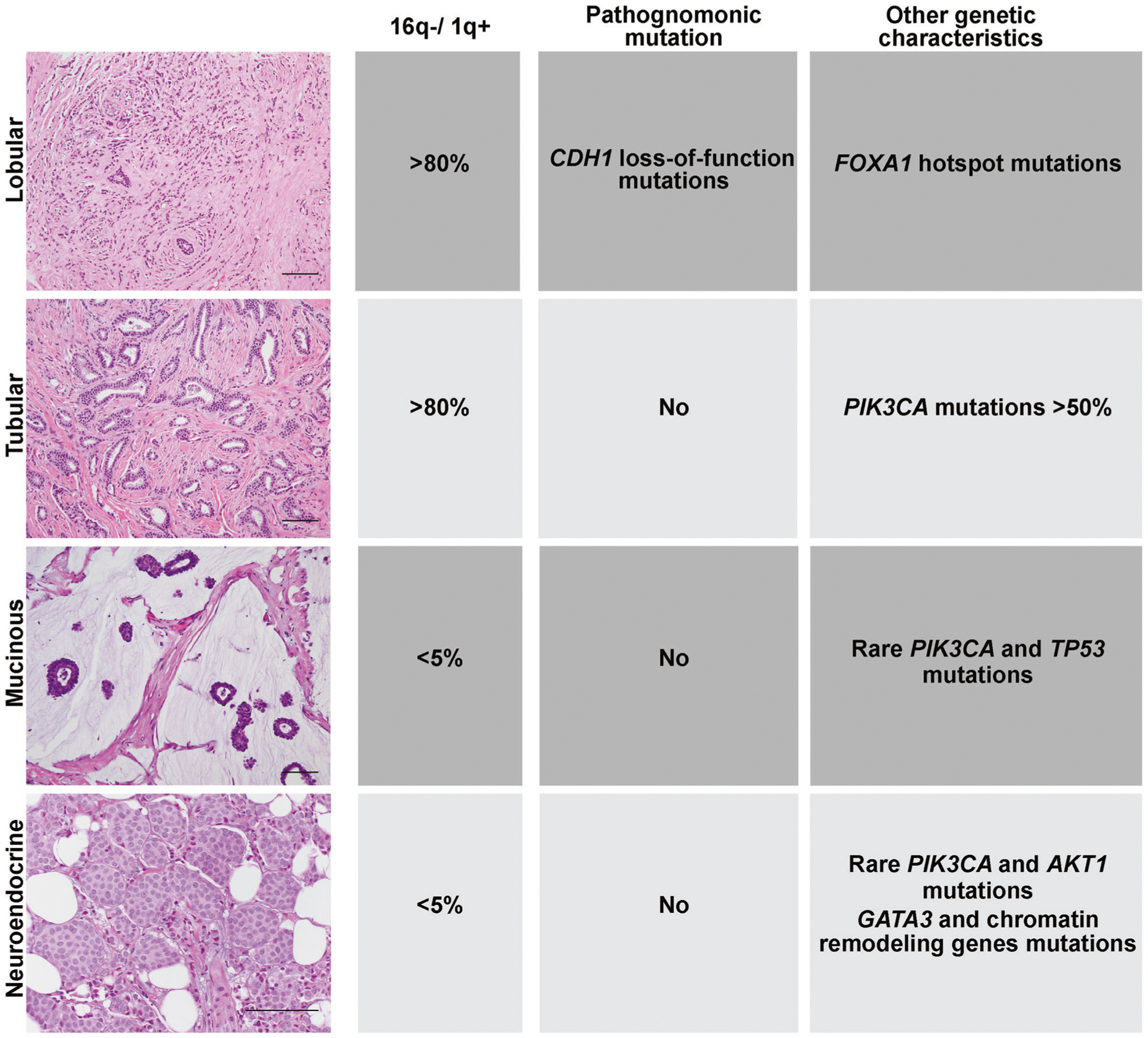
Representative micrographs and genetic features of invasive lobular carcinoma, tubular carcinoma, mucinous carcinoma and neuroendocrine tumors of the breast. Scale bars, 50 μm. Diagram summarizing the genomic features, including 16q losses (16q−) and 1q gains (1q+), hallmark genomic features of ER-positive/HER2-negative IDC-NSTs, the existence of pathognomonic genetic alterations that define each special histologic type and other genomic features of each subtype that differ from those found in ER-positive/HER2-negative IDC-NSTs.
Triple-negative breast cancer
TNBC is an operational term encompassing entities defined by the absence of ER, progesterone receptor (PR) and HER2 expression. As a group, the common forms of TNBCs largely encompass IDC-NSTs characterized by a constellation of histologic features associated with an aggressive clinical behavior and/or a BRCA1-associated phenotype, including high histologic grade, conspicuous nuclear pleomorphism, central necrosis, high mitotic activity, and brisk lymphocytic infiltrate [12, 13]. From a genetic standpoint, common forms of TNBC, despite the high mutational burden [14], have a limited number of highly recurrent driver genetic alterations. TP53 mutations are present in ~80% of these cancers [15–18], followed by somatic genetic alterations affecting PIK3CA in 10% of cases [15–18]. Other potentially clinically actionable mutations documented in TNBCs include BRCA1 germline and somatic mutations which are observed in up to 16% of TNBCs [19–21], PTEN losses and mutations, RB1 losses and mutations [16], and EGFR and FGFR2 amplifications [15, 22] in small subgroups (~5%) of the disease. Transcriptomically, given the phenotypic diversity of TNBCs, it is now understood that least four molecular subtypes exist, namely basal-like immune activated, basal-like immune depleted, mesenchymal-like, and luminal androgen receptor [23, 24].
A much rarer and less characterized group of TNBCs, however, is the subset of low-grade TNBCs [25]. These cancers, despite lacking ER, PR, and HER2, differ fundamentally from the common form of TNBCs at the phenotypic, transcriptomic, and genomic levels. Research on genotypic–phenotypic correlations in breast cancer has proven to be immensely fruitful in the context of a subset of low-grade forms of TNBC, the so-called salivary gland-like tumors of the breast (Fig. 2). These tumors have several characteristics in common: (i) despite being of triple-negative phenotype in the vast majority of cases, they display a rather indolent clinical behavior, (ii) they can originate not only in the breast, but also in the salivary glands and the lungs, (iii) they lack the typical genomic instability and high frequency of TP53 mutations found in common forms of TNBC and, instead, harbor highly recurrent, if not pathognomonic genetic alterations identical to those found in the respective salivary gland counterparts. This group includes secretory carcinoma, adenoid cystic carcinoma (AdCC), mucoepidermoid carcinoma, pleomorphic adenoma, polymorphous adenocarcinoma, and adenomyoepitheliomas (AMEs).
Fig. 2. Genotypic–phenotypic correlations in low-grade tumors of triple-negative phenotype.
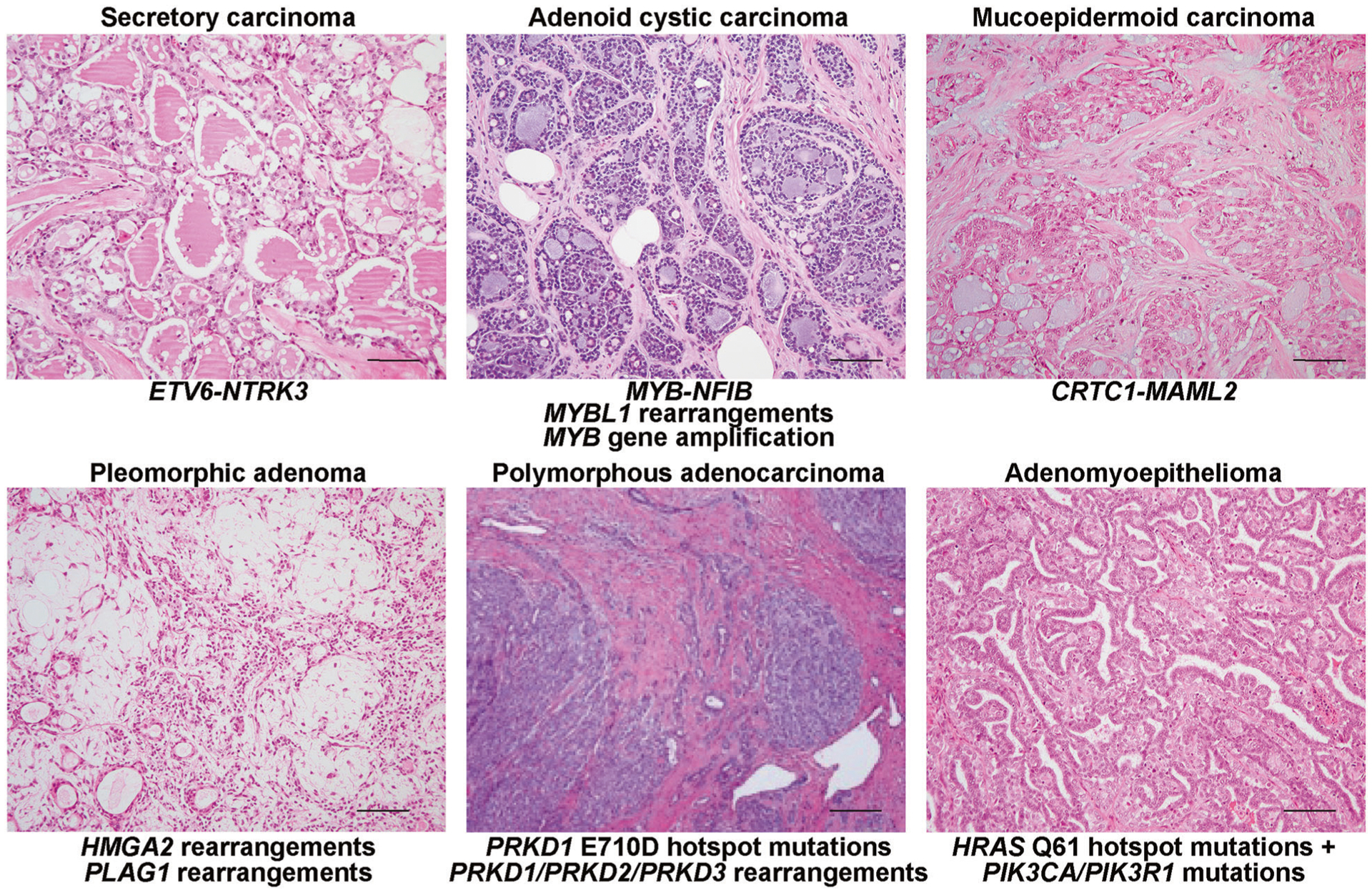
Representative micrographs and pathognomonic genetic alterations of secretory carcinoma, adenoid cystic carcinoma, mucoepidermoid carcinoma, pleomorphic adenoma, polymorphous adenocarcinoma, and estrogen receptor-negative adenomyoepithelioma. Scale bars, 50 μm.
Secretory carcinoma
Although originally described in children and named ‘juvenile carcinoma’, secretory carcinoma may occur at any age [26]. The clinical course of secretory carcinoma is usually excellent, even in the presence of nodal involvement or metastasis [26]. Secretory carcinomas display a peculiar phenotype, being characterized by a tubular, solid and/or microcystic architecture and abundant intra- and extracellular colloid-like secretions [3]. The hallmark genetic alteration of this entity is the ETV6-NTRK3 fusion gene [27], also identified in the mammary-analog secretory carcinoma, its salivary gland counterpart [28]. Consistent with the observations from other tumor types driven by highly recurrent or pathognomonic fusion genes or mutations and unlike common forms of TNBC, secretory carcinomas have simple genomes with quiet copy number profiles and a paucity of pathogenic mutations in cancer genes [29] in addition to the ETV6-NTRK3 fusion gene. In the context of primary breast cancers, this chimeric gene appears to be restricted to secretory carcinomas. Given that NTRK3 protein expression is not detected in breast epithelial cells physiologically, immunohistochemical analysis using pan-TRK antibodies has been shown to constitute a sensitive and specific approach for the detection of NTRK3 rearrangements in breast secretory carcinomas [30].
Rendering a diagnosis of secretory carcinoma is not an academic exercise of minimal import to oncologists and now has profound clinical implications. Patients with recurrent or metastatic secretory carcinomas are now eligible to be treated with one of the TRK inhibitors [31–35] approved for clinical use by the Food and Drug Administration (FDA), which have been shown to result in profound and long-lasting responses, even in the context of an aggressive clinical behavior.
Adenoid cystic carcinoma
Breast AdCC is rare histologic subtype of breast cancer, accounting for less than 0.1% of breast carcinomas [36]. These tumors are characterized by a dual cell population, including a myoepithelial and an epithelial component [3]. In contrast to AdCCs arising in the salivary glands, which oftentimes display a poor clinical outcome, breast AdCCs usually have an indolent clinical course [36], however distant metastases, in particular to the lungs, are on record [37, 38]. Akin to AdCC of the salivary glands and lungs, AdCCs arising in the breast are underpinned by rearrangements involving MYB, which most frequently result in the MYB-NFIB fusion gene [39, 40], which are present in 22.6–100% of cases [41–44]. Interestingly, breast AdCCs lacking the MYB-NFIB fusion gene have been shown to harbor MYBL1 rearrangements, including the MYBL1-ACTN1 and MYBL1-NFIB fusion genes, or MYB amplification [45], supporting the notion that AdCCs are a convergent phenotype driven by activation of MYB signaling via different molecular mechanisms. It should be noted, however, that for a subset of these tumors, the driver genetic alteration has yet to be identified.
A solid variant of AdCC featuring a basaloid morphology and composed predominantly of epithelial cells is recognized [46]. In contrast to conventional AdCC, solid type AdCC harbor MYB rearrangements in less than 20% of cases [47], display alterations affecting the NOTCH pathway-related genes, and appear to display a more aggressive clinical behavior than the classical form of AdCC. Further genomics studies are warranted to define whether solid-basaloid AdCCs constitute a single entity, and in a way akin to classical AdCCs would be driven by MYB pathway activation through different molecular mechanisms, or if these tumors would constitute a collection of different entities displaying a solid-basaloid histology.
Although AdCCs display rather simple genomes and lack TP53 mutations, progression to a high-grade form of TNBC has been reported. At variance with common forms of high-grade TNBC, high-grade tumors originating in AdCCs harbor the MYB-NFIB fusion gene, but also display additional genetic alterations, including mutations affecting NOTCH pathway-related genes, tyrosine kinase receptors, and chromatin remodelers [48].
Mucoepidermoid carcinoma
Mucoepidermoid carcinoma, a common salivary gland tumor, may rarely arise in the breast [49, 50], and due to its rarity, it often goes undiagnosed as such, being at times misclassified as metaplastic carcinomas with squamous differentiation. These tumors are histologically indistinguishable regardless of their anatomic origin, and are composed of various cell types, including mucinous, squamous, and intermediate cells [49].
Mucoepidermoid carcinomas arising in the salivary gland or lung are underpinned by the t(11;19) (q14–21; p12–13) translocation, resulting in the CRTC1-MAML2 fusion gene [51, 52], which has been shown to result in activation of Notch signaling [53]. Breast mucoepidermoid carcinomas have recently been found to harbor the hallmark CRTC1-MAML2 fusion gene [54, 55], and unlike common forms of TNBC, display a low mutation burden, lack mutations in commonly altered cancer genes, and have simple gene copy number profiles [54].
Pleomorphic adenoma
Pleomorphic adenomas are common salivary gland tumors [56], and may arise in other anatomic locations, such as breast [49, 57]. Pleomorphic adenomas are composed of by an admixture of epithelial and myoepithelial components in myxochondroid stroma. Pleomorphic adenomas arising in the salivary glands or soft tissue are characterized by recurrent rearrangements involving the transcription factor encoding genes HMGA2 or PLAG1 [58, 59]. Breast pleomorphic adenomas are rare and are also underpinned by fusion genes [55]. Indeed, we identified a breast pleomorphic adenoma harboring an HMGA2-WIF1 fusion gene and another case harboring a PLAG1 rearrangement [55].
Polymorphous adenocarcinoma
Polymorphous adenocarcinoma, previously known as polymorphous low-grade adenocarcinoma, is the second most frequent type of malignant tumors of the salivary gland and has been reported in the lungs; in the breast, however, this entity appears to be vanishingly rare and its existence is a matter of contention [3, 60]. Polymorphous adenocarcinomas were originally described as infiltrative tumors characterized by cytologic uniformity, with tumor cells displaying bland nuclei and scanty to moderate cytoplasm, and a panoply of architectural patterns. Although the majority of polymorphous adenocarcinomas follow an indolent course, some may progress to high-grade tumors and/or display a more aggressive clinical behavior, even with the development of distant metastases.
Our group has previously demonstrated that polymorphous adenocarcinomas are driven by a pathognomonic PRKD1 E710D hotspot mutation, which has been shown to be activating and oncogenic [61]. Polymorphous adenocarcinomas lacking this oncogenic hotspot mutation have been shown to harbor rearrangements involving genes of the PRKD gene family, including PRKD1, PRKD2, or PRKD3 [62, 63]. In a way akin to AdCCs, which can progress to high-grade TNBCs maintaining the original pathognomonic genetic alteration, polymorphous adenocarcinomas that progress to high-grade forms maintain the PRKD1 hotspot mutation or the rearrangement affecting a PRKD gene, but also acquire additional copy number alterations and/or mutations affecting genes related to the NOTCH pathway, chromatin remodelers, tyrosine kinase receptors, and the PI3K pathway [64].
Adenomyoepitheliomas
Breast AMEs are rare biphasic tumors with dual epithelial and myoepithelial differentiation, which may be ER-positive or ER-negative [3]. Although AMEs usually follow a benign course, recurrences and distal metastasis have been reported [3, 65]. The repertoire of genetic alterations of AMEs varies according to their ER status [66]. ER-positive AMEs harbor recurrent and mutually exclusive PIK3CA and AKT1 mutations, whereas ER-negative AMEs harbor recurrent HRAS Q61R/K hotspot mutations, which frequently coexist with mutations affecting PI3K-AKT genes, such as PIK3CA and PIK3R1 [66]. None of the AMEs analyzed were found to harbor TP53 somatic mutations. The immunohistochemical assessment of RAS Q61R has been shown to be highly specific and moderately sensitive for the detection of HRAS Q61R mutations in AMEs [67] (Fig. 3). Functional analyses have provided evidence that the combination of HRAS Q61 and PIK3CA hotspot mutations may not only constitute the drivers of ER-negative AMEs, but also result in the acquisition of the cardinal phenotypic features of these tumors. Forced expression of HRAS Q61R in MCF12A cells and in MCF10A cells with and without a PIK3CA H1047R somatic knock-in were found to result not only in an oncogenic phenotype, but also in the acquisition of myoepithelial differentiation [66].
Fig. 3. Immunohistochemical assessment of pathognomonic genetic alterations in rare breast tumors.
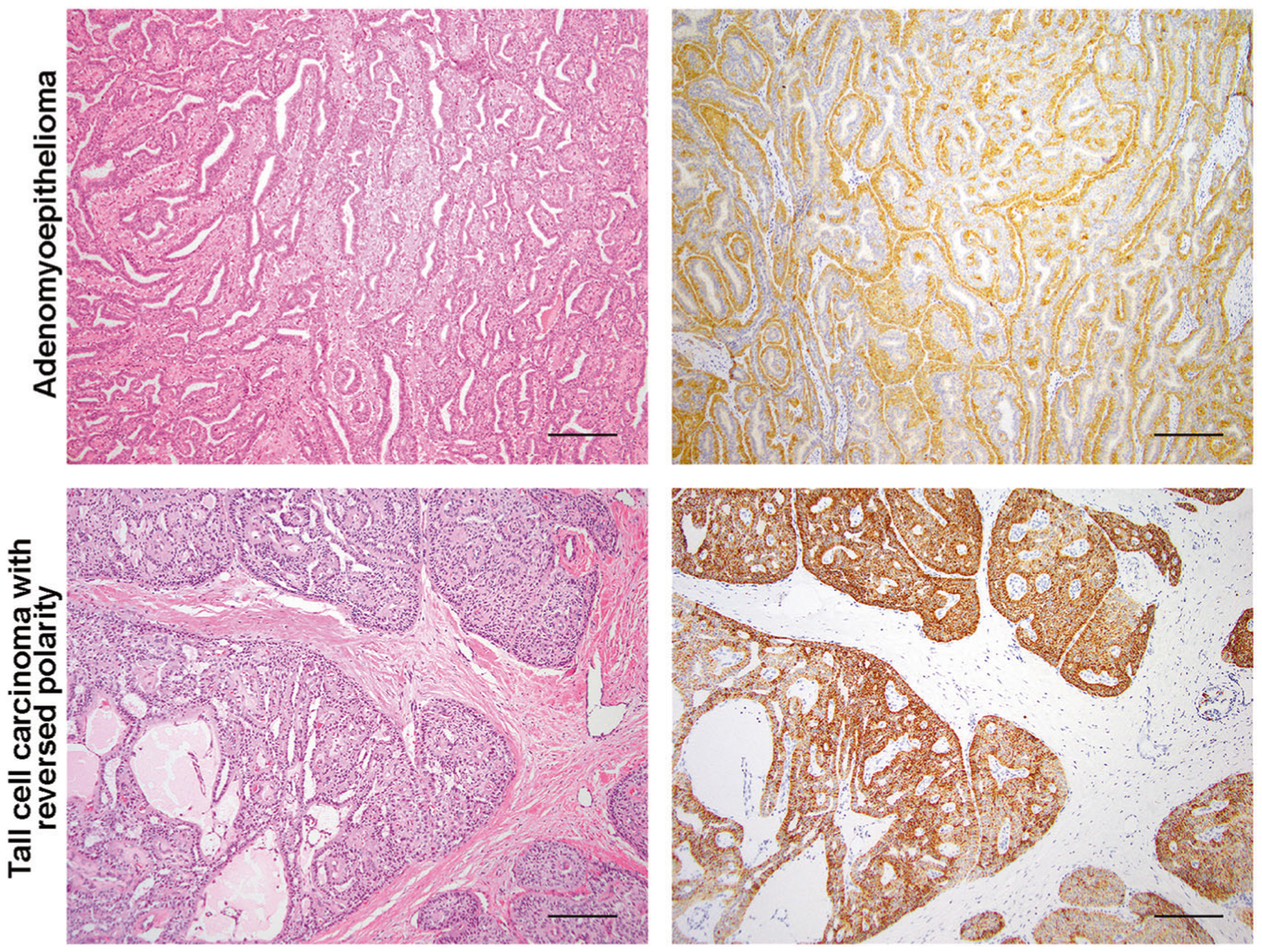
Representative micrographs of an estrogen receptor-negative adenomyoepithelioma harboring an HRAS Q61R mutation and of the immunohistochemical assessment of HRAS Q61R protein expression (upper panels), and of a tall cell carcinoma with reversed polarity harboring an IDH2 R172T mutation and of the immunohistochemical assessment of IDH2 R172 expression (lower panels). Scale bars, 100 μm.
It should be noted that consistent with other salivary gland-like tumors of the breast, ER-negative AMEs closely recapitulate histologically and genetically epithelial-myoepithelial tumors of the salivary glands, which also harbor recurrent and concurrent HRAS Q61 and PIK3CA hotspot mutations. On the other hand, another subset of AMEs display phenotypic similarities with pleomorphic adenomas, such as the presence of myxochondroid matrix [68]. Interestingly, an HMGA2-WIF1 fusion gene, previously described in pleomorphic adenomas of the salivary gland, has been recently described identified in an ER-positive AME lacking mutations in HRAS, PIK3CA, or AKT1 [69].
Progression of AMEs to high-grade TNBCs have been reported. In this context, the TNBCs developing synchronously with AMEs have been shown to be clonally related to the AMEs and, at variance with high-grade common forms of TNBCs, lack TP53 mutations and may harbor additional PI3K pathway gene mutations and TERT promoter hotspot mutations [66].
Acinic cell carcinoma and microglandular adenosis
Breast acinic cell carcinoma (ACC) is an exceedingly rare subtype of breast cancer [70] that usually shows a benign course, although may occasionally progress to high-grade TNBC [71]. Although being characterized by the presence of cells with diffuse serous differentiation and having the same name of a salivary gland cancer, breast ACCs differ histologically, and molecularly from their salivary gland counterparts. In fact, breast ACCs are composed of cytologically bland cells with abundant eosinophilic cytoplasm containing coarse or fine zymogen-like cytoplasmic granules, which are arranged in a mixture of infiltrative microglandular and nested growth patterns [3]. In fact, the histologic features of ACCs are more reminiscent of microglandular adenosis (MGA), a neoplastic lesion composed of a haphazard proliferation of small glands infiltrating adipose and collagenous tissue devoid of desmoplastic reaction [60], than of their namesake in the salivary glands. Albeit regarded by some as a benign hyperplastic lesion [72], MGA is now considered to be clonal and neoplastic, and its spectrum now includes pure MGA without atypia, atypical MGA (AMGA), and MGA associated with invasive carcinoma [73–76], which not uncommonly is in the form of an ACC. Notably, MGA, AMGA ACC, and other forms of MGA or ACC associated high-grade TNBCs display a similar immunophenotype, including the expression of S100 protein [73].
Genomic analyses have corroborated the notion that breast ACCs are not related to their salivary gland counterparts, but rather to MGA/AMGA. ACCs arising in the salivary glands harbor the t(4;9)(q13;q31) translocation resulting in rearrangements involving the nuclear transcription factor NR4A3 [77] and lack TP53 mutations [78], whereas breast ACCs lack a pathognomonic somatic mutation or fusion gene and harbor TP53 mutations [71, 79]. Conversely, ACCs and MGA display a remarkably similar pattern of complex gene copy number alterations, highly recurrent TP53 mutations and occasional PIK3CA hotspot mutations, akin to the repertoire of somatic genetic alterations of common forms of TNBC [71, 74–76, 78–80]. Consistent with these observations, a subset of ACCs has been shown to display genomic features of homologous recombination DNA repair defects, in a way akin to BRCA1 mutant TNBCs. Indeed, a subset of breast ACCs have been diagnosed in BRCA1 germline mutations carriers or display BRCA1 loss-of-function genetic alterations [71, 79, 80]. In addition, we have recently reported a bona fide example of ACC in a patient harboring an MLH1 pathogenic germline variant; the ACC lacked expression of MLH1 and displayed microsatellite instability, suggesting that these tumors may also arise in the setting of microsatellite instability [79].
Both ACCs and MGA/AMGA have a proclivity to progress to high-grade TNBCs. There are now several lines of evidence to demonstrate that ACCs and MGA/AMGA may constitute the substrate from which common forms of TNBC may develop. Recent studies have demonstrated that when synchronously present, the high-grade TNBC is clonally related to the MGA, AMGA, and/or ACCs [71, 75]. Interestingly, metaplastic TNBC has been reported in association with ACC and MGA/AMGA [71, 75]. The genetic alterations driving the progression from these lesions to high grade TNBC have yet to be identified.
It should be noted, however, that MGAs/AMGAs constitute a heterogeneous group of lesions. The genomic features discussed above have been primarily described in MGA/AMGA associated with invasive carcinoma. In fact, the majority of pure MGAs (i.e., devoid of invasive carcinoma) appear to differ from carcinoma-associated MGA/AMGA based on the TP53 mutations and copy number alterations affecting genomic regions commonly altered in TNBCs. This observation suggests that the earliest drivers of the development of MGA have yet to be identified and that the acquisition of TP53 mutations may occur relatively late in the development of these lesions. Alternatively, one could also contend the hypothesis that pure MGA and carcinoma-associated MGA might evolve through fundamentally distinct genetic pathways.
Tall cell carcinomas with reversed polarity
Tall cell carcinoma with reversed polarity (TCCRP), also known as solid papillary carcinoma with reverse polarity or breast tumor resembling the tall cell variant of papillary thyroid carcinoma, is a rare histologic subtype of breast cancer with a unique histology [81–84]. TCCRPs display a phenotype reminiscent to that of the tall cell variant of papillary thyroid carcinoma, as they feature a solid, papillary and follicular architecture and are composed of tall cells with apically located nuclei (i.e., reverse polarization) showing grooves and optical clearing [85]. TCCRPs harbor recurrent IDH2 R172 hotspot mutations, which frequently co-occur with genetic alterations in genes of the PI3K pathway [86–89]. Forced expression of IDH2 R172S in PIK3CA H1047R knock-in MCF10A cell models resulted in the recapitulation of the reverse polarization phenotype in vitro [86], supporting the notion that co-existing IDH2 and PIK3CA hotspot mutations are drivers of TCCRPs and result in the acquisition of the reverse polarization that defines this tumor type. Notably, IDH2 R172 mutations can be detected by immunohistochemical analysis using either IDH2 R172S or IDH1/2 (R132/172) mutation-specific antibodies in a sensitive and specific manner in both biopsy and excision specimens (Fig. 3) [90, 91].
Conclusions
The systematic genomic and transcriptomic analyses of special histologic types of breast cancer has been proven to be more than a mere exercise of academic philately; rather, these studies have helped refine the taxonomy of breast cancers, identify novel pathognomonic genetic alterations (e.g., PRKD1 E710D hotspot mutations), define new genotypic–phenotypic correlations (e.g., MYB pathway activation as a common denominator of AdCCs) and contextualize hotspot mutations affecting known oncogenes that are rarely altered in breast cancer (e.g., HRAS and IDH2). Most importantly, these findings have now highlighted the importance of histologic typing TNBCs, given that low-grade forms of TNBC have a distinct clinical behavior from the ‘garden variety’ of TNBCs and a diagnosis of secretory carcinoma now constitutes an indication for the use of TRK inhibitors in the advanced/metastatic setting.
We would contend that the genotypic–phenotypic associations identified so far represent only the extremes of phenotypes, which could be investigated through diligent histologic analyses of breast cancers in conjunction with state-of-the-art sequencing approaches. With the advent of artificial intelligence (AI) methods applied to whole-slide hematoxylin-and-eosin images, which allow for the extraction of phenotypic features above and beyond those that are currently systematically reported in diagnostic histopathology, combined with sequencing approaches, additional genotypic–phenotypic correlations are likely to be identified. We anticipate that the marriage of traditional pathology with cutting edge AI, genomics, transcriptomic, and epigenomics analyses of special histologic types of breast cancer will result in a taxonomy of breast cancers that more accurately recapitulates the biologic diversity of this panoply of neoplasms.
Acknowledgements
JSR-F is funded in part by the Breast Cancer Research Foundation. Research reported in this publication was funded in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No P30CA008748). FP is partially funded by a National Institutes of Health/National Cancer Institute K12 CA184746 grant and BW by Cycle for Survival and Stand Up To Cancer grants. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest JSR-F reports receiving personal/consultancy fees from Goldman Sachs and REPARE Therapeutics, membership of the scientific advisory boards of VolitionRx and Paige.AI, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro, outside the scope of this study. The remaining authors have no conflicts of interest to declare.
References
- 1.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–30. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Classification of Tumors Editorial Board. Breast tumours. WHO Classification of Tumors. 5th ed. Lyon: IARC; 2019. [Google Scholar]
- 4.Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–50. [DOI] [PubMed] [Google Scholar]
- 5.Longacre TA, Ennis M, Quenneville LA, Bane AL, Bleiweiss IJ, Carter BA, et al. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Mod Pathol. 2006;19: 195–207. [DOI] [PubMed] [Google Scholar]
- 6.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast. Cancer Cell. 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, et al. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34:1872–81. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen B, Veys I, Leduc S, Bareche Y, Majjaj S, Brown DN, et al. Genomic, transcriptomic, epigenetic, and immune profiling of mucinous breast cancer. J Natl Cancer Inst. 2019;111:742–6. [DOI] [PubMed] [Google Scholar]
- 9.Pareja F, Lee JY, Brown DN, Piscuoglio S, Gularte-Merida R, Selenica P, et al. The genomic landscape of mucinous breast cancer. J Natl Cancer Inst. 2019;111:737–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–8. [DOI] [PubMed] [Google Scholar]
- 11.Lee JY, Schizas M, Geyer FC, Selenica P, Piscuoglio S, Sakr RA, et al. Lobular carcinomas in situ display intralesion genetic heterogeneity and clonal evolution in the progression to invasive lobular carcinoma. Clin Cancer Res. 2019;25:674–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner NC, Reis-Filho JS. Tackling the diversity of triple-negative breast cancer. Clin Cancer Res. 2013;19:6380–8. [DOI] [PubMed] [Google Scholar]
- 13.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–32. [DOI] [PubMed] [Google Scholar]
- 14.Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. J Natl Cancer Inst. 2015; 107:1–13. [DOI] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young SR, Pilarski RT, Donenberg T, Shapiro C, Hammond LS, Miller J, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer. 2009;9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fostira F, Tsitlaidou M, Papadimitriou C, Pertesi M, Timotheadou E, Stavropoulou AV, et al. Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: implications for genetic screening selection criteria: a Hellenic Cooperative Oncology Group Study. Breast Cancer Res Treat. 2012;134:353–62. [DOI] [PubMed] [Google Scholar]
- 22.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Investig. 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pareja F, Geyer FC, Marchio C, Burke KA, Weigelt B, Reis-Filho JS. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016;2:16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horowitz DP, Sharma CS, Connolly E, Gidea-Addeo D, Deutsch I. Secretory carcinoma of the breast: results from the survival, epidemiology and end results database. Breast. 2012;21:350–3. [DOI] [PubMed] [Google Scholar]
- 27.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76. [DOI] [PubMed] [Google Scholar]
- 28.Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. [DOI] [PubMed] [Google Scholar]
- 29.Krings G, Joseph NM, Bean GR, Solomon D, Onodera C, Talevich E, et al. Genomic profiling of breast secretory carcinomas reveals distinct genetics from other breast cancers and similarity to mammary analog secretory carcinomas. Mod Pathol. 2017;30:1086–99. [DOI] [PubMed] [Google Scholar]
- 30.Harrison BT, Fowler E, Krings G, Chen YY, Bean GR, Vincent-Salomon A, et al. Pan-TRK immunohistochemistry: a useful diagnostic adjunct for secretory carcinoma of the breast. Am J Surg Pathol. 2019;43:1693–700. [DOI] [PubMed] [Google Scholar]
- 31.Marchio C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019;30:1417–27. [DOI] [PubMed] [Google Scholar]
- 32.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I Trials (ALKA-372–001 and STARTRK-1). Cancer Discov. 2017; 7:400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19:705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchio C, Weigelt B, Reis-Filho JS. Adenoid cystic carcinomas of the breast and salivary glands (or ‘The strange case of Dr Jekyll and Mr Hyde’ of exocrine gland carcinomas). J Clin Pathol. 2010;63:220–8. [DOI] [PubMed] [Google Scholar]
- 37.Slodkowska E, Xu B, Kos Z, Bane A, Barnard M, Zubovits J, et al. Predictors of outcome in mammary adenoid cystic carcinoma: a multi-institutional study. Am J Surg Pathol. 2020;44:214–23. [DOI] [PubMed] [Google Scholar]
- 38.Righi A, Lenzi M, Morandi L, Flamminio F, De Biase D, Farnedi A, et al. Adenoid cystic carcinoma of the breast associated with invasive duct carcinoma: a case report. Int J Surg Pathol. 2011;19:230–4. [DOI] [PubMed] [Google Scholar]
- 39.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei J, Flieder DB, Patchefsky A, Talarchek JN, Cooper HS, Testa JR, et al. Detecting MYB and MYBL1 fusion genes in tracheobronchial adenoid cystic carcinoma by targeted RNA-sequencing. Mod Pathol. 2019;32:1416–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreasen S, Tan Q, Agander TK, Steiner P, Bjorndal K, Hogdall E, et al. Adenoid cystic carcinomas of the salivary gland, lacrimal gland, and breast are morphologically and genetically similar but have distinct microRNA expression profiles. Mod Pathol. 2018; 31:1211–25. [DOI] [PubMed] [Google Scholar]
- 42.Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Alfonso TM, Mosquera JM, MacDonald TY, Padilla J, Liu YF, Rubin MA, et al. MYB-NFIB gene fusion in adenoid cystic carcinoma of the breast with special focus paid to the solid variant with basaloid features. Hum Pathol. 2014;45:2270–80. [DOI] [PubMed] [Google Scholar]
- 44.Wetterskog D, Lopez-Garcia MA, Lambros MB, A’Hern R, Geyer FC, Milanezi F, et al. Adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J Pathol. 2012;226:84–96. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Geyer FC, Martelotto LG, Ng CK, Lim RS, Selenica P, et al. MYBL1 rearrangements and MYB amplification in breast adenoid cystic carcinomas lacking the MYB-NFIB fusion gene. J Pathol. 2018;244:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin SJ, Rosen PP. Solid variant of mammary adenoid cystic carcinoma with basaloid features: a study of nine cases. Am J Surg Pathol. 2002;26:413–20. [DOI] [PubMed] [Google Scholar]
- 47.Masse J, Truntzer C, Boidot R, Khalifa E, Perot G, Velasco V, et al. Solid-type adenoid cystic carcinoma of the breast, a distinct molecular entity enriched in NOTCH and CREBBP mutations. Mod Pathol. 2020;33:1041–55. [DOI] [PubMed] [Google Scholar]
- 48.Fusco N, Geyer FC, De Filippo MR, Martelotto LG, Ng CK, Piscuoglio S, et al. Genetic events in the progression of adenoid cystic carcinoma of the breast to high-grade triple-negative breast cancer. Mod Pathol. 2016;29:1292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foschini MP, Morandi L, Asioli S, Giove G, Corradini AG, Eusebi V. The morphological spectrum of salivary gland type tumours of the breast. Pathology. 2017;49:215–27. [DOI] [PubMed] [Google Scholar]
- 50.Basbug M, Akbulut S, Arikanoglu Z, Sogutcu N, Firat U, Kucukoner M. Mucoepidermoid carcinoma in a breast affected by burn scars: comprehensive literature review and case report. Breast Care. 2011;6:293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunt JL. An update on molecular diagnostics of squamous and salivary gland tumors of the head and neck. Arch Pathol Lab Med. 2011;135:602–9. [DOI] [PubMed] [Google Scholar]
- 52.Salem A, Bell D, Sepesi B, Papadimitrakopoulou V, El-Naggar A, Moran CA, et al. Clinicopathologic and genetic features of primary bronchopulmonary mucoepidermoid carcinoma: the MD Anderson Cancer Center experience and comprehensive review of the literature. Virchows Arch. 2017;470:619–26. [DOI] [PubMed] [Google Scholar]
- 53.Komiya T, Park Y, Modi S, Coxon AB, Oh H, Kaye FJ. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25:6128–32. [DOI] [PubMed] [Google Scholar]
- 54.Bean GR, Krings G, Otis CN, Solomon DA, Garcia JJ, van Zante A, et al. CRTC1-MAML2 fusion in mucoepidermoid carcinoma of the breast. Histopathology. 2019;74:463–73. [DOI] [PubMed] [Google Scholar]
- 55.Pareja F, Da Cruz Paula A, Gularte-Merida R, Vahdatinia M, Li A, Geyer FC, et al. Pleomorphic adenomas and mucoepidermoid carcinomas of the breast are underpinned by fusion genes. NPJ Breast Cancer. 2020;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffith CC, Schmitt AC, Little JL, Magliocca KR. New developments in salivary gland pathology: clinically useful ancillary testing and new potentially targetable molecular alterations. Arch Pathol Lab Med. 2017;141:381–95. [DOI] [PubMed] [Google Scholar]
- 57.Pia-Foschini M, Reis-Filho JS, Eusebi V, Lakhani SR. Salivary gland-like tumours of the breast: surgical and molecular pathology. J Clin Pathol. 2003;56:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenman G Fusion oncogenes in salivary gland tumors: molecular and clinical consequences. Head Neck Pathol. 2013;7:S12–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO Classification of Breast Tumors. Lyon: IARC; 2012. [Google Scholar]
- 61.Weinreb I, Piscuoglio S, Martelotto LG, Waggott D, Ng CK, Perez-Ordonez B, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sebastiao APM, Xu B, Lozada JR, Pareja F, Geyer FC, Da Cruz Paula A, et al. Histologic spectrum of polymorphous adenocarcinoma of the salivary gland harbor genetic alterations affecting PRKD genes. Mod Pathol. 2020;33:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu B, Barbieri AL, Bishop JA, Chiosea SI, Dogan S, Di Palma S, et al. Histologic classification and molecular signature of polymorphous adenocarcinoma (PAC) and cribriform adenocarcinoma of salivary gland (CASG): an International Interobserver Study. Am J Surg Pathol. 2020;44:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sebastiao APM, Pareja F, Kumar R, Brown DN, Silveira C, da Silva EM, et al. Genomic analysis of recurrences and high-grade forms of polymorphous adenocarcinoma. Histopathology. 2019;75:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nadelman CM, Leslie KO, Fishbein MC. “Benign,” metastasizing adenomyoepithelioma of the breast: a report of 2 cases. Arch Pathol Lab Med. 2006;130:1349–53. [DOI] [PubMed] [Google Scholar]
- 66.Geyer FC, Li A, Papanastasiou AD, Smith A, Selenica P, Burke KA, et al. Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas. Nat Commun. 2018;9:1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pareja F, Toss MS, Geyer FC, da Silva EM, Vahdatinia M, Sebastiao APM, et al. Immunohistochemical assessment of HRAS Q61R mutations in breast adenomyoepitheliomas. Histopathology. 2020;76:865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLaren BK, Smith J, Schuyler PA, Dupont WD, Page DL. Adenomyoepithelioma: clinical, histologic, and immunohistologic evaluation of a series of related lesions. Am J Surg Pathol. 2005; 29:1294–9. [DOI] [PubMed] [Google Scholar]
- 69.Pareja F, Geyer FC, Brown DN, Sebastiao APM, Gularte-Merida R, Li A, et al. Assessment of HMGA2 and PLAG1 rearrangements in breast adenomyoepitheliomas. NPJ Breast Cancer. 2019;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roncaroli F, Lamovec J, Zidar A, Eusebi V. Acinic cell-like carcinoma of the breast. Virchows Arch. 1996;429:69–74. [DOI] [PubMed] [Google Scholar]
- 71.Guerini-Rocco E, Hodi Z, Piscuoglio S, Ng CK, Rakha EA, Schultheis AM, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavassoli FA, Norris HJ. Microglandular adenosis of the breast. A clinicopathologic study of 11 cases with ultrastructural observations. Am J Surg Pathol. 1983;7:731–7. [PubMed] [Google Scholar]
- 73.Wen YH, Weigelt B, Reis-Filho JS. Microglandular adenosis: a non-obligate precursor of triple-negative breast cancer? Histol Histopathol. 2013;28:1099–108. [DOI] [PubMed] [Google Scholar]
- 74.Geyer FC, Kushner YB, Lambros MB, Natrajan R, Mackay A, Tamber N, et al. Microglandular adenosis or microglandular adenoma? A molecular genetic analysis of a case associated with atypia and invasive carcinoma. Histopathology. 2009;55:732–43. [DOI] [PubMed] [Google Scholar]
- 75.Geyer FC, Lacroix-Triki M, Colombo PE, Patani N, Gauthier A, Natrajan R, et al. Molecular evidence in support of the neoplastic and precursor nature of microglandular adenosis. Histopathology. 2012;60:E115–30. [DOI] [PubMed] [Google Scholar]
- 76.Geyer FC, Berman SH, Marchio C, Burke KA, Guerini-Rocco E, Piscuoglio S, et al. Genetic analysis of microglandular adenosis and acinic cell carcinomas of the breast provides evidence for the existence of a low-grade triple-negative breast neoplasia family. Mod Pathol. 2017;30:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haller F, Bieg M, Will R, Korner C, Weichenhan D, Bott A, et al. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun. 2019;10:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piscuoglio S, Hodi Z, Katabi N, Guerini-Rocco E, Macedo GS, Ng CK, et al. Are acinic cell carcinomas of the breast and salivary glands distinct diseases? Histopathology. 2015;67:529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beca F, Lee SSK, Pareja F, Da Cruz Paula A, Selenica P, Ferrando L, et al. Whole-exome sequencing and RNA sequencing analyses of acinic cell carcinomas of the breast. Histopathology. 2019; 75:931–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ripamonti CB, Colombo M, Mondini P, Siranoush M, Peissel B, Bernard L, et al. First description of an acinic cell carcinoma of the breast in a BRCA1 mutation carrier: a case report. BMC Cancer. 2013;13:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colella R, Guerriero A, Giansanti M, Sidoni A, Bellezza G. An additional case of breast tumor resembling the tall cell variant of papillary thyroid carcinoma. Int J Surg Pathol. 2015;23:217–20. [DOI] [PubMed] [Google Scholar]
- 82.Masood S, Davis C, Kubik MJ. Changing the term “breast tumor resembling the tall cell variant of papillary thyroid carcinoma” to “tall cell variant of papillary breast carcinoma”. Adv Anat Pathol. 2012;19:108–10. [DOI] [PubMed] [Google Scholar]
- 83.Eusebi V, Damiani S, Ellis IO, Azzopardi JG, Rosai J. Breast tumor resembling the tall cell variant of papillary thyroid carcinoma: report of 5 cases. Am J Surg Pathol. 2003;27:1114–8. [DOI] [PubMed] [Google Scholar]
- 84.Cameselle-Teijeiro J, Abdulkader I, Barreiro-Morandeira F, Ruiz-Ponte C, Reyes-Santias R, Chavez E, et al. Breast tumor resembling the tall cell variant of papillary thyroid carcinoma: a case report. Int J Surg Pathol. 2006;14:79–84. [DOI] [PubMed] [Google Scholar]
- 85.Foschini MP, Asioli S, Foreid S, Cserni G, Ellis IO, Eusebi V, et al. Solid papillary breast carcinomas resembling the tall cell variant of papillary thyroid neoplasms: a unique invasive tumor with indolent behavior. Am J Surg Pathol. 2017;41:887–95. [DOI] [PubMed] [Google Scholar]
- 86.Chiang S, Weigelt B, Wen HC, Pareja F, Raghavendra A, Martelotto LG, et al. IDH2 mutations define a unique subtype of breast cancer with altered nuclear polarity. Cancer Res. 2016;76:7118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lozada JR, Basili T, Pareja F, Alemar B, Paula ADC, Gularte-Merida R, et al. Solid papillary breast carcinomas resembling the tall cell variant of papillary thyroid neoplasms (solid papillary carcinomas with reverse polarity) harbour recurrent mutations affecting IDH2 and PIK3CA: a validation cohort. Histopathology. 2018;73:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong E, Scognamiglio T, D’Alfonso T, Song W, Tran H, Baek I, et al. Breast tumor resembling the tall cell variant of papillary thyroid carcinoma: molecular characterization by next-generation sequencing and histopathological comparison with tall cell papillary carcinoma of thyroid. Int J Surg Pathol. 2019;27:134–41. [DOI] [PubMed] [Google Scholar]
- 89.Bhargava R, Florea AV, Pelmus M, Jones MW, Bonaventura M, Wald A, et al. Breast tumor resembling tall cell variant of papillary thyroid carcinoma: a solid papillary neoplasm with characteristic immunohistochemical profile and few recurrent mutations. Am J Clin Pathol. 2017;147:399–410. [DOI] [PubMed] [Google Scholar]
- 90.Pareja F, da Silva EM, Frosina D, Geyer FC, Lozada JR, Basili T, et al. Immunohistochemical analysis of IDH2 R172 hotspot mutations in breast papillary neoplasms: applications in the diagnosis of tall cell carcinoma with reverse polarity. Mod Pathol. 2020;33:1056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alsadoun N, MacGrogan G, Truntzer C, Lacroix-Triki M, Bedgedjian I, Koeb MH, et al. Solid papillary carcinoma with reverse polarity of the breast harbors specific morphologic, immunohistochemical and molecular profile in comparison with other benign or malignant papillary lesions of the breast: a comparative study of 9 additional cases. Mod Pathol. 2018;31:1367–80. [DOI] [PubMed] [Google Scholar]


