Abstract
Background
Conventional HER2-targeting therapies improve outcomes for patients with HER2-positive breast cancer (BC), defined as tumors showing HER2 protein overexpression by immunohistochemistry and/or ERBB2 gene amplification determined by in situ hybridization (ISH). Emerging HER2-targeting compounds show benefit in some patients with neither HER2 protein overexpression nor ERBB2 gene amplification, creating a need for new assays to select HER2-low tumors for treatment with these compounds. We evaluated the analytical performance of a targeted mass spectrometry-based assay for quantifying HER2 protein in formalin-fixed paraffin-embedded (FFPE) and frozen BC biopsies.
Methods
We used immunoaffinity-enrichment coupled to multiple reaction monitoring-mass spectrometry (immuno-MRM-MS) to quantify HER2 protein (as peptide GLQSLPTHDPSPLQR) in 96 frozen and 119 FFPE BC biopsies. We characterized linearity, lower limit of quantification (LLOQ), and intra- and inter-day variation of the assay in frozen and FFPE tissue matrices. We determined concordance between HER2 immuno-MRM-MS and predicate immunohistochemistry and ISH assays and examined the benefit of multiplexing the assay to include proteins expressed in tumor subcompartments (e.g., stroma, adipose, lymphocytes, epithelium) to account for tissue heterogeneity.
Results
HER2 immuno-MRM-MS assay linearity was ≥103, assay coefficient of variation was 7.8% (FFPE) and 5.9% (frozen) for spiked-in analyte, and 7.7% (FFPE) and 7.9% (frozen) for endogenous measurements. Immuno-MRM-MS-based HER2 measurements strongly correlated with predicate assay HER2 determinations, and concordance was improved by normalizing to glyceraldehyde-3-phosphate dehydrogenase. HER2 was quantified above the LLOQ in all tumors.
Conclusions
Immuno-MRM-MS can be used to quantify HER2 in FFPE and frozen BC biopsies, even at low HER2 expression levels.
Keywords: HER2, breast cancer, multiple reaction monitoring, formalin-fixed paraffin-embedded (FFPE), targeted mass spectrometry, immuno-MRM-MS, immunopeptide enrichment, tissue heterogeneity
Introduction
In ∼15% of breast cancers (BC), erb-b2 receptor tyrosine kinase 2 (ERBB2) amplification leads to overexpression of human epidermal growth factor receptor 2 (HER2) protein (1), which portends a worse prognosis unless treated with HER2-targeted therapy. Thus, accurate assessment of HER2 status is critical for optimal treatment of these patients. Under current guidelines (2), BCs are considered HER2-positive if the immunohistochemistry (IHC) assay score is 3+, or if ERBB2 gene amplification is observed on an in situ hybridization (ISH) assay. Harmonization of these assays has been challenging, due to a combination of preanalytical and analytical variables (1).
The 40%–50% of BCs demonstrating low HER2 expression and no detectable ERBB2 amplification (HER2-low: IHC 1+ or IHC 2+ with negative ISH) are considered “HER2-negative” and are not currently treated with HER2-targeted therapy (3) (e.g., trastuzumab, ado-trastuzumab emtansine). However, new anti-HER2 compounds such as antibody–drug conjugates are showing significant clinical benefit in the HER2-low subset of BCs, with one study showing a confirmed response rate of 37.0% (3). Thus, laboratory methods that are more quantitative and objective than IHC are needed to quantify HER2 expression in these traditionally “HER2 negative” BCs, to facilitate patient selection for the new generation of HER2-targeting therapies.
Prior studies (4–7) demonstrate the feasibility of using multiple-reaction monitoring mass spectrometry (MRM-MS) to quantify HER2 from protein extracted from formalin-fixed paraffin-embedded (FFPE) BC tissue, showing strong agreement between MRM results and IHC/ISH, and also demonstrating correlation between MRM-based measurements and clinical response to HER2-targeted therapy (6). One study demonstrated that amounts of 6 HER2 peptides were highly correlated, validating their use as surrogates of protein amounts in FFPE tissues (4). These studies quantified HER2 without enrichment, and HER2 expression levels were either below the lower limit of quantification (LLOQ) of the assay for a high percentage of the HER2-low tumors or required a large amount of input for the protein to be detected. Thus, higher assay sensitivity will be necessary to stratify patients with tumors expressing low levels of HER2 who may benefit from emerging therapies.
To address this unmet need, we tested the feasibility of using immuno-affinity enrichment of the HER2 proteotypic peptide GLQSLPTHDPSPLQR upstream of MRM (immuno-MRM-MS), hypothesizing that this would provide sufficient assay sensitivity to quantify HER2 in HER2-low tumors. The ability to differentiate HER2-low from HER2-negative cases could allow for better selection of patients for HER2-targeting antibody–drug conjugates. We characterized the analytical performance of the novel immuno-MRM-MS assay in both FFPE and frozen BC matrices and assessed concordance between the immuno-MRM-MS and predicate IHC/ISH assay results across a set of 215 BCs representing a wide range of tumor cellularity. We also considered effects of tissue heterogeneity on MRM-based HER2 determinations, by exploring the incorporation into the immuno-MRM-MS assay of additional tumor compartment biomarkers (e.g., stroma, adipose, epithelium, lymphocytes) in multiplex with HER2.
Materials and Methods
An expanded Materials and Methods section is available in the Supplemental Data.
Standards and Reagents
Custom anti-peptide monoclonal antibodies (mAbs) for immuno-MRM-MS assays were generated by Epitomics, an Abcam company (Burlingame, CA), and Excel BioPharm (Burlingame, CA) using hybridoma screening (8) or recombinant B-cell technology (9). A custom hybridoma mouse mAb (to PECAM1) was generated by the Fred Hutchinson Cancer Research Center Antibody Development Facility (Seattle, WA). Purified and crude heavy stable isotope-labeled standard (SIS) peptides were obtained from New England Peptide (Gardner, MA) and ThermoFisher Scientific (Waltham, MA). SIS peptides and matched light versions were handled according to published recommendations (10).
Study Specimens
The 119 archived FFPE BC biospecimens (53 HER2+, 64 HER2-, 2 equivocal) and the associated pathology reports were obtained from Stanford University Medical Center and NWBioTrust under approval and consistent with IRB reviews (Stanford University Medical Center IRB #348 and Fred Hutchinson Cancer Research Center IRB #7077), with no specific selection criteria. The 96 frozen BC biospecimens (13 HER2+, 77 HER2-, 6 equivocal or not available) and the associated pathology reports were collected as part of the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) program under the CPTAC2 breast procurement protocol (11) under IRB approval (at the collection site). All patients provided written informed consent. As previously described (12), samples were collected from a newly diagnosed patient with invasive breast adenocarcinoma undergoing surgical resection with no prior treatment for the disease and were selected for tumors with mass > 200 mg, percent necrosis ≤20%, and percent neoplastic cellularity ≥60%. Frozen tissue not needed for clinical management was divided into 2–4 segments. Hematoxylin and eosin (H&E) staining and imaging was performed for all FFPE and frozen samples. Tissue subcompartment cellularity (e.g., tumor, stroma, adipocytes, lymphocytes) was calculated using the HALO Tissue Classifier (Indica Labs, NM, USA) machine learning algorithm. Two pathologists, blinded to pathology results, independently trained algorithms on H&E-stained slides. One of the pathologists trained 2 algorithms separately on different subsets of the slides, so that 3 measurements were made from the different algorithms. Separate algorithms were trained for FFPE and frozen samples. A third pathologist scored the subcompartment cellularity of a subset of the slides to confirm the validity of the automated results.
Sample Preparation
FFPE samples were prepared according to American Society of Clinical Oncology/College of American Pathologists guidelines (13). 10 μm sections of the FFPE samples were cut using a microtome and mounted on glass slides. Every 10th FFPE slice was 4 μm and used for H&E and pathology review. Slide-mounted FFPE tissue sections were deparaffinized, rehydrated, and protein was extracted into a 0.1% RapiGest solution, as described previously (14). For frozen samples, a tissue segment was cryopulverized and the protein was extracted into a 6 M urea solution. Protein concentration was quantified by Micro BCA Assay (ThermoFisher). 20 to 50 µg protein from FFPE samples and 100 µg from frozen samples were reduced and alkylated, then incubated at 37 °C for 2 h after trypsin addition at 1:50 trypsin: protein ratio by mass and then overnight after trypsin addition at 1:100. Steady state digestion was confirmed for the HER2 peptide GLQSLPTHDPSPLQR at this incubation time and trypsin: protein ratio (see Supplemental Data). After digestion, a mixture of SIS peptides was spiked into the individual samples, and the samples were desalted.
Samples were processed in a blinded fashion, in triplicate when enough material was available (39 FFPE and 96 frozen samples were processed in triplicate). FFPE replicates consisted of independent sections that were processed independently (including deparaffinization, protein extraction, digestion, enrichment, and MRM) on different days; frozen replicates consisted of independent aliquots from a common lysate that underwent digestion and enrichment on different days.
Affinity Enrichment and Liquid Chromatography-Multiple-Reaction Monitoring-Mass Spectrometry (LC-MRM-MS)
Peptide immunoaffinity enrichment for all experiments was performed on ≥20 µg of digested tissue lysate and consisted of overnight incubations of the cross-linked mAbs using a mix of at least 1 µg of each of the 23 mAbs. A KingFisher platform was used to wash the beads as described previously (15) with modifications noted in the Supplemental Methods. For LC-MRM-MS analysis, 10 µL of a 26 µL eluate were injected on an ekspert nanoLC 425 with an NLC 400x AS autosampler (Eksigent Technologies, Dublin, CA) coupled to a 6500 QTRAP mass spectrometer (ABSciex, Foster City, CA). SIS peptides were obtained for each targeted peptide and used to optimize transitions and collision energies for MRM analysis (Supplemental Table 1). They were also used as internal calibrators to assign protein concentration per mg protein (see Supplemental Methods). The assay was multiplexed as described previously (15).
Assay Characterization
Performance figures of merit for the multiplex immuno-MRM-MS assay were determined according to best practices using a fit-for-purpose approach (16, 17). The quality of the analysis of peptides was assessed in a matrix composed of a mix of one-third of the individual samples from either FFPE (n = 40) or frozen (n = 34) breast tumor protein lysates. Response curves were used to characterize the linear range, upper and lower limits of detection (LLOQ and ULOQ). Sample processing repeatability (trypsin digestion through LC-MRM-MS) was characterized by measuring peptides spiked in the background matrix at 3 concentrations in triplicate over 5 separate days. The stability of the processed samples was validated using the background matrix processed in an identical manner and analyzed under several storage conditions in triplicate. The reproducibility of the endogenous measurements was characterized by measuring endogenous analyte peptides in background matrix in 5 process replicates over 5 separate days. Background noise and carryover signal was characterized by analyzing solvent blanks.
Clinical Study
The performance of the multiplex MRM assay for quantification of HER2 was retrospectively assessed in 215 BC biopsies (96 frozen, 119 FFPE). The clinical and demographic data for the patients are provided in Supplemental Table 2.
Sample Processing Quality Controls
Six process quality control (QC) samples consisting of a mix of protein lysates from multiple cell lines (T47D, adipocyte, fibroblast, and lymphoblastoid cell lines), as well as plasma, were included on every 96-well plate of samples and underwent the same workflow. Three predigested process QC samples were added to each plate after digestion and continued through the remainder of the workflow. Total variation was calculated from intra-plate and inter-plate variation.
Data Analysis and Statistics
MRM peak integration was performed in Skyline (18), with the sum of all transitions used for quantification. Peptide concentrations were calculated as the peak area ratio of the light and heavy peptides times the concentration of SIS peptide. Integration results were exported to the program R for machine learning modeling, correlations, and area under the curve (AUC) calculations.
Data Accessibility
Individual FFPE and frozen sample, and process QC MRM data are available (19). Characterization data for assays are available in the CPTAC Assay Portal (20). Expression level and gene copy number analysis of the frozen BCs has been released at the Genomic Data Commons and is accessible via the database of Genotypes and Phenotypes Study Accession phs000892 (21).
Results
The goals of the study were to i) evaluate immuno-MRM-MS for quantification of HER2 in BC biopsies (especially HER2-low), and ii) evaluate effects of tumor heterogeneity (measured by histological analysis and immuno-MRM-MS) on HER2 determinations using a bulk tissue MRM assay. The workflow is summarized in Fig. 1.
Fig. 1.
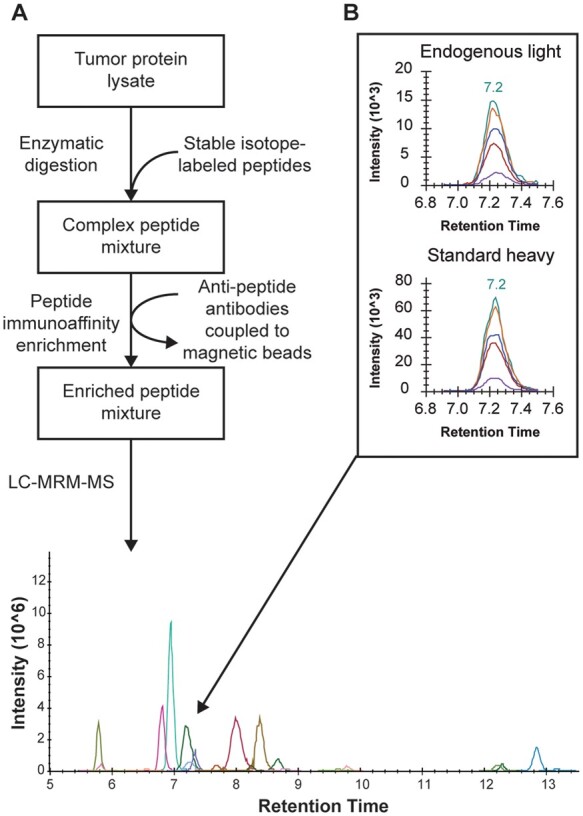
A multiplex immuno-MRM-MS workflow quantified targets in breast cancer tissue samples. (A), Schematic of sample preparation, from protein lysate through enzymatic digestion and addition of standards, to the enrichment of target peptides prior to targeted MS analysis. (B), HER2 levels quantified by the peak area ratio of light endogenous signal to heavy standard signal for the peptide GLQSLPTHDPSPLQR.
Fit-for-Purpose Assay Characterization
A 23-plex immuno-MRM-MS assay was developed to quantify HER2 protein (as peptide GLQSLPTHDPSPLQR) and 22 additional peptides (representing 13 proteins and pan-keratins, including housekeeping proteins, BC biomarkers, and tumor compartment-specific biomarkers; Supplemental Table 1). 21 peptides were successfully characterized in either FFPE or frozen sample by response curves (Supplemental Table 3, Supplemental Fig. 1), inter-day repeatability of the entire workflow (Supplemental Table 4, Supplemental Fig. 2), assay stability (Supplemental Table 5, Supplemental Fig. 3), and endogenous measurement reproducibility of the entire workflow (Supplemental Table 6, Supplemental Fig. 4). Background noise and carryover signal was also characterized (Supplemental Table 4). Assay response curves for HER2 are shown in Fig. 2 and a summary of the figures of merit can be found in Table 1.
Fig. 2.
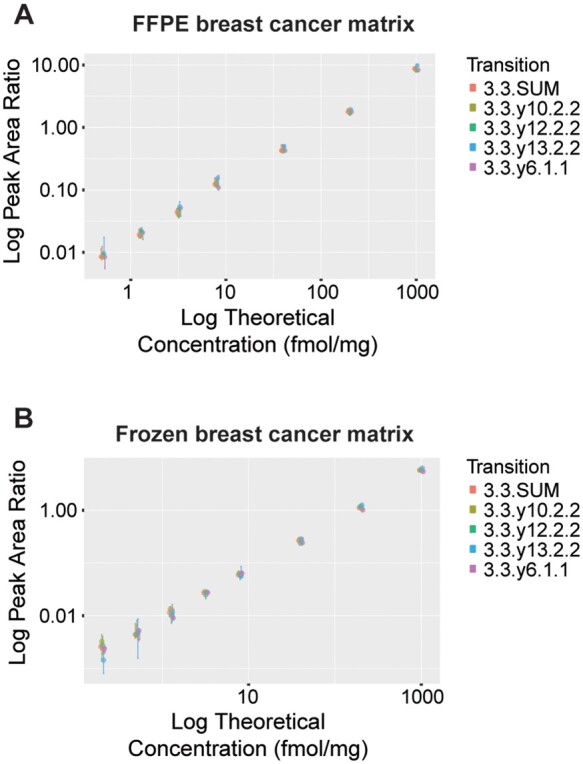
The assay targeting the HER2 peptide GLQSLPTHDPSPLQR was characterized by response curves. Response curves consist of 8 concentration points, covering almost 4 orders of magnitude, in (A), formalin-fixed paraffin-embedded (FFPE) and (B), frozen tumor tissue lysate. Error bars represent the standard deviation of the independent analysis of triplicate process replicates.
Table 1.
Assay characteristics for HER2 and the entire immuno-MRM-MS assay panel
| FFPE samples |
Frozen samples |
|||
|---|---|---|---|---|
| HER2 | All analytes | HER2 | All analytes | |
| LLOQ (fmol/mg protein) | 10 |
183 (10–12 000) |
2 |
111 (2–4400) |
| ULOQ (fmol/mg protein) | ≥ 19 719 |
≥ 20 708 (411–395 523) |
≥ 9985 |
≥ 10 503 (1079–198 230) |
| Inter-day total variability (CV%) | 7.8% |
9.2% (0.5%–52%) |
5.9% |
12.7% (4.9%–65%) |
| Endogenous measurement reproducibility (CV%) | 7.7% |
11.3% (3.1%–78%) |
7.9% |
12.4% (6.4%–54%) |
The multiplexed immuno-MRM-MS assay consisting of 23 peptides was characterized in formalin fixed paraffin embedded (FFPE) and frozen tumor tissue lysate matrix. Lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ) calculated from response curves (reported in fmol/mg protein), inter-day total variability and variability of endogenous signal measurement are reported (median and range are reported across all analytes).
Histological and Biochemical Characterization of Tissue Heterogeneity
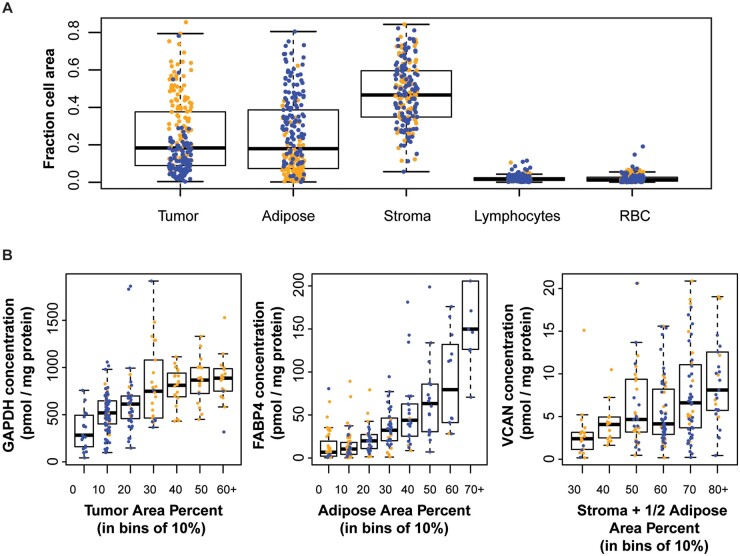
To study the impact of tumor heterogeneity on MRM-based analysis of bulk tissue to determine HER2 status, we quantified tumor composition using both histopathological analysis and tumor component biomarkers. Results of histopathological analyses from 3 independent machine learning algorithms (Supplemental Fig. 5) agreed for tissue components with area representation >5 mm2 (tumor, stroma, adipose; CVs 26%, 19%, and 28%, respectively; Supplemental Fig. 6, Supplemental Table 7) and agreed with manually-determined cellularity (median absolute difference between the average automated and manual measurement for tumor, stroma, and adipose is 7%, 12%, and 10%, respectively; Supplemental Fig. 7). This agreement is better than that reported when cellularity is estimated by independent pathologists (22). Histopathological quantification of tissue components with small area representation (≤5 mm2) in the clinical samples (lymphocytes, red blood cells) was not consistent, and thus these tissue components were not considered in further analyses. The results (Fig. 3, A and Supplemental Table 7) show a wide range of tumor cellularity (1%–80%) amongst study samples, which was on average lower in the FFPE (median 11%) than in the frozen (median 39%) samples, as expected based on inclusion criteria.
Fig. 3.
Tissue cellularity was characterized in the study samples. (A), Characterized by histopathology staining, a wide range of tumor (epithelium), adipocyte, stroma, lymphocyte and red blood cell cellularity was found across the 96 frozen (orange) and 119 FFPE (blue) samples. (B), Relationship between GAPDH measurements with tumor cellularity, FABP4 with adipose, and VCAN with stroma and adipose.
For biochemical characterization, we quantified expression levels of tissue component biomarkers, including epithelium (pan-keratin, KRT5 and KRT7), stroma (FBN1 and VCAN), blood vessels (PECAM1), lymphocytes (PTPRC), red blood cells (HBB), and adipocytes (FABP4 and PLIN4). Adipocytes did contribute to the stroma marker concentrations (Supplemental Fig. 8), likely due to the stromal vascular fraction (23). We also quantified ‘housekeeping’ proteins traditionally used to confirm protein loading for Western blot analyses [actin alpha1 (ACTA1), tubulin beta class (TUBB2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH)], although previous studies have shown higher GAPDH concentrations in cancer tissues compared with normal (24, 25). These biomarkers were quantified in multiplex with HER2 in the immuno-MRM-MS assay (Supplemental Table 8, A and B). Positive correlation was observed between expression levels of GAPDH with area percentages of tumor, FABP4 with adipose, and VCAN with stroma and adipose (Fig. 3, B), suggesting that these biomarkers might serve as biochemical signals to quantify these tissue compartments.
Quantification of HER2 in FFPE and Frozen Breast Cancer Biopsies
Protein lysates from the 215 study samples were prepared and analyzed in a blinded and randomized fashion in process replicates (depending on available material: 135 triplicate, 38 duplicate, and 42 singlicate samples). Replicates for the FFPE specimens were prepared from independent 10-micron sections. Endogenous concentrations of the HER2 peptide GLQSLPTHDPSPLQR and 22 other peptides were measured by immuno-MRM-MS and are shown in Supplemental Table 8, A and B.
Multiple QC samples (protein lysates from a mix of cell lines and plasma) and repeatability characterization samples (pooled tumor lysate) were run intermixed with the biopsies on each 96-well plate, including 3 samples that had been predigested with trypsin (“partial process QC”) and 6 that had not been (“full process QC”) (Supplemental Table 9, Supplemental Fig. 9). Three repeatability samples were prepared in complete process triplicate, including a low (8 fmol/sample), medium (80 fmol/sample), and high (800 fmol/sample) spike of heavy peptide mix, and a replicate was analyzed at the beginning, middle, and end of each plate (Supplemental Fig. 2, Supplemental Table 4). Across all 96-well plates, for the HER2 peptide GLQSLPTHDPSPLQR, total variation was 6.0% (5 FFPE sample plates) and 5.1% (6 frozen sample plates) for the partial process QC, and 7.3% (FFPE) and 7.6% (frozen) for the full process QC, indicating some variation introduced at the trypsin digestion step as reported previously (26). For the repeatability samples, total variation was 21.7% (low), 7.8% (medium), and 4.6% (high) for FFPE sample plates, and 15.7% (low), 5.9% (medium), and 4.6% (high) for frozen. For the entire assay panel targeting 23 peptides, the median total variation was 8.0% (FFPE) and 6.5% (frozen) for the partial process QC, and 14.0% (FFPE) and 11.2% (frozen) for the full process QC. For the repeatability samples, the median total variation was 20.8% (low), 9.2% (medium), and 8.1% (high) for FFPE, and 16.2% (low), 12.7% (medium), and 8.4% (high) for frozen.
Endogenous HER2 was detected above the LLOQ in all 215 biopsies, at a median concentration of 207 fmol/mg protein (range 21–8739) in FFPE samples (consistent with prior studies (5)) and 116 fmol/mg protein (range 19–8084) in frozen samples. Median CVs of the HER2 measurements in the 215 biopsies were 20% (FFPE) and 6% (frozen). HER2 determinations by predicate assays were available for 195 biopsies (105 FFPE, 90 frozen) from patients who did not receive neoadjuvant treatment (Supplemental Table 2), which has been shown to affect HER2 concentrations (27) (Supplemental Fig. 10).
Agreement between MRM and Predicate (IHC±ISH) Assays
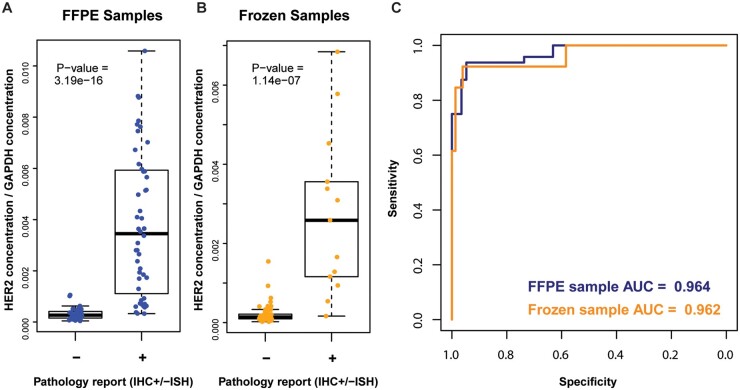
We evaluated the ability of immuno-MRM-MS measurements to differentiate HER2 status in FFPE and frozen tumor biopsies, using predicate (IHC±ISH) assay results as the gold standard to define the HER2 status. To account for the considerable variation in tumor cellularity in our study samples (Fig. 3, A), we investigated the use of tumor component biomarkers as a normalization factor for HER2 concentrations. Incorporating the immuno-MRM-MS measurements of tumor component biomarkers along with HER2, 3 machine learning models consistently found that measurements of HER2 in combination with GAPDH best predicts HER2 status in FFPE samples (Supplemental Fig. 11). Since GAPDH is positively correlated with tumor cellularity, we examined the agreement of HER2 normalized by GAPDH (as a surrogate for tumor cellularity) to HER2 status. Agreement between the GAPDH-normalized immuno-MRM-MS measurements of HER2 and HER2 status (by IHC±ISH) was excellent in both the FFPE (P-value = 1.09 e-16 by Mann-Whitney test; Fig. 4, A) and frozen (P-value = 1.14 e-7 by Mann-Whitney test; Fig. 4, B) biopsies. In addition, compared with standard of care, the immuno-MRM-MS assay had good sensitivity and specificity in both FFPE (AUC = 0.971) and frozen (AUC = 0.962) samples (Fig. 4, C). While normalizing HER2 to GAPDH signal improved sensitivity and specificity in the FFPE (P-value = 0.046) biopsies, normalization did not significantly improve assay performance in the frozen (P-value = 0.366) biopsies compared to HER2 immuno-MRM-MS measurements alone (Supplemental Fig. 12, A); this is likely due to higher tumor cellularity in the frozen vs FFPE samples (Fig. 3, A). Of note, although normalization to GAPDH did not improve concordance in the frozen samples, GAPDH normalization did significantly, but slightly, (P-value = 0.045) improve the correlation between HER2 messenger RNA (mRNA) and MRM measurements (R2 = 0.85 unnormalized, Supplemental Fig. 12, B; R2 = 0.90 after normalization, Supplemental Fig. 13, A), suggesting possible benefit to normalizing for quantitative molecular profiling.
Fig. 4.
Immuno-MRM-MS measurements can differentiate HER2 status in FFPE and frozen tumor biopsies. Agreement between MRM measurements of HER2 normalized to GAPDH and HER2 status as defined by predicate assays in (A), 105 FFPE and (B), 90 frozen samples. (C), Receiver operator curves for FFPE (blue) and frozen (orange) samples.
The observed discordant rate is consistent with the expected error rate (1%–8%) of the predicate HER2 clinical assays (28–30). Leveraging genomic data available for the frozen tumors (12), we examined HER2 mRNA expression level and ERBB2 gene copy number in the 3 tumors with discordant results between MRM vs predicate assays (1 putative false positive and 2 putative false negatives; Supplemental Fig. 13). HER2 positivity cutoffs were established by the accuracy of MRM, mRNA and copy number measurements to differentiate HER2 status (defined by predicate assays) (see Supplemental Methods). The putative false positive tumor (triangle in Supplemental Fig. 13) was scored IHC 0 but showed high level of HER2 mRNA expression, consistent with the immuno-MRM-MS data. The two putative false negative tumors (highlighted by the diamond and circle in Supplemental Fig. 13) were indeterminate by IHC (2+), and their HER2 status was determined by ISH. One of these tumors (circle in Supplemental Fig. 13) shows mRNA expression and copy number levels consistent with HER2- samples, corroborating the immuno-MRM-MS data. The other putative false negative tumor (diamond in Supplemental Fig. 13) showed ERBB2 copy number gain consistent with HER2+ samples and borderline mRNA expression level, and the MRM-based measurement of the HER2 protein was just below the cutoff for HER2 positivity. Of note, these comparisons are complicated by the preanalytical variable that different tumor sections went into each of the assays, which could contribute to observed discordances.
HER2 -Negative and -Low Tumors
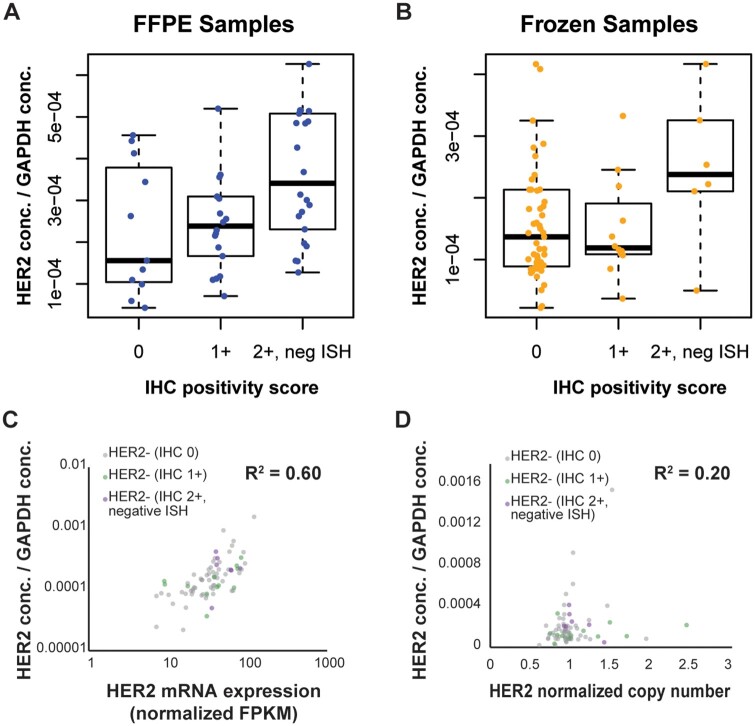
Immuno-enrichment of the HER2 peptide enabled detection above LLOQ in all study samples, including HER2-negative (IHC 0) and HER2-low tumors. The immuno-MRM-MS assay revealed a wide range of normalized HER2 expression levels amongst HER2-negative and -low tumors, spanning an approximate 70-fold range (Fig. 5, A & B). HER2 mRNA expression levels and ERBB2 copy number also spanned a wide range (17-fold and 4-fold, respectively) amongst HER2- negative and -low tumors (Fig. 5, C and D), and, as expected (31), mRNA expression levels showed higher correlation with MRM (R2 = 0.60) than did ERBB2 copy number (R2 = 0.20).
Fig. 5.
HER2 proteogenomic levels reveal a wide range of HER2 expression amongst HER2-negative and -low tumors. In HER2-negative and -low tumors, normalized HER2 MRM measurements compared to IHC positivity scores in (A), 57 FFPE and (B), 77 frozen samples. In the frozen samples, MRM measurements compared to (C), HER2 mRNA expression levels and (D), ERBB2 copy number.
Discussion
Interest in novel assays for quantifying HER2 protein is bolstered by recent data showing clinical activity of novel HER2-targeting therapies in nonbreast/gastric cancers (32) and in a subset of HER2-low BCs (1) that are scored HER2-negative using predicate assays. HER2-low and HER2-negative tumors are distinct and clinically relevant subtypes, but IHC and ISH assays have inherent discrepancies when delineating these IHC-negative subtypes (33). There is an emerging need for quantitative laboratory methods to measure tumor HER2 protein expression to facilitate patient selection for these novel treatments.
Prior studies have demonstrated the use of MRM to quantify HER2 protein in human cancers, showing correlation between MRM results and both predicate (IHC±ISH) assay results and clinical response to HER2-directed therapies (4–7). We confirm and extend these prior studies using a novel immuno-MRM-MS assay to HER2. The addition of immunoaffinity enrichment not only reduces the requirement of input material from precious clinical biospecimens, but also opens the possibility of multiplexing into the assay additional treatment response biomarker proteins that require enrichment. We demonstrate that our immuno-MRM-MS assay has acceptable analytical characteristics, high concordance with predicate assays, and enables precise, relative quantification of HER2 in HER2-low and HER2-negative tumors. The variability of the method was higher in FFPE tissues compared with frozen tumors, but this likely stems from the fact that different sections were used for the FFPE analysis compared with a single tissue fragment for frozen tumors. Steiner et al. demonstrated that amounts of 6 HER2 peptides, including GLQSLPTHDPSPLQR, were significantly correlated with each other, indicating that peptide concentrations can be used as surrogates of HER2 protein amounts (4).
LC-MRM-MS has been implemented in clinical laboratories for protein quantification in biofluids (34), but tissue applications are likely to be more complex due to intra- and inter-biopsy heterogeneity in tissue components (e.g., stroma, blood vessels, tumor cells). The reverse-transcriptase–polymerase-chain-reaction Oncotype DX assay (35) provides a precedent for clinical implementation of a bulk tissue assay, using a combination of 5 reference genes, a minimum tumor cellularity requirement, and macro-dissection to account for heterogeneity amongst BC biopsies.
We explored both histopathological and tissue component biomarkers to normalize bulk tissue MRM-based measurements for tissue heterogeneity. We describe novel immuno-MRM-MS assays for quantifying tumor component and housekeeping biomarkers KRT5, KRT7, FBN1, VCAN, PECAM1, PTPRC, HBB, FABP4, PLIN4, ACTA1 TUBB2, and GAPDH. We found that cellular, adipose, and stroma biomarkers positively correlated with their respective representation in the biopsies (Fig. 3, B), and that normalizing MRM results to the cellular biomarker protein GAPDH resulted in improved concordance between the immuno-MRM-MS and predicate assay tumor classification. Like Steiner et al. (4), we found that cytokeratins were not effective normalization markers, presumably due to variable expression amongst tumors (36).
The robust classification performance of the HER2 immuno-MRM-MS assay in the setting of substantial variability in tumor cellularity (Fig. 3, A) was initially surprising, but may be a reflection of the extremely large range of several orders of magnitude of HER2 protein expression between HER2-positive and -negative tumors (5) (Fig. 4, Supplemental Table 8). It will be interesting to explore the effects of tissue heterogeneity on a larger set of HER2-low tumors and on additional cancer biomarkers whose expression levels are not so wide-ranging, where normalization will likely play a larger role in obtaining accurate clinical classifications.
This study opens the door for studies of clinical validity of the quantitative HER2 immuno-MRM-MS bulk tissue assay for patient selection for novel HER2-targeted therapies, with enough sensitivity to allow for assaying material collected from FFPE blocks as well as core needle biopsies (e.g., a 1 cm-long core needle biopsy taken using an 18-gauge needle would have an estimated wet tissue weight of 5 mg and could generate 50 µg protein at 1% yield (typical yields are 2%–5%), sufficient for immuno-MRM analysis). Prior to clinical implementation, it would be beneficial to incorporate cleavable SIS peptide or isotope-labeled HER2 protein standards to control for trypsin digestion variation, since our QC samples demonstrate some variation in the digestion step (full vs partial process QC; Supplemental Table 9, Supplemental Fig. 9). Since the range of HER2 expression levels will be more compressed amongst HER2-low tumors (vs all BCs), it will be important in clinical validation studies to continue to explore the effects of tissue heterogeneity on the ability of MRM results to predict tumor response to therapy, as it may be necessary to refine the use of histopathology cutoffs (e.g., minimum tumor area) and biomarkers to normalize for tissue heterogeneity in this subset of tumors.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Nonstandard Abbreviations:
- BC
breast cancer
- HER2 or HER2-neu
human epidermal growth factor receptor 2
- IHC
immunohistochemistry
- ISH
in situ hybridization
- FFPE
formalin-fixed paraffin-embedded
- MRM
multiple reaction monitoring
- immuno-MRM-MS
peptide immunoaffinity enrichment coupled with quantitative multiple reaction monitoring-mass spectrometry
- SIS
heavy stable isotope-labeled standard peptides
- CPTAC
Clinical Proteomics Tumor Analysis Consortium
- H&E
hematoxylin and eosin staining
- mAbs
monoclonal antibodies
- LLOQ
lower limit of quantification
- ULOQ
upper limit of quantification
- CV
coefficient of variation
- QC
quality control
- AUC
area under the curve
- mRNA
messenger RNA
Human Genes
ERBB2, erb-b2 receptor tyrosine kinase 2; HER2, human epidermal growth factor receptor 2; ACTA1, actin alpha 1, skeletal muscle; FABP4, fatty acid binding protein 4; FBN1, fibrillin 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HBB, hemoglobin subunit beta; KRT5, keratin 5; KRT7, keratin 7; PECAM1, platelet and endothelial cell adhesion molecule 1; PLIN4, perilipin 4; PTPRC, protein tyrosine phosphatase receptor type C; TUBB, tubulin beta class I; VCAN, versican.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
P. Wang, G.S. Baird, J.J. Kennedy, J.R. Whiteaker, and A.G. Paulovich conceived and planned the experiments. J.J. Kennedy, D.E. Bosch, M.L. Lerch, R.M. Schoenherr, M.R. Kilgore, K.H. Allison, and L. Zhao contributed to identification of samples, assay characterization, and/or sample preparation. J.J. Kennedy, J.R. Whiteaker, C. Lin, and S. Chowdhury contributed to data acquisition and analysis. J.J. Kennedy, J.R. Whiteaker, L.C. Kennedy, D.E. Bosch, M.L. Lerch, M.R. Kilgore, P. Wang, A.N. Hoofnagle, and A.G. Paulovich contributed to interpretation of the results. J.J. Kennedy, A.G. Paulovich, and J.R. Whiteaker wrote the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
S. Chowdhury, Icahn School of Medicine at Mount Sinai; A.N. Hoofnagle, Clinical Chemistry, AACC; G.S. Baird, Avalon Healthcare Solutions; A.G. Paulovich, Precision Assays.
Consultant or Advisory Role
None declared.
Stock Ownership
G.S. Baird, Avalon Healthcare Solutions; A.G. Paulovich, Precision Assays.
Honoraria
None declared.
Research Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers R33CA173300 (to A.G. Paulovich), U24CA160034 (to A.G. Paulovich), U01CA214114 (to A.G. Paulovich), R01CA235575 (to A.G. Paulovich), and R50CA211499 (to J.R. Whiteaker). Histology and Image Analysis was done by Fred Hutchinson Experimental Histopathology Shared Resource.
Expert Testimony
None declared.
Patents
None declared.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
References
- 1. Tarantino P, Hamilton E, Tolaney SM, Cortes J, Morganti S, Ferraro E, et al. HER2-low breast cancer: pathological and clinical landscape. J Clin Oncol 2020;38:1951–62. [DOI] [PubMed] [Google Scholar]
- 2. Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline focused update. Arch Pathol Lab Med 2018;142:1364–82. [DOI] [PubMed] [Google Scholar]
- 3. Modi S, Park H, Murthy RK, Iwata H, Tamura K, Tsurutani J, et al. Antitumor activity and safety of trastuzumab deruxtecan in patients with HER2-Low-Expressing Advanced Breast Cancer: Results From a Phase Ib Study. J Clin Oncol 2020;38:1887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steiner C, Tille JC, Lamerz J, Kux van Geijtenbeek S, McKee TA, Venturi M, et al. Quantification of HER2 by targeted mass spectrometry in formalin-fixed paraffin-embedded (FFPE) breast cancer tissues. Mol Cell Proteomics 2015;14:2786–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hembrough T, Thyparambil S, Liao WL, Darfler MM, Abdo J, Bengali KM, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. J Mol Diagn 2013;15:454–65. [DOI] [PubMed] [Google Scholar]
- 6. Nuciforo P, Thyparambil S, Aura C, Garrido-Castro A, Vilaro M, Peg V, et al. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Mol Oncol 2016;10:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Do M, Kim H, Yeo I, Lee J, Park IA, Ryu HS, et al. Clinical application of multiple reaction monitoring-mass spectrometry to human epidermal growth factor receptor 2 measurements as a potential diagnostic tool for breast cancer therapy. Clin Chem 2020;66:1339–48. [DOI] [PubMed] [Google Scholar]
- 8. Schoenherr RM, Zhao L, Whiteaker JR, Feng LC, Li L, Liu L, et al. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring-mass spectrometry. J Immunol Methods 2010;353:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schoenherr RM, Saul RG, Whiteaker JR, Yan P, Whiteley GR, Paulovich AG.. Anti-peptide monoclonal antibodies generated for immuno-multiple reaction monitoring-mass spectrometry assays have a high probability of supporting Western blot and ELISA. Mol Cell Proteomics 2015;14:382–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoofnagle AN, Whiteaker JR, Carr SA, Kuhn E, Liu T, Massoni SA, et al. Recommendations for the generation, quantification, storage, and handling of peptides used for mass spectrometry-based assays. Clin Chem 2016;62:48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Cancer Institute. Cancer Proteomics Tumor Analysis Consortium Prospective Biospecimen Collection Protocol Breast Cancer v1.9. https://brd.nci.nih.gov/brd/sop/download-pdf/301 (Accessed 2017).
- 12. Krug K, Jaehnig EJ, Satpathy S, Blumenberg L, Karpova A, Anurag M, et al. ; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy [English]. Cell 2020;183:1436–56.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. ; College of American Pathologists. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013. [DOI] [PubMed] [Google Scholar]
- 14. Kennedy JJ, Whiteaker JR, Schoenherr RM, Yan P, Allison K, Shipley M, et al. Optimized protocol for quantitative multiple reaction monitoring-based proteomic analysis of Formalin-Fixed, Paraffin-embedded tissues. J Proteome Res 2016;15:2717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schoenherr RM, Whiteaker JR, Zhao L, Ivey RG, Trute M, Kennedy J, et al. Multiplexed quantification of estrogen receptor and HER2/Neu in tissue and cell lysates by peptide immunoaffinity enrichment mass spectrometry. Proteomics 2012;12:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics 2014;13:907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grant RP, Hoofnagle AN.. From lost in translation to paradise found: enabling protein biomarker method transfer by mass spectrometry. Clin Chem 2014;60:941–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.University of Washington. Panorama: Repository Software for Targeted Mass Spectrometry Assays from Skyline. https://panoramaweb.org/HER2_cellularity_breast_cancer_tissue.url (Accessed 2021).
- 20.National Cancer Institute. Clinical Proteomic Tumor Analysis Consortium Assay Portal. https://proteomics.cancer.gov/assay-portal (Accessed 2020).
- 21.National Center for Biotechnology Information. Genomic Data Commons database of Genotypes and Phenotypes. https://www.ncbi.nlm.nih.gov/gap/?term=phs000892 (Accessed 2020).
- 22. Smits AJ, Kummer JA, de Bruin PC, Bol M, van den Tweel JG, Seldenrijk KA, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014;27:168–74. [DOI] [PubMed] [Google Scholar]
- 23. Avram AS, Avram MM, James WD.. Subcutaneous fat in normal and diseased states: 2. Anatomy and physiology of white and brown adipose tissue. J Am Acad Dermatol 2005;53:671–83. [DOI] [PubMed] [Google Scholar]
- 24. Giricz O, Lauer-Fields JL, Fields GB.. The normalization of gene expression data in melanoma: investigating the use of glyceraldehyde 3-phosphate dehydrogenase and 18S ribosomal RNA as internal reference genes for quantitative real-time PCR. Anal Biochem 2008;380:137–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim JW, Kim SJ, Han SM, Paik SY, Hur SY, Kim YW, et al. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human cervical cancers. Gynecol Oncol 1998;71:266–9. [DOI] [PubMed] [Google Scholar]
- 26. Abbatiello SE, Schilling B, Mani DR, Zimmerman LJ, Hall SC, MacLean B, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics 2015;14:2357–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mittendorf EA, Wu Y, Scaltriti M, Meric-Bernstam F, Hunt KK, Dawood S, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res 2009;15:7381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfitzner BM, Lederer B, Lindner J, Solbach C, Engels K, Rezai M, et al. Clinical relevance and concordance of HER2 status in local and central testing—an analysis of 1581 HER2-positive breast carcinomas over 12 years. Mod Pathol 2018;31:607–15. [DOI] [PubMed] [Google Scholar]
- 29. Jorns JM, Healy P, Zhao L.. Review of estrogen receptor, progesterone receptor, and HER-2/neu immunohistochemistry impacts on treatment for a small subset of breast cancer patients transferring care to another institution. Arch Pathol Lab Med 2013;137:1660–3. [DOI] [PubMed] [Google Scholar]
- 30. Liu ZH, Wang K, Lin DY, Xu J, Chen J, Long XY, et al. Impact of the updated 2018 ASCO/CAP guidelines on HER2 FISH testing in invasive breast cancer: a retrospective study of HER2 fish results of 2233 cases. Breast Cancer Res Treat 2019;175:51–7. [DOI] [PubMed] [Google Scholar]
- 31. Myhre S, Lingjaerde OC, Hennessy BT, Aure MR, Carey MS, Alsner J, et al. Influence of DNA copy number and mRNA levels on the expression of breast cancer related proteins. Mol Oncol 2013;7:704–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsurutani J, Iwata H, Krop I, Jänne PA, Doi T, Takahashi S, et al. Targeting HER2 with trastuzumab deruxtecan: a dose-expansion, phase I study in multiple advanced solid tumors. Cancer Discov 2020;10:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lambein K, Van Bockstal M, Vandemaele L, Geenen S, Rottiers I, Nuyts A, et al. Distinguishing score 0 from score 1+ in HER2 immunohistochemistry-negative breast cancer: clinical and pathobiological relevance. Am J Clin Pathol 2013;140:561–6. [DOI] [PubMed] [Google Scholar]
- 34. Netzel BC, Grant RP, Hoofnagle AN, Rockwood AL, Shuford CM, Grebe SK.. First steps toward harmonization of LC-MS/MS thyroglobulin assays. Clin Chem 2016;62:297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. [DOI] [PubMed] [Google Scholar]
- 36. Vranes V, Vujasinović T, Rajković N, Kanjer K, Milošević NT, Radulovic M. Analysis of Spatial Distribution and Prognostic Value of Different Pan Cytokeratin Immunostaining Intensities in Breast Tumor Tissue Sections. Int J Mol Sci 2020;21:4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual FFPE and frozen sample, and process QC MRM data are available (19). Characterization data for assays are available in the CPTAC Assay Portal (20). Expression level and gene copy number analysis of the frozen BCs has been released at the Genomic Data Commons and is accessible via the database of Genotypes and Phenotypes Study Accession phs000892 (21).