Abstract
Structurally complex natural products have been a fruitful source for the discovery and development of new drugs. In an effort to construct a compound collection populated by architecturally complex members with unique scaffolds, we have used the natural product limonin as a starting point. Limonin is an abundant triterpenoid natural product and, through alteration of its heptacyclic core ring system using short synthetic sequences, a collection of 98 compounds was created, including multiple members with novel ring systems. The reactions leveraged in the construction of these compounds include novel ring cleavage, rearrangements, and cyclizations, and this work is highlighted by the discovery of a novel B-ring cleavage reaction, a unique B/C-ring rearrangement, an atypical D-ring cyclization, among others. Computational analysis shows that 52 different scaffolds/ring systems were produced during the course of this work, of which 36 are unprecedented. Phenotypic screening and structure–activity relationships identified compounds with activity against a panel of cancer cell lines.
Keywords: limonin, scaffold diversity, ring distortion, natural products, anticancer
Graphical Abrstract
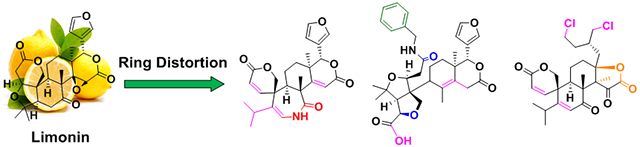
Many compound collections used in high-throughput screening are composed of members whose structural complexity is considerably lower than that of natural products. Here the natural product limonin was leveraged to generate 98 architecturally complex compounds with significant scaffold diversity. Phenotypic screening of the collection led to the discovery of novel anticancer compounds.
Introduction
High-throughput screening (HTS) is a cornerstone of drug discovery, routinely used to identify lead compounds that become first-in-class drugs.[1-2] As such, the type of drugs entering the market in part reflects the composition of screening libraries. Most compound collections are biased by medicinal chemistry conventions, such as Lipinski’s rule of five for oral bioavailability,[3] and thus are largely populated by relatively planar molecules, with functional groups arrayed around a small number of common skeletons.[4-5] The vast majority of such compounds would be considered structurally simple relative to natural products, and as screening these collections has increased and as the identified leads have progressed forward to approval, the resulting drugs tend to also have a low number of stereogenic centers and a low percentage of sp3-hybridized carbons.[6-7] Such collections have been outstanding for the discovery of biologically active compounds for targets with binding sites that favor flat molecules (e.g., kinase inhibitors), but less valuable in providing hits for other types of targets (e.g., transcription factors[8-9] and protein-protein interactions[10-11]) and complex multifactorial disorders (e.g., antibiotic-resistant bacteria).[12-13] Another drawback associated with the use of commercially available libraries in screening experiments is that such widely distributed collections are likely to have been thoroughly scoured for biologically active constituents.[14]
Scaffold diversity is a feature of compound collections that can have an impact on the success of biological screening.[15] In addition to providing compounds of different shapes and flexibilities, the scaffold (defined as the minimum core framework of a given compound)[16] also places peripheral functional groups in various positions of three-dimensional space, facilitating differential interaction with biological targets.[17] As such, compound collections composed of members that are complex and include members with scaffold diversity would be welcome.
Natural products are a source of inspiration in drug discovery with unsurpassed biological activity and scaffold diversity. Over 66% of all small-molecule approved drugs between 1981 and 2019 trace their structural origins back to natural products.[18] Natural products are typically characterized by complex scaffolds with considerable three-dimensionality, a high number of stereogenic centers and a high fraction of sp3–hybridized carbons (Fsp3),[3] and such parameters have been associated with improved target selectivity and successful progression through clinical trials.[4, 19-20] However, given the significant challenges with the isolation and purification of novel natural products, they are usually under-represented in screening collections.
To fill this gap, several creative strategies have been developed to rapidly generate complex compounds, including construction of natural product-inspired scaffolds,[21-25] diversity-oriented synthesis (DOS)[26-29] and biology-oriented synthesis (BIOS).[30-32] Inspired by nature, we have reported a complementary strategy named Complexity-to-Diversity (CtD). The CtD approach takes advantage of the structural complexity inherent within readily available natural products to generate unique complex compounds through ring distortion reactions (i.e., ring expansion, contraction, cleavage, fusion, and rearrangement).[33-34] This method has been applied to several natural products, including adrenosterone,[35] gibberellic acid,[35] quinine,[35] abietic acid,[36-37] sinomenine,[38] lycorine,[39] pleuromutilin,[40-42] yohimbine,[43] haemanthamine,[44] ilimaquinone[45] and steroids such as dutasteride and abiraterone acetate[46]. The resulting collections of natural product-like compounds have been instrumental in developing predictive guidelines for small molecule accumulation in E.coli,[47] and led to G protein-coupled receptor antagonists,[48] a new class of antiplasmodial agents,[49] and the discovery of a novel and potent inducer of ferroptotic cell death.[42]
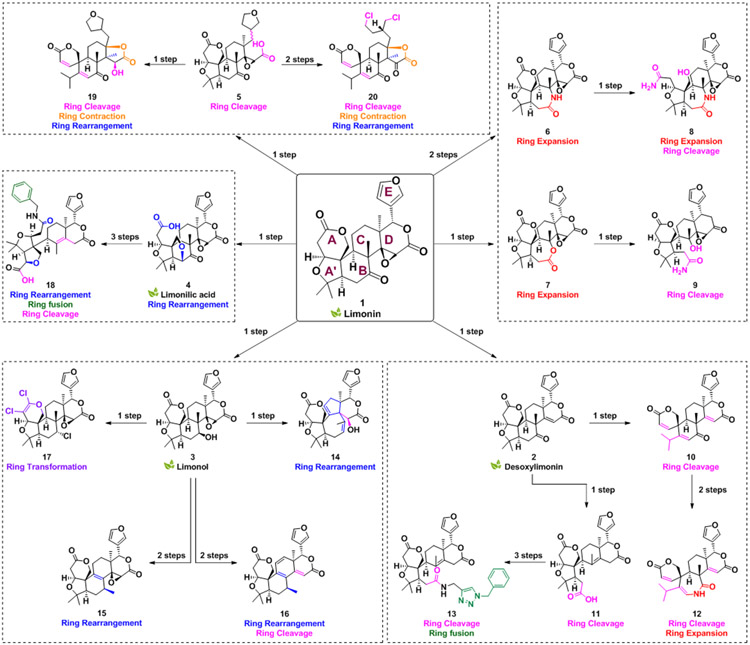
Nature offers a multitude of natural products that are exceptional starting points for the CtD strategy. The limonoids are a valuable source of biologically active natural products.[50] Although more than 300 limonoids have been isolated to date, their occurrence in the plant kingdom is confined mainly to the order Rutales, which includes the families Meliaceae and Rutaceae.[51] Limonin (1, Scheme 1) was the first member of this structural class to be identified, isolated from citrus fruit in 1841.[52] Limonin is a highly oxygenated modified triterpenoid with a dense interlocking ring system, consisting of seven rings (including the epoxide ring) and nine stereogenic centers, and it is characterized by a high Fsp3 (0.73) and an unusually high number of heterocycles (lean ring index, LRI = 2.5, defined as the number of heterocycles divided by the number of carbocycles in a given compound). Limonin, other limonoids, and their metabolites have demonstrated a broad range of biological activities in mammalian systems, including anticancer,[53] antibacterial,[54] anti-inflammatory,[55-56] and antiviral.[57] With the objective of constructing a variety of complex compounds with novel ring systems/scaffolds in a limited number of steps, herein we apply the CtD strategy to limonin (1), generating a compound collection with numerous complex and differentially decorated molecular frameworks; the pathways to key compounds 2-20 are overviewed in Scheme 1. As an initial evaluation, all compounds were tested for their ability to induce death of cancer cells in culture, but numerous other applications of this and analogous compound collections can be envisioned.
Scheme 1.
Overview of the scaffolds generated from the triterpenoid limonin.
Results
Design of compound collection
The limonin heptacyclic skeleton and functional group composition provide outstanding starting points for the development of novel and complex compounds, as overviewed in Scheme 1, and highlighted by the generation of multiple unique core ring systems/scaffolds. In designing synthetic routes to complex compounds from limonin, several known transformations were first leveraged to convert 1 into previously synthesized citrus limonoids 2-4 (Scheme S1) and the ring cleavage analogs 5 and S1 (Scheme S1).[58-61] During the construction of new compounds, several unprecedented reactions were discovered to alter the overall ring systems, including cleavage reactions of the B-ring, rearrangement of the B/C-ring system, and opening and novel cyclization of the D-ring. The ketone on the B-ring of 1 provides a suitable handle to generate scaffold diversity through ring expansion (6-7) and subsequent ring cleavage reactions (8-9). A’- and B-ring cleaved versions of 2 (10-11) can be further exploited to achieve both skeletal and functional group diversity (12-13). The B-ring-alcohol of limonol (3) and the carboxylic acid of limonolic acid (4) were leveraged to generate unknown ring systems via ring rearrangement (14-16), transformation (17), and cleavage (18). Finally, ring manipulations via D-ring contraction (19-20) were devised using 5 as a starting point.
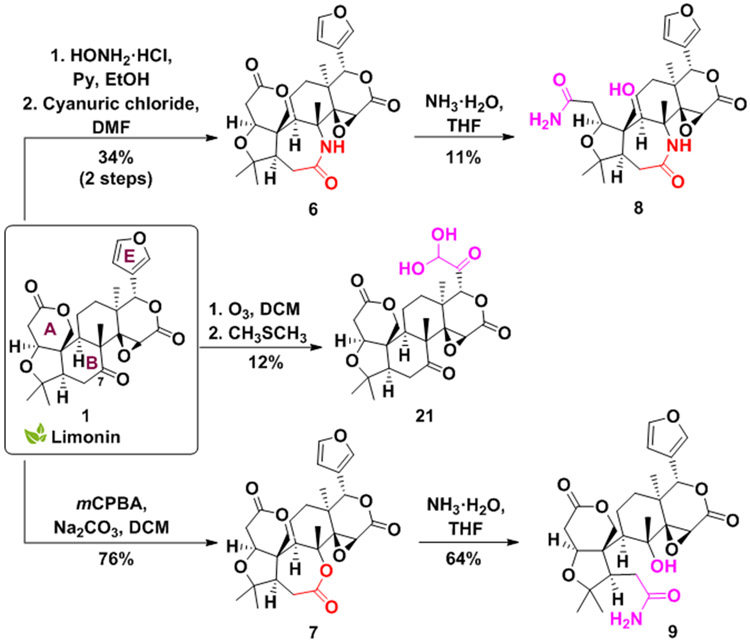
Diversifying limonin through B-ring expansion and E-ring opening
The B-ring of limonin presents the opportunity for the construction of new compounds with unexplored ring systems through ring expansions (Scheme 2). Seven-membered rings are medicinally important molecular frameworks commonly found in diverse bioactive natural products, but they are under-represented in compound screening collections and marketed drugs due to their challenging syntheses.[62] Attempts to generate the seven-member ring derivative 6 via a Schmidt reaction resulted in compound degradation. However, the known E-oxime intermediate (S2, Schemes S1 and S2) was synthesized[58] and utilized as the substrate for a Beckmann rearrangement using cyanuric chloride, yielding lactam 6 as a single constitutional isomer. Under these conditions, only the selective migration of the most substituted anti-group was observed. Baeyer-Villiger oxidation of 1 resulted in the analogous ring expanded lactone 7.
Scheme 2.
Limonin B-ring expansion, E-ring opening, and further manipulations of the A and B rings.
Further modifications of 6 and 7 were realized by subjecting the two compounds to limonin A-ring cleavage conditions (Scheme 2).[61] The reaction carried out on lactam 6 follows the classical limonin reactivity,[61] with the A-ring opening to derivative 8. However, the same conditions applied to 7 lead to selective cleavage of the newly expanded B-ring (9).
Many naturally occurring citrus limonoids contain a furan ring attached to the D-ring, designated as the E-ring. To generate a compound with alterations in the E-ring, 1 was subjected to ozone-mediated oxidative cleavage. While furans are known to generate carboxylic acid derivative via ozonolysis,[63] oxidation of the furan ring of 1 with ozone gives rise to novel dihydroxy ketone 21 (Scheme 2). The synthesis of this highly oxygenated product likely occurs by the formation of an α-ketoacid intermediate that converts to the dihydroxy analog 21.
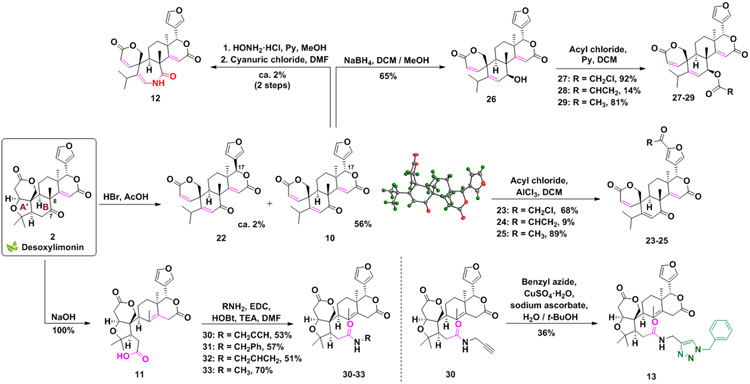
Diversifying desoxylimonin
Further skeleton manipulations of the A’ and B rings starting from desoxylimonin (2) are shown in Scheme 3. The synthesis of 10 was reported by Geissman and Tulagin in 1946, by refluxing 2 with red phosphorus.[64] To avoid the use of this toxic reagent, a more practical protocol utilizing hydrobromic acid was developed and optimized (see Scheme S3 for proposed mechanism). Under these conditions 10 and a small quantity of its C17-epimer (22) were isolated. The spirocyclic compound 10, whose structure was confirmed by X-ray crystallography, was further altered using AlCl3-catalyzed electrophilic aromatic substitution (using conditions previously developed on limonin)[65] to obtain a small series of novel acylated derivatives (23-25).
Scheme 3.
Synthesis and functionalization of the ring-opened derivatives 10 and 11.
Based on the success of Beckmann rearrangement carried out on 1 (in Scheme 2), a B-ring expansion of the spirocycle 10 was envisioned. Thus, 10 was converted to the corresponding oxime (S7, Scheme S4) and subsequently treated with cyanuric chloride generating lactam 12 with a regioselective NH incorporation between the carbonyl and the olefin. Although both S7 formation and Beckman rearrangement occur with low yields (25 and 8% respectively), these transformations enabled the generation of a novel molecular skeleton. The different migratory aptitudes observed in Beckmann rearrangement products of 6 and 12 rely on the different configurations assumed by the hydroxyl group of the two corresponding oximes (Z and E respectively). Sodium borohydride reduction of the B-ring ketone of 10 yielded β-alcohol 26 as a single diastereoisomer, further converted into the acylated derivatives 27-29 by reacting with the suitable acyl chloride in dichloromethane and pyridine (Scheme 3).
Treatment of 2 with sodium hydroxide at high temperature gave an unusual B-ring cleavage reaction, forming carboxylic acid 11 in quantitative yield. Compound 11 is a known limonin metabolite and natural product, first isolated from Pseudomonas in 1974 [66], but to our knowledge, it has never been synthesized before this work. The course of the reaction, which requires the presence of a δ-ketone α,β-unsaturated lactone, can be envisioned by hydroxyl anion attack on the ketone carbonyl and subsequent ring cleavage of the C7-C8 bond and olefin migrations (refer to Scheme S5 for proposed mechanism). Compound 11 afforded the opportunity to generate a small set of B-ring cleaved analogs via amide coupling reactions (30-33), and a subsequent copper-catalyzed azide-alkyne reaction to form triazole 13 (Schemes 3 and S6).
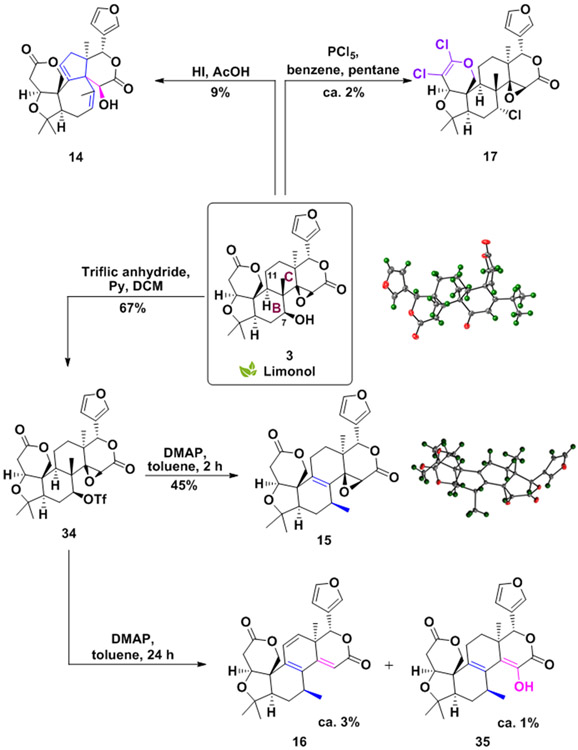
Novel ring rearrangements of limonol
Carbocation rearrangements are common reactions employed by both nature and synthetic chemists to generate complex molecules with new scaffolds.[67-68] Thus, we envisioned using the C7-alcohol of limonol (3) to trigger a skeleton rearrangement through carbocations, hydride shift, and alkyl shift, as shown in Schemes 4 and S7. Indeed, treatment of 3 with hydroiodic acid led to an unexpected ring system, compound 14. This ring rearrangement likely occurs through the displacement of C7-hydroxyl by iodine, which after C11-hydride abstraction initiates a rearrangement cascade culminating in the formation of compound 14 (refer to Scheme S8 for proposed mechanism).
Scheme 4.
Rearrangement of the B and C rings of limonol (3).
The fused ring systems within complex terpenoids are poised for manipulation through Wagner-Meerwein rearrangement, yielding complex and structurally divergent scaffolds.[69-70] Therefore, we sought to apply classical Wagner-Meerwein skeletal rearrangement to 3. Treatment of 3 with triflic anhydride provides activated alcohol 34, which can undergo a 1,2-shift to generate 15 as a single diastereoisomer (Scheme 4). After the C7 carbonium ion formation, the 1,2-sigmatropic shift of the methyl group could readily occur suprafacially with retention of configuration. The absolute configuration of 15 was confirmed by X-ray crystallography (Scheme 4).
By extending the reaction time, structural elaboration occurs via epoxide ring-opening and elimination to form 16 and 35 in low yields. Interestingly, in situ activations of the hydroxy function of 3, using phosphorus pentachloride, disfavors the Wagner-Meerwein rearrangement, and triggers a new unexpected ring A transformation (17, Scheme 4). After the displacement of the secondary alcohol by a chlorine ion, A-ring lactone conversion to a 5,6-dichloro-3,4-dihydro-2H-pyran ring occurs.
This unprecedented reaction allowed the generation of an unusual 5,6-dichloro-3,4-dihydro-2H-pyran moiety in one step. The formation of 17 likely occurs by phosphorus pentachloride-mediated A-lactone activation, followed by chloride ion addition to the ipso carbon of the lactone carbonyl. The formed intermediate can then further undergo an elimination reaction and a subsequent second chloride ion incorporation at the alpha carbon (see Scheme S9 for the proposed mechanism).
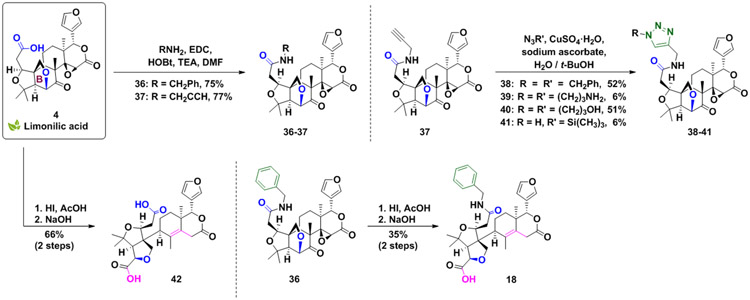
Limonilic acid-based ring distortion strategy
Limonilic acid (4) served as a key substrate to construct a subset of compounds with the rearranged A and B ring systems (Scheme 5). Coupling of the carboxylic acid of 4 with suitable amines provided amide derivatives (36-37). Compound 37 was modified using the azide-alkyne cycloaddition to build a small set of ring fusion analogs (38-41).
Scheme 5.
Manipulation of the ring-rearranged derivative 4.
The versatility of the novel ring B cleavage reaction discovered in the construction of 11 (in Scheme 3) was further demonstrated through the application of the developed conditions to the ring systems of 4 and 36 (Scheme 5). The suitable δ-ketone α,β-unsaturated lactone starting points S22 and S20 were generated from 4 and 36 via epoxide treatment with hydroiodic acid (Schemes S10 and S11). Compounds S22 and S20 were then submitted to sodium hydroxide ring cleavage yielding the new scaffolds 42 and 18.
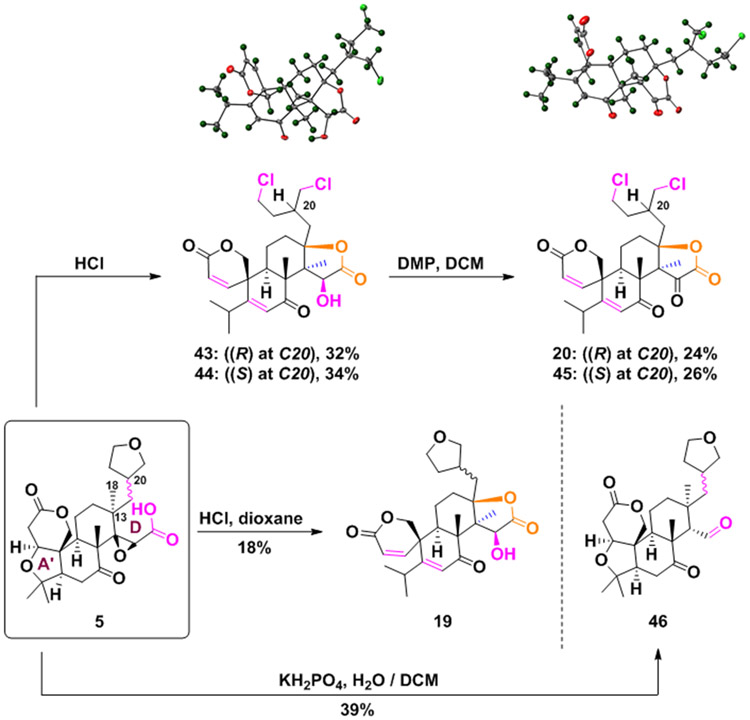
Ring D-distortion strategy
Finally, compound 5 also served as a starting point to build new scaffolds, as shown in Schemes 6 and S12. Key to this diversification strategy was the use of brønsted acids, which have had a broad utility in natural product chemistry.[71-73] Treatment of 5 with hydrochloric acid in dioxane at high temperature leads to two simultaneous transformations: the cleavage of the A’ ring (in a similar manner observed for compound 10 in Scheme 3) and an atypical D ring cyclization, forming compound 19 (Scheme 6). This heretofore-unknown ring cyclization occurs in a stereoselective fashion. The mechanistic underpinnings of this transformation are envisioned to proceed by epoxide opening to form the tertiary carbocation, which triggers the carboxylic acid attack to C13 and subsequent suprafacial methyl 18 migration (refer to Scheme S13 for proposed mechanism). Notably, increasing the acid concentration generates the dichlorinated derivatives 43 and 44 via tetrahydrofuran ring-opening, as previously observed by treating ethers with strong acids (see Scheme S14 for proposed mechanism).[74] The stereoselective nature of the novel D-ring cyclization was further confirmed by X-ray crystallography of the C20 (S)-diastereoisomer 43 (Scheme 6). Oxidation of 43 and 44 with Dess-Martin periodinane (DMP) generated the two diastereomeric ketones 20 (also confirmed by X-ray) and 45. Lastly, intermediate 5 was decarboxylated to generate aldehyde 46.
Scheme 6.
Manipulation of the D-ring cleaved derivative 5.
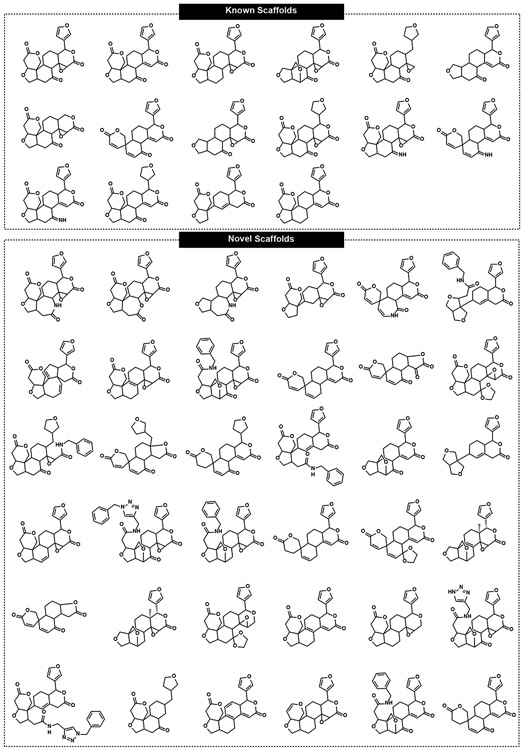
Computational analysis of limonin-derived complex molecules
Herein, the limonin scaffold was utilized to construct a collection of 98 complex compounds (compounds 2-47 and S1-S52 in Figures S1 and S2); 15 of these were previously known in the literature, while 83 are new compounds. Besides the 46 compounds described here, the synthetic route to the other 52 compounds are available in the Supporting Information (Schemes S15-S19). All compounds were isolated at >95% purity, and >20 mg quantities were produced for most compounds (including many of those produced through low-yielding routes, such as 14 and 17, Table S1). All the new CtD compounds accessed from limonin were submitted to chemoinformatic complexity analyses and compared to the physiochemical properties of existing screening libraries, natural products, and approved drugs (Figures S3-S16). The set of 99 compounds (including limonin) shows an average of Fsp3 = 0.66, average number of stereogenic centers = 7.77, ring fusion density = 0.27, and ring complexity index = 1.30, values markedly higher compared to those of standard screening collections (see Figures S3-S16).
Scaffold diversity was also assessed for these compounds generated from limonin. Several computational methods have been developed to analyze scaffold diversity, including the derivation of Murcko scaffold (defined as the union of ring systems and linkers in a given compound)[75] and the scaffold tree methodology (which describes the ring systems arranged in a hierarchical tree by removing rings one by one according to a set of prioritization rules).[76] By removing all the appendages from the set of 99 compounds, 52 unique Murcko scaffolds were identified (Figure 1 and Table S2). Among these, 16 are known (present in existing natural products or synthesized derivatives) while 36 are novel. Based on the Murcko fragments, structural features and molecular diversity was evaluated by the generation of a library scaffold tree (Figures S17-S27). In addition, a Tanimoto coefficient matrix (a measure of the similarity of different fingerprints) was also generated for all compounds (Figure S28), further highlighting the balance between scaffold diversity and compound density of the synthesized limonin collection.
Figure 1.
Scaffold collection derived from limonin (1).
Assessment of limonin CtD collection against cancer cells in culture
Molecular scaffolds play a pivotal role in the navigation of biologically relevant chemical space, thus we hypothesized that the variety of molecular frameworks generated herein from limonin could enhance the likelihood of identifying new bioactive agents relative to traditional screening collections.
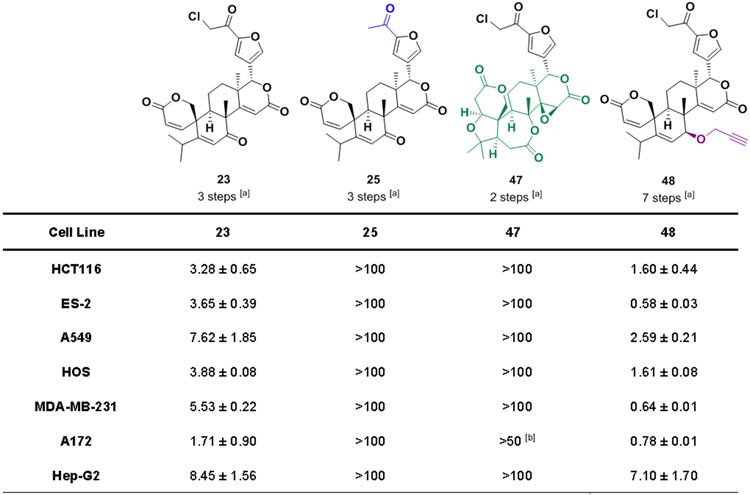
Phenotypic screening is a powerful strategy for the identification of bioactive molecules capable of modulating complex diseases.[77] The wide utility of this screening approach relies on its ability to identify new bioactive compounds before full elucidation of targets or mechanisms of action. With the objective of identifying new anticancer compounds and new chemical scaffolds with anticancer activity, all the synthesized compounds were assessed for their ability to kill cancer cell lines in culture, with initial screening using the ES-2 (ovarian carcinoma) and HCT116 (colorectal carcinoma) cell lines (Figure S29).
Cell viability was assessed after 72 hours treatment with compounds at 20 μM. From these assessments, compound 23 emerged as a promising anticancer agent, inducing over 85% cell death (at 20 μM) against both cancer cell lines, and was thus further evaluated against a panel of cancer cell lines with a full concentration range such that dose-response curves could be generated and half-maximal inhibitory concentrations (IC50s) values calculated. As shown in Figure 2, compound 23 has broad activity across cell lines from different cancer types with an IC50 range of 1.7–8.4 μM and near 100% Emax (maximum achievable effect,[78] see dose-response curves in Supporting Information Figure S30). One feature of 23 is the presence of an electrophilic group on the furan. α-chlorocarbonyl electrophiles are reactive with nucleophilic substrates such as cysteine thiols but can exhibit surprisingly low off-target reactivity and minimal activity towards glutathione, and thus can be suitable warheads for covalent drugs.[79] The α-chloroketone of 23 was systematically modified, revealing its importance to anticancer activity; for example, acylated compound 25 is inactive, and other electrophiles at this position also result in a significant drop in anticancer activity (Figure S31). The presence of the electrophilic warhead is not sufficient for anticancer activity, however, as compounds carrying the α-chlorocarbonyl electrophiles on the same position of the furan ring, but with different chemical skeletons, did not show any activity (47 and S39). Structure-activity relationship (SAR) studies also revealed that inversion of configuration at the furan (S57) results in loss of potency and that other functional groups (B-ring ketone and A-ring unsaturation, compounds S59, S67, S69 and S60, S64) could be removed without diminishment of activity (for full SAR refer to Supporting Information Figures S31-S35, and Schemes S20 and S21). Importantly, propargyl compound 48 retains excellent activity against cancer cells in culture, even more potent than 23 (Figures 2 and S36), setting the stage for future target identification efforts. Additional data suggest that compounds 23 and 48 induce death in cancer cells via apoptosis (Figures S37 and S38), and these compounds do not induce lysis of red blood cells (Figure S39).
Figure 2.
Biological evaluation of key compounds against cancer cells in culture. Bioactivity is expressed as a 72-hour IC50 value (in μM) against a panel of cell lines, as measured by Alamar Blue assay. Data represent mean ± SEM, Raptinal (50 μM) was used as dead control, n = 3 biological replicates; [a] number of steps from limonin, [b] not soluble at 100 μM.
Discussion and Conclusion
For high-throughput screens in drug discovery, structural diversity of the compound collection can be critical for success.[14, 80] Diverse scaffolds can place functionality in different positions of three-dimensional space, thus allowing for interaction with diverse biological targets. As such, the generation of multiple core ring systems/scaffolds is an efficient method for increasing the overall structural diversity of a compound collection.
Ring distortion of the heptacyclic scaffold of limonin allowed not only the generation of a variety of natural product-like scaffolds (119 compounds, considering 98 CtD compounds and 21 SAR analogs) and the discovery of novel reactions (cleavage of δ-ketone α,β-unsaturated lactone functional groups, unique B/C-rings rearrangement triggered by hydroiodic acid, unprecedented D-ring cyclization) but also afforded the opportunity for facile synthesis of a natural product. Indeed, nature takes advantage of divergent pathways from a common intermediate to generate multiple different natural products. This “natural products from natural products” result can also be a fortuitous consequence of applying new transformations to natural products, seen herein in the construction of compound 11 from limonin, and observed previously with sinomenine, converted into the natural product sinoracutine in only five steps[38] (compared to the twelve steps required for its total synthesis).[81] We anticipate that the compounds disclosed herein, and others generated from different natural products, will find utility in a wide variety of biological investigations.
Supplementary Material
Acknowledgements
We are grateful to the University of Illinois and the NIH (R01GM118575) for funding this work. We thank T. Woods and D. Gray (UIUC) for collection and analysis of X-ray data and F. Nocera (UIUC) for assistance in chemoinformatic analysis.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Data availability
The X-ray crystallography data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) using the following identifiers (www.ccdc.cam.ac.uk/structures/): 2082412 (compound 3), 2082413 (compound 10), 2082419 (compound 15), 2082414 (compound 20), 2082415 (compound 26), 2082416 (compound 44), 2082417 (compound S36) and 2082418 (compound S42).
References
- [1].Brown DG, Bostrom J, J. Med. Chem 2018, 61, 9442–9468. [DOI] [PubMed] [Google Scholar]
- [2].Macarron R, Banks M, Bojanic D, Burns D, Cirovic D, Garyantes T, Green D, Hertzberg R, Janzen W, Paslay J, Schopfer U, Sittampalam G, Nat. Rev. Drug Discov 2011, 10, 188–195. [DOI] [PubMed] [Google Scholar]
- [3].Stratton CF, Newman DJ, Tan DS, Bioorg. Med. Chem. Lett 2015, 25, 4802–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lovering F, Bikker J, Humblet C, J. Med. Chem 2009, 52, 6752–6756. [DOI] [PubMed] [Google Scholar]
- [5].Tajabadi FM, Campitelli MR, Quinn RJ, Springer Sci. Rev 2013, 1, 141–151. [Google Scholar]
- [6].Shultz MD, J. Med. Chem 2019, 62, 1701–1714. [DOI] [PubMed] [Google Scholar]
- [7].Stratton CF, Newman DJ, Tan DS, Bioorg. Med. Chem. Lett 2015, 25, 4802–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grivas PD, Kiaris H, Papavassiliou AG, Trends Mol. Med 2011, 17, 537–538. [DOI] [PubMed] [Google Scholar]
- [9].Grivas PD, Papavassiliou AG, Cancer 2013, 119, 1120–1128. [DOI] [PubMed] [Google Scholar]
- [10].Domling A, Curr. Opin. Chem. Biol 2008, 12, 281–291. [DOI] [PubMed] [Google Scholar]
- [11].Villoutreix BO, Kuenemann MA, Poyet JL, Bruzzoni-Giovanelli H, Labbe C, Lagorce D, Sperandio O, Miteva MA, Mol. Inform 2014, 33, 414–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL, Nat. Rev. Drug Discov 2007, 6, 29–40. [DOI] [PubMed] [Google Scholar]
- [13].Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP, Nat. Rev. Drug Discov 2017, 16, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Galloway WRJD, Spring DR, Exp. Opin. Drug Discov 2009, 4, 467–472. [DOI] [PubMed] [Google Scholar]
- [15].Shelat AA, Guy RK, Nat. Chem. Biol 2007, 3, 442–446. [DOI] [PubMed] [Google Scholar]
- [16].Nathan B, Edgar J, Mini Rev. Med. Chem 2006, 6, 1217–1229. [DOI] [PubMed] [Google Scholar]
- [17].Garcia-Castro M, Zimmermann S, Sankar MG, Kumar K, Angew. Chem. Int. Ed 2016, 55, 7586–7605. [DOI] [PubMed] [Google Scholar]
- [18].Newman DJ, Cragg GM, J. Nat. Prod 2020, 83, 770–803. [DOI] [PubMed] [Google Scholar]
- [19].Clemons PA, Bodycombe NE, Carrinski HA, Wilson JA, Shamji AF, Wagner BK, Koehler AN, Schreiber SL, PNAS 2010, 107, 18787–18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Talele TT, J. Med. Chem 2020, 63, 13291–13315. [DOI] [PubMed] [Google Scholar]
- [21].McLeod MC, Singh G, Plampin JN 3rd, Rane D, Wang JL, Day VW, Aubé J, Nat. Chem 2014, 6, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grenning AJ, Snyder JK, Porco JA, Org. Lett 2014, 16, 792–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Weber M, Owens K, Masarwa A, Sarpong R, Org. Lett 2015, 17, 5432–5435. [DOI] [PubMed] [Google Scholar]
- [24].McLeod MC, Aubé J, Tetrahedron 2016, 72, 3766–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Firth JD, Craven PG, Lilburn M, Pahl A, Marsden SP, Nelson A, Chem Commun (Camb) 2016, 52, 9837–9840. [DOI] [PubMed] [Google Scholar]
- [26].Choi Y, Kim H, Park SB, Chem. Sci 2019, 10, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kato N, Comer E, Sakata-Kato T, Sharma A, Sharma M, Maetani M, Bastien J, Brancucci NM, Bittker JA, Corey V, Clarke D, Derbyshire ER, Dornan GL, Duffy S, Eckley S, Itoe MA, Koolen KM, Lewis TA, Lui PS, Lukens AK, Lund E, March S, Meibalan E, Meier BC, McPhail JA, Mitasev B, Moss EL, Sayes M, Van Gessel Y, Wawer MJ, Yoshinaga T, Zeeman AM, Avery VM, Bhatia SN, Burke JE, Catteruccia F, Clardy JC, Clemons PA, Dechering KJ, Duvall JR, Foley MA, Gusovsky F, Kocken CH, Marti M, Morningstar ML, Munoz B, Neafsey DE, Sharma A, Winzeler EA, Wirth DF, Scherer CA, Schreiber SL, Nature 2016, 538, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim J, Jung J, Koo J, Cho W, Lee WS, Kim C, Park W, Park SB, Nat. Commun 2016, 7, 13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Galloway WRJD, Isidro-Llobet A, Spring DR, Nat. Commun 2010, 1, 80. [DOI] [PubMed] [Google Scholar]
- [30].Laraia L, Ohsawa K, Konstantinidis G, Robke L, Wu Y-W, Kumar K, Waldmann H, Angew. Chem. Int. Ed 2017, 56, 2145–2150. [DOI] [PubMed] [Google Scholar]
- [31].Švenda J, Sheremet M, Kremer L, Maier L, Bauer JO, Strohmann C, Ziegler S, Kumar K, Waldmann H, Angew. Chem. Int. Ed 2015, 54, 5596–5602. [DOI] [PubMed] [Google Scholar]
- [32].Liu J, Cremosnik GS, Otte F, Pahl A, Sievers S, Strohmann C, Waldmann H, Angew. Chem. Int. Ed 2021, 60, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morrison KC, Hergenrother PJ, Nat. Prod. Rep 2014, 31, 6–14. [DOI] [PubMed] [Google Scholar]
- [34].Motika SE, Hergenrother PJ, Nat. Prod. Rep 2020, 37, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huigens RW 3rd, Morrison KC, Hicklin RW, Flood TA Jr., Richter MF, Hergenrother PJ, Nat. Chem 2013, 5, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rafferty RJ, Hicklin RW, Maloof KA, Hergenrother PJ, Angew. Chem. Int. Ed 2014, 53, 220–224. [DOI] [PubMed] [Google Scholar]
- [37].Rafferty RJ, Hicklin RW, Maloof KA, Hergenrother PJ, Angew. Chem 2014, 126, 224–228. [DOI] [PubMed] [Google Scholar]
- [38].Garcia A, Drown BS, Hergenrother PJ, Org. Lett 2016, 18, 4852–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tasker SZ, Cowfer AE, Hergenrother PJ, Org. Lett 2018, 20, 5894–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hicklin RW, Lopez Silva TL, Hergenrother PJ, Angew. Chem. Int. Ed 2014, 53, 9880–9883. [DOI] [PubMed] [Google Scholar]
- [41].Hicklin RW, López Silva TL, Hergenrother PJ, Angew. Chem 2014, 126, 10038–10041. [DOI] [PubMed] [Google Scholar]
- [42].Llabani E, Hicklin RW, Lee HY, Motika SE, Crawford LA, Weerapana E, Hergenrother PJ, Nat. Chem 2019, 11, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Paciaroni NG, Ratnayake R, Matthews JH, Norwood V. M. t., Arnold AC, Dang LH, Luesch H, Huigens RW 3rd, Chemistry 2017, 23, 4327–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Govindaraju K, Masi M, Colin M, Mathieu V, Evidente A, Hudnall TW, Kornienko A, Molecules 2018, 23, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Evanno L, Lachkar D, Lamali A, Boufridi A, Séon-Méniel B, Tintillier F, Saulnier D, Denis S, Genta-Jouve G, Jullian J-C, Leblanc K, Beniddir MA, Petek S, Debitus C, Poupon E, Eur. J. Org. Chem 2018, 2486–2497. [Google Scholar]
- [46].Charaschanya M, Aube J, Nat. Commun 2018, 9, 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, Hergenrother PJ, Nature 2017, 545, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Paciaroni NG, Norwood VM, Ratnayake R, Luesch H, Huigens RW, Bioorg. Med. Chem 2020, 28, 115546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Paciaroni NG, Perry DL, Norwood VM, Murillo-Solano C, Collins J, Tenneti S, Chakrabarti D, Huigens RW, ACS Infect. Dis 2020, 6, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Y, Xu H, RSC Adv. 2017, 7, 35191–35220. [Google Scholar]
- [51].Roy A, Saraf S, Biol. Pharm. Bull 2006, 29, 191–201. [DOI] [PubMed] [Google Scholar]
- [52].Bernay S, Annalen 1841, 40, 317–319. [Google Scholar]
- [53].Spradlin JN, Hu X, Ward CC, Brittain SM, Jones MD, Ou L, To M, Proudfoot A, Ornelas E, Woldegiorgis M, Olzmann JA, Bussiere DE, Thomas JR, Tallarico JA, McKenna JM, Schirle M, Maimone TJ, Nomura DK, Nat. Chem. Bio 2019, 15, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vikram A, Jayaprakasha GK, Jesudhasan PR, Pillai SD, Patil BS, Food Control 2012, 26, 427–438. [Google Scholar]
- [55].Mahmoud MF, Gamal S, El-Fayoumi HM, Eur. J. Pharmacol 2014, 740, 676–682. [DOI] [PubMed] [Google Scholar]
- [56].Wang S, Han X, Yang Y, Zhou C, Luo D, He W, Zhu Q, Xu Y, Bioorg. Chem 2020, 100, 103886. [DOI] [PubMed] [Google Scholar]
- [57].Balestrieri E, Pizzimenti F, Ferlazzo A, Giofre SV, Iannazzo D, Piperno A, Romeo R, Chiacchio MA, Mastino A, Macchi B, Bioorg. Med. Chem 2011, 19, 2084–2089. [DOI] [PubMed] [Google Scholar]
- [58].Xu Y, Yang Y, Zhu Q, Wang X, Gong G, Yang L, China Pharmaceutical University, Peop. Rep. China 2014, p. 16pp. [Google Scholar]
- [59].Ruberto G, Renda A, Tringali C, Napoli EM, Simmonds MSJ, Agric J. Food Chem. 2002, 50, 6766–6774. [DOI] [PubMed] [Google Scholar]
- [60].Emerson OH, J. Am. Chem 1952, 74, 688–693. [Google Scholar]
- [61].Xu Y, Yang Y, Zhu Q, Wang S, Han X, Jia C, China Pharmaceutical University, Peop. Rep China: . 2018, p. 22pp. [Google Scholar]
- [62].Clarke AK, Unsworth WP, Chem. Sci 2020, 11, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Roydhouse M, Motherwell W, Constantinou A, Gavriilidis A, Wheeler R, Down K, Campbell I, RSC Adv. 2013, 3, 5076–5082. [Google Scholar]
- [64].Geissman TA, Tulagin V, J. Org. Chem 1946, 11, 760–770. [DOI] [PubMed] [Google Scholar]
- [65].Xu Y, He G, Jia C, Zhu Q, Chu Z, Ji Y, Gong G, China Pharmaceutical University, Peop. Rep China; Hefei Industrial Pharmaceutical Co., Ltd. . 2017, p. 26pp. [Google Scholar]
- [66].Hasegawa S, Maier VP, King AD, J. Agric. Food Chem 1974, 22, 523–526. [DOI] [PubMed] [Google Scholar]
- [67].O'Connor SE, Maresh JJ, Nat. Prod. Rep 2006, 23, 532–547. [DOI] [PubMed] [Google Scholar]
- [68].Boyce JH, Reisman BJ, Bachmann BO, Porco JA Jr., Angew. Chem. Int. Ed 2021, 60, 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Salvador J, Pinto R, Santos R, Le Roux C, Beja A, Paixão J, Org. Biomol. Chem 2009, 7, 508–517. [DOI] [PubMed] [Google Scholar]
- [70].Tkachenko IM, Mankova PA, Rybakov VB, Golovin EV, Klimochkin YN, Org. Biomol. Chem 2020, 18, 465–478. [DOI] [PubMed] [Google Scholar]
- [71].Boyce JH, Reisman BJ, Bachmann BO, Porco JA Jr., Angew. Chem. Int. Ed 2021, 60, 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gerard B, Sangji S, O'Leary DJ, Porco JA, J. Am. Chem 2006, 128, 7754–7755. [DOI] [PubMed] [Google Scholar]
- [73].Merad J, Lalli C, Bernadat G, Maury J, Masson G, Chem. – Eur. J 2018, 24, 3925–3943. [DOI] [PubMed] [Google Scholar]
- [74].Guo Q, Miyaji T, Hara R, Shen B, Takahashi T, Tetrahedron 2002, 58, 7327–7334. [Google Scholar]
- [75].Bemis GW, Murcko MA, J. Med. Chem 1996, 39, 2887–2893. [DOI] [PubMed] [Google Scholar]
- [76].Schuffenhauer A, Ertl P, Roggo S, Wetzel S, Koch MA, Waldmann H, J. Chem. Inf. Model 2007, 47, 47–58. [DOI] [PubMed] [Google Scholar]
- [77].Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M, Nat. Rev. Drug Discov 2017, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- [78].Holford N, Transl Clin Pharmacol 2017, 25, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jöst C, Nitsche C, Scholz T, Roux L, Klein CD, J. Med. Chem 2014, 57, 7590–7599. [DOI] [PubMed] [Google Scholar]
- [80].Follmann M, Briem H, Steinmeyer A, Hillisch A, Schmitt MH, Haning H, Meier H, Drug Discov. Today 2019, 24, 668–672. [DOI] [PubMed] [Google Scholar]
- [81].Volpin G, Vepřek NA, Bellan AB, Trauner D, Angew. Chem. Int. Ed 2017, 56, 897–901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray crystallography data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) using the following identifiers (www.ccdc.cam.ac.uk/structures/): 2082412 (compound 3), 2082413 (compound 10), 2082419 (compound 15), 2082414 (compound 20), 2082415 (compound 26), 2082416 (compound 44), 2082417 (compound S36) and 2082418 (compound S42).