Abstract
Human endogenous retroviruses (HERV) have been implicated in a variety of diseases including cancers. Recent research implicates HERVs in epigenetic gene regulation. Here we utilize a recently developed bioinformatics tool for identifying HERV expression at the locus-specific level to identify differential expression of HERVs in matched tumor-normal RNA-seq data from The Cancer Genome Atlas (TCGA). Data from 52 prostate cancer, 111 breast cancer, and 24 colon cancer cases were analyzed. Locus-specific analysis identified active HERV elements and differentially expressed HERVs in prostate cancer, breast cancer, and colon cancer. Additionally, differentially expressed host genes were identified across prostate, breast, and colon cancer datasets, respectively, including several involved in demethylation and antiviral response pathways, supporting previous findings regarding the pathogenic mechanisms of HERVs. A majority of differentially expressed HERVs intersected protein coding genes or lncRNAs in each dataset, and a subset of differentially expressed HERVs intersected differentially expressed genes in prostate, breast, and colon cancers, providing evidence towards regulatory function. Lastly, patterns in HERV expression were identified in multiple cancer types, with 155 HERVs differentially expressed in all three cancer types. This analysis extends previous results identifying HERV transcription in cancer RNA-seq datasets to a locus-specific level, and in doing so provides a foundation for future studies investigating the functional role of HERV in cancers and identifies a number of novel targets for cancer biomarkers and immunotherapy.
Keywords: endogenous retroviruses, breast cancer, colon cancer, prostate cancer, transcriptomics
Introduction
Prostate, breast, and colon cancers together account for over 500,000 new cancer cases in the United States each year (1). Recently, human endogenous retroviruses (HERV) – a class of transposable elements resulting from the integration of ancient exogenous retroviruses into the human genome (2) – have been implicated in cancer mechanisms. HERVs comprise approximately 8-9% of the human genome and have been incorporated into primate genomes via retrotransposition for at least 40 million years (2). Sequence degradation over time has rendered the majority of protein-coding HERV sequences inactive, however HERV transcription has been identified in all commonly studied human tissue types (3). While HERV nomenclature varies in the literature, HERVs separate phylogenetically into over 40 distinct groups, the most commonly studied being the HERV-K group and its subgroup HML-2.
Previous in vitro work in tumor cell lines and tissue samples has identified evidence of HERV transcription in prostate, breast, and colon cancers. Individual HERV-K elements including HERV-K102 and HERV-K118, as well as solo LTRs, are transcribed in prostate cancer cell lines (4). Expression of an antigen related to the HERV-K gag gene has also been found in prostate tumors (5). Additionally, HERVK expression has been found in other urologic cancers (6,7), and the HERV-K(HML-2) protein Rec has been suggested to be oncogenic in these cancers by de-repressing the androgen receptor (8). In breast cancers, HERV transcription has been found in non-malignant breast tissue, as well as lower overall HERV transcription in tumor tissues, with the latter being heterogeneous across patients (9). Further studies have identified evidence of HERV-K transcripts including HERV-K102 and HERV-K(HML-2) loci 1q22 and 3q21.2 (10-12) in breast cancers, as well as evidence of HERV-K env transcription (13,14). In colorectal cancers, several HERV-H elements are expressed in both tumor and normal tissues, particularly at the locus Xp22.32 (15,16), as well as the ERV3-1 Env protein (17).
Unveiling HERV expression patterns across multiple cancer types is important to understanding the broader role of these elements in cancer progression. Towards this aim, large biomedical databases have been utilized to conduct pan-cancer studies of transposable element expression (18-21). However, identifying locus-specific HERV expression in transcriptomics data remains challenging from a bioinformatics standpoint due to high sequence similarity among HERV groups, and therefore, the vast majority of these studies report group-level expression patterns. As such, novel computational frameworks are needed in tandem with large -omics datasets to identify locus-specific HERV expression patterns across human cancers.
While HERV transcription signatures are useful as prognostic biomarkers (22-24), elucidating the underlying mechanisms of HERV activation and function in the context of cancer development is also essential to treatment applications. As methylation is a key mechanism in silencing HERV function, hypomethylation patterns observed in tumor cells may induce HERV expression. HERV expression has also been shown to contribute to immunomodulation of host cells and HERVs have been implicated in the regulation of innate immunity (25,26). Furthermore, a viral mimicry phenotype has been identified in subsets of metastatic tumors which corresponds to increased HERV expression, further suggesting the contribution of HERVs to inflammatory immune responses supporting tumor survival (27). Thus, an understanding of host cell factors including epigenetics and immunogenicity, in addition to HERV expression, is highly relevant to translational applications.
Here, we utilize Telescope (28) to present a locus-specific characterization of differential HERV expression in paired tumor-normal prostate, breast, and colon cancer RNA-Seq data from The Cancer Genome Atlas (TCGA) (29). We then combine these results with differential expression analysis of host genes to identify potential host genomic targets of HERV expression and regulatory mechanisms. Lastly, we analyze broader patterns in dysregulated HERV elements across these three cancer types to identify potential mechanisms of HERV activation associated with cancers.
Materials and Methods
Data
Paired-end RNA-Seq data (Illumina) were downloaded in BAM format (*.bam) from TCGA (29) using the Genomic Data Commons (GDC) Data Transfer Tool (30). In total, RNA-Seq files from paired primary tumor (non-metastatic) and solid tissue normal (non-cancerous tissues adjacent to the tumor) samples from 52 prostate cancer (TCGA-PRAD) cases, 111 breast cancer (TCGA-BRCA) cases, and 24 colon cancer (TCGA-COAD) cases were pulled. FFPE and metastatic samples were excluded from further analysis, and in cases where multiple tumor files remained for a single case, per-base sequence quality (evaluated using FastQC v0.11.9 (31)) was used to select one file for use in this study. Corresponding clinical and demographic metadata were downloaded using TCGABiolinks v3.11 (32) and all related analyses were performed in R v4.0.0 (33) (Supplemental Table S1).
Quality Control and Trimming
Downloaded BAM files were converted to FASTQ format using SAMtools v. 1.10 (34). FASTQ files were then trimmed using Trimmomatic v. 0.39 (35) with a leading/trailing threshold of 3, sliding window of 4bp with a threshold of 14, minimum read length of 36 bp, and Illumina adapters removed. In TCGA-PRAD data, the percentage of reads remaining after trimming ranged from 71.30 to 97.50% (average 95.15% +/− 2.71) and the number of trimmed reads per file ranged from 24,556,156 to 112,956,172 (average 72,099,135 +/− 12,007,080). In TCGA-BRCA data, the percentage of reads remaining after trimming ranged from 65.02 to 97.08% (average 93.41% +/− 3.24) and the number of trimmed reads per file ranged from 28,327,165 to 150,387,689 (average 70,310,030 +/− 17,254,417). In TCGA-COAD data, the percentage of reads remaining after trimming ranged from 88.44 to 96.85% (average 94.02% +/− 2.57) and the number of trimmed reads per file ranged from 28,901,359 to 101,130,017 (average 56,122,694.8 +/− 13,567,413.5).
Retrotranscriptome and Transcriptome Quantification
Trimmed reads were aligned to Hg38 using Bowtie2 v2.3.5.1 (36) with a very sensitive local alignment search, up to 100 alignments per read, and with fragments having at least 95% identity reported. Alignment rates averaged 92.9% (+/− 0.94%) for TCGA-PRAD, 96.0% (+/− 0.025%) for TCGA-BRCA, and 92.1% (+/− 0.72%) for TCGA-COAD. The Telescope v1.0.3-0 (28) assign module was then used for TE quantification using the HERV annotation described in (28) and a theta prior of 200,000. The Telescope algorithm is unique in identifying locus-specific HERV expression, as opposed to groups of HERVs, as is common in other studies. Additionally, Telescope uses a novel Bayesian reassignment method to map fragments onto transcripts, allowing for the utilization of more data than other approaches which either are limited to mapping just unique reads, thereby, using a very limited data set or incorporate all reads, but without the reassignment step resulting in low resolution to differentiate among alternative mapping options.
Kallisto v0.46.2 (37) was used to quantify gene expression data using default parameters and the Ensembl v96 human transcriptome assembly indexed with default settings. Tximport v1.16.1 (38) was used to read abundance files into R. Average pseudoalignment rate was 92.2% (+/− 2.3%) for TCGA-PRAD data, 90.5% (+/− 5.3%) for TCGA-BRCA data, and 90.9% (+/− 7.5%) for TCGA-COAD data.
Differential Expression Analysis
Differential expression analysis of retrotranscriptomic and transcriptomic data between tumor and normal samples was performed using DESeq2 v1.28.1 (39) using a negative binomial model and Wald test with a significance cutoff of p=0.05, using the patient identifier and condition (tumor or normal tissue) as factors in the DESeq model ( ~ patient + condition). Variance stabilizing transformation was used prior to principal component analysis (PCA) and hierarchical clustering. The R packages ggplot2 v3.2.2 (40) and EnhancedVolcano v1.6.0 (41) were used in addition to DESeq2 functions to visualize PCA and volcano plots, respectively, and the circlize v0.4.10 (42) and GenomicRanges v1.40.0 (43) packages were used to construct circos plots of differential gene and HERV expression patterns. Lastly, the pheatmap v1.0.12 (44) package was used to visualize sample-wise expression and perform hierarchical clustering for top differentially expressed HERVs.
Gene Set Enrichment Analysis
Gene set enrichment analyses (GSEA) of differentially expressed HERVs and differentially expressed genes were performed using GSEAPreranked as implemented in GSEA v4.0.3 (45). HERVs and genes, as applicable, were ranked by sign(log2(fold change))*−log10(adjusted p-value), using fold change metrics and p-values from DESeq2. For HERVs, a custom gene set was generated from the HERV annotation described in (28). For genes, gene ontology sets from the Molecular Signatures Database (MSigDB; (45,46)), as provided by GSEA, were used.
Survival Analysis
Survival analysis was performed using the survival R package v. 3.2.3 (47) using the log-rank test for by-group comparisons. Kaplan-Meier survival curves were visualized using ggfortify v. 0.4.10 (48). Survival times were censored at 5 years, with patients listed as alive censored at the date of the last follow up.
HERV Meta-Annotations
Annotations of the intersected and nearby genes for the TE reference utilized here were constructed using the ENSEMBL Hg38 release 99. Annotations of enhancer regions were constructed using FANTOM5 (49).
Results
Prostate Cancer
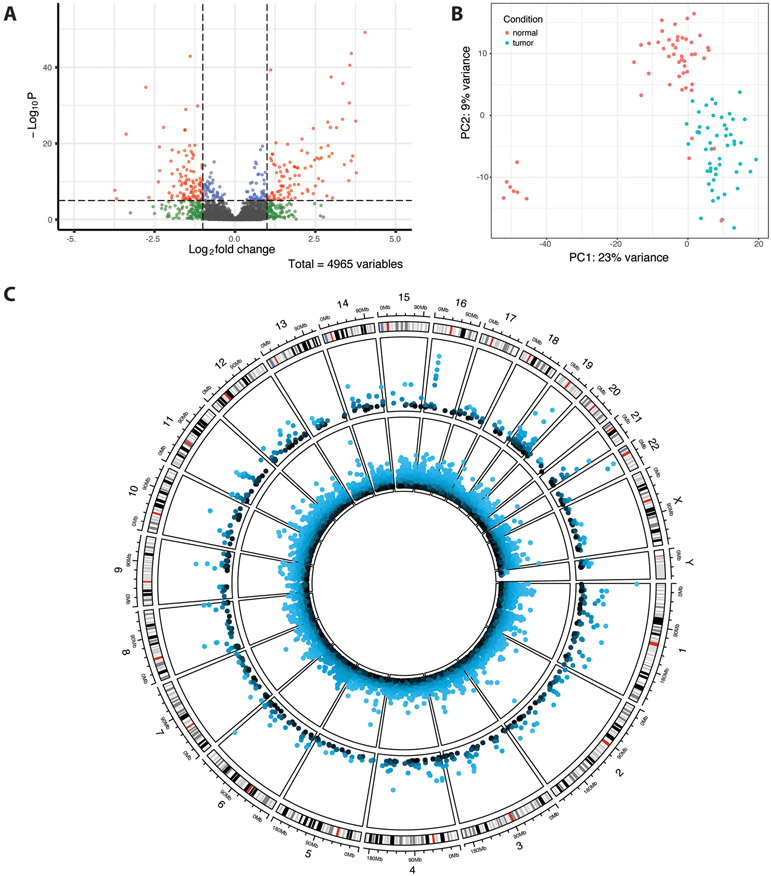
In total, out of 4,965 HERVs identified, 720 were differentially expressed between tumor and normal prostate cancer samples, with 361 upregulated in tumor tissue and 259 downregulated (Figure 1A, Table 1). A full list of differentially expressed HERVs is available in the supplemental data. The HERV groups MER4, HERVL/ERVL, HERVK, and HERVH were most prevalent among both up- and downregulated HERV elements (Supplemental Data, Table S2), however only HERVE was significantly enriched in downregulated HERVs (q<0.25), and no groups were significantly enriched in upregulated HERVs (Supplemental Data, Table S3). PCA of the 500 most variable HERVs shows separation of tumor and normal HERV expression data in the majority of samples, and also a group of seven normal tissue samples which cluster separately (Figure 1B). These seven samples additionally cluster together in hierarchical clustering analysis of the top 100 most significantly differentially expressed HERV loci (Supplemental Data; Figure S1), which shows notable upregulation of HML3 10q11.21a, HARLEQUIN 1q32.1, HERVIP10F 19p13.2b, and PABLA 14q11.2 and downregulation of HERVL18 2q31.1 in these samples compared to other normal tissue samples. Secondary Gleason grade, age at diagnosis, and race were not significantly different between cases corresponding to each group (Chi-square test, p-values in [0.21,0.79]). Survival data were not available for this dataset.
Figure 1.
TCGA-PRAD differential HERV expression results. (A) Volcano plot of differentially expressed HERVs. Shrunken log fold changes are shown. (B) Principal component analysis of the 500 most variably expressed HERVs. Data were transformed using the variance stabilizing transformation. (C) Circos plot showing differentially expressed (p<0.05) host genes (inner ring) and HERVs (outer ring) across the human genome. In both rings, −log10(p-value) is plotted, with outermost points as well as lighter colors corresponding to greater significance. The outermost ring displays cytoband data for each chromosome.
Table 1.
Top 10 up- and downregulated differentially expressed HERVs in TCGA-PRAD data, ranked by adjusted p-value.
| HERV | Locus | Chromosome | Start | End | Log Fold Change | Adjusted p-value | Group | |
|---|---|---|---|---|---|---|---|---|
| Upregulated HERVs | ||||||||
| 1 | HERVK11 | 22q11.21 | 22 | 19934438 | 19940687 | 4.36908947 | 1.50E-46 | HERVK |
| 2 | HML2 | 22q11.23 | 22 | 23536062 | 23546900 | 3.93780804 | 2.48E-41 | HERVK |
| 3 | HML1 | 16p12.3a | 16 | 20641357 | 20642589 | 3.80177955 | 1.54E-38 | HERVK |
| 4 | HERVS71 | 19q13.12a | 19 | 36324873 | 36331043 | 1.11668712 | 2.09E-37 | HERVS |
| 5 | ERVLB4 | 6q27d | 6 | 168400842 | 168407700 | 3.22389969 | 1.32E-35 | ERVL |
| 6 | HERVIP10FH | 16p12.3 | 16 | 20642834 | 20644949 | 3.55247703 | 5.39E-34 | HERVI |
| 7 | HERVW | 16p12.3 | 16 | 20644952 | 20646904 | 3.81852268 | 5.89E-29 | HERVW |
| 8 | MER4B | 16p12.3b | 16 | 20655409 | 20660157 | 3.64956313 | 8.46E-25 | MER4 |
| 9 | HERVIP10F | 8p11.22 | 8 | 39848200 | 39853109 | 4.31450739 | 2.35E-24 | HERVI |
| 10 | LTR57 | 9q22.32 | 9 | 94577573 | 94578558 | 3.13229217 | 4.14E-24 | LTR |
| Downregulated HERVs | ||||||||
| 1 | HARLEQUIN | 1p36.33 | 1 | 1412252 | 1418852 | −1.4072252 | 8.79E-41 | HARLEQUIN |
| 2 | ERV316A3 | 21q22.3b | 21 | 42598743 | 42600906 | −2.8617665 | 4.99E-33 | ERV3 |
| 3 | HERVH | 19p13.2d | 19 | 9741826 | 9751972 | −1.1657101 | 3.19E-28 | HERVH |
| 4 | HML3 | 19q13.2 | 19 | 38935164 | 38941835 | −1.5507787 | 2.36E-27 | HERVK |
| 5 | HARLEQUIN | 10q23.1 | 10 | 84167186 | 84172465 | −2.3486084 | 8.82E-23 | HARLEQUIN |
| 6 | HERVK11 | 1p13.3 | 1 | 109702615 | 109709643 | −1.594515 | 2.95E-22 | HERVK |
| 7 | HUERSP3B | 1q21.3 | 1 | 153603714 | 153606012 | −1.5841396 | 3.66E-22 | HUERS |
| 8 | PABLA | 14q11.2 | 14 | 21003288 | 21010322 | −3.8428954 | 3.94E-21 | PAB |
| 9 | HML2 | 4p16.1b | 4 | 9657956 | 9667550 | −1.2569849 | 2.78E-18 | HERVK |
| 10 | HML3 | 1p32.3 | 1 | 54630287 | 54636120 | −1.4478303 | 3.19E-18 | HERVK |
In addition, 12,861 genes were identified as differentially expressed in tumor samples as compared to normal samples, with 6,423 being upregulated in tumor samples and 6,438 downregulated (Figure 1C; Supplemental Data, Figure S2A). Gene ontology enrichment analysis identified the enrichment of multiple processes relevant to HERV expression in upregulated genes, including viral protein expression (e.g., GO:0019080, q<0.05), epigenetic gene regulation (e.g., GO:0016458, GO:0045814, GO:0040029; q<0.05), and immune response, including innate immune response (e.g., GO:0002251, GO:0002227; q<0.25; Supplemental Data). Genes related to immunoglobulin secretion were also enriched in downregulated genes (e.g., q<0.25), as well as interferon response (e.g., GO:0035455, q<0.25) and inflammation (GO:0002269, q<0.05). Additionally, two TET genes (TET1 and TET3), which are associated with demethylation and are frequently mutated in cancer cells, were significantly upregulated in tumor samples. Several APOBEC genes, which are also involved in DNA demethylation pathways, were also differentially expressed, including upregulation of APOBEC3B and downregulation of APOBEC3C, APOBEC3G, APOBEC3F, APOBEC3D, APOBEC3H, and APOBEC2 in tumor samples (p<0.05). Several of these genes are also involved in RNA editing and can act as viral restriction factors. Furthermore, the antiviral signaling gene MAVS, interferon regulating factor IFN7, and two interferon genes (IFNAR1 and IFNGR2) were upregulated (p<0.05). The seven normal tissue samples which clustered separately by HERV expression also clustered separately by host gene expression in PCA and hierarchical clustering (Supplemental Data, Figure S2B, Figure S3). AOX1, which has been associated with prostate cancer survival times, displays higher expression levels in this subgroup, as well as GSTP1, the methylation of which has been associated with prostate tumor carcinogenesis. GCNT4, a colon cancer biomarker, additionally shows higher expression. Furthermore, the two most significantly upregulated HERVs in PRAD – both of which are HERV-K elements – are located on the q arm of chromosome 22, which is also the site of a TMPRSS2 and ERG gene fusion leading to overexpression of ERG, an oncogene. This fusion is among the most common in prostate cancer, and ERG was additionally found to be significantly overexpressed in tumor samples (p<0.001).
We then used HERV meta-annotations provided by Telescope to examine functional properties of differentially expressed elements, as well as nearby or intersected genes. Of 720 differentially expressed HERVs, 262 are located in exonic regions, 232 are located in intronic regions, and 226 are located in intergenic regions. A total of 67 are located in enhancer regions. Furthermore, these HERVs intersect 271 protein coding genes and 242 long noncoding RNAs (lncRNAs). Several of these regions correspond to known gene expression patterns in prostate and other cancers. Specifically, among upregulated HERVs, HERVK11 22q11.21 intersects TXNRD2, a protein-coding gene which was also significantly upregulated in tumor samples (p<0.001). TXNRD2 expression has been linked with tumor growth and suggested as a drug target. Additionally, HML2 22a11.23 intersects PCAT14, a lncRNA which has been identified as a prostate cancer biomarker. HML1 16p12.3a intersects the genes ACSM3 and ACSM1, both of which have been suggested as clinical biomarkers for prostate cancer. ACSM1 was significantly upregulated in tumor samples (p<0.001). Among downregulated HERVs, PABLA 14q11.2 intersects AL161668.3, a lncRNA which has been shown to be downregulated in head and neck cancer. HARLEQUIN 10q23.1 intersects CERNA2, a lncRNA associated with late stage and metastatic cervical cancers and malignant gastric cancers. In total, 155 differentially expressed HERVs intersected a gene that was also differentially expressed, with 142/155 being differentially expressed in the same direction. Furthermore, several differentially expressed HERVs are proximal to relevant genes. Notably, PABLB 4p15.2, which was upregulated in tumor samples (p<0.001), is 15.3 kb 3’ of DHX15, the expression of which is associated with androgen receptor activation and prostate cancer progression. Additionally, HML3 8p11.22, also upregulated (p<0.001) is 26.3 kb 5’ of IDO1, the upregulation of which is implicated in a mechanism for epithelial-mesenchymal transition in prostate cancer cells. In total, there were 75 differentially expressed HERVs for which the closest 5’ gene was also differentially expressed, and there were 77 differentially expressed HERVs for which the closest 3’ gene was also differentially expressed.
Breast Cancer
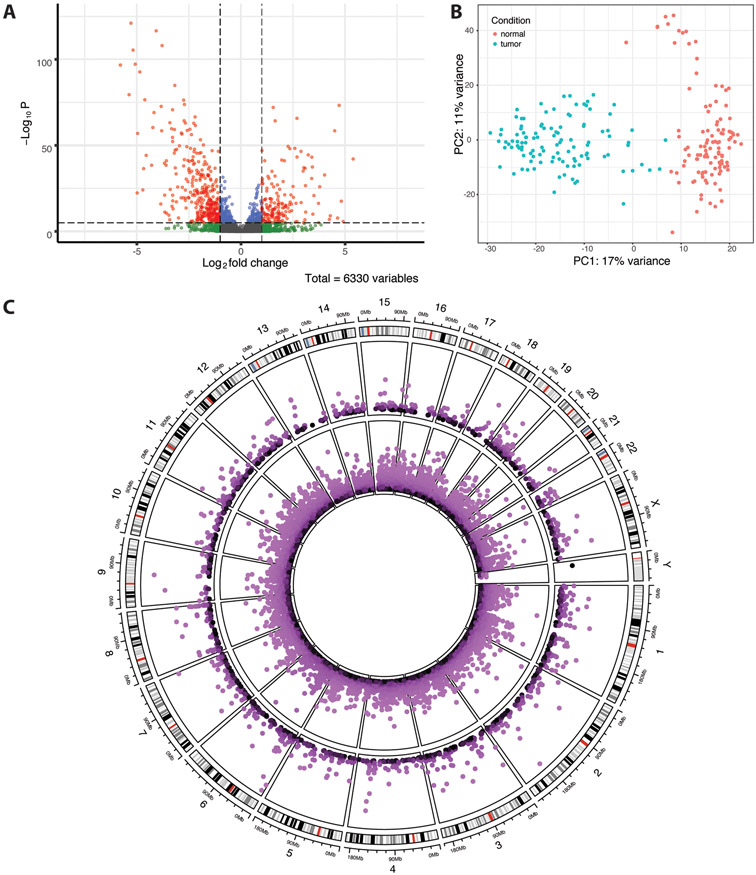
In total, of 6,330 HERVs identified, 1,547 HERVs were differentially expressed in tumor versus normal TCGA-BRCA samples (p<0.05), including 640 upregulated and 907 downregulated HERVs (Table 2, Figure 2A). A full list of differentially expressed HERVs is available in the supplemental data. MER4, HERVL/ERVL, HERVH, and HERVK were most prevalent among both up- and down-regulated HERVs (Supplemental Data, Table S4). GSEA results indicate that groups HERVL and ERV1 are enriched among upregulated HERVs, while HERVH is enriched in downregulated HERVs (false discovery rate q<0.25; Supplemental Data, Table S5). PCA of the 500 most variable HERVs shows separation of tumor and normal samples, with some separation of normal samples along principal component 2 (Figure 2B). This separation corresponds with the clustering of 14 normal samples separated from others in hierarchical clustering of the 100 most variable HERVs (Supplemental Data, Figure S4), in which decreased expression of several HERVs relative to other normal samples is visible. Survival analysis revealed no significant difference in survival times of these 14 cases versus others (p=0.1; Supplemental Data, Figure S5). Tumor stage, menopause stage, age at diagnosis, and race were not significantly different between cases corresponding to each group (Chi-square test, p-values in [0.17,1]). However, a greater proportion of patients in the smaller subset had received hormone therapy as a part of treatment (Chi-squared test, p=0.03).
Table 2.
Top 10 up- and downregulated differentially expressed HERVs in TCGA-BRCA data, ranked by adjusted p-value.
| HERV | Locus | Chromosome | Start | End | Log Fold Change | Adjusted p-value | Group | |
|---|---|---|---|---|---|---|---|---|
| Upregulated HERVs | ||||||||
| 1 | ERVLE | 18q21.1b | 18 | 48459770 | 48463770 | 4.76229862 | 1.48E-71 | ERVL |
| 2 | HERVS71 | 19q13.12a | 19 | 36324873 | 36331043 | 1.53951594 | 2.08E-70 | HERVS |
| 3 | ERVLB4 | 8q24.3d | 8 | 143744356 | 143745392 | 2.73317918 | 3.45E-64 | ERVL |
| 4 | ERV316A3 | 8q13.3a | 8 | 70099953 | 70104701 | 1.63373913 | 9.21E-63 | ERV3 |
| 5 | HERV3 | 21q22.13 | 21 | 38224043 | 38229805 | 4.61421579 | 3.58E-57 | ERV1 |
| 6 | HERVL | 5q12.3 | 5 | 65486807 | 65491213 | 2.27725807 | 3.28E-48 | HERVL |
| 7 | HERVH | 11q14.1b | 11 | 78080836 | 78087032 | 2.51036203 | 4.21E-47 | HERVH |
| 8 | HERV3 | 19p13.3 | 19 | 566440 | 568668 | 2.31841646 | 6.06E-47 | ERV1 |
| 9 | MER4 | Xq21.31c | X | 88094741 | 88098939 | 1.00744161 | 7.11E-46 | MER4 |
| 10 | PABLA | 2p25.1 | 2 | 10199041 | 10202321 | 2.85683078 | 2.08E-45 | PAB |
| Downregulated HERVs | ||||||||
| 1 | HERVH | 6p25.1 | 6 | 6027810 | 6033600 | −6.0124842 | 3.09E-118 | HERVH |
| 2 | ERVLB4 | 2q32.3 | 2 | 192042435 | 192046614 | −4.280831 | 4.48E-114 | ERVL |
| 3 | ERV316A3 | 6p25.2c | 6 | 3252869 | 3256853 | −3.9394259 | 1.13E-105 | ERV3 |
| 4 | HERVH | 9q21.12 | 9 | 69404587 | 69410267 | −6.0219238 | 3.51E-103 | HERVH |
| 5 | HERVH | 6p22.3c | 6 | 18754144 | 18759870 | −6.1987908 | 4.47E-95 | HERVH |
| 6 | MER4 | 4q32.1b | 4 | 155120046 | 155123356 | −7.6576203 | 1.15E-94 | MER4 |
| 7 | HML3 | 7q36.3 | 7 | 155238195 | 155244187 | −5.2711014 | 9.74E-91 | HERVK |
| 8 | MER41 | 9p13.3a | 9 | 33703617 | 33708862 | −3.2855468 | 5.96E-83 | MER4 |
| 9 | HERVH | 4q32.1b | 4 | 155108278 | 155114237 | −7.0346713 | 1.37E-77 | HERVH |
| 10 | HML2 | 3p25.3 | 3 | 9847662 | 9854552 | −4.7081634 | 1.11E-74 | HERVK |
Figure 2.
TCGA-BRCA differential HERV expression results. (A) Volcano plot of differentially expressed HERVs. Shrunken log fold changes are shown. (B) Principal component analysis of the 500 most variably expressed HERVs. Data was transformed using the variance stabilizing transformation. (C) Circos plot showing differentially expressed (p<0.05) host genes (inner ring) and HERVs (outer ring) across the human genome. In both rings, −log10(p-value) is plotted, with outermost points as well as lighter colors corresponding to greater significance. The outermost ring displays cytoband data for each chromosome.
In total, 18,425 genes were identified as differentially expressed in the TCGA-BRCA data, including 8,609 upregulated and 9,816 downregulated in primary tumors (Figure 2C; Supplemental Data, Figure S6A). Similar to the prostate cancer data, pathways related to epigenetic gene regulation (e.g., GO:0016458, GO:0045815, GO:0045814, GO:0040029; q<0.05) and innate immune response (e.g., GO:0045089, GO:0002218; q<0.25) were significantly enriched in upregulated genes, as is consistent with proposed mechanisms of HERV activation (Supplemental Data). BRCA1 and BRCA2 were both significantly upregulated in tumor samples. Several methylation-associated genes were also dysregulated, including upregulation of TET3, APOBEC3B, APOBEC3A, APOBEC3H, and APOBEC4 as well as downregulation of APOBEC3C, APOBEC2, and APOBEC3G (p<0.05). Additionally, IRF7 and several interferon genes (IFN1, IFNL3P1, IFNGR2, and IFNG) were overexpressed (p<0.05). The clustering of normal samples in PCA is consistent with that found in HERV expression PCA, and a majority of the 14 samples in the separate sub-group cluster together in hierarchical clustering (Supplemental Data, Figure S6B, Figure S7). The samples in the sub-group and those which cluster with them display a notable increase in LEP expression, which is known to be pro-carcinogenic in breast cancers.
Of the 1,547 differentially expressed HERVs identified, 497, 554, and 496 are located in exonic, intronic, and intergenic regions, respectively. Additionally, 133 are located in enhancer regions. The identified HERVs intersect 651 protein coding genes and 443 lncRNAs. Of note in upregulated HERVs, HARLEQUIN 17q21.31 intersects BRCA1. LTR25 7q21.12 and MER61 7q21.12 both intersect TP53TG1, a lncRNA and component of the p53 pathway which inhibits tumor growth and is inactivated in human cancer cells via methylation. PABLA 2p35.1 intersects RRM2, which promotes breast cancer metastasis and was upregulated in tumor samples (p<0.001). Additionally, HERV3 21q22.13 intersects KCNJ15, which has been associated with esophageal and renal cancers. KCNJ15 was also upregulated in tumor samples (p<0.001). ERV316A3 8q23.1c, which was downregulated in tumor samples, intersects PKHD1L1, which is commonly mutated in triple-negative breast cancers. PKHD1L1 was also downregulated in tumor samples (p<0.001). In total, there were 387 differentially expressed HERVs for which the intersected gene was also differentially expressed, with 336/387 being differentially expressed in the same direction. HERVH 6p25.1, which was downregulated in tumor samples, is 20.2 kb 5’ of NRN1, which is known to be hypermethylated in breast cancer and was also downregulated in tumor samples (p<0.001). In total, there were 84 differentially expressed HERVs for which the closest 5’ gene was also differentially expressed and 73 differentially expressed HERVs for which the closest 3’ gene was also differentially expressed.
Colon Cancer
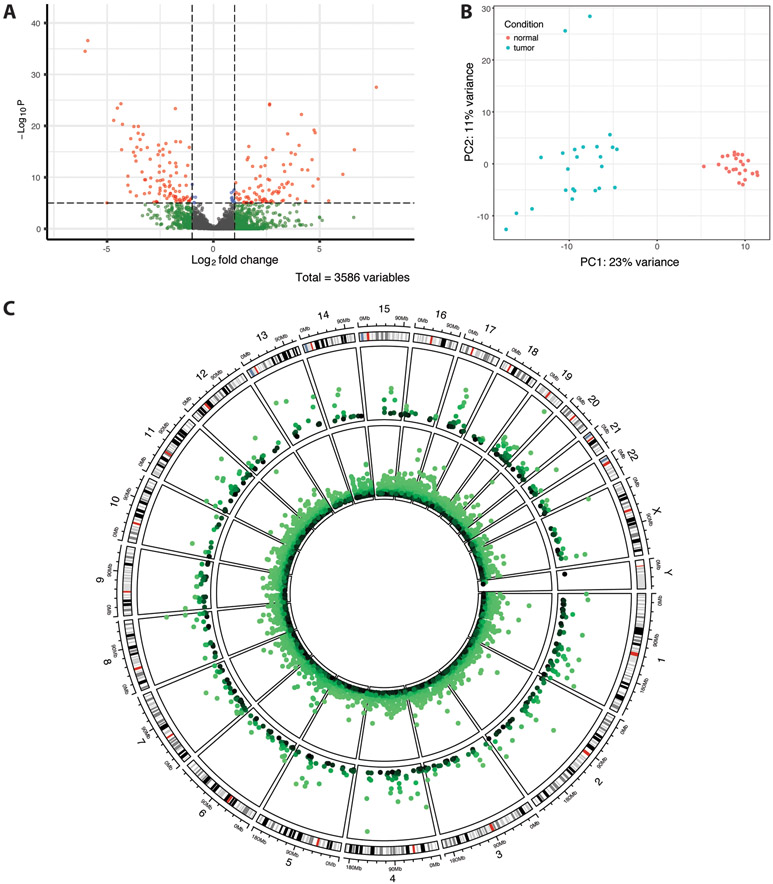
In the TCGA-COAD data, of 3,586 HERVs identified, 575 were differentially expressed in tumor versus normal samples, with 283 upregulated and 292 downregulated (p<0.05) (Table 3, Figure 3A). A full list of differentially expressed HERVs is available in the supplemental data. The HERV groups MER4, HERVL/ERVL, HERVH, and HERVK were most prevalent among both up- and downregulated HERVs (Supplemental Data, Table S6). GSEA results indicate enrichment of PAB and ERV1 groups in upregulated HERVs (Supplemental Data, Table S7). PCA of HERV expression data shows a clear distinction between tumor and normal samples, with additional separation of two tumor samples along PC2 (Figure 3B). These two samples, along with two others, cluster with normal samples in hierarchical clustering, though separation between tumor and normal samples is otherwise distinct (Supplemental Data, Figure S8). Overall differences in HERV expression data are less apparent than in PRAD or BRCA datasets, though notable overexpression of two HERVH elements (Xp22.32a and 20p11.23b) is seen in several tumor samples.
Table 3.
Top 10 up- and downregulated differentially expressed HERVs in TCGA-COAD data, ranked by adjusted p-value.
| HERV | Locus | Chromosome | Start | End | Log Fold Change | Adjusted p-value | Group | |
|---|---|---|---|---|---|---|---|---|
| Upregulated HERVs | ||||||||
| 1 | HERVH | Xp22.32a | X | 4540474 | 4546320 | 7.41804354 | 2.17E-25 | HERVH |
| 2 | ERVLB4 | 8q24.3d | 8 | 143744356 | 143745392 | 2.65863273 | 2.37E-22 | ERVL |
| 3 | HML3 | 13q12.11a | 13 | 19626322 | 19631230 | 2.66118817 | 3.00E-22 | HERVK |
| 4 | HERVH | 13q14.11b | 13 | 43059672 | 43061602 | 4.13841523 | 1.42E-20 | HERVH |
| 5 | HERV9 | 5q31.2 | 5 | 139502943 | 139512402 | 4.72808626 | 9.94E-18 | HERVW |
| 6 | HERVH | 13q33.3 | 13 | 109265090 | 109271116 | 4.95089626 | 3.02E-17 | HERVH |
| 7 | MER61 | 4q21.1a | 4 | 76230620 | 76234657 | 3.7165833 | 4.43E-17 | MER4 |
| 8 | MER4B | Xq26.2a | X | 131689719 | 131692783 | 4.09495849 | 5.17E-15 | MER4 |
| 9 | HARLEQUIN | 7q22.1 | 7 | 100838700 | 100847185 | 1.67979636 | 8.12E-15 | HARLEQUIN |
| 10 | ERV316A3 | 11q13.3 | 11 | 69383332 | 69384472 | 2.92229141 | 2.72E-14 | ERV3 |
| Downregulated HERVs | ||||||||
| 1 | ERVLE | 19q13.31a | 19 | 43364281 | 43366105 | −6.5117269 | 5.56E-34 | ERVL |
| 2 | ERVLE | 4q31.23c | 4 | 148613113 | 148614226 | −6.222073 | 3.39E-32 | ERVL |
| 3 | ERVLE | 17q21.2b | 17 | 41557971 | 41561541 | −4.5728292 | 2.37E-22 | ERVL |
| 4 | HERVL74 | 2q11.2 | 2 | 100276117 | 100280003 | −4.8984686 | 1.07E-21 | HERVL |
| 5 | MER41 | 4p16.1b | 4 | 8358459 | 8364122 | −1.8001679 | 1.17E-21 | MER4 |
| 6 | HML3 | 17q21.32 | 17 | 49170645 | 49176690 | −5.9316334 | 1.81E-19 | HERVK |
| 7 | ERV316A3 | 21q21.1f | 21 | 22032698 | 22035130 | −4.9657293 | 9.67E-19 | ERV3 |
| 8 | ERVLB4 | 11q23.3a | 11 | 114648240 | 114649258 | −4.0180887 | 2.19E-18 | ERVL |
| 9 | ERV316A3 | 1q43e | 1 | 240485614 | 240489056 | −3.6953533 | 2.19E-18 | ERV3 |
| 10 | PRIMA4 | 14q22.1 | 14 | 52280360 | 52286612 | −3.8018022 | 3.50E-17 | PRIMA |
Figure 3.
TCGA-COAD differential HERV expression results. (A) Volcano plot of differentially expressed HERVs. Shrunken log fold changes are shown. (B) Principal component analysis of the 500 most variably expressed HERVs. Data was transformed using the variance stabilizing transformation. (C) Circos plot showing differentially expressed (p<0.05) host genes (inner ring) and HERVs (outer ring) across the human genome. In both rings, −log10(p-value) is plotted, with outermost points as well as lighter colors corresponding to greater significance. The outermost ring displays cytoband data for each chromosome.
Additionally, 12,701 genes were differentially expressed in the TCGA-COAD data, including 5,851 upregulated and 6,850 downregulated genes (Figure 3C; Supplemental Data, Figure S9A). As before, upregulated genes are enriched for pathways relevant to epigenetic gene regulation (e.g., GO:0016458, GO:0045815, GO:0045814, GO:0040029; q<0.25). Notably, four of the top ten most significantly enriched pathways in downregulated genes are related to immune response and immunoglobulin circulation (GO:0019814, GO:0034987, GO:0042571, GO:0002455; q<0.05; Supplemental Data). TET2 was significantly downregulated in tumor samples (p<0.001), which would promote DNA methylation, as well as APOBEC3C, APOBEC2, and APOBEC3G (p<0.05). Several interferon genes (IFNWP19, IFNLP31, and IFNG) and interferon regulatory factor IRF7 were also significantly overexpressed (p<0.05). Hierarchical clustering and PCA of gene expression data show clear separation of tumor and normal samples, as well as distinct differences in gene expression patterns (Supplemental Data, Figure S9B, Figure S10).
The 575 differentially expressed HERVs are distributed across exonic, intronic, and intergenic regions with 218, 172, and 185 per region, respectively. Of these, 63 are located in enhancer regions. These HERVs intersect 231 protein coding genes and 178 lncRNAs. Included among these, as in the breast cancer data, is MER61 7q21.12, which intersects TP53TG1. Additionally, HERVH 4q22.1c, which was downregulated in tumor samples, intersects ABCG2, which was also significantly downregulated (p<0.001) and has been linked to treatment resistance in colon cancer (50,51). HERVH 4q22.3a, which was also downregulated in tumor samples, intersects the downregulated gene (p<0.001) HPGDS, which has been shown to inhibit intestinal tumors in a mouse model. In total, there were 119 differentially expressed HERVs for which the intersected gene was also differentially expressed, with 114/119 being differentially expressed in the same direction. Additionally, there were 70 differentially expressed HERVs for which the closest 5’ gene was also differentially expressed and 58 for which the closest 3’ gene was also differentially expressed. Included among these is HERVL 4q32.1a, which was downregulated in tumor samples and is 1.1 kb 5’ of TDO2, which was upregulated in tumor samples (p<0.01) and has been associated with advanced stage colorectal cancers.
Comparisons Across Cancer Types
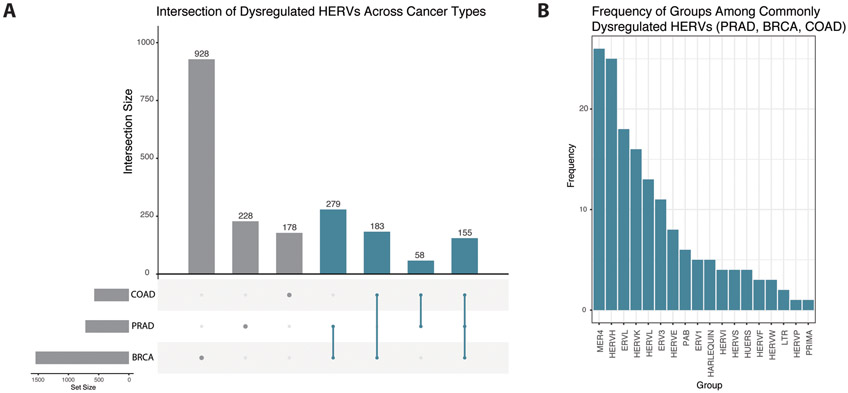
Many HERV loci were identified as differentially expressed in more than one of prostate, breast, and colon cancers, including 155 loci which were differentially expressed in all three cancer types (Figure 4A). Of these, 114 were differentially expressed in the same direction in all three cancer types. The majority were of the groups MER4 (26/155), HERVH (25/155), ERVL (18/155), HERVK (16/155), HERVL (13/155), and ERV3 (11/155) (Figure 4B). Of these HERV loci, 72 are located in exonic regions, 43 are located in intronic regions, and 40 are located in intergenic regions. These elements intersect 77 protein coding genes and 53 lncRNAs. Notably, one HERV which is differentially expressed in all three cancer types (HARLEQUIN 1q32.1) intersects the tumor suppressor gene BRCA1. This element was upregulated in PRAD and COAD samples, but downregulated in BRCA samples. Furthermore, 279 HERV loci were differentially expressed in both PRAD and BRCA datasets, 183 were differentially expressed in both BRCA and COAD datasets, and 58 were differentially expressed in both COAD and PRAD datasets. Patterns in the prevalence of HERV groups among up- and downregulated HERVs are similar across all three cancer types, with MER4, HERVL, HERVK, HERVH, and ERVL being the five most frequently dysregulated groups in all datasets.
Figure 4.
Comparison of dysregulated HERVs across multiple cancer types. (A) Upset plot displaying frequencies of individual HERV loci differentially regulated in one or more of prostate, breast, and colon cancers. Bars colored blue indicate HERV loci dysregulated in at least two cancer types. (B) Frequencies of HERV groups among 155 HERVs dysregulated in all three cancer types (PRAD, BRCA, and COAD).
Discussion
Here, we present locus-specific differential HERV expression patterns in prostate, breast, and colon cancers. In general, MER4, HERVK, and HERVL/ERVL elements were most commonly dysregulated in tumor tissues across breast, prostate, and colon cancers, with over 600 unique elements being dysregulated in more than one cancer type. Additionally, we identified host gene expression and analyzed the intersection sites of dysregulated HERV loci in order to address potential pathogenic mechanisms.
In the TCGA-PRAD dataset, a total of 720 differentially expressed HERVs were identified, as well as 12,861 host genes. A majority of the differentially expressed HERVs were upregulated vs. downregulated elements (361 and 359, respectively), as has been shown to be common in many cancer types (19). Gene set enrichment analysis found that no HERV groups were enriched in upregulated HERVs and only HERVE was enriched in downregulated HERVs (q<0.25), likely owing to the prevalence of more frequently dysregulated HERV groups (e.g., MER4, HERVL/ERVL, HERVK, and HERVH) in both up- and downregulated loci. Two distinct clusters of normal samples were observed in both HERV and host gene expression data, which did not correlate with sequencing batches or clinical variables. This clustering was consistent in PCA and hierarchical clustering of both HERVs and host genes. Additionally, several cancer-associated genes show overexpression in the smaller (n=7) subset, including AOX1, GSTP1, and GCNT4, with these findings together suggesting that the clustering pattern has biological significance. This observation is consistent with previous empirical studies which found subgroups of patients with significantly different HERV expression profiles, including in normal tissue samples (9), and those which have identified prognostic markers in tumor-adjacent normal tissues (52). Furthermore, another recent study of TCGA data identified heterogeneity in HERV expression across patients, as is consistent with this observation (21). The correspondence between HERV and gene expression profiles (Figure 1C) – as well as the proximity to and/or intersection of differentially expressed, cancer-associated genes by the identified HERVs (152 and 155 unique HERVs, respectively) – further suggests a functional link between HERV expression and cancer pathways. A majority (513/720) of differentially expressed HERVs intersect either protein coding genes or lncRNAs; the latter being typically associated with HERVs and activated in cancers (53). Several known prostate cancer-associated genes and lncRNAs were intersected by or located immediately 5’ or 3’ to a differentially expressed HERV, including TXNRD2, PCAT14, ACSM1/3, AL161668.3, CERNA2, DHX15, and IDO1.
In the TCGA-BRCA dataset, a total of 1,547 differentially expressed HERVs were identified, as well as 18,425 genes. The most significantly differentially expressed HERVs are predominantly downregulated (907/1,546; Figure 2B), as is consistent with previous studies [22,31]. HERVL and ERV1 were enriched in upregulated HERVs, while HERVH was enriched in downregulated HERVs (q<0.25). As in the TCGA-PRAD data, a subgroup (n=14) of normal samples was identified by PCA and hierarchical clustering that was consistent between HERV and gene expression data. The presence of a subset of samples with different HERV expression profiles is consistent with a previous empirical study in breast cancer cell lines (9). Here, this subgroup also displays a clear increase in LEP expression, which has been implicated in obesity-related breast cancers (54). While clinical data relating to BMI or obesity status was not available for these samples, it is possible that this clustering could indicate obesity-related cases. Notably, the smaller subset of normal tissues corresponds to a significantly greater proportion of patients having received hormone therapy for breast cancer treatment (p<0.05), indicating a potential association between such therapies and HERV activation. This is consistent with previous work in which HERV-K has been shown to be hormonally activated in breast cancers. There was no significant difference in other clinical variables between the two clusters, nor was there a difference in survival probabilities (p=0.1), though this analysis may have been limited by the small sample size. As in prostate cancer, many of the differentially expressed genes intersect or are located in close proximity to differentially expressed genes (387 and 157, respectively), and several breast cancer-associated genes were found to be differentially expressed in these data, including BRCA1 and BRCA2. As in prostate cancer, several TET and APOBEC genes were also differentially expressed. One upregulated HERV (HARLEQUIN 17q21.31) intersects BRCA1, which was also upregulated in tumor samples (p<0.05). Additionally, NRN1 – which is typically hypermethylated in breast tumors – is 3’ of downregulated element HERVH 6p25.1 (p<0.01). In total, 1,094/1,547 differentially expressed HERVs intersected either a protein coding gene or lncRNA.
In the TCGA-COAD dataset, 575 differentially expressed HERVs and 12,701 differentially expressed genes were identified. PAB and ERV1 groups were enriched among upregulated HERVs (q<0.25). The separation of normal samples into two groups that was found in prostate and breast cancers was not seen in colon cancer, though this is possibly a result of the much lower sample size. Two tumor samples do separate along PC2 in HERV expression data, though this is not matched in hierarchical clustering or in PCA of gene expression data. Overall, differences in HERV expression between tumor and normal samples are less pronounced than in the other two datasets. Still, many differentially expressed HERVs intersect differentially expressed genes (119 HERVs) or are immediately 5’ or 3’ of a differentially expressed gene (128 HERVs). Many genes previously associated with colon cancer were identified as differentially expressed in these samples. As in the other cancer types, several differentially expressed HERVs intersect clinically relevant genes, and a majority (409/575) of HERVs intersect a protein coding gene or lncRNA.
In total, 675 unique HERV loci were differentially expressed in more than one of the cancer types studied, suggesting that a significant amount of HERV expression in cancer is not disease or tissue specific. Moreover, the same HERV groups – MER4, HERVH, ERVL/HERVL, and HERVK – were most prevalent among dysregulated HERVs in all three cancer types, which is consistent with other studies identifying overlap in expressed HERV groups across TCGA cancer types (21). As the same groups were predominant among both up- and downregulated HERVs, collapsing HERV transcripts at the gene level may mask differential expression patterns. HERVK and HERVH were found to be two of the most frequently overexpressed HERVs in a previous study as well, where it was postulated that their overexpression was a result of being more recently integrated into the human genome and thus more functional (19). However, MER4 elements, which are older and thus no longer functional, were most prevalent. These results suggest that other HERV groups in addition to HERV-K should also be a focus of disease studies. Interestingly, the highest overlap of differentially expressed HERV loci is seen between prostate and breast cancers. These cancers commonly are associated with sex hormone dependence and mutation of BRCA1, suggesting that these two factors may also be linked to HERV expression. Furthermore, the subgroup ERV316A3 was repeatedly implicated in top differentially expressed HERVs in all three cancer types, motivating future study of these elements. Across all three datasets, many differentially expressed HERVs intersect genes relevant to disease phenotypes, and many are located in enhancer regions, motivating further studies into the functional effects of HERV expression in cancer tissues.
Demethylation is a key mechanism of HERV expression. Here, TET and APOBEC genes – which are important to DNA demethylation pathways – were differentially expressed in all three cancer types. Previous work has associated TET2 overexpression with increased HERV expression and antiviral immune response signatures (27), suggesting a link between demethylation and HERV activation in cancer. TET2 was downregulated in COAD, while TET3 was upregulated in BRCA and PRAD, with TET1 also being upregulated in PRAD. Thus, these results add to existing evidence suggesting changes to DNA demethylation activity contribute to HERV activation in tumor cells. Interestingly, both TET and APOBEC genes were both up- and downregulated in tumor samples, suggesting that epigenetic alterations in normal tissues are of importance, as well as tumor samples. Moreover, several genes associated with viral mimicry and host antiviral response – including MAVS, IRF7, and several interferon genes – were significantly overexpressed in tumor tissues (p<0.05), which is consistent with previous work establishing demethylation of HERVs as a precursor to an immunogenic response (55). Additionally, gene ontology enrichment analysis identified enrichment of several relevant biological pathways in dysregulated genes across cancer types, including epigenetic gene regulation and immune response. Therefore, gene expression data support the roles of both demethylation and viral mimicry pathways in contributing to HERV pathogenesis in cancer.
While further empirical studies will be required to validate the functional effects of HERV expression patterns identified here, significant empirical evidence exists for the immunogenic effects of HERV expression. In fact, HERVL/ERVL, HERVK, and ERV1 – three HERV classes which were repeatedly identified in our analyses – were recently implicated in biochemical experiments linking HERV expression to interferon response in ovarian cancer cell lines (56,57). Furthermore, HERV elements at several loci identified in the top significantly differentially expressed HERVs in our analyses have been identified in previous empirical studies, including the 21q22.3 locus (55). Lastly, a recent study found evidence of HERV-induced transcriptional modulation of zinc-finger proteins in lung cancer cell lines, many of which were differentially regulated across all three cancer types studied here (21). Thus, previous empirical work supports the utility of our bioinformatics analyses in identifying targets of future validation studies.
The primary limitations of this study stem from low sample size. While paired tumor-normal tissue samples are necessary to identify dysregulated elements while controlling for heterogeneity among patients, normal solid tissue samples are difficult to obtain and thus are not commonly included in sequencing databases. In the TCGA-PRAD data in particular, survival data were not available, and thus the impact of HERV expression clustering on survival probability was not able to be determined. Also, only one patient in the TCGA-PRAD dataset had received hormone therapy, and so the effects of such treatment on HERV activity could not be evaluated for this dataset. In the TCGA-BRCA data, while survival data were available, the cluster of interest was small (n=14), and so re-evaluation with a larger sample set would be beneficial. Moreover, all tumor samples included in this study were primary tumors. Additional samples from metastatic tumors would be useful to determine differential expression of HERVs throughout cancer progression. Lastly, methylation data were not incorporated into this analysis as the methylation arrays used in the TCGA project primarily cover genic regions, however further studies incorporating appropriate methylation data would be of great benefit towards uncovering causal relationships in HERV transcription pathways. While other studies have addressed HERV expression in cancer RNA-Seq data, our study is unique in identifying locus-specific expression, as opposed to more general HERV groups or subgroups. Additionally, the Telescope algorithm utilizes a greater proportion of reads by not requiring reads to be uniquely mapped via its Bayesian re-assignment approach (28).
In conclusion, by utilizing RNA-Seq data from matched samples, we identified differentially expressed HERVs in both tumor and normal tissue in prostate, breast, and colon cancers. Our work is novel in providing locus-specific characterization of these elements, which has previously been unattainable due to bioinformatics challenges. We found that HERV expression in normal tissue samples from prostate and breast cancers differentiates samples into two distinct groups, and that in breast cancer, this clustering was associated with hormone therapy. Furthermore, we found that a sizable number of HERV loci are differentially expressed across multiple cancer types, particularly between prostate and breast cancers, suggesting that the pathogenic role of HERVs in cancer may not be entirely disease-specific. By pairing HERV expression results with an analysis of host genes, we found that many DNA demethylation and antiviral response related genes were also differentially regulated in tumor tissues, providing evidence towards demethylation and viral mimicry as driving factors for HERV expression in cancers. Future work validating these findings using empirical data and determining causal relationships would be of great benefit, as such knowledge would elucidate regulatory mechanisms of disease-associated HERVs towards the development of novel cancer therapies.
Supplementary Material
Statement of Significance:
Expressed human endogenous retroviruses are mapped at locus-specific resolution and linked to specific pathways to identify potential biomarkers and therapeutic targets in prostate, breast, and colon cancers.
Acknowledgements
This work was supported by the National Institutes of Health, grant numbers CA206488 (D.F.N. and K.A.C.), UL1TR000075 (K.A.C.), and a Medical Scientist Training Program grant to the Weill Cornell-Rockefeller-Sloan Kettering Tri-Institutional MD-PhD Program T32GM007739 (J.L.M.). The work also benefited from support from a Research Innovation Award (02RIA032020) from the Milken Institute School of Public Health (K.A.C., K.B.C.). The results shown here are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thank Miguel Branco and an anonymous reviewer for helpful comments to improve our manuscript.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.American Cancer Society. Cancer Facts & Figures 2019. Atlanta; 2019. [Google Scholar]
- 2.Bannert N, Kurth R. The Evolutionary Dynamics of Human Endogenous Retroviral Families. Annu Rev Genomics Hum Genet. 2006;7:149–73. [DOI] [PubMed] [Google Scholar]
- 3.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, et al. Comprehensive Analysis of Human Endogenous Retrovirus Transcriptional Activity in Human Tissues with a Retrovirus-Specific Microarray. J Virol. 2005;79:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol. 2013;3:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 2008;8:1–10. [PMC free article] [PubMed] [Google Scholar]
- 6.Boller K, Konig H, Sauter M, Mueller-Lantzsch N, Lower R, Lower J, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–53. [DOI] [PubMed] [Google Scholar]
- 7.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, et al. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanke K, Chudak C, Kurth R, Bannert N. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer. 2013;132:556–67. [DOI] [PubMed] [Google Scholar]
- 9.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, et al. Variable Transcriptional Activity of Endogenous Retroviruses in Human Breast Cancer. J Virol. 2008;82:1808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, et al. Characterization of Human Endogenous Retroviral Elements in the Blood of HIV-1-Infected Individuals. J Virol. 2012;86:262–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, et al. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–60. [PubMed] [Google Scholar]
- 13.Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-k envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012;104:189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhyu DW, Kang YJ, Ock MS, Eo JW, Choi YH, Kim WJ, et al. Expression of human endogenous retrovirus env genes in the blood of breast cancer patients. Int J Mol Sci. 2014;15:9173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves PMS, Lévy N, Stevenson BJ, Bouzourene H, Theiler G, Bricard G, et al. Identification of tumor-associated antigens by large-scale analysis of genes expressed in human colorectal cancer. Cancer Immun. 2008;8:1–11. [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS One. 2012;7:e29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Kang YJ, Jo JO, Ock MS, Baek KW, Eo J, et al. Elevation of human ERV3-1 env protein expression in colorectal cancer. J Clin Pathol. 2014;67:840–4. [DOI] [PubMed] [Google Scholar]
- 18.Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M, et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong Y, Rose CM, Cass AA, Williams AG, Darwish M, Lianoglou S, et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun. 2019;10:5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolbe AR, Bendall ML, Pearson AT, Paul D, Nixon DF, Pérez-Losada M, et al. Human Endogenous Retrovirus Expression Is Associated with Head and Neck Cancer and Differential Survival. Viruses. 2020;12:956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito J, Kimura I, Soper A, Coudray A, Koyanagi Y, Nakaoka H, et al. Endogenous retroviruses drive KRAB zinc-finger family protein expression for tumor suppression. Sci Adv. 2020;6:eabc3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CC, Beckermann KE, Bortone DS, Cubas AA, Bixby LM, Lee SJ, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128:4804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang-Johanning F, Li M, Esteva FJ, Hess KR, Yin B, Rycaj K, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer. 2014;134:587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Radvanyi L, Yin B, Li J, Chivukula R, Lin K, et al. Downregulation of human endogenous retrovirus type K (HERV-K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 2017;23:5892–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morozov VA, Dao Thi VL, Denner J. The Transmembrane Protein of the Human Endogenous Retrovirus - K (HERV-K) Modulates Cytokine Release and Gene Expression. PLoS One. 2013;8:e70399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topham JT, Titmuss E, Pleasance ED, Williamson LM, Karasinska JM, Culibrk L, et al. Endogenous retrovirus transcript levels are associated with immunogenic signatures in multiple metastatic cancer types. Mol Cancer Ther. 2020;19:1889–97. [DOI] [PubMed] [Google Scholar]
- 28.Bendall ML, De Mulder M, Iñiguez LP, Lecanda-Sánchez A, Pérez-Losada M, Ostrowski MA, et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput Biol. 2019;15:e1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol. 2015;19:A68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Internet]. 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 32.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria; 2017. Available from: https://www.r-project.org/ [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:535–7. [DOI] [PubMed] [Google Scholar]
- 38.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickham H. ggplot2: Elegant Graphics for Data Analysis [Internet]. Springer-Verlag; New York; 2016. Available from: https://ggplot2.tidyverse.org [Google Scholar]
- 41.Blighe K, Rana S, Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling [Internet]. 2020. Available from: https://github.com/kevinblighe/EnhancedVolcano [Google Scholar]
- 42.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolde R. pheatmap : Pretty Heatmaps [Internet]. 2015. Available from: https://cran.r-project.org/package=pheatmap [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thurneau T. survival: A Package for Survival Analysis in R [Internet]. 2020. Available from: https://cran.r-project.org/package=survival [Google Scholar]
- 48.Tang Y, Horikoshi M, Li W. Ggfortify: Unified interface to visualize statistical results of popular r packages. R J. 2016;8.2:478–89. [Google Scholar]
- 49.Abugessaisa I, Shimoji H, Sahin S, Kondo A, Harshbarger J, Lizio M, et al. FANTOM5 transcriptome catalog of cellular states based on Semantic MediaWiki. Database (Oxford). 2016;2016:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westover D, Li F. New trends for overcoming ABCG2/BCRP-mediated resistance to cancer therapies. J Exp Clin Cancer Res. Journal of Experimental & Clinical Cancer Research; 2015;34:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Candeil L, Gourdier I, Peyron D, Vezzio N, Copois V, Bibeau F, et al. ABCG2 overexpression in colon cancer cells resistant to SN38 and in irinotecan-treated metastases. Int J Cancer. 2004;109:848–54. [DOI] [PubMed] [Google Scholar]
- 52.Huang X, Stern DF, Zhao H. Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival - Evidence from TCGA Pan-Cancer Data. Sci Rep. 2016;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibb EA, Warren RL, Wilson GW, Brown SD, Robertson GA, Morin GB, et al. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Surmacz E. Obesity hormone leptin: A new target in breast cancer? Breast Cancer Res. 2007;9:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natoli M, Gallon J, Lu H, Amgheib A, Pinato DJ, Mauri FA, et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J Immunother Cancer. 2021;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H, Ning S, Nolley R, Scicinski J, Oronsky B, Knox SJ, et al. The immunomodulatory anticancer agent, RRx-001, induces an interferon response through epigenetic induction of viral mimicry. Clin Epigenetics. 2017;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.