Abstract
Adoptive cell therapy with virus-specific T cells has been used successfully to treat life-threatening viral infections, supporting application of this approach to coronavirus disease 2019 (COVID-19). We expand severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) T cells from the peripheral blood of COVID-19-recovered donors and non-exposed controls using different culture conditions. We observe that the choice of cytokines modulates the expansion, phenotype, and hierarchy of antigenic recognition by SARS-CoV-2 T cells. Culture with interleukin (IL)-2/4/7, but not under other cytokine-driven conditions, results in more than 1,000-fold expansion in SARS-CoV-2 T cells with a retained phenotype, function, and hierarchy of antigenic recognition compared with baseline (pre-expansion) samples. Expanded cytotoxic T lymphocytes (CTLs) are directed against structural SARS-CoV-2 proteins, including the receptor-binding domain of Spike. SARS-CoV-2 T cells cannot be expanded efficiently from the peripheral blood of non-exposed controls. Because corticosteroids are used for management of severe COVID-19, we propose an efficient strategy to inactivate the glucocorticoid receptor gene (NR3C1) in SARS-CoV-2 CTLs using CRISPR-Cas9 gene editing.
Keywords: SARS-CoV-2, CTL expansion, glucocorticoid receptor, adoptive cell therapy, CRISPR-Cas9, convalescent plasma
Graphical abstract
Basar et al. demonstrate that SARS-CoV-2 T cells can be efficiently generated from COVID19-recovered donors with specificity against multiple structural SARS-CoV-2 proteins, including the Spike protein. Moreover, these cells can be genetically modified to render them resistant to corticosteroids, making their application clinically feasible.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019 marks the third and most devastating large-scale epidemic of coronavirus infection (known as coronavirus disease 2019 [COVID-19]) in recent times. A number of potential treatment options for SARS-CoV-2 are under investigation, including use of convalescent plasma, remdesivir, lopinavir/ritonavir, and interferon-beta (Beigel et al., 2020; Cao et al., 2020; Casadevall and Pirofski, 2020; Hung et al., 2020; ClinicalTrials.gov: NCT04315948). So far, none appear to be curative, making it critical to develop novel therapeutic strategies.
SARS-CoV-2 infection is characterized by profound T-lymphopenia associated with a dysregulated/excessive innate response, thought to be the underlying mechanism of acute respiratory distress syndrome (ARDS), the major cause of morbidity and mortality with this virus (Fathi and Rezaei, 2020; Mehta et al., 2020). Recent studies of individuals with COVID-19 point to an important role of T cell adaptive immunity in protection and clearance of the virus (Braun et al., 2020), with T cell responses documented against the structural SARS-CoV-2 viral proteins Spike (S), Membrane (M), and Nucleocapsid (N) (Grifoni et al., 2020; Ni et al., 2020; Sekine et al., 2020; Thieme et al., 2020). Indeed, in preclinical models of SARS-CoV-1 infection, adoptive transfer of virus-specific T cells (VSTs) has been shown to be curative in infected mice, supporting use of adoptive cell therapy (ACT) in coronavirus-related infection. ACT with allogeneic cytotoxic T lymphocytes (CTLs) has been used successfully to treat other severe viral infections, such as cytomegalovirus (CMV), adenovirus, BK virus (BKV), Epstein-Barr virus, and human herpes virus 6 in immunosuppressed individuals, with responses ranging from 60%–100% (Haque et al., 2007; Muftuoglu et al., 2018; O’Reilly et al., 2016; Tzannou et al., 2017). Thus, ACT may be an attractive approach for management of COVID-19-related disease. However, many individuals with severe COVID-19 receive corticosteroids, which, because of their lymphocytotoxic effects, limit the efficacy of ACT. Here we describe a novel approach for generation of highly functional and steroid-resistant SARS-CoV-2-reactive T cells for immunotherapy of individuals with COVID-19.
Results
Expansion of SARS-CoV-2-reactive T cells from COVID-19-recovered donors
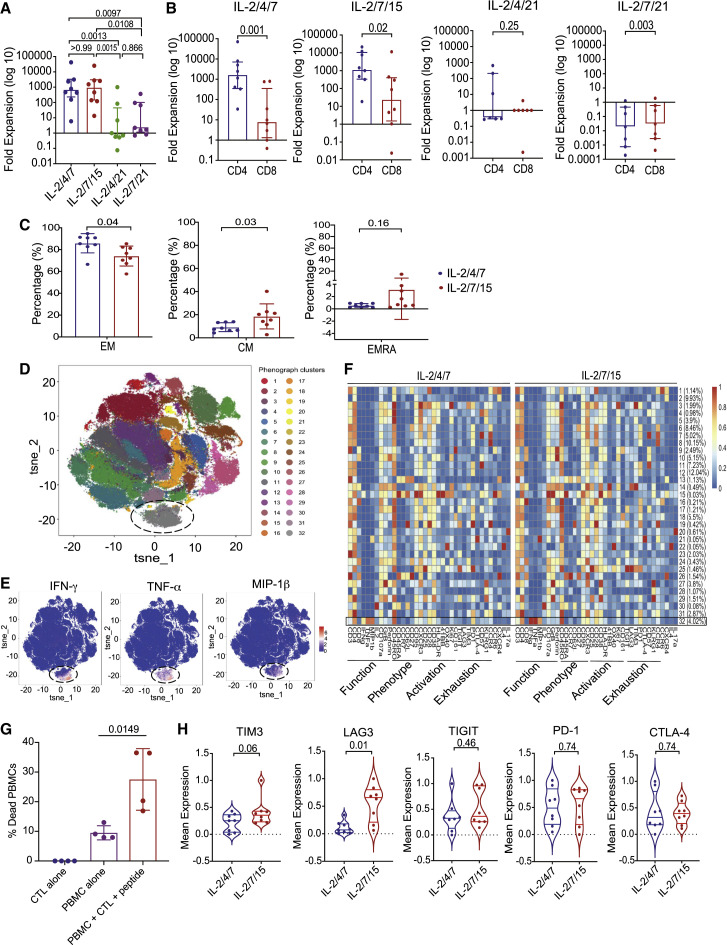
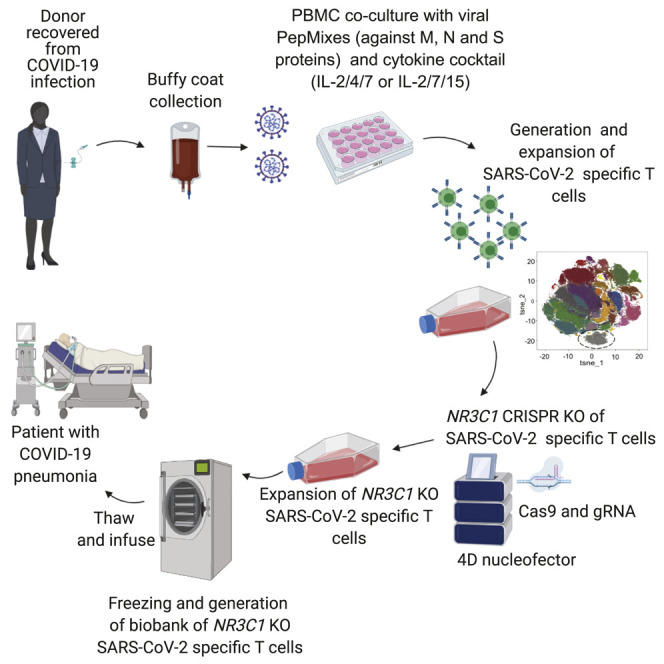
Our group has previously reported the feasibility of generating VSTs from the peripheral blood (PB) of healthy donors for ACT (Muftuoglu et al., 2018). Here we utilized this approach to derive and expand SARS-CoV-2-specific T cells. Briefly, peripheral blood mononuclear cells (PBMCs) from 10 CoV19-recovered donors (RDs) were cultured with 11 different peptide libraries (15-mers overlapping by 11 amino acids) spanning the entire sequence of the SARS-CoV-2 antigens, including the structural (S, M, N, and E) and non-structural (AP3A, Y14, NS6, NS7a, NS7B, NS8, ORF9B, and ORF10) proteins in the presence of interleukin (IL)-2/4/7, IL-2/7/15, IL-2/4/21, or IL-2/7/21 for 14 days. At the end of the culture period, SARS-CoV-2-reactive T cells were enumerated based on their ability to produce interferon-gamma (IFN-γ) in response to ex vivo stimulation with the viral antigens. When cultured in the presence of IL-2/4/7 or IL-2/7/15, expansion was successful in 8 of 10 cases, with a median fold expansion of 719.14 (range, 7.16–45572.50) and 1138.41 (range, 15.97–27716.61), respectively. However, expansion using IL-2/4/21 or IL-2/7/21 was suboptimal, with a median fold expansion of only 0.71 (range, 0.08–996.18) and 2.72 (range, 0.85–415.98), respectively (Figure 1 A; Tables 1 , S2, and S3). IL-2/4/7 and IL-2/7/15 culture conditions supported expansion of CD4+ and CD8+ SARS-CoV-2-specific T cells with a predominance of CD4+ T cells, whereas expansion with IL-2/4/21 and IL-2/7/21 failed to result in significant expansion of SARS-CoV-2 CD4+ or CD8+ T cells (Figure 1B).
Figure 1.
Successful expansion of SARS-CoV-2 T cells from COVID-19-RDs
(A) Bar graph showing the log 10-fold expansion of SARS-CoV-2 T cells cultured with different cytokine cocktails: IL-2/4/7 (blue), IL-2/7/15 (red), IL-2/4/21 (green), and IL-2/7/21 (purple).
(B) Bar graphs showing the log 10-fold expansion of the CD4+ (blue) and CD8+ (red) subsets of SARS-CoV-2 T cells cultured under the different cytokine stimulation conditions.
(C) Quantification of EM (CD45RO+ CD45RA− CD62L−, left panel), CM (CD45RO+ CD45RA− CD62+, center panel), and EMRA (CD45RO− CD45RA+ CD62L−, right panel) cells among SARS-CoV-2 T cells expanded with IL-2/4/7 (blue) or IL-2/7/15 (red) (n = 8 samples per group); bars represent median values with interquartile range. p values are indicated at the top of each graph.
(D) Mass cytometry analysis of T cells (gated on CD45+CD3+) expanded from 8 RDs using a combination of M, N, and S peptide libraries and cytokine cocktails (IL-2/4/7 and IL-27/15 conditions are overlapped in this phenograph). The t-distributed neighbor embedding (tSNE) map shows the 32 clusters obtained, each highlighted in a corresponding color. Cluster 32 (circled) represents polyfunctional SARS-CoV-2 T cells.
(E) Individual tSNE maps showing expression of IFN-γ, TNF-α, and MIP-1β mostly restricted to cluster 32. Expression levels are indicated by a color scale ranging from blue, representing low expression, to red, representing high expression.
(F) Cluster identity and frequency are summarized in heatmaps showing marker expression levels (x axis) for T cell populations (y axis) expanded with the two different cytokine cocktails IL-2/4/7 (left heatmap) or IL-2/7/15 (right heatmap). Markers associated with function, phenotype, activation, or exhaustion are indicated below each heatmap. Expression level is indicated by a color scale ranging from low (blue) to high (red). Cluster 32 is indicated with a black rectangle.
(G) Bar graph showing the percentage of dead (Annexin V+ and live/dead+) cells in autologous PBMCs pre-loaded with SARS-CoV-2 PepMix (M, N, and S proteins) cultured alone (purple) or in the presence of expanded CTLs (red) for 16 h at a 1:1 ratio. The percentage of dead cells in CTLs cultured alone (blue) is also plotted as a negative control.
(H) Violin plots comparing expression of TIM3, LAG3, T cell immunoreceptor with Ig and ITIM domains (TIGIT), PD-1, and CTLA-4 and between the two cytokine stimulation conditions IL-2/4/7 (blue) and IL-2/7/15 (red); n = 8 samples per group. p values are indicated at the top of each graph.
The bars represent median values with interquartile ranges. Statistical analysis by one-way ANOVA with Tukey’s correction for multiple comparisons (A), paired t test (B, C, and G), or Wilcoxon signed-rank test (H).
Table 1.
Cytokine production of SARS-CoV-2 T cells from RDs expanded with different cytokine cocktails against the M, N, and S structural proteins
| CD3% |
CD4% |
CD8% |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M | N | S | M | N | S | M | N | S | ||
| IL-2/4/7 | IFN-γ median | 4.27 | 4.98 | 10.60 | 4.24 | 5.88 | 10.65 | 2.42 | 4.81 | 0.82 |
| IFN-γ min | 0.11 | 0.12 | 0.21 | 0.14 | 0.14 | 0.22 | 0.10 | 0.05 | 0.25 | |
| IFN-γ max | 33.60 | 17.10 | 14.80 | 33.40 | 18.90 | 20.10 | 42.00 | 19.40 | 3.99 | |
| IL-2 median | 0.68 | 1.01 | 0.96 | 0.41 | 0.70 | 0.90 | 0.42 | 0.42 | 0.27 | |
| IL-2 min | 0.14 | 0.23 | 0.20 | 0.14 | 0.18 | 0.16 | 0.00 | 0.16 | 0.18 | |
| IL-2 max | 1.41 | 1.85 | 2.31 | 1.44 | 2.09 | 1.82 | 1.20 | 0.71 | 0.79 | |
| TNF-α median | 2.80 | 2.35 | 3.99 | 2.64 | 2.09 | 4.59 | 1.45 | 0.97 | 0.61 | |
| TNF-α min | 0.19 | 0.22 | 0.25 | 0.05 | 0.04 | 0.07 | 0.10 | 0.00 | 0.09 | |
| TNF-α max | 12.90 | 7.50 | 7.86 | 12.90 | 8.41 | 7.90 | 5.65 | 3.70 | 2.42 | |
| IL-2/7/15 | IFN-γ median | 6.05 | 3.60 | 4.47 | 6.00 | 3.61 | 4.55 | 0.33 | 3.13 | 1.15 |
| IFN-γ min | 0.15 | 0.17 | 0.48 | 0.13 | 0.21 | 0.42 | 0.09 | 0.04 | 0.12 | |
| IFN-γ max | 20.80 | 15.50 | 26.00 | 34.20 | 16.10 | 29.40 | 47.30 | 20.50 | 2.65 | |
| IL-2 median | 1.03 | 0.53 | 0.79 | 0.68 | 0.58 | 0.80 | 0.37 | 0.49 | 0.41 | |
| IL-2 min | 0.15 | 0.32 | 0.31 | 0.20 | 0.28 | 0.22 | 0.20 | 0.23 | 0.27 | |
| IL-2 max | 2.55 | 1.56 | 6.59 | 1.64 | 0.91 | 6.52 | 2.24 | 1.16 | 1.75 | |
| TNF-α median | 4.18 | 1.70 | 2.37 | 3.90 | 1.46 | 2.74 | 0.69 | 1.29 | 0.76 | |
| TNF-α min | 0.24 | 0.38 | 0.78 | 0.10 | 0.48 | 0.33 | 0.00 | 0.10 | 0.24 | |
| TNF-α max | 7.06 | 4.20 | 10.10 | 12.60 | 4.50 | 11.20 | 18.50 | 7.27 | 3.61 | |
| IL-2/4/21 | IFN-γ median | 0.30 | 0.20 | 0.36 | 0.51 | 0.29 | 0.16 | 0.00 | 0.00 | 0.00 |
| IFN-γ min | 0.03 | 0.14 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| IFN-γ max | 2.46 | 0.64 | 2.53 | 4.99 | 1.08 | 3.63 | 3.37 | 0.22 | 0.97 | |
| IL-2 median | 0.16 | 0.21 | 0.16 | 0.13 | 0.20 | 0.16 | 0.00 | 0.00 | 0.00 | |
| IL-2 min | 0.10 | 0.09 | 0.09 | 0.00 | 0.09 | 0.06 | 0.00 | 0.00 | 0.00 | |
| IL-2 max | 0.40 | 0.84 | 1.02 | 0.52 | 0.33 | 0.81 | 0.24 | 0.48 | 2.17 | |
| TNF-α median | 0.40 | 0.15 | 0.14 | 0.34 | 0.20 | 0.14 | 0.00 | 0.00 | 0.00 | |
| TNF-α min | 0.10 | 0.09 | 0.00 | 0.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| TNF-α max | 0.93 | 0.47 | 1.66 | 1.95 | 0.50 | 2.36 | 3.70 | 7.69 | 0.00 | |
| IL-2/7/21 | IFN-γ median | 0.74 | 0.25 | 0.29 | 1.27 | 0.16 | 0.18 | 0.00 | 0.33 | 0.41 |
| IFN-γ min | 0.14 | 0.06 | 0.11 | 0.07 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| IFN-γ max | 0.97 | 0.38 | 0.50 | 2.93 | 0.81 | 1.36 | 1.52 | 5.82 | 1.11 | |
| IL-2 median | 0.09 | 0.07 | 0.11 | 0.20 | 0.22 | 0.14 | 0.10 | 0.13 | 0.34 | |
| IL-2 min | 0.03 | 0.05 | 0.07 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| IL-2 max | 0.73 | 0.66 | 0.31 | 1.27 | 1.10 | 0.57 | 1.75 | 0.60 | 0.83 | |
| TNF-α median | 0.19 | 0.19 | 0.20 | 0.45 | 0.33 | 0.59 | 0.39 | 0.53 | 0.10 | |
| TNF-α min | 0.13 | 0.07 | 0.14 | 0.17 | 0.19 | 0.18 | 0.00 | 0.00 | 0.00 | |
| TNF-α max | 1.70 | 0.31 | 0.26 | 2.55 | 1.01 | 2.58 | 0.94 | 0.72 | 0.93 | |
Shown are percent IFN-γ, IL-2, and TNF-α production (median, minimum [min], and maximum [max] values) from the CD3+, CD4+, and CD8+ compartments of SARS-CoV-2 T cells stimulated with the peptide libraries derived from the M, N, and S structural proteins after expansion under the different culture conditions with the four different cytokine cocktails IL-2/4/7, IL-2/7/15, IL-2/4/21, and IL-2/7/21.
SARS-CoV-2-reactive T cells generated from CoV-RDs are polyfunctional
We next interrogated the functional phenotype of the ex-vivo-expanded SARS-CoV-2 CTLs. Because IL-2/4/7 and IL-2/7/15 resulted in the best cell expansion, we focused our analysis on SARS-CoV-2 CTLs generated using these two conditions. Both culture conditions supported expansion of effector memory (EM) and central memory (CM) T cells, although use of IL-2/7/15 resulted in expansion of CM T cells (Figure 1C).
Previous studies of individuals with severe COVID-19 have reported the presence of T cells with an exhausted phenotype and reduced polyfunctionality (Chen et al., 2020; Zheng et al., 2020a, 2020b). Thus, we performed a comprehensive single-cell analysis of expanded SARS-CoV-2 CTLs from 8 RDs using mass cytometry. Phenotypic assessment of SARS-CoV-2-reactive T cells expanded with IL-2/4/7 or IL-2/7/15 (identified based on their ability to produce IFN-γ in response to ex vivo stimulation with a mixture of S, M, and N peptide libraries) revealed that SARS-CoV-2-specific CTLs are polyfunctional based on their ability to secrete multiple cytokines, including IFN-γ, tumor necrosis factor alpha (TNF-α), and MIP-1β (cluster 32; Figures 1D and 1E) and chemokine receptors that support trafficking to the lungs, such as CXCR3 and CCR4 (Figure S1). Moreover, ex-vivo-expanded SARS-CoV-2 CTLs did not express high levels of inhibitory/checkpoint molecules, arguing against an exhausted phenotype (cluster 32; Figure 1F). Indeed, analysis of functional markers revealed a cytotoxic Th1 phenotype, characterized by expression of IFN-γ, TNF-α, CD107a, and granzyme B (GrB) (Figure 1F) and the ability to kill SARS-CoV-2 PepMix-loaded autologous PBMCs by Annexin V killing assay (Figure 1G). These data indicate a direct antiviral killing capacity of our CTLs, although we are unable to discriminate between TNF-α- or perforin-dependent killing, given the longer duration of the cytotoxicity assay used in our experiment. SARS-CoV-2 CTLs did not produce significant amounts of IL-2 in response to antigenic stimulation. Single-cell phenotypic comparison of SARS-CoV-2 CTLs expanded using the two different culture conditions did not reveal major differences in the expression patterns of activation and functional markers between these two groups (Figure 1F). However, cells expanded in the presence of IL-2/4/7 expressed lower levels of some exhaustion markers, such as TIM3 and LAG3, compared with cells expanded with IL-2/7/15 (Figure 1H). These data support the notion that polyfunctional, non-exhausted T cells capable of reacting against SARS-CoV-2 antigens can be expanded from the PB of CoV-RDs.
We also performed a multiplex analysis to measure cytokines in supernatants collected from cultures of SARS-CoV-2 CTLs with SARS-CoV-2 antigens (n = 4 samples for each of the culture conditions: IL-2/4/7 and IL-2/7/15). As expected, the expanded SARS-CoV-2 CTLs released effector cytokines such as IFN-γ, TNF-α, and MIP-1β in response to antigenic stimulation. Notably, they did not produce cytokines, such as IL-6, IL-1α, or IL-10, that could contribute to a higher risk of toxicity or cytokine release syndrome (CRS) (Figure S2).
Expanded SARS-CoV-2 CTLs from CoV-RDs are directed against structural proteins, including the C and N termini of the S protein
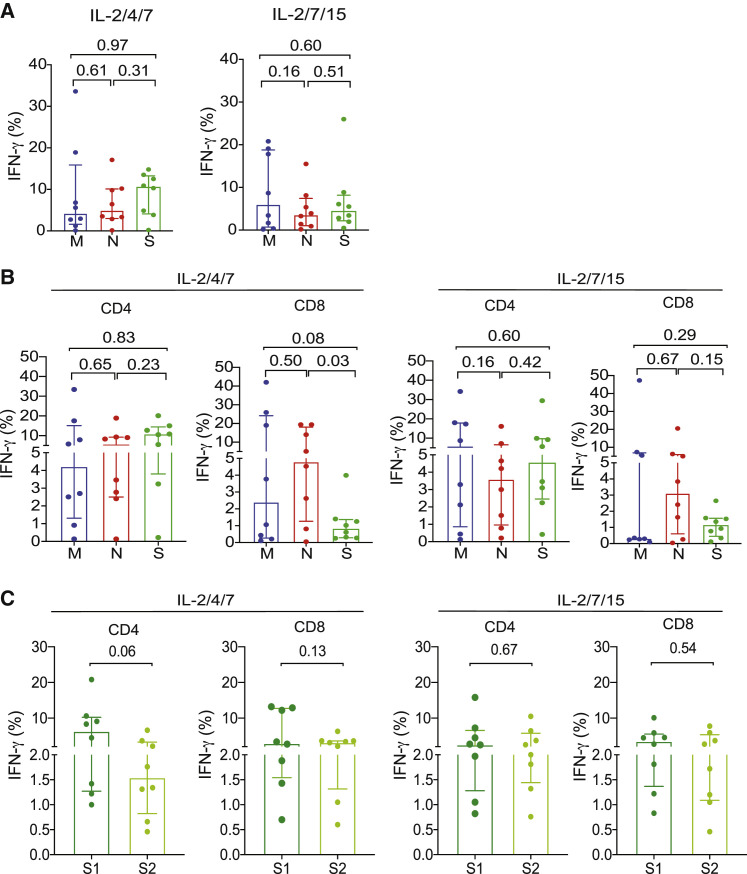
To identify the dominant antigen(s) driving expansion of SARS-CoV-2 CTLs, the expanded cells were stimulated ex vivo with peptide libraries derived from M, N, S, or E (structural proteins) or AP3A, Y14, NS6, NS7a, NS7B, NS8, ORF9B, or ORF10 (non-structural proteins). Analysis of IFN-γ production showed that, for the CTL lines expanded with IL-2/4/7, the antiviral response was mostly directed against S (median, 10.60%; range, 0.21%–4.8%), with the remaining cells responding to M (median, 4.27%; range, 0.11%–33.6%) or N (median, 4.98%; range, 0.12%–17.10%) (Figure 2 A). For the lines expanded with IL-2/7/15, the antiviral response favored M (median, 6.05%; range, 0.15%–20.80%), followed by S (median, 4.47%; range, 0.48%–26.00%) and N (median, 3.60%; range, 0.17%–15.50%) (Figure 2A). In sum, for the IL-2/4/7 and IL-2/7/15 culture settings, seven lines were directed against M, N, and S; one line was directed against S and N; and no line reacted to M and N in the absence of S. There was no significant expansion of CTLs in response to the non-structural proteins or the structural E protein.
Figure 2.
Expanded SARS-CoV-2 CTLs are directed against structural proteins, including the C and N termini of the S protein
(A) Bar graphs showing the percentage of total IFN-γ (+) SARS-CoV-2 CD3+ T cells stimulated with the peptide libraries derived from the different structural proteins M (blue), N (red), and S (green) cultured with different cytokine cocktails: IL-2/4/7 (left panel) or IL-2/7/15 (right panel).
(B) Bar graphs showing IFN-γ (+) expression in CD4+ and CD8+ T cell subsets of SARS-CoV-2 T cells stimulated with the peptide libraries derived from the different structural proteins M (blue), N (red), and S (green) cultured with IL-2/4/7 (left panels) or IL-2/7/15 (right panels).
(C) Quantification of IFN-γ (+) SARS-CoV-2 T cells subsets (CD4+ or CD8+) directed against the N terminus (S1, dark green) or C terminus (S2, light green) of the S protein under IL-2/4/7 (left panels) and IL-2/7/15 (right panels) stimulation conditions (n = 8 samples per group).
The bars represent median values with interquartile range. p values are indicated at the top of each graph. Statistical analysis by paired t test.
When we considered the CD4+ and CD8+ T cell responses separately, we found that the response of CD4+ T cells to individual SARS-CoV-2 antigens followed a pattern similar to that observed in the overall CD3+ T cell population. Interestingly, however, CD8+ T cell responses were mostly directed against the N protein, irrespective of the cytokine cocktail used for SARS-CoV-2 CTL expansion (Figure 2B; Table 1).
Peptides derived from the C terminus of the S protein have higher homology with the S glycoprotein of human endemic “common cold” coronaviruses; in contrast, the N terminus of the S protein includes peptides from the receptor-binding domain (the target of neutralizing antibodies) that are more specific to SARS-CoV-2 (Braun et al., 2020; Walls et al., 2020). Our expanded CD4+ and CD8+ SARS-CoV-2 CTLs were capable of reacting to epitopes found on the N or C terminus or the S protein (peptide pools 1 and 2, respectively), indicating their specificity for the receptor binding domain (RBD) of SARS-CoV-2 (Figure 2C; Tables 2 , S4, and S5).
Table 2.
Cytokine production of SARS-CoV-2 T cells from RDs expanded with different cytokine cocktails against S1 and S2 (N and C termini of the S protein)
| CD3% |
CD4% |
CD8% |
|||||
|---|---|---|---|---|---|---|---|
| S1 | S2 | S1 | S2 | S1 | S2 | ||
| IL-2/4/7 | IFN-γ median | 5.81 | 3.88 | 6.40 | 1.55 | 3.02 | 3.26 |
| IFN-γ min | 1.33 | 0.71 | 1.00 | 0.46 | 0.70 | 0.60 | |
| IFN-γ max | 26.50 | 8.54 | 20.80 | 6.60 | 13.20 | 6.29 | |
| IL-2 median | 0.53 | 0.51 | 0.41 | 0.38 | 1.78 | 2.87 | |
| IL-2 min | 0.14 | 0.24 | 0.06 | 0.07 | 0.00 | 0.15 | |
| IL-2 max | 5.03 | 4.48 | 2.55 | 2.72 | 6.15 | 6.47 | |
| TNF-α median | 3.24 | 1.41 | 2.56 | 0.87 | 5.51 | 1.80 | |
| TNF-α min | 0.45 | 0.72 | 0.33 | 0.51 | 0.00 | 0.00 | |
| TNF-α max | 9.47 | 2.63 | 12.80 | 1.60 | 27.30 | 14.70 | |
| IL-2/7/15 | IFN-γ median | 5.94 | 2.99 | 2.61 | 2.27 | 3.59 | 2.22 |
| IFN-γ min | 1.73 | 1.61 | 0.82 | 0.76 | 0.83 | 0.46 | |
| IFN-γ max | 18.00 | 14.60 | 15.80 | 10.50 | 10.10 | 7.77 | |
| IL-2 median | 0.80 | 0.90 | 0.22 | 0.27 | 3.03 | 1.87 | |
| IL-2 min | 0.12 | 0.05 | 0.05 | 0.03 | 0.14 | 0.40 | |
| IL-2 max | 4.37 | 4.97 | 3.02 | 2.34 | 4.13 | 4.48 | |
| TNF-α median | 2.20 | 2.02 | 2.62 | 1.36 | 10.27 | 11.38 | |
| TNF-α min | 1.57 | 0.95 | 0.84 | 0.38 | 0.36 | 0.00 | |
| TNF-α max | 10.50 | 4.14 | 13.90 | 3.05 | 26.20 | 22.60 | |
| IL-2/4/21 | IFN-γ median | 0.17 | 0.14 | 0.14 | 0.22 | 0.00 | 0.00 |
| IFN-γ min | 0.03 | 0.08 | 0.00 | 0.05 | 0.00 | 0.00 | |
| IFN-γ max | 4.15 | 0.26 | 6.21 | 0.60 | 0.67 | 0.00 | |
| IL-2 median | 0.17 | 0.16 | 0.14 | 0.16 | 0.00 | 0.00 | |
| IL-2 min | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | |
| IL-2 max | 0.40 | 0.57 | 0.68 | 0.44 | 0.66 | 0.61 | |
| TNF-α median | 0.23 | 0.14 | 0.29 | 0.18 | 0.00 | 0.00 | |
| TNF-α min | 0.07 | 0.05 | 0.08 | 0.00 | 0.00 | 0.00 | |
| TNF-α max | 1.78 | 0.72 | 2.69 | 0.30 | 0.00 | 1.59 | |
| IL-2/7/21 | IFN-γ median | 0.20 | 0.12 | 0.23 | 0.10 | 0.43 | 0.00 |
| IFN-γ min | 0.05 | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | |
| IFN-γ max | 0.89 | 0.63 | 2.22 | 1.24 | 0.63 | 0.47 | |
| IL-2 median | 0.08 | 0.08 | 0.17 | 0.39 | 0.00 | 0.35 | |
| IL-2 min | 0.06 | 0.04 | 0.09 | 0.11 | 0.00 | 0.00 | |
| IL-2 max | 0.11 | 0.21 | 0.37 | 1.50 | 0.54 | 0.87 | |
| TNF-α median | 0.20 | 0.09 | 0.35 | 0.21 | 0.00 | 0.00 | |
| TNF-α min | 0.08 | 0.06 | 0.20 | 0.07 | 0.00 | 0.00 | |
| TNF-α max | 0.37 | 0.31 | 1.18 | 1.50 | 0.63 | 0.83 | |
Shown are percent IFN-γ, IL-2, and TNF-α production (median, minimum [min], and maximum [max] values) from the CD3+, CD4+, and CD8+ compartments of SARS-CoV-2 T cells stimulated with the peptide libraries derived from S1 and S2 (N and C-terminals of the S protein) after expansion under different cytokine stimulation conditions (IL-2/4/7, IL-2/7/15, IL-2/4/21 and IL-2/7/21).
Culture with IL-2/4/7 results in preferential expansion of T cells against the N terminus of the S protein
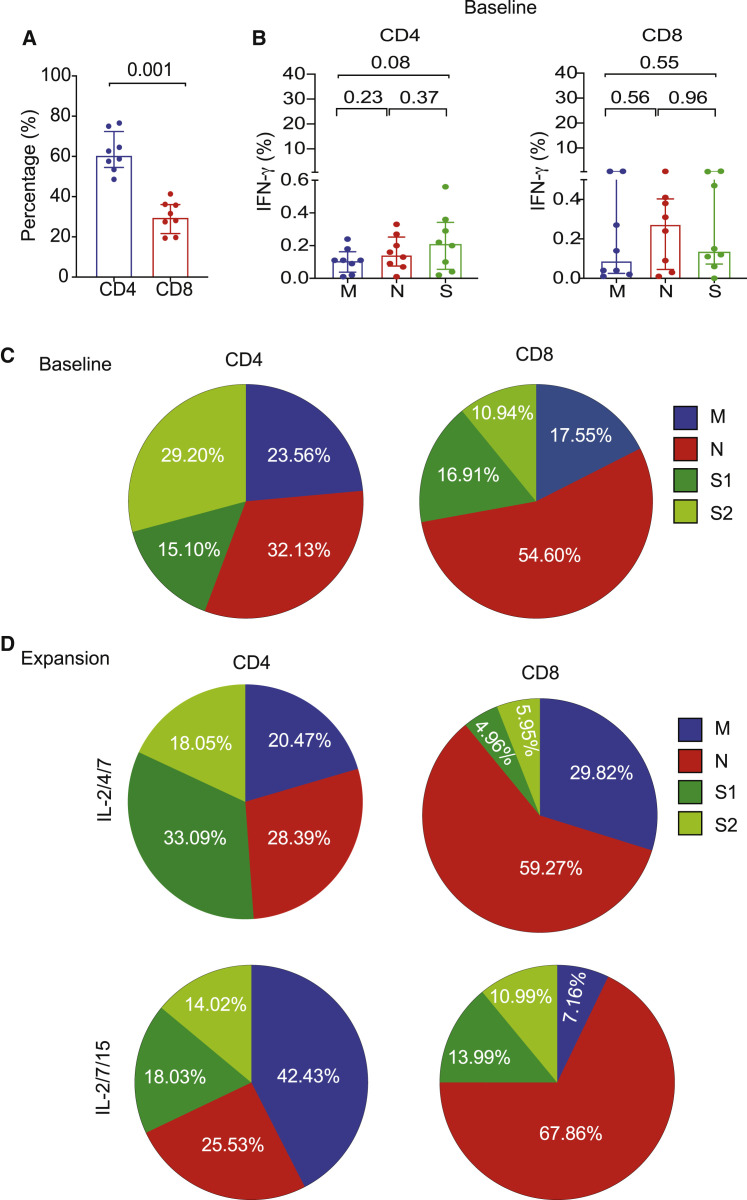
Because the choice of cytokines used to expand SARS-CoV-2 T cells modulates the hierarchy of antigenic response, we compared the T cell response against S (S1 and S2), M, and N proteins of SARS-CoV-2 in PB samples at baseline (prior to expansion) with that observed in paired expanded CTLs from CoV-RDs.
At baseline (pre-expansion), the responses were mostly CD4 dominant (Figure 3 A) and directed against the S protein (median, 0.21%; range, 0.02%–0.56%), followed by N (median, 0.15%; range, 0.01%–0.33%) and M (median, 0.11%; range, 0.01%–0.24%) (Figure 3B), indicating an immunogenic dominance for S protein human leukocyte antigen (HLA) class II epitopes. We did not detect measurable responses against non-structural proteins (data not shown). Following expansion, culture with IL-2/4/7 maintained the hierarchy of CD4+ T cell response toward the S protein (Figures 2B, 3C, and 3D), with a greater proportion of T cells directed against S1, whereas culture with IL-2/7/15 favored a response toward the M protein (Figures 2B, 3C, and 3D). Expansion with IL-2/4/21 and IL-2/7/21 yielded very low numbers of CTLs, most of which were CD4+. Interestingly, the pattern of antigenic response for cells cultured using these two conditions differed from that observed for IL-2/4/7 and was similar to IL-2/7/15 in that the majority of CD4+ CTLs reacted instead to the M and N proteins (Figure S3A). For CTLs that did show a response to the S protein, further analysis detected no particular pattern of reactivity to either terminus of the protein (Figure S3B).
Figure 3.
Pattern of antigenic responses after expansion of SARS-CoV-2 T cells from COVID-19-RDs compared with baseline
(A) Bar graph showing the percentages of CD4+ and CD8+ subsets of SARS-CoV-2 T cells at baseline.
(B) Graphical analysis showing the percentages of IFN-γ (+) SARS-CoV-2 T cells from RDs at baseline in the CD4+ compartment (left panel) or CD8+ compartment (right panel) when stimulated with the peptide libraries derived from the different structural proteins M (blue), N (red), and S (green).
(C and D) Pie charts illustrating the percent distribution of IFN-γ (+) CD4 and CD8 T cells reactive to M (blue), N (red), S1 (dark green), and S2 (light green) peptide libraries at baseline (C) or following expansion with IL-2/4/7 (top panel) or IL-2/7/15 (bottom panel) cytokine cocktails (D).
The bars represent median values with interquartile ranges. Statistical analysis by paired t test (A and B).
Furthermore, we found a significant correlation (p = 0.002, R2 = 0.82) between the S protein immunoglobulin G (IgG) antibody titer measured in plasma from RDs and the absolute number of SARS-CoV-2-specific T cells following expansion with IL-2/4/7 but not with the IL-2/7/15 cytokine cocktail (Figure S4A). However, there was no correlation between the S protein IgG antibody titer and the absolute number of SARS-CoV-2 T cells prior to ex vivo expansion at baseline (p = 0.52, R2 = 0.07) (Figure S4B). Nonetheless, we recognize that the correlation observed following expansion could be skewed by data from a single individual who represents an outlier and that further studies with larger cohorts are needed to confirm this trend.
These data indicate that antigenic skewing can be driven by the immunodominance of the protein and by the culture conditions and support use of IL-2/4/7 for expansion of SARS-CoV-2 T cells for clinical use.
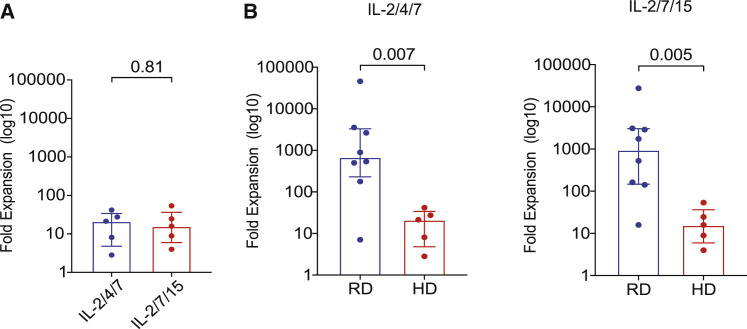
SARS-CoV-2 T cells can be expanded from the PB of healthy donors but at lower frequencies than for CoV-RDs
Recent reports indicate the presence of SARS-CoV-2 T cells in the PB of healthy donors (HDs) not exposed to COVID-19 (Braun et al., 2020; Pia, 2020). Thus, we wanted to find out whether SARS-CoV-2 T cells can be expanded from the PB of HDs and whether they have a similar pattern of SARS-CoV-2 recognition as those generated from CoV-RDs. PBMCs from 5 HDs were expanded using the same protocol as for those from CoV-RDs. We achieved only modest expansion of CTLs recognizing SARS-CoV-2 antigens over a 14-day culture period, with a median 20.37-fold increase (range, 2.85–41.84) for the IL-2/4/7 culture condition and 21.49-fold increase (range, 4.00–53.95) for the IL-2/7/15 culture condition (Figure 4 A). The frequencies of SARS-CoV-2 CTLs from the PB of HDs after 14 days of culture were significantly lower than those achieved with PB from CoV-RDs (Figure 4B). At baseline (pre-expansion), assessment of IFN-γ production and CTL frequencies suggested that, in HDs, responses were directed mostly against the N protein (median, 0.13%; range, 0.03%–0.37%), followed by S (median, 0.10%; range, 0.04%–0.12%) and M (median, 0.09%; range, 0.01%–0.75%) (Figure S5A). Culture in the presence of either cytokine cocktail could skew the response toward S, albeit at much lower frequencies than that observed with CoV-RDs (Figure S5B; Tables S6 and S7). We did not find any particular pattern of antigenic response in the CD4 and CD8 compartments (Figure S5B; Tables S6 and S7). Expansion was not successful under IL-2/4/21 or IL-2/7/21 stimulation conditions. Thus, the different patterns of antigen recognition and the weaker expansion of SARS-CoV-2 T cells from HDs support use of buffy coats from CoV-RDs as starting material for generation of SARS-CoV-2-reactive T cells for ACT.
Figure 4.
SARS-CoV-2 CTLs can be expanded from the PB of HDs but at lower frequencies compared with RDs
(A) Graphical representation of the log 10-fold expansion of SARS-CoV-2 T cells derived from HDs cultured with different cytokine cocktails: IL-2/4/7 (blue) and IL-2/7/15 (red).
(B) Comparison of SARS-CoV-2 T cell expansion between recovered donors (RDs; blue) and healthy donors (HDs; red) cultured under the different cytokine stimulation conditions IL-2/4/7 (left panel) and IL-2/7/15 (right panel).
The bars represent median values with interquartile ranges. Statistical analysis by paired t test (A) or unpaired t test (B).
Expanded SARS-CoV-2 T cells can be genetically modified to render them steroid resistant
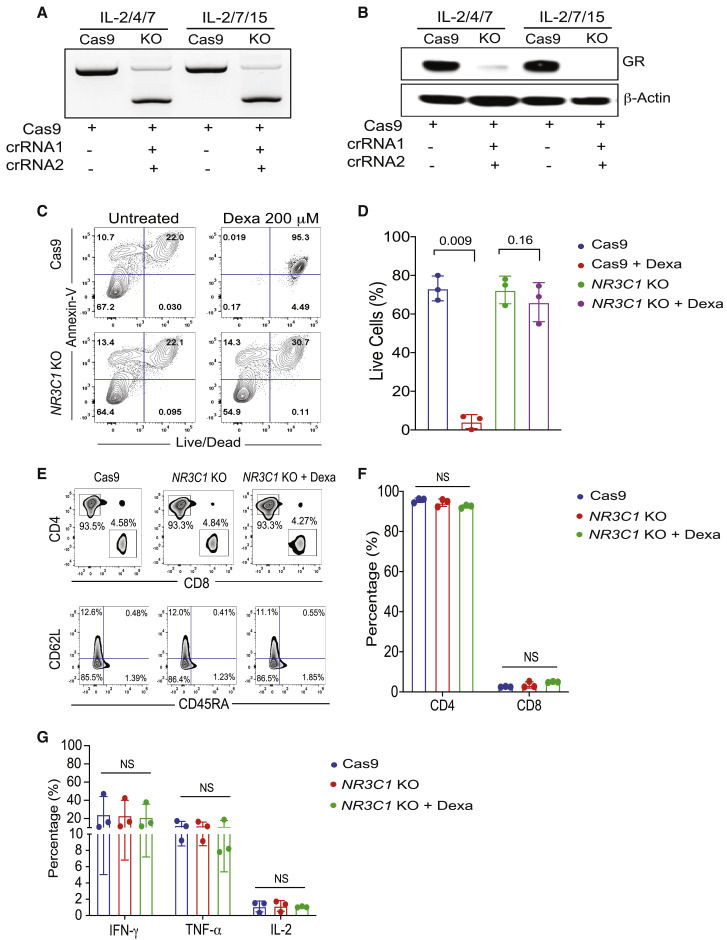
Corticosteroids are used for treatment of individuals with COVID-19-related ARDS to reduce mortality associated with this condition. SARS-CoV-2-specific T cell therapy is not an option in these individuals because corticosteroids induce apoptosis of adoptively transferred T cells, significantly limiting the efficacy of this approach. To address this challenge, we used CRISPR-Cas9 gene editing to knock out the glucocorticoid receptor gene (nuclear receptor subfamily 3 group C member 1 [NR3C1]) in SARS-CoV-2 CTLs and confirmed high efficiency of deletion (>90%), as determined by PCR and western blot analysis (Figures 5A and 5B). An Annexin V apoptosis assay confirmed that the viability of NR3C1 knockout (KO) CTLs treated with dexamethasone was similar to that of control CTLs (defined as CTLs electroporated with Cas9 alone) (Figures 5C and 5D). Moreover, NR3C1 KO SARS-CoV-2 CTLs maintained a similar phenotype and distribution of CD4+ and CD8+ T cell subsets compared with control SARS-CoV-2 CTLs and retained their effector functions (Figures 5E–5G).
Figure 5.
Expanded SARS-CoV-2 CTLs can be genetically modified to become steroid resistant
(A and B) NR3C1 KO efficiency shown by PCR gel electrophoresis (A) and western blot (B) in SARS-CoV-2 CTLs expanded with IL-2/4/7 or IL-2/7/15, after electroporation with Cas9 alone or Cas9 complexed with crRNA 1 and crRNA 2 targeting exon 2 of the NR3C1 gene. SARS-CoV-2 CTLs electroporated with Cas9 alone were used as controls. β-Actin was used as loading control for the western blot.
(C) Representative fluorescence-activated cell sorting (FACS) plots showing the percentage of apoptotic cells (Annexin V+) and live or dead cells (live/dead stain) in control Cas9 versus NR3C1 KO SARS-CoV-2 CTLs after culture with or without dexamethasone (Dexa; 200 μM) for 72 h. Inset values indicate the percentage of Annexin V and live/dead cells from each group.
(D) Bar graph summarizing the percentage of live cells between control Cas9 and NR3C1 KO SARS-CoV-2 CTLs treated with or without 200 μM Dexa for 72 h (n = 3 samples). Bars represent median values with interquartile range. p values are indicated above the graphs.
(E) Representative FACs plots showing the distribution of CD4+ and CD8+ T cells (top panel) and phenotype based on CD62L and CD45RA expression (bottom panel) in Cas9-only or NR3C1 KO SARS-CoV-2 CTLs with or without 200 μM Dexa.
(F) Percentage of CD4+ and CD8+ T cells within SARS-CoV-2 CTLs treated with control Cas9 (blue), NR3C1 KO (red), or NR3C1 KO plus Dexa (200 μM, green).
(G) Frequency of SARS-CoV-2 CTLs producing IFN-γ, TNF-α, or IL-2 in control Cas9 (blue), NR3C1 KO (red), or NR3C1 KO plus Dexa (200 μM, green) in response to 6 h of stimulation with viral PepMix (n = 3 samples). Functional analysis of the Cas9 + Dexa group was not performed because of the absence of viable cells resulting from the lymphocytotoxic effect of steroids.
The bars represent mean values with SD. NS, not significant. Statistical analysis by paired t test (D) or two-way ANOVA with Tukey’s correction for multiple comparisons (F and G).
Discussion
Here we show that large numbers of SARS-CoV-2 T cells can be generated from buffy coats of convalescent individuals with specificity directed against multiple structural proteins of this virus, including the RBD of the S protein. These cells can be genetically modified to render them resistant to the lymphocytotoxic effect of corticosteroids, making their application clinically feasible.
We performed single-cell analysis of ex-vivo-expanded SARS-CoV-2 CTLs to better understand their phenotypic and functional properties. SARS-CoV-2 T cells were classified based on their state of differentiation into naive, CM, EM, or terminally differentiated EM (TEMRA) cells. SARS-CoV-2 T cells were CD4+ dominant, comprising mostly EM and CM cells. The effector phenotype predicts the capacity to mediate early viral clearance, whereas CM T cells persist and provide long-term immunity after adoptive transfer (Powell et al., 2005). Indeed, our group and others have confirmed that adoptive infusion of VSTs with a predominantly CD4+ EM phenotype results in successful eradication of severe viral infections such as JCV, BKV, and adenovirus (Heslop et al., 2010; Leen et al., 2010; Muftuoglu et al., 2018; Olson et al., 2021). We also investigated the functional state of SARS-CoV-2 T cells based on their ability to produce one or more cytokines in response to ex vivo stimulation with SARS-CoV-2 antigens. Polyfunctionality is defined as production of multiple cytokines by T cells and is associated with protective immune responses to viruses and vaccines (Minton, 2014). We confirmed that SARS-CoV-2 T cells were polyfunctional with a predominantly Th1 phenotype and capable of killing antigen-loaded targets.
T cells derived from individuals with severe COVID-19 have been reported to express multiple inhibitory molecules (Song et al., 2020), raising concerns that, following ex vivo expansion, they may have an exhausted phenotype with poor effector function and replicative senescence. In our study, T cells from COVID-19-recovered individuals did not have an exhaustion phenotypic signature following in vitro expansion and retained their functional phenotype.
SARS-CoV-2 neutralizing antibodies are directed against the RBD within the N terminus of the S glycoprotein. Peptides derived from this region of the protein are believed to be specific to SARS-CoV-2, whereas peptides from the C terminus are shared with other betacoronaviruses (Braun et al., 2020). We showed that expanded SARS-CoV-2 T cells from CoV-RDs were capable of recognizing epitopes from the N and C terminus of the S glycoprotein, indicating specificity for the SARS-CoV-2 virus. In addition, expanded CTLs reacted against N and M proteins, which are reportedly also shared among different betacoronaviruses (Patrick et al., 2006). The specific antigens that drive an effective and protective T cell response against SARS-CoV-2 are not yet known. They may be proteins that are shared with other betacoronaviruses, or they may be unique to SARS-CoV-2 (e.g., RBD) or may likely be a combination of both.
Cytokines can modulate the phenotype of T cells by activating different signaling pathways. We tested different combinations of cytokines, including IL-2, IL-4, IL-7, IL-15, and IL-21 to identify the optimal conditions to promote expansion of SARS-CoV-2 T cells with a memory phenotype and without evidence of exhaustion. Although IL-2, IL-4, IL-7, and IL-15 support expansion of central and EM T cells (Clénet et al., 2017; Geginat et al., 2001), with IL-2, IL-7, and IL-15 driving proliferation through STAT5 signaling (Drake et al., 2016), in vitro culture with IL-21 facilitates generation of memory stem-like T cells (Chen et al., 2018). Because CD4 and CD8 T cells are involved in successful antiviral response, we also investigated whether cytokines would support expansion of both subsets. We observed that cocktails including IL-2/4/7 or IL-2/7/15 resulted in expansion of clinically relevant doses of polyfunctional SARS-CoV-2 T cells with a CM and EM phenotype. Interestingly, the combination of IL-2/4/7 preferentially supported expansion of T cells against S and, in particular, the RBD-containing S1 region of the protein, making this the cytokine cocktail of choice for production of SARS-CoV-2 T cells for clinical use. Inclusion of IL-21 in the cytokine cocktail resulted in poor expansion of SARS-CoV-2 T cells, in line with previous studies reporting that the effect of IL-21 to augment the antigen-specific T cell response is limited to the naive and not the memory T cell population (Li et al., 2005).
CRS is a major complication of COVID-19 (Moore and June, 2020) and is caused by production of inflammatory cytokines such as IL-6 by virus-infected myeloid cells. The inflammatory milieu overstimulates cells of the innate and adaptive immune system, which, in turn, contributes to the observed cytokine storm (Kang et al., 2019). Therefore, a legitimate concern with our approach is that the adoptively infused SARS-CoV-2 T cells, which also include a small percentage of Th2 and Th17 subsets, could amplify CRS and worsen the condition (Hotez et al., 2020; Wu and Yang, 2020). However, we believe that ACT for COVID-19 is unlikely to worsen CRS because the adoptively infused CTLs will target and kill SARS-CoV-2-infected myeloid cells, breaking the vicious cycle driving the cytokine storm. Furthermore, CRS is not unique to betacoronavirus infections and has been reported with other viral infections, such as CMV, EBV, and adenovirus (Humar et al., 1999; McLaughlin et al., 2018; Ramos-Casals et al., 2014), where ACT with virus-specific CTLs has been used to treat hundreds of individuals with severe infections effectively and with minimal complications (Bollard and Heslop, 2016; McLaughlin et al., 2018; Muftuoglu et al., 2018; Tzannou et al., 2017).
Our approach allows cryopreservation and banking of SARS-CoV-2 T cells, facilitating rapid identification and selection of off-the-shelf VSTs for immediate ACT based on the most closely HLA-matched third-party donor, as published by our group and others for other severe viral infections (Eiz-Vesper et al., 2013; Haque et al., 2007; Leen et al., 2010; Muftuoglu et al., 2018; O’Reilly et al., 2016). Thus, we initiated a phase I/II clinical trial to assess the feasibility, safety, and efficacy of off-the-shelf SARS-CoV-2 T cells in individuals with mild to moderate COVID-19 disease (ClinicalTrials.gov: NCT04742595). Because there is increasing evidence that use of corticosteroids may reduce mortality in individuals with COVID-19-related ARDS (Horby et al., 2021), we also developed an efficient and novel strategy to inactivate the glucocorticoid receptor in SARS-CoV-2 T cells using RNA-guided endonuclease CRISPR. In the event of unanticipated toxicities, corticosteroid-resistant SARS-CoV-2 T cells can be eliminated easily using T cell-directed antibodies such as anti-thymocyte globulin (ATG) or alemtuzumab.
Adoptive transfer of SARS-CoV-2 T cells may be a suitable therapeutic strategy for treatment of individuals with COVID-19.
Limitations of the study
Our study evaluated the cytotoxic effect of SARS-CoV-2 T cells using well-validated in vitro assays. We could not test the efficacy of our expanded SARS-CoV-2 T cells in vivo because a robust mouse model of human COVID-19 is not readily available. We also recognize that our study lacks data from affected individuals; however, a phase I/II clinical trial is currently underway to assess the feasibility, safety, and efficacy of our off-the-shelf SARS-CoV-2 T cells in individuals with mild to moderate COVID-19 (ClinicalTrials.gov: NCT04742595).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 APC Cy7 (HIT3A) | Biolegend | Cat# 300318; RRID: AB_314054 |
| CD4 APC (SK3) | BD Biosciences | Cat# 565994; RRID: AB_2739445 |
| CD8 PerCP Cy5.5 (SK1) | Biolegend | Cat# 344710; RRID: AB_2044010 |
| CD95 BV785 (DX2) | Biolegend | Cat# 305646; RRID: AB_2629742 |
| CD45RO BV650 (UCHL1) | BD Biosciences | Cat# 563749; RRID: AB_2744412 |
| CD45RA PECy7 (HI100) | Biolegend | Cat# 304126; RRID: AB_10708879 |
| CD27 BV711 (O323) | Biolegend | Cat# 302834; RRID: AB_2563809 |
| CCR7 FITC (150503) | BD Biosciences | Cat# 561271; RRID: AB_10561679 |
| CD62L BV605 (DREG56) | Biolegend | Cat# 304834; RRID: AB_2562130 |
| IL-2 PE (Clone MQ1-17H12) | BD Biosciences | Cat# 554566; RRID: AB_395483 |
| IFN-γ BV450 (BD Biosciences, Clone B27) | BD Biosciences | Cat# 560371; RRID: AB_1645594 |
| TNF-α AF700 (Biolegend, Clone MAB11) | Biolegend | Cat# 502928; RRID: AB_2561315 |
| Annexin V (V500; BD Biosciences) | BD Biosciences | Cat# 560506; RRID: AB_10694254 |
| live/dead viability dye (efluor 660; Invitrogen) | Invitrogen | Cat# 65-0864-14 |
| LIVE/DEAD Fixable Aqua | Invitrogen | Cat# L34957 |
| GR (clone D6H2L) XP rabbit monoclonal antibody | Cell Signaling Technology | Cat# 12041S; RRID:AB_11179215 |
| β-actin antibody (clone 8H10D10) | Cell Signaling Technology | Cat# 3700S; RRID:AB_2242334 |
| Biological samples | ||
| Convalescent donor blood samples | MD Anderson Cancer Center Blood Bank | N/A |
| Healthy donor blood samples | MD Anderson Cancer Center Blood Bank | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| PepMix Peptide Pools | JPT Peptide Technologies | https://www.jpt.com |
| Bovine Serum Albumin | Millipore Sigma | Cat# A9418 |
| Brefeldin A (BFA) | Millipore Sigma | Cat# B7651 |
| Phorbol 12-myristate 13-acetate (PMA) | Millipore Sigma | Cat# P8139 |
| Ionomycin | Millipore Sigma | Cat# I0634 |
| Proleukin® Aldesleukin (IL-2) | Prometheus | Cat# 798818 |
| Recombinant Human IL-4 Protein | R&D Systems | Cat# 204-IL |
| Recombinant Human IL-7 Protein | R&D Systems | Cat# 207-IL |
| Recombinant Human IL-15 Protein | R&D Systems | Cat# 247-ILB |
| Recombinant Human IL-21 Protein | PeproTech | Cat# 200-21 |
| Critical commercial assays | ||
| Milliplex® MAP Human Cytokine/Chemokine panel | Millipore Sigma | https://www.emdmillipore.com/US/en |
| Fixation/permeabilization kit | BD Biosciences | https://www.bdbiosciences.com/en-us |
| Oligonucleotides | ||
| Primer Exon 2 GR gene, Fwd: GGACTCCAAAGAATCATTAACTCCTGG | This paper | N/A |
| Primer Exon 2 GR gene, Rev: AATTACCCCAGGGGTGCAGA | This paper | N/A |
| Software and algorithms | ||
| R package cytofkit (v1.11.3) | (Chen et al., 2016) | https://github.com/JinmiaoChenLab/cytofkit |
| R package Rtsne (v0.15) | Krijthe (2015) | https://cran.r-project.org/web/packages/Rtsne/index.html |
| R package Rphenograph (v0.99.1) | Levine et al. (2015) | https://github.com/JinmiaoChenLab/Rphenograph |
| R package ggplot2 (v3.3.2) | Wickham (2016) | https://cran.r-project.org/web/packages/ggplot2/index.html |
| R package pheatmap (v1.0.12) | Kolde (2019) | https://cran.r-project.org/web/packages/pheatmap/index.html |
| R package ggpubr (v0.4.0) | Kassambara (2020) | https://cran.r-project.org/web/packages/ggpubr/index.html |
| GeneSys software | Syngene | https://www.syngene.com/software/genesys-rapid-gel-image-capture/ |
| Helios 6.5.358 acquisition software | Fluidigm | https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/cytof-software-6.7-rl-400314/cytof-software-6.7-rl-400314/fluidigm%3Afile |
| FlowJo 10 | BD Biosciences | https://www.flowjo.com |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests may be directed to and will be fulfilled by the Lead Contact Katayoun Rezvani (KRezvani@mdanderson.org).
Materials availability
All requests for data and materials will be reviewed by MD Anderson Cancer Center to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared by the corresponding author will be released freely or via a Material Transfer Agreement if deemed necessary.
Data and code availability
-
•
The data will be available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
COVID-19 recovered donors and healthy donors
Buffy coat units were processed from 500mL of whole blood collected from each of the 10 COVID-19 recovered donors (CoV-RD) and 20 mL of peripheral blood from 5 healthy donors were collected under local Institutional Review Board approved protocols (Lab02-0630 and PA13-0647) and following informed consent. All donors were 18 years or older and were recruited without consideration of disease severity, race, ethnicity or gender. All CoV-RD had recovered from proven symptomatic COVID-19 confirmed by a positive test for SARS-CoV-2. At the time of blood collection, all were asymptomatic for at least 14 days and had a negative PCR test, confirming full recovery.
Blood from Cov-RD was collected in heparin-coated blood bags and stored at room temperature prior to processing for peripheral blood mononuclear cell (PBMC) isolation. PBMCs were isolated by density-gradient sedimentation using Ficoll-Paque (Lymphoprep, Oslo, Norway). Isolated PBMCs were either used fresh for ex vivo expansion of SARS-CoV-2 specific T cells (SARS-CoV-2 CTLs) or cryopreserved in freezing media containing 10% DMSO (GIBCO), supplemented with 10% heat inactivated Human Serum AB (Gemini Bio) and stored in liquid nitrogen until used for phenotypic and functional assays.
Method details
Functional assessment of SARS-CoV-2 reactive T cells
For intracellular assessment of cytokine production, cells were stimulated ex vivo with 15-mer PepMixes overlapping by 11 amino acids derived from SARS-CoV-2 Spike (S) (peptide pool 1 or 2), Membrane (M), Nucleocapsid (N), Envelope (E), or the non-structural proteins (AP3A, Y14, NS6, NS7a, NS7B, NS8,ORF9B and ORF10) (JPT, Germany) [1μg /ml per peptide] for 4 hours. Stimulation with an equimolar amount of DMSO was performed as negative control and with Phorbol 12-myristate 13-acetate (PMA)- Ionomycin (1.25ng/ul and 0.05ng/μl, respectively) as positive control. Brefeldin A (Millipore Sigma, St. Louis, MO) was added into the culture for 4 hours. Cells were stained with an antibody cocktail containing live/dead viability dye Aqua (Invitrogen), CD3 APC Cy7 (Biolegend, Clone HIT3A), CD4 APC (E Biosciences, Clone SK3), CD8 PerCP Cy5.5 (Biolegend, Clone SK1), CD95 BV785 (Biolegend, Clone DX2), CD45RO BV650 (BD Biosciences, Clone UCHL1), CD45RA PECy7 (Biolegend, Clone HI100), CD27 BV711 (Biolegend, Clone O323), CCR7 FITC (BD Biosciences, Clone 150503) and CD62L BV605 (Biolegend, Clone DREG56) for 30 minutes on ice, then fixed and permeabilized using the BD fixation/permeabilization kit (BD Biosciences, San Diego, CA) according to manufacturer’s protocol. Cells were subsequently stained with antibodies against IL-2 PE (BD Biosciences, Clone MQ1-17H12), IFN-γ BV450 (BD Biosciences, Clone B27), and TNF-α AF700 (Biolegend, Clone MAB11) for 30 mins. Following a final wash, cells were re-suspended in FACS buffer and data were acquired on a BD LSRFortessa (BD Biosciences). Data analysis was performed using Flowjo (Tree Star, Ashland, OR). The gates applied for the identification of IFN-γ, IL-2, and TNF-α on the total population of CD4+ and CD8+ T cells were defined according to the negative control for each individual. Similar functional assays were performed for NR3C1 knockout (KO) CTLs.
SARS-CoV-2 antibody assay
IgM and IgG responses against Nucleocapsid, S1 receptor-binding domain (RBD), S1S2, S2, S1, OC43, HKU1, NL63 Nucleoprotein, and 229E Spike derived from SARS-CoV-2 and other human coronaviruses were performed at Genalyte (Austin, TX) CLIA-certified laboratory using plasma from convalescent patients.
Cytokine and chemokine measurement
Cells were stimulated ex vivo with 15-mer PepMixes from S, M and N for 24 hours at 37°C and 5% CO2. Supernatants were collected and assayed with the Milliplex® MAP Human Cytokine/Chemokine panel (EMD Millipore Corporation, Burlington, MA) following the manufacturer’s instructions.
Generation of SARS-CoV-2 specific T cells
Isolated PBMC from CoV-RD and HD were pulsed with a SARS-CoV-2 PepMix (JPT, Germany) comprising the entire length of the structural (S, M, N, E) and non-structural (AP3A, Y14, NS6, NS7a, NS7B, NS8, ORF9B and ORF10) proteins at a concentration of 1 μg/ml per peptide. Cells were cultured in complete media with 5% human AB serum and supplemented with four different cytokine cocktails: IL-2 (50 IU/ml), IL-4 (60 ng/ml) and IL-7 (10 ng/ml) versus IL-2 (50 IU/ml), IL-7 (10 ng/ml) and IL-15 (10 ng/ml) versus IL-2 (50 IU/ml), IL-4 (60 ng/ml) and IL-21 (30 ng/ml) versus IL-2 (50 IU/ml), IL-7 (10 ng/ml) and IL-21 (30 ng/ml) every 3 days. After 14 days of expansion, the frequencies of SARS-CoV-2 specific T cells were determined by intracellular cytokine staining.
Mass Cytometry
A panel of 40 metal-tagged antibodies was used for the in-depth characterization of SARS-CoV-2 reactive T cells (Table S1). All unlabeled antibodies were purchased in carrier-free form (Fluidigm) and conjugated in-house with the corresponding metal tag using Maxpar X8 polymer per the manufacturer’s instructions (Fluidigm) and as previously described (Muftuoglu et al., 2018). Briefly, thawed PBMCs were rested overnight at 37°C / 5% CO2 and stained with a freshly prepared antibody mix against cell surface markers for 30 minutes at room temperature on a shaker (100 rpm). For the last 3 minutes of incubation, cells were incubated with 2.5 μM cisplatin (Pt198, Fluidigm) for viability assessment, washed twice with cell staining buffer and fixed/permeabilized using BD fixation/permeabilization solution for 30 minutes in dark at 4°C. Cells were washed twice with perm/wash buffer, stained with antibodies directed against intracellular markers and after an additional wash step, stored overnight in 500 μl of 1.6% paraformaldehyde (EMD Biosciences)/PBS with 125 nM iridium nucleic acid intercalator (Fluidigm). Samples were supplemented with EQ calibration beads (Fluidigm) and acquired at 300 events/second on a Helios instrument (Fluidigm) using the Helios 6.5.358 acquisition software (Fluidigm).
Mass cytometry data were normalized based on EQ™ four element signal shift over time using Fluidigm normalization software 2. Initial data processing was performed using Flowjo version 10.2. Calibration beads were gated out and singlets were chosen based on iridium 193 staining and event length. Dead cells were excluded by the Pt198 channel and manual gating was performed to select the CD45+CD3+ population which was subsequently exported for downstream analyses. A total of 156,384 cells were evenly sampled from 16 samples derived from 8 patients to perform automated clustering analysis. The data were processed using the R package cytofkit (v1.11.3) (Chen et al., 2016). Expression values for each marker were arcsine transformed with a cofactor of 5. Data dimensionality reduction was performed using the R package Rtsne (v0.15) (Krijthe, 2015) for t-Distributed Neighbor Embedding (tSNE) analysis. The R package Rphenograph (v0.99.1) (Levine et al., 2015) was used to cluster all cells into 32 clusters. Both the R package Rstne (v0.15) and the R package Rphenograph (v0.99.1) were implemented in the R package cytofkit (v1.11.3). The t-SNE plots were generated using the R package ggplot2 (v3.3.2) (Wickham, 2016). Normalized mean values of marker expressions in each cluster were plotted as heatmap using the function “pheatmap” from R package pheatmap (v1.0.12) (Kolde, 2019). Min-max normalization was used to scale each marker’s mean expressions range to [0,1]. The normalized mean values of marker expressions were plotted as boxplots using the function “ggpaired” from R package ggpubr (v0.4.0) (Kassambara, 2020). The mean comparison p values of Wilcoxon signed-rank test were added to the plots using the function “stat_compare_means” from R package ggpubr (v0.4.0).
CRISPR-Cas9 gene editing of the glucocorticoid receptor
Knockout (KO) of NR3C1 (the glucocorticoid receptor gene) was performed on day 7 of T cell expansion using ribonucleoprotein (RNP) complex. We used two crRNAs targeting exon 2 of the human NR3C1 gene: crRNA #1 TGAGAAGCGACAGCCAGTGA, crRNA#2 GGCCAGACTGGCACCAACGG as previously described (Basar et al., 2020) Briefly, Cas9 protein (IDT) and gRNA (crRNA + tracrRNA combination) were complexed and electroporated into 1 million SARS-CoV-2 specific T cells using the Neon transfection system (Thermo Fisher Scientific).
Annexin V apoptosis assay
Annexin V apoptosis assay was performed to evaluate the effect of dexamethasone on the viability of CTLs from Cas9 control and NR3C1 KO groups. CTLs from both groups were treated with 200 μM dexamethasone (Sigma) for 72 hours. Cells were then collected, washed with Annexin V buffer, and stained with Annexin V (V500; BD Biosciences) and live/dead viability dye (efluor 660; Invitrogen) in addition to CD3 APC Cy7 (Biolegend, Clone HIT3A), CD4 APC (E Biosciences, Clone SK3), and CD8 PerCP Cy5.5 (Biolegend, Clone SK1). The proportion of apoptotic (positive for Annexin V) and dead CTLs (positive for live/dead stain) was determined by flow cytometry.
To confirm the ability of SARS-CoV-2 CTLs to mediate cytotoxicity, autologous PBMCs were labeled with carboxyfluorescein succinimidyl ester (CFSE) per manufacturer’s recommendation and loaded with SARS-CoV-2 Spike (S) (peptide pool 1 or 2), Membrane (M) and Nucleocapsid (N) PepMix (JPT, Germany) [1μg /ml per peptide] overnight. The next day, the SARS-CoV-2 PepMix-loaded PBMCs were co-cultured with the expanded CTLs at 1:1 ratio for 16 hours followed by Annexin V staining as described above. The proportion of dead/apoptotic PBMCs (Live/Dead+ and Annexin V+) was determined by flow cyometry.
PCR gel electrophoresis
DNA was extracted and purified (QIAamp DNA Blood Mini Kit; QIAGEN Inc) from SARS-CoV-2 specific T cells (control and NR3C1 KO conditions). We used the Platinum SuperFi Green PCR Master Mix from Invitrogen for polymerase chain reaction (PCR) amplification using the following PCR primers spanning the Cas9–single-guide RNA cleavage site of exon 2 of the GR gene: exon 2 forward primer, GGACTCCAAAGAATCATTAACTCCTGG; exon 2 reverse primer, AATTACCCCAGGGGTGCAGA. DNA bands were separated by agarose gel electrophoresis prepared with SYBR-safe DNA gel stain in 0.5 × Tris/Borate/EDTA. Gel images were obtained using GeneSys software in a G:BOX gel documentation system (Syngene).
Western blot
To detect GR protein expression, CTLs were lysed in lysis buffer (IP Lysis Buffer; Pierce Biotechnology Inc) supplemented with protease inhibitors (Complete Mini, EDTA-free Cocktail tablets; Roche Holding) and incubated for 30 minutes on ice. Protein concentration was determined by the bicinchoninic acid (BCA) assay (Pierce Biotechnology Inc). The following primary antibodies were used: GR (clone D6H2L) XP rabbit monoclonal antibody and β-actin antibody (clone 8H10D10); both antibodies were obtained from Cell Signaling Technology. Blots were imaged using a G:BOX gel documentation system and GeneSys software (Syngene).
Quantification and statistical analysis
Statistical significance was assessed with Prism 9.0 software (GraphPad Software, Inc.). Means were compared using unpaired t test, paired t test, one-way ANOVA, or two-way ANOVA as needed. The Tukey correction was used for correction of multiple comparisons. For correlation analysis, R squared simple linear regression was used. Graphs represent mean and standard deviation (SD). A p ≤ 0.05 was considered to be statistical significance. When analyzing variables with more than 2 categories, P values were adjusted for multiple comparisons. The statistical details are found in the figure legends.
Acknowledgments
Supported in part by a generous donation from Lyda Hill Philanthropies; generous philanthropic contributions to The University of Texas MD Anderson Cancer Center AML Moonshot Program; National Institute of Health, National Cancer Institute 5R01CA211044-04; and a Cancer Center Support (CORE) grant (CA016672) that supports the Flow Cytometry and Cellular Imaging Facility and RNA sequencing core facility at MD Anderson Cancer Center.
Author contributions
Conceptualization, R.B. and K.R.; methodology, R.B., N.U., E.E., and M.D.; investigation, N.U., E.E., M.S., B.H., E.G., A.K.N.C., M.M., F.R.S., S.A., S.L., L.M.G., and P.L.; writing, K.R., R.B., N.U., E.E., M.D., T.L., L.M.F., and D. Marin; funding acquisition, K.R.; resources, K.K., F.M., F.A., H.S., S.G.Y., Q.M., J.D., K.C., D. Mak, and A.L.; supervision, K.R., P.P.B., R.E.C., and E.J.S.
Declaration of interests
R.B., D. Marin, E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Affimed GmbH. Because MD Anderson is committed to the protection of human subjects and the effective management of its financial conflicts of interest in relation to its research activities, MD Anderson is implementing an Institutional Conflict of Interest Management and Monitoring Plan to manage and monitor the conflict of interest with respect to MD Anderson’s conduct of any other ongoing or future research related to this relationship. R.B., M.D., P.P.B., D. Marin, R.E.C., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical for the licensing of the technology related to CAR-NK cell research. MD Anderson has implemented an Institutional Conflict of Interest Management and Monitoring Plan to manage and monitor the conflict of interest with respect to MDACC’s conduct of any other ongoing or future research related to this relationship. K.R. participates on the Scientific Advisory Board for GemoAb, AvengeBio, Virogin, GSK, and Bayer.
Published: July 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109432.
Supplemental information
References
- Basar R., Daher M., Uprety N., Gokdemir E., Alsuliman A., Ensley E., Ozcan G., Mendt M., Hernandez Sanabria M., Kerbauy L.N. Large-scale GMP-compliant CRISPR-Cas9-mediated deletion of the glucocorticoid receptor in multivirus-specific T cells. Blood Adv. 2020;4:3357–3367. doi: 10.1182/bloodadvances.2020001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the Treatment of Covid-19 — Preliminary Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard C.M., Heslop H.E. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood. 2016;127:3331–3340. doi: 10.1182/blood-2016-01-628982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. Presence of SARS-CoV-2 reactive T cells in COVID-19 patients and healthy donors. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yu F., Jiang Y., Chen J., Wu K., Chen X., Lin Y., Zhang H., Li L., Zhang Y. Adoptive Transfer of Interleukin-21-stimulated Human CD8+ T Memory Stem Cells Efficiently Inhibits Tumor Growth. J. Immunother. 2018;41:274–283. doi: 10.1097/CJI.0000000000000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lau M.C., Wong M.T., Newell E.W., Poidinger M., Chen J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clénet M.-L., Gagnon F., Moratalla A.C., Viel E.C., Arbour N. Peripheral human CD4+CD8+ T lymphocytes exhibit a memory phenotype and enhanced responses to IL-2, IL-7 and IL-15. Sci. Rep. 2017;7:11612. doi: 10.1038/s41598-017-11926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake A., Kaur M., Iliopoulou B.P., Phennicie R., Hanson A., Chen J. Interleukins 7 and 15 Maintain Human T Cell Proliferative Capacity through STAT5 Signaling. PLoS ONE. 2016;11:e0166280. doi: 10.1371/journal.pone.0166280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiz-Vesper B., Maecker-Kolhoff B., Blasczyk R. Adoptive T-cell immunotherapy from third-party donors: Characterization of donors and set up of a T-cell donor registry. Front. Immunol. 2013;3:410. doi: 10.3389/fimmu.2012.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi N., Rezaei N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020;44:1792–1797. doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat J., Sallusto F., Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells. J. Exp. Med. 2001;194:1711–1719. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque T., Wilkie G.M., Jones M.M., Higgins C.D., Urquhart G., Wingate P., Burns D., McAulay K., Turner M., Bellamy C. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood. 2007;110:1123–1131. doi: 10.1182/blood-2006-12-063008. [DOI] [PubMed] [Google Scholar]
- Heslop H.E., Slobod K.S., Pule M.A., Hale G.A., Rousseau A., Smith C.A., Bollard C.M., Liu H., Wu M.F., Rochester R.J. Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood. 2010;115:925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P.J., Bottazzi M.E., Corry D.B. The potential role of Th17 immune responses in coronavirus immunopathology and vaccine-induced immune enhancement. Microbes Infect. 2020;22:165–167. doi: 10.1016/j.micinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humar A., St Louis P., Mazzulli T., McGeer A., Lipton J., Messner H., MacDonald K.S. Elevated serum cytokines are associated with cytomegalovirus infection and disease in bone marrow transplant recipients. J. Infect. Dis. 1999;179:484–488. doi: 10.1086/314602. [DOI] [PubMed] [Google Scholar]
- Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Tanaka T., Narazaki M., Kishimoto T. Targeting Interleukin-6 Signaling in Clinic. Immunity. 2019;50:1007–1023. doi: 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- Kassambara, A. (2020). Package ‘ggpubr’: “ggplot2” Based Publication Ready Plots. R Packag. Version 0.4.0. https://cran.r-project.org/web/packages/ggpubr/index.html.
- Kolde, R. (2019). pheatmap: Pretty Heatmaps version 1.0.12 from CRAN. https://rdrr.io/cran/pheatmap/.
- Krijthe (2015). Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. https://github.com/jkrijthe/Rtsne.
- Leen A.M., Bollard C.M., Mendizabal A., Shpall E.J., Szabolcs P., Antin J.H., Kapoor N., Pai S.-Y., Grilley B., Gee A.P. Most Closely HLA-Matched Allogeneic Virus Specific Cytotoxic T-Lymphocytes (CTL) to Treat Persistent Reactivation or Infection With Adenovirus, CMV and EBV After Hemopoietic Stem Cell Transplantation (HSCT) Blood. 2010;116:829. [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bleakley M., Yee C. IL-21 influences the frequency, phenotype, and affinity of the antigen-specific CD8 T cell response. J. Immunol. 2005;175:2261–2269. doi: 10.4049/jimmunol.175.4.2261. [DOI] [PubMed] [Google Scholar]
- McLaughlin L.P., Bollard C.M., Keller M.D. Adoptive T Cell Therapy for Epstein-Barr Virus Complications in Patients With Primary Immunodeficiency Disorders. Front. Immunol. 2018;9:556. doi: 10.3389/fimmu.2018.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. Mechanisms of T cell polyfunctionality. Nat. Rev. Immunol. 2014;14:7. [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M., Olson A., Marin D., Ahmed S., Mulanovich V., Tummala S., Chi T.L., Ferrajoli A., Kaur I., Li L. Allogeneic BK Virus-Specific T Cells for Progressive Multifocal Leukoencephalopathy. N. Engl. J. Med. 2018;379:1443–1451. doi: 10.1056/NEJMoa1801540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly R.J., Prockop S., Hasan A.N., Koehne G., Doubrovina E. Virus-specific T-cell banks for ‘off the shelf’ adoptive therapy of refractory infections. Bone Marrow Transplant. 2016;51:1163–1172. doi: 10.1038/bmt.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A., Lin R., Marin D., Rafei H., Bdaiwi M.H., Thall P.F., Basar R., Abudayyeh A., Banerjee P., Aung F.M. Third-party BK virus specific cytotoxic T lymphocyte therapy for hemorrhagic cystitis following allotransplantation. J. Clin. Oncol. 2021 doi: 10.1200/JCO.20.02608. Published online April 30, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J. An Outbreak of Human Coronavirus OC43 Infection and Serological Cross-reactivity with SARS Coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006;17:330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pia L. SARS-CoV-2-reactive T cells in patients and healthy donors. Nat. Rev. Immunol. 2020;20:353. doi: 10.1038/s41577-020-0333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D.J., Dudley M.E., Robbins P.F., Rosenberg S.A. Transition of late-stage effector T cells to CD27+ CD28 + tumor-reactive effector memory T cells in humans after adoptive cell transfer therapy. Blood. 2005;105:241–250. doi: 10.1182/blood-2004-06-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Casals M., Brito-Zerón P., López-Guillermo A., Khamashta M.A., Bosch X. Adult haemophagocytic syndrome. Lancet. 2014;383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Straling K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., Cao W.J., Yang T., Dai X.P., Wang S.Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme C., Anft M., Paniskaki K., Blázquez Navarro A., Doevelaar A., Seibert F., Hölzer B., Konik M.J., Brenner T., Tempfer C. The SARS-COV-2 T-Cell Immunity is Directed Against the Spike, Membrane, and Nucleocapsid Protein and Associated with COVID 19 Severity. Cell Rep. Med. 2020;1:100092. doi: 10.1016/j.xcrm.2020.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzannou I., Papadopoulou A., Naik S., Leung K., Martinez C.A., Ramos C.A., Carrum G., Sasa G., Lulla P., Watanabe A. Off-the-Shelf Virus-Specific T Cells to Treat BK Virus, Human Herpesvirus 6, Cytomegalovirus, Epstein-Barr Virus, and Adenovirus Infections After Allogeneic Hematopoietic Stem-Cell Transplantation. J. Clin. Oncol. 2017;35:3547–3557. doi: 10.1200/JCO.2017.73.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data will be available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.