Abstract
Introduction
In humans and companion animals, obesity is accompanied by metabolic derangements. Studies have revealed differences in the composition of the fecal microbiome between obese dogs and those with an ideal body weight.
Objectives
We have previously reported that the fecal microbiome in obese dogs changes after controlled weight reduction, induced by feeding a diet high in fiber and protein. Despite these findings, it is unclear if taxonomic differences infer differences at the functional level between obese dogs and those with an ideal body weight.
Methodology
Untargeted fecal metabolome analysis was performed on dogs with obesity before and after weight loss achieved by feeding a high-fiber-high-protein diet.
Results
Fecal metabolome analysis revealed a total of 13 compounds that changed in concentration in obese dogs after weight loss. Of these compounds, metabolites associated with bacterial metabolism decreased after weight loss including purine, L-(-)-methionine, coumestrol, and the alkaloids 1-methylxanthine and trigonelline. Conversely, the polyphenols (-)-epicatechin and matairesinol and the quinoline derivatives 1,5-isoquinolinediol and 2-hydroxiquinoline increased after weight loss.
Conclusion
These results suggest differences in intestinal microbiome at the functional level after weight loss, but further studies are needed to determine the role of these compounds in the etiology of obesity and weight loss.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11306-021-01815-1.
Keywords: Canine obesity, Fecal microbiota, Fecal metabolome, Weight loss, Untargeted metabolome analysis
Introduction
Canine obesity has recently been characterized as a disease by many international veterinary organizations (Ward et al., 2019), on account of its significant impact on health and wellbeing (German, 2006; German et al., 2010). Recent studies have identified metabolic changes associated with canine obesity, including insulin resistance, decreased adiponectin concentrations and markers of renal function or disease (German et al., 2009; Tvarijonaviciute et al., 2012, 2013). Other obesity-related metabolic changes have been identified using proteomic analysis, including altered expression of proteins involved in lipid metabolism, immune response, and antioxidant status (Tvarijonaviciute et al., 2016).
Obesity is associated with alterations of the gut microbiota (Ridaura et al., 2013; Turnbaugh et al., 2009). The first studies that compared the fecal microbiome of obese dogs and those with an ideal body weight showed only minor differences in their fecal microbiome (Handl et al., 2013; Park et al., 2015). However, subsequent studies evaluating the fecal microbiota of obese dogs before and after weight loss, reported an increase in the bacterial biodiversity and a decrease of the Firmicutes/Bacteroidetes ratio after the dogs completed their weight loss program (Bermudez Sanchez et al., 2020; Salas-Mani et al., 2018).
Gut bacteria are known to have important functions affecting host physiology (Sittipo et al., 2019) and maintaining gut homeostasis, including development and modulation of the immune system, the gut epithelium, bile acid metabolism, xenobiotic degradation, and biosynthesis of vitamins (Jandhyala et al., 2015). In addition, the gut microbiota harbors an expansive number of genes that confer upon bacteria a wide variety of digestive functions, associated with dietary and host metabolites (Kho & Lal, 2018).
Studies using animal models that focused on defining the role of the gut microbiota in the etiology of obesity, point to a direct interaction with the host through bacterial metabolites (Boulangé et al., 2016). In this respect, the gut microbiota are thought to modulate host gut microenvironment and host energy homeostasis by metabolizing nutrients from the diet (Arora et al., 2011). Examples include synthesis of short-chain fatty acids (SCFAs) from undigested polysaccharides and bile acid interconversion (Rowland et al., 2018). These microbial processes are thought to contribute to the etiology of obesity by modulation of energy generation, storage, and expenditure, in addition to intestinal inflammation mediated by LPS (Harakeh et al., 2016).
The analysis of the fecal metabolome is a non-invasive method of characterizing functional differences of the gut microbiome between individuals, with results depending upon the composition of the overall gut microbiota (Zierer et al., 2018). However, the vast majority of metabolites remain unknown with undefined functions. Therefore, to better understand associations between the gut microbiota and obesity in dogs, we evaluated the composition of the fecal metabolome in 20 client-owned dogs with obesity, that had recently participated in a study examining the composition of the fecal microbiota (Bermudez Sanchez et al., 2020). Untargeted fecal metabolome analysis was performed on fecal samples of obese dogs before and after controlled weight reduction by feeding them a diet rich in protein and fiber.
Material and methods
Study animals, eligibility criteria, and ethical considerations
Dogs with obesity enrolled into this study had been referred to the Royal Canin Weight Management Clinic at the University of Liverpool, UK, between June 2009 and August 2017. All dogs had completed their weight loss program between November 2009 and August 2018, and eligibility criteria were the same as those described previously (Bermudez Sanchez et al., 2020), including having a body condition score (BCS) > 6 on a 9-point scale, no antimicrobial usage in the past month (Igarashi et al., 2014), and no signs of gastrointestinal disease. The studied group comprised 25 dogs with obesity. Five dogs stopped the study prematurely and were excluded from the analysis; the remaining 20 dogs completed the weight loss programme, and fecal samples were available before dietary change (T0) and after a period of controlled weight reduction with a therapeutic diet (see below) to reach a pre-defined target weight (T1).
The study protocol was reviewed and approved by the University of Liverpool Veterinary Research Ethics Committee (Approval reference: RETH000353 and VREC793), the Royal Canin ethical review committee, and the WALTHAM ethical review committee. Owners of dogs with obesity gave informed consent in writing.
Weight loss regimen
Full details of the weight loss regimen have been previously described (German et al., 2015). Briefly, dogs were first determined to be systemically well (apart from their obesity), based on physical examination findings, complete blood count, serum biochemical analysis, serum free thyroxine concentration (by equilibrium dialysis), and urinalysis. Body condition score (BCS) was estimated using a 9-integer scale (Laflamme, 1997) by the attending clinician (AJG). Body composition was measured by dual-energy x-ray absorptiometry (DEXA), as previously described (Raffan et al., 2006), the results of which were used to determine the ideal body weight of each dog (German et al., 2015). Body composition measurements were available for 19 of the 20 dogs, from both before and after weight reduction.
All dogs were fed either a dry or wet high-protein high-fiber therapeutic weight reduction diet (Canine Satiety® diet, Royal Canin), with an increased protein and fiber content as previously described (German et al., 2015). Advice was also given to owners about lifestyle and activity alterations that could help their dog lose weight. For each dog, a target weight was also set, which was individually tailored for each dog, based upon expected health benefits, once again, as described (German, 2016). The weight reduction process was monitored through regular weight checks using calibrated electronic scales.
Fecal collection
As previously described (Bermudez Sanchez et al., 2020), fecal samples were collected after spontaneous defecation and stored at − 20 °C, before being shipped to the Gastrointestinal Laboratory at Texas A&M University in February 2019.
Sample preparation for metabolome analysis
Samples were lyophilized, weighed, and extracted with a methanol:chloroform:water based extraction method. Briefly 800 µL ice cold methanol:chloroform (1:1, v:v) was added to each sample in a bead based lysis tube (Bertin, Rockville, MD). Samples were extracted on a Precyllys 24 (Bertin) tissue homogenizer for 30 s at a speed of 6000 rpm. The supernatant was collected and samples were homogenized a second time with 800 µL ice cold methanol:chloroform. Then 600 µL ice cold water was added to the combined extract, vortexed, and centrifuged to separate the phases. The upper aqueous layer was passed through a 0.2 µm nylon filter (Merck Millipore, Burlington, MA). Then 500 µL of the filtered aqueous phase was passed through a 3 kDa cutoff column (Thermo Scientific) and the flow through was collected for analysis.
Metabolome analysis by HPLC–MS
Untargeted liquid chromatography high resolution accurate mass spectrometry (LC-HRAM) analysis was performed on a Q Exactive Plus orbitrap mass spectrometer (Thermo Scientific, Waltham, MA) coupled to a binary pump HPLC (UltiMate 3000, Thermo Scientific). Full MS spectra were obtained at 70,000 resolution (200 m/z) with a scan range of 50–750 m/z. Full MS followed by ddMS2 scans were obtained at 35,000 resolution (MS1) and 17,500 resolution (MS2) with a 1.5 m/z isolation window and a stepped NCE (20, 40, 60). Samples were maintained at 4 °C before injection. The injection volume was 10 µL. Chromatographic separation was achieved on a Synergi Fusion 4 µm, 150 mm × 2 mm reverse phase column (Phenomenex, Torrance, CA) maintained at 30 °C using a solvent gradient method. Solvent A was water (0.1% formic acid). Solvent B was methanol (0.1% formic acid). The gradient method used was 0–5 min (10% B to 40% B), 5–7 min (40% B to 95% B), 7–9 min (95% B), 9–9.1 min (95% B to 10% B), 9.1–13 min (10% B). The flow rate was 0.4 mL/min. Sample acquisition was performed by the Xcalibur software package (Thermo Scientific). Data analysis was performed with the Compound Discoverer software package 2.1 (Thermo Scientific).
Data analysis (Compound Discoverer 2.1)
Background was excluded by setting the minimal peak intensity as greater than 169,436.00. As inclusion criteria for the analysis, we applied at least an 80% of match score with the compound of reference provided by the MzCloud library, which considers the mass spectrum of a catalog of the compounds. Unnamed and repeated compounds were excluded. Resulting peaks were manually curated, and the peak intensity for each sample and compound was exported into an excel file (Microsoft Excel; Supplementary data 1). A total of 134 named compounds were identified in the fecal samples of the dogs enrolled.
Statistical analysis
Normality was tested using Shapiro–Wilk tests for all continuous variables. Results were reported as median (range) when data were non-parametric, and as mean (± SD) when data passed normality testing. Differences in dog characteristics between groups were compared using t-tests or Wilcoxon tests for paired analyses and Mann–Whitney tests for unpaired analyses. Metabolomic data were analyzed with the web-based free tool MetaboAnalyst 5.0.; http://www.metaboanalyst.ca) (Chong & Xia, 2020; Xia et al., 2015). Data were normalized by pareto scaling and log transformation. MetaboAnalyst was used to generate Principal Coordinate Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) plots in 2D to display metabolic communities for all groups. Heatmap visualization was performed based on Wilcoxon tests for paired analyses and reorganization of metabolites to show contrast between the groups. Correction for multiple testing was done by calculating the false discovery rate (FDR). Normalized data were used with a statistical software package, GraphPad Prism version 8.0 (La Jolla, California, USA), to generate graphs for individual values of fecal metabolites for each dog.
Results
Animal population characteristics and details of the study period
At enrollment, the median age of the dogs was 63 months (range 16 to 135 months). The breeds of the dogs enrolled in the study were: Labrador Retriever (6), Golden Retriever (2), Cavalier King Charles spaniel (2), Border Collie (1), Lhasa Apso (1), American Bulldog (1), Dachshund (1), Rottweiler (1), Newfoundland (1), Bichon Frise (1), and mixed breed (3). Ten dogs were neutered female and 10 were neutered male. Prior to weight reduction, the median BCS was 8 (range 6 to 9). Mean body fat mass before weight loss was 45% (SD 5.2%).
The median time between fecal sampling (T0 to T1) was 271 (range 112 to 768) days during which time dogs had lost a mean of 26% (SD 8.6%) of their starting bodyweight, equating to a rate of 0.7% (SD 0.30%) per week. Body condition score changed significantly (T0 median BCS, 8 [range 6–9]; T1 median BCS, 5 [range 4–7], P < 0.001). Body fat mass decreased significantly after weight loss (T0 mean, 45% [SD 5.2%]; T1 mean, 30% [SD 6.7%], P < 0.001). The mean daily energy intake during the weight loss period was 60.8 kcal/kg0.75 (SD 5.6 kcal/kg0.75) calculated based on the ideal body weight (Bermudez Sanchez et al., 2020).
No significant associations between baseline fecal metabolites and sex as well as age were found (Supplementary data 2).
The fecal metabolome of dogs with obesity changes after a period of controlled weight reduction
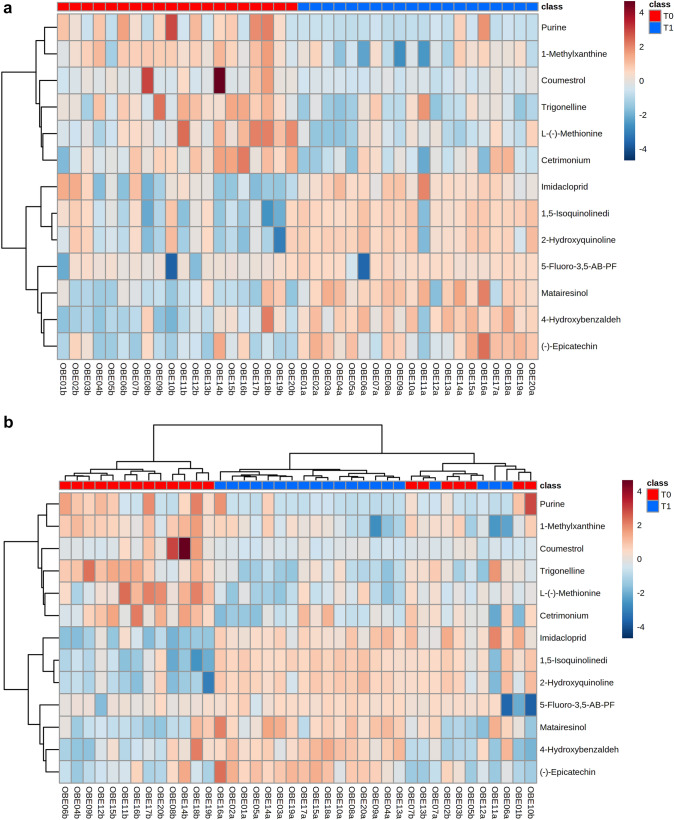
A total of 134 compounds were identified in the feces of the dogs, of which, 13 metabolites changed significantly between T0 and T1, based on adjusted P-values (Fig. 1).
Fig. 1.
A Reorganized and B not reorganized hierarchical clustering and heatmaps of the 13 metabolites that were different in their peak intensity before and after weight reduction. Ordered columns represent the fecal metabolite profile for each dog before (T0) and after a period of weight reduction (T1) using a therapeutic diet and sorted by group. Colored bars on the x-axis represent group of dogs (red, obese dogs before weight loss; blue, obese dogs after weight loss). Rows represent the 13 metabolites that differed, highlighting two clusters of metabolites: the first comprised metabolites that decreased after weight reduction (i.e., purine, 1-methylxanthine, coumestrol, trigonelline, L-(-)-methionine, and cetrimonium); the second comprised metabolites that increased after weight reduction (i.e., imidacloprid, 1,5-isoquinolinediol, 2-hydroxiquinoline, 5-fluoro-3,5-AB-PFUPPYCA, matairesinol, 4-hydroxybenzaldehido, and (-)-epicatechin)
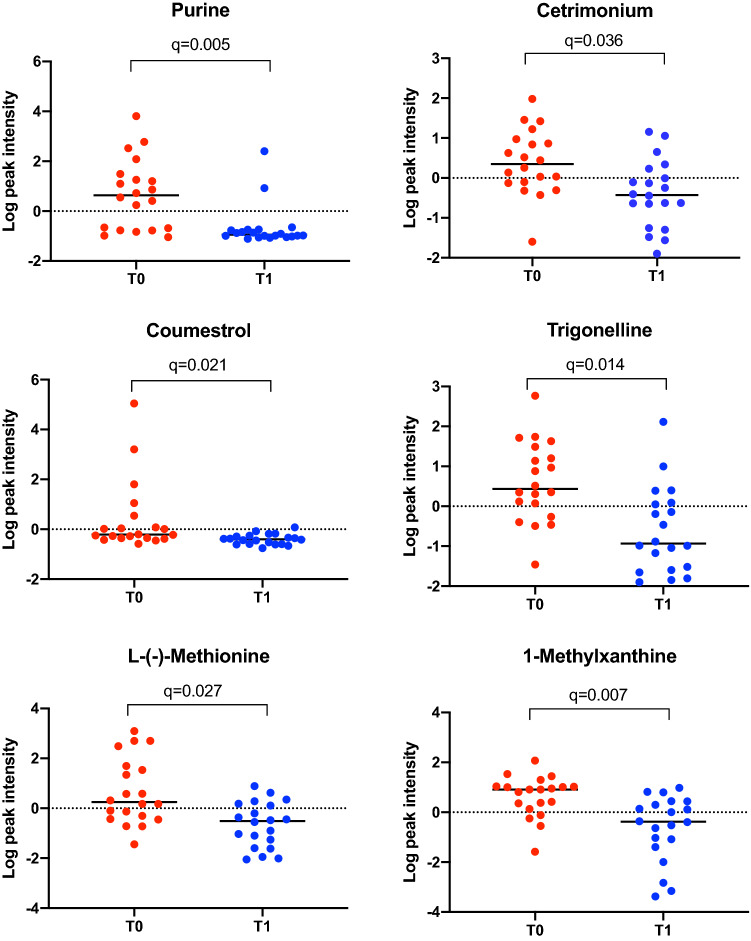
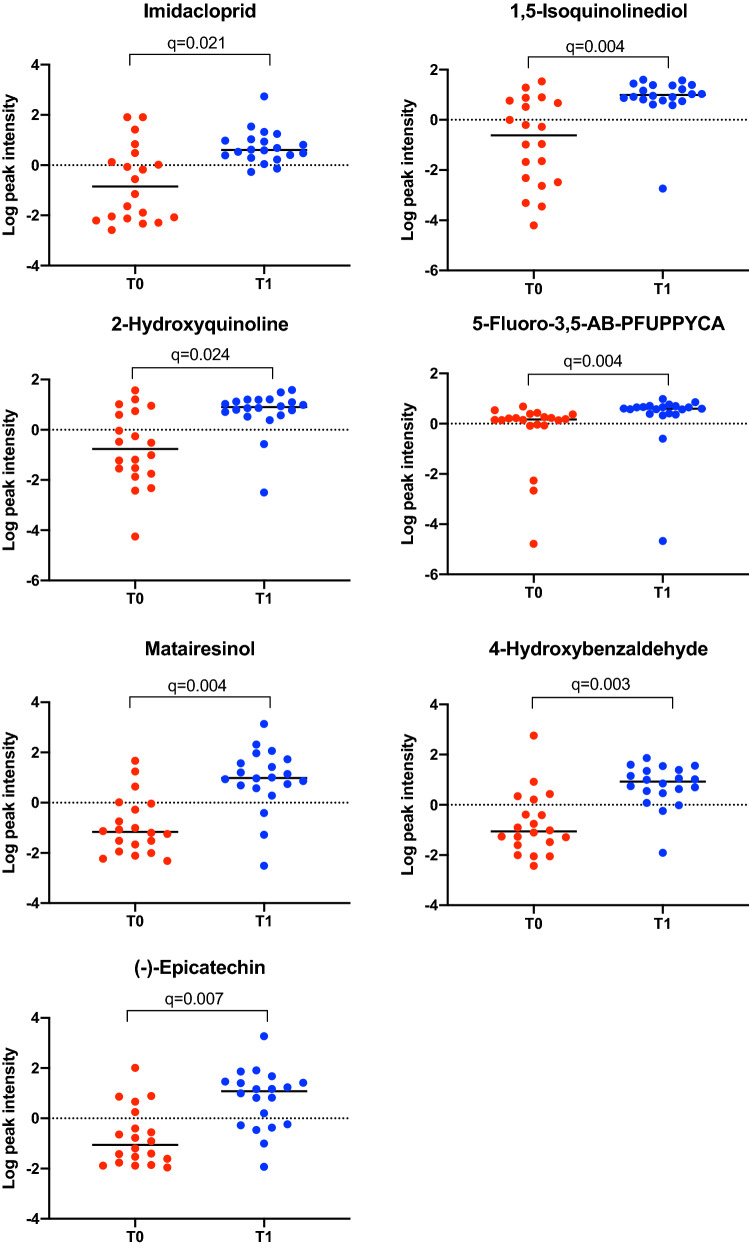
Purine, 1-methylxanthine, cetrimonium, coumestrol, trigonelline, and L-(-)-methionine decreased after weight loss (Fig. 2). Conversely, imidacloprid, 1,5-isoquinolinediol, 2-hydroxyquinoline, 5-fluoro-3,5-AB-PFUPPYCA, matairesinol, 4-hydroxybenzldehyde and (-)-epicatechin increased after weight loss (Fig. 3).
Fig. 2.
Normalized data were used to create individual value graphs of the fecal metabolites that decreased after weight reduction in dogs. The bar represents the median for each group, (T0) before, (T1) after a period of weight reduction. Wilcoxon tests were used for statistical comparison and significance was based on FDR values
Fig. 3.
Normalized data were used to create individual value graphs of the fecal metabolites that increased after weight reduction in dogs. The bar represents the median for each group, (T0) before, (T1) after a period of weight reduction. Wilcoxon tests were used for statistical comparison and significance was based on FDR values
Based on PCA, there was no separation of the overall metabolic profile of dogs before (T0) and after (T1) weight reduction (Fig. 4A). However, clear separation was observed on the PLS-DA plot (Supplementary Fig. 1). In addition, when the 13 compounds which showed a significant different concentration after weight loss were plotted on the PCA, separated clusters were shown for each group of dogs (Fig. 4B).
Fig. 4.
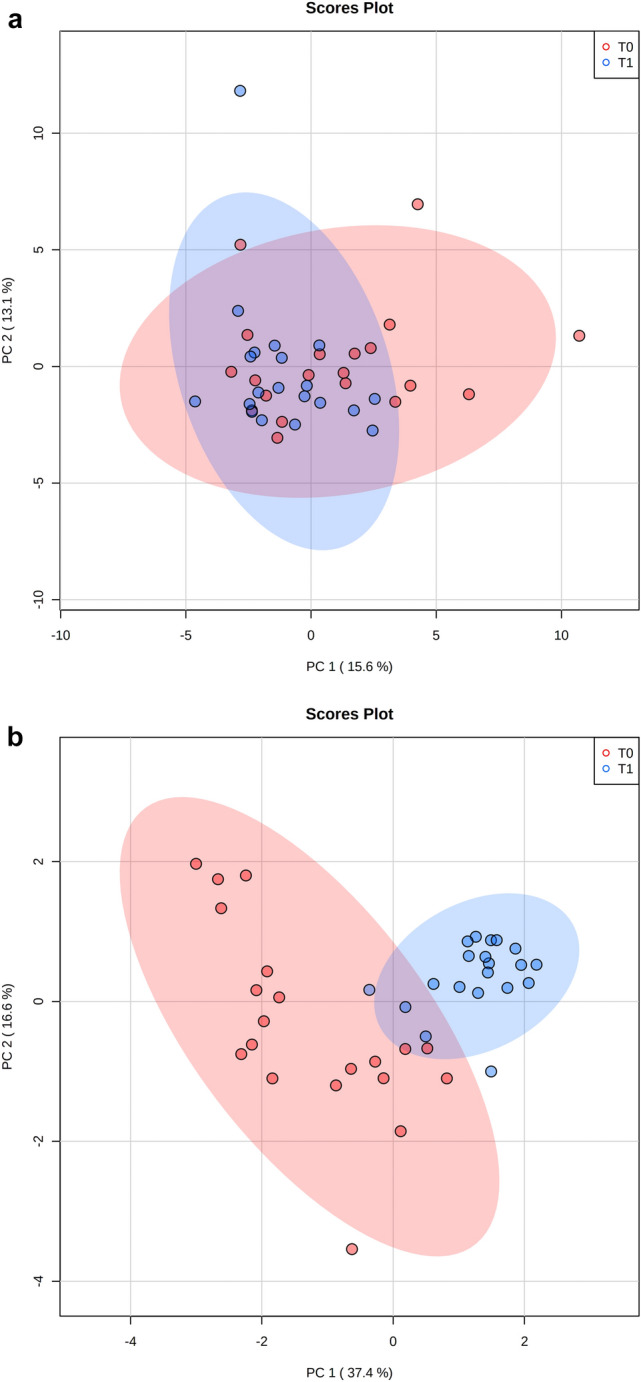
A 2D PCA score plot displaying the fecal metabolomics data from dogs before (T0, red) and after (T1, blue) weight reduction by feeding a therapeutic diet. B 2D PCA score plot displaying the data from the 13 metabolites which were significantly different in their peak intensity before (T0, red) and after (T1, blue) weight loss reduction. Ellipses represent the 95% confidence interval of the metabolite profile for each group
Discussion
In this study, we investigated the fecal metabolome of obese dogs following a controlled weight reduction program that utilized a diet rich in fiber and protein. In a previous study involving this cohort, significant changes in the composition of the fecal microbiome composition had been observed after weight reduction (Bermudez Sanchez et al., 2020). However, given that many different bacterial species can occupy the same niche, changes in microbiome composition do not necessarily result in changes in microbiome function. Therefore, to understand whether the changes in the fecal metabolome may have an impact on intestinal health, we chose to analyze the fecal metabolome of the same twenty obese dogs before and after weight loss in the current study. Overall, we were able to show significant changes in the concentration of 13/134 fecal metabolites. Whilst notable, the magnitude of these differences was insufficient to create a separation in the metabolome profiles on PCA plot (Fig. 4A). The reason for this is not known, but might be related to the variety of diets that dogs were fed before the start of the study, and this variability might have obscured the shifts seen with weight reduction as a result of the therapeutic diet fed. Furthermore, the dogs studied were diverse in terms of their signalment and, being pet dogs, were exposed to different environmental factors. Additional work would be required to examine this possibility further.
The specific changes seen in the fecal metabolome profile in the current study include decreases in the concentrations of purine, 1-methylxhantine, cetrimonium, coumestrol, trigonelline, and L(-)-methionine, and an increase in the concentrations of imidacloprid, 1,5-isoquinolinediol, 2-hydroxyquinoline, 5-fluoro-3,5-AB-PFUPPYCA, matairesinol, 4-hydroxybenzaldehyde, and (-)-epicatechin (Figs. 2 and 3).
Purine is a metabolic substrate of intestinal bacteria, which has been reported to be high in people ingesting high-protein diets (Guo et al., 2016). Thus, a decrease in purine concentration is contrary to expectations, given that the therapeutic diet fed was rich in protein. Possible reasons for this include the reduced food intake during the weight reduction period, an increase in purine requirement by the host, or differences in the composition of the microbiota, leading to a change in the metabolic syntrophy of the bacteria (Bermudez Sanchez et al., 2020). This has been observed for the alkaloids 1-methylxanthine, which is a substrate of different species of Pseudomonas (Woolfolk & Downard, 1977; Yu et al., 2009), and trigonelline, which modulates the intestinal microbiota in animal models (Anwar et al., 2018; Bruce-Keller et al., 2020), and plays a role in the metabolism of choline, indirectly affecting lipid accumulation as well as improving specific markers of health status during ingestion of a high-fat diet in mice (Knott et al., 2017; Salek et al., 2007; Zhou et al., 2012).
Another compound that decreased in fecal samples after weight loss was the amino acid L-(-)-methionine, which is usually present in the diet of dogs (Harrison et al., 2020; Middelbos et al., 2006). It has been reported that an increased intake of this amino acid is associated with adverse effects on host metabolism and the gut microbiome in mice (Miousse et al., 2017). Therefore, dietary methionine restriction has a protective role on health, extending life expectancy, reducing the weight gain, and regulating insulin in mice (Ables et al., 2012; Yang et al., 2018). Conversely, methionine supplementation is associated with improved intestinal integrity and oxidative status in piglets (Su et al., 2018; Yang & Liao, 2019). Interestingly, L-(-)-methionine is a substrate of gut bacteria, including members of the Proteobacteria phylum, the Bacilli class and the Clostridium and Bifidobacterium genera (Portune et al., 2016).
Dietary polyphenols are bioactive compounds with antioxidant activity, and are commonly found in fruits, vegetables, seed, and beverages such as coffee, tea, and wine (Cardona et al., 2013). Polyphenols in fecal samples arise directly from the diet; however, some polyphenols are also subject to metabolism by gut bacteria (Gaya et al., 2016), as is the case for (-)-epicatechin (Tzounis et al., 2011). Consumption of polyphenols has been associated with a modulation of the intestinal microbiota (Lee et al., 2006), maintenance of host intestinal barrier integrity, and regulation of immune functions (Kolátorová et al., 2018). Potential anti-obesogenic effects have also been attributed to some polyphenols, including catechins (Wang et al., 2014), with possible mechanisms including suppression of fat oxidation, inhibition of adipocyte differentiation, inhibition of fat absorption, and induction of adipocyte apoptosis (Hursel & Westerterp-Plantenga, 2010; Lin et al., 2005). Intestinal bacteria can also transform matairesinol into mammalian lignan enterolactone, which can undergo colonic absorption (Jansen et al., 2005) and is also known to possess estrogenic, and antioxidative functions (Borriello et al., 1985; Bowey et al., 2003; Heinonen et al., 2001). In the current study, various polyphenolic compounds were detected in the fecal samples of dogs with obesity. Given that (-)-epicatechin and matairesinol concentrations were increased, but coumestrol concentration was decreased, our results did not demonstrate a consistent effect of weight reduction on the fecal polyphenolic profile in dogs with obesity. Further studies would be required to determine the significance of these findings.
Concentrations of quinolines 2-hydroxyquinoline and 1–5, isoquinolinediol decreased after weight reduction in dogs with obesity. Quinoline derivates are products of bacterial metabolism with quorum-sensing functions (Hubbard et al., 2019) that can be metabolized by some bacteria, such as Rhodococcus sp., Pseudomonas (Sun et al., 2009), Thauera, and Bacillus (Tuo et al., 2012). Interestingly, in humans, 2-hydroxyquinoline inhibits PON1 (paraoxonase-1), an antioxidant enzyme involved in lipid metabolism (Graves & Scott, 2008). Furthermore, some quinoline derivates have been proposed as Aryl hydrocarbon receptor (AhR) ligands in humans (Hubbard et al., 2019). AhR signaling contributes to the immune homeostasis by suppressing inflammation but also modulating the maturation of immune cells involved in the mucosal immunity and the defense against pathogens by xenobiotic metabolism and epithelial renewal in the intestine (Agus et al., 2018; Lamas et al., 2018; Stockinger et al., 2014; Zhou, 2016).
We also detected changes in the concentrations of the synthetic compounds imidacloprid, cetrimonium, and 5-fluoro-3,5-AB-PFUPPYCA. Imidacloprid is an antiparasitic drug widely used in companion animals (Jeschke & Nauen, 2008), but it is also used as an insecticide in agriculture (Badgujar et al., 2013). Administering this compound together with a high fat diet is associated with greater adiposity and insulin resistance in mice (Sun et al., 2016), but the effects in dogs have not previously been reported. Cetrimonium is a disinfectant whereas 5-fluoro-3,5-AB-PFUPPYCA is a synthetic cannabinoid derivative but, to our knowledge, the relevance of the changes in the concentration of the rest of these synthetic compounds after weight loss and during health in general has not yet been studied.
A limitation of the current study is that it is unclear whether the observed changes in the fecal metabolome were the result of the weight reduction, or whether they were simply the result of the dietary change, as demonstrated by a recent study on the fecal metabolome of dogs (Jackson & Jewell, 2019). A second limitation was that only two fecal samples were collected from each dog, meaning that sequential changes in the intestinal microbiome could not be elucidated. This was further complicated by the fact that follow-up samples were taken after a variable time, dictated by the duration of weight loss, rather than being taken at specific time points after the start of the study.
In conclusion, although preliminary, the findings of the current study suggest that controlled weight reduction in obese dogs using a protein and fiber rich therapeutic diet is associated with alterations in the fecal metabolome, characterized by increased concentrations of polyphenols and quinoline derivatives. However, further studies are necessary to clarify the significance of these findings, and if they are a consequence of the therapeutic diet, a direct effect of weight reduction, or are an indirect effect of changes in the composition of the fecal microbiome.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary data 1. Spreadsheet of the data from peak intensity for each sample and compound imported from Compound discoverer 2.1. (XLSX 341 kb)
Supplementary data 2. Association between baseline fecal metabolites and sex, and between baseline fecal metabolites and age. (XLSX 102 kb)
Supplementary figure 1. 2D PLS-DA score plot displaying the fecal metabolomics data from dogs before (T0, red) and after (T1, blue) weight reduction by feeding a therapeutic diet. Ellipses represent the 95% confidence interval of the metabolite profile for each group. (PNG 213 kb)
Acknowledgements
The authors wish to acknowledge the referring veterinarians for referring cases and the owners of all dogs for enrolling them.
Author contributions
SBS conducted the experiments, analyzed the data, performed the statistical analysis, prepared the figures and wrote the first draft of the paper. BS performed the data analysis. RP designed the study, performed the data analysis, and revised the work critically for important content. AJG designed the study, contributed to the clinical assessment and management of patients, sample acquisition and storage, collection and collation of clinical data, contributed to writing the manuscript, and revised the work critically for important content. GRTW revised the work critically for important content, contributed to the clinical assessment and management of patients, sample acquisition and storage, collection and collation of clinical data. JSS, FM, AG, JAL and JMS designed the study and revised the work critically for important content. All authors read and approved the manuscript. The journal’s authorship policy was followed.
Funding
This work has received financial support from Mars Petcare (VCR10030) and from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement (No. 713714) to ESR 09 (Sandra Bermudez Sanchez).
Data availability
The metabolomics data reported in this paper are available via supplementary material.
Declarations
Conflict of interest
The diets used in this study were manufactured by Royal Canin. Alexander J. German and Georgiana R.T. Woods are employees of the University of Liverpool but his academic post is funded by Royal Canin, part of Mars Petcare. Both have received financial remuneration and gifts for providing educational material, speaking at conferences, and consultancy work. Alexander J. Germans’s position at the University of Liverpool is funded by Royal Canin; Alexander J. German has also received financial remuneration and gifts for providing educational material, speaking at conferences, and consultancy work. Rachel Pilla, Joerg M. Steiner, Jonathan A. Lidbury and Jan S. Suchodolski are employed by the Gastrointestinal Laboratory at Texas A&M University, which provides assay for intestinal function and microbiota analysis on a fee-for-service basis. Jan S. Suchodolski have also received consulting fees from Royal Canin.
Ethical approval
The study protocol was reviewed and approved by the University of Liverpool Veterinary Research Ethics Committee (Approval reference: RETH000353 and VREC793), the Royal Canin ethical review committee, and the WALTHAM ethical review committee.
Informed consent
Informed consent was obtained from all owners of the dogs included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ables GP, Perrone CE, Orentreich D, Orentreich N. Methionine-restricted C57BL/6J mice are resistant to diet-induced obesity and insulin resistance but have low bone density. PLoS ONE. 2012;7:e51357. doi: 10.1371/journal.pone.0051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host & Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Anwar S, Bhandari U, Panda BP, Dubey K, Khan W, Ahmad S. Trigonelline inhibits intestinal microbial metabolism of choline and its associated cardiovascular risk. Journal of Pharmaceutical and Biomedical Analysis. 2018;159:100–112. doi: 10.1016/j.jpba.2018.06.027. [DOI] [PubMed] [Google Scholar]
- Arora T, Sharma R, Frost G. Propionate. Anti-obesity and satiety enhancing factor? Appetite. 2011;56:511–515. doi: 10.1016/j.appet.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Badgujar PC, Jain SK, Singh A, Punia JS, Gupta RP, Chandratre GA. Immunotoxic effects of imidacloprid following 28 days of oral exposure in BALB/c mice. Environmental Toxicology and Pharmacology. 2013;35:408–418. doi: 10.1016/j.etap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Bermudez Sanchez S, Pilla R, Sarawichitr B, Gramenzi A, Marsilio F, Steiner JM, Lidbury JA, Woods GRT, German AJ, Suchodolski JS. Fecal microbiota in client-owned obese dogs changes after weight loss with a high-fiber-high-protein diet. PeerJ. 2020;8:e9706. doi: 10.7717/peerj.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello SP, Setchell KD, Axelson M, Lawson AM. Production and metabolism of lignans by the human faecal flora. Journal of Applied Bacteriology. 1985;58:37–43. doi: 10.1111/j.1365-2672.1985.tb01427.x. [DOI] [PubMed] [Google Scholar]
- Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Medicine. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: A study in germ-free and human flora associated rats. Food and Chemical Toxicology. 2003;41:631–636. doi: 10.1016/S0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Richard AJ, Fernandez-Kim SO, Ribnicky DM, Salbaum JM, Newman S, Carmouche R, Stephens JM. Fenugreek counters the effects of high fat diet on gut microbiota in mice: Links to metabolic benefit. Science and Reports. 2020;10:1245. doi: 10.1038/s41598-020-58005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. Journal of Nutritional Biochemistry. 2013;24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Chong J, Xia J. Using metaboanalyst 4.0 for metabolomics data analysis, interpretation, and integration with other omics data. Methods in Molecular Biology. 2020;2104:337–360. doi: 10.1007/978-1-0716-0239-3_17. [DOI] [PubMed] [Google Scholar]
- Gaya P, Medina M, Sánchez-Jiménez A, Landete JM. Phytoestrogen metabolism by adult human gut microbiota. Molecules. 2016;21:1034. doi: 10.3390/molecules21081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German AJ. The growing problem of obesity in dogs and cats. Journal of Nutrition. 2006;136:1940S–S1946. doi: 10.1093/jn/136.7.1940S. [DOI] [PubMed] [Google Scholar]
- German AJ. Weight management in obese pets: The tailoring concept and how it can improve results. Acta Veterinaria Scandinavica. 2016;58:57. doi: 10.1186/s13028-016-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German AJ, Hervera M, Hunter L, Holden SL, Morris PJ, Biourge V, Trayhurn P. Improvement in insulin resistance and reduction in plasma inflammatory adipokines after weight loss in obese dogs. Domestic Animal Endocrinology. 2009;37:214–226. doi: 10.1016/j.domaniend.2009.07.001. [DOI] [PubMed] [Google Scholar]
- German AJ, Holden SL, Bissot T, Morris PJ, Biourge V. A high protein high fibre diet improves weight loss in obese dogs. The Veterinary Journal. 2010;183:294–297. doi: 10.1016/j.tvjl.2008.12.004. [DOI] [PubMed] [Google Scholar]
- German AJ, Titcomb JM, Holden SL, Queau Y, Morris PJ, Biourge V. Cohort study of the success of controlled weight loss programs for obese dogs. Journal of Veterinary Internal Medicine. 2015;29:1547–1555. doi: 10.1111/jvim.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves TL, Scott JE. A high throughput serum paraoxonase assay for discovery of small molecule modulators of PON1 activity. Current Chemical Genomics. 2008;2:51–61. doi: 10.2174/1875397300802010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, Su X, Qiao J, Zheng Y, Wang L, Koh E, Danliang H, Xu J, Lee YK, Zhang H. Intestinal microbiota distinguish gout patients from healthy humans. Science and Reports. 2016;6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl S, German AJ, Holden SL, Dowd SE, Steiner JM, Heilmann RM, Grant RW, Swanson KS, Suchodolski JS. Faecal microbiota in lean and obese dogs. FEMS Microbiology Ecology. 2013;84:332–343. doi: 10.1111/1574-6941.12067. [DOI] [PubMed] [Google Scholar]
- Harakeh SM, Khan I, Kumosani T, Barbour E, Almasaudi SB, Bahijri SM, Alfadul SM, Ajabnoor GM, Azhar EI. Gut microbiota: A contributing factor to obesity. Frontiers in Cellular and Infection Microbiology. 2016;6:95. doi: 10.3389/fcimb.2016.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M, Thomas G, Gilham M, Gray K, Colyer A, Allaway D. Short-term determination and long-term evaluation of the dietary methionine requirement in adult dogs. British Journal of Nutrition. 2020;123:1333–1344. doi: 10.1017/S0007114520000690. [DOI] [PubMed] [Google Scholar]
- Heinonen S, Nurmi T, Liukkonen K, Poutanen K, Wähälä K, Deyama T, Nishibe S, Adlercreutz H. In vitro metabolism of plant lignans: New precursors of mammalian lignans enterolactone and enterodiol. Journal of Agriculture and Food Chemistry. 2001;49:3178–3186. doi: 10.1021/jf010038a. [DOI] [PubMed] [Google Scholar]
- Hubbard TD, Liu Q, Murray IA, Dong F, Miller C, Smith PB, Gowda K, Lin JM, Amin S, Patterson AD, Perdew GH. Microbiota metabolism promotes synthesis of the human ah receptor agonist 2,8-dihydroxyquinoline. Journal of Proteome Research. 2019;18:1715–1724. doi: 10.1021/acs.jproteome.8b00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hursel R, Westerterp-Plantenga MS. Thermogenic ingredients and body weight regulation. International Journal of Obesity. 2010;34:659–669. doi: 10.1038/ijo.2009.299. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Maeda S, Ohno K, Horigome A, Odamaki T, Tsujimoto H. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLoS ONE. 2014;9:e107909. doi: 10.1371/journal.pone.0107909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MI, Jewell DE. Balance of saccharolysis and proteolysis underpins improvements in stool quality induced by adding a fiber bundle containing bound polyphenols to either hydrolyzed meat or grain-rich foods. Gut Microbes. 2019;10:298–320. doi: 10.1080/19490976.2018.1526580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World Journal of Gastroenterology. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen GH, Arts IC, Nielen MW, Müller M, Hollman PC, Keijer J. Uptake and metabolism of enterolactone and enterodiol by human colon epithelial cells. Archives of Biochemistry and Biophysics. 2005;435:74–82. doi: 10.1016/j.abb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Jeschke P, Nauen R. Neonicotinoids-from zero to hero in insecticide chemistry. Pest Management Science. 2008;64:1084–1098. doi: 10.1002/ps.1631. [DOI] [PubMed] [Google Scholar]
- Kho ZY, Lal SK. The human gut microbiome—A potential controller of wellness and disease. Frontiers in Microbiology. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott EJ, Richard AJ, Mynatt RL, Ribnicky D, Stephens JM, Bruce-Keller A. Fenugreek supplementation during high-fat feeding improves specific markers of metabolic health. Science and Reports. 2017;7:12770. doi: 10.1038/s41598-017-12846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolátorová, L., Lapčík, O., & Stárka, L. (2018). Phytoestrogens and the intestinal microbiome. Physiological Research,67(Suppl 3), S401–S408. [DOI] [PubMed]
- Laflamme D. Development and validation of a body condition score system for dogs. Canine Practice. 1997;22:10–15. [Google Scholar]
- Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunology. 2018;11:1024–1038. doi: 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
- Lee HC, Jenner AM, Low CS, Lee YK. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Research in Microbiology. 2006;157:876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Della-Fera MA, Baile CA. Green tea polyphenol epigallocatechin gallate inhibits adipogenesis and induces apoptosis in 3T3-L1 adipocytes. Obesity Research. 2005;13:982–990. doi: 10.1038/oby.2005.115. [DOI] [PubMed] [Google Scholar]
- Middelbos IS, Karr-Lilienthal LK, Folador J, Vazquez-Anon M, Yi G, Fahey GC. Effects of dietary supplementation of DL-methionine or 2-hydroxy-4-(methylthio)-butanoic acid on food intake, nutrient digestibility, nitrogen balance, and urinary and blood metabolites in healthy, growing dogs. Archives of Animal Nutrition. 2006;60:301–316. doi: 10.1080/17450390600785483. [DOI] [PubMed] [Google Scholar]
- Miousse IR, Pathak R, Garg S, Skinner CM, Melnyk S, Pavliv O, Hendrickson H, Landes RD, Lumen A, Tackett AJ, Deutz NEP, Hauer-Jensen M, Koturbash I. Short-term dietary methionine supplementation affects one-carbon metabolism and DNA methylation in the mouse gut and leads to altered microbiome profiles, barrier function, gene expression and histomorphology. Genes & Nutrition. 2017;12:22. doi: 10.1186/s12263-017-0576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Lee SE, Kim HB, Isaacson RE, Seo KW, Song KH. Association of obesity with serum leptin, adiponectin, and serotonin and gut microflora in beagle dogs. Journal of Veterinary Internal Medicine. 2015;29:43–50. doi: 10.1111/jvim.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portune KJ, Beaumont M, Davila A-M, Tomé D, Blachier F, Sanz Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends in Food Science & Technology. 2016;57:19. doi: 10.1016/j.tifs.2016.08.011. [DOI] [Google Scholar]
- Raffan E, Holden SL, Cullingham F, Hackett RM, Rawlings JM, German AJ. Standardized positioning is essential for precise determination of body composition using dual-energy x-ray absorptiometry in dogs. Journal of Nutrition. 2006;136:1976S–S1978. doi: 10.1093/jn/136.7.1976S. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: Metabolism of nutrients and other food components. European Journal of Nutrition. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Mani A, Jeusette I, Castillo I, Manuelian CL, Lionnet C, Iraculis N, Sanchez N, Fernández S, Vilaseca L, Torre C. Fecal microbiota composition changes after a BW loss diet in Beagle dogs. Journal of Animal Science. 2018;96:3102–3111. doi: 10.1093/jas/sky193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, Nunez D, Sweatman BC, Haselden JN, Cox RD, Connor SC, Griffin JL. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiological Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- Sittipo P, Shim JW, Lee YK. Microbial metabolites determine host health and the status of some diseases. International Journal of Molecular Sciences. 2019;20:5296. doi: 10.3390/ijms20215296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: Multitasking in the immune system. Annual Review of Immunology. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- Su W, Zhang H, Ying Z, Li Y, Zhou L, Wang F, Zhang L, Wang T. Effects of dietary L-methionine supplementation on intestinal integrity and oxidative status in intrauterine growth-retarded weanling piglets. European Journal of Nutrition. 2018;57:2735–2745. doi: 10.1007/s00394-017-1539-3. [DOI] [PubMed] [Google Scholar]
- Sun Q, Bai Y, Zhao C, Xiao Y, Wen D, Tang X. Aerobic biodegradation characteristics and metabolic products of quinoline by a Pseudomonas strain. Bioresource Technology. 2009;100:5030–5036. doi: 10.1016/j.biortech.2009.05.044. [DOI] [PubMed] [Google Scholar]
- Sun Q, Xiao X, Kim Y, Kim D, Yoon KS, Clark JM, Park Y. Imidacloprid promotes high fat diet-induced adiposity and insulin resistance in male C57BL/6J mice. Journal of Agriculture and Food Chemistry. 2016;64:9293–9306. doi: 10.1021/acs.jafc.6b04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuo BH, Yan JB, Fan BA, Yang ZH, Liu JZ. Biodegradation characteristics and bioaugmentation potential of a novel quinoline-degrading strain of Bacillus sp. isolated from petroleum-contaminated soil. Bioresource Technology. 2012;107:55–60. doi: 10.1016/j.biortech.2011.12.114. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarijonaviciute A, Ceron JJ, de Torre C, Ljubić BB, Holden SL, Queau Y, Morris PJ, Pastor J, German AJ. Obese dogs with and without obesity-related metabolic dysfunction—A proteomic approach. BMC Veterinary Research. 2016;12:211. doi: 10.1186/s12917-016-0839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarijonaviciute A, Ceron JJ, Holden SL, Biourge V, Morris PJ, German AJ. Effect of weight loss in obese dogs on indicators of renal function or disease. Journal of Veterinary Internal Medicine. 2013;27:31–38. doi: 10.1111/jvim.12029. [DOI] [PubMed] [Google Scholar]
- Tvarijonaviciute A, Ceron JJ, Holden SL, Cuthbertson DJ, Biourge V, Morris PJ, German AJ. Obesity-related metabolic dysfunction in dogs: A comparison with human metabolic syndrome. BMC Veterinary Research. 2012;8:147. doi: 10.1186/1746-6148-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. American Journal of Clinical Nutrition. 2011;93:62–72. doi: 10.3945/ajcn.110.000075. [DOI] [PubMed] [Google Scholar]
- Wang S, Moustaid-Moussa N, Chen L, Mo H, Shastri A, Su R, Bapat P, Kwun I, Shen CL. Novel insights of dietary polyphenols and obesity. Journal of Nutritional Biochemistry. 2014;25:1–18. doi: 10.1016/j.jnutbio.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, Z. J., Bleich, S. N., Cradock, A. L., Barrett, J. L., Giles, C. M., Flax, C., Long, M. W., & Gortmaker, S. L. (2019). Projected U.S. State-Level Prevalence of Adult Obesity and Severe Obesity. New England Journal of Medicine,381, 2440–2450. [DOI] [PubMed]
- Woolfolk CA, Downard JS. Distribution of xanthine oxidase and xanthine dehydrogenase specificity types among bacteria. Journal of Bacteriology. 1977;130:1175–1191. doi: 10.1128/jb.130.3.1175-1191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Liao SF. Physiological effects of dietary amino acids on gut health and functions of swine. Frontiers in Veterinary Science. 2019;6:169. doi: 10.3389/fvets.2019.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang J, Wu G, Sun J, Wang Y, Guo H, Shi Y, Cheng X, Tang X, Le G. Dietary methionine restriction regulated energy and protein homeostasis by improving thyroid function in high fat diet mice. Food & Function. 2018;9:3718–3731. doi: 10.1039/C8FO00685G. [DOI] [PubMed] [Google Scholar]
- Yu CL, Louie TM, Summers R, Kale Y, Gopishetty S, Subramanian M. Two distinct pathways for metabolism of theophylline and caffeine are coexpressed in Pseudomonas putida CBB5. Journal of Bacteriology. 2009;191:4624–4632. doi: 10.1128/JB.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. AHR function in lymphocytes: Emerging concepts. Trends in Immunology. 2016;37:17–31. doi: 10.1016/j.it.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chan L, Zhou S. Trigonelline: A plant alkaloid with therapeutic potential for diabetes and central nervous system disease. Current Medicinal Chemistry. 2012;19:3523–3531. doi: 10.2174/092986712801323171. [DOI] [PubMed] [Google Scholar]
- Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves CJ, Valdes AM, Spector TD, Menni C. The fecal metabolome as a functional readout of the gut microbiome. Nature Genetics. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1. Spreadsheet of the data from peak intensity for each sample and compound imported from Compound discoverer 2.1. (XLSX 341 kb)
Supplementary data 2. Association between baseline fecal metabolites and sex, and between baseline fecal metabolites and age. (XLSX 102 kb)
Supplementary figure 1. 2D PLS-DA score plot displaying the fecal metabolomics data from dogs before (T0, red) and after (T1, blue) weight reduction by feeding a therapeutic diet. Ellipses represent the 95% confidence interval of the metabolite profile for each group. (PNG 213 kb)
Data Availability Statement
The metabolomics data reported in this paper are available via supplementary material.