Abstract
The novel betacoronavirus severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) causes a form of severe pneumonia disease called coronavirus disease 2019 (COVID-19). To develop human neutralizing anti-SARS-CoV-2 antibodies, antibody gene libraries from convalescent COVID-19 patients were constructed and recombinant antibody fragments (scFv) against the receptor-binding domain (RBD) of the spike protein were selected by phage display. The antibody STE90-C11 shows a subnanometer IC50 in a plaque-based live SARS-CoV-2 neutralization assay. The in vivo efficacy of the antibody is demonstrated in the Syrian hamster and in the human angiotensin-converting enzyme 2 (hACE2) mice model. The crystal structure of STE90-C11 Fab in complex with SARS-CoV-2-RBD is solved at 2.0 Å resolution showing that the antibody binds at the same region as ACE2 to RBD. The binding and inhibition of STE90-C11 is not blocked by many known emerging RBD mutations. STE90-C11-derived human IgG1 with FcγR-silenced Fc (COR-101) is undergoing Phase Ib/II clinical trials for the treatment of moderate to severe COVID-19.
Keywords: SARS-CoV-2, RBD, spike protein, neutralizing antibody, recombinant antibody, phage display, immune library, in vivo neutralization, coronavirus, SARS-CoV-2 variants
Graphical abstract
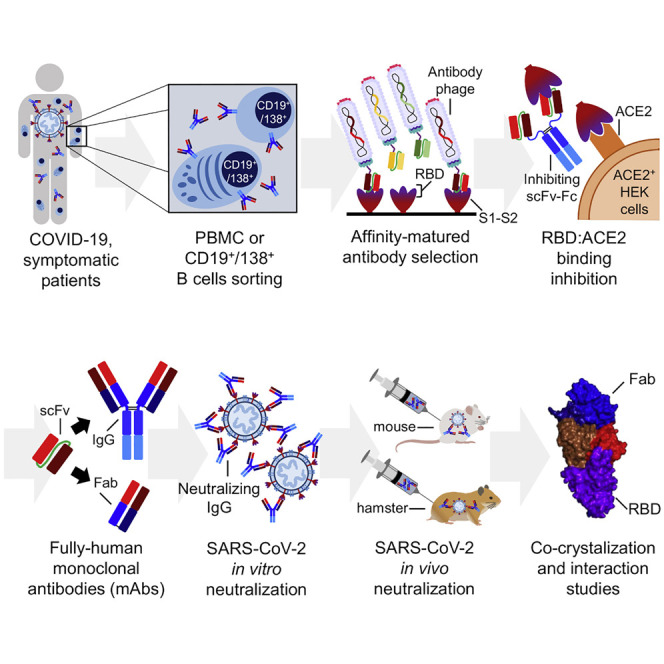
Bertoglio et al. describe the anti-SARS-CoV-2 antibody generation by phage display and development of the human antibody COR-101. In vitro and in vivo neutralization in two animal models is demonstrated. The structure is solved in complex with RBD. COR-101 with silenced Fc part is in clinical Phase Ib/II trial.
Introduction
The severe pneumonia COVID-19 (coronavirus disease 2019) is a disease caused by the novel coronavirus severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) and was described at the end of 2019 in Wuhan, China (Lu et al., 2020; Zhou et al., 2020). This new human pathogenic coronavirus is closely related to the bat coronavirus RATG13, indicating an animal-to-human transition (Shang et al., 2020a). The Spike (S) protein of SARS-CoV-2 binds to the human zinc peptidase angiotensin-converting enzyme 2 (ACE2) as the main receptor for cell entry. The S protein has two subunits: the N-terminal S1, harboring the receptor binding domain (RBD), and the viral membrane-anchored C-terminal S2 subunit, which is required for trimerization and fusion of the virus and host membrane for viral entry (Shang et al., 2020b; Starr et al., 2020; Walls et al., 2020; Wang et al., 2020b; Wrapp et al., 2020; Yan et al., 2020). Blocking of the RBD-ACE2 interaction by therapeutic antibodies as a strategy to treat COVID-19 (Zhou and Zhao, 2020) is a promising approach since the successful generation of neutralizing antibodies against S1 and RBD was already demonstrated for SARS-CoV and Middle East respiratory syndrome (MERS) (Coughlin and Prabhakar, 2012; Widjaja et al., 2019; Zhu et al., 2007). Some of the anti-SARS-CoV antibodies also showed cross-reaction and cross-neutralization of SARS-CoV-2 (Huo et al., 2020; Tian et al., 2020; Wang et al., 2020a).
Antibody phage display is a widely used in vitro technology to select human antibody fragments for the development of therapeutic antibodies (Frenzel et al., 2016). Currently, 14 European Medicines Agency- (EMA) or US Food and Drug Administration- (FDA) approved antibodies were generated by antibody phage display (Alfaleh et al., 2020). Human antibodies can be selected from two kinds of antibody gene libraries: universal libraries and immune libraries. Universal libraries, which have a naive, semi-synthetic, or synthetic antibody gene repertoire, allow, in theory, the selection of antibodies against any kind of molecule, while immune libraries derived from immunized donors typically facilitate the antibody generation against the pathogen that affected the donors (Bradbury et al., 2011; Frenzel et al., 2017; Kretzschmar and von Rüden, 2002; Kügler et al., 2018; Kuhn et al., 2016). Immune phage display libraries from convalescent patients or immunized human donors were used for the successful generation of antibodies against infectious diseases (Duan et al., 2009; Rahumatullah et al., 2017; Trott et al., 2014; Wenzel et al., 2020a; Zhang et al., 2003). Human antibodies were also selected directly against SARS-CoV-2 using different approaches, including single B cell cloning or antibody phage display (Baum et al., 2020; Bertoglio et al., 2021; Kreer et al., 2020; Robbiani et al., 2020; Shi et al., 2020; Zeng et al., 2020).
Recently, several SARS-CoV-2 variants with mutations in the RBD emerged. Here, most prominent are the B.1.1.7 (“UK,” RBD mutation N501Y), B.1.351 (“Southafrica,” K417N, E484K, N501Y), and P1 (B.1.1.28.1) (“Brazil,” K417T, E484K, N501Y) (Rees-Spear et al., 2021). More recently other variants such as B.1.429+B.1.427 (“Southern California,” L452R) (Zhang et al., 2021), B.1.526 (“New York,” E484K, in some variants S477N instead of E484K) (Annavajhala et al., 2021), B.1.258Δ (“Czech,” N439K) (Brejová et al., 2021; Surleac et al., 2021), P2 (B.1.1.28.2) (E484K) (Nonaka et al., 2021), P3 (B.1.1.28.3) (E484K, N501Y) (Tablizo et al., 2021), B.1.1.33 (E484K) (Resende et al., 2021), B.1.617 (“India,” L452R, E484Q) (Ranjan et al., 2021), and other variants such as B.1.525 (E484K) (Hodcroft et al., 2021) occurred.
In this work, immune phage display libraries from six COVID-19 convalescent patients were constructed and RBD-binding antibodies have been selected, resulting in SARS-CoV-2-inhibiting and -neutralizing antibodies. The crystal structure of the best neutralizing candidate STE90-C11 in complex with wild-type (WT) RBD was elucidated. Together with its unique binding pattern that tolerates a large number of RBD mutations, its properties suggest its use as a therapeutic agent against SARS-CoV-2 infections.
Results
Immune library construction
For the immune library construction, we collected blood samples of 16 qRT-PCR-confirmed COVID-19 convalescent patients. To analyze the immune answer, the sera of all patients were titrated on RBD (Data S1A). Patients 1, 2, 5, 6, 9, and 14 showed the highest immunoglobulin G (IgG) titers against RBD and were chosen for library construction. Peripheral blood mononuclear cells (PBMCs) were isolated and CD19+/CD138+ plasma cells were enriched by fluorescence-activated cell sorting (FACS) from some of these samples. Both PBMCs and plasma cells were used for library construction. Six individual libraries were constructed from the PBMCs of patients 2, 6, and 14, while 4 additional libraries were made from CD19+/CD138+ plasma cell pools (pool 1: patients 2, 6, and 14; pool 2: patients 1, 5, and 9). All of the libraries were constructed cloning kappa and lambda antibody genes separately, resulting in diversities ranging from 0.7 × 107 to 1.7 × 108 estimated independent clones.
Antibody fragments were selected against SARS-CoV-2
Antibodies were selected by panning in microtiter plates against SARS-CoV-2 S1 subunit using S1-S2 in the first panning round, RBD in the second panning round, and S1-S2 in the third panning round. This approach focused on the antibody selection on epitopes located within the RBD while being presented in the correct conformation of the intact S protein trimer. The two plasma cell-derived immune libraries were combined for kappa, and lambda, respectively, and the panning was performed separately for lambda and kappa libraries. All kappa and lambda libraries originating from the PBMCs were pooled together. The following single-clone screening was performed by antigen ELISA in 96-well microtiter plates (MTPs), using soluble monoclonal single-chain variable fragment (scFv) produced by Escherichia coli. A total of 542 monoclonal hits were identified. DNA sequencing revealed 197 unique antibodies. The V gene combinations for kappa and lambda (Data S1B) antibodies were analyzed. The most abundantly used V genes were VH3-66, VK1D-39, and VL3-21. Antibody sequences that showed potential developability liabilities (i.e., glycosylation sites or single cysteines in the complementarity-determining regions [CDRs]) were excluded from further analysis. A total of 135 antibodies were re-cloned into the bivalent scFv-Fc format and produced in Expi293F cell in 5-mL culture scales, with yields ranging from 70 to 260 mg/L.
Screening for inhibition in a cell-based assay
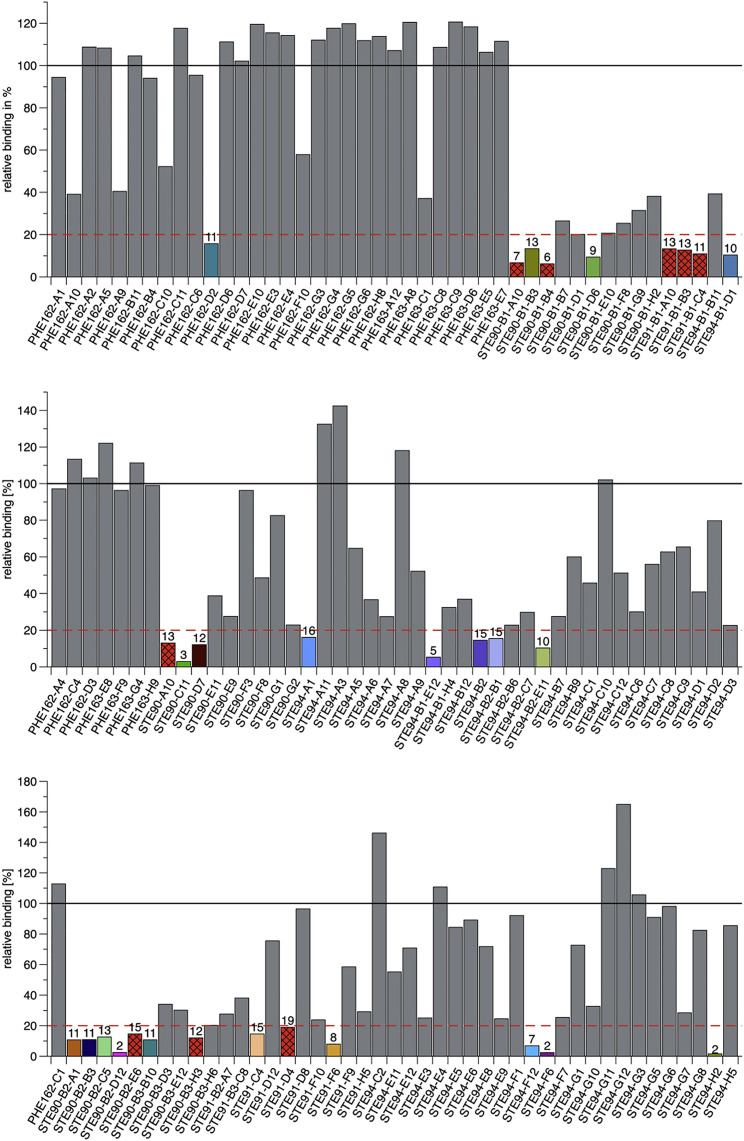
To screen for SARS-CoV-2-blocking antibodies, an inhibition assay was performed by flow cytometry on ACE2-expressing EXPI293F cells, measuring direct competition between S1-S2 trimer and scFv-Fc antibodies. The entire S protein ectodomain was used for this inhibition assay for optimal representation of viral binding. In a first screening, the 135 antibodies were tested at 1,500 nM S1-S2 (molar ratio antibody:S1-S2 30:1) (Figure 1 ). Thirty antibodies that inhibited S1-S2 binding >80% were selected for further analysis. The germline genes of these inhibiting scFv-Fcs are given in Table 1 . VH3-66 was the most abundantly used heavy-chain V gene of the well-inhibiting scFv-Fc. All VH3-66 derived antibodies had closely related variable domains of heavy chains (VH) CDR amino acid (aa) sequence, while the corresponding light chains were different.
Figure 1.
Inhibition of SARS-CoV-2 spike protein binding cells (flow cytometry)
Inhibition prescreen of 135 scFv-Fc antibodies on ACE2+ cells using 1,500 nM antibody and 50 nM Spike protein (30:1 ratio). The antibodies selected for detailed analysis are marked in colors. Bars with a red background and crossed stripes indicate scFv-Fcs not analyzed further. Data show single measurements.
Table 1.
Overview on inhibiting antibodies
| Antibody name | VH | Germinality index VH (%) | VL | Germinality index VL (%) | EC50 ELISA [nM] |
Flow cytometry Spike binding inhibition assay |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RBD | S1 | S1-S2 | IC50 (nM) with 50 nM Spike | Molar ratio antibody arm: Spike | IC50 (nM) with 10 nM RBD | Molar ratio antibody arm: RBD | |||||
| PHE162-D2 | IGHV4-31 | 98.9 | IGLV1-40 | 98.5 | 3.3 | 3.9 | 7.5 | 160 | 3.20 | 34.0 | 3.40 |
| STE90-A10 | IGHV3-66 | 98.6 | IGKV1-5 | 94.4 | – | – | – | – | – | – | – |
| STE90-C11 | IGHV3-66 | 98.6 | IGKV1-9 | 100 | 0.2 | 0.2 | 0.3 | 2.56a | 0.05a | 0.99a | 0.03a |
| STE90-D7 | IGHV3-66 | 98.6 | IGKV1-12 | 98.1 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| STE94-A1 | IGHV3-66 | 98.6 | IGLV1-40 | 100 | n.d. | n.d. | n.d. | 460 | 9.20 | 31.3 | 3.13 |
| STE90-B1-A10 | IGHV3-66 | 98.6 | IGKV1-5 | 98.1 | – | – | – | – | – | – | – |
| STE90-B1-B3 | IGHV3-66 | 98.6 | IGKV1-9 | 96.0 | – | – | – | 21.1 | 0.42 | 4.56 | 0.46 |
| STE90-B1-B4 | IGHV3-66 | 98.6 | IGKV1D-39 | 92.5 | – | – | – | – | – | – | – |
| STE90-B1-D6 | IGHV3-53 | 97.7 | IGKV1D-39 | 96.4 | – | – | – | 80.6 | 1.61 | 81.6 | 0.82 |
| STE90-B2-A1 | IGHV3-66 | 98.6 | IGKV1D-39 | 92.8 | 1.4 | 2.6 | 5.2 | 620 | 12.4 | 19.3 | 1.93 |
| STE90-B2-B3 | IGHV3-66 | 98.6 | IGKV1-5 | 100 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| STE90-B2-C5 | IGHV3-66 | 98.6 | IGKV1-5 | 100 | n.d. | n.d. | n.d. | 420 | 8.40 | 640 | 64.0 |
| STE90-B2-D12 | IGHV3-66 | 98.6 | IGKV1-9 | 100 | 0.3 | 0.4 | 0.4 | 27.1 | 0.54 | 4.49 | 0.45 |
| STE90-B2-E6 | IGHV3-53 | 100 | IGKV1D-39 | 98.1 | – | – | – | n.d. | n.d. | n.d. | n.d. |
| STE90-B3-B10 | IGHV3-66 | 98.6 | IGKV1-5 | 100 | n.d. | n.d. | n.d. | 250 | 5.0 | 490 | 49.0 |
| STE90-B3-H3 | IGHV3-66 | 98.6 | IGKV1D-12 | 100 | – | – | – | – | – | – | – |
| STE91-B1-A10 | IGHV3-66 | 98.6 | IGLV3-1 | 89.4 | – | – | – | – | – | – | – |
| STE91-B1-B3 | IGHV3-66 | 98.6 | IGLV3-21 | 100 | – | – | – | – | – | – | – |
| STE91-B1-C4 | IGHV3-66 | 98.6 | IGLV3-1 | 85.9 | – | – | – | – | – | – | – |
| STE91-C4 | IGHV3-66 | 98.6 | IGLV3-21 | 91.6 | 4 | 7.9 | n.d. | 36.4 | 0.73 | 7.15 | 0.72 |
| STE91-D4 | IGHV3-66 | 98.6 | IGLV3-1 | 89.4 | – | – | – | – | – | – | – |
| STE91-F6 | IGHV3-66 | 98.6 | IGLV1-40 | 100 | 2.3 | 3.8 | 5.9 | 30.0 | 0.60 | 5.99 | 0.60 |
| STE94-B1-D1 | IGHV3-66 | 98.6 | IGLV3-21 | 95.0 | 1 | 0.9 | 2.5 | 15.5 | 0.31 | 3.09 | 0.31 |
| STE94-B1-E12 | IGHV3-66 | 98.6 | IGLV3-1 | 85.0 | 0.4 | 0.7 | 1.6 | 11.5 | 0.23 | 2.49 | 0.25 |
| STE94-B2 | IGHV3-66 | 98.6 | IGLV3-21 | 98.3 | 5.1 | 4.8 | n.d. | n.d. | n.d. | 5.55 | 0.56 |
| STE94-B2-B1 | IGHV3-66 | 98.6 | IGLV1-40 | 100 | 1.2 | 2 | 7.5 | 140 | 2.80 | 3.35 | 0.34 |
| STE94-B2-E11 | IGHV3-66 | 98.6 | IGLV1-51 | 91.3 | 4.7 | 7.7 | n.d. | 39.1 | 0.78 | 6.25 | 0.63 |
| STE94-F6 | IGHV1-69 | 96.3 | IGKV1D-39 | 96.2 | 0.2 | 0.3 | 0.3 | 10.3 | 0.21 | 2.06 | 0.21 |
| STE94-F12 | IGHV3-66 | 98.6 | IGLV3-21 | 93.3 | 0.7 | 1.2 | 2.4 | 43.7 | 0.87 | 8.74 | 0.87 |
| STE94-H2 | IGHV3-66 | 98.6 | IGKV1-9 | 100 | 0.3 | 0.4 | 0.5 | 5.55 | 0.11 | 1.11 | 0.11 |
The EC50s were determined using 30 ng immobilized RBD-mFc, S1-mFc, and S1-S2-His (trimer) by ELISA. The IC50 was determined by flow cytometry using 50 nM (in relation to monomer) S1-S2 trimer, respectively 10 nM RBD and ACE2+ cells. The molar ratio of antibody binding site:S1–S2 or RBD is given for 50% inhibition. EC50 were calculated with GraphPad Prism version 6.1, fitting to a 4-parameter logistic curve. IC50s were calculated with OriginPro using the Logistic5 Fit. –, not applicable. n.d., not determined.
The IC50 data of STE90-C11 were generated in the validation experiment shown in Figure S6.
Antigen binding and cell-based inhibition by IgG
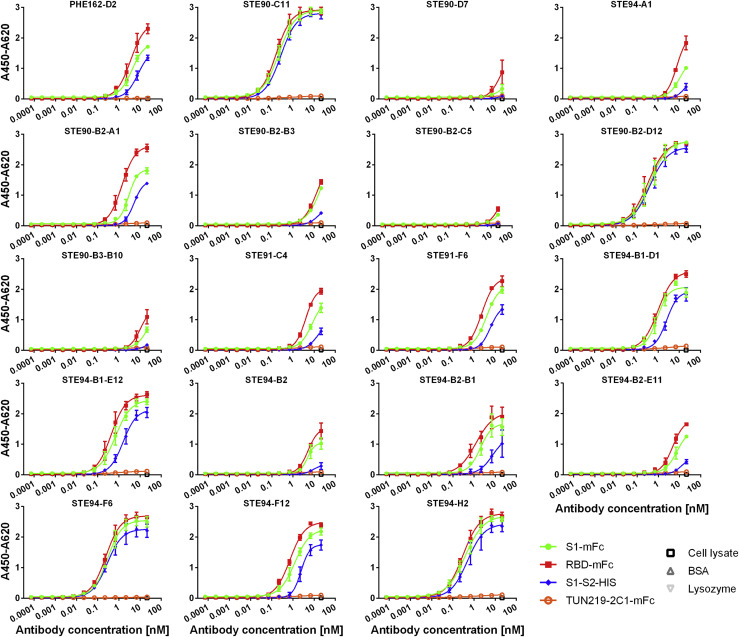
The 30 antibodies were screened in a cytopathic effect (CPE)-based neutralization screening assay (data not shown) to select antibodies for further characterization as IgG. This assay was performed with 250 plaque-forming units (pfu)/well SARS-CoV-2 and 1 μg/mL (∼10 nM) scFv-Fc. CPE is characterized by rounding and detachment clearly visible in phase contrast microscopy upon SARS-CoV-2 infection within 4 days, while uninfected cells maintained an undisturbed confluent monolayer. The best neutralizing 19 scFv-Fc were re-cloned and produced as human IgG in 50-mL culture scales, with yields ranging from 12 to 93 mg/L. Half-maximal effective concentration (EC50) values of binding to RBD, S1, or S1-S2 were determined (Figure 2 ) and are shown in Table 1. Antibodies STE90-C11, STE90-B2-D12, STE94-F6, and STE94-H2 showed EC50 values ranging between 0.2 and 0.5 nM on all of the antigens tested. Inhibition of ACE2 binding was assessed at concentrations from 100 to 0.3 nM IgG using 10 nM RBD (Data S2A) or 500–1.5 nM IgG using 50 nM S1-S2 Spike (Data S2B) in the previously described cell-based inhibition assay (Bertoglio et al., 2021). The extent of inhibition by STE90-C11 of both RBD and S1-S2 was validated in a further cellular assay with ACE2-expressing cells (Data S3). The best antibodies showed half-maximal inhibitory concentration (IC50) values of 2.6–15 nM IgG for 50 nM S1-S2 Spike, respectively, and 1–3 nM IgG for 10 nM RBD (Table 1). For 50% inhibition, the best molar ratios were from 0.02 to 0.3:1 (antibody binding site:antigen).
Figure 2.
Determination of EC50 in ELISA
Binding in titration ELISA of the IgGs to RBD (fusion protein with murine Fc part), S1 (fusion protein with murine Fc part), or S1-S2 (fusion protein with His tag). An unrelated antibody with murine Fc part (TUN219-2C1), human HEK293 cell lysate, BSA, and lysozyme were used as controls. Experiments were performed in duplicate and means ± SEMs are given. EC50 were calculated with GraphPad Prism Version 6.1, fitting to a 4-parameter logistic curve.
Antibodies from COVID-19 patient libraries neutralize active SARS-CoV-2 virus
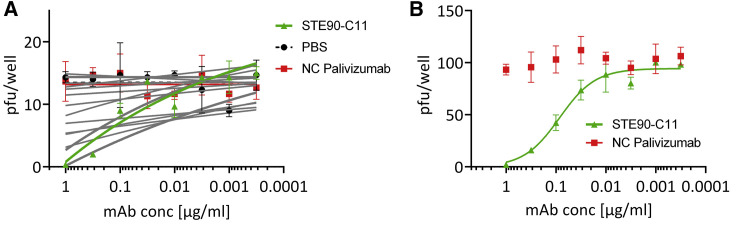
To analyze the neutralization capacity of the antibodies, the 19 IgGs were first screened in a plaque titration assay using active ∼15 pfu SARS-CoV-2 virus. The antibody palivizumab was used as a control antibody (Figure 3 A). The antibody STE90-C11 was chosen to be further analyzed. To confirm its neutralizing activity, a plaque assay using ∼150 pfu was performed and the determined IC50 was 0.56 nM in the IgG format (Figure 3B).
Figure 3.
In vitro neutralization of authentic SARS-CoV-2
(A) Neutralization screening of ~15 plaque-forming units (pfu) SARS-CoV-2 by 19 anti-RBD SARS-CoV-2 IgGs.
(B) STE90-C11 titration on ~150 pfu SARS-CoV-2 to determine the IC50. Neutralization assays were performed in triplicate; mean ± SEMs are given. Palivizumab was used as isotype control in both assays. IC50s were calculated with OriginPro using the Logistic5 Fit.
STE90-C11 is specific for SARS-CoV-2 and binds most RBD mutations, including B.1.617
A detailed characterization of the best neutralizer was performed in regard to cross-reaction to other coronaviruses, binding to known RBD mutations, and biochemical and biophysical properties. The binding of STE90-C11 to other coronaviruses SARS-CoV-1, MERS-CoV, HCov-HKU1, HCoV-229E, and HCoV-NL63 S proteins was tested by titration ELISA (Data S4A). STE90-C11 bound specifically SARS-CoV-2 and did not show any cross-reactions to other human-pathogenic coronaviruses.
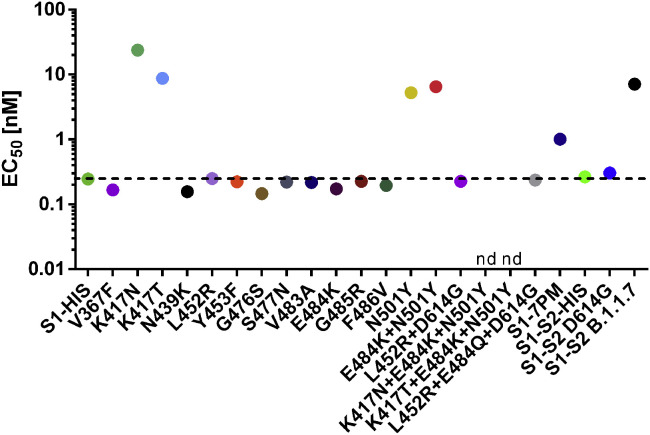
Because the first SARS-CoV-2 mutants in patients are being identified (https://www.gisaid.org and Shi et al., 2020) and more mutations within the RBD have been characterized to arise under antibody selection pressure (Baum et al., 2020), S1 subunits harboring single-point mutations (PMs) described in GISAID in the RBD were produced and the binding of STE90-C11 to those mutants was tested. In addition, a variant with 7 PMs (V367F, N439K, G476, V483A, E484K, G485R and F486V) was analyzed (S1-7PM). The EC50 values of STE90-C11 are given in Figure 4 , and the corresponding ELISAs are given in Data S4B–S4E, including further binding assays comparing STE90-C11 to REGN10933, REGN10987, CR3022, and CB6. All S1 or S1-S2 constructs were still able to bind recombinant ACE2 (Data S4F), indicating correct folding. The antibody STE90-C11 showed reduced binding to N501Y and lost binding to K417N/K417T, but it was able to bind to all of the other investigated RBD mutations, including a S1-7PM mutant and more relevant to E484K, N439K, and L452R, which are the RBD mutations in the emerging B.1.525, B.1.526, B.1.1.33, B.1.258, and B.1.429/B.1.427 variants. Most important is the binding to B.1.617 (L452R+E484Q), which is emerging in India and other parts of the world. Subsequently, the inhibition of S1 binding to ACE2 for the RBD mutants bound best by STE90-C11 was confirmed in the cell-based inhibition assays (Data S5). The inhibition of B.1.617 was slightly better than the inhibition of the WT.
Figure 4.
Cross-reactivity analysis of STE90-C11
Binding to S1 or S1-S2 of different SARS-CoV-2 RBD mutations identified in virus variants from COVID-19 patients analyzed by ELISA. The EC50 values are given for each analyzed mutant. The corresponding ELISAs are given in Data S4B–S4D. nd, not determinable. S1-7PM contains the 7 mutations V367F, N439K, G476, V483A, E484K, G485R, and F486V.
STE90-C11 is protective in vivo in the hamster challenge model
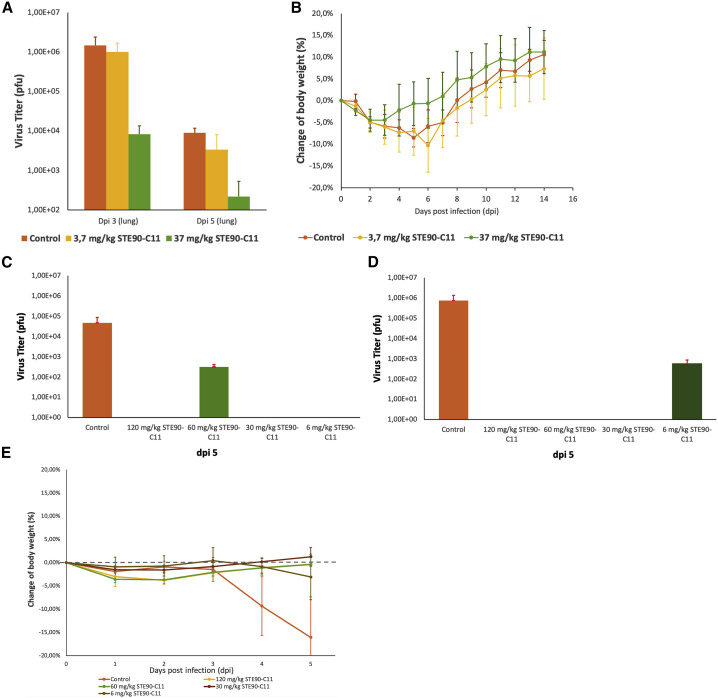
The SARS-CoV-2 neutralizing effect of STE90-C11 was tested in vivo in a Syrian hamster challenge model. In this model, hamsters were intranasally infected with 1 × 105 pfu genuine SARS-CoV-2 and treated 2 h later with 3.7 or 37 mg/kg STE90-C11 or with PBS in the control group. The virus titer in the lung of treated animals showed a dose-dependent reduction of viral load on days 3 and 5 after infection (Figure 5 A). The measured mean viral load was 8.3 × 103 pfu with 37 mg/kg IgG compared to 1.5 × 106 pfu in the PBS control at day 3. The hamster model is further characterized by a rapid weight loss in the first days after SARS-CoV-2 infection but also rapid recovery of body weight from ∼1 week post-infection on. Animals treated with the higher STE90-C11 dose showed a reduced loss of weight and a faster weight recovery compared to the PBS control (Figure 5B).
Figure 5.
In vivo protection of STE90-C11 in a Syrian hamster challenge model and in the transgenic mice model
(A and B) Hamster model. Challenging of 9 animals per group with authentic SARS-CoV-2 and treatment with 3.7 mg/kg, 37 mg/kg STE90-C11, or PBS.
(A) Quantification of SARS-CoV-2 pfu from lung homogenates after SARS-CoV-2 challenge 3 days after infection (3 dpi) or 5 days (5 dpi).
(B) Body weight of hamsters after SARS-CoV-2 challenge from days 0 to 14. The mean values ± SEMs from 9 animals per group are given.
(C–E) K18hACE2 mice model: 2–4 animals per group were treated with 6, 30, 60, or 120 mg/kg STE90-C11 or with PBS as control and 1 h later infected with authentic SARS-CoV-2.
(C and D) Quantification of SARS-CoV-2 pfu from lung homogenates after SARS-CoV-2 challenge 5 days post-infection (5 dpi). Two independent experiments are shown. Experiment 1 (C) was performed with 2,000 pfu and experiment 2 (D) with 1,000 pfu SARS-CoV-2.
(E) Body weight of the mice after SARS-CoV-2 challenge from day 0 to 5 from 3 independent experiments. The mean values ± SEMs from 2 to 4 animals per group are given.
STE90-C11 is protective in vivo in the transgenic ACE2 mice challenge model
The SARS-CoV-2 neutralizing effect of STE90-C11 was further tested in vivo in the transgenic human ACE2 mice model (K18hACE2). Mice were first treated with 6, 30, 60, or 120 mg/kg STE90-C11 or with PBS in the control group. After 1 h, they were inoculated intranasally with 1,000 or 2,000 pfu genuine SARS-CoV-2 or with PBS in the control group. The virus titer was measured with 4.7 × 104 pfu in the first experiment with 1,000 pfu and 7.4 × 105 pfu in the experiment with 2,000 pfu. Only in the two experiments, one with 60 mg/kg and one with 6 mg/kg, was a remaining viral load of 5.6 × 102 and 3.1 × 102 pfu, respectively, detected. In all other mice treated with STE90-C11, the virus was completely removed from the lungs (Figures 5C and 5D). In the control group, a significant weight loss was measured 4 days post-infection, which was not observed in the groups with STE90-C11 treatment (Figure 5E).
SARS-CoV-2 escape mutant screening leads to increased K417x but not N501x mutations
An in vivo escape mutant screening experiment was performed according to Baum et al. (2020) to determine RBD mutants that are not neutralized by STE90-C11. Here, serial dilutions of STE90-C11 and palivizumab as the control antibody were used to infect Calu-3 cells with active WT SARS-CoV-2. The supernatant with the propagated virus particles were used for the second round of neutralization and infection. After the second passage, the viruses were analyzed by DNA sequencing. In the experiment with STE90-C11, 4.7% of the remaining viruses were WT, 81.9% had the K417T mutations, 3.35 K417T+A475V, 0.3% A475V, 0.3% N481H, 0.12% N481Y, and 0.11% N501T. In the control experiment with a non-related antibody and no selection pressure, 89.9% of the remaining viruses were WT, 0.32% N481H, 0.14% K417T, 0.13% N501T, and 0.1% Q414K. This experiment shows that the K417x mutation was not neutralized by STE90-C11, but no selection pressure was found on the N501x mutations, indicating a sufficient neutralization of these variants by STE90-C11.
Biochemical characterization of STE90-C11
Aggregation propensity is an important quality parameter in therapeutic antibody development. STE90-C11 showed no relevant aggregation under normal conditions (pH 7.4, 4°C in PBS), heat stress conditions (pH 7.4, 45°C, 24 h in PBS), and pH stress (pH 3, 24 h, room temperature [RT]) (Data S6A). A further test to exclude polyreactivity was performed on different antigens (DNA, lipopolysaccharide [LPS], lysozyme, and EXPI cell lysate) with the FDA-approved antibody avelumab as control (Data S6B). Here, STE90-C11 showed equal or less unspecificity compared to avelumab. The monovalent affinity of STE90-C11 was determined by bio-layer interferometry (BLI) in three different setups (Data S6C–S6E). In the first setup, RBD-mFc was immobilized using anti-Fc capture tips, and the KD of STE90-C11 in monovalent Fab format was determined to be 8.1 nM. In the second setup, the IgG was immobilized using a Fab2G capture tip, and the KD of monovalent S1-His was determined to be 1.6 nM. In the third assay, the IgG was immobilized using a protein A capture tip, resulting in a measured KD of 6.5 nM to S1-His, confirming that the affinity of STE90-C11 is in the low nanomolar range. STE90-C11 did not bind to RBD on immunoblots after treatment at 95°C and SDS-gel separation under reducing conditions, while it was able to detect RBD when separated under non-reducing conditions heated either at 95°C or 56°C (Data S6F). These data suggest that the antibody recognizes a conformational epitope.
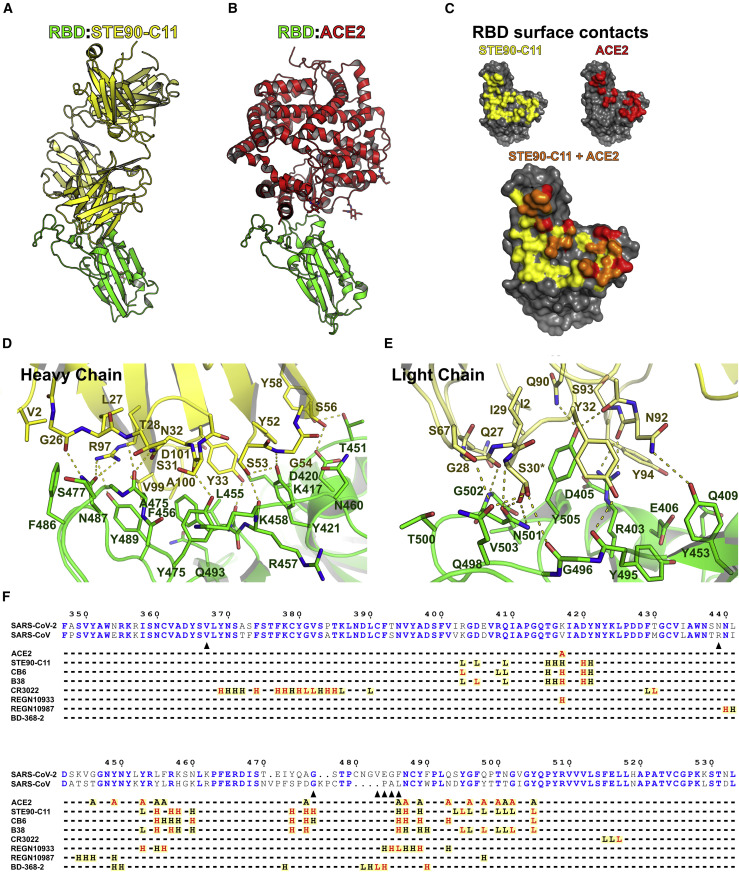
STE90-C11 binds at the ACE2-RBD interface
To get further insight into the neutralizing mechanism of STE90-C11, a complex of STE90-C11 Fab and SARS-CoV-2 RBD (22-kDa fragment) was prepared and subjected to crystallization screening. X-ray diffraction images collected from the resulting crystals yielded a dataset to an overall resolution limit of 2.0 Å (Data S7A). After solving the structure by molecular replacement, a model was built into the electron density (Figure 6 ). Figure 6A shows the binding of STE90-C11 to RBD and Figure 6B shows the binding of ACE2 to RBD. STE90-C11 binds SARS-CoV2 RBD with an interface area of 1,113 Å2 (Figure 6C; Data S7B). Roughly 60% of this area can be contributed to the VH segment, forming up to 10 hydrogen bonds at the same time and the region dominated by hydrophobic interaction (V99, A100, Y33, and Y52) (Figure 6D). The remaining 40% are provided by the variable domain of light chain (VL) segment contributing 8 additional hydrogen bonds to stabilize the interaction (Figure 6E). All 6 CDR loops contribute to the interaction between STE90-C11 Fab and SARS-CoV-2 RBD. The superposition of the RBDs of the STE90-C11:SARS-CoV-2 RBD complex with a ACE2:SARS-CoV-2 RBD complex (Lan et al., 2020) resulted in a low Cα root mean square distance of 0.532 Å (183 atoms), indicating that the binding of STE90-C11 did not induce substantial changes in the RBD. The superposition further revealed that the neutralization mechanism of STE90-C11 is based on directly competing for the ACE2 binding side, as the interaction interfaces on the RBD of both molecules almost completely overlap. Further superposition of the STE90-C11:SARS-CoV-2 RBD complex model onto cryoelectron microscopy (cryo-EM) models of the whole SARS-CoV-2 S protein (Walls et al., 2020) indicate that due to steric hindrance, STE90-C11-like ACE2 is expected to bind only to the open conformation of the S protein (Data S7C).
Figure 6.
The crystal structure of STE90-C11 in complex with SARS-CoV-2-RBD
(A and B) The structure of the complex between RBD (green) and STE90-C11 (A; heavy chain: yellow; light chain: pale yellow) or ACE2 (B; red; PDB: 6M0J; Lan et al., 2020) depicted in illustrative form after superposition of the RBD.
(C) Surface representation of the RBD with the binding surface (cutoff 4 Å) of STE90:C11 in yellow and of ACE2 in red. The competitive binding surface is in orange.
(D and E) Detailed view of the interactions between RBD and the heavy chain (D) and light chain (E) of STE90-C11. Possible hydrogen bonds are displayed as dashed yellow lines and residues with alternate conformations are marked with an asterisk.
(F) Sequence alignment of RBD. Depicted is a part of the sequence alignment of the SARS-CoV-2 and SARS-CoV S-protein (UniProt: P0DTC2; P59594). Identical residues are depicted as bold blue letters. Under the alignment contacts between residues of the S protein and ACE2 (PDB: 6M0J; Lan et al., 2020), STE90-C11 (PDB: 7B3O), CB6 (PDB: 7C01; Shi et al., 2020), B38 (PDB: 7BZ5; Wu et al., 2020), CR3022 (PDB: 6W41; Yuan et al., 2020), REGN10933, REGN10987 (PDB: 6XDG; Hansen et al., 2020), and BD-368-2 (PDB: 7CHF; Cao et al., 2020) are marked by an A (ACE2), H (heavy chain), or L (light chain). A black letter indicates a distance under 4 Å and a red letter indicates a distance under 3.2 Å between non-hydrogen atoms of the RBD and the respective protein. Black arrows indicate residues often exchanged in the RBD of SARS-CoV-2, which were tested for their influence on STE90-C11 binding in this work. Distances were calculated with CNS (Brünger et al., 1998) and the representation was prepared using ESPript (Robert and Gouet, 2014).
Discussion
For the treatment of COVID-19 but also to protect risk groups, human anti-SARS-CoV-2 antibodies are a promising therapeutic option. Human recombinant antibodies were successfully used for the treatment of other viral diseases. The antibody mAb114 (ansuvimab-zykl) (Corti et al., 2016) and three-antibody cocktail REGN-EB3 (atoltivimab/maftivimab/odesivimab) (Pascal et al., 2018) were approved by the FDA in 2020 and showed a good efficiency in clinical trials against the Ebola virus, especially in comparison to remdesivir (Mulangu et al., 2019). For the treatment of a severe respiratory infection of infants caused by the respiratory syncytial virus (RSV), the antibody palivizumab is approved by both EMA and FDA (van Mechelen et al., 2016; Subramanian et al., 1998). These anti-viral antibodies can be used as a blueprint to develop therapeutic antibodies against SARS-CoV-2. Currently, monoclonal antibodies against SARS-CoV-2 are selected by rescreening memory B cells from a SARS patient (Pinto et al., 2020), selected from COVID-19 patients by single B cell PCR (Cao et al., 2020; Shi et al., 2020) or FACS sorting (Kreer et al., 2020), selected from transgenic mice and from patients by single B cell FACS sorting (Hansen et al., 2020) or using phage display with universal or patient libraries, including different antibody formats (Bertoglio et al., 2021; Chi et al., 2020; Li et al., 2020; Liu et al., 2020; Ma et al., 2021; Noy-Porat et al., 2020; Parray et al., 2020; Zeng et al., 2020).
In this work, antibody phage display immune libraries were constructed using lymphocytes from six local convalescent COVID-19 patients. A total of 197 unique antibodies were selected against RBD. We intently focused on RBDs, aiming to select antibodies that directly interfere with the interaction between S protein and ACE2, to avoid potential antibody-dependent enhancement (ADE), especially in patients with severe symptoms. ADE during coronavirus entry has been described for both MERS (Wan et al., 2020) and SARS (Wang et al., 2014). It is defined as “enhancement of disease severity in an infected person or animal when an antibody against a pathogen…worsens its virulence by a mechanism that is shown to be antibody-dependent” (Arvin et al., 2020). Furthermore, immune dysregulation and lung inflammation was also caused by anti-Spike antibodies in an acute SARS-CoV infection (Liu et al., 2019). However, Quinlan et al. (2020) reported that animals immunized with RBD SARS-CoV-2 did not mediate ADE. A therapeutic antibody directed against RBD and the usage of a Fc part that is not binding to Fc-γ receptors (“silent Fc”) would reduce the risk of ADE, in particular because ADE cannot be fully predicted from in vitro studies or from animal models (Arvin et al., 2020). Winkler et al. (2021) argue that an intact Fc part is needed for optimal therapeutic protection. The ACTIV-3/TICO LY-CoV555 Study Group et al. (2021) described no efficacy of LY-COV-555 in hospitalized patients and also discussed ADE as a potential explanation for the lacking efficacy. Because the role of ADE is not fully deciphered in the case of SARS-CoV-2 (Lee et al., 2020) and LY-CoV555 was not effective in patients with severe symptoms, it is advisable that a silent Fc part be used to address hospitalized patients with moderate to severe symptoms. Therefore, all assays with STE90-C11 IgGs were performed with a silenced Fc part.
The selected anti-RBD antibodies were analyzed for RBD::ACE2 inhibition in the scFv-Fc format. The advantage of the IgG-like bivalent scFv-Fc format is that it speeds up analysis by requiring only one cloning step and provides high expression yields in a small-scale format (Bertoglio et al., 2021; Jäger et al., 2013; Wenzel et al., 2020a). A total of 30 scFv-Fc antibodies inhibited the binding of the S protein to ACE2-expressing living cells. Interestingly, the majority of the VH V genes of the selected anti-RBD antibodies and 28 of these 30 inhibiting antibodies is the VH3-66 V gene showed strong enrichment of this V gene in the immune response of COVID-19 patients against RBD. This is in accordance with Cao et al. (2020), who found enriched use of VH3-53 or VH3-66. VH3-53 and VH3-66 V-genes are closely related. Robbiani et al. (2020) showed an enrichment of VH3-30, VH3-53, and VH3-66 and Hansen et al. (2020) reported a bias toward VH3-53. The VH3-53 and VH3-66 V-genes are closely related. In contrast, Ju et al. (2020) identified mainly VH1-2, VH3-48, and VH3-9 V-genes for their selected anti-RBD antibodies from one patient and Brouwer et al. (2020) identified a strong enrichment of VH1-69, VH3-30 and VH1-24 in three patients, but also an enrichment of VH3-53 and VH3-66. VH3-66 antibodies against RBD were also selected from the naive HAL9/10 antibody gene libraries made long before the SARS-CoV-2 outbreak (Kügler et al., 2015) but not significantly enriched (Bertoglio et al., 2021). The selected inhibiting antibodies in our study (Table 1) have a high germinality index and are therefore very similar to the not hypermutated human germline, promising low immunogenicity when used as a therapeutic agent. Only some light chains showed a major difference to their closest related V gene. Kreye et al. (2020) also noted that the antibodies they selected from COVID-19 patients were very close to the germline genes. Interestingly, the VH V genes of these immune libraries are closer to their germline compared to the anti-RBD antibodies selected from the naive HAL9/10 antibody gene libraries (Bertoglio et al., 2021).
In the cell-based inhibition assay, some of the antibodies showed a molar antibody:spike monomer ratio lower than 1:1. Similar behavior was also found by Bertoglio et al., (2020) and may be explained by the observation that three RBDs on the same S trimer can be in different conformations (“up” and “down”), while only the RBDs in “up” positions are able to bind ACE2 (Walls et al., 2020), and therefore, the “down” RBDs are not fully accessible.
The monovalent affinity of STE90-C11 was measured with 1.6–8.1 nM, depending on the BLI setup. This is in the same range as BD-368-2, with 0.82 nM affinity to RBD (Cao et al., 2020), 3.3 nM for REGN10933 in the setup with the monovalent RBD (Hansen et al., 2020), 70 nM for B38 (Wu et al., 2020), or 0.8–4.7 nM for CB6 (Shi et al., 2020).
The analysis of the binding STE90-C11 to other human coronaviruses showed specificity for SARS-CoV-2. Comparison between the epitope of STE90-C11 and other published antibodies (Figure 5F) revealed that it binds in the same region as CB6 (Shi et al., 2020) and B38 (Wu et al., 2020). The selectivity of STE90-C11 and CB6 for SARS-CoV-2 over SARS-CoV RBD can mainly be attributed to the interaction with residues 473–476, as they are not involved in ACE2 binding, and SARS-CoV RBD folds into a different conformation of this loop, causing steric hindrance for antibody binding (Lan et al., 2020).
In addition to the in vitro neutralization of authentic SARS-CoV-2, the in vivo efficacy of STE90-C11 was demonstrated in the Syrian hamster challenge model (Kreye et al., 2020) and in the model using transgenic mice with hACE2 receptor (Wu et al., 2020) in a dose-dependent manner.
The binding of STE90-C11 to S1 containing RBD mutations observed in strains from COVID-19 patients was analyzed, showing binding to most variants. As the cell-based inhibition analysis demonstrates, STE90-C11 is able to inhibit most analyzed mutants, validating a tolerance for RBD mutations. REGN10933 (Hansen et al., 2020), which is also binding at the RBD-ACE2 interface, showed a loss of neutralization in an assay using pseudoviral particles for the F486V and a reduced neutralization for both G485D and E484K mutations (Baum et al., 2020). Loss of binding to F486A is also described for VH-Fc ab8 (Li et al., 2020). In the epitope analysis, REGN10933 has more molecular interactions within the region aa483–aa486 compared to STE90-C11. Currently, the variant B.1.1.7 is widespread and STE90-C11 has reduced binding to N501Y and lost binding to mutants with the K417N/T mutations in combination with N501Y, which occur in B.1.351 and B.1.1.28.1 variants. The structure suggests that the exchange N501Y may push the light chain CDR1 loop of the antibody further away, while the exchange of K417N/T leads to an abolishment of a salt bridge to D101 of the heavy chain, similar to observations done for COVOX-269 and COVOX-222, respectively (Dejnirattisai et al., 2021; Supasa et al., 2021). In COVOX-269, binding to the RBD-N501Y leads to a conformational change of light-chain CDR3 loop around Y94. In the escape mutant experiment, no N501x mutants were enriched, indicating a sufficient neutralization of the N501x but not K417x mutation.
On the other hand, STE90-C11 binds strongly to the RBD mutations in the emerging SARS-CoV-2 variants B.1.429/B.1.427 (L452R), B.1.526 (E484K or S477N), B1.258Δ (N439K), B.1.525 (E484K), B.1.1.28.2 (E484K), B.1.1.33 (E484K), and B1.617 (L452R, E484Q). These variants are emerging; for example, the frequency of B.1.429+B.1.427 reached 40% in January 2021 (Zhang et al., 2021) or B.1.258 with 59% in the samples sequenced in Czech in the last 3 months of 2020 (Brejová et al., 2021). B1.617 (L452R, E484Q) and derivates have superseded B.1.1.7 in India, and the prevalence of this variant was the majority of all of the sequenced viruses at the end of April 2021 (https://outbreak.info/location-reports?loc=IND). We hypothesize that STE90-C11 still binds to most analyzed RDB mutants because of both the wide interface area of 1,133 Å2 and the extensive light-chain contacts that may compensate the exchange of several amino acids. The calculated interface area of STE90-C11 VL is more than twice the area of REGN10933 and REGN10987 and slightly larger compared to CB6 or CR3022 (Data S7B).
In summary, the patient-derived SARS-CoV-2 neutralizing anti-RBD antibody STE90-C11 binding to the RBD-ACE2 interface maintains high similarity to the human germline V genes VH3-66, the same family of many isolated anti-SARS-CoV-2 neutralizing antibodies. STE90-C11 is tolerant to most known RBD mutants, especially those of the mutants B.1.429/B.1.427, B.1.526, B1.258Δ, B.1.535, B.1.617, and B.1.1.33, which are currently emerging. A Phase Ib/II clinical trial with a full human IgG1 variant with FcγR-silenced Fc STE90-C11 (COR-101) was started in April 2021 (ClinicalTrials.gov ID: NCT04674566) to assess the safety and efficacy of COR-101 in hospitalized patients with moderate to severe COVID-19.
STAR★Methods
Key resources table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| goat-anti-hIgG(Fc)-HRP | Sigma | Cat#A0170; RRID:AB_257868 |
| murine mAb 9E10 | Expressed for this paper in-house, original (Chan et al., 1987) | N/A |
| goat-anti-mIgG(Fc)-HRP | Sigma | Cat#A0168; RRID:AB_257867 |
| Hyper-Myc (TUN219-2C1-mFc) | Abcalis | Cat#ABC0503 |
| Avelumab | Expressed for this paper in-house, original (Boyerinas et al., 2015) | N/A |
| Palivizumab | Expressed for this paper in-house, original (Subramanian et al., 1998) | N/A |
| STE90-C11 | This paper | N/A |
| mouse anti-His epitope tag | Dianova | Cat#DIA-900-200 |
| mounse anti-penta His | QIAGEN | Cat#34660; RRID:AB_2619735 |
| goat anti-hIgG(Fc)-AP | Jackson ImmunoResearch | Cat#AB_2337599; RRID:AB_2337599 |
| goat anti-mIgG-AP | Dianova | Cat#115-055-071; RRID:AB_2338535 |
| mouse α-CD138 (FITC conjugated) | BioLegend | Cat#352304; RRID:AB_10900441 |
| Bacterial and virus strains | ||
| E. coli XL1 Blue MRF’ | Stratagene | Cat#200230 |
| E. coli TG1 | Lucigen | Cat#605021 |
| SARS-CoV-2/Münster/FI110320/1/2020 | University of Münster, Germany | N/A |
| SARS-CoV-2 BetaCoV/Germany/BavPat1/2020 | Charité, Germany | N/A |
| Hyperphage | Produced in-house | N/A |
| M13KO7 helperphage | Produced in-house | N/A |
| Experimental model: cell lines | ||
| Expi293F | Thermo Fisher Scientific | Cat#A14527 |
| VeroE6 | ATCC | ATCC CRL-1586 |
| High Five | Thermo Fisher Scientific | Cat#B85502 |
| Biological samples | ||
| COVID-19 patient samples | local hospitals | N/A |
| COVID-19 immune libraries | This paper | N/A |
| Healthy adult control samples (from pre-COVID-19 time) | LADR GmbH Medizinisches Versorgungszentrum Braunschweig | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Spike protein (RBD; S1; S1-S2; N) | This paper | GenBank: MN908947 |
| ACE2 | This paper | GenBank: NM021804.3 |
| PEI 40 kDa | Polysciences | Cat#24765-1 |
| Ficoll | GE Healthcare | Cat#45-001-749 |
| TRIzol LS | Life Technologies | Cat#10296028 |
| Superscript IV | Thermo Fisher Scientific | Cat#18090010 |
| Trypsin | Sigma | Cat#9002-07-7 |
| IPTG (50 μM) | Sigma | Cat#367-93-1 |
| GIBCO FreeStyle F17 expression media | Thermo Fisher Scientific | Cat#10388033 |
| HyClone SFM4Transfx-293 media | GE Healthcare | Cat#10015813 |
| HyClone Boost 6 supplement | GE Healthcare | Cat#11576341 |
| Laemmli sample buffer | Laemmli, 1970 | N/A |
| 5-Brom-4-Chlor-3-Indolyl-Phosphat/Nitro Tetrazolium Blue Chloride | Thermo Fisher Scientific | Cat#B6149 |
| CHO CD efficient feed A | Thermo Fisher Scientific | Cat#A1023401 |
| ExCell 405 serum-free medium | Sigma | Cat#14405C |
| 2,3-(R,R)-butandiol | Alfa Aeser | Cat#513-85-9 |
| Critical commercial kits | ||
| Direct-zol RNA Miniprep Plus Kit | Zymo Research | Cat#R2072 |
| NucleoBond Xtra Midi Kit | Macherey-Nagel | Cat#12798402 |
| NucleoSpin Gel and PCR Clean-up Kit | Macherey-Nagel | Cat#11992242 |
| NucleoSpin Plasmid EasyPure | Macherey-Nagel | Cat#15162537 |
| NEB Luna Universal Probe One-Step RT-qPCR kit | NEB | Cat#E3006S |
| Deposited data | ||
| Crystal structure of STE90-C11 in complex with SARS-CoV-2-RBD | This paper | PDB 7B3O |
| Oligonucleotides | ||
| Primer set for VH and VL amplification | (Wenzel et al., 2020b) | N/A |
| Recombinant DNA | ||
| Spike protein (RBD; S1; S1-S2; N) | Thermo Fisher/Gene Art | GenBank: MN908947 |
| ACE2 | Thermo Fisher/Gene Art | GenBank: NM021804.3 |
| Software and algorithms | ||
| OrginPro (2019) | OrginLab | N/A |
| GraphPad Prism 6.1 | GraphPad | https://www.graphpad.com |
| VBASE | MRC Centre for Protein Engineering | https://www2.mrc-lmb.cam.ac.uk/vbase/ |
| VBASE2 | (Mollova et al., 2010) | www.vbase2.org |
| DataGraph (4.5.1) | DataGraph | https://www.visualdatatools.com/DataGraph/ |
| IncuCyte S3 GUI | Sartorius | N/A |
| Phenix | (Liebschner et al., 2019) | www.phenix-online.org |
| BD FACS Chorus | BD | N/A |
| PyMol | Schrödinger LLC | www.pymol.org/2/ |
| PISA | European Bioinformatics Institute | https://www.ebi.ac.uk/pdbe/prot_int/pistart.html |
| Octet qKe Data Analysis HT 11.0 | Fortebio/Sartorius | N/A |
| Other | ||
| Äkta Go/Äkta Pure | Cytiva | N/A |
| Profinia System | BIO-RAD | N/A |
| Precision XS microplate sample processor | BioTek | N/A |
| Sartorius IncuCyte S3 | Sartorius | N/A |
| BD FACSMelody Cell Sorter | BD | N/A |
| Octet qKe | Fortebio/Sartorius | N/A |
| PETRA III | DESY Hamburg | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Hust (m.hust@tu-bs.de)
Materials availability
All requests for resources and reagents should be directed to the Lead Contact author. This includes antibodies, plasmids and proteins. All reagents are available on request after completion of a Material Transfer Agreement.
Data and code availability
The structure data and the antibody sequence of COR-101 is available at PDB 7B3O. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Additional resources
The clinical trial Ib/II studies are registered at ClinicalTrials.gov (ID: NCT04674566).
Experimental model and subject details
For the generation of human immune antibody libraries against SARS-CoV-2, blood from COVID-19 convalescent patients were collected from local hospitals. This was performed in accordance with the Declaration of Helsinki. All the voluntary donors were informed about the project and gave their consent. The use of blood samples for the development of antibody phage display libraries was approved by the ethical committee of the Technische Universität Braunschweig (Ethik-Kommission der Fakultät 2 der TU Braunschweig, approval number FV-2020-02). The donors have given their consent for study publication. The donors included both sex and were older than 18 years. A control serum was obtained from LADR Braunschweig.
Method details
Production of antigens in insect cells
The antigens were produced and purified as described before (Bertoglio et al., 2021; Korn et al., 2020). In brief, different domains or subunits of the Spike protein (GenBank: MN908947) were produced Baculovirus-free in High Five cells (Thermo Fisher Scientific, Schwerte, Germany) by transient transfection. High Five cells were cultivated at 27°C, 110 rpm in EX-CELL 405 media (Sigma Aldrich, Munich, Germany) and kept at a cell density between 0.3 – 5.5 x106 cells/mL. For transfection cells were centrifuged and resuspended in fresh media to a density of 4x106 cells/mL and transfected with 4 μg plasmid/mL and 16 μg/mL of PEI 40 kDa (Polysciences). After 4 h to 24 h after transfection cells were diluted to a final density of 1x 106 cells/mL. At 48 h after transfection, the culture volume was doubled. Cell supernatant was harvested five days after transfection in a two-step centrifugation (4 min at 180xg and 20 min at above 3500xg) and 0.2 μm filtered for purification.
Protein purification
Protein purification was performed as described before (Bertoglio et al., 2021) depending on the production scale in either 24 well filter plate with 0.5 mL resin (10 mL scale) or 1 mL column on Äkta go (Cytiva), Äkta Pure (Cytiva) or Profinia System (BIO-RAD). MabSelect SuRe or HiTrap Fibro PrismA (Cytiva) was used as resin for Protein A purification. For His-tag purification of Expi293F supernatant HisTrap FF Crude column (Cytiva) and for His-tag purification of insect cell supernatant HisTrap excel column (Cytiva) was used. All purifications were performed according to the manufacturer’s manual. Indicated antigens were further purified by size exclusion chromatography by a 16/600 Superdex 200 kDa pg (Cytiva). All antigens, antibodies and scFv-Fc were run on Superdex 200 Increase 10/300GL (Cytiva) on Äkta or HPLC (Techlab) on an AdvanceBio SEC 300Å 2.7 μm, 7.8x300 mm (Agilent) for quality control.
Serum ELISA
For COVID-19 convalescent serum analysis, sera were titrated in 11 steps (dilution ratio 1:√3) on 100 ng/well of RBD-His. As unspecificity controls, sera were tested on 100 ng/well BSA. All immobilization steps were performed in 0.05 M carbonate buffer (pH 9.6). Serum IgGs were detected using goat-anti-hIgG(Fc)-HRP (1:70,000, A0170, Sigma). Titration assays were performed in 96 well microtiter plates (Costar).
COVID-19 convalescent patient library construction
For the generation of human immune antibody libraries against SARS-CoV-2, blood from 6 donors showing a good antibody titer against RBD was used (Data S1A). The library construction was performed as described previously with minor modifications (Kügler et al., 2018). In brief, the peripheral blood mononuclear cells (PBMC) were extracted from the blood via Ficoll (GE Healthcare, Freiburg, Germany) and RNA isolated with TRIzol LS reagent (Life Technologies, Carlsbad, USA) and Direct-zol RNA Miniprep Plus kit (Zymo Research, Freiburg, Germany). For the plasma B cell sorted library, plasma B cells were double-stained with mouse α-CD19 APC-conjugated (MHCD1905, Thermo Fisher Scientific, Schwerte, Germany) and mouse α-CD138 (FITC conjugated) antibody (DL-101, BioLegend, San Diego, USA), sorted via FACSMelody (BD, Franklin Lakes, USA). Sorted cells were directly collected in TRIzol LS reagent and the RNA was extracted as indicated above. RNA was converted into cDNA using Superscript IV (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer’s instructions. Library construction was achieved as described previously (Wenzel et al., 2020a) with slight modifications. In brief, specific primers for the VH chain, kappa light chain and lambda light chain were used to amplify antibody genes from the cDNA. These resulting PCR products were purified and again amplified with primers adding specific restriction sites for further cloning into the E. coli expression vector pHAL52. The vector pHAL52 is derived from the vector pHAL30 (Kügler et al., 2015) with an AscI restriction site in the VL stuffer and a SalI restriction site in the VH stuffer for removal of uncut vector backbone. First, the light chain was cloned between the restriction sites MluI-HF and NotI-HF. In a second step the VH chain was cloned with the restriction sites HindIII-HF and NcoI-HF into previously cloned pHAL52-VL, additionally digested with AscI. The efficiency of library cloning was tested by colony PCR and the rate of complete scFv insertion was determined. The libraries were packaged with Hyperphage (Rondot et al., 2001; Soltes et al., 2007). Antibody phage were precipitated with PEG-NaCl and resuspended in phage dilution buffer (10 mM TrisHCl pH7,5, 20 mM NaCl, 2 mM EDTA). Resulting phage titer was determined by infection of E. coli XL1 blue MRF’.
Antibody selection using phage display
Antibody selection was performed as described previously with modifications (Russo et al., 2018). In brief, 5 μg of of S1-S2-His or RBD-His (produced in High Five cells) was diluted in carbonate buffer (50 mM NaHCO3/Na2CO3, pH 9.6) and coated onto the wells of a High binding 96 well microtiter plate (High Binding, Costar) at 4°C overnight. Next, the wells were blocked with 350 μL 2% MBPST (2% (w/v) milk powder in PBS; 0.05% Tween20) for 1 h at RT and then washed 3 times with PBST (PBS; 0.05% Tween20). Before adding the libraries to the coated wells, the libraries (5x1010 phage particles) were preincubated with 5 μg of an unrelated scFv-Fc and 2% MPBST on blocked wells for 1 h at RT, to deprive libraries of human Fc fragment binders. The libraries were transferred to the antigen coated wells, incubated for 2 h at RT. After 10 washes, bound phage were eluted with 150 μL trypsin (10 μg/mL) at 37°C, 30 minutes and used for the next panning round. The eluted phage solution was transferred to a 96 deep well plate (Greiner Bio-One, Frickenhausen, Germany) and incubated with 150 μL E. coli TG1 (OD600 = 0.5) first for 30 min at 37°C, then 30 min at 37°C and 650 rpm to infect the phage particles. 1 mL 2xYT-GA (1.6% (w/v) Tryptone; 1% (w/v) Yeast extract; 0.5% (w/v) NaCl (pH 7.0), 100 mM D-Glucose, 100 μg/mL ampicillin) was added and incubated for 1 h at 37°C and 650 rpm, followed by addition of 1x1010 cfu M13KO7 helper phage. Subsequently, the infected bacteria were incubated 30 min at 37°C followed by 30 min at 37°C and 650 rpm before centrifugation for 10 min at 3220xg. The supernatant was discarded and the pellet resuspended in fresh 2xYT-AK (1.6% (w/v) Tryptone; 1% (w/v) Yeast extract; 0.5% (w/v) NaCl (pH 7.0), 100 μg/mL ampicillin, 50 μg/mL kanamycin). The antibody phage were amplified overnight at 30°C and 650 rpm and used for the next panning round. In total four panning rounds were performed. In each round, the stringency of the washing procedure was increased (20x in panning round 2, 30x in panning round 3) and the amount of antigen was reduced (2.5 μg in panning round 2, 1.5 μg in panning round 3). After third panning round single clones were analyzed for production of RBD specific scFv by screening ELISA.
Screening of monoclonal recombinant binders using E. coli scFv supernatant
Soluble antibody fragments (scFv) were produced in 96-well polypropylene MTPs (U96 PP, Greiner Bio-One) as described before (Russo et al., 2018; Wenzel et al., 2020a). Briefly, 150 μL 2xYT-GA was inoculated with the bacteria bearing scFv expressing phagemids. MTPs were incubated overnight at 37°C and 800 rpm in a MTP shaker (Thermoshaker PST-60HL-4, Lab4You, Berlin, Germany). A volume of 180 μL 2xYT-GA in a PP-MTP well was inoculated with 20 μL of the overnight culture and grown at 37°C and 800 rpm for 90 minutes (approx. OD600 of 0.5). Bacteria were harvested by centrifugation for 10 min at 3220xg and the supernatant was discarded. To induce expression of the antibody genes, the pellets were resuspended in 200 μL 2xYT supplemented with 100 μg/mL ampicillin and 50 μM isopropyl-beta-D-thiogalacto-pyranoside (IPTG) and incubated at 30°C and 800 rpm overnight. Bacteria were pelleted by centrifugation for 20 min at 3220xg and 4°C.
For the ELISA, 100 ng of antigen was coated on 96 well microtiter plates (High Binding, Costar) in PBS (pH 7.4) overnight at 4°C. After coating, the wells were blocked with 2% MPBST for 1 h at RT, followed by three washing steps with H2O and 0.05% Tween20. Supernatants containing secreted monoclonal scFv were mixed with 2% MPBST (1:2) and incubated onto the antigen coated plates for 1 h at 37°C followed by three H2O and 0.05% Tween20 washing cycles. Bound scFv were detected using murine mAb 9E10 which recognizes the C-terminal c-myc tag (1:50 diluted in 2% MPBST) and a goat anti-mouse serum conjugated with horseradish peroxidase (HRP) (A0168, Sigma) (1:42000 dilution in 2% MPBST). Bound antibodies were visualized with tetramethylbenzidine (TMB) substrate (20 parts TMB solution A (30 mM Potassium citrate; 1% (w/v) Citric acid (pH 4.1)) and 1 part TMB solution B (10 mM TMB; 10% (v/v) Acetone; 90% (v/v) Ethanol; 80 mM H2O2 (30%)) were mixed). After stopping the reaction by addition of 1 N H2SO4, absorbance at 450 nm with a 620 nm reference was measured in an ELISA plate reader (Epoch, BioTek). Monoclonal binders were sequenced and analyzed using VBASE2 (www.vbase2.org) (Mollova et al., 2010) and possible glycosylation positions in the CDRS were analyzed according Lu et al. (2019.
ScFv-Fc and IgG production
Unique scFv sequences isolated by antibody-phage display were subcloned into pCSE2.7-hIgG1-Fc-XP using NcoI/NotI (New England Biolabs, Frankfurt, Germany) for mammalian production in Expi293F cells as scFv-Fc (Wenzel et al., 2020a). For IgG production, the variable domains were recloned into the IgG vectors human IgG1 format by subcloning of VH in the vector pCSEH1c (heavy chain) and VL in the vector pCSL3l/pCSL3k (light chain lambda/kappa) (Steinwand et al., 2014) adapted for Golden Gate Assembly procedure with Esp3I restriction enzyme (New England Biolabs, Frankfurt, Germany). For the antibodies CB6 (pdb 7C01), CR3022 (pdb 6W41) and REGN10933 and REGN10987 (pdb 6XDG) the public available amino acid sequences were used and the V-Genes were ordered as GeneArt Strings DNA fragments (Thermo Fisher Scientific, Schwerte, Germany) and recloned in the above indicated vectors. A “silenced” Fc part with following point mutations described Armour et al. (1999) and Shields et al. (2001) were used: E233P, L234V, L235A, deletion of G236, D265G, A327Q and A330S. Expi293F cells were cultured at 37°C, 110 rpm and 5% CO2 in GIBCO FreeStyle F17 expression media (Thermo Fisher Scientific) supplemented with 8 mM Glutamine and 0.1% Pluronic F68 (PAN Biotech). At the day of transfection cell density was between 1.5 - 2x106 cells/mL and viability at least above 90%. For formation of DNA:PEI complexes 1 μg DNA/mL transfection volume and 5 μg of 40 kDa PEI (Polysciences) were first diluted separately in 5% transfection volume in supplemented F17 media. DNA (1:1 ratio of the vectors for IgG production) and PEI was then mixed and incubated ∼25 min at RT before addition to the cells. 48 h later the culture volume was doubled by feeding HyClone SFM4Transfx-293 media (GE Healthcare) supplemented with 8 mM Glutamine. Additionally, HyClone Boost 6 supplement (GE Healthcare) was added with 10% of the end volume. One week after transfection supernatant was harvested by 15 min centrifugation at 1500xg.
Inhibition of S1-S2 and RBD binding to ACE2 expressing cells using flow cytometry
The inhibition tests in flow cytometry on EXPI293F cells were performed based on a previously published protocol (Bertoglio et al., 2021). Briefly, Expi293F cells were transfected according to the protocol above using pCSE2.5-ACE2fl-His and 5% eGFP plasmid. Two days after transfection, purified S1-S2-His was labeled using Monolith NTTM His-Tag Labeling Kit RED-tris-NTA (Nanotemper) according to the manufacturer’s protocol. In this setup 50 nM antigen was incubated with min. 1 μM of different scFv-Fc and the ACE2 expressing cells. The resulting median antigen fluorescence of GFP positive living single cells was measured. For comparison of the different scFv-Fc first the median fluorescence background of cells without antigen was subtracted, second it was normalized to the antigen signal where no antibody was applied. ScFv-Fc showing an inhibition in this first setup were further titrated as IgGs (max. 500 nM- 0.5 nM) on S1-S2-His, S1-His or on RBD-mFc (max. 100 nM-0.1 nM). S1-His and the corresponding mutants were detected with mouse anti-penta His (QIAGEN) and goat anti-mFc APC-conjugated antibody (Dianova). RBD-mFc was detected directed with the goat anti-mFc APC-conjugated antibody (Dianova). Measurements were performed with MACSQuant Analyzer (Milteny Biotech) (Data S2). The IC50 was calculated using the equation f(x) = Amin+(Amax-Amin)/(1+(x0/x)ˆh)ˆs and parameters from OriginPro (2019).
Dose dependent binding of IgG in titration ELISA
For titration ELISA, purified IgGs were titrated from 3.18 μg/mL- 0.001 ng/mL on 30 ng/well of the following antigens: S1-S2-His (High Five cell produced), RBD-mFc (High Five cell produced), S1-mFc (High Five cell produced) and TUN219-2C1-mFc (as control for unspecific Fc binding). In addition, all scFv-hFc were also tested only at the highest concentration (3.18 μg/mL) for unspecific cross-reactivity on Expi293F cell lysate (104 cells/well), BSA (1% w/v) and lysozyme. IgGs were detected using goat-anti-hIgG(Fc)-HRP (1:70000, A0170, Sigma). Titration assays were performed using 384 well or 96 well microtiter plates (Greiner Bio-One) using Precision XS microplate sample processor (BioTek), EL406 washer dispenser (BioTek) and BioStack Microplate stacker (BioTek). EC50 were calculated with by GraphPad Prism Version 6.1, fitting to a four-parameter logistic curve. Titration ELISAs on other coronaviruses and S1-HIS mutants were performed as described above.
Screening and titrating monoclonal antibodies for SARS-CoV-2 neutralization in cell culture
VeroE6 cells (ATCC CRL-1586) were seeded at a density of 6∗104/well onto cell culture 96-well plates (Nunc, Cat.#167008). Two days later, cells reached 100% confluence.
For titration, antibodies were diluted in 1/√10 steps and mixed with a fixed inoculum of SARS-CoV-2/Münster/FI110320/1/2020 (kind gift of Stephan Ludwig, University of Münster, Germany) (10-20, respectively 100-150 pfu) in a total volume of 500 μl of Vero E6 medium (DMEM, 10% FCS, 2 mM glutamine, penicillin, streptomycin). After one hour incubation at 37°C, cells were infected with the antibody/virus mix, incubated for one hour and then overlaid with Vero E6 medium containing 1.5% methyl-cellulose. Three days postinfection, wells were imaged using a Sartorius IncuCyte S3 (4x objective, whole-well scan) and plaques were counted from these images. Image data was quantified with the IncuCyte S3 GUI tools measuring the decrease of confluence concomitant with the cytopathic effect of the virus in relation to uninfected controls and controls without antibody and analyzed with Origin using the Logistic5 fit.
Specificity assay
To test specificity of the antibody candidates an ELISA on DNA, LPS, lysozyme and cell lysate was performed under standard conditions (see above). In brief, 10 μg/mL of the respective antigen was immobilized on 96 well microtiter plates (High binding, Costar) in PBS (pH 7.4) overnight at 4°C. After blocking 10 μg/mL of STE90-C11 IgG, Avelumab, Palivizumab and IVIG respectively were incubated and later detected using goat-anti-hIgG(Fc)-HRP (1:70000, A0170, Sigma). TMB reaction took place for 30 min and absorbance at 450 nm with a 620 nm reference was measured in an ELISA plate reader (Epoch, BioTek). All signals were normalized to the absorbance of Avelumab.
Analytical size exclusion chromatography (SEC)
All purified antigens and indicated antibodies were run on Superdex 200 Increase 10/300GL column (Cytiva) on Äkta pure system (Cytiva) according to the manufactures protocol.
Affinity measurement by Bio-Layer Interferometry
The affinity was measured by Bio-Layer Interferometry in three different assays using the Octet qKe (Fortebio/Sartorius GmbH, Göttingen, Germany).
In the first assay, anti-Mouse Fc-Capture (AMC) sensors were activated for 10 min in PBS. After that, the sensors were equilibrated in assay buffer (PBS containing 1% BSA and 0.05% Tween 20) for 60 s before RBD-mFc (Sino Biologicals) was loaded onto the sensors at 10 μg/ml for 180 s. After a stable baseline measurement was established (60 s), antigen-loaded sensors were transferred to an 8-point antibody dilution series (500, 150, 50, 15, 5, 1.5, 0.5 and 0 nM). Association of the Fab antibody to the antigen was measured for 300 s. After that, the sensors were transferred into assay buffer were the dissociation was measured for 900 s. Significant binding of the antibody to an unloaded sensor was not detected. For data analysis, the reference measurement (0 nM) was subtracted from the other measurements and data traces ranging from 150 to 1.5 nM were used for modeling of the kinetic data using a 1:1 binding model (Data S6C).
In the second assay, anti-human Fab (FAB2G) sensors were activated for 10 min in PBS. After that, the sensors were equilibrated in assay buffer (PBS containing 1% BSA and 0.05% Tween 20) for 60 s before the IgG antibody was loaded onto the sensors at 2.5 μg/ml for 180 s. After a stable baseline measurement was established (60 s), antibody-loaded sensors were transferred to an 8-point S1-HIS antigen dilution series (500, 150, 50, 15, 5, 1.5, 0.5 and 0 nM). Association of the S1 antigen to the antibody was measured for 300 s. After that, the sensors were transferred into assay buffer were the dissociation was measured for 900 s. Significant binding of the antigen to an unloaded sensor was not detected. For data analysis, the reference measurement (0 nM) was subtracted from the other measurements and data traces ranging from 50 to 5 nM were used for modeling of the kinetic data using a 1:1 binding model in the Data Analysis HT 11.0 software tool (Data S6D).
In the third assay, protein A sensors were activated for 10 min in PBS. Before use, the sensors were regenerated for 5 cycles in 10 mM Glycine buffer (pH2.0) followed by neutralization in PBS. Each step was performed for 5 s. After that, the regenerated sensors were equilibrated in assay buffer (PBS containing 1% BSA and 0.05% Tween 20) for 60 s before the IgG antibody was loaded onto the sensors at 2.5 μg/ml for 180 s. After a stable baseline measurement was established (60 s), antibody-loaded sensors were transferred to an 8-point S1-HIS antigen dilution series (500, 150, 50, 15, 5, 1.5, 0.5 and 0 nM). Association of the S1 antigen to the antibody was measured for 300 s. After that, the sensors were transferred into assay buffer were the dissociation was measured for 900 s. Significant binding of the antigen to an unloaded sensor was not detected. For data analysis, the reference measurement (0 nM) was subtracted from the other measurements and data traces ranging from 50 to 0.5 nM were used for modeling of the kinetic data using a 1:1 binding model (Data S6E).
Immunoblot analysis
For the immunoblot analysis RBD-His was disrupted either by incubation for 10 minutes at 56°C without β-Mercaptoethanol, at 95°C without β-Mercaptoethanol or at 95°C in Laemmli sample buffer (Laemmli, 1970) with β-Mercaptoethanol. RBD was separated by 12% SDS-PAGE and blotted onto a nitrocellulose membrane (Amersham Protan 0.2 μm NC, GE HealthCare). The membrane was blocked with 2% M-PBST for 1 h at RT. For the detection of RBD-His, 10 μg/mL STE90-C11 IgG was used. For control, RBD-His (95°C + β-Mercaptoethanol) was detected with 2μg/mL mouse anti-His (Dia-900-200, Dianova, Hamburg, Germany) for 1.5 h at RT. After 3x washing with PBST the secondary antibody goat anti-human Fc AP conjugated (1:20,000, Jackson ImmunoResearch, Cambridge House, UK) was used for the detection of STE90-C11 and goat anti-mouse IgG AP conjugated (1:30,000, 115-055-071, Dianova) was used for the anti-His antibody and incubated for 1 h. Finally, the membrane was washed 2x with PBST and 4x with PBS. Staining and visualization of specific proteins was performed with 5-Brom-4-Chlor-3-Indolyl-Phosphat/Nitro Tetrazolium Blue Chloride (NBT-BCIP, Thermo Fisher Scientific GmbH, Dreieich, Germany) according to standard protocols (Data S6F).
Hamster model of SARS-CoV-2 infection
Animal procedures were performed according to the European Guidelines for Animal Studies after approval by the Institutional Animal Care Committee and the relevant state authority (Landesamt für Gesundheit und Soziales, Berlin, Permit number 0086/20).
SARS-CoV-2 isolate BetaCoV/Germany/BavPat1/2020 (Wölfel et al., 2020) was used as challenge virus for hamster experiments. The virus was propagated and titrated on Vero E6 cells (ATCC CRL-1586) in minimal essential medium (MEM; PAN Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (PAN Biotech), 100 IU/ml penicillin G and 100 μg/ml streptomycin (Carl Roth, Karlsruhe, Germany) and stored at −80°C prior to experimental infections.
Per group, nine male and female Syrian hamsters (Mesocricetus auratus) strain RjHAN:AURA (Janvier, Le Genest-Saint-Isle, France) were used. Animals were housed in GR-900 IVC cages (Tecniplast, Buguggiate, Italy) and provided with food ad libidum and bountiful enrichment and nesting materials (Carfil, Oud-Turnhout, Belgium). Hamsters were randomly distributed into experimental groups and treated intraperitoneally with 3.7 mg/kg or 37 mg/kg STE90-C11 in a total volume of 1 mL PBS, two hours post infection, the control group received 1 mL PBS only at the same time-point.
SARS-CoV-2 infection was performed as previously described (Osterrieder et al., 2020). Briefly, anaesthetized hamsters received 1x105 pfu SARS-CoV-2 in 60 μL MEM intranasally two hours before treatment. Following infection, the clinical presentation of all animals was monitored twice a day, body weight of all hamsters was recorded daily. On days 3, 5 and 14 post infection, three randomly assigned hamsters per group were euthanized. Euthanasia was applied by exsanguination under general anesthesia as described (Nakamura et al., 2017). Oropharyngeal swabs and lungs were collected for virus titrations, RT-qPCR and/or histopathological examinations. All organs were immediately frozen at −80°C or preserved in 4% formaldehyde for subsequent in-depth histopathological investigations.
To assess virus titers from 50 mg lung tissue, tissue homogenates were prepared using a bead mill (Analytic Jena) and 10-fold serial dilutions were prepared in MEM, and plated on Vero E6 cells in 12-well-plates. The dilutions were removed after 2 h and cells were overlaid with 1.25% microcrystalline cellulose (Avicel) in MEM supplemented with 10% FBS and penicillin/streptomycin. Three days later, cells were formalin-fixed, stained with crystal violet, and plaques were counted.
Transgenic mice model of SARS-CoV-2 infection
All animal experiments were performed in compliance with the German Animal Welfare Act (TierSchG BGBl. I S. 1206, 1313; May 18, 2006) and Directive 2010/63/EU. The mice were handled in accordance with good animal practice as defined by the Federation for Laboratory Animal Science Associations and Gesellschaft für Versuchstierkunde/Society of Laboratory Animal Science. All animal experiments were approved by the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety) under permits number 20_3567. K18hACE2 mice (B6.Cg-Tg(K18-ACE2)2Prlmn/J) were purchased from Charles River (Sulzfeld, Germany), Mice were housed at the animal facility of the Helmholtz Centre for Infection Research under pathogen-free conditions.
Mice were fixed in a restrainer before injection and the lateral tail veins were hyperaemized. Different antibody concentrations were diluted in 100ul of PBS and injected intravenous into the lateral tail vein.
Female and male at least 6-wk-old mice were infected with tissue culture– derived virus and housed in specific pathogen-free conditions throughout the experiment. Mice were anesthetized with Ketamin/Xylazin and inoculated intranasally with 2,000 PFU of virus in 20 ul of Phosphate buffered saline (PBS).
The mice were sacrificed by CO2 asphyxiation on day 5. Lungs were collected aseptically, homogenized in 500 μL PBS and stored at −80◦C. Part of the organ homogenates were used for titration cells and the other part for qPCR Analysis. Organ homogenates were serially diluted 1:10 – 1:105 in medium (DMEM supplemented with 5% FCS, 2 mM glutamine, 100 IU/mL penicillin and 100 μg/mL streptomycin). Vero E-6 cells were then inoculated with 200ul of diluted homogenates and incubated for 1h, 37°C, CO2 incubator. Cells were then coverd with 1.75% Carboxymethyl-cellulose and incubated at 37°C, CO2 incubator for 3-5 days. Plates were then fixed with 6% Paraformaldehyde for 1h and then stained with 1% Crystal violet. Plaques were then read under microscope.
In vitro evolution of SARS-CoV-2 by antibody co-cultivation
This assay was performed with STE90-C11 and Palivizumab as described by Baum et al. (2020).
Fab and RBD22 production for co-crystallization
The production of STE90-C11 Fab fragment was done by transient co-transfection of plasmids encoding the heavy and the light chain in Expi293F cells cultivated at 37°C, 5% CO2 and 100rpm in Expi Expression Medium. The transfection was performed at a density of 3∗10ˆ6 cells/ml by adding 1μg/ml culture mixed plasmids and 4 μg/ml culture PEI 40 kDa (Polyscience). The culture was incubated for 72 hours according to the protocol. The supernatant was harvested by centrifugation (30min, 3000 g) and sterile filtration (0.2μm). The Fab-fragment was purified by affinity chromatography using 1ml HisTrap Excel column (GE Healthcare) followed by a size exclusion chromatography on a 10/300 Superdex200 Increase column (GE Healthcare) according to manufactures manual.
For the production of RBD22 High Five cells grown in EX-CELL 405 serum-free medium (Sigma) were transiently transfected with 5μg/mL plasmid and 20μg/mL PEI 40 kDa (Polyscience) at a cell density of 5∗10ˆ6 cells/ml. After 4h incubation at 27°C and 100rpm the cells were diluted 5-fold with EX-CELL medium to a density of 1∗10ˆ6 cells/ml and further incubated for three more days. Following centrifugation (5000xg 30min) and sterile filtration (0.2μm) the supernatant was loaded on a 5ml HisTrap Excel column (GE Healthcare). After washing and elution with imidazole the RBD22 containing fractions were further purified by size exclusion chromatography on a 26/600 Superdex 200 pg using 20mM Tris-HCl pH8 and 150mM NaCl as a buffer. Aliquots were snap-frozen in liquid nitrogen and stored at −80°C till further use.
Crystallization, data collection and structure determination
The Fab fragment of STE90-C11 was incubated overnight with a 1.2 molar excess of purified RBD22 at 4°C. The complex was isolated by size exclusion chromatography on a Superdex 200 Increase 10/300GL (GE Healthcare) with 20mM Tris-HCl pH8 and 150mM NaCl as a running buffer. Fractions containing the complex were concentrated utilizing a Vivaspin2 ultrafiltration unit (10,000 MWCO; Sartorius). Crystallizations trails were set up in 96-well sitting-drop vapor diffusion plates (Intelli 96-3 plates, Art Robbins Instruments) with a pipetting robot (Crystal Gryphon, Art Robbins Instruments) mixing 200nl reservoir solution with 200nl of protein solution (14mg/mL, 7mg/mL) and equilibrated against 60 μL of reservoir solution. As initial screens the Cryos Suite and the JCSGplus Suite (QIAGEN) were chosen and crystal growth was monitored in a crystal hotel (RockImager, Formulatrix). Initial hits were further optimized by a random screen assembled with a Formulator pipetting robot (Formulatrix). Best diffracting crystals grew in 13.3% (w/v) polyethylene glycol 6,000, 0.1M MES pH 5.6 and 0.24M tri sodium citrate. Crystals were harvested with nylon loops and soaked in reservoir solution mixed with 2,3-(R,R)-butandiol (Alfa Aeser) to a final concentration of 10% (v/v) prior to flash cooling in liquid nitrogen.
3600 diffraction images with an oscillation angle of 0.1° per image were collected at the beamline P11 at PETRA III (DESY, Hamburg, Germany) (Burkhardt et al., 2016) on a Pilatus 6M fast detector (Dectris) and processed with XDS (Kabsch, 2010) and Aimless (Evans and Murshudov, 2013) yielding a dataset with a resolution cut off of 2.0 Å based on a CC1/2 value greater than 0.5. Initial phases were determined by molecular replacement with Phaser (McCoy et al., 2007). As a search model the coordinates of a Fab fragment and the RBD was used (PDB: 7BWJ) (Ju et al., 2020). The model was further improved by manual rebuilding in Coot (Emsley et al., 2010) and computational refinement with phenix. refine (Afonine et al., 2012) including placement of water, TLS refinement and riding hydrogens in the final steps of the procedure. Depictions of the model were generated with PyMol molecular graphics system (Schrödinger LLC; version 2.3.2). Data processing and model refinement statistics can be found in Data S7A. The final model can be accessed under the PDB code 7B3O.
Quantification and statistical analysis
The quantification and statistical analysis is indicated for each individual experiment in the figure legends and the material and methods.
Acknowledgments
We kindly acknowledge the financial support of MWK Niedersachsen (14-76103-184 CORONA-2/20) for the projects “Antibody generation,” “Neutralization experiments” and “Structure-based analysis of antiviral strategies against CoV-2 target proteins.” We also acknowledge support of the European Union for the ATAC (“antibody therapy against corona,” Horizon2020 number 101003650) consortium and the Deutsche Herzstiftung (“Menschliche monoklonale Antikörper gegen SARS-CoV2 zur Prophylaxe gegen COVID-19 bei vorerkrankten Risikopatienten – Unterstützung der Entwicklung”). We acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities. Parts of this research were carried out at PETRAIII, and we would like to thank Johanna Hakanpää for assistance in using the P11 beamline. We are deeply grateful to Adelheid Langner, Andrea Walzog, Bettina Sandner, Cornelia Oltmann, and Wolfgang Grassl for constant help and support during the pandemic.
Author contributions
F.B., A.F., T.S., S.D., J.V.d.H., L.Č.-Š., M. Schubert, and M.H. conceptualized the study. F.B., V.F., M.R., P.A.H., L.A., U.R., T.K., D.M., N.L., S. Steinke, R.B., K.-T.S., K.D.R.R., P.R., D.S., A.K., S.Z.-E., K.E., Y.K., M.B., M. Scholz, G.M.S.G.M., E.V.W., G. Roth, J.A., J.T., J.K., M.Z.C., and M. Schubert performed and designed the experiments. F.B., D.M., N.L., S. Steinke, U.R., P.K., J.V.d.H., J.T., A.F., L.Č.-Š., M. Schubert, and M.H. analyzed the data. H.S.P.G., S.C., and A.G. provided the patient samples and support. S.D., L.Č.-Š., and M.H. conceived the funding. P.K., E.V.W., G. Russo, A.H., V.F., S.D., G. Roth, A.F., J.V.d.H., M. Schubert, and M.H. advised on the experimental design and data analysis. F.B., T.K., S.D., L.Č.-Š., M. Schubert, and M.H. wrote the manuscript.
Declaration of interests
F.B., D.M., N.L., S. Steinke, P.A.H., R.B., M.R., K.-T.S., K.D.R.R., S.Z.-E., M.B., V.F., S.D., M. Schubert, and M.H. are inventors on a patent application on blocking antibodies against SARS-CoV-2. A.H., A.F., and T.S. are officers of CORAT Therapeutics GmbH, a company founded by YUMAB GmbH for the clinical and regulatory development of STE90-C11 (development name COR-101) (ClinicalTrials.gov ID: NCT04674566 for safety and efficacy of COR-101 in hospitalized patients with moderate to severe COVID-19). A.H. is a shareholder of CORAT Therapeutics GmbH. S.D. and M.H. are advisors to Corat Therapeutics GmbH. A.F., T.S., S.D., and M.H. are shareholders of YUMAB GmbH.
Published: July 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109433.
Supplemental information
References
- ACTIV-3/TICO LY-CoV555 Study Group. Lundgren J.D., Grund B., Barkauskas C.E., Holland T.L., Gottlieb R.L., Sandkovsky U., Brown S.M., Knowlton K.U., Self W.H., et al. A Neutralizing Monoclonal Antibody for Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384:905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaleh M.A., Alsaab H.O., Mahmoud A.B., Alkayyal A.A., Jones M.L., Mahler S.M., Hashem A.M. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 2020;11:1986. doi: 10.3389/fimmu.2020.01986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.-C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 2021 doi: 10.1101/2021.02.23.21252259. [DOI] [Google Scholar]
- Armour K.L., Clark M.R., Hadley A.G., Williamson L.M. Recombinant human IgG molecules lacking Fcgamma receptor I binding and monocyte triggering activities. Eur. J. Immunol. 1999;29:2613–2624. doi: 10.1002/(SICI)1521-4141(199908)29:08<2613::AID-IMMU2613>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoglio F., Meier D., Langreder N., Steinke S., Rand U., Simonelli L., Heine P.A., Ballmann R., Schneider K.-T., Roth K.D.R., et al. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat. Commun. 2021;12:1577. doi: 10.1038/s41467-021-21609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B., Jochems C., Fantini M., Heery C.R., Gulley J.L., Tsang K.Y., Schlom J. Antibody-Dependent Cellular Cytotoxicity Activity of a Novel Anti-PD-L1 Antibody Avelumab (MSB0010718C) on Human Tumor Cells. Cancer Immunol. Res. 2015;3:1148–1157. doi: 10.1158/2326-6066.CIR-15-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A.R.M., Sidhu S., Dübel S., McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 2011;29:245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejová B., Hodorová V., Boršová K., Čabanová V., Reizigová L., Paul E.D., Čekan P., Klempa B., Nosek J., Vinař T. B.1.258$∖Delta$, a SARS-CoV-2 variant with ΔH69/ΔV70 in the Spike protein circulating in the Czech Republic and Slovakia. arXiv. 2021 doi: 10.1007/s11262-021-01866-5. http://arxiv.org/abs/2102.04689v1 2102.04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burkhardt A., Pakendorf T., Reime B., Meyer J., Fischer P., Stübe N., Panneerselvam S., Lorbeer O., Stachnik K., Warmer M., et al. Status of the crystallography beamlines at PETRA III. Eur. Phys. J. Plus. 2016;131:56. [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Gabra H., Hill F., Evan G., Sikora K. A novel tumour marker related to the c-myc oncogene product. Mol. Cell. Probes. 1987;1:73–82. doi: 10.1016/0890-8508(87)90008-9. [DOI] [PubMed] [Google Scholar]
- Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., Ren L., Jin Q., Wang J., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11:4528. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Misasi J., Mulangu S., Stanley D.A., Kanekiyo M., Wollen S., Ploquin A., Doria-Rose N.A., Staupe R.P., Bailey M., et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Coughlin M.M., Prabhakar B.S. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev. Med. Virol. 2012;22:2–17. doi: 10.1002/rmv.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan T., Ferguson M., Yuan L., Xu F., Li G. Human Monoclonal Fab Antibodies Against West Nile Virus and its Neutralizing Activity Analyzed in Vitro and in Vivo. J. Antivir. Antiretrovir. 2009;1:36–42. doi: 10.4172/jaa.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A., Schirrmann T., Hust M. Phage display-derived human antibodies in clinical development and therapy. MAbs. 2016;8:1177–1194. doi: 10.1080/19420862.2016.1212149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A., Kügler J., Helmsing S., Meier D., Schirrmann T., Hust M., Dübel S. Designing Human Antibodies by Phage Display. Transfus. Med. Hemother. 2017;44:312–318. doi: 10.1159/000479633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Domman D.B., Snyder D.J., Oguntuyo K., Van Diest M., Densmore K.H., Schwalm K.C., Femling J., Carroll J.L., Scott R.S., et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. MedRxiv. 2021 [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger V., Büssow K., Wagner A., Weber S., Hust M., Frenzel A., Schirrmann T. High level transient production of recombinant antibodies and antibody fusion proteins in HEK293 cells. BMC Biotechnol. 2013;13:52. doi: 10.1186/1472-6750-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn J., Schäckermann D., Kirmann T., Bertoglio F., Steinke S., Heisig J., Ruschig M., Rojas G., Langreder N., Wenzel E.V., et al. Baculovirus-free insect cell expression system for high yield antibody and antigen production. Sci. Rep. 2020;10:21393. doi: 10.1038/s41598-020-78425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T., von Rüden T. Antibody discovery: phage display. Curr. Opin. Biotechnol. 2002;13:598–602. doi: 10.1016/s0958-1669(02)00380-4. [DOI] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler J., Wilke S., Meier D., Tomszak F., Frenzel A., Schirrmann T., Dübel S., Garritsen H., Hock B., Toleikis L., et al. Generation and analysis of the improved human HAL9/10 antibody phage display libraries. BMC Biotechnol. 2015;15:10. doi: 10.1186/s12896-015-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler J., Tomszak F., Frenzel A., Hust M. Construction of Human Immune and Naive scFv Libraries. Methods Mol. Biol. 2018;1701:3–24. doi: 10.1007/978-1-4939-7447-4_1. [DOI] [PubMed] [Google Scholar]
- Kuhn P., Fühner V., Unkauf T., Moreira G.M.S.G., Frenzel A., Miethe S., Hust M. Recombinant antibodies for diagnostics and therapy against pathogens and toxins generated by phage display. Proteomics Clin. Appl. 2016;10:922–948. doi: 10.1002/prca.201600002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Li W., Schäfer A., Kulkarni S.S., Liu X., Martinez D.R., Chen C., Sun Z., Leist S.R., Drelich A., Zhang L., et al. High Potency of a Bivalent Human VH Domain in SARS-CoV-2 Animal Models. Cell. 2020;183:429–441.e16. doi: 10.1016/j.cell.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gao F., Gou L., Chen Y., Gu Y., Ao L., Shen H., Hu Z., Guo X., Gao W. Neutralizing Antibodies Isolated by a site-directed Screening have Potent Protection on SARS-CoV-2 Infection. bioRxiv. 2020 doi: 10.1101/2020.05.03.074914. [DOI] [Google Scholar]
- Lu X., Nobrega R.P., Lynaugh H., Jain T., Barlow K., Boland T., Sivasubramanian A., Vásquez M., Xu Y. Deamidation and isomerization liability analysis of 131 clinical-stage antibodies. MAbs. 2019;11:45–57. doi: 10.1080/19420862.2018.1548233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., Meng X., Huang X., Yang Y., Zhao D., Zhou P., Wang X., Zhao C., Sun Y., et al. Potent Neutralization of SARS-CoV-2 by Hetero-bivalent Alpaca Nanobodies Targeting the Spike Receptor-Binding Domain. J. Virol. 2021;95:e02438-20. doi: 10.1128/JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova S., Retter I., Hust M., Dübel S., Müller W. In: Dübel S., Kontermann R., editors. Springer Verlag; 2010. Analysis of single chain antibody sequences using the VBASE2 Fab analysis tool; pp. 3–10. (Antibody Engineering). [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., et al. PALM Writing Group. PALM Consortium Study Team A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Karakida N., Dantsuka A., Ichii O., Elewa Y.H.A., Kon Y., Nagasaki K.-I., Hattori H., Yoshiyasu T. Effects of a mixture of medetomidine, midazolam and butorphanol on anesthesia and blood biochemistry and the antagonizing action of atipamezole in hamsters. J. Vet. Med. Sci. 2017;79:1230–1235. doi: 10.1292/jvms.17-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka C.K.V., Franco M.M., Gräf T., de Lorenzo Barcia C.A., de Ávila Mendonça R.N., de Sousa K.A.F., Neiva L.M.C., Fosenca V., Mendes A.V.A., de Aguiar R.S., et al. Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil. Emerg. Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parray H.A., Chiranjivi A.K., Asthana S., Yadav N., Shrivastava T., Mani S., Sharma C., Vishwakarma P., Das S., Pindari K., et al. Identification of an anti-SARS-CoV-2 receptor-binding domain-directed human monoclonal antibody from a naïve semisynthetic library. J. Biol. Chem. 2020;295:12814–12821. doi: 10.1074/jbc.AC120.014918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal K.E., Dudgeon D., Trefry J.C., Anantpadma M., Sakurai Y., Murin C.D., Turner H.L., Fairhurst J., Torres M., Rafique A., et al. Development of Clinical-Stage Human Monoclonal Antibodies That Treat Advanced Ebola Virus Disease in Nonhuman Primates. J. Infect. Dis. 2018;218(Suppl 5):S612–S626. doi: 10.1093/infdis/jiy285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., Parcells M.S., Luo G., Li W., Zhong G., et al. Social Science Research Network; 2020. The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response Without Antibody-Dependent Enhancement. [Google Scholar]
- Rahumatullah A., Abdul Karim I.Z., Noordin R., Lim T.S. Antibody-Based Protective Immunity against Helminth Infections: Antibody Phage Display Derived Antibodies against BmR1 Antigen. Int. J. Mol. Sci. 2017;18:2376. doi: 10.3390/ijms18112376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P., Neha, Devi C., Das P. Bioinformatics analysis of SARS-CoV-2 RBD mutant variants and insights into antibody and ACE2 receptor binding. bioRxiv. 2021 doi: 10.1101/2021.04.03.438113. [DOI] [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. SAFER Investigators The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Gräf T., Paixão A.C.D., Appolinario L., Lopes R.S., Mendonça A.C. da F., da Rocha A.S.B., Motta F.C., Neto L.G.L., Khouri R., et al. A potential SARS-CoV-2 variant of interest (VOI) harboring mutation E484K in the Spike protein was identified within lineage B.1.1.33 circulating in Brazil. Viruses. 2021;13:724. doi: 10.3390/v13050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent Antibody Responses to SARS-CoV-2 Infection in Convalescent Individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondot S., Koch J., Breitling F., Dübel S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001;19:75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- Russo G., Meier D., Helmsing S., Wenzel E., Oberle F., Frenzel A., Hust M. Parallelized Antibody Selection in Microtiter Plates. Methods Mol. Biol. 2018;1701:273–284. doi: 10.1007/978-1-4939-7447-4_14. [DOI] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shields R.L., Namenuk A.K., Hong K., Meng Y.G., Rae J., Briggs J., Xie D., Lai J., Stadlen A., Li B., et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J. Biol. Chem. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- Soltes G., Hust M., Ng K.K.Y., Bansal A., Field J., Stewart D.I.H., Dübel S., Cha S., Wiersma E.J. On the influence of vector design on antibody phage display. J. Biotechnol. 2007;127:626–637. doi: 10.1016/j.jbiotec.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwand M., Droste P., Frenzel A., Hust M., Dübel S., Schirrmann T. The influence of antibody fragment format on phage display based affinity maturation of IgG. MAbs. 2014;6:204–218. doi: 10.4161/mabs.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K.N., Weisman L.E., Rhodes T., Ariagno R., Sánchez P.J., Steichen J., Givner L.B., Jennings T.L., Top F.H., Jr., Carlin D., Connor E. Safety, tolerance and pharmacokinetics of a humanized monoclonal antibody to respiratory syncytial virus in premature infants and infants with bronchopulmonary dysplasia. MEDI-493 Study Group. Pediatr. Infect. Dis. J. 1998;17:110–115. doi: 10.1097/00006454-199802000-00006. [DOI] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surleac M., Casangiu C., Banica L., Milu P., Florea D., Sandulescu O., Streinu-Cercel A., Vlaicu O., Tudor A., Hohan R., et al. Evidence of Novel SARS-CoV-2 Variants Circulation in Romania. AIDS Res Hum Retroviruses. AIDS Res Hum Retroviruses. 2021;37:329–332. doi: 10.1089/AID.2021.0009. [DOI] [PubMed] [Google Scholar]
- Tablizo F.A., Kim K.M., Lapid C.M., Castro M.J.R., Yangzon M.S.L., Maralit B.A., Ayes M.E.C., la Paz E.M.C., Guzman A.R.D., Yap J.M.C., et al. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. medRxiv. 2021 doi: 10.1101/2021.03.03.21252812. [DOI] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott M., Weiß S., Antoni S., Koch J., von Briesen H., Hust M., Dietrich U. Functional characterization of two scFv-Fc antibodies from an HIV controller selected on soluble HIV-1 Env complexes: a neutralizing V3- and a trimer-specific gp41 antibody. PLoS ONE. 2014;9:e97478. doi: 10.1371/journal.pone.0097478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mechelen L., Luytjes W., de Haan C.A.M., Wicht O. RSV neutralization by palivizumab, but not by monoclonal antibodies targeting other epitopes, is augmented by Fc gamma receptors. Antiviral Res. 2016;132:1–5. doi: 10.1016/j.antiviral.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020;94:e02015-19. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-F., Tseng S.-P., Yen C.-H., Yang J.-Y., Tsao C.-H., Shen C.-W., Chen K.-H., Liu F.-T., Liu W.-T., Chen Y.-M.A., Huang J.C. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel E.V., Bosnak M., Tierney R., Schubert M., Brown J., Dübel S., Efstratiou A., Sesardic D., Stickings P., Hust M. Human antibodies neutralizing diphtheria toxin in vitro and in vivo. Sci. Rep. 2020;10:571. doi: 10.1038/s41598-019-57103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel E.V., Roth K.D.R., Russo G., Fühner V., Helmsing S., Frenzel A., Hust M. Antibody Phage Display: Antibody Selection in Solution Using Biotinylated Antigens. Methods Mol. Biol. 2020;2070:143–155. doi: 10.1007/978-1-4939-9853-1_8. [DOI] [PubMed] [Google Scholar]
- Widjaja I., Wang C., van Haperen R., Gutiérrez-Álvarez J., van Dieren B., Okba N.M.A., Raj V.S., Li W., Fernandez-Delgado R., Grosveld F., et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes Infect. 2019;8:516–530. doi: 10.1080/22221751.2019.1597644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Li L., Lin J., Li X., Liu B., Kong Y., Zeng S., Du J., Xiao H., Zhang T., et al. Isolation of a human monoclonal antibody specific for the receptor binding domain of SARS-CoV-2 using a competitive phage biopanning strategy. Antib. Ther. 2020;3:95–100. doi: 10.1093/abt/tbaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.-Y., Shu Y., Phogat S., Xiao X., Cham F., Bouma P., Choudhary A., Feng Y.-R., Sanz I., Rybak S., et al. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods. 2003;283:17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16:1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A., et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure data and the antibody sequence of COR-101 is available at PDB 7B3O. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.