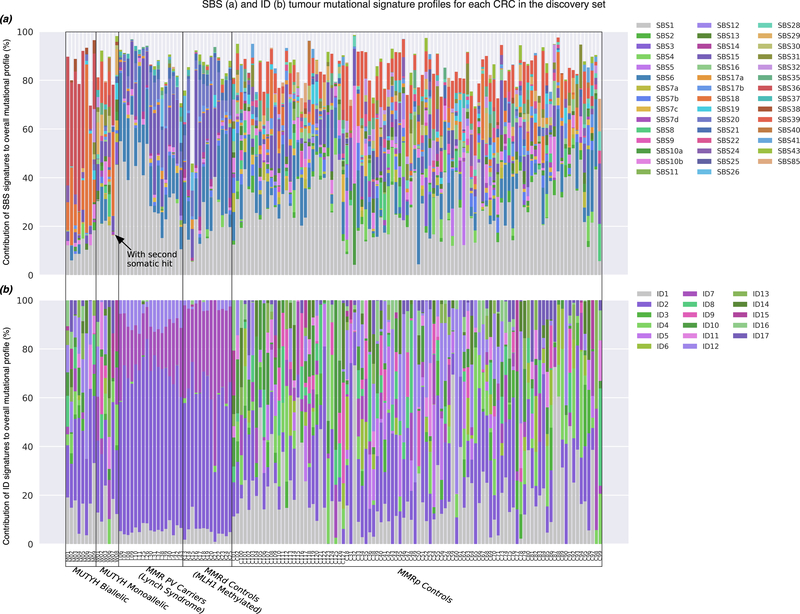
Figure 2.
The tumour mutational signature (TMS) profiles based on the COSMIC v3 signature set for each of the 142 CRCs tested by whole exome sequencing and included in the discovery group: (a) Single base substitution (SBS)-derived TMS and (b) insertion and deletion (ID)-derived TMS profiles. CRCs were grouped by subtype: (i) biallelic MUTYH pathogenic variant carriers, (ii) monoallelic MUTYH pathogenic variant carriers, (iii) mismatch repair (MMR) gene pathogenic variant carriers (Lynch syndrome), (iv) MMR-deficient (MMRd) controls related to MLH1 gene promoter hypermethylation and (v) MMR-proficient (MMRp) controls. Individual SBS or ID TMS with proportional values below 5% across all the CRC samples were excluded.