Abstract
Odorant-binding proteins (OBPs) are prevalent in the antennal transcriptomes of different orders of insects. Studies on OBPs have focused on their role in the insect chemosensory system, but knowledge of their functions in the insect testis is limited. We sequenced the transcriptomes of the Athetis dissimilis reproductive organs and analyzed the expression of AdisOBP genes in different tissues. We identified 23 OBPs in the testis and ovaries and 31 OBPs in antennal transcriptomes. The results of real-time quantitative PCR revealed that 23 of the 54 OBP genes were highly expressed in both female and male antennae, including three that exhibited male-biased expression and 15 that exhibited female-biased expression. A total of 24 OBPs were highly expressed in the testis of A. dissimilis, while expression of OBPs in the ovaries was very low. These findings highlight the functional diversity of OBPs in insects and can facilitate further studies on the OBPs in A. dissimilis and lepidopteran species.
Subject terms: Genetics, Molecular biology, Zoology
Introduction
The olfactory system in insects regulates their intersex communication, host-plant interactions, oviposition, foraging, escape from predators and reproduction1–5. Insects have a complex chemosensory system in which pheromones and plant odors are initially recognized by odorant-binding proteins (OBPs) expressed in the antennal sensilla lymph that transfer the odorants to membrane-bound olfactory receptors (ORs) to activate olfactory receptor neurons (ORNs) and stimulate behavioral responses6–11.
Understanding the molecular mechanisms of olfaction is essential for better using olfactory-based pest management strategies and the development of novel strategies. OBPs are more accessible targets for research, considering they are small, soluble, stable and easier to manipulate and modify. OBPs are small water soluble proteins that have six positionally conserved cysteines to form three interlocking disulphide bridges that stabilize the protein’s three-dimensional structure12–19. OBPs were first discovered in the antenna of Antheraea polyphemus, where they distinguish and bind to lipophilic odorant compounds20–25. However, emerging data suggests that OBPs are not restricted to the sensory organs of insect and show expression in non-sensory organs including reproductive organs26,27. Li et al. showed that AaegOBP22 was highly expressed in the male reproductive organs of Aedes aegypti and transfers to females during mating. This suggests an additional function for this protein as pheromone carrier, analogously to vertebrates’ urinary and salivary proteins as well as some insect chemosensory proteins26. Sun et al. also found that HarmOBP10 and HassOBP10 is highly abundant in seminal fluid of Helicoverpa armigera and H. assulta and transfers to female during mating. HarmOBP10 and HassOBP10 also bind 1-dodecene, a known insect repellent27.
Athetis dissimilis Hampson (Lepidoptera: Noctuidae) is an important agricultural pest and mainly distributed in Asian countries including China, Japan, Philippines, Korea, Indonesia and India, causing serious damages to maize, wheat, peanut, soybean and sweet potato28–30. Because of the fact that larvae of A. dissimilis live under plant residues, it is difficult to control the spread of the pest with chemical pesticides. Therefore, novel control managements are urgently needed to mitigate crop damage. We first sequenced the antennal transcriptomes of A. dissimilis31 and characterized 5 OBPs that showed tissue-specific expression patterns32. Of note, AdisOBP6 was highly expressed in the testes of A. dissimilis32. We reasoned that the testis of insects possess a defined set of OBPs in a manner comparable to the antenna. In this study, we reanalyzed the previous antennal transcriptome data and identified 31 OBP genes. We also sequenced the transcriptomes of the A. dissimilis reproductive organs, and studied the expression of the OBPs in the antennae, testis and ovaries. Our study provides a new reference for studying the function of OBP genes.
Results
Illumina sequencing and assembly
A total of 34,565,866, 32,154,799, and 26,952,526 clean reads containing 10.35, 9.63, and 8.07 giga base (Gb) pairs of clean nucleotides respectively, were obtained from the three replicates of the A. dissimilis ovaries. A total of 27,752,168, 28,900,040, and 30,838,686 clean reads containing 8.29, 8.65 and 9.23 giga base (Gb) pairs of clean nucleotides respectively, were obtained from the three replicates of A. dissimilis testes. The quality of the transcriptome sequences was high, with Q30 percentages of 94.03%, 94.36%, 94.21%, 94.42%, 94.27% and 94.01% for the three replicates of A. dissimilis ovaries and testes, with a GC content of ~ 50% (Table 1). Then 221,074 transcripts and 82,016 unigenes with N50 length of 1350 and 1243 were obtained from assembled using Trinity (Table 2).
Table 1.
Summary of the sequence assemblies according to the RNA-seq data of the A. dissimilis.
| Sample name | Clean reads | Clean bases | GC content (%) | Q30 (%) |
|---|---|---|---|---|
| Ovaries | ||||
| Repeat 1 | 34,565,866 | 10.35 G | 48.00 | 94.03 |
| Repeat 2 | 32,154,799 | 9.63 G | 48.35 | 94.36 |
| Repeat 3 | 26,952,526 | 8.07 G | 48.27 | 94.21 |
| Testis | ||||
| Repeat 1 | 27,752,168 | 8.29 G | 48.85 | 94.42 |
| Repeat 2 | 28,900,040 | 8.65 G | 47.20 | 94.27 |
| Repeat 3 | 30,838,686 | 9.23 G | 46.65 | 94.01 |
Table 2.
Summary of de novo assembly of the A. dissimilis transcriptomes.
| Length range | Transcript | Rate% | Unigene | Rate% |
|---|---|---|---|---|
| < 300 | 0 | 0 | 0 | 0 |
| 300–500 | 83,670 | 37.85 | 37,104 | 45.24 |
| 500–1000 | 70,088 | 31.70 | 24,792 | 30.23 |
| 1000–2000 | 44,935 | 20.33 | 12,864 | 15.68 |
| > 2000 | 22,381 | 10.12 | 7256 | 8.85 |
| Total number | 221,074 | 82,016 | ||
| Total length | 216,261,287 | 73,549,396 | ||
| N50 length | 1350 | 1243 | ||
| Mean length | 978.23 | 896.77 |
Functional annotation
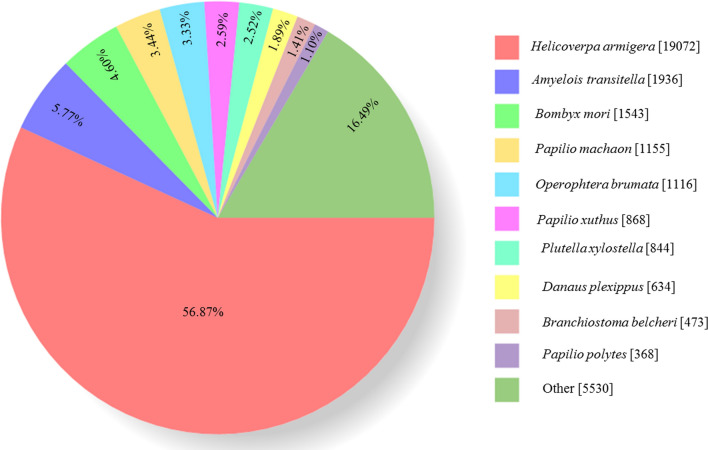
Significant matches of 33,587 unigenes (96.91%) in the NR; 29,936 (86.38%) in the eggnog; 20,134 (58.09%) in the Pfam; 15,174 (43.78%) in the Swissprot database; 14,775 (42.63%) in the KEGG; 7797 (22.50%) in the GO; and 6712 (19.37%) in the COG were observed. As a result, up to 34,658 putative coding sequences were identified (Table 3). NR database queries revealed a high percentage of A. dissimilis sequences that closely matched to sequences of H. armigera (19,072, 56.87%), Amyelois transitella (1936, 5.77%), Bombyx mori (1543, 4.60%), Papilio machaon (1155, 3.44%), Papilio xuthus (868, 2.59%), Plutella xylostella (844, 2.52%), Danaus plexippus (634, 1.89%), Branchiostoma belcheri (473, 1.41%), and Papilio polytes (368, 1.10%) (Fig. 1).
Table 3.
Functional annotation of the A. dissimilis transcriptomes.
| Database | Number | Rate (%) | 300 ≦ length < 1000 | Length ≧ 1000 |
|---|---|---|---|---|
| COG | 6712 | 19.37 | 2638 | 4074 |
| GO | 7797 | 22.50 | 4453 | 3344 |
| KEGG | 14,775 | 42.63 | 8205 | 6570 |
| Pfam | 20,134 | 58.09 | 8577 | 11,557 |
| Swissprot | 15,174 | 43.78 | 6987 | 8187 |
| eggNOG | 29,936 | 86.38 | 16,283 | 13,653 |
| NR | 33,587 | 96.91 | 18,939 | 14,648 |
| All | 34,658 | 19,914 | 14,744 |
Figure 1.
The Blastx results of Athetis dissimilis reproductive organs unigenes in NR database.
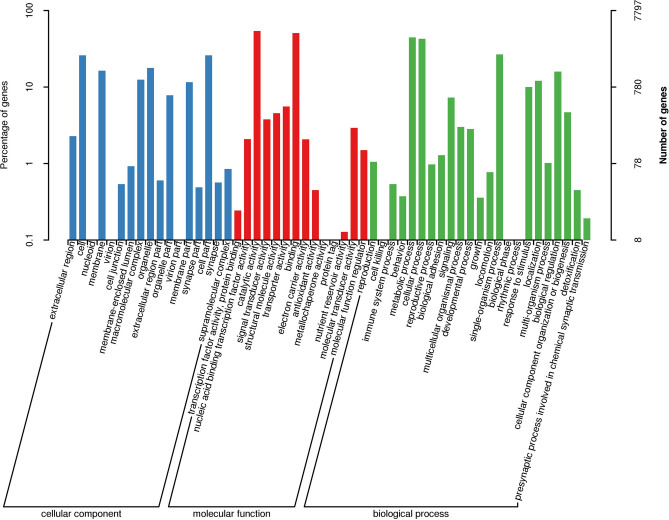
For GO analysis, 7797 unigenes (22.50%) could be assigned to three GO terms including: cellular components, molecular functions and biological process (Fig. 2). For the “molecular functions” ontology, catalytic activity (4227, 42.19%) and binding (3972, 39.64%) were most prevalent.
Figure 2.
Gene Ontology (GO) classifications of Athetis dissimilis reproductive organs unigenes according to their involvement in biological processes, cellular component and molecular function.
Identification of putative odorant-binding proteins
In the A. dissimilis antennal and reproductive organ transcriptome, we identified 54 candidate OBPs (Genbank accession number: KR780027–KR780030, MH900289–MH900338), 31 of which were from the antennae (through the analysis of previous A. dissimilis antennal transcriptomes) and 23 from the testis and ovaries transcriptomes of A. dissimilis (Table 4). A total of 44 AdisOBP sequences had full-length ORFs. Their cDNAs encoded protein of 131–293 amino acids with molecular weights of 11.6–33.2 kDa and isoelectric points of 4.44–9.74. Excluding 7 AdisOBPs (AdisOBP28, 30, 31, 35, 36, 41, 42, 52, 53 and 54) signal peptides were predicted at the N-terminus. AdisOBPs had 39–99% sequence homology with previously identified OBPs from other insect species, displaying a high level of sequence similarity. For example, AdisOBP13 had a 95% identity with Spodoptera exigua OBP9 (Table 4). There was 11.87% the lowest identity level in a pairwise comparison of AdisOBPs.
Table 4.
The characteristic of candidate OBP genes in the antennae and reproductive organs of A. dissimilis.
| Order | Gene name | GenBank accession no. | ORF (aa) | Molecular weight (kD) | Isoelectric point | Signal peptide | Full length | Homology search with the known proteins | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene annotation | Species | Protein ID | Score | E-value | Identity (%) | ||||||||
| c69042 | AdisPBP1 | KR780029 | 166 | 17.32 | 5.19 | Yes | Yes | PBP1 | Mamestra brassicae | AAC05702 | 266 | 3e−89 | 79 |
| c65047 | AdisPBP2 | KR780030 | 162 | 18.08 | 5.30 | Yes | Yes | PBP2 | Mamestra brassicae | AAC05701 | 281 | 4e−95 | 81 |
| c65143 | AdisPBP3 | MH900289 | 164 | 18.71 | 5.25 | Yes | Yes | PBP3 | Agrotis ipsilon | AFM36758 | 292 | 1e−99 | 82 |
| c47645 | AdisGOBP1 | KR780027 | 163 | 18.89 | 5.19 | Yes | Yes | GOBP1 | Sesamia inferens | AGS36742 | 289 | 3e−98 | 99 |
| c60029 | AdisGOBP2 | KR780028 | 161 | 18.09 | 5.09 | Yes | Yes | GOBP2 | Agrotis ipsilon | AFM36760 | 297 | 2e−101 | 88 |
| c68783 | AdisOBP1 | MH900290 | 293 | 33.20 | 5.76 | Yes | Yes | OBP | Bombyx mori | NP_001153663 | 264 | 1e−84 | 51 |
| c69959 | AdisOBP2 | MH900291 | 246 | 27.36 | 5.40 | Yes | Yes | OBP10 | Ostrinia furnacalis | BAV56797 | 310 | 4e−104 | 66 |
| c60098 | AdisOBP3 | MH900292 | 145 | 16.22 | 8.37 | Yes | Yes | OBP | Spodoptera exigua | ADY17886 | 251 | 5e−84 | 79 |
| c65852 | AdisOBP5 | MH900293 | 242 | 26.78 | 6.33 | Yes | Yes | OBP35 | Dendrolimus punctatus | ARO70194 | 215 | 2e−66 | 46 |
| c72710 | AdisOBP8 | MH900294 | 240 | 27.01 | 6.53 | Yes | Yes | OBP25 | Spodoptera exigua | AKT26502 | 305 | 3e−102 | 63 |
| c61153 | AdisOBP9 | MH900295 | 167 | 18.50 | 4.51 | Yes | Yes | OBP10 | Sesamia inferens | AGS36751 | 233 | 3e−76 | 79 |
| c60049 | AdisOBP11 | MH900296 | 141 | 16.38 | 4.47 | Yes | Yes | OBP8 | Spodoptera exigua | AGH70104 | 232 | 2e−76 | 86 |
| c65401 | AdisOBP13 | MH900297 | 133 | 15.14 | 9.01 | Yes | Yes | OBP9 | Spodoptera exigua | AGH70105 | 261 | 6e−88 | 95 |
| c58306 | AdisOBP14 | MH900298 | 185 | 20.13 | 6.04 | Yes | Yes | OBP1 | Agrotis ipsilon | AGR39564 | 279 | 1e−93 | 74 |
| c64058 | AdisOBP15 | MH900299 | 146 | 16.43 | 6.29 | Yes | Yes | OBP6 | Agrotis ipsilon | AGR39569 | 238 | 4e−79 | 88 |
| c53621 | AdisOBP16 | MH900300 | 118 | – | – | – | Internal | OBP18 | Spodoptera exigua | AKT26496 | 124 | 2e−33 | 48 |
| c68160 | AdisOBP17 | MH900301 | 252 | 28.95 | 6.19 | Yes | Yes | OBP23 | Spodoptera exigua | AKT26500 | 442 | 7e−156 | 81 |
| c67912 | AdisOBP18 | MH900302 | 203 | 22.50 | 5.69 | Yes | Yes | OBP19 | Helicoverpa assulta | AGC92793 | 245 | 9e−80 | 62 |
| c60881 | AdisOBP19 | MH900303 | 139 | 14.55 | 8.58 | Yes | Yes | OBP5 | Agrotis ipsilon | AGR39568 | 168 | 4e−51 | 62 |
| c71719 | AdisOBP20 | MH900304 | 139 | 15.69 | 7.52 | Yes | Yes | OBP8 | Spodoptera litura | AKI87969 | 257 | 2e−86 | 87 |
| c65033 | AdisOBP21 | MH900305 | 147 | 15.65 | 4.90 | Yes | Yes | OBP5 | Helicoverpa armigera | AEB54581 | 221 | 6e−72 | 75 |
| c63129 | AdisOBP22 | MH900306 | 146 | 15.92 | 7.53 | Yes | Yes | OBP23 | Spodoptera litura | XP_022826767 | 238 | 2e−78 | 77 |
| c57331 | AdisOBP23 | MH900307 | 149 | 15.96 | 5.03 | Yes | Yes | OBP26 | Spodoptera exigua | AKT26503 | 233 | 1e−76 | 76 |
| c64709 | AdisOBP24 | MH900308 | 148 | 16.77 | 5.45 | Yes | Yes | OBP7 | Helicoverpa armigera | AEB54591 | 187 | 5e−57 | 57 |
| c81048 | AdisOBP25 | MH900309 | 71 | – | – | – | Internal | OBP22 | Spodoptera exigua | AKT26499 | 130 | 3e−37 | 87 |
| c53707 | AdisOBP26 | MH900310 | 134 | 14.28 | 4.51 | Yes | Yes | OBP34 | Helicoverpa assulta | ASA40070 | 225 | 9e−74 | 86 |
| c28876 | AdisOBP27 | MH900311 | 124 | – | – | – | Internal | OBP11 | Spodoptera exigua | AGP03457.1 | 219 | 3e−71 | 81 |
| c67118 | AdisOBP28 | MH900312 | 236 | 27.80 | 4.90 | No | Yes | OBP9 | Spodoptera litura | ALD65883 | 383 | 1e−131 | 82 |
| c57589 | AdisOBP29 | MH900313 | 129 | – | – | – | 5ʹ lose | OBP33 | Helicoverpa assulta | ASA40072 | 208 | 5e−67 | 76 |
| c62521 | AdisOBP30 | MH900314 | 180 | 20.26 | 4.84 | No | Yes | OBP9 | Helicoverpa armigera | AEB54592 | 167 | 3e−50 | 54 |
| c63839 | AdisOBP31 | MH900315 | 116 | 12.77 | 6.12 | No | Yes | OBP14 | Spodoptera exigua | AGP03460 | 199 | 7e−64 | 83 |
| Gene.53346 | AdisOBP32 | MH900316 | 184 | 20.65 | 6.32 | Yes | Yes | GOBP70 | Helicoverpa armigera | XP_021188671 | 375 | 1e−131 | 98 |
| Gene.77161 | AdisOBP33 | MH900317 | 207 | 23.94 | 9.19 | Yes | Yes | OBP19 | Helicoverpa assulta | AGC92793 | 151 | 1e−42 | 39 |
| Gene.60926 | AdisOBP34 | MH900318 | 193 | 22.42 | 5.48 | Yes | Yes | OBP9 | Cnaphalocrocis medinalis | ALT31639 | 289 | 5e−97 | 70 |
| Gene.32069 | AdisOBP35 | MH900319 | 137 | 15.34 | 8.85 | No | Yes | OBP | Helicoverpa armigera | AEX07279 | 238 | 7e−79 | 88 |
| Gene.44893 | AdisOBP36 | MH900320 | 143 | 15.92 | 5.57 | No | Yes | OBP19 | Helicoverpa assulta | AGC92793 | 187 | 1e−57 | 66 |
| Gene.35132 | AdisOBP37 | MH900321 | 102 | – | – | – | 5ʹ lose | OBP24 | Cnaphalocrocis medinalis | ALT31654 | 182 | 7e−58 | 86 |
| Gene.54044 | AdisOBP38 | MH900322 | 141 | 15.05 | 8.77 | Yes | Yes | OBP5 | Agrotis ipsilon | AGR39568 | 155 | 6e−46 | 57 |
| Gene.7082 | AdisOBP39 | MH900323 | 156 | 17.94 | 4.86 | Yes | Yes | PBP1 | Helicoverpa armigera | XP_021192649 | 129 | 1e−34 | 39 |
| Gene.113597 | AdisOBP40 | MH900324 | 166 | 19.09 | 8.61 | Yes | Yes | OBP38 | Dendrolimus punctatus | ARO70197 | 157 | 7e−46 | 63 |
| Gene.77158 | AdisOBP41 | MH900325 | 141 | 16.29 | 9.12 | No | Yes | OBP19 | Helicoverpa assulta | AGC92793 | 115 | 2e−29 | 44 |
| Gene.14505 | AdisOBP42 | MH900326 | 102 | 11.15 | 5.44 | No | Yes | OBP23 | Spodoptera litura | ALD65897 | 98.6 | 3e−24 | 49 |
| Gene.54039 | AdisOBP43 | MH900327 | 76 | – | – | – | 5ʹ lose | OBP | Helicoverpa armigera | AEX07280 | 87.8 | 1e−20 | 59 |
| Gene.58201 | AdisOBP44 | MH900328 | 76 | – | – | – | 5ʹ lose | OBP23 | Spodoptera litura | ALD65897 | 71.6 | 6e−14 | 48 |
| Gene.32531 | AdisOBP45 | MH900329 | 150 | 16.43 | 4.77 | Yes | Yes | OBP2 | Agrotis ipsilon | AGR39565 | 119 | 1e−31 | 42 |
| Gene.5319 | AdisOBP46 | MH900330 | 70 | – | – | – | 5ʹ lose | OBP14 | Spodoptera exigua | AGP03460 | 117 | 2e−32 | 81 |
| Gene.86678 | AdisOBP47 | MH900331 | 120 | – | – | – | 5ʹ lose | OBP13 | Sesamia inferens | AGS36753 | 137 | 8e−39 | 53 |
| Gene.141496 | AdisOBP48 | MH900332 | 106 | 12.10 | 6.95 | No | Yes | OBP39 | Dendrolimus punctatus | ARO70198 | 183 | 4e−57 | 82 |
| Gene.142856 | AdisOBP49 | MH900333 | 157 | 17.96 | 9.74 | Yes | Yes | OBP18 | Dendrolimus punctatus | ARO70177 | 119 | 3e−31 | 51 |
| Gene.17592 | AdisOBP50 | MH900334 | 144 | 16.21 | 4.44 | Yes | Yes | OBP9 | Helicoverpa armigera | AEB54592 | 163 | 5e−49 | 54 |
| Gene.54647 | AdisOBP51 | MH900335 | 84 | – | – | – | 5ʹ lose | OBP39 | Dendrolimus punctatus | ARO70198 | 140 | 1e−40 | 86 |
| Gene.76032 | AdisOBP52 | MH900336 | 105 | 11.60 | 4.71 | No | Yes | OBP | Spodoptera litura | ALD65897 | 111 | 4e−29 | 52 |
| Gene.111996 | AdisOBP53 | MH900337 | 105 | 12.28 | 8.21 | No | Yes | OBP | Operophtera brumata | KOB73304 | 194 | 1e−61 | 88 |
| Gene.158529 | AdisOBP54 | MH900338 | 131 | 14.34 | 4.86 | No | Yes | OBP11 | Spodoptera exigua | AGP03457 | 226 | 3e−74 | 79 |
Genes beginning with the lowercase letter “c” came from the identification of antenna transcriptome, and genes beginning with “Gene” came from testis and ovary transcriptome identification.
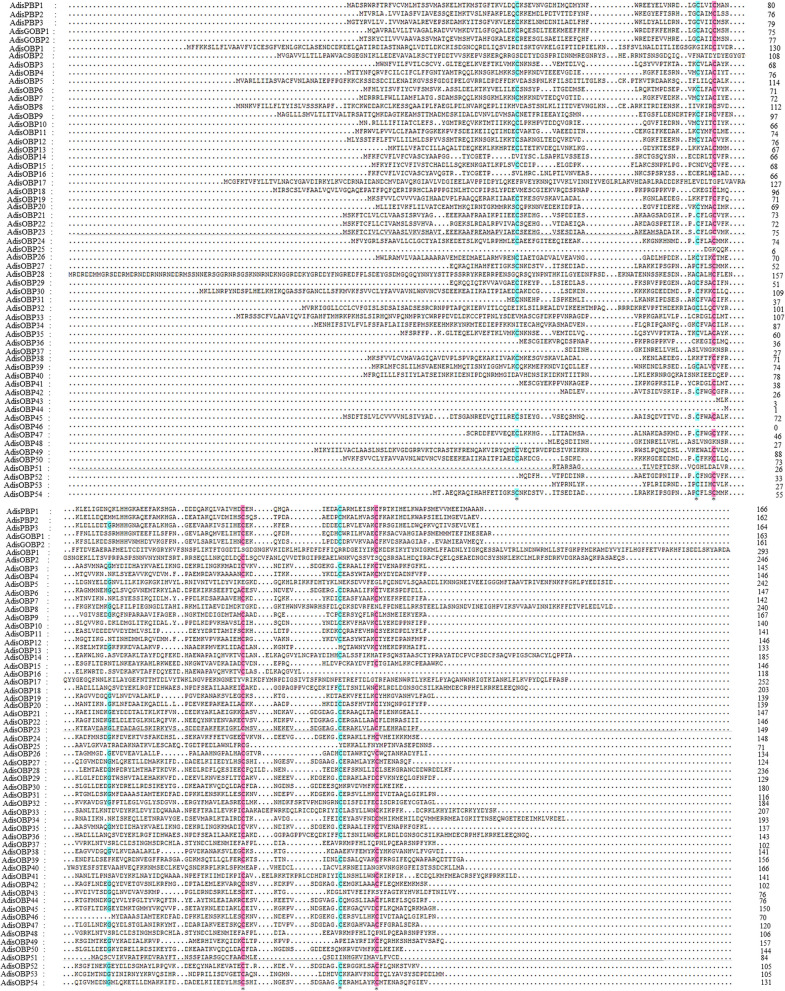
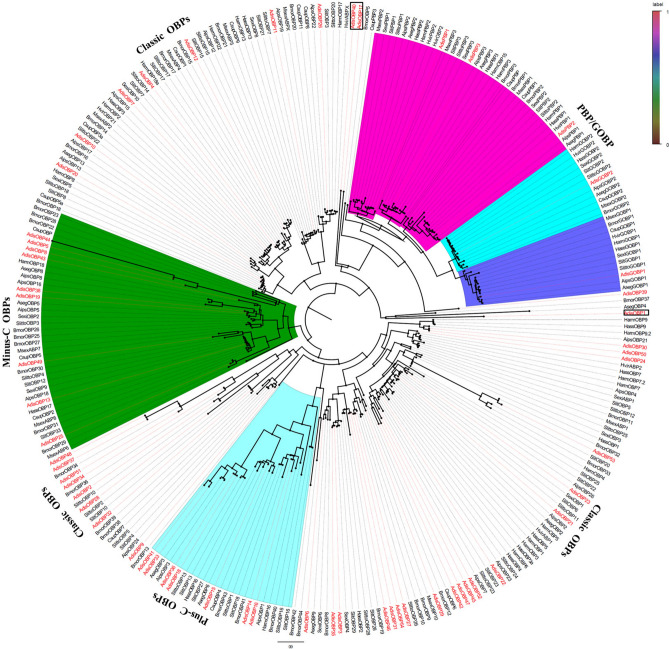
Multiple sequence alignments of the A. dissimilis OBPs revealed the presence of expected conserved cysteines (Fig. 3). The phylogenetic tree of A. dissimilis and other lepidopteran OBPs constructed using the neighbor-joining method, indicated five clades that contained four possible subclass OBPs (Fig. 4). In addition, the tree showed low levels of clustering highlighting the diversity of the lepidopteran OBPs. Five AdisOBPs (AdisPBP1-3, GOBP1-2) belonged to PBP/GOBP. A total of 30 OBPs (AdisOBP2-3, 9, 11, 20–24, 26–32, 34–35, 37, 39, 42, 45–48, 50–54) were ‘Classic’ OBPs that contained six positionally-conserved cysteine residues. Seven OBPs (AdisOBP14-16, 18, 33, 36 and 41) belonged to ‘Plus-C’ subclass OBP genes with more cysteines in addition to those of the conserved motif. Nine OBPs belonged to ‘Minus-C’ subclass OBP genes with only four cysteines. Interestingly, AdisOBP1, AdisOBP17 and AdisOBP40 did not belong to any of the four subclass OBPs (Fig. 4). However, according to BLAST results these three genes were homologous with OBP genes of Bombyx mori, Spodoptera exigua and Dendrolimus punctatus (Table 4). The transcription abundance of A. dissimilis OBPs in antennae of female and males, ovary and testis are profiled in Fig. 5.
Figure 3.
Sequence alignments of Athetis dissimilis OBPs. The six conserved cysteine residues are indicated with the asterisks under the sequence.
Figure 4.
Phylogenetic relationships of candidate OBP proteins (including 5 OBPs identified in a previous study) from Athetis dissimilis and 33 Lepidoptera species.
Figure 5.
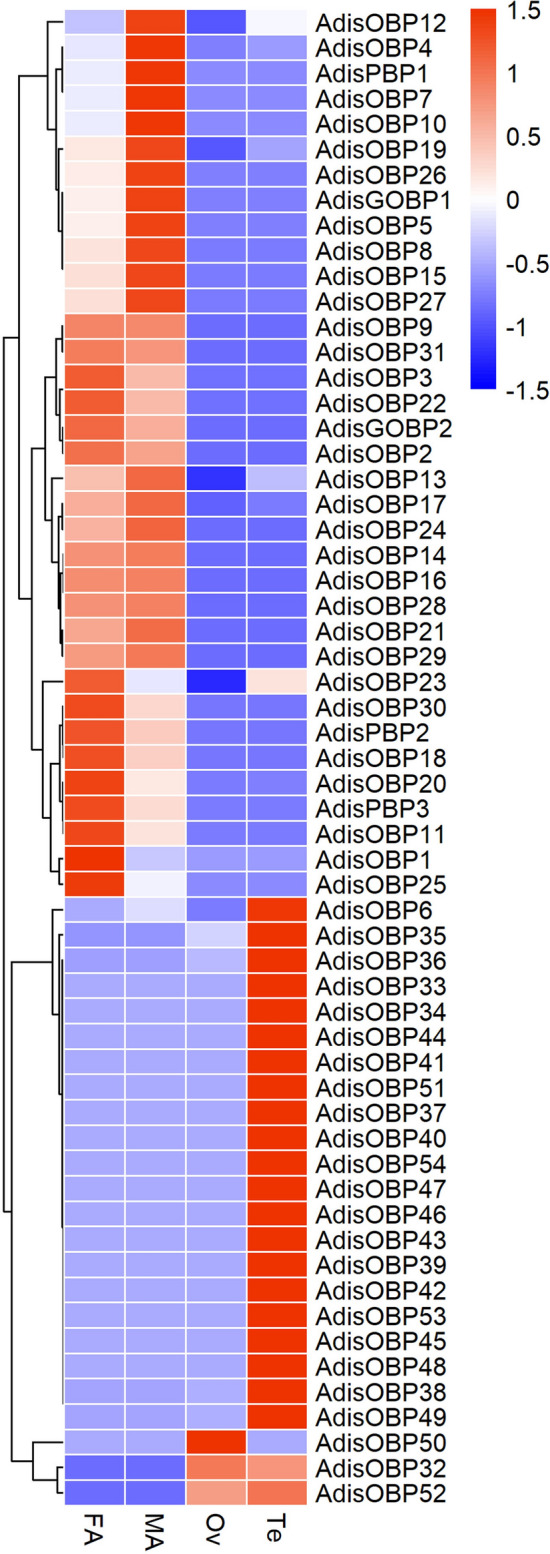
Heat map showing the abundance of unigenes encoding OBPs (including 5 OBPs identified in a previous study) in the Athetis dissimilis different tissues transcriptomes presented as normalized reads in reads per kilobase per million mapped reads (RPKM). In the figure each column represents 1 samples; each line represents 1 OBP gene. The color depth represents the number of reads contained in OBPs; red means more; blue means less. FA female antennae, MA male antennae, Ov ovaries, Te testis.
Expression of the OBPs in the antennae, ovaries and testis of A. dissimilis
Next, we measured the relative expression levels of the identified OBPs in different tissues of A. dissimilis via fluorescence qRT-PCR (Fig. 6). A total of 23 OBPs (AdisGOBP1-2, PBP1-3, OBP1-2, 8–9, 11, 17, 20–22, 24, 26–31, 50 and 54) were highly expressed in the antennae compared to the reproductive organs, including three OBPs (AdisPBP1, OBP17 and OBP26) that exhibited male-biased expression, 15 OBPs (AdisGOBP2, PBP2-3, OBP1-2, 11, 20–22, 27–28, 30–31, 50 and 54) that exhibited female-biased expression, and five OBPs (Adis GOBP1, OBP8-9, 24 and 29) that showed comparable expression in the male and female antennae of A. dissimilis.
Figure 6.
Expression profiles of the candidate OBPs in different tissues of Athetis dissimilis. FA female antennae, MA male antennae, Ov ovaries, Te testis. The standard errors are represented by the error bars; different lowercase letters (a–c) above the bars denote significant differences at p ˂ 0.05.
A total of 24 OBPs (AdisOBP3, 5, 15, 18–19, 23, 25, 33–41, 44–45, 47–49 and 51–53) were highly expressed in the testis of A. dissimilis compared to other tissues. The expression of the OBPs was low in the ovaries of A. dissimilis.
Discussion
In this study, we identified 31 novel OBPs through the analysis of A. dissimilis antennal transcriptomes, except for 5 AdisOBP genes identified in a previous study32. The number of OBPs in A. dissimilis antennae were similar to those in the antennal transcriptomes of S. litura (33)17 and S. littoralis (36)33 but more abundant than S. exigua (11)34, M. sexta (18)35 and H. armigera (26)36. We additionally sequenced the transcriptomes of A. dissimilis ovaries and testis. The alignments against the Nr database showed that 56.87% of the A. dissimilis unigenes were comparable to H. armigera sequences. A total of 23 OBPs were identified in the transcriptomes of A. dissimilis reproduction organs.
Based on sequence alignments and the cluster analysis of the phylogenetic trees, five PBP/GOBP genes, 35 Classic genes, 7 Plus-C genes and 9 Minus-C genes were obtained from the A. dissimilis antennal library. These results were similar to the classifications of most insect OBPs17,27,37. Interestingly, AdisOBP1, AdisOBP17 and AdisOBP40 could not be clustered into any subfamilies, and multiple sequence alignments of all AdisOBP genes revealed that the three OBPs contain no conserved cysteines. The phylogenetic tree supports a highly dynamic evolutionary process for the lepidopteran OBP family and a high degree of OBP sequence divergence. The diversification of OBPs might be the result of multiple and late independent gene duplications. In addition, they might be derived from a common ancestor and later diverged into different subfamilies by different selection pressures, which has been evidenced by evolutionary selection analysis in several insect species38–40.
OBPs are expressed specifically in the antennae and other parts associated with olfactory organs15,19,31,41–43. Our comprehensive expression analysis revealed that 23 AdisOBPs were found to be restricted to the antenna. It is worth noting that only 3 AdisOBPs had male-biased expression pattern in the antennae, suggesting that females require more abundant OBPs for spawning. It is interesting to note 24 AdisOBPs showed significant expression in the testis of A. dissimilis compared to other tissues, but the expression of AdisOBPs in the ovaries was low. The expression of OBPs in reproduction has also been reported in some literature44–46. It was previously speculated that OBPs expressed in the testis deliver compounds to the females during mating26,27. Hence, it is understandable to presume that such stable proteins could be used in the testis of insect where there is need for transportation of hydrophobic molecules in aqueous media or protection of chemicals from degradation, as well as to assure a gradual release of semiochemicals in the environment. So these proteins have been named for ‘‘encapsulins”, to imply the common role of encapsulating small ligands47. qRT-PCR was conducted on 53 candidate genes, and the expression level of most genes were consistent with the variation of RPKM values.
Like the OBP families of insect antennae, insect testes contain a large number of OBP genes. The functions of these genes is unclear, and they need us to further study. Our results provide a reference for the study of these genes.
Materials and methods
Insect rearing and sample preparation
The A. dissimilis strain was collected from Luoyang (province of Henan, China) corn fields (112° 26′ E, 34° 43′ N) in 2014 and maintained at the Henan Science and Technology University. Colonies were reared on an artificial diet at 25 ± 1 °C, 80 ± 5% relative humidity and a 16-h/8-h light/dark cycle.
Based on preliminary data, we found that the A. dissimilis sperm and eggs began to mature 3 days after emergence. We respectively collected the ovaries and testes of 3-day old virgin females and male adults (n = 40 per treatment) from three biological replications. Dissections were performed in sterile PBS-DEPC and immediately frozen in liquid nitrogen until RNA isolation.
cDNA library preparation and sequencing
Total RNA from the A. dissimilis ovaries and testis tissues were extracted using RNAiso Plus kit (TaKaRa, Dalian, China) and treated with DNase I (TaKaRa, Dalian, China) as per the manufacturer’s protocols. RNA was assessed through 1% agarose gel electrophoresis and Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA), Qubit 2.0 (Life Technologies, Carlsbad, CA, USA) and Agilent 2100 (Agilent, Santa Clara, CA, USA) analysis.
Following the TruSeq RNA Sample Preparation Guide v2 (Illumina, San Diego, CA, USA), mRNA was enriched using magnetic beads crosslinked with Oligo (dT). Enriched RNA was then fragmented using fragmentation buffer and first-strand cDNA synthesis was used to produce small mRNA fragments, random primers, reverse transcriptase, and second-strand cDNA synthesis through the addition of dNTPs, DNA polymerase I, and RNase H. Double-stranded cDNA was purified with AMPure XP beads (Beckman Coulter, Brea, CA, USA) and treated to repair ends, remove poly(-A) tails, and link sequencing adapters. Fragment sizes were selected using AMPure XP beads and cDNA libraries were constructed through PCR amplification (Veriti 96-Well Thermal Cycle, Applied Biosystems, Foster City, USA). The concentration and insert size of the cDNA libraries were detected using Qubit 2.0 and Agilent 2100 and quantified via q-PCR (CFX-96, Bio-Rad, Hercules, CA, USA).
Finally, sequencing was performed using the Illumina HiSeq 4000 platform to generate 150-bp paired-end reads. Sequencing analyses were performed by the Genomics Services of the Beijing Biomarker Technologies Co., Ltd. (Beijing, China). Raw data processing and base calling were performed using Illumina software.
Assembly and functional annotation
Raw data (raw reads) in the FASTQ format were first modified into clean data (clean reads) through Perl scripts. This was performed through the removal of reads containing adapter sequences, > 10% unknown nucleotides and quality values ≤ 20. The Q20, Q30, and GC content were then calculated using high-quality data.
Transcriptomes were assembled using Trinity (version trinityrnaseq_r20131110) with default settings, except for min_kmer_cov set to 248. Unigene functions were annotated based on NCBI non-redundant protein sequences (NR, NCBI blast 2.2.28+, e-value = 1e−5), NCBI nucleotide sequences (NT, NCBI blast 2.2.28+, e-value = 1e−5), Protein family (Pfam, HMMER 3.0 package, hmmscan, e-value = 0.01), eukaryotic Ortholog Groups (KOG, NCBI blast 2.2.28+, e-value = 1e−3), SwissProt (NCBI blast 2.2.28+, e-value = 1e−5), the Kyoto Encyclopedia of Genes and Genomes (KEGG; KEGG Automatic Annotation Server [KASS], e-value = 1e−10) and Gene Ontology (GO, Blast2GO v2.5, e-value = 1e−6). Coding sequences (CDS) were predicted through aligning transcriptome sequences to the Nr and Swiss-Prot database or using estscan 3.0.349. The read count for each gene was obtained by mapping clean reads to the assembled transcriptome using RSEM (bowtie2 parameters: mismatch 0). The final read count was calculated as Fragments Per Kilobase of transcript per Million mapped reads (FPKM)50.
Sequence and phylogenetic analysis
Sequence similarities were assessed using the NCBI-Blast network server (http://blast.ncbi.nlm.nih.gov/). The signal peptides of OBPs were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/)51. Multiple sequence alignments were assessed using DNAMAN 6.0. Sequence alignments of the candidate OBPs were performed using ClustalX 2.152 and used to construct phylogenetic trees with PhyML in Seaview v.4 based on the Jones–Taylor–Thormton (JTT) model with nearest-neighbor interchanges. Trees were viewed and edited using FigTree v.1.3.1. Amino acid sequences of OBPs in phylogenetice tree were listed in Supplementary File 1.
Expression analysis through quantitative real-time polymerase chain reaction
Male antennae (100), female antennae (100), ovaries (80) and testes (150) tissue from adults at 3 post-eclosion were excised and frozen in liquid nitrogen. Total RNA was extracted using RNAiso Plus kits (TaKaRa, Dalian, China) and isolated RNA was transcribed to first-strand cDNA using PrimeScript RT reagent with gDNA Eraser (TaKaRa, Dalian, China) following the manufacturer’s protocols. Real-time quantitative PCR (RT-qPCR) was performed with SYBR® Premix Ex Taq II (TaKaRa). The A. dissimilis GAPDH gene was used as an endogenous control to correct for sample-to-sample variations. A 200 ng/μL cDNA sample was used for per tissue. Primers were designed using Primer Premier 5.0 software and are listed in Supplementary File 2. RT-qPCR reactions contained: 10 μL of SYBR Premix Ex Taq II, 20 ng of cDNA template, 0.2 μM of each primer and nuclease-free water. The cycling conditions were 1 cycle of 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 s and 55 °C for 30 s. Melt curve conditions were 95 °C for 10 s and 65 °C for 30 s. No-template controls (NTC) were included to detect possible contamination. Three biological replicates were analyzed and the relative expression of the OBP genes was measured using the 2−∆∆CT method53. Expression was calculated relative to levels in the female antennae, which were arbitrarily set to 1. Differences in the expression of AdisOBP genes between the different tissues were compared using a one-way nested analysis of variance (ANOVA), followed by a Tukey’s honestly significance difference (HSD) test using SPSS (SPSS Institute 17.0, SPSS Inc, Chicago, IL, USA).
Supplementary Information
Acknowledgements
This study is supported by the National Natural Science Foundation of China (31701788) and Science and Technology Project in Henan Province (202102110069).
Author contributions
H.Z. and Y.Q. conceived this project and analyzed the data. Z.Y., Q.H., Q.X. and J.F. assisted the preparation of samples and operated experiment. Y.Q. wrote the main manuscript text. H.Z. edited the manuscript. All the authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-93423-1.
References
- 1.Field LM, Pickett JA, Wadhams LJ. Molecular studies in insect olfaction. Insect Mol. Biol. 2000;9:545–551. doi: 10.1046/j.1365-2583.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh E, Bohbot J, Zwiebel LJ. Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 2014;6:86–92. doi: 10.1016/j.cois.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L, et al. Perception of potential sex pheromones and host-associated volatiles in the cotton plant bug, Adelphocoris fasciaticollis (Hemiptera: Miridae): Morphology and electrophysiology. Appl. Entomol. Zool. 2014;49:43–57. doi: 10.1007/s13355-013-0223-1. [DOI] [Google Scholar]
- 5.Zhang J, Walker WB, Wang G. Pheromone reception in moths: From molecules to behaviors. Prog. Mol. Biol. Transl. Sci. 2015;130:109–128. doi: 10.1016/bs.pmbts.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 7.Pelosi P, Maida R. Odorant-binding proteins in insects. Comp. Biochem. Phys. B. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- 8.Angeli S, et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999;262:745–754. doi: 10.1046/j.1432-1327.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- 9.Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464:85–90. doi: 10.1016/S0014-5793(99)01683-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhou JJ. Odorant-binding proteins in insects. Vitamins Hormones. 2010;83:241–272. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- 11.Lagarde A, et al. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J. Mol. Biol. 2011;414:401–412. doi: 10.1016/j.jmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Pelosi P, Zhou JJ, Ban L, Calvello M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leal WS. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Smagghe G, Lei ZR, Wang JJ. Identification of male-and female-specific olfaction genes in antennae of the oriental fruit fly (Bactrocera dorsalis) PLoS ONE. 2016;11:e0147783. doi: 10.1371/journal.pone.0147783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao H, et al. Discrimination of alarm pheromone (E)-beta-farnesene by aphid odorant-binding proteins. Insect Biochem. Mol. 2009;39:414–419. doi: 10.1016/j.ibmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhou JJ, et al. Characterisation of Bombyx mori Odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009;389:529–545. doi: 10.1016/j.jmb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 17.Gu SH, et al. Identification and comparative expression analysis of odorant binding protein genes in the tobacco cutworm Spodoptera litura. Sci. Rep. 2015;5:13800. doi: 10.1038/srep13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larter NK, Sun JS, Carlson JR. Organization and function of Drosophila odorant binding proteins. Elife. 2016;5:e20242. doi: 10.7554/eLife.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X, Zhang X, Liu H, Wang R, Zhang T. Identification of chemosensory genes from the antennal transcriptome of Indianmeal moth Plodiainterpunctella. PLoS ONE. 2018;13:e0189889. doi: 10.1371/journal.pone.0189889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao DP, et al. Identification of candidate olfactory genes in Chilosuppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 2014;10:846. doi: 10.7150/ijbs.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang SF, Zhang Z, Wang HB, Kong XB. Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae) Insect Biochem. Mol. 2014;52:69–81. doi: 10.1016/j.ibmb.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Jia XJ, et al. Antennal transcriptome and differential expression of olfactory genes in the yellow peach moth, Conogethes punctiferalis (Lepidoptera: Crambidae) Sci. Rep. 2016;6:29067. doi: 10.1038/srep29067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng WN, Zhang YD, Liu W, Li GW, Zhu-Salzman K. Molecular and functional characterization of three odorant-binding proteins from the wheat blossom midge, Sitodiplosis mosellana. Insect Sci. 2019;98:e21456. doi: 10.1111/1744-7917.12677. [DOI] [PubMed] [Google Scholar]
- 24.Pregitzer P, et al. Expression of odorant-binding proteins in mouthpart palps of the desert locust Schistocerca gregaria. Insect Mol. Biol. 2019;28:264–276. doi: 10.1111/imb.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin J, et al. Functional characterization of odorant-binding proteins from the scarab beetle Holotrichia oblita based on semiochemical-induced expression alteration and gene silencing. Insect Biochem. Mol. 2019;104:11–19. doi: 10.1016/j.ibmb.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Li S, et al. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Co. 2008;372:464–468. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- 27.Sun YL, Huang LQ, Pelosi P, Wang CZ. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS ONE. 2012;7:e30040. doi: 10.1371/journal.pone.0030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JW, et al. Morphologically a like species of Athetis lepigone (Möschler)—A. dissimilis (Hampson) found in Shandong Province of China. Plant Prot. 2014;40:193–195. [Google Scholar]
- 29.Takahashi M. Athetis dissimilis Hampson, a new nuisance? Jpn. Soc. Med. Entomol. Zool. 1975;26:66. doi: 10.7601/mez.26.66_1. [DOI] [Google Scholar]
- 30.Li NG. Relationships between cold hardiness and ice nucleating activity, glycerol and protein contents in the hemolymph of caterpillars, Aporia crataegi L. CryoLetters. 2012;33:134–142. [PubMed] [Google Scholar]
- 31.Dong J, Song Y, Li W, Shi J, Wang Z. Identification of putative chemosensory receptor genes from the Athetis dissimilis antennal transcriptome. PLoS ONE. 2016;11:e0147768. doi: 10.1371/journal.pone.0147768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun HZ, Song YQ, Du J, Wang XD, Cheng ZJ. Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Athetis dissimilis (Lepidoptera: Noctuidae) Appl. Entomol. Zool. 2016;51:409–420. doi: 10.1007/s13355-016-0413-8. [DOI] [Google Scholar]
- 33.Poivet E, et al. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE. 2013;8:e60263. doi: 10.1371/journal.pone.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu JY, Zhang LF, Ze SZ, Wang DW, Yang B. Identification and tissue distribution of odorant binding protein genes in the beet armyworm, Spodoptera exigua. J. Insect Physiol. 2013;59:722–728. doi: 10.1016/j.jinsphys.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Grosse-Wilde E, et al. Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. 2011;108:7449–7454. doi: 10.1073/pnas.1017963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Gu SH, Zhang YJ, Guo YY, Wang GR. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS ONE. 2012;7:e48260. doi: 10.1371/journal.pone.0048260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H, et al. Candidate odorant binding proteins and chemosensory proteins in the larval chemosensory tissues of two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS ONE. 2017;12:e0179243. doi: 10.1371/journal.pone.0179243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forêt S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera) Genome Res. 2006;16:1404–1413. doi: 10.1101/gr.5075706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou JJ, et al. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2010;19:113–122. doi: 10.1111/j.1365-2583.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang GH, Li YP, Xu XL, Chen H, Wu JX. Identification and characterization of two general odorant binding protein genes from the oriental fruit moth, Grapholita molesta (Busck) J. Chem. Ecol. 2012;38:427–436. doi: 10.1007/s10886-012-0102-1. [DOI] [PubMed] [Google Scholar]
- 41.Feng B, Guo QS, Zheng KD, Qin YX, Du YJ. Antennal transcriptome analysis of the piercing moth Oraesia emarginata (Lepidoptera: Noctuidae) PLoS ONE. 2017;12:e0179433. doi: 10.1371/journal.pone.0179433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei HS, Li KB, Zhang S, Cao YZ, Yin J. Identification of candidate chemosensory genes by transcriptome analysis in Loxostege sticticalis Linnaeus. PLoS ONE. 2017;12:e0174036. doi: 10.1371/journal.pone.0174036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian ZQ, et al. Antennal transcriptome analysis of the chemosensory gene families in Carposina sasakii (Lepidoptera: Carposinidae) BMC Genomics. 2018;19:544. doi: 10.1186/s12864-018-4900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang TT, et al. Male-and female-biased gene expression of olfactory-related genes in the antennae of Asian corn borer, Ostrinia furnacalis (Guenee) (Lepidoptera: Crambidae) PLoS ONE. 2015;10:e0128550. doi: 10.1371/journal.pone.0128550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song LM, et al. Male tarsi specific odorant-binding proteins in the diving beetle Cybister japonicus sharp. Sci. Rep. 2016;6:31848. doi: 10.1038/srep31848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen XF, et al. Genome-wide identification and expression profiling of odorant-binding proteins in the oriental fruit fly, Bactrocera dorsalis. Comp. Biochem. Phys. D. 2019;31:100605. doi: 10.1016/j.cbd.2019.100605. [DOI] [PubMed] [Google Scholar]
- 47.Leal WS. Molecular-based chemical propecting of mosquito attractants and repellents. In: Debboun MF, Strickman D, editors. Insect Repellents: Principles, Methods, and Uses. CRC Press; 2006. pp. 229–244. [Google Scholar]
- 48.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iseli C, Jongeneel CV, Bucher P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999;7:138–148. [PubMed] [Google Scholar]
- 50.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomesby RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 51.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 52.Larkin M, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-ΔΔC) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.