Abstract
The tea-oil tree (Camellia oleifera Abel.) is a commercial edible-oil tree in China, and anthracnose commonly occurs in its plantations, causing great losses annually. We have previously revealed that CfSnf1 is essential for pathogenicity in Colletotrichum fructicola, the major pathogen of anthracnose on the tea-oil tree. Here, we identified CfGcn5 as the homolog of yeast histone acetyltransferase ScGcn5, which cooperates with ScSnf1 to modify histone H3 in Saccharomyces cerevisiae. Targeted gene deletion revealed that CfGcn5 is important in fungi growth, conidiation, and responses to environmental stresses. Pathogenicity assays indicated that CfGcn5 is essential for C. fructicola virulence both in unwounded and wounded tea-oil tree leaves. Further, we found that CfGcn5 is localized to the nucleus and this specific localization is dependent on both NLS region and HAT domain. Moreover, we provided evidence showing that the nuclear localization is essential but not sufficient for the full function of CfGcn5, and the NLS, HAT, and Bromo domains were proven to be important for normal CfGcn5 functions. Taken together, our studies not only illustrate the key functions of CfGcn5 in growth, development, and pathogenicity but also highlight the relationship between its locations with functions in C. fructicola.
Keywords: histone acetyltransferase, conidiation, pathogenicity, nucleus, localization, C. fructicola
Introduction
The tea-oil tree (Camellia oleifera Abel.) is a commercial shrub native to China and has been widely grown in southern China for over 2000 years (Chen et al., 2015). The tea oil extracted from its seed, similar to olive oil, is an excellent edible oil for humans and popular in Chinese cooking (Feas et al., 2013; Di et al., 2018). Though plantations have increased to more than 3.7 million hectares and tea oil yield ranges from 450 to 750 kg/ha, it is still not enough for the demand (Chen et al., 2015). Unfortunately, the tea-oil tree can be infected by numerous serious diseases which limit its cultivation.
Anthracnose is the most devastating disease afflicting the tea-oil tree and commonly occurs in plantations (Li et al., 2016). All tea-oil tree tissues are susceptible to the disease, resulting in the constant dropout of leaves and fruit. Anthracnose is currently causing a 10–30% reduction in tea oil each year, and some areas often incur over 50% tea oil losses. Colletotrichum, the causal agent of anthracnose, is one of the top 10 plant-pathogenic fungi and almost every crop is susceptible to one or more species of such a genus (Dean et al., 2012). For a long time, the Colletotrichum gloeosporioides complex has been considered as the pathogen of anthracnose on the tea-oil tree. Whereas, we recently revealed that its pathogens include at least C. gloeosporioides, C. fructicola, C. siamense, C. karstii, and C. camelliae, and that C. fructicola, the widest distributed and strongest pathogenic specie, was the major pathogen (Li et al., 2016; Jiang and Li, 2018).
C. fructicola was first identified in coffee berries in 2009 and was distributed all over the world, which resulted in the infection of over 50 plants, such as apples, pears and strawberries, among others (Prihastuti et al., 2009; Weir et al., 2012; Rockenbach et al., 2016; Liang et al., 2018). Despite its economic and ecological importance, its molecular pathogenesis is unclear. Previous studies revealed that C. fructicola contains a large amount of candidate virulence genes and many of them are upregulated during early plant colonization (Baroncelli et al., 2016; Liang et al., 2018), but the functions of these putative virulence genes remain unknown.
We recently demonstrated that CfSnf1, a key component of AMPK (AMP-activated protein kinase), regulates the utilization of specific carbon sources, appressorium formation, and pathogenicity in C. fructicola (Zhang et al., 2019). In Saccharomyces cerevisiae, ScSnf1 cooperates with the histone acetyltransferase ScGcn5 (general control non-derepressible 5) to regulate transcription (Lo et al., 2001; Abate et al., 2012). ScGcn5 was the first HAT (histone acetyltransferase) identified in yeast and contains a HAT domain and a Bromo domain (Haynes et al., 1992; Candau et al., 1997). It mainly targets specific lysine residues in histones H3 and H2B, and thus epigenetically regulates global gene transcription to environmental adaption (Zhang et al., 1998; Suka et al., 2001; Gaupel et al., 2015; Hu et al., 2015). There is little evidence supporting the importance of Gcn5 in the fungal development and virulence of phytopathogens. In Magnaporthe oryzae, MoGcn5 regulates the pathogenicity through acetylating the autophagy protein MoAtg7 (Zhang et al., 2017). Gcn5 proteins are also required for the development and virulence of Ustilago maydis and Fusarium graminearum (Gonzalez-Prieto et al., 2014; Kong et al., 2018). Despite this, the function of such a protein in forest fungal pathogens including C. fructicola is unknown. Here, we characterized the roles of CfGcn5 in forest pathogens for the first time. And we also provided evidence demonstrating the contributions of the NLS region, HAT, and Bromo domains to localization and function by domain deletion.
Materials and Methods
Strains and Culture Conditions
The C. fructicola CFLH16 strain was used as wide-type (WT) for transformation. The WT, ΔCfgcn5 mutant, complemented strain ΔCfgcn5/CfGCN5, and domain deleted strains (ΔNLS, ΔHAT, and ΔBromo) were cultured on Potato Dextrose Agar (PDA: 200 g peeled potato, 20 g dextrose, and 15 g agar in 1 L ddH2O) or Minimal Medium (MM: 0.52 g KCl, 6 g NaNO3, 1.52 g KH2PO4, 0.152 g MgSO4⋅7H2O, 0.01 g VB1, 10 g Glucose, 1 ml 1,000 × trace elements, and 15 g agar in 1 L ddH2O) plates at 28°C in the dark. The strains cultured in liquid PDB (Potato Dextrose Broth) for 2 or 3 days were used for DNA extraction and conidiation analysis.
Phylogenetic Tree Construction and Domain Prediction
The Gcn5 proteins in F. fujikuroi, F. graminearum, Trichoderma reesei, C. fructicola, C. gloeosporioides, M. oryzae, Neurospora crassa, Aspergillus nidulans, and S. cerevisiae were obtained from the NCBI database1. The MEGA 7.0 program with the neighbor-joining method was used to construct the phylogenetic tree. The domains and NLS region of CfGcn5 were predicted by the SMART2 and cNLS Mapper3 websites, respectively.
Gene Deletion, Complementation, and Domain Deletion Assays
Targeted CfGCN5 gene deletion was carried out by the one-step replacement strategy as per our previous description (Zhang et al., 2019). The two ∼1.0 kb sequences flanking CfGCN5 were overlapped to the two sides of the HPH (hygromycin resistance cassette) gene, respectively. Then, the ∼3.4 kb fragments were transformed into protoplasts of CFLH16 for gene deletion. For the complementation and domain deletion assays, the ∼1.5 kb native promoter and the full-length or truncated fragments (ΔNLS, or ΔHAT, or ΔBromo) of CfGCN5 were inserted into the pYF11 (bleomycin resistance) vector as described previously (Bruno et al., 2004). After sequencing, the fused-pYF11 plasmids were transformed into protoplasts of the ΔCfgcn5 mutant to acquire the complemented and domain deleted strains.
Growth, Conidiation, Germination, and Appressoria Formation Assays
The strains were cultured on PDA and MM agar plates at 28°C in the darkness for 4 days, and the colony diameters were measured and statistically analyzed. For conidiation assays, the strains were cultured in 200 mL liquid shaking PDB for 3 days, then were filtered with three layers of lens paper and the conidia were collected and quantified on a microscope. For germination and appressoria formation assays, the collected conidia were washed by ddH2O twice and resuspended to a concentration of 3 × 105 spores/mL and inoculated onto hydrophobic artificial surfaces or an onion epidermis for germination and appressorium formation.
Stress Response Assays
The strains were cultured on PDA and PDA plus with various stresses, including osmotic stress (1 M NaCl and 1 M KCl), cell wall integrity stress (400 μg/ml CFW, 800 μg/ml CR and 0.01% SDS), oxidative stress (2.5 mM and 5.0 mM H2O2), and ER stress (2.5 mM and 5.0 Mm DTT). After 4 days of incubation, the colony diameters were measured and the inhibition rates were statistically analyzed.
Pathogenicity Assays
The mycelia or conidial suspensions (3 × 105) of the strains were inoculated onto the edge of unwounded and wounded tea-oil leaves. For the pathogenicity on wounded apples, the apples were first punched into several 8-mm-diameter holes and the peel tissue was removed from the punched area, then 3 × 3 mm mycelia plugs were inoculated onto it. The inoculated leaves or apples were kept in a high humidity conditions at room temperature. After incubation for 4∼5 days, the lesions were observed and photographed.
Protoplast Release Assays
The mycelia were incubated in liquid PDB for 2 days, followed by filtration and drying. For each sample, 1.0 g of dried biomass was resuspended in 0.7 M NaCl with lysing enzyme and incubated for 2 h at 30°C by gentle shaking (70 rpm). The protoplasts were observed under microscope.
Localization Observation and DAPI Staining
The full-length and truncated CfGcn5 were fused with a green fluorescent protein (GFP) tag and transformed into the ΔCfgcn5 mutant. The hyphae and conidia of the strains were further stained with DAPI (50 μg/ml) for 5 min. After being washed twice with ddH2O, the green and blue fluorescent signals were observed under microscope.
Statistical Analysis
All statistical data were expressed as mean ± SD and analyzed by a one-way ANOVA (Analysis of Variance) with Duncan’s new multiple range test, P < 0.01.
Results
Identification and Phylogenetic Analysis of CfGcn5 in C. fructicola
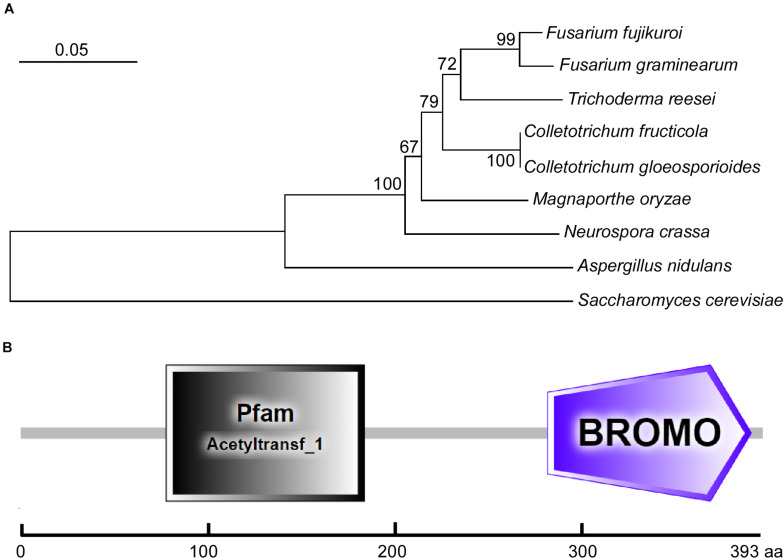
Using the S. cerevisiae ScGcn5 sequence as the trace, we identified its single homolog in the C. fructicola genome database by a BLAST_P search. Then, we submitted its sequence to the NCBI database (see text footnote 1) (GenBank accession number MW701426) and named it CfGcn5. We also collected the sequence of its homologs in other filamentous fungi from the NCBI database and constructed the phylogenetic tree. The phylogenetic dendrogram revealed that CfGcn5 shows sequence conservation among other fungi Gcn5 proteins; with CfGcn5 being most similar to that of C. gloeosporioides (94% identities) and most distant to that of S. cerevisiae (still 66% identities; Figure 1A). The domain prediction by the SMART website2 suggested that CfGcn5 contains a histone acetyltransferase acetyltransf_1 (HAT) domain and a protein binding Bromo domain (Figure 1B).
FIGURE 1.
Phylogenetic analysis and domain prediction of CfGcn5. (A) The Gcn5 proteins from diverse fungi were aligned using CLUSTAL_W, and the phylogenetic tree was constructed using MEGA 7.0 and the neighbor-joining method with 1000 bootstrap replicates. The sequences were collected from the NCBI database and the GenBank accession numbers are shown as follows: F. fujikuroi (XP_023425542.1), F. graminearum (XP_011324943.1), T. reesei (XP_006966860.1), C. fructicola (MW701426), C. gloeosporioides (EQB51886.1), M. oryzae (XP_003716207.1), N. crassa (XP_001728480.2), A. nidulans (XP_661225.1), and S. cerevisiae (NP_011768.1). (B) The domain prediction of CfGcn5. The gray rectangle (77∼182 amino acids) indicates the acetyltransf_1 (HAT) domain and the purple pentagon (279∼387 amino acids) refers to the Bromo domain.
Targeted Deletion of the CfGCN5 Gene in C. fructicola
To characterize the functions of CfGcn5, the coding region of CfGCN5 was replaced with the HPH gene according to the homologous recombination principle (Supplementary Figure 1A). Putative transformants were screened on hygromycin media and verified by PCR amplification. We thus acquired the CfGCN5 gene deletion mutant ΔCfgcn5 (Supplementary Figure 1B). Moreover, the mutant was also complemented with the wild-type CfGCN5 gene that restored all defects.
CfGcn5 Is Involved in Vegetative Growth and Conidiation
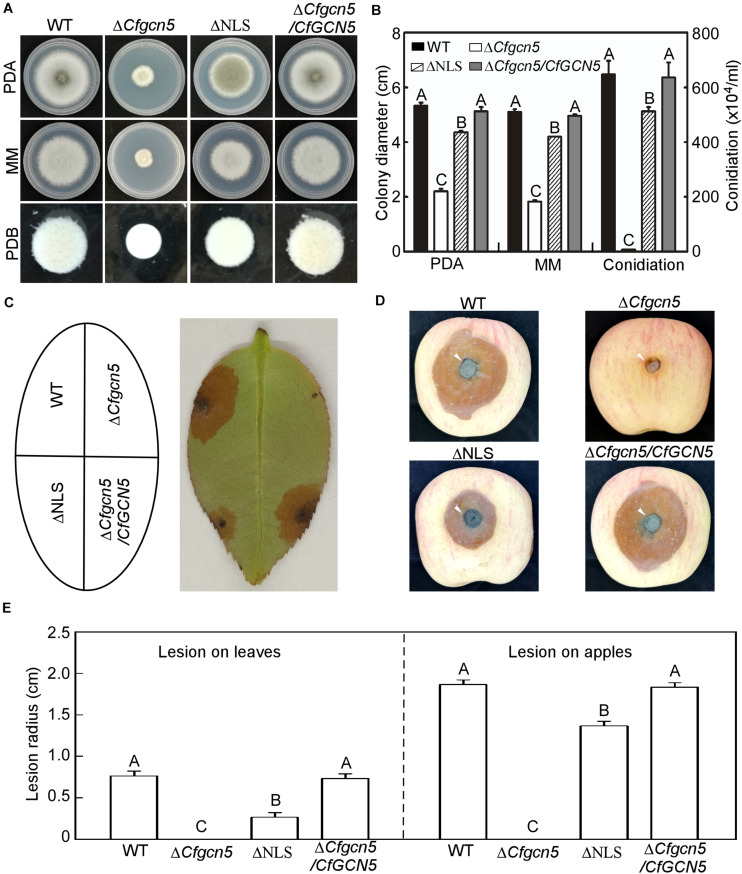
To test growth, wide-type (WT), ΔCfgcn5 mutant, and complemented strain ΔCfgcn5/CfGCN5 were cultured on PDA and MM plates. After 4 days, ΔCfgcn5 showed significant reduced growth rates compared with WT and ΔCfgcn5/CfGCN5 (Figures 2A,B). Similar results were observed following incubation in liquid PDB for 2 days (Figure 2C). Moreover, we also found that the ΔCfgcn5 mutant showed reduced aerial hyphal growth, which exhibited a flat colony compared with the fluffy colony of WT and ΔCfgcn5/CfGCN5 (Figure 2D). Meanwhile, we found that the ΔCfgcn5 mutant produced significantly fewer conidia than WT and ΔCfgcn5/CfGCN5 (Figure 2B).
FIGURE 2.
CfGcn5 is involved in vegetative growth and conidiation. (A) Growth of wide-type (WT), ΔCfgcn5 mutant, and complemented strain ΔCfgcn5/CfGCN5 on PDA and MM plates for 4 days. (B) Colony diameters and conidia were measured and statistically analyzed by Duncan analysis. Asterisks indicate significant differences (p < 0.01). (C) Mycelial growth following incubation in liquid PDB for 2 days. (D) Aerial hyphae growth is observed. Strains were cultured in PDA plates for 4 days and colony side views are displayed.
CfGcn5 Is Essential for Pathogenicity
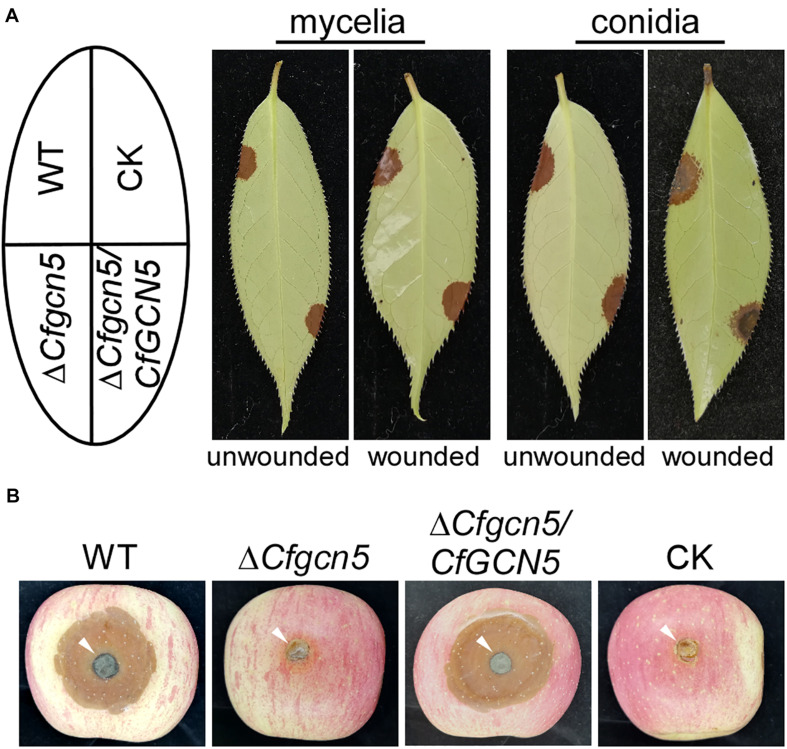
To characterize the roles of CfGcn5 in pathogenicity, mycelial plugs or conidial suspensions of WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were inoculated on unwounded tea-oil tree leaves. After 4 days, ΔCfgcn5 showed no lesions compared with the large and typical lesions caused by WT and ΔCfgcn5/CfGCN5 (Figure 3A). Moreover, we also conducted a pathogenicity assay on wounded tea-oil tree leaves. We found that ΔCfgcn5 still produced no lesions, in contrast to the typical lesions caused by WT and ΔCfgcn5/CfGCN5 (Figure 3A). Further, to test whether the non-pathogenicity of ΔCfgcn5 was specific to the tea-oil tree, mycelial plugs were inoculated on wounded apples, which resulted in ΔCfgcn5 producing no lesions and WT and ΔCfgcn5/CfGCN5 producing typical lesions (Figure 3B). The above observations indicate that CfGcn5 is essential for pathogenicity.
FIGURE 3.
CfGcn5 is essential for pathogenicity. (A) Unwounded and wounded tea-oil tree leaves were inoculated with mycelial plugs or conidial suspensions of WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5, respectively. Diseased symptoms were photographed at 4 days post inoculation (dpi). (B) Diseased symptoms of wounded apples inoculated with different mycelial plugs at 5 dpi. CK: compared control, the agar plug or ddH2O was inoculated onto it.
CfGcn5 Is Involved in Germination and Appressoria Formation
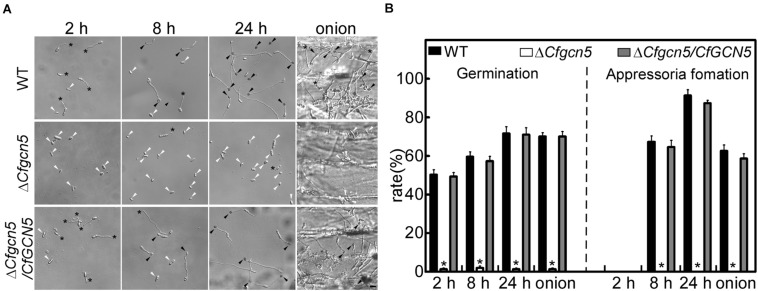
To explore the underlying mechanism of the pathogenicity defects of the ΔCfgcn5 mutant, we tested conidial germination and appressoria development. After inoculation on artificial hydrophobic surfaces for 2 h, approximately 50% of conidia were germinated in WT and ΔCfgcn5/CfGCN5, compared with 3% in the ΔCfgcn5 mutant (Figures 4A,B). At 8 h and 24 h, ΔCfgcn5 still showed less than 5% germination rates and its germ tubes did not form any appressoria, compared with more than 60% appressoria formation for WT and ΔCfgcn5/CfGCN5 (Figures 4A,B). A similar result was observed when inoculations were made on an onion epidermis, the natural appressorium-inducing hydrophobic surface. ΔCfgcn5 showed less than 5% germination rates and no appressoria formation at 24 h, compared with more than 60% of that for WT and ΔCfgcn5/CfGCN5 (Figures 4A,B). These results illustrate the importance of CfGcn5 in germination and appressoria formation.
FIGURE 4.
CfGcn5 is involved in germination and appressoria formation. (A) The conidia of WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were inoculated on artificial hydrophobic surfaces and onion epidermis, respectively. Germination and appressoria formation were observed after inoculation on hydrophobic surfaces at different time points (2, 8, and 24 h), and on onions epidermis (24 h). Asterisks indicate germ tubes, black arrows indicates appressoria, and white arrows indicates conidia. Bar = 10 μm. (B) Statistical analysis of germination and appressoria formation rates. Error bars represent standard deviations (SD) of three replicates and asterisks indicate significant differences (p < 0.01). The experiments were repeated three times with three replicates each time, and more than 100 conidia were counted per replicate.
CfGcn5 Contributes to the Osmotic Stresses Tolerance
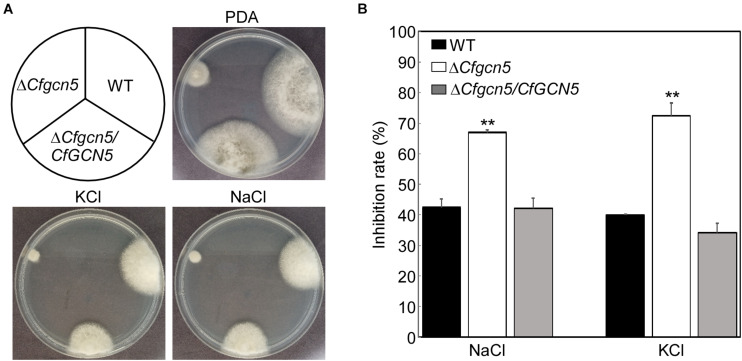
For normal growth and infection, pathogenic fungi must undergo many types of stress in nature. To address the role of CfGcn5 in environmental adaptation, WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were incubated on PDA and PDA added with osmotic stresses (1 M NaCl, and 1 M KCl) for 4 days. ΔCfgcn5 showed higher inhibition rates than WT and ΔCfgcn5/CfGCN5 both in NaCl and KCl (Figures 5A,B). This result indicates that CfGcn5 contributes to osmotic stresses tolerance.
FIGURE 5.
CfGcn5 contributes to the osmotic stresses tolerance. (A) The WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were incubated on PDA plates with various osmotic stresses of NaCl and KCl at 28°C for 4 days. (B) Statistical analysis of inhibition rates of the strains to osmotic stresses against untreated control. Error bars represent SD of three replicates and asterisks indicate significant differences (p < 0.01).
CfGcn5 Plays Roles in Cell Wall Integrity
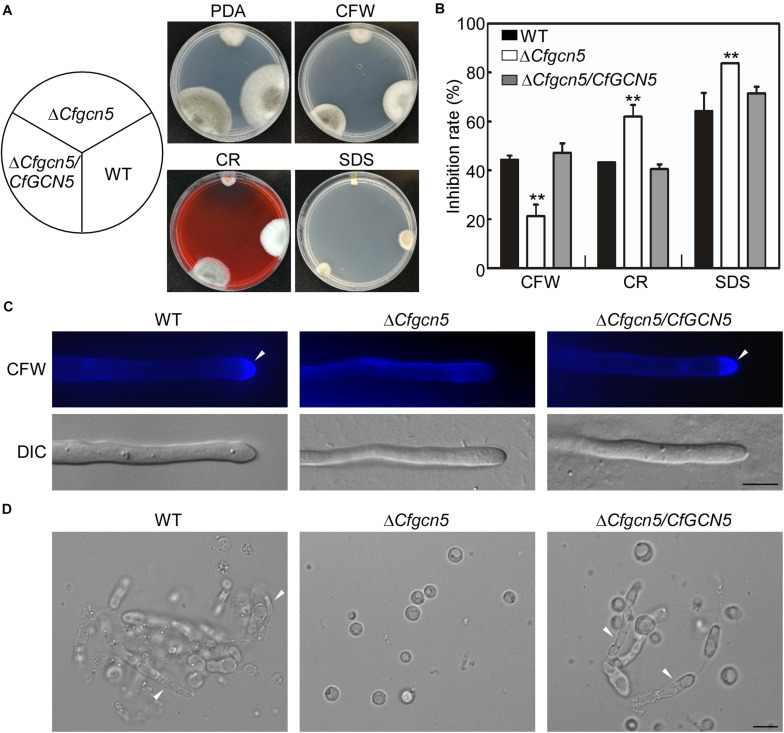
We further tested the roles of CfGcn5 in the responses to cell wall stresses, namely Calcofluor white (CFW), Congo red (CR), and sodium dodecylsulfate (SDS), the results showed that ΔCfgcn5 exhibited lower inhibition rates in CFW and higher inhibition rates in CR and SDS than WT and ΔCfgcn5/CfGCN5 (Figures 6A,B). As CFW and CR bind to nascent chitin chains and inhibit the assembly enzymes that connect chitin to β-1,3-glucan and β-1,6-glucan (Ram and Klis, 2006), we hypothesized that CfGcn5 contributes to chitin properties. To test this, we used the CFW as a chitinous fluorochrome to stain the hyphae. In WT and ΔCfgcn5/CfGCN5, CFW fluorescence was mostly distributed at the growing apices, where chitin, one of the main components of the fungal cell wall, was actively synthesized, while fluorescence was distributed evenly at the cell wall in ΔCfgcn5 (Figure 6C). We further treated the mycelia of the above strains with lytic enzymes. Mycelial fragments were still observed in WT and ΔCfgcn5/CfGCN5 after 2-h treatment, but all mycelia were degraded to protoplasts in ΔCfgcn5 (Figure 6D). These results revealed that CfGcn5 plays roles in maintaining cell wall integrity.
FIGURE 6.
CfGcn5 plays roles in cell wall integrity. (A) The WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were incubated on PDA plates with various cell wall stresses of CFW, CR, and SDS at 28°C for 4 days. (B) Statistical analysis of inhibition rates of the strains to cell wall stresses and asterisks indicate significant differences (p < 0.01). (C) The mycelia of the strains were stained with 10 μg/mL CFW for 5 min without light before being photographed, arrows indicates the stained hyphal tips. The experiment was repeated three times with triplicates, which showed the same results. DIC, differential interference contrast image. (D) Light microscopic observation of protoplast release after treatment with cell wall-degrading enzymes for 2 h at 30°C, arrows indicates the non-degraded hypha. Bar = 10 μm.
CfGcn5 Participates in the Responses to Oxidative and ER Stresses
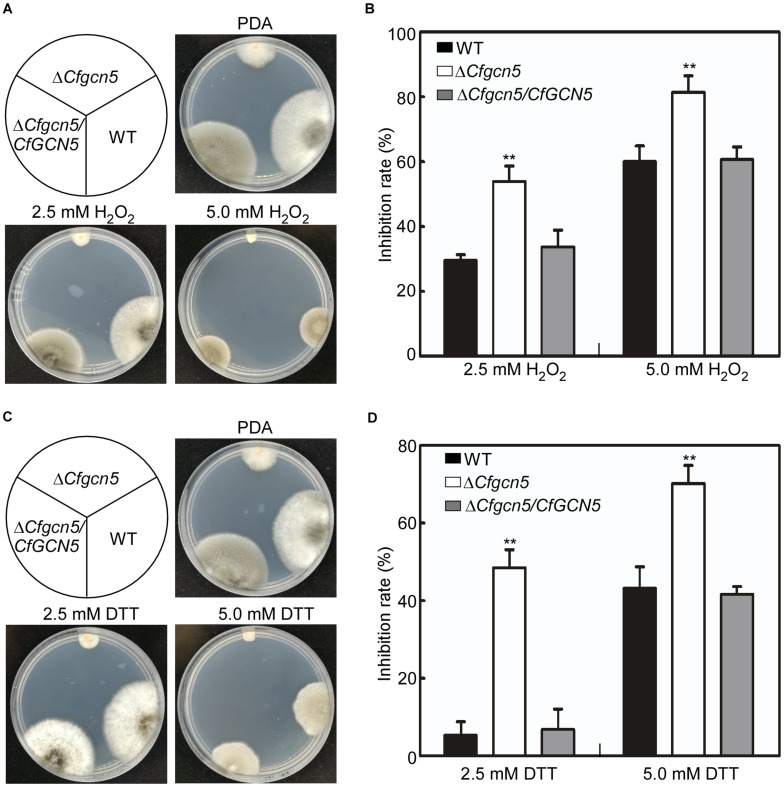
Apart from the environmental stresses, pathogenic fungi are also confronted with stresses from their host. The reactive oxygen species (ROS) is a common defense response for plants in pathogen–plant interactions, and pathogens must counteract host-derived ROS for successful infection (Hemetsberger et al., 2012; Liu et al., 2020). Thus, we used H2O2 to mimic host-derived ROS stress. When the strains were treated with 2.5 mM H2O2, ΔCfgcn5 showed an inhibition rate of 53.9%, in contrast to that of 29.5 and 33.4% in WT and ΔCfgcn5/CfGCN5, respectively (Figures 7A,B). As the concentration of H2O2 increased, the inhibition rates of all the strains were also increased, but that of ΔCfgcn5 was still significantly higher than that of WT and ΔCfgcn5/CfGCN5 (Figures 7A,B). The recent studies showed that pathogens also face host-derived ER stress during infection (Tang et al., 2020; Yin et al., 2020). We thus used DTT to mimic ER stress derived from plants. When treated with 2.5 mM DTT, ΔCfgcn5 showed an inhibition rate of 48.5%, in contrast to that of 5.3 and 6.8% in WT and ΔCfgcn5/CfGCN5, respectively (Figures 7C,D). A similar result was observed when the concentration of DTT was increased to 5.0 mM (Figures 7C,D). These observations revealed that CfGcn5 participates in the responses to ER stress.
FIGURE 7.
CfGcn5 participates in the responses to oxidative and ER stresses. (A) The WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were incubated on PDA plates with 2.5 and 5.0 mM H2O2 at 28°C for 4 days. (B) Statistical analysis of inhibition rates of the strains to H2O2 stress. (C) The WT, ΔCfgcn5, and ΔCfgcn5/CfGCN5 were incubated on PDA plates with 2.5 mM DTT and 5.0 mM DTT at 28°C for 4 days. (D) Statistical analysis of inhibition rates of the strains to DTT stress. Error bars represent SD of three replicates and asterisks indicate significant difference (P < 0.01).
CfGcn5 Is Localized to the Nucleus
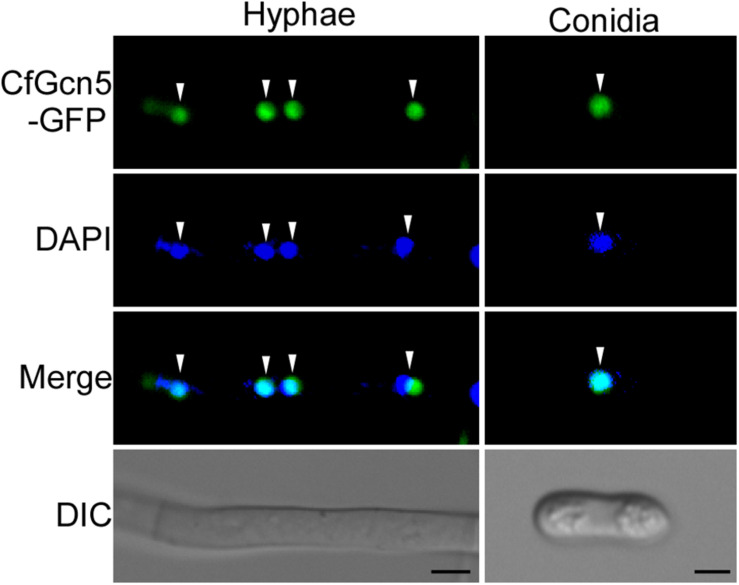
To further dissect the function of CfGcn5, a green fluorescent protein (GFP) tag was fused to the C-terminus of CfGcn5 and its cellular localization was observed. Spot green fluorescent signals were observed in vegetative hyphae and conidia. Further observation by 4, 6-diamidino-2-phenylindole (DAPI, a nucleus dye) staining showed that the spot green fluorescent signals co-localized well with the blue fluorescent signals (Figure 8), indicating that CfGcn5 is localized to the nucleus.
FIGURE 8.
CfGcn5 is localized to the nucleus. The nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI). CfGcn5-GFP co-localized with DAPI in hyphae and conidia. Arrowheads show the representative co-localized areas. Bar = 5 μm.
The NLS Contributes to the Nuclear Localization of CfGcn5
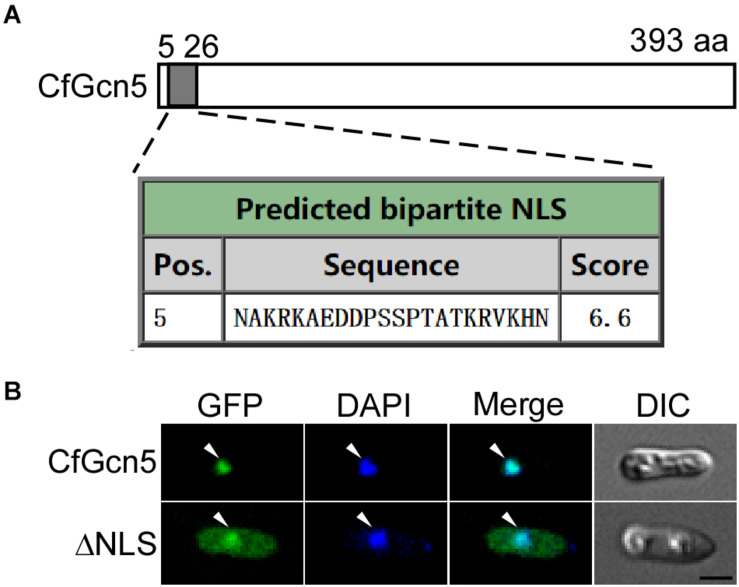
We next predicted that CfGcn5 contains an NLS region (score > 6.0) of 5∼26 amino acids by the cNLS Mapper website (Figure 9A). We hypothesized that the localization of CfGcn5 might be mediated by the NLS region. To demonstrate this, we deleted the NLS region, and found that the truncated CfGcn5 (ΔNLS) was distributed in the cytoplasm though some still localized to the nucleus (Figure 9B). This result suggests that the NLS region mediates the nuclear localization of CfGcn5.
FIGURE 9.
The NLS contributes to the nuclear localization of CfGcn5. (A) The NLS prediction of CfGcn5 with the cNLS Mapper website. (B) The deleted NLS region of CfGcn5 (ΔNLS) showed cytoplasmic and nuclear localization. Arrowheads show the representative co-localized areas. Bar = 5 μm.
The NLS Region Is Important for the Function of CfGcn5
In addition to examining the role of the NLS region in localization, we examined its contribution to function. In a growth and conidiation test, ΔNLS showed moderate growth and conidiation rates between that of ΔCfgcn5 and WT (Figures 10A,B). In wounded tea-oil tree leaves, ΔNLS showed some lesions but fewer than WT, while ΔCfgcn5 showed no lesions (Figures 10C,E). Similar results were observed in wounded apples where ΔNLS showed some lesions but still fewer than WT (Figures 10D,E). These results suggest that the NLS region is important for the function of CfGcn5.
FIGURE 10.
The NLS region is important for the function of CfGcn5. (A) Growth of WT, ΔCfgcn5, ΔNLS, and ΔCfgcn5/CfGCN5 on PDA and MM plates for 4 days or in liquid PDB for 2 days. (B) Colony diameters and conidiation were measured and statistically analyzed by Duncan analysis. Different letters indicate significant difference (P < 0.01). (C) Diseased symptoms of wounded tea-oil tree leaves inoculated with related mycelial plugs at 4 dpi. (D) Diseased symptoms of wounded apples inoculated with related mycelial plugs at 5 dpi. Arrows indicate the infection area. (E) Statistical analysis of lesion radius on tea-oil tree leaves and apples. Error bars represent SD of three replicates and different letters indicate significant difference (P < 0.01).
The HAT Domain Is Required for the Localization and Function of CfGcn5 and Bromo Domain Contributes to Its Function
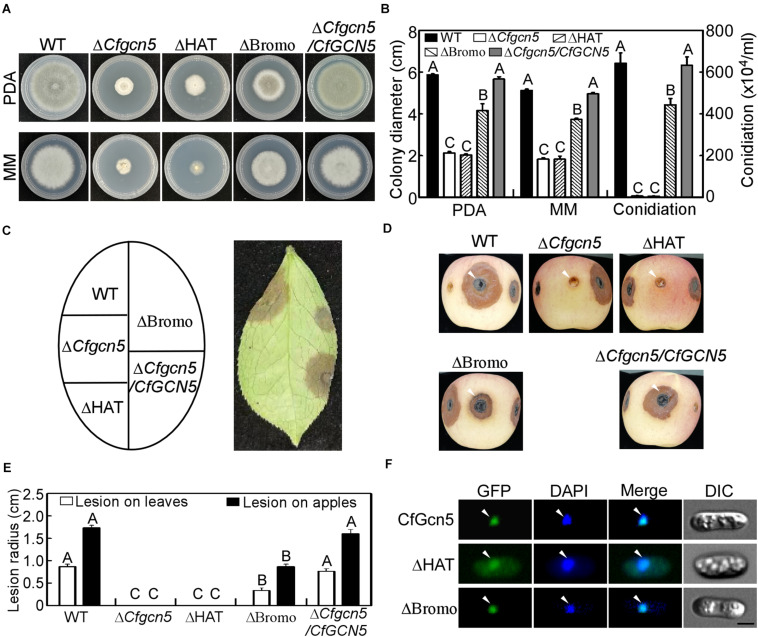
Except the NLS region, CfGcn5 contains the HAT and Bromo domains. To test their contribution to function, we deleted the HAT and Bromo domains and introduced them into the ΔCfgcn5 mutant. We found that ΔHAT failed to restore the ΔCfgcn5 mutant in growth and conidiation, while ΔBromo partially restored these phenotypes (Figures 11A,B). In wounded tea-oil tree leaves, ΔHAT produced no lesions like ΔCfgcn5, while ΔBromo produced some lesions but fewer than WT (Figures 11C,E). A similar pathogenic result was also observed in wounded apples (Figures 11D,E). In the localization assay, ΔHAT was distributed in the cytoplasm though some still localized to the nucleus, while ΔBromo still localized to the nucleus (Figure 10F). These results suggested that both HAT and Bromo domains contribute to the function of CfGcn5, but only the HAT domain is required for subcellular localization.
FIGURE 11.
Functional and localized analysis of HAT and Bromo domains. (A) Growth of WT, ΔCfgcn5, ΔHAT, ΔBromo, and ΔCfgcn5/CfGCN5 on PDA and MM plates for 4 days. (B) Colony diameters and conidiation were measured and statistically analyzed by Duncan analysis for different strains. Different letters indicate significant difference (P < 0.01). (C) Diseased symptoms of wounded tea-oil tree leaves inoculated with related mycelial plugs at 4 dpi. (D) Diseased symptoms of wounded apples inoculated with related mycelial plugs at 5 dpi. Arrows indicate the infection area. (E) Statistical analysis of lesion radius on tea-oil tree leaves and apples. Error bars represent SD of three replicates and different letters indicate significant difference (P < 0.01). (F) The localization of deleted HAT domain (ΔHAT) and Bromo domain (ΔBromo). Arrows show the representative co-localized areas. Bar = 5 μm.
Discussion
In the present study, we identified a yeast histone acetyltransferase ScGcn5 homolog CfGcn5 and characterized its functions. We found that the nucleus-localized CfGcn5 is important in growth, conidiation, stress responses, and pathogenicity in C. fructicola.
In U. maydis and A. nidulans, targeted GCN5 gene deletion mutants showed a slight reduction in growth rate (Canovas et al., 2014; Gonzalez-Prieto et al., 2014), while that of mutants exhibited severe growth defects in F. graminearum and M. oryzae (Zhang et al., 2017; Kong et al., 2018). Similar to the growth phenotypes of these fungi, we found that the ΔCfgcn5 mutant showed a significantly decreased colony size and aerial hyphal growth. These observations indicate that Gcn5 proteins share conserved roles in growth, to a distinct extent.
There are some contradictions about the roles of such proteins in conidiation in other systems. In M. oryzae, MoGcn5 repressed conidia and the MoGCN5 deletion mutant showed largely increased conidiation both in the light and dark conditions (Zhang et al., 2017), while such proteins were essential for conidiation in T. reesei and F. graminearum, which resulted in the mutants producing no conidia (Xin et al., 2013; Kong et al., 2018). The strongly decreased conidiation of the ΔCfgcn5 mutant in our study indicated that CfGcn5 is important but not essential for conidiation, which is also supported by the finding in A. nidulans (Reyes-Dominguez et al., 2008). These diversities in conidiation by the GCN5 mutants reflect different mechanisms of Gcn5 proteins during asexual development in distinct fungi.
A number of external stresses that mimic the diverse micro-environments on the tea-oil tree were adapted in this study. The ΔCfgcn5 mutant does not only affect the resistance to osmotic stresses and cell wall integrity stresses, but is also sensitive to mimic host-derived ROS and ER stresses. The important role of CfGcn5 in the responses to environmental stress might foretell its function in virulence. Indeed, the ΔCfgcn5 mutant showed non-pathogenicity on healthy tea-oil leaves, which is mainly caused by the decreased conidial germination rate and no appressoria formation. Furthermore, the lack of lesions of the mutant on wounded tea-oil trees and apples indicate that there are other underlying mechanisms besides the appressorium formation, and further studies are warranted to confirm this.
Previous studies revealed that Gcn5 proteins target histones in the nucleus to epigenetically regulate global gene transcription (Chang et al., 2015; Gaupel et al., 2015; Zhang et al., 2017). We also found that CfGcn5 is localized to the nucleus both in hyphae and conidia, suggesting that nucleus localization is critical for CfGcn5 function. Both cytoplasm and nucleus localization of the deleted N-terminal NLS region indicate its contributions to localization, supported by the studies in Candida albicans (Chang et al., 2015). Again, the HAT domain-deleted CfGcn5 also localized in both the cytoplasm and nucleus, which shows some differences to cytoplasm but not nucleus localization of such truncated proteins in Candida albicans. One reason for this is that the importance of HAT in localization is distinct in different cells, and a similar result was observed in human cells when HAT-deleted Gcn5 proteins were distributed between the nucleus and cytoplasm at varying levels in different cell types (Blanco-Garcia et al., 2009). Taken together, we concluded that the HAT domain also contributes to the localization of CfGcn5, but the Bromo domain plays no role in its localization.
Combining the localization and phenotype together, we revealed that nucleus localization is essential but not sufficient for the full function of CfGcn5. The deleted NLS and HAT affected its localization, and these truncated proteins did not fully rescue the defects of the ΔCfgcn5 mutant when reintroduced. The deleted Bromo did not affect nucleus localization and also could not fully restore the defects of the ΔCfgcn5 mutant when reintroduced. These findings demonstrated that the NLS region and HAT and Bromo domains are all required for the normal function of CfGcn5. Among them, HAT may be the most important domain, as the HAT-deleted mutant showed similar defects to the ΔCfgcn5 mutant. This might be because HAT is essential for histone acetyltransferase activity and auto-acetylation (Candau et al., 1997; Santos-Rosa et al., 2003). It might be very interesting to investigate how acetylation occurs and how this post-translation modification regulates the pathogenicity in C. fructicola and other phytopathogens, such further studies are highly warranted.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s. The data presented in the study are deposited in the NCBI (accession number MW701426).
Author Contributions
HL and SZ conceived and designed the study. YG, SC, and SZ performed the experiments. SZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was funded by the National Natural Science Foundation of China (32071765) and the Natural Science Foundation of Hunan Province (2020JJ5979).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.680415/full#supplementary-material
Generation of the CfGCN5 gene deletion mutant in C. fructicola. (A) Schematic illustration for deletion strategy of CfGCN5 gene. (B) Validation of the gene deletion mutant by PCR amplified with primers 1 (NBF/NBR) and primers 2 (BWF/HPHR). M: marker; +: positive control; -: negative control; #6: mutant.
Primers used in this study.
References
- Abate G., Bastonini E., Braun K. A., Verdone L., Young E. T., Caserta M. (2012). Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochim. Biophys. Acta 1819 419–427. 10.1016/j.bbagrm.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroncelli R., Amby D. B., Zapparata A., Sarrocco S., Vannacci G., Le Floch G., et al. (2016). Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics 17:555. 10.1186/s12864-016-2917-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Garcia N., Asensio-Juan E., de la Cruz X., Martinez-Balbas M. A. (2009). Autoacetylation regulates P/CAF nuclear localization. J Biol. Chem. 284 1343–1352. 10.1074/jbc.m806075200 [DOI] [PubMed] [Google Scholar]
- Bruno K. S., Tenjo F., Li L., Hamer J. E., Xu J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryotic Cell. 3 1525–1532. 10.1128/ec.3.6.1525-1532.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R., Zhou J. X., Allis C. D., Berger S. L. (1997). Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16 555–565. 10.1093/emboj/16.3.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas D., Marcos A. T., Gacek A., Ramos M. S., Gutierrez G., Reyes-Dominguez Y., et al. (2014). The histone acetyltransferase GcnE (GCN5) plays a central role in the regulation of Aspergillus asexual development. Genetics 197 1175–1189. 10.1534/genetics.114.165688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Fan X., Chen J. (2015). Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet. Biol. 81 132–141. 10.1016/j.fgb.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang B., Chen J., Wang X., Wang R., Peng S., et al. (2015). Identification of rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front. Plant Sci. 6:189. 10.3389/fpls.2015.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J. A., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di T. M., Yang S. L., Du F. Y., Zhao L., Li X. H., Xia T., et al. (2018). Oleiferasaponin A(2), a novel saponin from Camellia oleifera Abel. seeds, inhibits lipid accumulation of HepG2 cells through regulating fatty acid metabolism. Molecules 23:3296. 10.3390/molecules23123296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feas X., Estevinho L. M., Salinero C., Vela P., Sainz M. J., Vazquez-Tato M. P., et al. (2013). Triacylglyceride, antioxidant and antimicrobial features of virgin Camellia oleifera, C. reticulata and C. sasanqua oils. Molecules 18 4573–4587. 10.3390/molecules18044573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupel A. C., Begley T. J., Tenniswood M. (2015). Gcn5 modulates the cellular response to oxidative stress and histone deacetylase inhibition. J. Cell. Biochem. 116 1982–1992. 10.1002/jcb.25153 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Prieto J. M., Rosas-Quijano R., Dominguez A., Ruiz-Herrera J. (2014). The UmGcn5 gene encoding histone acetyltransferase from Ustilago maydis is involved in dimorphism and virulence. Fungal Genet. Biol. 71 86–95. 10.1016/j.fgb.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. (1992). The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603. 10.1093/nar/20.10.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemetsberger C., Herrberger C., Zechmann B., Hillmer M., Doehlemann G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathogens 8:e1002684. 10.1371/journal.ppat.1002684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. R., Song N., Zheng M., Liu X. Y., Liu Z. S., Xing J. W., et al. (2015). Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 84 1178–1191. 10.1111/tpj.13076 [DOI] [PubMed] [Google Scholar]
- Jiang S. Q., Li H. (2018). First report of leaf anthracnose caused by Colletotrichum karstii on tea-oil trees (Camellia oleifera) in China. Plant Dis. 102 674–675. 10.1094/pdis-08-17-1195-pdn [DOI] [Google Scholar]
- Kong X., van Diepeningen A. D., van der Lee T. A. J., Waalwijk C., Xu J., Zhang H., et al. (2018). The Fusarium graminearum histone acetyltransferases are important for morphogenesis, DON biosynthesis, and pathogenicity. Front. Microbiol. 9:654. 10.3389/fmicb.2018.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhou G. Y., Liu J. A., Xu J. (2016). Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PloS One 11:e0156841. 10.1371/journal.pone.0156841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Shang S., Dong Q., Wang B., Zhang R., Gleason M. L., et al. (2018). Transcriptomic analysis reveals candidate genes regulating development and host interactions of Colletotrichum fructicola. BMC Genomics 19:557. 10.1186/s12864-018-4934-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhou Q., Guo Z., Liu P., Shen L., Chai N., et al. (2020). A self-balancing circuit centered on MoOsm1 kinase governs adaptive responses to host-derived ROS in Magnaporthe oryzae. eLife 9:e61605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., Shiekhattar R., et al. (2001). Snf1–a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293 1142–1146. 10.1126/science.1062322 [DOI] [PubMed] [Google Scholar]
- Prihastuti H., Cai L., Chen H., McKenzie E. H. C., Hyde K. D. (2009). Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers 39 89–109. [Google Scholar]
- Ram A. F., Klis F. M. (2006). Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protocols 1 2253–2256. 10.1038/nprot.2006.397 [DOI] [PubMed] [Google Scholar]
- Reyes-Dominguez Y., Narendja F., Berger H., Gallmetzer A., Fernandez-Martin R., Garcia I., et al. (2008). Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryotic Cell. 7 656–663. 10.1128/ec.00184-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenbach M. F., Velho A. C., Goncalves A. E., Mondino P. E., Alaniz S. M., Stadnik M. J. (2016). Genetic structure of Colletotrichum fructicola associated to apple bitter rot and glomerella leaf spot in southern Brazil and Uruguay. Phytopathology 106 774–781. 10.1094/phyto-09-15-0222-r [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Valls E., Kouzarides T., Martinez-Balbas M. (2003). Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 31 4285–4292. 10.1093/nar/gkg655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M. (2001). Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8 473–479. 10.1016/s1097-2765(01)00301-x [DOI] [PubMed] [Google Scholar]
- Tang W., Jiang H., Aron O., Wang M., Wang X., Chen J., et al. (2020). Endoplasmic reticulum-associated degradation mediated by MoHrd1 and MoDer1 is pivotal for appressorium development and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 22 4953–4973. 10.1111/1462-2920.15069 [DOI] [PubMed] [Google Scholar]
- Weir B. S., Johnston P. R., Damm U. (2012). The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73 115–180. 10.3114/sim0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Q., Gong Y., Lv X., Chen G., Liu W. (2013). Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr. Microbiol. 67 580–589. 10.1007/s00284-013-0396-4 [DOI] [PubMed] [Google Scholar]
- Yin Z., Feng W., Chen C., Xu J., Li Y., Yang L., et al. (2020). Shedding light on autophagy coordinating with cell wall integrity signaling to govern pathogenicity of Magnaporthe oryzae. Autophagy 16 900–916. 10.1080/15548627.2019.1644075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Guo Y., Li S., Zhou G., Liu J., Xu J., et al. (2019). Functional analysis of CfSnf1 in the development and pathogenicity of anthracnose fungus Colletotrichum fructicola on tea-oil tree. BMC Genetics 20:94. 10.1186/s12863-019-0796-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liang M., Naqvi N. I., Lin C., Qian W., Zhang L. H., et al. (2017). Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae. Autophagy 13 1318–1330. 10.1080/15548627.2017.1327103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Bone J. R., Edmondson D. G., Turner B. M., Roth S. Y. (1998). Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17 3155–3167. 10.1093/emboj/17.11.3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of the CfGCN5 gene deletion mutant in C. fructicola. (A) Schematic illustration for deletion strategy of CfGCN5 gene. (B) Validation of the gene deletion mutant by PCR amplified with primers 1 (NBF/NBR) and primers 2 (BWF/HPHR). M: marker; +: positive control; -: negative control; #6: mutant.
Primers used in this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s. The data presented in the study are deposited in the NCBI (accession number MW701426).