Abstract
Ubiquitously distributed microorganisms are natural decomposers of environmental pollutants. However, because of continuous generation of novel recalcitrant pollutants due to human activities, it is difficult, if not impossible, for microbes to acquire novel degradation mechanisms through natural evolution. Synthetic biology provides tools to engineer, transform or even re-synthesize an organism purposefully, accelerating transition from unable to able, inefficient to efficient degradation of given pollutants, and therefore, providing new solutions for environmental bioremediation. In this review, we described the pipeline to build chassis cells for the treatment of aromatic pollutants, and presented a proposal to design microbes with emphasis on the strategies applied to modify the target organism at different level. Finally, we discussed challenges and opportunities for future research in this field.
Keywords: Aromatic compounds, Synthetic biology, Bioremediation, Environmental pollution, Microorganisms
1. Introduction
With rapid growth of the global population and material consumption, discharge of various pollutants continues to increase, and environmental pollution has become one of the most severe issues affecting human health [1]. Pollutants refer to substances that can cause environmental pollution and have an adverse effect on the environment if discharged into the atmosphere, water or soil during human daily life [2]. Environmental pollution have direct damage to the ecological systems, such as water deterioration, forest destruction, and desertification, or indirect damage to human [3]. Among the major environmental pollutants, aromatic compounds are of great concern because they will be persistent in the environment due to the high thermodynamic stability of the benzene group. How to degrade these pollutants is currently a key challenge in environmental pollution control. Thanks to diverse types of microbial metabolism, most pollutants can be degraded or transformed by certain microorganisms [4]. The microbial degradation of aromatic pollutants has been developed for 40 years and has always been a hot topic in environmental protection (see Fig. 1). Naturally genes involved in aromatic pollutants degradation generally exist in clusters and are often located on the plasmids with low copy numbers and large sizes [5]. The gene clusters comprise catabolic genes encoding enzymes, transport genes encoding proteins for uptake of the aromatic compounds, and regulatory genes responsible for regulating the expression of both catabolic and transport genes. Gene clusters from Comamonas sp. strain E6 can degrade o-phthalate, terephthalate, and isophthalate via the protocatechuate 4,5-cleavage pathway [6,7]. Microbes generally only contain the catabolic genes for a single compound, and the degradation of multiple compound pollutants by a single strain is currently not well resolved [8]. Microbial remediation has been drawing increasing attention in the recent years, especially in the rising era of synthetic biology. Synthetic biology provides a strategy to construct engineered microorganisms that can monitor, aggregate and degrade environmental pollutants, with the aim to eliminate water pollution, remove garbage, and reduce air pollution [9]. So far, there are only limited reports on the construction of microorganism to degrade aromatic compounds using synthetic biology [10]. However, many efforts have been made to degrade 1,2,3-trichloropropane via synthetic biology [11], and synthetic biosensors for rapid detection of water contaminants [12], these successful cases bring us the hope to the degradation of aromatic compounds. Here, we review current progresses in aromatic compounds degradation using microbes and present a proposal for the rational design and construction of microbial strains to degrade aromatic compounds (see Fig. 2). Such a proposal usually follows the Chassis selection-Pathway design-Metabolism optimization-Tolerance engineering cycle, where iteration of each cycle leads to the improvement of the microbes.
Fig. 1.
Development of microbial degradation of aromatic pollutants.
In the 1980s, it was an exciting era of microbe discovery; In the 1990s, naturally occurring microbes already have considerable ability to remove many environmental pollutants; In the 2000s, sanger sequencing leads to the discovery of microbial degradation gene clusters; In the future, the emerging of synthetic biology technologies brings a new artificial microorganism for pollutants degradation.
Fig. 2.
Schematic overview of synthetic biology strategies applying to microbial degradation of aromatic pollutants (Naphthalene, Toluene, and Phenanthrene). The workflow includes chassis selection, pathway design, metabolism optimization, and tolerance engineering. (A) Not just model microbes but also some nonconventional microbes can serve as a chassis cell for the degradation of pollutants, such as Naphthalene, Toluene, and Phenanthrene. When selecting a host, consideration should be given to the characteristics of the pollutant, the chassis's genetic manipulation tools, genetic databases, and growth characteristics. (B) Biodegradation pathways containing gene clusters can be integrated into the chromosome or plasmid, and pathway design rely on genome data (gene clusters), mining tools (KEGG and MRE), and engineering tools (DNA assembly, CRISPR/Cas editing and Enzyme engineering). (C) Recently developed synthetic biology tools will accelerate the optimization of catabolism pathways for pollutants (AI-based design parts). (D) Most of the efforts in tolerance engineering have relied on improving the native gene function (nah,tmo, xyl and phn) and capabilities of a chassis cell.
2. Synthetic biology conception of microbial remediation
In the field of synthetic genomics, the de novo design and synthesis capabilities of DNA sequences have evolved from a single gene to the entire microbial genome. Viruses, prokaryotic genomes and eukaryotic chromosomes have been successfully synthesized in the past research studies [[13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]]. At the same time, the existing enabling technology allows introduction of complex exogenous metabolic pathways into specific microbial hosts with particular modifications to achieve specific goals. In recent years, with the rapid development of synthetic biology, microbial remediation has been drawing increased attention and shown the great potential in degrading pollutants compared to traditional physical and chemical methods. In the last decade, by the means of “top-down” and “bottom-up” engineering approaches, synthetic biology has been proven to be a very powerful tool in utilization and modification of existing genetic materials to redesign organisms with desired abilities. Taking “Design-Build-Test-Learn (DBTL)” biological engineering cycle, the standardized and universal biological component modules could be designed, constructed, integrated, tested and optimized in simple chassis cells to achieve efficient operation of a quantitative and controllable platform-based new living system [29]. The reconstruction of synthetic pathways in dedicated chassis cells for natural product production (e.g. artemisinin, avermectin, resveratrol, or penicillin) has been successfully conducted in this strategy [30]. These results have not only achieved great academic and commercial success, so that more and more researchers are focusing on the synthesis of medicines and raw materials (see Table 1). However, synthetic biology is not yet widely applied in engineering the catabolism pathways in microbes so far. Therefore, the field of synthetic biology is increasingly expanding from a focus on natural products to pollutants degradation by engineering microbes, and artificially synthesized catabolic pathways provide a new approach for environmental bioremediation [33].
Table 1.
Progress of synthetic biomanufacturing of aromatic compounds.
| Compound | Production host | Titer (g/L) | Refs |
|---|---|---|---|
| Salicylate | E.coli | 11.5 | [31] |
| 4-hydroxybenzoate | E.coli | 1.82 | [31] |
| 3-hydroxybenzoate | E.coli | 2.18 | [31] |
| 4-aminobenzoate | E.coli | 2.88 | [31] |
| 2-aminobenzoate | E.coli | 1.83 | [31] |
| l-tyrosine | E.coli | 1.62 | [31] |
| Phenol | E.coli | 1.1 | [31] |
| Muconic acid | E.coli | 3.1 | [31] |
| Cinnamaldehyde | S. cerevisiae | 0.0003 | [32] |
| Cinnamyl alcohol | S. cerevisiae | 0.0278 | [32] |
| Hydrocinnamyl alcohol | S. cerevisiae | 0.1131 | [32] |
2.1. Designing and constructing microbes
Aromatic pollutants may be selected as a degradation target for their various toxic effects on humans. For a given aromatic pollutant, the first step towards heterologous degradation is selection of an appropriate host species in which to engineer the pathway. Within a species, use of previously developed strains that efficiently degrade pollutants can greatly accelerate progress. And lastly, within a given strain, preliminary engineering of the host prior to incorporation of heterologous enzymes can facilitate implementation of the non-native pathway in a new context.
2.1.1. Choosing a suitable chassis
The construction of microbial chassis and their application as artificial cell factories are important factors in microbial degradation [34], that is to identify a suitable host organism. Many microbes have been used as chassis for natural product production, whereas most studies focused on model organisms such as Escherichia coli and Saccharomyces cerevisiae [35]. There is still an urgent need for identification of new chassis. In nature, microbial strains capable of degrading persistent organic pollutants include Pseudomonas [36], Bacillus [37], Sphingomonas [38], Rhodococcus [39], Mycobacterium [40], and Dehalococcoides [41]. Table 2 provides a partial list of microbial strains containing certain gene clusters degrading aromatic pollutants. Using a host which naturally is capable of degrading the target pollutant, even not so efficiently, could greatly accelerate the engineering progress. Fortunately, Pseudomonas putida KT2440 belongs to this type of starting chassis strains. Pseudomonas putida is a gram-negative bacterium that can be encountered in diverse ecological habitats. This ubiquity is traced to its remarkably versatile metabolism, adapted to withstand physicochemical stress, and the capacity to thrive in harsh environments in which pollutants often exist. The genome-editing methods have been well established in Pseudomonas putida KT2440, which has been considered as a robust metabolic chassis for catabolic pathway assembly. To date, many biodegradation pathways have been integrated into the chromosome of Pseudomonas putida KT2440 [[53], [54], [55]]. In addition, V. natriegens recently emerges as a promising chassis for aromatic pollutants degradation because of its fastest-growing and non-pathogenic nature [56,57]. A number of tools for genetic manipulation have been established in V. natriegens. These methods comprise DNA transformation methods, such as electroporation, heat-shock transformation and conjugation, as well as genome engineering techniques, such as recombineering, multiplexed genome editing via natural transformation and CRISPR-Cas9. Clearly, V. natriegens could reduce the cycle time drastically, thereby accelerating the optimization of catabolic pathways and protein expression (enzymes).
Table 2.
Function gene clusters in the aromatic pollutants degradation of microbes.
| Substrates | Degrading genes | strains | Refs |
|---|---|---|---|
| Benzene | bnz | Rhodococcus opacus | [42] |
| Toluene | tmo, xyl | Pseudomonas stutzeri | [43] |
| Xylene | xylCMABN | Halioxenophilus aromaticivorans | [44] |
| Phenylpropanoid | phd | Corynebacterium glutamicum | [45] |
| Phenol | phe | Pseudomonas pseudoalcaligenes C70 | [46] |
| Chlorophenol | cph | Arthrobacter chlorophenolicus | [47] |
| Nitrophenol | hnp, mnp, pnp | Cupriavidus sp. strain CNP-8, Burkholderia sp. Strain SJ98 | [[48], [49], [50]] |
| Naphthalene | nah | Pseudomonas putida | [51] |
| Phenanthrene | phn | Burkholderia sp. strain RP007 | [52] |
2.1.2. Engineering a strain with known heterologous pathways
Following selection of a suitable host, engineering a strain with the degradation pathways will be designed and executed. When designing the suitable degradation pathway for a specific pollutant, the process of engineering microbial host would be enhanced if we could identify all involved enzymes in the natural degradation pathway. The natural microbial degradation pathways of aromatic compounds mainly involve anaerobic reduction dehalogenation, aerobic dehalogenation, ring-opening mineralization and other co-metabolic steps. Certain anaerobic microbes can obtain energy to support their growth and metabolism through the reductive dehalogenation process using dehalogenase in dehalogenation respiratory reaction, which is an exothermic reaction [58]. Generally, the most common hexachlorobenzene degradation pathway is anaerobic reduction due to halogenated aromatic compounds resistance to aerobic degradation. DDT is converted into DDD (Dichlorodiphenyldichloroethane) through removing one chlorine atom by the microbes, which requires transition metal and metal complexes as reducing agents [59]. DDD undergoes reductive dechlorination to generate dichlorodiphenylmethane, and then the benzene ring is cracked under aerobic conditions. The biodegradation of hexachlorocyclohexane also occurs under anaerobic conditions [60]. The anaerobic dehalogenation reaction of aromatic compounds catalyzed by microbes is a relatively slow process, which takes up to 7–12 weeks. After reductive dehalogenation by microbes, the compounds with increased hydrophilicity can become electron donors and enter the microbial aerobic degradation process. Followed further dehalogenation and ring-opening decomposition into small molecules, they subsequently enter the tricarboxylic acid cycle and are oxidized into water and carbon dioxide [61,62]. In contrast, the microbial aerobic degradation reaction of aromatic compounds is a relatively short-period process and does not produce toxic byproducts, which is the major way of aromatic compounds mineralization. The microbial aerobic degradation pathway of organochloride pesticides (OCPs) is to hydroxylate OCPs to form the intermediate product chlorinated catechol, which ring-opening and dechlorinating by lactonization [63,64]. Naphthalene is catalyzed by dioxygenase and dehydrogenase to produce 1, 2-dihydroxynaphthalene, and further generate salicylic acid, which is then converted to catechol under the action of salicylic acid hydroxylase, or converted into gentisic acid under monooxygenase, and finally undergoes ring-opening degradation [65]. To access degradation potential, an innovative discovery pipeline was developed to systematically annotate the degradation abilities of microbes using comparative metabolomics and heterologous gene expression. With this platform, microbial genomic DNA fragments containing intact biodegradation gene clusters are inserted into yeast artificial chromosomes (YACs) and are used to transform a yeast host to discover new degradation products. For example, we can design a naphthalene metabolism pathway from catabolic gene clusters through synthetic biology methods, and reconstructed, cloned, and heterologously expressed a naphthalene gene clusters using YACs. Then we employ the YAC-metabolite scoring strategy to identify the degradation product of this gene clusters and probe its biodegradation pathway. The core metabolic networks of model organisms (yeast) are well-characterized and can be used to guide overexpression and knockout modifications for metabolic burden and to address common challenges (e.g., feedback inhibition or other metabolic regulation). The opd gene and the p-nitrophenol degradative operons were introduced into P. putida KT2442 to construct a parathion-degrading pathway [66]. In the practical application example of synthetic biology for the degradation of 1,2,3-trichloropropane, a complete, artificial five-step catabolic pathway has been engineered into Escherichia coli, which assembled haloalkane dehalogenase from Rhodococcus rhodochrous, haloalcohol dehalogenase and epoxide hydrolase from Agrobacterium radiobacter [67]. Then computational models was used to identify bottlenecks in the catabolic pathway and employed forward engineering to improve 1,2,3-trichloropropane degradation [68].
2.1.3. Identifying enzymes and redesigning novel metabolic pathways
A novel candidate degradation pathway is first outlined through selection of stepwise chemical intermediates leading from host metabolism to the target compound (e.g., H2O, CO2), followed by selection of enzymes to carry out each specified reaction. For certain pollutants, detailed knowledge of the native catabolic pathway is available and can be used to outline all intermediates and enzymes in a pathway, facilitating pathway engineering into a heterologous host. In such cases, candidate pathway design, enzyme selection, and pathway testing all offer distinct challenges which can be solved by computational tools. At the design stage, mining the gene clusters involved in aromatic compounds degradation is the first step in the construction of engineering microbes. Many degradation gene clusters have been identified in known metabolic pathways (see Table 2), such as nah, xyl, tmo, phn, bnz, phe etc. In addition, some computational tools, KEGG and MRE, can be used for genome mining and pathway prediction involved in aromatic compounds decomposition. After chosen degrading enzymes, the enzyme gene clusters are synthesized in the form of functional clusters and introduced into a suitable chassis to achieve the goal of degradation of pollutants. Therefore, there are growing interests in engineering microbes through molecular biology manipulation to construct novel pathways with improved catalyzing activities for pollution removal. With recent advances in high-throughput screening methods, decomposition enzymes receive increased attention in environmental remediation. Enzymes are very promising in the degradation of pollutants (see Table 3). Among them, oxidoreductases and hydrolases are most studied enzymes for degrading pollutants owing to their high catalytic activity and ability to target a broad range of organic pollutants [83]. Oxidoreductases typically include peroxidases, laccases, and oxygenases. Peroxidases, such as manganese peroxidases, horseradish peroxidases, chloroperoxidases, and lignin peroxidases have been used in environmental bioremediation [84,85]. Laccase, which is a copper-containing oxidase, is capable of exercising one-electron oxidation of a broad range of pollutants using oxygen as the electron acceptor, and can degrade various persistent organic pollutants such as endocrine-disrupting chemicals, pesticides, and drugs [86,87]. In order to adapt to natural environment, Phanerochaete chrysosporium secretes lignin degrading enzymes, including laccase, lignin peroxidase, and manganese peroxidase, which can oxidize and degrade refractory organic pollutants [88]. In addition, the monooxygenase cytochrome P450 in Pleurotus ostreatus also plays a role in degrading pollutants [89]. Hydrolases usually break the large molecule into two small ones, which include proteases, esterases, amylases, lipases, acylases, and phosphatases [90]. These enzymes with high potential in degradation of pollutants have been applied in engineering microbes. Recently, Tournier V et al. reported that engineered enzyme was able to break down 200 g of polyethylene terephthalate in 10 h [91]. In engineered Pseudomonas putida KT2440 for 1,2,3-trichloropropane bioremediation, a catabolic pathway composed of haloalkane dehalogenase, haloalcohol dehalogenase and epoxide hydrolase was integrated into the chromosome for the conversion of 1,2,3-trichloropropane into glycerol. In addition, combinatorial engineering strategies were implemented to improve 1,2,3-trichloropropane mineralization, such as enhancing the carbon flux by deleting the glpR gene, improving the oxygen sequestering capacity through the heterologous expression of hemoglobin, and further increasing intracellular energy charge (ATP/ADP ratio) and reducing power (NADPH/NADP + ratio) by deleting flagella-related genes [92].
Table 3.
Enzymes for contaminant degradation.
| Enzymes | Species | Substrate | Refs |
|---|---|---|---|
| Oxidoreductases | |||
| Peroxidases | Ganoderma lucidum IBL-05 | Dye | [69] |
| Trametes pubescens strain i8 | Lignin | [70] | |
| Phanerochaete chrysosporium | Tetracycline and Oxytetracycline | [71] | |
| Ganoderma lucidum | Phenol | [72] | |
| Oxygenases | Pseudomonas putida G786 | Chlorofluorocarbons | [73] |
| Laccases |
Trametes villosa | Bisphenol | [74] |
| Coriolus versicolor | Lignin | [75] | |
| Trametes versicolor | Dye | [76] | |
|
Bacillus subtilis |
Polycyclic aromatic hydrocarbons |
[77] |
|
| Hydrolases | |||
| Lipase | Candida rugosa | Polyurethane | [78] |
| Rhizomucor miehei | Slop oil | [79] | |
| Lactobacillus sps. | Poly (ε-caprolactone) | [80] | |
| Cellulase | Bacillus megaterium | Cellulose | [81] |
| Protease | Myrothecium verrucaria | Poultry feather | [82] |
2.2. Optimizing the degradation system to boost efficiency by biological parts
Many microorganisms often have only one or none catabolic pathways, so they cannot completely mineralize aromatic pollutants. To solve this problem, one strategy in synthetic biology is to construct engineered strains which could be used as a bioremediation agent for aromatic compounds under complex environmental conditions. For example, one uses computational tools to analyze the genes, enzymes and metabolic pathways of degrading microorganism in different aromatic pollutants, and artificially synthesizes various biological parts [93], such as regulatory elements, transporters and stress-resistant elements (Table 4), and then integrate them into a microbial chassis to from a new metabolic network to achieve complete mineralization of refractory aromatic pollutants. When a fine-tuning of gene expression is required, the best approach is synthetic biology, which is the reconstruction, rewiring, and complete de novo design of transcriptional networks. Applications range from enhancing the understanding of gene regulatory and transcriptional mechanisms to the highly controlled expression of complete metabolic pathways. Orthogonal systems for heterologous protein expression as well as for the engineering of synthetic gene regulatory circuits in hosts depend on synthetic transcription factors and corresponding cis-regulatory binding sites. Synthetic promoters and corresponding synthetic transcription factors can be used to regulate the expression of heterologous genes without extensively relying on endogenous host transcription factors. A tunable and reasonable range of expression strengths is desired, especially when novel catabolic pathways are to be implemented. For example, precise control of key pathway enzymes can help maximize pollutant degradation, and synthetic promoters, terminators and ribosome-binding site can be inserted into the gene clusters in the right place to drive gene expression in host (Fig. 2B).
Table 4.
Boosting degradation efficiency by biological parts.
| Name | Description | Refs |
|---|---|---|
| promoters | An engineered haloalkane dehalogenase with the constitutive dhlA promoter for improving 1,2,3-Trichloropropane degradation activity in Pseudomonas putida MC4. | [94] |
| The AI-designed promoters are experimentally demonstrated to be functional in E. coli. | [95] | |
| Efficient keratinase expression via promoter engineering strategies for degradation of feather wastes. | [96] | |
| terminators | A panel of short (35–70 bp) synthetic terminators can be used for modulating gene expression in yeast. | [97] |
| Synthetic terminator performs the same function as natural terminator. | [98] | |
| RBS | Automated design of synthetic ribosome binding sites to control protein expression. | [99] |
| RBS optimization of the key enzymes was used for improving the synthesis of natural product. | [100,101] | |
| transport proteins | Transporters for benzoic acid, 4-hydroxybenzoic acid, protocatechuic acid and vanillic acid. | [[102], [103], [104]] |
| mobile genetic elements | Many catabolic genes have been found adjacent to mobile genetic elements. | [105] |
| genomic islands | The genomic islands-deleted Pseudomonas putida KT2440 was an optimum chassis for improving the γ-hexachlorocyclohexane and 1,2,3-trichloropropane biodegradation pathways. | [106] |
| plasmid | Catabolic plasmids that encode genes for the degradation of contaminants such as toluene, naphthalene, phenol, and nitrobenzene. | [107] |
| transposons | Transposons for the catabolism of toluene (Tn4651, Tn4653, Tn4656), chlorobenzoate (Tn5271), chlorobenzene (Tn5280), benzene (Tn5542) and naphthalene (Tn4655). | [108] |
2.2.1. Regulatory elements
Synthetic biology builds artificial biological systems, and regulatory elements are the basis in this system, such as promoters, terminators, operators, open reading frame and ribosome-binding site (RBS). Gene expression is the basis for achieving gene function, especially for enzyme. The classic strategy to regulate this process is to construct an inducible expression system using the repressor protein and operon, such as IPTG system with lac operon in Escherichia coli. However, artificial systems constructed directly from natural elements often have the disadvantages of higher leakage expression, low induction efficiency, and poor stability for protein [109]. To overcome these bottlenecks, redesign artificial biological elements is highly desired [110]. We can develop high-performance biological elements, and standardize them to improve gene expression strength and stability, and achieve ‘plug and play’ goal. Samin et al. [94] reported that an engineered haloalkane dehalogenase with the constitutive dhlA promoter for improving 1,2,3-trichloropropane degradation activity in Pseudomonas putida MC4. Gong et al. [96] improved keratinase expression via promoter engineering strategies for degradation of feather wastes. In addition, the AI-assistant designed promoters have already been experimentally demonstrated to be functional in Escherichia coli [95]. Synthetic terminator also performs the same function as natural terminator [98]. A native promoter of the bph operon was replaced by a constitutive promoter through homologous recombination, which greatly improved the biphenyl and polychlorinated biphenyl degradation activity of Pseudomonas sp. strain KKS102 [111]. Curran [97] et al. synthesized a panel of short terminators with stronger termination efficiency, which have been applied in modulating gene expression in yeast.
2.2.2. Transporting elements
In the previous studies, there are few reports on the transporting elements involved in microbial remediation process. The transport proteins HBT1/HBT2, as a benzoic acid transporter, involved in the catabolic degradation of hydroxyaromatic compounds in the pathogenic yeast Candida parapsilosis [102]. Xu et al. reported that the gentisate transporter GenK was carried in the metabolism of gentisic acid by Corynebacterium glutamicum [103]. In addition, the transport proteins, which involve in the bioremediation process, have been found from Corynebacterium glutamicum. It also contains 4-hydroxybenzoic acid, protocatechuic acid, vanillic acid, and benzoic acid transporters [104].
2.2.3. Other elements
In addition to regulatory elements and transporters, there are other biological parts including mobile genetic elements, plasmids, genomic islands, and transposons. In order to quickly adapt to the stimuli and changes in the environment, microorganisms have gradually evolved the ability to obtain mobile genetic elements from surroundings [105], so that microbes have more and stronger catabolism capabilities for pollutants. For example, the resulting catabolic plasmids obtained certain genes encoding enzyme for the degradation of contaminants such as toluene, naphthalene, phenol, and nitrobenzene [107]. Deletion of some genes can also improve the degradation rate, and the genomic islands-deleted Pseudomonas putida KT2440 was an optimum chassis for improving the γ-hexachlorocyclohexane and 1,2,3-trichloropropane biodegradation pathways [106]. Transposons are small pieces of DNA that can transpose through either RNA or DNA intermediates, and used for the catabolism of toluene (Tn4651, Tn4653, Tn4656), chlorobenzoate (Tn5271), chlorobenzene (Tn5280), benzene (Tn5542) and naphthalene (Tn4655) [108]. Though microbes showed the advantages in naturally degrading pollutants, they have intrinsic disadvantage that they usually possess the metabolic gene only for one specific compound. Although their advantages are obvious and proof-of-concept have been demonstrated successful in the laboratory, construction of engineered strains is time consuming, and it is difficult for a single strain to degrade compound pollutants. Thus, few products have entered the market so far. However, taking advantages of synthetic biology, we are confident that current technical challenges in microbial remediation will be overcome in the future.
2.3. Strategies to improve the chassis tolerance
The discovery and application of stress-resistant elements is an effective method to enhance the viability of microbes under stress conditions and thereby improve microbe's pollutant degradation capacity [112]. The long-term domestication of environmental microbes under natural conditions has a special and efficient stress response mechanism. Therefore, the tolerance elements in such microbes have wider application. However, information related to the unique stress response mechanism in environmental microbes remains unclear. Although the microbial degradation mechanism of organic pollutants in the environment has been studied, such as nicotine and biphenyl [[113], [114], [115]], it is still unknown why natural microbes usually appear in harsh environments with high temperature, high salt, strong acid and alkali, or hypertonicity, which exhibit increased metabolic burden and inhibited growth of natural microbes. In practical, engineered strains would also encounter various stresses, such as extreme temperature and pH, oxidative pressure, organic solvents, osmotic pressure, high concentrations of substrates, toxic products or byproducts etc. The metabolic activity of these strains under such stressful growth conditions is severally inhibited or even completely lost, causing dramatically dropping pollutants degradation efficiency. In the case of pollutants, the harsh environment toxicity is one of the main bottlenecks in achieving optimal pollutant degradation. Since overcoming sensitivity to the environment often requires engineering tolerance mechanisms, this specific area of host optimization has recently been termed tolerance engineering. Prominent categories of genes used in tolerance engineering include chaperones, membrane-modifying enzymes, redox enzymes, transport pumps, and transcriptional factors. For instance, chaperones specifically have emerged as a powerful category of proteins that bestow tolerance and often improve degradation efficiency of engineering strains. CRISPRi and Red/ET recombineering allow us to target multiple genes simultaneously and provide powerful new approaches in tolerance engineering. Genome shuffling across multiple strains with desirable phenotypes, coupled with strong screens, can be a potential approach to obtain degrading strains. Adaptive evolution is also an effective method to obtain engineering strains under specific environmental conditions, which is used to screen microorganisms resistant to environmental stresses.
The tolerance of chassis cells against the harsh environments can be rationally enhanced if the tolerance elements present in natural microbes are well understood. Tolerance elements are closely related to the cell structure, physiological properties, metabolic pathways and regulatory processes of cells, such as cell wall and cell membrane, efflux pump, heat shock proteins, and compatible solutes (Table 5).
Table 5.
Examples of tolerance engineering.
| Tolerance mechanism | Proteins | Species | Stress resistance | Refs |
|---|---|---|---|---|
| chaperones | GroESL | Clostridium acetobutylicum | n-Butanol | [116] |
| transporters | RbsB MsmK | Lactococcus lactis | Acid | [117] |
| membrane | Med2 | Candida glabrata | Acid | [118] |
| MgtA | E. coli | Succinic acid | [119] | |
| efflux pumps | RcdA | E. coli | Limonene | [120] |
| regulators | MetR | E. coli | 3-Methyl | [121] |
| heat shock proteins | HspX, Y, Z | P. putida | Phenol | [122] |
| compatible solute | RHD | Martelella AD-3 | saline | [123] |
2.3.1. Cell wall and cell membrane
The cell wall is an important biological barrier separating microorganisms from the external environment. The murA gene encoding UDP-N-acetylglucosamine enolpyruvyl transferase, is known to catalyze the biosynthesis of peptidoglycan, which is an important cellular component of cell wall in prokaryotes [124]. Yuan et al. reported that overexpression of peptidoglycan biosynthesis murA2 gene from the Lactobacillus plantarum could enhance Escherichia coli's tolerance to alcohols [125]. On the other hand, cell membrane is the major target under external stress conditions, and engineering membrane and cell-wall is a good strategy for developing industrial strains with increased stress tolerance [126,127]. Cellular analysis and comparative transcriptomics revealed that Candida tropicalis raised the tolerance to phenol through improvement of cell wall resistance [128].
2.3.2. Efflux pump
Efflux pumps are membrane transporters localized in the cytoplasmic membrane of various cells, such as TtgABC, TtgDEF, TtgGHI and SrpABC in Pseudomonas Putida and AcrAB-TolC in E. coli. AcrAB-TolC is involved in the tolerance towards olefin compounds in Escherichia coli [129]. The EmhABC in Pseudomonas fluorescens LP6a effluxes phenanthrene and anthracene, and the presence of EmhABC decreased the efficiency of phenanthrene biodegradation. However, the EmhABC is involved in naphthalene tolerance and increases the efficiency of naphthalene metabolism in Pseudomonas fluorescens LP6a [130].
2.3.3. Heat shock proteins
Heat shock proteins, as chaperones, play an important role in stress tolerance, and are induced or overexpressed for degradation and reactivation of damaged proteins. Suo et al. reported that overexpression of GroESL increased the butyric acid tolerance of Clostridium tyrobutyricum ATCC 25755 [131]. The genes groES and groEL from Clostridium acetobutylicum ATCC 824 were introduced into Clostridium beijerinckii NCIMB 8052, which enhanced its tolerance to ferulic acid [132]. Overexpressing three heat shock protein genes hsp18.5, hsp18.55 and hsp19.3 in Lactobacillus plantarum strain WCFS1 improved adaptation to heat, cold, and solvent tolerance [133]. Growth competition experiments showed that HspX, Y and Z of Pseudomonas putida are involved in tolerance against the toxic effects of phenol, and the novel heat shock operon hspXYZ, which is a part of the chaperone network could mediate stress tolerance in the natural environment [122].
2.3.4. Compatible solute
Accumulation of compatible solutes such as trehalose, proline, and betaine in the cell can balance intracellular and extracellular osmotic pressure, and maintain the metabolic activity of the cell to adapt to stress environment. Betaine, a regulator of osmosis, is synthesized by aldehyde dehydrogenase, and heterologous expression of the betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus facilitated engineered Escherichia coli to confer high heat and salt tolerance under high temperature at 55 °C and 700 mM NaCl [134]. Intracellular proline protects yeast from damage caused by freezing and ethanol, and is important for breeding stress-tolerant industrial yeast strains [135]. A phenanthrene-degrading strain, Martelella AD-3, was isolated from high salt environments. Label-free proteomics revealed that Martelella relied on aromatic ring-hydroxylating dioxygenase and dihydrodiol dehydrogenase to degrade phenanthrene, and accumulated compatible solutes for resistance to salt stress [123].
3. Challenges and future directions
Microbial remediation plays a critical role in the treatment of environmental pollution. Due to the diversity and complexity of environmental pollution, the tolerance of microbes under pollution condition has become the bottleneck for bioremediation. The application of synthetic biology in microbial remediation is still in its infancy but already offers exciting possibilities for creating a cleaner, safer, healthier environment. Many challenges remain to be resolved before the engineered cells can be used to treat environmental pollution in practice. For example, there may be multiple pollutants in the environment, which makes microbes unable to execute the expected degradation function. Since after a microorganism targeting a single pollutant is released into the environment, other pollutants and environmental factors may affect the growth and degradation speed and efficiency of this microorganism. We need to create a new design model that takes uncertainty into account and allows us to gradually increase the complexity when mimicking the natural environment. In order to solve the problem of in-situ treatment of aromatic pollutants, we proposed the following technical routes: (i) Mine genes in microbial systems, such as degradation genes, transport genes, molecular switches, and stress resistance genes; (ii) Rationally design degradation pathways and systematically optimize high-efficiency degradation components; (iii) Improve the movement, aggregation, interaction of synthetic microorganisms and the ability to adapt to complex environments; (iv) Design, assemble multifunctional metabolic networks, and use artificial intervention to build a synthetic biological system for the aromatic pollutants degradation; (v) Develop artificial degradation metabolic systems in in-situ treatment of pollutants and apply it in actual industrial application.
The more enzymatic steps in a heterologous pathway, the more formidable the challenge for construction of the pathway, and discovery of the requisite enzymatic components. Advances in synthetic genomics, DNA sequencing, synthesis and assembly make it possible to redesign and artificially synthesize whole genomes, and these enabling technologies have allowed the discovery and engineering of increasingly long catabolism pathways. In the future, the fast development of synthetic biology has stimulated the reconstruction of cellular components to create synthetic microorganisms for solving the problem of pollutants (Fig. 3). By complying with the central dogma, which operates microbial catabolism, synthetic biology researchers have reprogramed some organelles, such as nucleus, ribosomes, and mitochondria, and have created synthetic microorganisms which can serve as smart chassis for pollutant degradation.
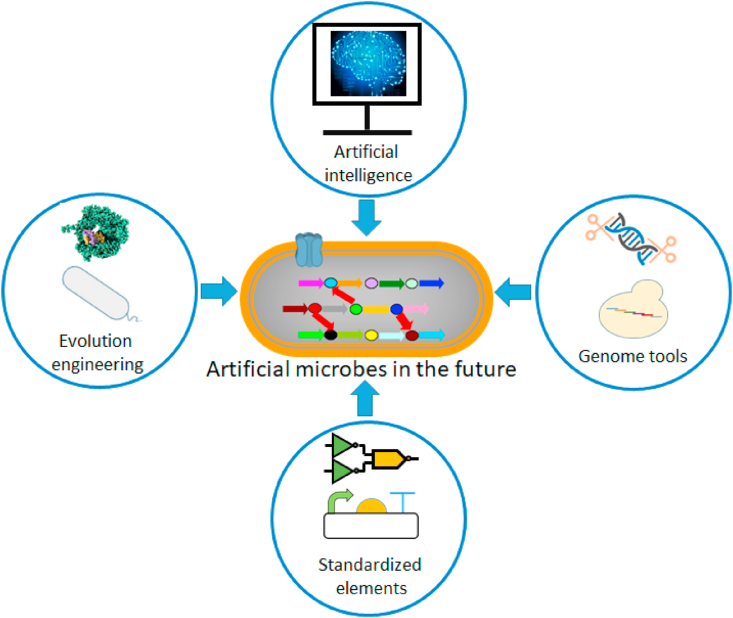
Fig. 3.
Future perspectives on the construction of artificial microbes.
In the future, artificial intelligence should be used to assist in construction of artificial microbes, especially in genetic model building and protein and metabolic pathway design. Genome tools should be developed to enable efficient genetic manipulation of artificial microbes as well as natural decomposer. The construction of standardized elements will shorten the cycle of complex genetic circuit design. Finally, protein directed evolution and strain adaptive evolution should be established to accelerate the efficiency of pollutant degradation.
However, commercial applications remain limited due to complicated biodegradation processes, and synthetic biology also brings many uncertainties to microbes. First, existing synthetic biology tools may not be suitable for new chassis and need to be further optimized. Second, considering that bioremediation will ultimately be performed in an open environment, the ecological safety of engineered bacteria must be considered, such as possibility of introducing mobile genetic elements and antibiotic resistance marker genes into the environment. Third, the change in microbial metabolic pathway, will release unknown toxic products for the environment, indirectly acting as opposition microbial candidates.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31971347, 32030004 and 31725002), Shenzhen Science and Technology Program (KQTD20180413181837372), Guangdong Provincial Key Laboratory of Synthetic Genomics (2019B030301006) and Shenzhen Outstanding Talents Training Fund.
CRediT authorship contribution statement
Liang Xiang: Writing – original draft, Writing – review & editing, writing of the original draft, all authors: review and editing, All authors have read and agreed to the published version of the manuscript. Hongzhi Tang: Conceptualization, Writing – review & editing, all authors: review and editing, All authors have read and agreed to the published version of the manuscript. Junbiao Dai: Conceptualization, Writing – review & editing, all authors: review and editing, All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors indicate that they have no conflict of interest.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Hongzhi Tang, Email: tanghongzhi@sjtu.edu.cn.
Junbiao Dai, Email: junbiao.dai@siat.ac.cn.
References
- 1.Dvořák P., Nikel P.I., Damborský J., de Lorenzo V. Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol Adv. 2017;35(7):845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Padhye L.P. Fate of environmental pollutants. Water Environ Res. 2016;88(10):1619–1636. doi: 10.2175/106143016X14696400495578. [DOI] [PubMed] [Google Scholar]
- 3.Sana B. Bioresources for control of environmental pollution. Adv Biochem Eng Biotechnol. 2015;147:137–183. doi: 10.1007/10_2014_276. [DOI] [PubMed] [Google Scholar]
- 4.Che S., Men Y. Synthetic microbial consortia for biosynthesis and biodegradation: promises and challenges. J Ind Microbiol Biotechnol. 2019;46(9–10):1343–1358. doi: 10.1007/s10295-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 5.Rucká L., Nešvera J., Pátek M. Biodegradation of phenol and its derivatives by engineered bacteria: current knowledge and perspectives. World J Microbiol Biotechnol. 2017;33(9):174. doi: 10.1007/s11274-017-2339-x. Published 2017 Sep. 6. [DOI] [PubMed] [Google Scholar]
- 6.Fukuhara Y., Inakazu K., Kodama N. Characterization of the isophthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microbiol. 2010;76(2):519–527. doi: 10.1128/AEM.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimodaira J., Kamimura N., Hosoyama A., Yamazoe A., Fujita N., Masai E. Draft genome sequence of Comamonas sp. strain E6 (NBRC 107749), a degrader of phthalate isomers through the protocatechuate 4,5-cleavage pathway. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00643-15. e00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawson C.E., Harcombe W.R., Hatzenpichler R. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol. 2019;17(12):725–741. doi: 10.1038/s41579-019-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V. Systems biology approaches to bioremediation. Curr Opin Biotechnol. 2008;19(6):579–589. doi: 10.1016/j.copbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 10.McCarty N.S., Ledesma-Amaro R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019;37(2):181–197. doi: 10.1016/j.tibtech.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurumbang N.P., Dvorak P., Bendl J., Brezovsky J., Prokop Z., Damborsky J. Computer-assisted engineering of the synthetic pathway for biodegradation of a toxic persistent pollutant. ACS Synth Biol. 2014;3(3):172–181. doi: 10.1021/sb400147n. [DOI] [PubMed] [Google Scholar]
- 12.Jung J.K., Alam K.K., Verosloff M.S. Cell-free biosensors for rapid detection of water contaminants. Nat Biotechnol. 2020;38(12):1451–1459. doi: 10.1038/s41587-020-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith H.O., Hutchison C.A., 3rd, Pfannkoch C., Venter J.C. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A. 2003;100(26):15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson D.G., Benders G.A., Andrews-Pfannkoch C. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319(5867):1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 15.Gibson D.G., Glass J.I., Lartigue C. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 16.Hutchison C.A., 3rd, Chuang R.Y., Noskov V.N. Design and synthesis of a minimal bacterial genome [published correction appears in ACS Chem Biol. 2016 May 20;11(5):1463] Science. 2016;351(6280):aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 17.Dymond J.S., Richardson S.M., Coombes C.E. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477(7365):471–476. doi: 10.1038/nature10403. Published 2011 Sep. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annaluru N., Muller H., Mitchell L.A. Total synthesis of a functional designer eukaryotic chromosome [published correction appears in Science. 2014 May 23;344(6186):816] Science. 2014;344(6179):55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercy G., Mozziconacci J., Scolari V.F. 3D organization of synthetic and scrambled chromosomes. Science. 2017;355(6329) doi: 10.1126/science.aaf4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell L.A., Wang A., Stracquadanio G. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355(6329) doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 21.Richardson S.M., Mitchell L.A., Stracquadanio G. Design of a synthetic yeast genome. Science. 2017;355(6329):1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y., Wang Y., Chen T. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355(6329) doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Li B.Z., Zhao M. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355(6329) doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z.X., Li B.Z., Mitchell L.A. Perfect" designer chromosome V and behavior of a ring derivative. Science. 2017;355(6329) doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W., Zhao G., Luo Z. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355(6329) doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 26.Shao Y., Lu N., Wu Z. Creating a functional single-chromosome yeast. Nature. 2018;560(7718):331–335. doi: 10.1038/s41586-018-0382-x. [DOI] [PubMed] [Google Scholar]
- 27.Luo J., Sun X., Cormack B.P., Boeke J.D. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature. 2018;560(7718):392–396. doi: 10.1038/s41586-018-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredens J., Wang K., de la Torre D. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569(7757):514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F., Ellis T. The second decade of synthetic biology: 2010-2020. Nat Commun. 2020;11(1):5174. doi: 10.1038/s41467-020-19092-2. Published 2020 Oct 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Liu Y., Du G., Ledesma-Amaro R., Liu L. Microbial chassis development for natural product biosynthesis. Trends Biotechnol. 2020;38(7):779–796. doi: 10.1016/j.tibtech.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Noda S., Shirai T., Oyama S., Kondo A. Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab Eng. 2016;33:119–129. doi: 10.1016/j.ymben.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Gottardi M., Knudsen J.D., Prado L., Oreb M., Branduardi P., Boles E. De novo biosynthesis of trans-cinnamic acid derivatives in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2017;101(12):4883–4893. doi: 10.1007/s00253-017-8220-x. [DOI] [PubMed] [Google Scholar]
- 33.Rylott E.L., Bruce N.C. How synthetic biology can help bioremediation. Curr Opin Chem Biol. 2020;58:86–95. doi: 10.1016/j.cbpa.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Gu Y., Xu X., Wu Y. Advances and prospects of Bacillus subtilis cellular factories: from rational design to industrial applications. Metab Eng. 2018;50:109–121. doi: 10.1016/j.ymben.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Xiang M., Kang Q., Zhang D. Advances on systems metabolic engineering of Bacillus subtilis as a chassis cell. Synth Syst Biotechnol. 2020;5(4):245–251. doi: 10.1016/j.synbio.2020.07.005. Published 2020 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin J., Liu X., Liu W., Zheng X. Effects of biochar-BDE-47 interactions on BDE-47 bioaccessibility and biodegradation by Pseudomonas putida TZ-1. Ecotoxicol Environ Saf. 2014;106:27–32. doi: 10.1016/j.ecoenv.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 37.Li J., Luo C., Zhang D., Cai X., Jiang L., Zhang G. Stable-isotope probing-enabled cultivation of the indigenous bacterium Ralstonia sp. strain M1, capable of degrading phenanthrene and biphenyl in industrial wastewater. Appl Environ Microbiol. 2019;85(14) doi: 10.1128/AEM.00511-19. e00511-19. Published 2019 Jul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y.M., Nam I.H., Murugesan K., Schmidt S., Crowley D.E., Chang Y.S. Biodegradation of diphenyl ether and transformation of selected brominated congeners by Sphingomonas sp. PH-07. Appl Microbiol Biotechnol. 2007;77(1):187–194. doi: 10.1007/s00253-007-1129-z. [DOI] [PubMed] [Google Scholar]
- 39.Robrock K.R., Coelhan M., Sedlak D.L., Alvarez-Cohent L. Aerobic biotransformation of polybrominated diphenyl ethers (PBDEs) by bacterial isolates. Environ Sci Technol. 2009;43(15):5705–5711. doi: 10.1021/es900411k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dogra C., Raina V., Pal R. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J Bacteriol. 2004;186(8):2225–2235. doi: 10.1128/jb.186.8.2225-2235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Field J.A., Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollut. 2008;155(1):1–12. doi: 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Na K.S., Kuroda A., Takiguchi N., Ikeda T., Ohtake H., Kato J. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains. J Biosci Bioeng. 2005;99(4):378–382. doi: 10.1263/jbb.99.378. [DOI] [PubMed] [Google Scholar]
- 43.Heinaru E., Naanuri E., Grünbach M., Jõesaar M., Heinaru A. Functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene. 2016;589(1):90–98. doi: 10.1016/j.gene.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Iwaki H., Yamamoto T., Hasegawa Y. Isolation of marine xylene-utilizing bacteria and characterization of Halioxenophilus aromaticivorans gen. nov., sp. nov. and its xylene degradation gene cluster. FEMS Microbiol Lett. 2018;365(7) doi: 10.1093/femsle/fny042. [DOI] [PubMed] [Google Scholar]
- 45.Kallscheuer N., Vogt M., Kappelmann J. Identification of the phd gene cluster responsible for phenylpropanoid utilization in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2016;100(4):1871–1881. doi: 10.1007/s00253-015-7165-1. [DOI] [PubMed] [Google Scholar]
- 46.Jõesaar M., Viggor S., Heinaru E. Strategy of Pseudomonas pseudoalcaligenes C70 for effective degradation of phenol and salicylate. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173180. Published 2017 Mar 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordin K., Unell M., Jansson J.K. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl Environ Microbiol. 2005;71(11):6538–6544. doi: 10.1128/AEM.71.11.6538-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min J., Chen W., Hu X. Biodegradation of 2,6-dibromo-4-nitrophenol by Cupriavidus sp. strain CNP-8: kinetics, pathway, genetic and biochemical characterization. J Hazard Mater. 2019;361:10–18. doi: 10.1016/j.jhazmat.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 49.Min J., Chen W., Wang J., Hu X. Genetic and biochemical characterization of 2-chloro-5-nitrophenol degradation in a newly isolated bacterium, cupriavidus sp. strain CNP-8. Front Microbiol. 2017;8:1778. doi: 10.3389/fmicb.2017.01778. Published 2017 Sep. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Min J., Lu Y., Hu X., Zhou N.Y. Biochemical characterization of 3-Methyl-4-nitrophenol degradation in Burkholderia sp. strain SJ98. Front Microbiol. 2016;7:791. doi: 10.3389/fmicb.2016.00791. Published 2016 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izmalkova T.Y., Sazonova O.I., Nagornih M.O., Sokolov S.L., Kosheleva I.A., Boronin A.M. The organization of naphthalene degradation genes in Pseudomonas putida strain AK5. Res Microbiol. 2013;164(3):244–253. doi: 10.1016/j.resmic.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Laurie A.D., Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181(2):531–540. doi: 10.1128/JB.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong T., Liu R., Che Y. Engineering Pseudomonas putida KT2440 for simultaneous degradation of carbofuran and chlorpyrifos. Microb Biotechnol. 2016;9(6):792–800. doi: 10.1111/1751-7915.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong T., Xu X., Dang Y. An engineered Pseudomonas putida can simultaneously degrade organophosphates, pyrethroids and carbamates. Sci Total Environ. 2018;628–629:1258–1265. doi: 10.1016/j.scitotenv.2018.02.143. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y., Che Y., Zhang F. Development of an efficient pathway construction strategy for rapid evolution of the biodegradation capacity of Pseudomonas putida KT2440 and its application in bioremediation. Sci Total Environ. 2021;761:143239. doi: 10.1016/j.scitotenv.2020.143239. [DOI] [PubMed] [Google Scholar]
- 56.Hoff J., Daniel B., Stukenberg D., Thuronyi B.W., Waldminghaus T., Fritz G. Vibrio natriegens: an ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ Microbiol. 2020;22(10):4394–4408. doi: 10.1111/1462-2920.15128. [DOI] [PubMed] [Google Scholar]
- 57.Tschirhart T., Shukla V., Kelly E.E. Synthetic biology tools for the fast-growing marine bacterium Vibrio natriegens. ACS Synth Biol. 2019;8(9):2069–2079. doi: 10.1021/acssynbio.9b00176. [DOI] [PubMed] [Google Scholar]
- 58.Popat S.C., Deshusses M.A. Reductive dehalogenation of trichloroethene vapors in an anaerobic biotrickling filter. Environ Sci Technol. 2009;43(20):7856–7861. doi: 10.1021/es901305x. [DOI] [PubMed] [Google Scholar]
- 59.Kengara F.O., Doerfler U., Welzl G., Munch J.C., Schroll R. Evidence of non-DDD pathway in the anaerobic degradation of DDT in tropical soil. Environ Sci Pollut Res Int. 2019;26(9):8779–8788. doi: 10.1007/s11356-019-04331-x. [DOI] [PubMed] [Google Scholar]
- 60.Bumpus J.A., Aust S.D. Biodegradation of DDT [1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane] by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1987;53(9):2001–2008. doi: 10.1128/AEM.53.9.2001-2008.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S., Wondrousch D., Cooper M., Zinder S.H., Schüürmann G., Adrian L. Anaerobic dehalogenation of chloroanilines by Dehalococcoides mccartyi strain CBDB1 and dehalobacter strain 14DCB1 via different pathways as related to molecular electronic structure. Environ Sci Technol. 2017;51(7):3714–3724. doi: 10.1021/acs.est.6b05730. [DOI] [PubMed] [Google Scholar]
- 62.Smidt H., de Vos W.M. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- 63.Ito K., Takagi K., Matsushima Y. Identification of the novel hcbB operon catalyzing the dechlorination of pentachlorophenol in the Gram-positive bacterium Nocardioides sp. strain PD653. J Pestic Sci. 2018;43(2):124–131. doi: 10.1584/jpestics.D17-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito K., Takagi K., Iwasaki A. Identification of the hcb gene operon involved in catalyzing aerobic hexachlorobenzene dechlorination in Nocardioides sp. strain PD653 [published correction appears in Appl Environ Microbiol. 2017 Dec 15;84(1):] Appl Environ Microbiol. 2017;83(19) doi: 10.1128/AEM.00824-17. e00824-17. Published 2017 Sep. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seo J.S., Keum Y.S., Li Q.X. Bacterial degradation of aromatic compounds. Int J Environ Res Publ Health. 2009;6(1):278–309. doi: 10.3390/ijerph6010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker A.W., Keasling J.D. Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol Bioeng. 2002;78(7):715–721. doi: 10.1002/bit.10251. [DOI] [PubMed] [Google Scholar]
- 67.Kurumbang N.P., Dvorak P., Bendl J., Brezovsky J., Prokop Z., Damborsky J. Computer-assisted engineering of the synthetic pathway for biodegradation of a toxic persistent pollutant. ACS Synth Biol. 2014;3(3):172–181. doi: 10.1021/sb400147n. [DOI] [PubMed] [Google Scholar]
- 68.Demko M., Chrást L., Dvořák P., Damborský J., Šafránek D. Computational modelling of metabolic burden and substrate toxicity in Escherichia coli carrying a synthetic metabolic pathway. Microorganisms. 2019;7(11):553. doi: 10.3390/microorganisms7110553. Published 2019 Nov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaheen R., Asgher M., Hussain F., Bhatti H.N. Immobilized lignin peroxidase from Ganoderma lucidum IBL-05 with improved dye decolorization and cytotoxicity reduction properties. Int J Biol Macromol. 2017;103:57–64. doi: 10.1016/j.ijbiomac.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 70.Rekik H., Zaraî Jaouadi N., Bouacem K. Physical and enzymatic properties of a new manganese peroxidase from the white-rot fungus Trametes pubescens strain i8 for lignin biodegradation and textile-dyes biodecolorization. Int J Biol Macromol. 2019;125:514–525. doi: 10.1016/j.ijbiomac.2018.12.053. [DOI] [PubMed] [Google Scholar]
- 71.Wen X., Jia Y., Li J. Degradation of tetracycline and oxytetracycline by crude lignin peroxidase prepared from Phanerochaete chrysosporium--a white rot fungus. Chemosphere. 2009;75(8):1003–1007. doi: 10.1016/j.chemosphere.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 72.Xu H., Guo M.Y., Gao Y.H., Bai X.H., Zhou X.W. Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol. BMC Biotechnol. 2017;17(1):19. doi: 10.1186/s12896-017-0338-5. Published 2017 Feb 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hur H.G., Sadowsky M.J., Wackett L.P. Metabolism of chlorofluorocarbons and polybrominated compounds by Pseudomonas putida G786(pHG-2) via an engineered metabolic pathway. Appl Environ Microbiol. 1994;60(11):4148–4154. doi: 10.1128/AEM.60.11.4148-4154.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukuda T., Uchida H., Takashima Y., Uwajima T., Kawabata T., Suzuki M. Degradation of bisphenol A by purified laccase from Trametes villosa. Biochem Biophys Res Commun. 2001;284(3):704–706. doi: 10.1006/bbrc.2001.5021. [DOI] [PubMed] [Google Scholar]
- 75.Kajita S., Sugawara S., Miyazaki Y. Overproduction of recombinant laccase using a homologous expression system in Coriolus versicolor. Appl Microbiol Biotechnol. 2004;66(2):194–199. doi: 10.1007/s00253-004-1663-x. [DOI] [PubMed] [Google Scholar]
- 76.Legerská B., Chmelová D., Ondrejovič M. Decolourization and detoxification of monoazo dyes by laccase from the white-rot fungus Trametes versicolor. J Biotechnol. 2018;285:84–90. doi: 10.1016/j.jbiotec.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Zeng J., Zhu Q., Wu Y., Lin X. Oxidation of polycyclic aromatic hydrocarbons using Bacillus subtilis CotA with high laccase activity and copper independence. Chemosphere. 2016;148:1–7. doi: 10.1016/j.chemosphere.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 78.Gautam R., Bassi A.S., Yanful E.K. Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium. Biotechnol Lett. 2007;29(7):1081–1086. doi: 10.1007/s10529-007-9354-1. [DOI] [PubMed] [Google Scholar]
- 79.Marchut-Mikolajczyk O., Drożdżyński P., Struszczyk-Świta K. Biodegradation of slop oil by endophytic Bacillus cereus EN18 coupled with lipase from Rhizomucor miehei (Palatase®) Chemosphere. 2020;250:126203. doi: 10.1016/j.chemosphere.2020.126203. [DOI] [PubMed] [Google Scholar]
- 80.Khan I., Ray Dutta J., Ganesan R. Lactobacillus sps. lipase mediated poly (ε-caprolactone) degradation. Int J Biol Macromol. 2017;95:126–131. doi: 10.1016/j.ijbiomac.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 81.Kalbarczyk K.Z., Mazeau E.J., Rapp K.M., Marchand N., Koffas M.A.G., Collins C.H. Engineering Bacillus megaterium strains to secrete cellulases for synergistic cellulose degradation in a microbial community. ACS Synth Biol. 2018;7(10):2413–2422. doi: 10.1021/acssynbio.8b00186. [DOI] [PubMed] [Google Scholar]
- 82.Moreira-Gasparin F.G., de Souza C.G., Costa A.M. Purification and characterization of an efficient poultry feather degrading-protease from Myrothecium verrucaria. Biodegradation. 2009;20(5):727–736. doi: 10.1007/s10532-009-9260-4. [DOI] [PubMed] [Google Scholar]
- 83.Zhu B., Chen Y., Wei N. Engineering biocatalytic and biosorptive materials for environmental applications. Trends Biotechnol. 2019;37(6):661–676. doi: 10.1016/j.tibtech.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 84.Rao M.A., Scelza R., Acevedo F., Diez M.C., Gianfreda L. Enzymes as useful tools for environmental purposes. Chemosphere. 2014;107:145–162. doi: 10.1016/j.chemosphere.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 85.Dragana R., Nikola G., Željko D., Gordana A., Olgica N. Separation of peroxidases from Miscanthus x giganteus, their partial characterisation and application for degradation of dyes. Plant Physiol Biochem. 2017;120:179–185. doi: 10.1016/j.plaphy.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Macellaro G., Pezzella C., Cicatiello P., Sannia G., Piscitelli A. Fungal laccases degradation of endocrine disrupting compounds. BioMed Res Int. 2014;2014:614038. doi: 10.1155/2014/614038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim Y., Yeo S., Song H.G., Choi H.T. Enhanced expression of laccase during the degradation of endocrine disrupting chemicals in Trametes versicolor. J Microbiol. 2008;46(4):402–407. doi: 10.1007/s12275-007-0236-y. [DOI] [PubMed] [Google Scholar]
- 88.Bumpus J.A., Tien M., Wright D., Aust S.D. Oxidation of persistent environmental pollutants by a white rot fungus. Science. 1985;228(4706):1434–1436. doi: 10.1126/science.3925550. [DOI] [PubMed] [Google Scholar]
- 89.Bezalel L., Hadar Y., Fu P.P., Freeman J.P., Cerniglia C.E. Initial oxidation products in the metabolism of pyrene, anthracene, fluorene, and dibenzothiophene by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1996;62(7):2554–2559. doi: 10.1128/AEM.62.7.2554-2559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharma B., Dangi A.K., Shukla P. Contemporary enzyme based technologies for bioremediation: a review. J Environ Manag. 2018;210:10–22. doi: 10.1016/j.jenvman.2017.12.075. [DOI] [PubMed] [Google Scholar]
- 91.Tournier V., Topham C.M., Gilles A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580(7802):216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- 92.Gong T., Xu X., Che Y. Combinatorial metabolic engineering of Pseudomonas putida KT2440 for efficient mineralization of 1,2,3-trichloropropane. Sci Rep. 2017;7(1):7064. doi: 10.1038/s41598-017-07435-x. Published 2017 Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen B., Lee H.L., Heng Y.C. Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnol Adv. 2018;36(7):1870–1881. doi: 10.1016/j.biotechadv.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 94.Samin G., Pavlova M., Arif M.I., Postema C.P., Damborsky J., Janssen D.B. A Pseudomonas putida strain genetically engineered for 1,2,3-trichloropropane bioremediation. Appl Environ Microbiol. 2014;80(17):5467–5476. doi: 10.1128/AEM.01620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y., Wang H., Wei L., Li S., Liu L., Wang X. Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res. 2020;48(12):6403–6412. doi: 10.1093/nar/gkaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong J.S., Ye J.P., Tao L.Y. Efficient keratinase expression via promoter engineering strategies for degradation of feather wastes. Enzym Microb Technol. 2020;137:109550. doi: 10.1016/j.enzmictec.2020.109550. [DOI] [PubMed] [Google Scholar]
- 97.Curran K.A., Morse N.J., Markham K.A., Wagman A.M., Gupta A., Alper H.S. Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synth Biol. 2015;4(7):824–832. doi: 10.1021/sb5003357. [DOI] [PubMed] [Google Scholar]
- 98.Chen Y.J., Liu P., Nielsen A.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10(7):659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 99.Salis H.M., Mirsky E.A., Voigt C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen J., Zhu X., Tan Z. Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl Microbiol Biotechnol. 2014;98(5):2197–2205. doi: 10.1007/s00253-013-5387-7. [DOI] [PubMed] [Google Scholar]
- 101.Li M., Chen H., Liu C. Improvement of isoprene production in Escherichia coli by rational optimization of RBSs and key enzymes screening. Microb Cell Factories. 2019;18(1):4. doi: 10.1186/s12934-018-1051-3. Published 2019 Jan 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cillingová A., Zeman I., Tóth R. Eukaryotic transporters for hydroxyderivatives of benzoic acid. Sci Rep. 2017;7(1):8998. doi: 10.1038/s41598-017-09408-6. Published 2017 Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu Y., Wang S.H., Chao H.J., Liu S.J., Zhou N.Y. Biochemical and molecular characterization of the gentisate transporter GenK in Corynebacterium glutamicum. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0038701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chaudhry M.T., Huang Y., Shen X.H., Poetsch A., Jiang C.Y., Liu S.J. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology (Read) 2007;153(Pt 3):857–865. doi: 10.1099/mic.0.2006/002501-0. [DOI] [PubMed] [Google Scholar]
- 105.Rios Miguel A.B., Jetten M.S.M., Welte C.U. The role of mobile genetic elements in organic micropollutant degradation during biological wastewater treatment. Water Res X. 2020;9:100065. doi: 10.1016/j.wroa.2020.100065. Published 2020 Aug 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liang P., Zhang Y., Xu B. Deletion of genomic islands in the Pseudomonas putida KT2440 genome can create an optimal chassis for synthetic biology applications. Microb Cell Factories. 2020;19(1):70. doi: 10.1186/s12934-020-01329-w. Published 2020 Mar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Segura A., Molina L., Ramos J.L. Plasmid-Mediated tolerance toward environmental pollutants. Microbiol Spectr. 2014;2(6) doi: 10.1128/microbiolspec.PLAS-0013-2013. [DOI] [PubMed] [Google Scholar]
- 108.Tan H.M. Bacterial catabolic transposons. Appl Microbiol Biotechnol. 1999;51(1):1–12. doi: 10.1007/s002530051356. [DOI] [PubMed] [Google Scholar]
- 109.Liu L., Yang H., Shin H.D. How to achieve high-level expression of microbial enzymes: strategies and perspectives. Bioengineered. 2013;4(4):212–223. doi: 10.4161/bioe.24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu R., Liu L., Li X., Liu D., Yuan Y. Engineering yeast artificial core promoter with designated base motifs. Microb Cell Factories. 2020;19(1):38. doi: 10.1186/s12934-020-01305-4. Published 2020 Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohtsubo Y., Shimura M., Delawary M. Novel approach to the improvement of biphenyl and polychlorinated biphenyl degradation activity: promoter implantation by homologous recombination. Appl Environ Microbiol. 2003;69(1):146–153. doi: 10.1128/aem.69.1.146-153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y., Tang H., Lin Z., Xu P. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv. 2015;33(7):1484–1492. doi: 10.1016/j.biotechadv.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 113.Hu H., Wang L., Wang W. Regulatory mechanism of nicotine degradation in Pseudomonas putida. mBio. 2019;10(3) doi: 10.1128/mBio.00602-19. e00602-19. Published 2019 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang H., Wang L., Wang W. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 2013;9(10) doi: 10.1371/journal.pgen.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Q., Wang X., Yin G. New metabolites in dibenzofuran cometabolic degradation by a biphenyl-cultivated Pseudomonas putida strain B6-2. Environ Sci Technol. 2009;43(22):8635–8642. doi: 10.1021/es901991d. [DOI] [PubMed] [Google Scholar]
- 116.Tomas C.A., Welker N.E., Papoutsakis E.T. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl Environ Microbiol. 2003;69(8):4951–4965. doi: 10.1128/aem.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu Z., Yang J., Yang P., Wu Z., Zhang J., Du G. Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Microb Cell Factories. 2019;18(1):136. doi: 10.1186/s12934-019-1188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou P., Yuan X., Liu H., Qi Y., Chen X., Liu L. Candida glabrata Yap6 recruits Med2 to alter glycerophospholipid composition and develop acid pH stress resistance. Appl Environ Microbiol. 2020;86(24):e01915–e01920. doi: 10.1128/AEM.01915-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang J., Zhang B., Zhang J. Enhanced succinic acid production and magnesium utilization by overexpression of magnesium transporter mgtA in Escherichia coli mutant. Bioresour Technol. 2014;170:125–131. doi: 10.1016/j.biortech.2014.07.081. [DOI] [PubMed] [Google Scholar]
- 120.Frederix M., Hütter K., Leu J. Development of a native Escherichia coli induction system for ionic liquid tolerance. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Foo J.L., Jensen H.M., Dahl R.H. Improving microbial biogasoline production in Escherichia coli using tolerance engineering. mBio. 2014;5(6) doi: 10.1128/mBio.01932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Krajewski S.S., Joswig M., Nagel M., Narberhaus F. A tricistronic heat shock operon is important for stress tolerance of Pseudomonas putida and conserved in many environmental bacteria. Environ Microbiol. 2014;16(6):1835–1853. doi: 10.1111/1462-2920.12432. [DOI] [PubMed] [Google Scholar]
- 123.Chen X., Wang W., Hu H. Insights from comparative proteomic analysis into degradation of phenanthrene and salt tolerance by the halophilic Martelella strain AD-3. Ecotoxicology. 2020 doi: 10.1007/s10646-020-02310-4. 10.1007/s10646-020-02310-4. [DOI] [PubMed] [Google Scholar]
- 124.Du W., Brown J.R., Sylvester D.R. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J Bacteriol. 2000;182(15):4146–4152. doi: 10.1128/jb.182.15.4146-4152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yuan Y., Bi C., Nicolaou S.A., Zingaro K.A., Ralston M., Papoutsakis E.T. Overexpression of the Lactobacillus plantarum peptidoglycan biosynthesis murA2 gene increases the tolerance of Escherichia coli to alcohols and enhances ethanol production. Appl Microbiol Biotechnol. 2014;98(19):8399–8411. doi: 10.1007/s00253-014-6004-0. [DOI] [PubMed] [Google Scholar]
- 126.Sandoval N.R., Papoutsakis E.T. Engineering membrane and cell-wall programs for tolerance to toxic chemicals: beyond solo genes. Curr Opin Microbiol. 2016;33:56–66. doi: 10.1016/j.mib.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Qi Y., Liu H., Chen X., Liu L. Engineering microbial membranes to increase stress tolerance of industrial strains. Metab Eng. 2019;53:24–34. doi: 10.1016/j.ymben.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 128.Wang H., Li Q., Peng Y. Cellular analysis and comparative transcriptomics reveal the tolerance mechanisms of Candida tropicalis toward phenol. Front Microbiol. 2020;11:544. doi: 10.3389/fmicb.2020.00544. Published 2020 Apr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mingardon F., Clement C., Hirano K. Improving olefin tolerance and production in E. coli using native and evolved AcrB. Biotechnol Bioeng. 2015;112(5):879–888. doi: 10.1002/bit.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Adebusuyi A.A., Foght J.M. The EmhABC efflux pump in Pseudomonas fluorescens LP6a is involved in naphthalene tolerance but not efflux. Appl Microbiol Biotechnol. 2013;97(6):2587–2596. doi: 10.1007/s00253-012-4373-9. [DOI] [PubMed] [Google Scholar]
- 131.Suo Y., Luo S., Zhang Y., Liao Z., Wang J. Enhanced butyric acid tolerance and production by Class I heat shock protein-overproducing Clostridium tyrobutyricum ATCC 25755. J Ind Microbiol Biotechnol. 2017;44(8):1145–1156. doi: 10.1007/s10295-017-1939-7. [DOI] [PubMed] [Google Scholar]
- 132.Lee S., Lee J.H., Mitchell R.J. Analysis of Clostridium beijerinckii NCIMB 8052's transcriptional response to ferulic acid and its application to enhance the strain tolerance. Biotechnol Biofuels. 2015;8:68. doi: 10.1186/s13068-015-0252-9. Published 2015 Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fiocco D., Capozzi V., Goffin P., Hols P., Spano G. Improved adaptation to heat, cold, and solvent tolerance in Lactobacillus plantarum. Appl Microbiol Biotechnol. 2007;77(4):909–915. doi: 10.1007/s00253-007-1228-x. [DOI] [PubMed] [Google Scholar]
- 134.Yu H.Q., Wang Y.G., Yong T.M., She Y.H., Fu F.L., Li W.C. Heterologous expression of betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus confers high salt and heat tolerance to Escherichia coli. Gene. 2014;549(1):77–84. doi: 10.1016/j.gene.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 135.Takagi H., Taguchi J., Kaino T. Proline accumulation protects Saccharomyces cerevisiae cells in stationary phase from ethanol stress by reducing reactive oxygen species levels. Yeast. 2016;33(8):355–363. doi: 10.1002/yea.3154. [DOI] [PubMed] [Google Scholar]