Summary
While isolating immune cells from spleens and lungs is routinely achieved using flow cytometry, it is challenging to isolate viable immune cells from skin. Here, we describe a step-by-step protocol for skin digestion using a murine melanoma model, which is amenable for detection of low abundant immune cell populations including group 2 innate lymphoid cells.
Subject areas: Cancer, Cell isolation, Flow Cytometry/Mass Cytometry, Immunology, Model Organisms
Graphical abstract
Highlights
-
•
Optimized skin and melanoma digestion protocol for immune phenotyping
-
•
Detection of ILC2s and other immune cells isolated from skin and melanoma samples
-
•
Detailed gating strategies to identify ILC2s, T cells, and myeloid cell subsets
While isolating immune cells from spleens and lungs is routinely achieved using flow cytometry, it is challenging to isolate viable immune cells from skin. Here, we describe a step-by-step protocol for skin digestion using a murine melanoma model, which is amenable for detection of low abundant immune cell populations including group 2 innate lymphoid cells.
Before you begin
Melanoma induction using 4-hydroxytamoxifen
Timing: 30 min–1 h
BRAFV600E/PTENnull/null melanoma transgenic mouse model
The BRAFV600E/PTENnull/null melanoma transgenic mouse model faithfully recapitulates human cutaneous melanoma, with the presence of pigmented lesions (moles) which progress to metastatic melanomas (Dankort et al., 2009, Miller and Mihm, 2006). This melanoma model was designed to allow 4-hydroxytamoxifen (4-HT) inducible melanocyte-targeted BRAFV600E expression and simultaneous PTEN inactivation. This is made possible due to the presence of the 4-HT-inducible Cre recombinase-mutated estrogen receptor fusion transgene which is under the control of the tyrosinase promoter (Tyr::CreER). Given that this is an inducible melanoma model, not requiring injection of melanoma cells into the mouse, it can serve as a powerful tool to study the immune cells which are present as soon as the pigmented lesions are visible.
A challenge in immune phenotyping of melanomas early in the development of this disease is the digestion of the skin and melanoma using enzymes such as collagenase, dispase and liberase. Certain enzymes affect the expression of cell surface markers that decorate immune cells (Autengruber et al., 2012), and may reduce the viability of specific immune cell types in the skin. To overcome these challenges, we developed a protocol for skin tumor processing and digestion, which can be used for leukocyte phenotyping of skin and melanoma. The isolated single cells from the step-by-step digestion protocol presented here can be used for subsequent multiparameter flow cytometry to enumerate and define distinct functions of numerous cell types, including T cells and myeloid cells, but also for the detection of very low abundant cell types including ILC2s (Salimi et al., 2013).
-
1.
Sedate 6–8 week old male mice with 3% Isoflurane and oxygen (Figure 1A)
-
2.
While sedated, use an electric shaver to shave the fur off the lower back (Figure 1B)
-
3.
Using a cotton swab, apply hair depilatory cream on to the shaved skin and massage the cream for maximum contact. Leave it on for 2–3 min to remove the fur. (Figure 1C)
-
4.
Use a moist cotton gauze to completely remove the depilatory cream (Figure 1D)
-
5.
Use a dry cotton gauze to dry the skin (Figure 1E)
-
6.
Place mouse in cage under heating lamp to recover and proceed to work on the next mouse (Figure 1F)
-
7.
Allow mice to recover for at least 12 h
-
8.
The following morning, take the 4-HT aliquot (5 mM) out of the freezer to thaw on ice
-
9.
While handling the mouse by the base of the tail, allow the mouse to grab the cage grill
Note: The mouse may be sedated with 3% isoflurane if the operator is uncomfortable handling mice while awake
-
10.
Add 2 μL 4-HT (5 mM) to the shaved skin (Figure 1G)
-
11.
Use the side of the same pipette tip to spread the 4-HT around the exposed skin (Figure 1H)
-
12.
Allow the 4-HT to dry until no visible drug is left on the skin
-
13.
Place mouse back into the cage and work on the next mouse
-
14.
Repeat steps 8–13 for the next 2 days, such that the 4-HT is applied for three consecutive days
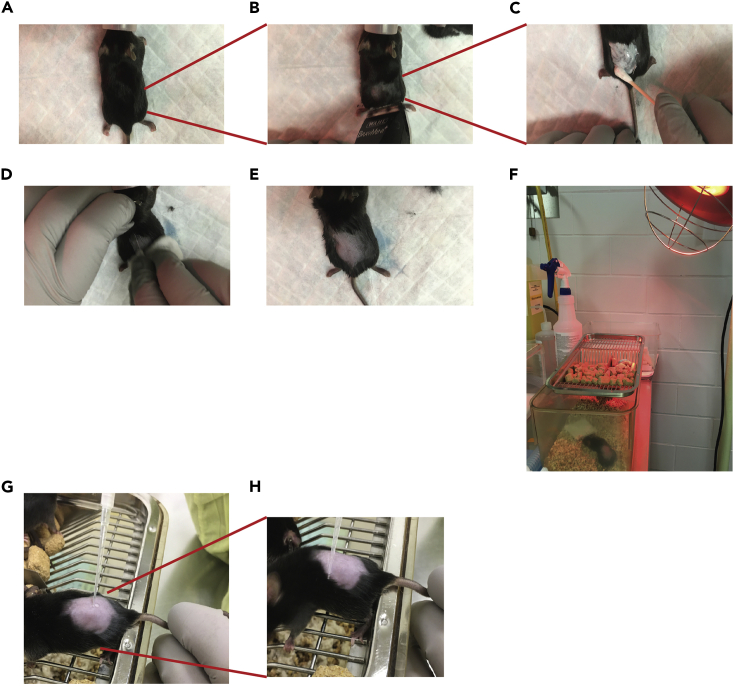
Figure 1.
Application of 4-Hydroxytamoxifen (4-HT) for the induction of Melanoma formation
(A) Mice are sedated with 3% Isoflurane.
(B) Mice are shaved at the lower back near the base of tail.
(C) Hair depilatory cream is applied for 2–3 min to remove residual fur.
(D) Wet swab used to remove the cream.
(E) Dry and inspect skin for any residual fur.
(F) Place mice back in the cage with a heating lamp to allow recovery.
(G) Using a pipette tip, add 2μl of 5mM 4-HT to the shaved, exposed skin.
(H) Use the side of a pipette tip to spread 4-HT on the skin and allow it to dry before placing the mouse back into the cage
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GATA3 BV711 (clone L50-823) | BD Biosciences | Cat #565449 |
| Mouse monoclonal anti-IgG1,κ Isotype Control BV711 (clone X40) | BD Biosciences | Cat #563044 |
| Mouse monoclonal anti-ST2 PerCP-eFluor 710 (clone RMST2-2) | eBioscience | Cat #46-9335 |
| Mouse monoclonal anti-IgG2,κ Isotype Control PerCP-eFluor 710 (clone eBR2a) | eBioscience | Cat #46-4321-80 |
| Mouse monoclonal anti-CD127 BV650 (clone A7R34) | BioLegend | Cat #135043 |
| Mouse monoclonal anti-IgG2α,κ Isotype Control BV650 (clone RTK2758) | BioLegend | Cat # 400542 |
| Mouse monoclonal anti-KLRG1 AF488 (clone 2F1) | BD Biosciences | Cat #561619 |
| Mouse monoclonal anti-CD45.2 BV785 (clone 30-F11) | BioLegend | Cat #103149 |
| Mouse monoclonal anti-ICOS APC (clone C398.4A) | eBioscience | Cat #17-9949-82 |
| Mouse monoclonal anti-Thy 1(CD90.2) BV510 (clone 53-2.1) | BioLegend | Cat #140319 |
| Mouse monoclonal anti-CD25 PE-Cy7 (clone PC61) | BD Biosciences | Cat #561780 |
| Mouse monoclonal anti-FcεR1 PE (clone MAR-1) | eBioscience | Cat #12-5898-83 |
| Mouse monoclonal anti-CD19 PE (clone eBioID3) | eBioscience | Cat #12-0193-82 |
| Mouse monoclonal anti-CD3ε PE (clone 145-2C11) | eBioscience | Cat #12-0031-85 |
| Mouse monoclonal anti-B220 PE (Clone RA3-6B2) | eBioscience | Cat #12-0452-83 |
| Mouse monoclonal anti-TCRβ PE (clone H57-597) | eBioscience | Cat #12-5961-83 |
| Mouse monoclonal anti-TCRγ/δ PE (clone EBioGL3) | eBioscience | Cat #12-5711-82 |
| Mouse monoclonal anti-NK1.1 PE (clone PK136) | eBioscience | Cat #12-5941-82 |
| Mouse monoclonal anti-Gr-1 eF450 (clone RB6-8C5) | eBioscience | Cat #48-5931-82 |
| Mouse monoclonal anti-TER-119 eF450 (clone TER-119) | eBioscience | Cat #48-5921-82 |
| Mouse monoclonal anti-CD11c eF450 (clone N418) | eBioscience | Cat #48-0114-82 |
| Mouse monoclonal anti-CD11b eF450 (clone M1/70) | eBioscience | Cat #48-0112-82 |
| Mouse monoclonal anti-CD45.2 APC-Cy7 (clone 30-F11) | BD Biosciences | Cat #561037 |
| Mouse monoclonal anti-CD3ε BV650 (clone 145-2C11) | BD Biosciences | Cat #563044 |
| Mouse monoclonal anti-CD8α PerCP-Cy5.5 (clone 53-6.7) | BD Biosciences | Cat #561109 |
| Mouse monoclonal anti-CD4 PE-Cy7 (clone GK1.5) | BioLegend | Cat #100-422 |
| Mouse monoclonal anti-CD11c BV786 (clone HL3) | BD Biosciences | Cat #563735 |
| Mouse monoclonal anti-F4/80 PE (clone BM8) | eBioscience | Cat #12-4801-80 |
| Mouse monoclonal anti-Ly-6G/Ly-6C AF488 (clone RB6-8C5) | eBioscience | Cat #53-5931-82 |
| Mouse monoclonal anti-Ly-6C APC (clone AL-21) | Cat #560595 | |
| Mouse monoclonal anti-FOXP3 PE-eF610 (clone FJK-16s) | eBioscience | Cat #61-5773 |
| Mouse monoclonal anti-Arginase-1 AF700 (clone AlexF5) | eBioscience | Cat #56-3697 |
| Mouse monoclonal anti-CD206 BV711 (clone C068C2) | BioLegend | Cat #141727 |
| Chemicals, peptides, and recombinant proteins | ||
| Isofluorane USP | Fresenius Kabi | Cat #CP0406V2 |
| 4-Hydroxytamoxifen (4-HT) | Millipore Sigma | Cat #H6278 |
| 1× Dulbecco’s phosphate-buffered saline (DPBS) | Wisent Bioproducts | Cat #311-425-CL |
| Dulbecco’s modified Eagle media (DMEM) | Wisent Bioproducts | Cat #319-005-CL |
| Collagenase P | Roche | Cat #11213865001 |
| Deoxyribonuclease I (DNase I) | Sigma-Aldrich | Cat #D5025-15KU |
| Fetal bovine serum | Wisent Bioproducts | Cat #080-150 |
| LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit | Invitrogen | Cat #L10119 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Invitrogen | Cat #L34965 |
| OneComp eBeadsTM Compensation Beads | Invitrogen | Cat ##01-1111 |
| UltraComp eBeadsTM Compensation Beads | Invitrogen | Cat #01-2222 |
| FOXP3 / Transcription Factor Staining Buffer | eBioscience | Cat #00-5523-00 |
| Mouse FcR Blocking Reagent | Miltenyi Biotec | Cat #130-092-575 |
| Experimental models: Organisms/strains | ||
| Male C57BL/6 mouse with the BRafV600E, Tyr::CreER and Ptenlox4-5 genotype (6 to 8 weeks old) | Dankort D. et al., 2009 | Gift from Dr Dankort |
| Software and algorithms | ||
| FlowJo™ v10.7 | Becton Dickinson | SCR_008520 |
| Other | ||
| Depilatory cream | Nair | Cat # 6170022261 |
| Electric shaver | Wahl | Cat # 1590-1300 |
| Cotton gauze | AMD Ritmed | Cat # A3007 |
| Cotton swab | Medicom | Cat # 807 |
| P2 micropipette | Gilson | Cat # F144801 |
| 2 μL Pipette tips | SARSTEDT | Cat # 70.113 |
| 15 mL Tubes | Falcon | Cat # 352096 |
| 50 mL Tubes | Falcon | Cat # 352070 |
| 100 mm Culture dishes | Ultident | Cat # 229621 |
| 6-Well plate | Corning | Cat # 353046 |
| Cell strainer 70 μM | Falcon | Cat # 352350 |
| Cell strainer 50 μM | Sysmex Celltrics | Cat # 04-0042-2317 |
| 5 mL Syringe | BD Biosciences | Cat # 309646 |
| Pipette-Aid | Mandel | Cat # TM-155000 |
| 25 mL Pipette | SARSTEDT | Cat # 86.1685.001 |
| 5 mL Round-bottom tube | Falcon | Cat # 352008 |
| U-bottom 96-well plate | SARSTEDT | Cat # 83.3925 |
| BD LSR Fortessa | BD Biosciences | N/A |
Materials and equipment
Note: We use 5% FBS for our FACS buffer, however other labs use 2% FBS for their staining, which is acceptable(Duerr, 2016). Furthermore, the amount of FACS buffer prepared can be scaled down according to your needs.
Flow cytometer
The panels used in this protocol were designed to be used on the BD LSRFortessa flow cytometer with the following configuration: 2 channels for the blue 488 nm laser (Alexa Fluor 488, PerCP-Cy5.5, and PerCP-eFluor710), 3 channels for the red 640 nm laser (APC, Alexa Fluor 700, APC-Cy7, and Near IR), 5 channels for the violet laser 405 nm (Aqua, eFluor450, Brilliant Violet 510, Brilliant Violet 650, Brilliant Violet 711, Brilliant Violet 785, and Brilliant Violet 786), 3 channels for the yellow-green 561 nm laser (PE, PE-eFluor610, and PE-Cy7).
Step-by-step method details
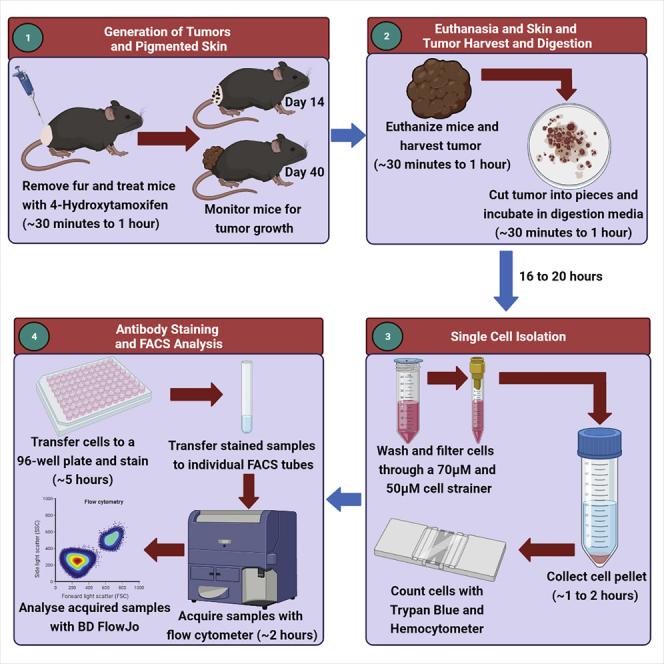
Tumor harvest, digestion, and single-cell isolation
Timing: 2 Days
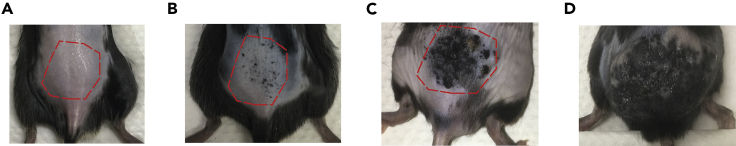
Pigmented lesions will start to form on the backs of mice between days 12–15 after the last 4-HT treatment (Figures 2A and 2B). The pigmented lesions will progress to a measurable melanoma between days 24–28 (Figure 2C). The humane endpoint is set at day 50 (Figure 2D). In our experience, 100% of the BRAFV600E/PTENnull/null mice painted with 4-HT develop melanoma and outgrow at a similar rate (Huang et al., 2021). An advantage of this optimized digestion protocol is that we can isolate viable immune cells on day 7, prior to the generation of pigmented lesions on the backs of mice (Figures 2A). The latter enables us to characterize potential rare immune cell populations such as ILC2s, which are known to be “first-responder” immune cells at the site of inflammation.
Figure 2.
Melanoma progression following 4-HT treatment
(A) Day 7 following 4-HT treatment. During this timepoint, no pigmented lesions yet visible.
(B) Day 12 – 14 following 4-HT treatment. During this timepoint, pigmented lesions will start to form.
(C) Day 24 – 28 following 4-HT treatment. During this timepoint, measurable tumors will start to form.
(D) Any time after Day 40 following 4-HT treatment, mice are monitored closely for human endpoints.
Prior to harvesting the skin or tumor, it is recommended to shave the mice. We determined that cutting the skin tumors with a razor into thin strips and then into 1–2 mm2 squares resulted in optimal cell viability and numbers.
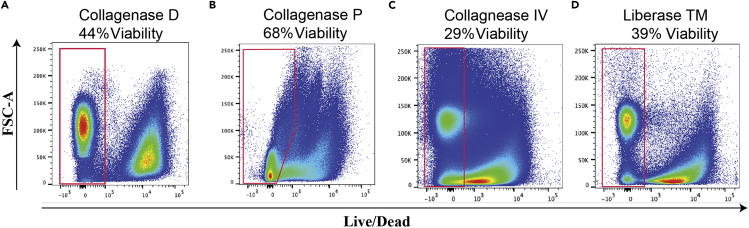
By optimizing the skin digestion protocol, we determined Collagenase P to be the optimal digestion enzyme. We also tested Collagenase IV, Collagenase D, Dispase I, and Liberase TM but found that the obtained cell viabilities and counts were very low (~30% viable cells). Cell viability and yield further improved when tissues were digested for 16–20 h at 4°C, compared to 1 h at 37°C (Figures 3A–3D). It is important to note that this is different from our routinely used lung digestion protocol which employs Liberase TM and DNase for 45 min at 37°C. We opted to not to include a red blood cell (RBC) lysis buffer, as we observed it to decrease cell viability. However, all of our staining procedures included intracellular protein detection using fixation and permeabilization reagents that lyse RBCs. As such, removal of RBC was ensured.
-
1.
Euthanize 4-HT treated mice using approved institutional guidelines, typically through 5% isoflurane and carbon dioxide asphyxiation and confirmation of death by cervical dislocation (Figure 4A)
-
2.
Shave off any hair from the tumor and peripheral area (Figure 4B)
-
3.
Using a pair of scissors and forceps, cut the tumor around the periphery, minimizing the amount of non-tumor bearing skin (Figure 4C)
Note: The tumour bearing skin will typically separate from the body easily. If not, place the scissors under the tumour and open the scissors repeatedly to separate the tumour from the body by removing the connective tissue
-
4.
Harvest the tumor bearing skin and weigh it
-
5.
Place the tumor in a 50 mL conical tube containing FACS buffer (Figure 4D) (Table 1)
-
6.
Place tube on ice and work on the next mouse
-
7.
Place tumor on a 100 mm dish on ice (Figure 4E)
-
8.
Using a single-use blade, cut the tumor into small strips (Figure 4F)
-
9.
From the strips, cut the tumor into approximately 1–2 mm2 squares (Figure 4G)
CRITICAL: We have tried mincing the tissues with scissors to form a paste but determined that this results in a great reduction in cell viability. Instead, we established that cutting tissues into strips and squares yields a cell preparation with significantly increased viability.
-
10.
Place the cut tumor in a 6-well plate with 5 mL tumor digestion media (Figure 4H) (Tables 2 and 3)
-
11.
Place the 6-well plate on a rocker in a cold room for 16–20 h (Figure 4I)
-
12.
The following morning transfer the contents of each well to an individual 50 mL conical tube overlaid with a 70 μM cell strainer (Figure 4J)
-
13.
Add 2 mL DMEM with 10% FBS to the wells to collect any remaining single cells and transfer to the same cell strainer (Table 2)
-
14.
Use a 5 mL syringe plunger to further crush the tissue pieces on the 70 μM strainer, 3 times (Figure 4K)
-
15.
Rinse the cell strainer with 5 mL DMEM plus 10% FBS between each tissue crush
-
16.
Place the 50 mL conical tube in an ice box while working on the next samples
-
17.
Once all samples have been processed as in steps 12–15, place the 50 mL tubes in a centrifuge and spin at 450 × g for 5 min at 4°C
-
18.
Decant the supernatant and resuspend the pellet with 5 mL DMEM with 10% FBS
-
19.
Transfer the resuspended contents to a 15 mL conical tube containing a 50 μM cell strainer (Figure 4L)
-
20.
Place the tubes in a centrifuge and spin at 450 × g for 5 min at 4°C
-
21.
Decant the supernatant and resuspend the pellet with 1 mL DMEM with 10% FBS
-
22.
Count cells using a hemocytometer and trypan blue
CRITICAL: We have observed cell viability between 60%–70% with our protocol. Having said that, we may occasionally observe viability between 40%–50% and we still acquired data from those samples. However, we do not recommend staining samples with viabilities lower than 40% viability, as we have experienced difficulties identifying rare cell populations such as ILC2s.
Figure 3.
Single cell suspensions of melanomas were prepared using distinct enzymatic treatments and cell viabilities were determined by flow cytometric analysis.
(A) Collagenase D, (B) Collagenase P, (C) Collagenase IV at 30°C for 1 h, (D) Liberase TM
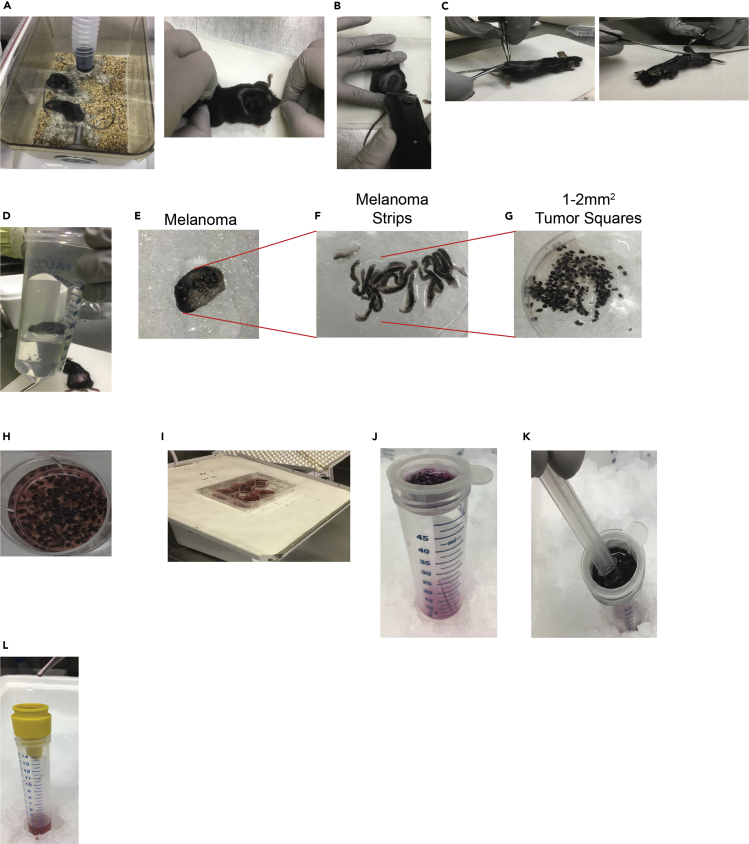
Figure 4.
Skin and melanoma harvest
(A) Euthanize mice according to institutional guidelines, typically by sedation with 5% Isoflurane, followed by asphyxiation and physical confirmation of death by cervical dislocation.
(B) Shave fur from tumor sites and peripheral area.
(C) Harvest tumors using scissors and forceps.
(D) Place tumors in cold PBS.
(E) Place tumors in 10cm dishes on ice.
(F) Cut tumors into strips.
(G) Cut strips into 1–2 mm2 squares.
(H) Place tumors squares into a 6-well plate and add 5 mL digestion media.
(I) Place plate containing tumor squares on a rocker in a cold room and incubate for 16–20 h.
(J) Place tumors and digestion media through a 70μM cell strainer.
(K) A 3 mL or 5 mL syringe plunger is used to mash the tumor on the strainer and wash the strainer twice with DMEM containing 5% FBS.
(L) Filter the tumor and media through a 50μM cell strainer, wash, and count cells with Trypan Blue and a hemocytometer.
Table 1. Fluorescence-Activated Cell Sorting (FACS) Buffer Recipe
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× Dulbecco’s Phosphate-Buffered Saline (DPBS) | N/A | 475 mL |
| Fetal Bovine Serum (FBS) | 5% | 25 mL |
| Total | n/a | 500 mL |
Store at 4°C for up to 1 month.
Table 2. Dulbecco’s Modified Eagle Media (DMEM) with 10% FBS Recipe
| Reagent | Final concentration | Amount |
|---|---|---|
| Dulbecco’s Modified Eagle Media (DMEM) | N/A | 450 mL |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 1 month.
Table 3. Tissue Digestion Media Recipe
| Reagent | Final concentration | Amount |
|---|---|---|
| Dulbecco’s Modified Eagle Media (DMEM) + 10% FBS | N/A | 5 mL |
| 50 mg/mL Collagenase P | 0.5 mg/mL | 50 μL |
| 1 mg/mL Deoxyribonuclease I (DNase I) | 10 μg/mL | 50 μL |
| Total | N/A | 5.1 mL |
Store at 4°C for up to 1 month
Characterization of ILC2 cells over the time course of melanoma progression
Timing: 5 h
After acquiring the single cells from the digested tumor, ILC2s are characterized by flow cytometry using specific markers to the receptors and transcription factors that they express such as CD45, ST2/IL-33R, CD127/IL-7Rα, and GATA3. In addition, ILC2s do not express lineage-specific markers such as TCRβ, TCRγ/δ, Gr-1 (myeloid differentiation marker), NK1.1 (marker of NK cells), FcεR1 (high affinity IgE receptor) and B220/CD45R (marker of B cells), and thus these are useful to discriminate between lineage-positive and lineage-negative populations. To effectively distinguish the various populations, Fluorescence Minus One (FMO) controls, isotype controls and an unstained control are used for proper gating. The titrations listed below are optimized for our institute’s flow cytometer (BD LSRFortessa) and further titration may be required for your institute’s flow cytometer.
Note: 1- Titration of antibodies will be required to determine optimal antibody dilutions for your institute’s flow cytometer. 2- ∗NK1.1 is used only when immune phenotyping tissue from C57BL/6 mice, while CD49b/DX5 is used for BALB/c mice.
-
23.
Prepare antibody lineage cocktail in FACS buffer
-
24.
Add ILC2 extracellular surface antibodies to the cocktail
-
25.
Prepare the GATA3 antibody dilution in FACS buffer in a separate tube for intracellular staining
-
26.
Aliquot 2–4 million cells per sample per well in a 96-well plate for staining with the ILC2 extracellular surface antibody cocktail
Note: Remaining cells from samples once allocated for ILC2 staining can be used for FMO and unstained controls
Note: 2 sets of cells per sample are required for test and isotype controls
-
27.
Wash cells with cold 1× DPBS to remove DMEM at 450 g at 4°C for 5 min.
-
28.
Decant the supernatant in a quick downward motion
-
29.
In the dark, add 50 μL of 1/2000 viability dye to each well, except to the unstained control well.
-
30.
Incubate for 30 min on ice in the dark
-
31.
Add 100 μL cold 1× DPBS to each well
-
32.
Centrifuge at 450 × g for 5 min at 4°C
-
33.
Decant the supernatant in a quick downward motion
-
34.
Add 10 μL of Fc receptor blocking reagent to each well
-
35.
Incubate for 30 min on ice in the dark
-
36.
Without washing the plate, add 25 μL of the ILC2 antibody cocktail directly to the wells
-
37.
Incubate for 30 min on ice in the dark
-
38.
Add 100 μL cold FACS buffer to each well
-
39.
Centrifuge at 450 × g for 5 min at 4°C
-
40.
Decant the supernatant in a quick downward motion
-
41.
Add 100 μL of FOXP3 Fixation/Permeabilization solution (1:4 dilution with Fixation/Permeabilization Diluent) to each well
-
42.
Incubate for 30 min on ice in the dark
-
43.
Add 100 μL cold 1× permeabilization buffer to each well
-
44.
Centrifuge at 600 × g for 5 min at 4°C
-
45.
Decant the supernatant in a quick downward motion
-
46.
Add 10 μL of Fc receptor blocking reagent to each well
-
47.
Incubate for 30 min on ice in the dark
-
48.
Without washing the plate, add 50 μL of the diluted GATA3 antibody directly to the wells
-
49.
Incubate for 30 min at room temperature (20°C–25°C) in the dark
-
50.
Add 150 μL cold 1× permeabilization buffer to each well
-
51.
Centrifuge at 600 × g for 5 min at 4°C
-
52.
Decant the supernatant in a quick downward motion
-
53.
Resuspend in 150 μL cold 1× permeabilization buffer to each well
-
54.
Incubate for 5 min at room temperature (20°C–25°C) in the dark
-
55.
Centrifuge at 600 × g for 5 min at 4°C
-
56.
Decant the supernatant in a quick downward motion
-
57.
Resuspend in 200 μL cold 1× FACS buffer to each well
-
58.
Wrap plate in aluminum foil to protect from light and keep at 4°C
CRITICAL: We optimized our staining panel according to the available fluorophores. If your lab or institute is in a region where it may be difficult to purchase the indicated markers with the above-mentioned fluorophores, we recommend using different fluorophores and adjust and optimize accordingly.
CRITICAL: If your institute’s flow cytometer has an UV laser, we recommend using BUV-conjugated antibodies (e.g.,: Mouse Anti-GATA3 BUV395, BD #565448) and titrate accordingly.
CRITICAL: For endpoint tumors (Day 40), we typically harvest at least 20 million cells. While 2–4 million cells are recommended for the staining, for the Day 14 immune phenotyped samples, no measurable tumor has yet formed. Thus, it is primarily pigmented skin and low cell numbers are recovered. We have added 200,000 cells for our staining and have acquired between 100,000 and 150,000 events via FACSDiva. The end-users of this protocol can pool the skin samples should they wish to stain an increased number of cells.
Characterization of T cells and myeloid cells over the time course of melanoma progression
Timing: 5 h
After acquiring the single cells from the digested tumor, T-cells and myeloid cells are characterized by flow cytometry using specific markers to the receptors and transcription factors that they express, such as CD45, CD3ε, CD11b, CD11c, F4/80 and FOXP3. In this panel we identify: CD3+ cells, CD4+ T-cells, CD8+ T-cells, regulatory T-cells (Tregs), monocytic-myeloid-derived suppressor cells (M-MDSCs) and granulocytic-MDSCs (G-MDSCs), non-lymphoid tissue dendritic cells, myeloid dendritic cells, and macrophages. To effectively distinguish the various populations, Fluorescence Minus One (FMO) controls, isotype controls and an unstained control are used for proper gating. This protocol applies to C57BL/6 mice, but as already specified in Major Step 2, it can be applied to BALB/c mice with one modification: instead of using the NK1.1 antibody, the CD49b/DX5 antibody is instead used for staining immune cells from BALB/c mice (Table 4). The titrations listed below are optimized for our institute’s flow cytometer (BD LSRFortessa) and further titration may be required for your institute’s flow cytometer.
CRITICAL: We optimized our staining panel according to the available fluorophores. If your lab or institute is in a region where it may be difficult to purchase those markers with the above-mentioned fluorophores, we recommend using different fluorophores and adjust and optimize accordingly.
CRITICAL: If your institute’s flow cytometer has an UV laser, we recommend using BUV-conjugated antibodies (e.g.,: CD45 BUV395, BD #264279) and titrate accordingly.
-
59.
Prepare extracellular antibody cocktail (Excluding FOXP3 and Arginase-1) in FACS buffer
-
60.
Prepare FOXP3 and Arginase-1 antibody dilutions in FACS buffer in a separate tube for intracellular staining
-
61.
Aliquot 2–4 million cells per sample per well in a 96-well plate
Note: Remaining cells from samples once allocated for staining can be used for FMO and unstained controls
Note: 2 sets of cells per sample are required for test and isotype controls
-
62.
Wash cells with cold 1× DPBS to remove DMEM at 450 g at 4°C for 5 min.
-
63.
Decant the supernatant in a quick downward motion
-
64.
In the dark, add 50 μL of 1/250 viability dye to each well, except to the unstained control well.
-
65.
Incubate for 30 min on ice in the dark
-
66.
Add 100 μL cold 1× DPBS to each well
-
67.
Centrifuge at 450 × g for 5 min at 4°C
-
68.
Decant the supernatant in a quick downward motion
-
69.
Add 10 μL of Fc receptor blocking reagent to each well
-
70.
Incubate for 30 min on ice in the dark
-
71.
Without washing the plate, add 25 μL of the antibody cocktail directly to the wells
-
72.
Incubate for 30 min on ice in the dark
-
73.
Add 100 μL cold FACS buffer to each well
-
74.
Centrifuge at 450 × g for 5 min at 4°C
-
75.
Decant the supernatant in a quick downward motion
-
76.
Add 100 μL of FOXP3 Fixation/Permeabilization solution (1:4 dilution with Fixation/Permeabilization Diluent) to each well
-
77.
Incubate for 30 min on ice in the dark
-
78.
Add 100 μL cold 1× permeabilization buffer to each well
-
79.
Centrifuge at 600 × g for 5 min at 4°C
-
80.
Decant the supernatant in a quick downward motion
-
81.
Add 10 μL of Fc receptor blocking reagent to each well
-
82.
Incubate for 30 min on ice in the dark
-
83.
Without washing the plate, add 50 μL of the diluted FOXP3 and Arginase-1 antibody directly to the wells
-
84.
Incubate for 30 min at room temperature (20°C–25°C) in the dark
-
85.
Add 150 μL cold 1× permeabilization buffer to each well
-
86.
Centrifuge at 600 × g for 5 min at 4°C
-
87.
Decant the supernatant in a quick downward motion
-
88.
Resuspend in 150 μL cold 1× permeabilization buffer to each well
-
89.
Incubate for 5 min at room temperature (20°C–25°C) in the dark
-
90.
Centrifuge at 600 × g for 5 min at 4°C
-
91.
Decant the supernatant in a quick downward motion
-
92.
Resuspend in 200 μL cold 1× FACS buffer to each well
-
93.
Wrap plate in aluminum foil and keep at 4°C
Pause point: At this step, the fixed and stained cells can be stored at 4°C for a week prior to moving onto the acquisition of samples by flow cytometry. However, it is preferable to acquire within 24–48 h.
Table 5.
List of antibodies used to characterize T-cells and Myeloid Cells
| Antibody | Clone | Fluorochrome | Dilution | Company and Catalog # |
|---|---|---|---|---|
| CD45.2 | 30-F11 | APC-Cy7 | 1/800 | BD #561037 |
| CD3ε | 145-2C11 | Brilliant Violet 650 | 1/12.5 | BD #563044 |
| CD8α | 53-6.7 | PerCP-Cy5.5 | 1/50 | BD #561109 |
| CD4 | GK1.5 | PE-Cy7 | 1/100 | BioLegend #100-422 |
| CD11b | M1/70 | eFluor450 | 1/2000 | eBioscience #48-0112-82 |
| CD11c | HL3 | Brilliant Violet 786 | 1/500 | BD # 563735 |
| F4/80 | BM8 | PE | 1/100 | eBioscience #12-4801-80 |
| Ly-6G/Ly-6C | RB6-8C5 | Alexa Fluor 488 | 1/2000 | eBioscience #53-5931-82 |
| Ly-6C | AL-21 | APC | 3/400 | BD #560595 |
| FOXP3 | FJK-16s | PE-eFluor610 | 1/100 | eBioscience #61-5773 |
| Arginase-1 | A1exF5 | Alexa Fluor 700 | 1/100 | eBioscience #56-3697 |
| CD206 | C068C2 | Brilliant Violet 711 | 1/200 | BioLegend #141727 |
| Viability Dye | - | Aqua | 1/250 | Invitrogen #L34965 |
(Note: Titration of antibodies required to determine an optimal dilution for your institute’s flow cytometer)
Table 4.
List of antibodies used to characterize ILC2s
| Antibody | Clone | Fluorochrome | Dilution | Company and Catalog # |
|---|---|---|---|---|
| ILC2-Specific Markers | ||||
| GATA3 | L50-823 | Brilliant Violet 711 | 1/100 | BD #565449 |
| IgG1, κ Isotype Control | X40 | Brilliant Violet 711 | 1/100 | BD #563044 |
| ST2 | RMST2-2 | PerCP-eFluor710 | 1/50 | eBioscience #46-9335 |
| IgG2, κ Isotype Control | eBR2a | PerCP-eFluor710 | 1/50 | eBioscience #46-4321-80 |
| CD127 | A7R34 | Brilliant Violet 650 | 1/200 | BioLegend #135043 |
| IgG2α, κ Isotype Control | RTK2758 | Brilliant Violet 650 | 1/200 | BioLegend # 400542 |
| KLRG1 | 2F1 | Alexa Fluor 488 | 1/50 | BD #561619 |
| CD45.2 | 30-F11 | Brilliant Violet 785 | 1/2000 | BioLegend #103149 |
| ICOS | C398.4A | APC | 1/50 | eBioscience #17-9949-82 |
| Thy 1 (CD90.2) | 53-2.1 | Brilliant Violet 510 | 1/400 | BioLegend #140319 |
| CD25 | PC61 | PE-Cy7 | 1/100 | BD #561780 |
| Viability Dye | - | Near IR | 1/2000 | Invitrogen #L10119 |
| Lineage Markers | ||||
| FcεR1 | MAR-1 | PE | 1/50 | eBioscience #12-5898-83 |
| CD19 | eBioID3 | PE | 1/100 | eBioscience #12-0193-82 |
| CD3ε | 145-2C11 | PE | 1/400 | eBioscience #12-0031-85 |
| B220 | RA3-6B2 | PE | 1/200 | eBioscience #12-0452-83 |
| TCRβ | H57-597 | PE | 1/200 | eBioscience #12-5961-83 |
| TCRγ/δ | EBioGL3 | PE | 1/400 | eBioscience #12-5711-82 |
| NK1.1∗ | PK136 | PE | 1/50 | eBioscience #12-5941-82 |
| Gr-1 | RB6-8C5 | eFluor450 | 1/500 | eBioscience #48-5931-82 |
| TER-119 | TER-119 | eFluor450 | 1/50 | eBioscience #48-5921-82 |
| CD11c | N418 | eFluor450 | 1/50 | eBioscience #48-0114-82 |
| CD11b | M1/70 | eFluor450 | 1/500 | eBioscience #48-0112-82 |
Lineage markers are included as these provide a lineage-negative population to discriminate ILC2s.
Flow cytometry acquisition
Timing: 2 h (For 5 samples)
This step details the acquisition of our samples using the BD LSRFortessa flow cytometer from our institution. You might have to modify this step to accommodate to your institute’s instruments and guidelines.
-
94.
As the samples are fixed, acquisition of the samples can be acquired within 2 days.
-
95.
Take the plate out of the fridge and transfer the samples to individual 5 mL FACS tubes.
Note: If your instrument has a plate reader attached, you may wish to use it and follow your institute’s core facility’s guidelines.
-
96.
Prepare compensation tubes with each fluorochrome listed above.
Note: For violet laser-excited fluorochromes e.g.,: eFluor450, BV711, BV510, UltraComp compensation beads are recommended due to background autofluorescence. OneComp compensation beads are typically used for blue, red, yellow-green and UV laser-excited fluorochromes.
-
97.
Perform the acquisition using the BD LSRFortessa
Note: You will require an instrument in your institution’s core facility that can accommodate up to a 15-color panel e.g., BD LSRFortessa, BD LSRII.
-
98.
Ensure there is enough sheath fluid in the tank and that the waste tank is not full.
-
99.
Using BD FACSDiva (or the available acquisition software in your institute’s core facility) create a new data acquisition matrix
-
100.
Ensure that all the forward scatter and side scatter parameters are selected (e.g., Area (A), Height (H), and Width (W))
-
101.
Assign each channel to your fluorochromes and delete the rest
-
102.
Prime the instrument twice to remove any debris that may block and clog the instrument during acquisition
-
103.
Perform (Cytometer Setup and Tracking) CST if your institute core facility requires the first user of the day to do so
-
104.
Use your unstained control to set the voltages of your FSC-A, SSC-A and fluorochrome by right-clicking on Cytometer Settings → Application Settings → Create Worksheet, which creates a new global worksheet
-
105.
In the SSC-A and FSC-A plot, draw a gate around the lymphocyte population. In the other plots showing the fluorochromes, make sure the negative population is on the “+” sign
-
106.
Perform compensation using your compensation tubes and set an interval gate for the negative population peak and move your P2 gate on the positive population peak
-
107.
Calculate compensation.
Note: In BD FACSDiva it is Instrument → Instrument Setup → Calculate Compensation. Click “Link & Save”.
-
108.
Click on the little sheet icon to get back to the analysis schematic and set up your gating strategy
-
109.
Acquire all samples, including FMO and unstained controls and ensure that the threshold rate is at 10,000 events/second
-
110.
Once acquisition is completed, export your data according to your institute’s core facility’s guidelines.
Note: Ensure that it is FCS3.0 to allow analysis on the BD FlowJo program.
-
111.
Perform cleaning and shutdown (if last person of the day) procedures according to your institute’s core facility guidelines.
CRITICAL: As ILC2s are a rare cell population, we acquire all the stained cells in the tube and do not set a pre-determined number of events to acquire. We then normalize to CD45+ cells, or as an absolute number of cells from the total cell count, tissue weight, and total cells acquired via FACSDiva.
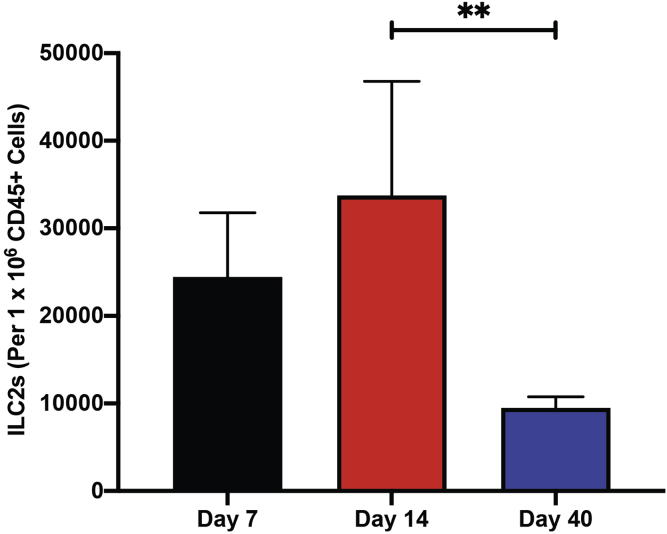
CRITICAL: Depending on the timepoint following 4-HT induction of melanomagenesis, we have a variable range of ILC2s acquired. For Day 7, our range is approximately 10,000–40,000 ILC2s per 1 million CD45+ cells, while at Day 14 it is approximately 3,000–60,000 ILC2s per 1 million CD45+ cells, and at Day 40 it is from 4,000–15,000 ILC2s per 1 million CD45+ cells.
Expected outcomes
From the digestion of the late-stage endpoint tumors, the number of cells will range from 8 million to 10 million cells. However, when working with skin samples from Day 7 and 14 mice (following the last day of 4-HT treatment), we typically count less than 2 million cells. We have included the gating strategies for ILC2s, T-Cells, dendritic cells, MDSCs, and macrophages (Figures 5, 6, 7, 8, 9, and 10 respectively). We have also included the levels of ILC2s, which is a rare population at 3 different timepoints during the course of melanoma development (Days 7, 14 and 40) (Figure 11).
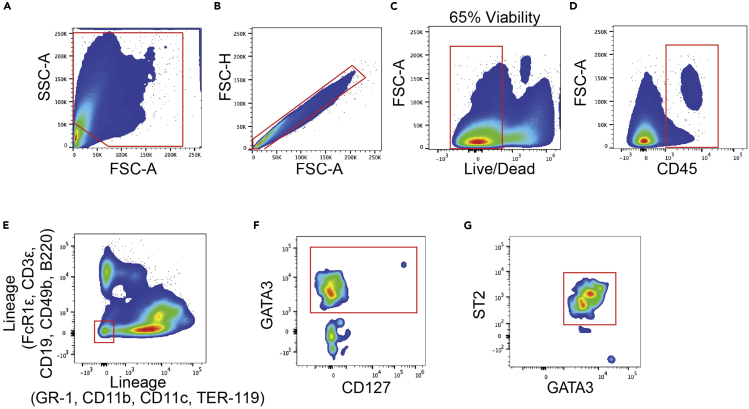
Figure 5.
ILC2 gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) Lineage-negative cell population gating.
(F) GATA3+ cell population gating.
(G) ILC2 were characterized as GATA3+ST2+ cell population.
Figure 6.
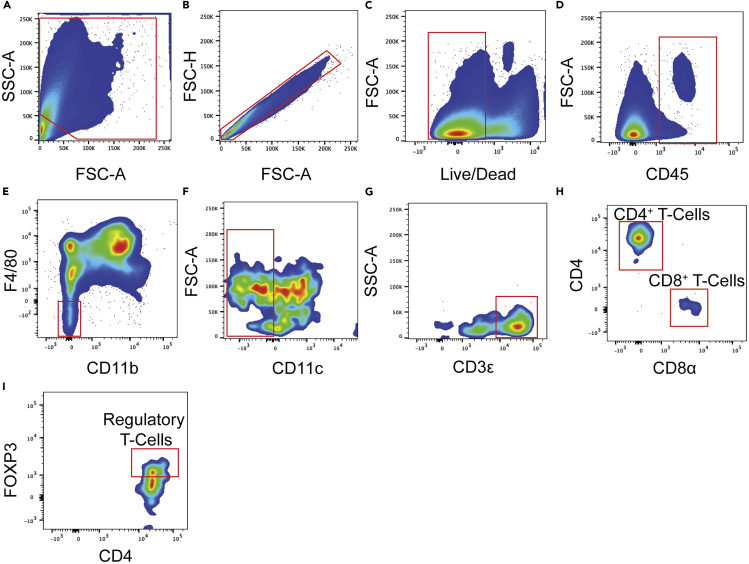
Regulatory T-cells (Tregs) gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) CD11b and F4/80 double-negative gating.
(F) CD11c negative gating.
(G) CD3ε+ T cell gating.
(H) CD4+ T cell and CD8+ T cell gating.
(I) CD4+FOXP3+ gating to identify Tregs
Figure 7.
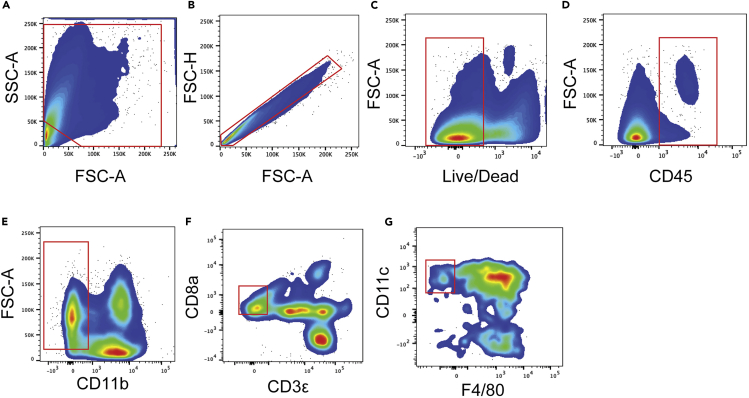
Non-lymphoid tissue dendritic cells (NLT DCs) gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) CD11b negative gating.
(F) CD3ε and CD8ε double negative gating.
(G) F4/80Low, CD11cMid gating to identify NLT DCs.
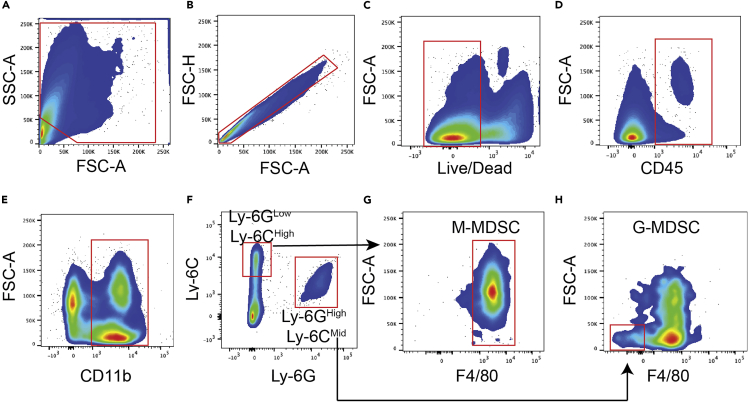
Figure 8.
Myeloid-derived suppressor cells (MDSC) gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) CD11b positive gating.
(F) Ly-6GLow and Ly-6CHigh gating for M-MDSCs and Ly-6GHigh and Ly-6CMid for G-MDSCs.
(G) F4/80High gating to identify M-MDSCs.
(H) F4/80Low gating to identify G-MDSCs
Figure 9.
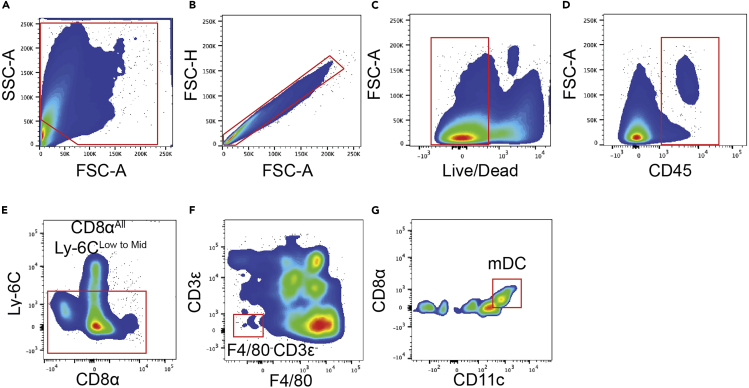
Monocytic tissue dendritic cells (mDCs) gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) CD8αAll and Ly-6CLow gating.
(F) F4/80-CD3ε- for mDCs.
(G) CD8αLow and CD11cMid to High to identify mDCs
Figure 10.
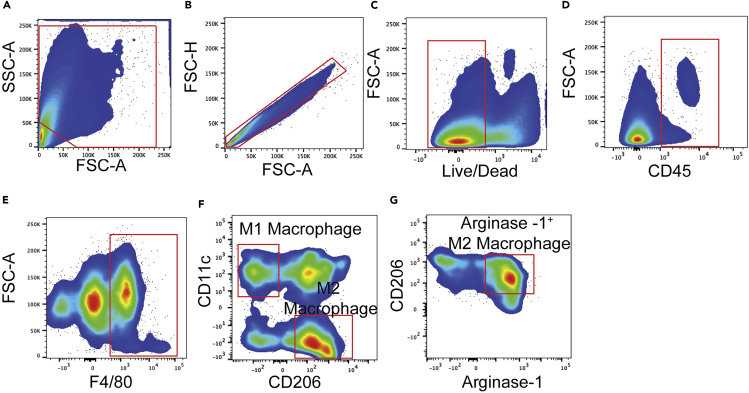
Macrophage gating strategy
(A) Forward and sideward scatter gating to exclude debris.
(B) Singlet cell gating.
(C) Live cell gating.
(D) CD45+ cell gating.
(E) F4/80High population gating.
(F) CD11cHigh and CD206Low gating for M1 Macrophages, and CD11cLow and CD206Mid to High gating for M2 Macrophages.
(G) M2 Macrophages express high levels of Arginase-1.
Figure 11.
Detection of ILC2s at 3 different timepoints during melanoma progression (mouse model shown in Figure 2)
ILC2s reduce in number in melanomas characterized on Day 40, compared to Days 7 and 14. One-way ANOVA. All values are represented as mean ± SD. ∗∗P ≤ 0.01.
Limitations
A limitation of the BRAFV600E/PTENnull/null melanoma mouse model is the development of spontaneous tumors in approximately 10%–15% of male mice per cohort in various anatomical locations. BRAFV600E/PTENnull/null female mice are not used for this protocol, as approximately 40% per cohort will develop spontaneous tumors. While the cell viability of digested late-stage endpoint tumors (i.e., Day 40 following the last day of 4-HT treatment) remained consistently high, one may encounter issues with viability when working with Day 24 tumors. While this protocol shows the markers used to identify skin and tumor ILC2s, care must be taken when attempting to identify ILC2s in the lung, gut, or other parts of the body. While ILC2s express the above-mentioned markers, ILC2s at various sites express different levels of markers (Ricardo-Gonzalez et al., 2018, Meininger et al., 2020, Simoni et al., 2017). Having said that, using a “base” of lineage negative, CD127 and GATA3 to identify total ILC2s can be used and then the other markers can be gated. An alternative is to use Mean Fluorescence Intensity to determine the levels of activation and/or inhibition markers at different timepoints.
Troubleshooting
Problem 1
Pigmented lesions and tumors not formed after 4-Hydroxytamoxifen treatment.
Potential solution 1
Backcrossing of mice with previous generations may be warranted due to genetic drift.
Re-genotype mice to ensure the presence of Tyr::CreER transgene, remove the breeder mice immediately if pubs were detected negative for Tyr::CreER. In addition, all mice used in this study were tested PTENflox/flox and BRAFCA/+. Primer sequences can be found at (Huang et al., 2021).
Problem 2
Low cell count after tumor digestion and single cell isolation.
Potential solution 2
This may be possible especially with early-timepoint tissues such as non-pigmented skin and pigmented skin. Pooling of cells may be warranted.
Problem 3
Low viability after tumor digestion and single cell isolation.
Potential solution 3
Reduce incubation in digestion media for 8–12 h.
Problem 4
No positive staining of markers by flow cytometry.
Potential solution 4
Repeat compensation with new set of single-stained, and unstained controls and adjust PMT voltages.
You may need to use a different fluorophore that might work on your institute’s flow cytometer.
Re-titration of antibodies may be warranted to suit your institute’s flow cytometer.
Problem 5
Intracellular stain cannot be detected.
Potential solution 5
Increase eBioscience FOXP3 Fixation/Permeabilization solution incubation to 1 h and at room temperature instead.
It is possible to use the BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (Catalog #554714) and to follow the manufacturer’s instructions.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sonia del Rincon PhD, soniavictoria.delrincon@mcgill.ca
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze [datasets/code].
Acknowledgments
This research is funded by the Canadian Institutes for Health Research (grant PJT-162260 to S.V.d.R. and J.H.F.). This work was also supported by a grant from McGill Interdisciplinary Initiative in Infection and Immunity (MI4) to S.V.d.R. and a Transition Award from the Cole Foundation to S.V.d.R. S.S.K. receives financial support from the Fonds de la Recherche en Sante du Quebec (FRQS), McGill University Department of Physiology, and the Estate of Max E. Childress (Max E. and Jane K. Childress Entrance Fellowship in Physiology). Animal experiments adhered to the regulations established by the Canadian Council of Animal Care, and protocols were approved by the McGill University Animal Care and Use Committee (2015-7672). We thank Julien Leconte and Camille Stegen from the Life Science Complex (LSC) Flow Cytometry and Cell Sorting Facility at McGill University, as well as Christian Young from the Lady Davis Institute for Medical Research Flow Cytometry Facility for excellent technical support. The LSC facility's infrastructure is supported by the Canada Foundation for Innovation (CFI). Graphical abstract was created with BioRender.com.
Author contributions
J.H.F. and S.V.d.R. designed the study and edited the manuscript. S.S.K. developed the staining panels. S.S.K. and C.G. developed the skin digestion protocol. N.G., F.H., and D.P. maintained the mouse colony and breeding. S.S.K. performed the sample collection, processing, and staining and flow cytometry analysis, drafted the manuscript, and designed the figures.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jörg H. Fritz, Email: jorg.fritz@mcgill.ca.
Sonia V. del Rincon, Email: soniavictoria.delrincon@mcgill.ca.
References
- Autengruber A., Gereke M., Hansen G., Hennig C., Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur. J. Microbiol. Immunol. (Bp) 2012;2:112–120. doi: 10.1556/EuJMI.2.2012.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D., Curley D.P., Cartlidge R.A., Nelson B., Karnezis A.N., Damsky W.E., Jr., You M.J., Depinho R.A., Mcmahon M., Bosenberg M. Braf(V600e) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr Claudia. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nature Immunology. 2016;17:65–75. doi: 10.1038/ni.3308. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Goncalves C., Bartish M., Remy-Sarrazin J., Issa M.E., Cordeiro B., Guo Q., Emond A., Attias M., Yang W., Plourde D. Inhibiting the Mnk1/2-Eif4e axis impairs melanoma phenotype switching and potentiates anti-tumor immune responses. J. Clin. Invest. 2021 doi: 10.1172/JCI140752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meininger I., Carrasco A., Rao A., Soini T., Kokkinou E., Mjosberg J. Tissue-specific features of innate lymphoid cells. Trends Immunol. 2020;41:902–917. doi: 10.1016/j.it.2020.08.009. [DOI] [PubMed] [Google Scholar]
- Miller A.J., Mihm M.C., Jr. Melanoma. N. Engl. J. Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Ricardo-Gonzalez R.R., Van Dyken S.J., Schneider C., Lee J., Nussbaum J.C., Liang H.E., Vaka D., Eckalbar W.L., Molofsky A.B., Erle D.J., Locksley R.M. Tissue signals imprint Ilc2 identity with anticipatory function. Nat. Immunol. 2018;19:1093–1099. doi: 10.1038/s41590-018-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.-C., Johnson D., Scanlon S.T., Mckenzie A.N.J., Fallon P.G., Ogg G.S. A role for Il-25 and Il-33–driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni Y., Fehlings M., Kloverpris H.N., Mcgovern N., Koo S.L., Loh C.Y., Lim S., Kurioka A., Fergusson J.R., Tang C.L. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze [datasets/code].