Abstract
PURPOSE
Variation in risk of adverse clinical outcomes in patients with cancer and COVID-19 has been reported from relatively small cohorts. The NCATS’ National COVID Cohort Collaborative (N3C) is a centralized data resource representing the largest multicenter cohort of COVID-19 cases and controls nationwide. We aimed to construct and characterize the cancer cohort within N3C and identify risk factors for all-cause mortality from COVID-19.
METHODS
We used 4,382,085 patients from 50 US medical centers to construct a cohort of patients with cancer. We restricted analyses to adults ≥ 18 years old with a COVID-19–positive or COVID-19–negative diagnosis between January 1, 2020, and March 25, 2021. We followed N3C selection of an index encounter per patient for analyses. All analyses were performed in the N3C Data Enclave Palantir platform.
RESULTS
A total of 398,579 adult patients with cancer were identified from the N3C cohort; 63,413 (15.9%) were COVID-19–positive. Most common represented cancers were skin (13.8%), breast (13.7%), prostate (10.6%), hematologic (10.5%), and GI cancers (10%). COVID-19 positivity was significantly associated with increased risk of all-cause mortality (hazard ratio, 1.20; 95% CI, 1.15 to 1.24). Among COVID-19–positive patients, age ≥ 65 years, male gender, Southern or Western US residence, an adjusted Charlson Comorbidity Index score ≥ 4, hematologic malignancy, multitumor sites, and recent cytotoxic therapy were associated with increased risk of all-cause mortality. Patients who received recent immunotherapies or targeted therapies did not have higher risk of overall mortality.
CONCLUSION
Using N3C, we assembled the largest nationally representative cohort of patients with cancer and COVID-19 to date. We identified demographic and clinical factors associated with increased all-cause mortality in patients with cancer. Full characterization of the cohort will provide further insights into the effects of COVID-19 on cancer outcomes and the ability to continue specific cancer treatments.
INTRODUCTION
COVID-19 rapidly emerged as a global pandemic affecting access and quality of care, as well as health outcomes.1,2 Patients with cancer were among vulnerable populations shown to be at increased risk of severe disease and death from COVID-19, which has been attributed to the manifestation of cancer itself, interaction with cancer therapies, and the hindrance of cancer care delivery by the pandemic.3-6 Compared with patients without cancer, patients with cancer had higher mortality rates despite receipt of more frequent antiviral and immune-related therapies.7
CONTEXT
Key Objective
Patients with cancer are among vulnerable populations at increased risk of severe outcomes and death from COVID-19. We used the National COVID Cohort Collaborative (N3C) to build a cohort of patients with cancer with and without COVID-19 and examined risk factors for overall mortality from COVID-19.
Knowledge Generated
Older age, male gender, increasing comorbidities, hematologic malignancy, and recent cytotoxic therapy were associated with higher mortality in COVID-19 patients with cancer. COVID-19–positive patients who received recent immunotherapies or targeted therapies did not have significantly higher risks of overall mortality from COVID-19.
Relevance
The N3C is a large-scale national Real-World Data resource that can be leveraged by clinicians and researchers to examine effects of COVID-19 on outcomes in patients with cancer. Further characterization of the curated cancer cohort will provide additional guidance on resource and clinical management of patients by cancer type.
The impact of COVID-19 on cancer outcomes and care delivery has been developing over time, with one of the earliest reports demonstrating more than five-fold increase in the likelihood of severe COVID-19 and death among patients with cancer.8 However, early studies might have suffered from small sample sizes and potential significant confounders.8-13 Subsequent studies reported increased risk of severe infection and death among patients with cancer but provided a more nuanced picture regarding COVID-19 severity, risk of death, and the impact of the cancer type and cancer therapy. For example, an analysis of the Lean European Open Survey on SARS-CoV-2 Infected Patients registry found minimal change in COVID-19 risk among patients with and without cancer after adjusting for age, sex, and comorbidity.14 Studies that have specifically assessed the impact of cancer-related therapy have demonstrated mixed results. The most recent report from the COVID-19 and Cancer Consortium (CCC-19) found a 28% increased risk of COVID-19 severity and 61% increased risk of 30-day mortality with cytotoxic agents; among specific therapies, rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone, platinum-etoposide, and DNA methyltransferase inhibitors were associated with increased 30-day all-cause mortality.15 Conversely, studies like the UK Coronavirus Cancer Monitoring Project and the Gustave Roussy cohort did not find an increased risk of death with immunotherapies, targeted therapies, and hormone therapies after adjusting for age, sex, and comorbidities.16,17 The inconsistencies in the findings could be due to relatively limited cohort sizes, geographical locations, or lack of a COVID-19–negative cancer control.
To address the lack of a national clinical patient registry that could be used to study COVID-19 at scale, the National COVID Cohort Collaborative (N3C) was developed as a centralized repository of electronic health record (EHR) data representing the largest multicenter cohort of COVID-19 cases and controls nationwide.18,19 As of March 25, 2021, 1,077,445 confirmed COVID-19 cases and 3,304,640 controls from 50 contributing sites were available in the N3C platform. Our cohort study involved 398,579 patients with cancer who had any encounter with the health system including the emergency department, outpatient, telehealth, home visits, or inpatient hospitalization, and among whom, 63,413 tested positive for COVID-19. In this report, we aimed to characterize the cohort of patients with cancer within N3C and to identify risk factors associated with all-cause mortality within our cohort.
METHODS
Study Cohort
The N3C clinical cohort was built using broad inclusion criteria to include COVID-19 cases and non-COVID-19 controls of both outpatients and inpatients from contributing sites. At each site, all patients with COVID-19 with any clinical encounter after January 1, 2020, were included in the overall N3C clinical cohort. All patients without COVID-19 were initially included from contributing sites and starting December 2020 were randomly selected from the corresponding site using a 2:1 ratio to match the overall prevalence of age, sex, and race of COVID-19 cases. Historical patient data dating back to January 1, 2018, were included to document existing health conditions within the same health system for both cases and controls. Institutions with multiple source data models (eg, PCORNet, Accruals to Clinical Trials, TriNetX, and Observational Medical Outcomes Partnership [OMOP]) have provided data to the consortium. All data were harmonized, mapped, and deposited into an OMOP common data model (CDM)20 (V5.3.1) following thorough data quality and harmonization checks by the N3C core teams.19
To construct a retrospective cohort of patients with cancer for this study within the larger N3C cohort, we used the Observational Health Data Sciences and Informatics ATLAS tool to search and navigate vocabularies and to build cohort definitions. Our study cohort was built using the Malignant Neoplastic Disease standard concept (SNOMED Code: 363346000). Within this cohort, 4,528 SNOMED Neoplasm Concepts were mapped to N3C Enclave Concepts; benign concepts listed in Appendix Table A1 (online only) were excluded.
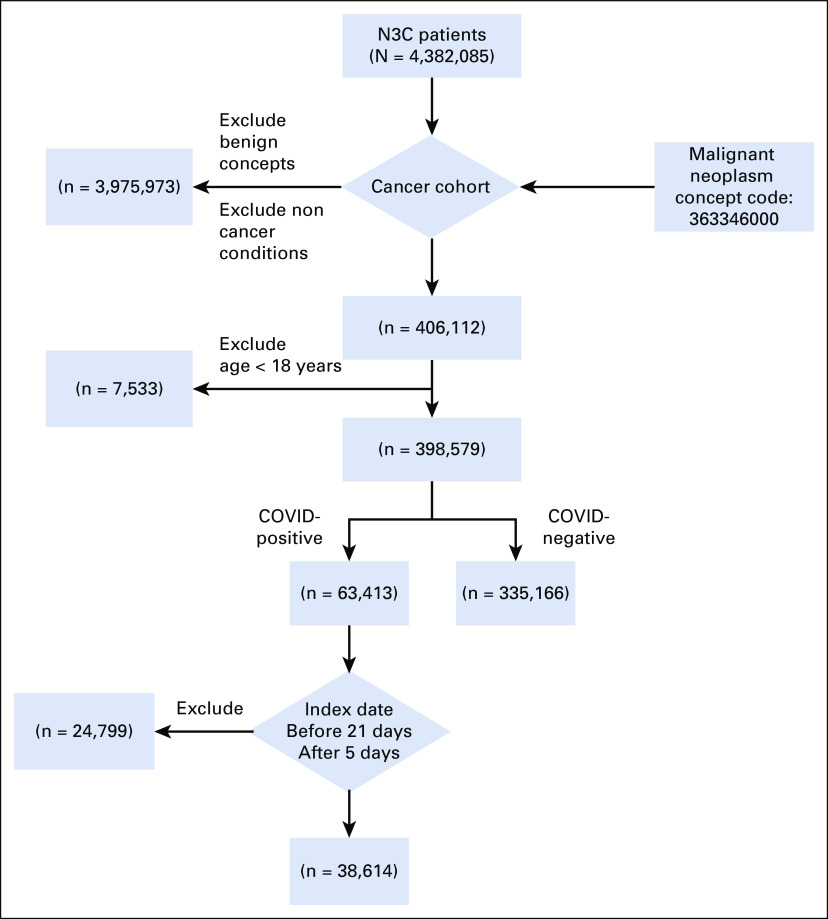
To define COVID-19 status, we followed the N3C phenotyping guidelines to define COVID-19 positivity on the basis of a COVID-19–positive polymerase chain reaction or antigen test, or an International Classification of Diseases (ICD)-10-CM diagnostic code for COVID-19 during the same single index encounter.19 We also used N3C guidelines to identify a single index encounter that represented the critical COVID-19–related visit for each laboratory-confirmed positive patient, hereafter referred to as the index encounter (Appendix 1, online only). We limited our analytic cohort to patients with COVID-19 who had their earliest COVID-19 diagnosis within 21 days before the start of the index encounter and up to 5 days after the start of the index encounter. We restricted the cancer cohort to adult patients ≥ 18 years old with a past or existing primary cancer diagnosis and an index encounter between January 1, 2020, and March 25, 2021 (Fig 1). Finally, because of Health Information Portability and Accountability Act limitations, all age groups > 89 years have been set to 90 years for our analysis.
FIG 1.
Flow diagram for study cohort. N3C, National COVID Cohort Collaborative.
Indicator Variables
The N3C clinical data set is a collection of limited data sets (ie, containing real dates and geographic location) with potential prognostic variables. In our analysis, we included data for age, sex, race and ethnicity, geographical location of patient residence, smoking status, and COVID-19 treatment received. In addition, we used available data to calculate indicator variables on Charlson Comorbidity Index (CCI)21 adjusted for a cancer diagnosis, primary cancer diagnosis, and cancer therapies.
Primary Diagnosis
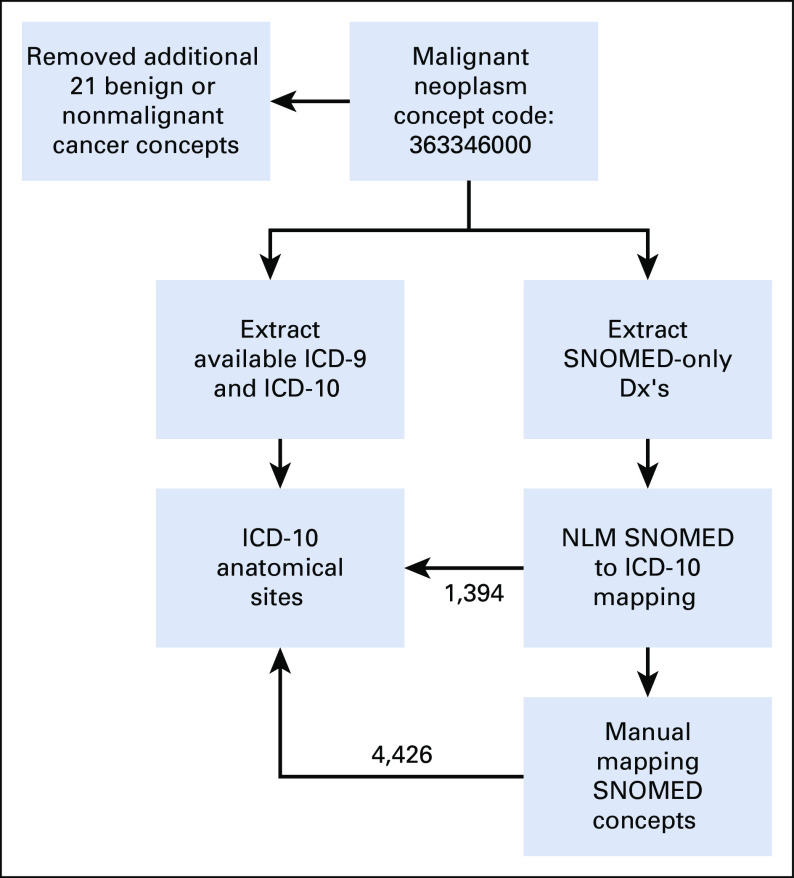
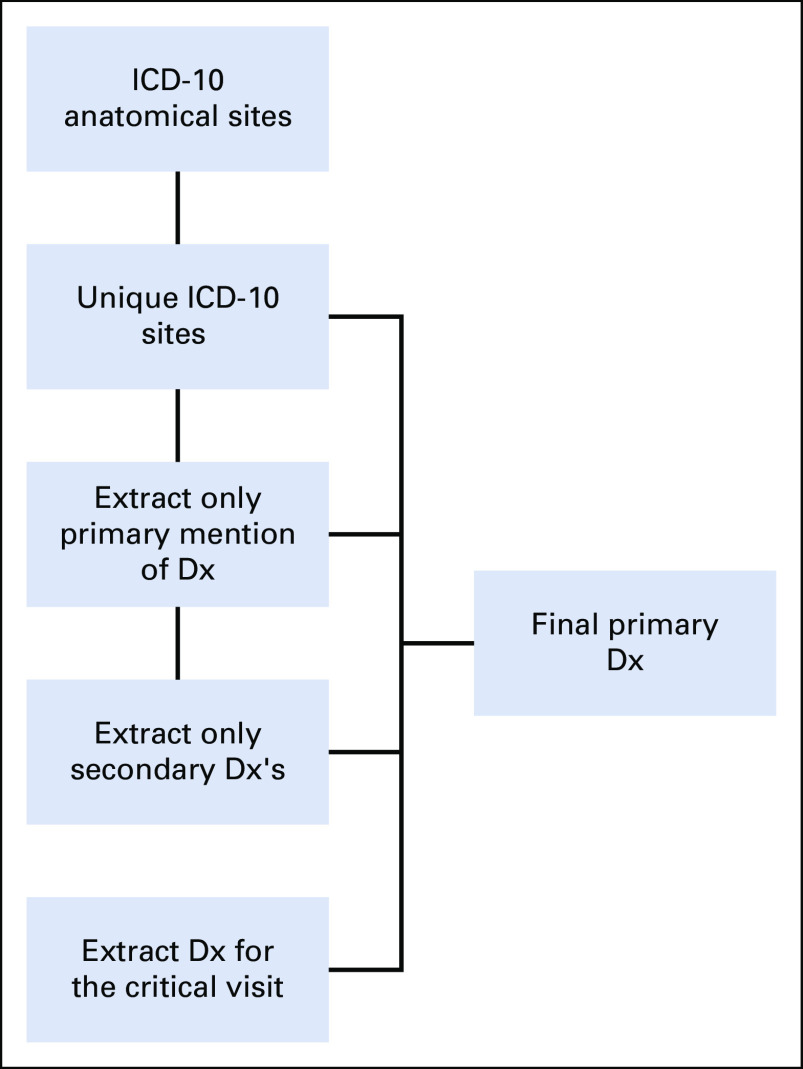
Primary cancer diagnosis and associated diagnostics features are challenging to determine from the CDMs because of the lack of contextual clinical features (eg, pathology). Additionally, limited historical data availability within the N3C Enclave (starting at January 1, 2018) precludes a conclusive determination of a first cancer diagnosis. Using a diagnosis mapping to ICD-10 process (Appendix Fig A1, online only), we were able to map 363,856 patients' diagnosis to one (or more) ICD-10 topography code(s). In 9,924 patients, either the SNOMED-reported cancer type could not be mapped to a single ICD-10 anatomical site or they had an ICD-10 code of C76 or C80 (code C76 corresponds to malignant neoplasm of other and ill-defined sites, and C80 corresponds to malignant neoplasm without specification of site). As such, we were not able to map those patients to a single primary diagnosis and were categorized as having unknown or undefined primary, respectively, in the analyses. As depicted in Appendix Figure A2 (online only), we employed a multistep process to identify a primary diagnosis for patients with a mapped diagnosis. Initially, we extracted the single-cancer ICD-10 topography-reported patients. Second, we searched the keyword primary in cancer type and corresponding ICD-10 topography if it is mapped to a single topography. Third, we extracted secondary-only diagnosis on the basis of ICD-10 topography and finally, identified unique occurrence of an ICD-10 topography within the index encounter. This process resulted in a total of 321,337 patients with a primary diagnosis. We were unable to conclusively associate 52,443 patients with a single primary diagnosis because of reporting of more than one cancer site for these patients and insufficient data to differentiate a primary versus subsequent malignancy vs metastasis.
Cancer Therapies
Exposure to systemic, nontopical cancer therapies were assessed for each person in our cohort using a string search for each cancer therapy in the drug concept name and manually reviewed for correctness. Although steroids are an integral portion of cancer therapy, steroids were excluded from our categorization as a sole indication of cancer therapy exposure. Each cancer therapy was categorized as cytotoxic, targeted, immunotherapy, or endocrine therapy on the basis of the drug's mechanism of action (Appendix Table A2, online only). The most recent cancer therapy received within 30 days of the index encounter was included in the analyses.
Outcomes
The primary outcome of interest was all-cause mortality during the index encounter. Secondary outcomes included indicators of clinical severity requiring hospitalization: use of mechanical ventilation and extracorporeal membrane oxygenation.
Statistical Analysis
We calculated frequencies of potential prognostic factors comparing COVID-19 cases with controls using chi-square tests. We calculated frequency of death from any cause in the entire cohort and death and clinical severity indicators in hospitalized patients. Survival probabilities for the study cohort from 3 days, 10 days, and up to 90 days, as well as their 95% CIs, were estimated using the Kaplan-Meier estimator for censored data. We also estimated the survival probability for the major cancer types in the COVID-19–positive patients. Kaplan-Meier curves were used to visualize the corresponding survival probability, and the log-rank test to test statistical differences in survival probability between COVID-19–positive and COVID-19–negative patients. Multivariable analyses were performed using Cox Proportional Hazard models to estimate hazard ratios (HRs) for association of potential risk factors with all-cause mortality within 1 year comparing COVID-19–positive and COVID-19–negative patients adjusting for age group, sex, race and ethnicity, smoking status, geographical location of patient residence, adjusted CCI index, primary cancer types, and cancer treatment at 30 days. All tests were 2-sided using a 5% significance threshold. We also fit a separate model including only COVID-19–positive patients adding COVID-19 treatments including azithromycin, hydroxychloroquine, remdesivir, systemic steroids, and antibiotics to the model.
Per N3C policy, exact counts that are 20 or less were not reported to protect the privacy of individuals. All analyses were performed in the N3C Data Enclave on the Palantir platform.
The Role of the Institutional Review Board
All authors who performed analyses and had access to N3C data in the Enclave obtained individual institutional review board approvals from their respective institutions for this project and were also approved to use a limited data set by the N3C Data Use Request Committee.
RESULTS
As of March 25, 2021, our N3C-derived cohort consists of 373,780 adults (≥ 18 years old) with a history of cancer (mean age 65 years, 51% female, 68% non-Hispanic White, and 39% with four or more comorbidities). The median observation time over the study period between January 1, 2018, and March 25, 2021, was 2.85 years (interquartile range: 1.2 years). Within the analytic cohort, 38,614 patients were COVID-19–positive and 335,166 patients were negative (Fig 1). COVID-19–positive and COVID-19–negative subgroups did not have significantly different proportions by sex, whereas more non-Hispanic White individuals were COVID-19–negative (69% v 62%; P < .001). COVID-19–negative individuals were more likely to have a higher CCI compared with COVID-19–positive patients (CCI ≥ 4 was observed in 41% v 28%, respectively; P < .001). Top four cancer subtypes were more prevalent in the COVID-19–positive patients compared with COVID-19–negative patients. Recent cancer-related therapies for both COVID-19–positive and COVID-19–negative patients were low (overall approximately 1% on any specific therapy) (Table 1).
TABLE 1.
Study Cohort Demographic, Clinical, and Tumor Characteristics
Among the 373,780 patients in the analytic cohort, the index encounter was a hospitalization visit in 204,503 patients (19,515 COVID-19–positive and 184,988 COVID-19–negative). The average length of stay in the hospital was 6.5 days (standard deviation 10.3 days). Among COVID-19–positive hospitalized patients, 14.8% of patients died and 8.2% required invasive ventilation during their index hospitalization compared with 12.5% and 5.2%, respectively, in COVID-19–negative patients (Table 2). Only 0.14% of COVID-19–positive hospitalized patients required extracorporeal membrane oxygenation.
TABLE 2.
Prevalence of Death and Invasive Ventilation in the Entire and Hospitalized Cohort by Risk Factor
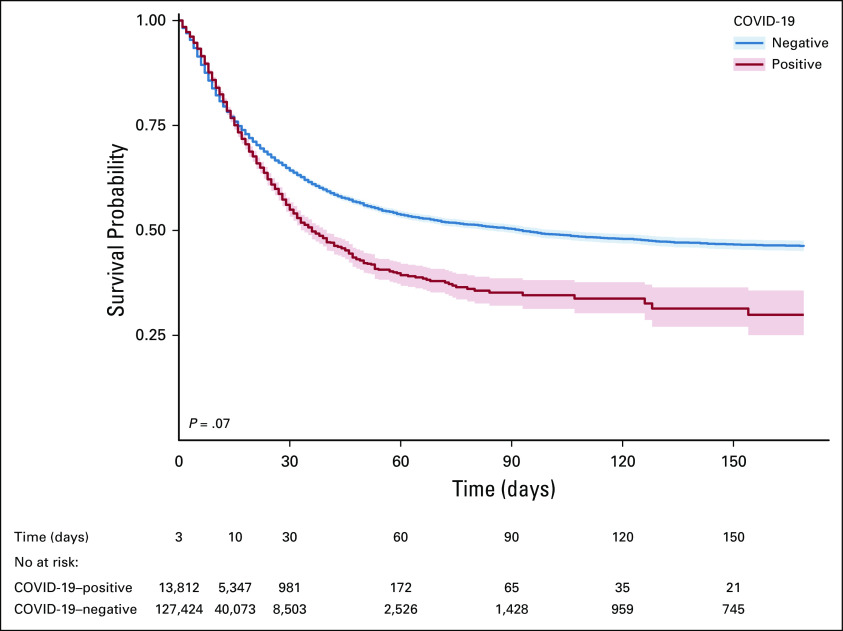
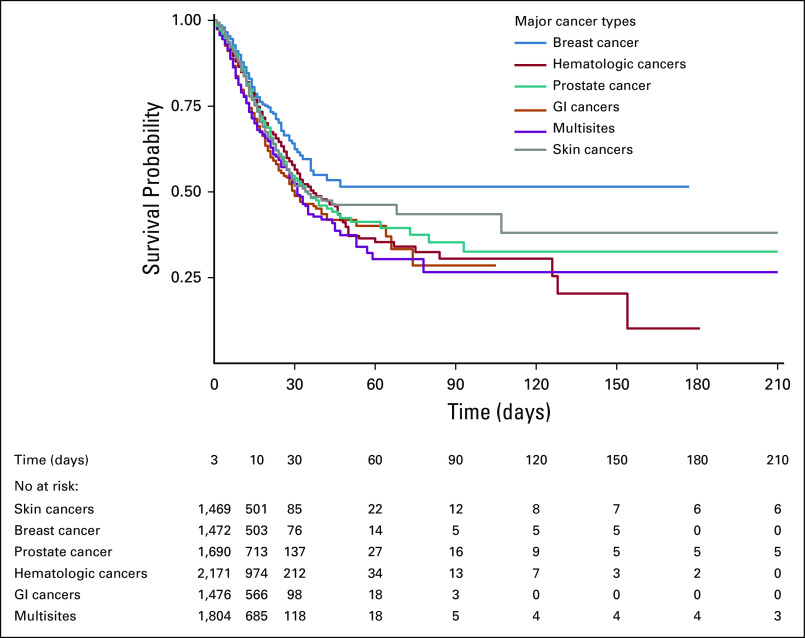
Among COVID-19–positive patients, survival probabilities were 84% at 10 days, 55% at 30 days, and 35% at 90 days, compared with 82%, 64%, and 50% in COVID-19–negative patients, respectively. Overall, there was no statistically significant difference in survival probabilities using log-rank test (P = .07). At 90 days survival, COVID-19–positive breast cancer showed better survival than other cancer types (51%; 95% CI, 44 to 61), whereas patients with multisite cancers showed lowest survival (26%; 95% CI, 18 to 39) (Figs 2 and 3; Table 3). Survival curves for the top cancer types by age group are shown in Appendix Figure A3 (online only).
FIG 2.
Survival probability curves for COVID-19–positive and COVID-19–negative patients.
FIG 3.
Survival probability curve by cancer type for COVID-19–positive patients.
TABLE 3.
Survival Probabilities for COVID-19–Positive Patients Over Time
COVID-19 positivity was significantly associated with increased risk of all-cause mortality (HR, 1.14; 95% CI, 1.1 to 1.2; P < .001) after adjusting for all other potential risk factors (Fig 4A). Among COVID-19–positive patients, older age more than 65 years (HR, 1.9; 95% CI, 1.3 to 3.1), male gender (HR, 1.11; 95% CI, 1.02 to 1.20), Southern and Western US regions (HR, 1.3; 95% CI, 1.1 to 1.6 and HR, 1.7; 95% CI, 1.3 to 2.2, respectively), higher number of comorbidities (HR, 2.0; 95% CI, 1.8 to 2.3), hematologic malignancies (HR, 1.2; 95% CI, 1.0 to 1.3) and multisite tumors (HR, 1.3; 95% CI, 1.1 to 1.4), and recent cytotoxic therapy (HR, 1.5; 95% CI, 1.1 to 2.1) were associated with increased risk of all-cause mortality (Fig 4B; Appendix Table A3, online only). Recent receipt of immunotherapies and targeted therapies were not associated with increased all-cause mortality. Non-Hispanic Black race (HR, 0.8; 95% CI, 0.7 to 0.9), recent hormonal therapy (HR, 0.5; 95% CI, 0.3 to 0.9), and treatment of COVID-19 with dexamethasone (HR, 0.8; 95% CI, 0.7 to 0.9) were associated with decreased risk of all-cause mortality (Fig 4B). Our analysis of the hematologic malignancy subcohort showed no significant statistical difference among myeloid and lymphoid malignancies in mortality risk (Appendix Table A4, online only).
FIG 4.
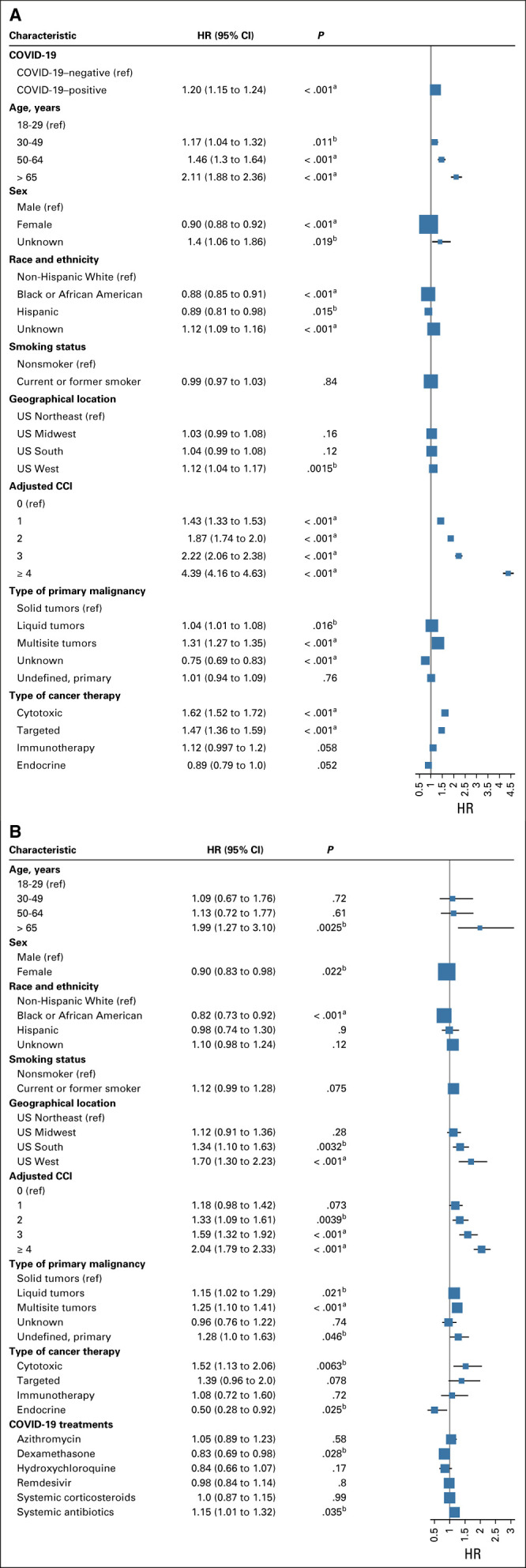
Adjusted HRs Cox proportional hazard model for association of potential risk factors with 1-year all-cause mortality in (A) entire cohort, and (B) COVID-19–positive patients only. CCI, Charlson Comorbidity Index; HR, hazard ratio; ref, reference. aP < .001. bP < .05.
DISCUSSION
We used the harmonized and integrated N3C clinical cohort, a patient registry that includes data on approximately 4.3 million COVID-19–tested patients with at least one clinical encounter after January 1, 2020 (inpatient or outpatient), at 50 US medical centers, to construct a cohort of patients with cancer. To the best of our knowledge, this is the first collaborative network study on patients with cancer and COVID-19 of this magnitude that demonstrates the feasibility of performing such large-scale observational research on the interaction of COVID-19 infection and cancer management across multiple healthcare sites nationwide.
The survival profile of patients with cancer infected by SARS-CoV-2 demonstrated characteristics related to both COVID-19 and cancer. Older age, male gender, and existing comorbidities are well-established mortality risk factors for COVID-19. The impact of age, sex, and comorbidity on survival remained prominent in our cohort with a pre-existing cancer diagnosis. Although some studies have shown racial disparities in mortality risk from COVID-19 between non-Hispanic Whites and Blacks,15,22,23 other large studies did not find that race significantly affected rates of hospitalization or all-cause mortality.24-26 Notably, our analysis showed that non-Hispanic Black and Hispanic patients with cancer had significantly lower risk of mortality. A more nuanced examination of neighborhood level characteristics, social determinants of health, and health literacy may provide better insights into the association of race and mortality risk from COVID-19.27,28
Although patients with cancer in our entire cohort who were on active cytotoxic therapies, targeted therapies, and immunotherapies were at increased mortality risk, consistent with previous studies,16,17,29-31 receipt of recent immunotherapy and targeted cancer therapy did not appear to have a significantly increased effect on the mortality risk in COVID-19–positive patients. On the other hand, recent cytotoxic therapy was associated with increased mortality risk,15 whereas hormonal therapy was associated with decreased mortality risk. In addition, patients with COVID-19 also demonstrated differential prognosis by cancer type, which is largely consistent with prior knowledge before the COVID-19 pandemic, with breast cancers having the best prognosis and hematologic and GI cancers showing poor survival. Despite the observed high mortality rate in our cohort, the cancer therapy exposure detected in the preceding 30 days of the index encounter was lower than expected in our cohort especially when compared with manually extracted registry data (eg, the CCC19 cohort).15,31 Our cohort consists of patients who are at varying stages in their cancer journey and could be on active treatment, in remission, or receiving end-of-life care. It is also possible that despite being on active cancer therapy, a COVID-19 diagnosis might have delayed or prevented a planned or ongoing cancer treatment.32,33 We defined our cohort by an index encounter within a specified timeframe, which might have inadvertently decreased the likelihood of capturing recent cancer therapy particularly if a patient has received care for COVID-19 diagnosis at an institution different from where they receive their usual cancer care. The N3C Real-World Data–based cohort was constructed from a wide range of institutions that included patients with and without cancer, as such certain health systems may be better at documenting cancer diagnosis or respective therapy in their data warehouse especially if there is an embedded or linked cancer center within the health system. We attempted to minimize potential therapy misclassification by excluding certain non-cancer–specific therapies such as steroids or certain hormonal therapies and by considering spelling variations of therapy names.
Strengths of this study are derived from the construction of a large-scale patient registry such as N3C.18 Through concept mapping and standardization, the OMOP CDM in the N3C platform facilitates collaboration and interoperability across heterogeneous EHR databases. As more high-quality extract, transform, and load tools are being developed in the Observational Health Data Sciences and Informatics community,20,34 the CDM holds promise to decrease barriers to open collaborative cancer research. With data originating from 50 sites via four different data models (ie, Accruals to Clinical Trials, OMOP, PCORNet, and TriNetX), we encountered data quality issues during the aggregation and mapping of the data. Identifying the primary cancer diagnosis from the structured EHR data is a known challenge. The limited historical data within the N3C platform further restricted our ability to identify a single primary diagnosis for all the patients within our study cohort. Some patients might be misclassified with cancer while they were in remission or they merely underwent workups for cancer rule-out. More than half of our study population were hospitalized patients, which may explain the poor long-term survival outcomes observed in our study cohort. The continuing growth of the N3C cohort will, however, provide further insight into the outcomes of hospitalized and nonhospitalized patients with COVID-19 and cancer. We opted to define our cohort using the index encounter (known as the critical visit within N3C).19 This allowed the identification of the most important or critical COVID-related encounter in a patient's clinical record and the most critical non-COVID–related visit for COVID-negative controls. Hence, an important strength of our study is the inclusion of non-COVID controls in our cohort, allowing us to estimate independent effects of COVID-19 infection on mortality risk in patients with cancer. The manner that the COVID-19–negative cohort was constructed, however, is not without limitations. Initially, all COVID-19–negative patients were included from the contributing sites. Starting December 2020, COVID-19–negative controls are being randomly selected from each of the data contributing sites using a 2:1 ratio to match the overall prevalence of age, sex, and race of COVID-19 cases from those sites. Future studies could explore more detailed and consistent matching strategies within the cohort. Data missingness from EHR is a well-known problem for studies using clinical records. Although some patient records may be completely captured by a given hospital system, other records are only partially captured, when a patient sought care at a different facility not affiliated with the given hospital system. This limitation extends to all clinical data domains including death information. Many hospital systems link their records with state death records to close this gap, but this is often done on a semiannual basis and it may not be done at every site. All-cause mortality, our primary study outcome, may be under-represented in the data; however, the scope and scale of the N3C data repository potentially overcome the limitations of individual sites.
In conclusion, through constructing the largest COVID-19 and cancer cohort within the United States, we examined risks of adverse outcomes associated with COVID-19–positive patients with cancer, particularly all-cause mortality. Despite the known limitations of large-scale data networks such as N3C, the consortium represents an unmatched resource for clinicians and researchers to examine the effects of cancer on COVID-19 outcomes and vice versa. Consistent with previous literature, older age, male gender, and increasing comorbidities were associated with higher mortality in patients with cancer and COVID-19. The N3C data set also confirmed that patients with cancer and COVID-19 who received recent immunotherapies or targeted therapies were not at higher risks of overall mortality. Future studies of the cohort will provide insights into the evolving effect of COVID-19 on patients with cancer and additional evidence to guide the clinical management of this patient population.
ACKNOWLEDGMENT
The authors would like to extend special thanks to Amin Mannaa for providing support on the analytic platform.
We gratefully acknowledge contributions from the following N3C core teams: Principal Investigators: Melissa A. Haendel*, Christopher G. Chute*, Kenneth R. Gersing, and Anita Walden; Workstream, Subgroup, and Administrative Leaders: Melissa A. Haendel*, Tellen D. Bennett, Christopher G. Chute, David A. Eichmann, Justin Guinney, Warren A. Kibbe, Hongfang Liu, Philip R.O. Payne, Emily R. Pfaff, Peter N. Robinson, Joel H. Saltz, Heidi Spratt, Justin Starren, Christine Suver, Adam B. Wilcox, Andrew E. Williams, and Chunlei Wu; Key liaisons at data partner sites; Regulatory staff at data partner sites; Individuals at the sites who are responsible for creating the data sets and submitting data to N3C; Data Ingest and Harmonization Team: Christopher G. Chute*, Emily R. Pfaff*, Davera Gabriel, Stephanie S. Hong, Kristin Kostka, Harold P. Lehmann, Richard A. Moffitt, Michele Morris, Matvey B. Palchuk, Xiaohan Tanner Zhang, and Richard L. Zhu; Phenotype Team (individuals who create the scripts that the sites use to submit their data, on the basis of the COVID and Long COVID definitions): Emily R. Pfaff*, Benjamin Amor, Mark M. Bissell, Marshall Clark, Andrew T. Girvin, Stephanie S. Hong, Kristin Kostka, Adam M. Lee, Robert T. Miller, Michele Morris, Matvey B. Palchuk, and Kellie M. Walters; Project Management and Operations Team: Anita Walden*, Usman Sheikh, Yooree Chae, Connor Cook, Alexandra Dest, Racquel R. Dietz, Thomas Dillon, Patricia A. Francis, Rafael Fuentes, Alexis Graves, Julie A. McMurry, Andrew J. Neumann, Shawn T. O'Neil, Andréa M. Volz, and Elizabeth Zampino; Partners From NIH and Other Federal Agencies: Christopher P. Austin*, Kenneth R. Gersing*, Samuel Bozzette, Mariam Deacy, Nicole Garbarini, Michael G. Kurilla, Sam G. Michael, Joni L. Rutter, and Meredith Temple-O'Connor; Analytics Team (individuals who build the Enclave infrastructure, help create codesets and variables, and help Domain Teams and project teams with their data sets): Benjamin Amor*, Mark M. Bissell, Katie Rebecca Bradwell, Andrew T. Girvin, Amin Manna, and Nabeel Qureshi; Publication Committee Management Team: Mary Morrison Saltz*, Christine Suver*, Christopher G. Chute, Melissa A. Haendel, Julie A. McMurry, Andréa M. Volz, and Anita Walden; Publication Committee Review Team: Carolyn Bramante, Jeremy Richard Harper, Wenndy Hernandez, Farrukh M. Koraishy, Federico Mariona, Saidulu Mattapally, Amit Saha, and Satyanarayana Vedula.
Stony Brook University—U24TR002306; University of Oklahoma Health Sciences Center—U54GM104938: Oklahoma Clinical and Translational Science Institute (OCTSI); West Virginia University—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI); University of Mississippi Medical Center—U54GM115428: Mississippi Center for Clinical and Translational Research (CCTR); University of Nebraska Medical Center—U54GM115458: Great Plains IDeA-Clinical & Translational Research; Maine Medical Center—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network; Wake Forest University Health Sciences—UL1TR001420: Wake Forest Clinical and Translational Science Institute; Northwestern University at Chicago—UL1TR001422: Northwestern University Clinical and Translational Science Institute (NUCATS); University of Cincinnati—UL1TR001425: Center for Clinical and Translational Science and Training; The University of Texas Medical Branch at Galveston—UL1TR001439: The Institute for Translational Sciences; Medical University of South Carolina—UL1TR001450: South Carolina Clinical & Translational Research Institute (SCTR); University of Massachusetts Medical School Worcester—UL1TR001453: The UMass Center for Clinical and Translational Science (UMCCTS); University of Southern California—UL1TR001855: The Southern California Clinical and Translational Science Institute (SC CTSI); Columbia University Irving Medical Center—UL1TR001873: Irving Institute for Clinical and Translational Research; George Washington Children's Research Institute—UL1TR001876: Clinical and Translational Science Institute at Children's National (CTSA-CN); University of Kentucky—UL1TR001998: Appalachian Translational Research Network (ATRN); University of Rochester—UL1TR002001: UR Clinical & Translational Science Institute; University of Illinois at Chicago—UL1TR002003: UIC Center for Clinical and Translational Science; Penn State Health Milton S. Hershey Medical Center—UL1TR002014: Penn State Clinical and Translational Science Institute; The University of Michigan at Ann Arbor—UL1TR002240: Michigan Institute for Clinical and Health Research; Vanderbilt University Medical Center—UL1TR002243: Vanderbilt Institute for Clinical and Translational Research; University of Washington—UL1TR002319: Institute of Translational Health Sciences; Washington University in St Louis—UL1TR002345: Institute of Clinical and Translational Sciences; Oregon Health & Science University—UL1TR002369: Oregon Clinical and Translational Research Institute; University of Wisconsin-Madison—UL1TR002373: Wisconsin Network For Health Research; Rush University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM); The University of Chicago—UL1TR002389: The Institute for Translational Medicine (ITM); University of North Carolina at Chapel Hill—UL1TR002489: North Carolina Translational and Clinical Science Institute; University of Minnesota—UL1TR002494: Clinical and Translational Science Institute; Children's Hospital Colorado—UL1TR002535: Colorado Clinical and Translational Sciences Institute; The University of Iowa—UL1TR002537: Institute for Clinical and Translational Science; The University of Utah—UL1TR002538: Uhealth Center for Clinical and Translational Science; Tufts Medical Center—UL1TR002544: Tufts Clinical and Translational Science Institute; Duke University—UL1TR002553: Duke Clinical and Translational Science Institute; Virginia Commonwealth University—UL1TR002649: C. Kenneth and Dianne Wright Center for Clinical and Translational Research; The Ohio State University—UL1TR002733: Center for Clinical and Translational Science; The University of Miami Leonard M. Miller School of Medicine—UL1TR002736: University of Miami Clinical and Translational Science Institute; University of Virginia—UL1TR003015: iTHRIVL Integrated Translational health Research Institute of Virginia; Carilion Clinic—UL1TR003015: iTHRIVL Integrated Translational health Research Institute of Virginia; University of Alabama at Birmingham—UL1TR003096: Center for Clinical and Translational Science; Johns Hopkins University—UL1TR003098: Johns Hopkins Institute for Clinical and Translational Research; University of Arkansas for Medical Sciences—UL1TR003107: Consortium of Rural States (CORES); Nemours—U54GM104941: Delaware CTR ACCEL Program; University Medical Center New Orleans—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center; University of Colorado Denver, Anschutz Medical Campus—UL1TR002535: Colorado Clinical and Translational Sciences Institute; Mayo Clinic Rochester—UL1TR002377: Mayo Clinic Center for Clinical and Translational Science (CCaTS); Tulane University—UL1TR003096: Center for Clinical and Translational Science; Loyola University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM); Advocate Health Care Network—UL1TR002389: The Institute for Translational Medicine (ITM); OCHIN—INV-018455: Bill and Melinda Gates Foundation grant to Sage Bionetworks.
The Rockefeller University—UL1TR001866: Center for Clinical and Translational Science; The Scripps Research Institute—UL1TR002550: Scripps Research Translational Institute; University of Texas Health Science Center at San Antonio—UL1TR002645: Institute for Integration of Medicine and Science; The University of Texas Health Science Center at Houston—UL1TR003167: Center for Clinical and Translational Sciences (CCTS); NorthShore University HealthSystem—UL1TR002389: The Institute for Translational Medicine (ITM); Yale New Haven Hospital—UL1TR001863: Yale Center for Clinical Investigation; Emory University—UL1TR002378: Georgia Clinical and Translational Science Alliance; Weill Medical College of Cornell University—UL1TR002384: Weill Cornell Medicine Clinical and Translational Science Center; Montefiore Medical Center—UL1TR002556: Institute for Clinical and Translational Research at Einstein and Montefiore; Medical College of Wisconsin—UL1TR001436: Clinical and Translational Science Institute of Southeast Wisconsin; University of New Mexico Health Sciences Center—UL1TR001449: University of New Mexico Clinical and Translational Science Center; George Washington University—UL1TR001876: Clinical and Translational Science Institute at Children's National (CTSA-CN); Stanford University—UL1TR003142: Spectrum: The Stanford Center for Clinical and Translational Research and Education; Regenstrief Institute—UL1TR002529: Indiana Clinical and Translational Science Institute; Cincinnati Children's Hospital Medical Center—UL1TR001425: Center for Clinical and Translational Science and Training; Boston University Medical Campus—UL1TR001430: Boston University Clinical and Translational Science Institute; The State University of New York at Buffalo—UL1TR001412: Clinical and Translational Science Institute; Aurora Health Care—UL1TR002373: Wisconsin Network For Health Research; Brown University—U54GM115677: Advance Clinical Translational Research (Advance-CTR); Rutgers, The State University of New Jersey—UL1TR003017: New Jersey Alliance for Clinical and Translational Science; Loyola University Chicago—UL1TR002389: The Institute for Translational Medicine (ITM); #N/A—UL1TR001445: Langone Health's Clinical and Translational Science Institute; Children's Hospital of Philadelphia—UL1TR001878: Institute for Translational Medicine and Therapeutics; University of Kansas Medical Center—UL1TR002366: Frontiers: University of Kansas Clinical and Translational Science Institute; Massachusetts General Brigham—UL1TR002541: Harvard Catalyst; Icahn School of Medicine at Mount Sinai—UL1TR001433: ConduITS Institute for Translational Sciences; Ochsner Medical Center—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center; HonorHealth—None (Voluntary); University of California, Irvine—UL1TR001414: The UC Irvine Institute for Clinical and Translational Science (ICTS); University of California, San Diego—UL1TR001442: Altman Clinical and Translational Research Institute; University of California, Davis—UL1TR001860: UC Davis Health Clinical and Translational Science Center; University of California, San Francisco—UL1TR001872: UCSF Clinical and Translational Science Institute; University of California, Los Angeles—UL1TR001881: UCLA Clinical Translational Science Institute; University of Vermont—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network; Arkansas Children's Hospital—UL1TR003107: UAMS Translational Research Institute.
Please see Appendix 2 (online only) for the full list of the National COVID Cohort Collaborative (N3C) core authors and affiliations.
This research was possible because of the patients whose information is included within the data from participating organizations (https://ncats.nih.gov/n3c/resources/data-contribution/data-transfer-agreement-signatories) and scientists (https://covid.cd2h.org/duas) who have contributed to the on-going development of this community resource doi.org/10.5281/zenodo.3979622.
The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol No. IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources. *Denotes N3C core team leads.
APPENDIX 1
Methods: Algorithm to Define a Single Index Encounter Representing the Critical COVID-19–Related Visit for Each Patient
We followed N3C definitions to select one single index encounter per person by COVID status and severity. Among multiple recorded encounters per patient, the most appropriate single encounter for analysis is chosen using the following procedure:
Select visits with an associated COVID-positive test result, if available
Select visits with an associated COVID-negative test result, if available
Select visits with a suspected COVID diagnosis, if available
Select inpatient visits, if available
Select emergency department visits, if available
Select hospital visits, if available
If the patient is recorded as dead, select the most recent visit
Select visits that included extracorporeal membrane oxygenation or mechanical ventilation, if available
Select the longest visit
Select the most recent visit
APPENDIX 2
Addendum of the National COVID Cohort Collaborative (N3C) core authors and affiliations
Amit Mitra,1 Ramakanth Kavuluru,2 Melissa A. Haendel,3 Christopher G. Chute4
1Amit Mitra, Drug Discovery and Development (DDD), Center for Pharmacogenomics and Single-Cell Omics (AUPharmGx), Auburn University, Auburn; e-mail: mitra79@gmail.com
I would like to be indexed in Medline including: data curation; critical revision of the manuscript for important intellectual content; N3C Phenotype definition; project evaluation.
2Ramakanth Kavuluru, University of Kentucky, Lexington, KY, USA; e-mail: ramakanth.sai@gmail.com
I would like to be indexed in Medline including: data analysis; critical revision of the manuscript for important intellectual content; statistical analysis.
3Melissa A. Haendel, Center for Health AI, University of Colorado Anschutz Medical Campus, Aurora, CO, USA; e-mail: melissa@tislab.org
I would like to be indexed in Medline including: funding acquisition; governance; critical revision of the manuscript for important intellectual content; project management.
4Christopher G. Chute, Schools of Medicine, Public Health, and Nursing, Johns Hopkins University, Baltimore, MD, USA; e-mail: chute@jhu.edu
I would like to be indexed in Medline including: clinical data model expertise; data curation; data integration; data quality assurance; funding acquisition; governance; critical revision of the manuscript for important intellectual content; N3C Phenotype definition; project management; regulatory oversight/admin.
FIG A1.
Primary cancer type mapping process. Dx, diagnosis; ICD, International Classification of Diseases.
FIG A2.
Primary cancer type identification process. Dx, diagnosis; ICD, International Classification of Diseases.
FIG A3.
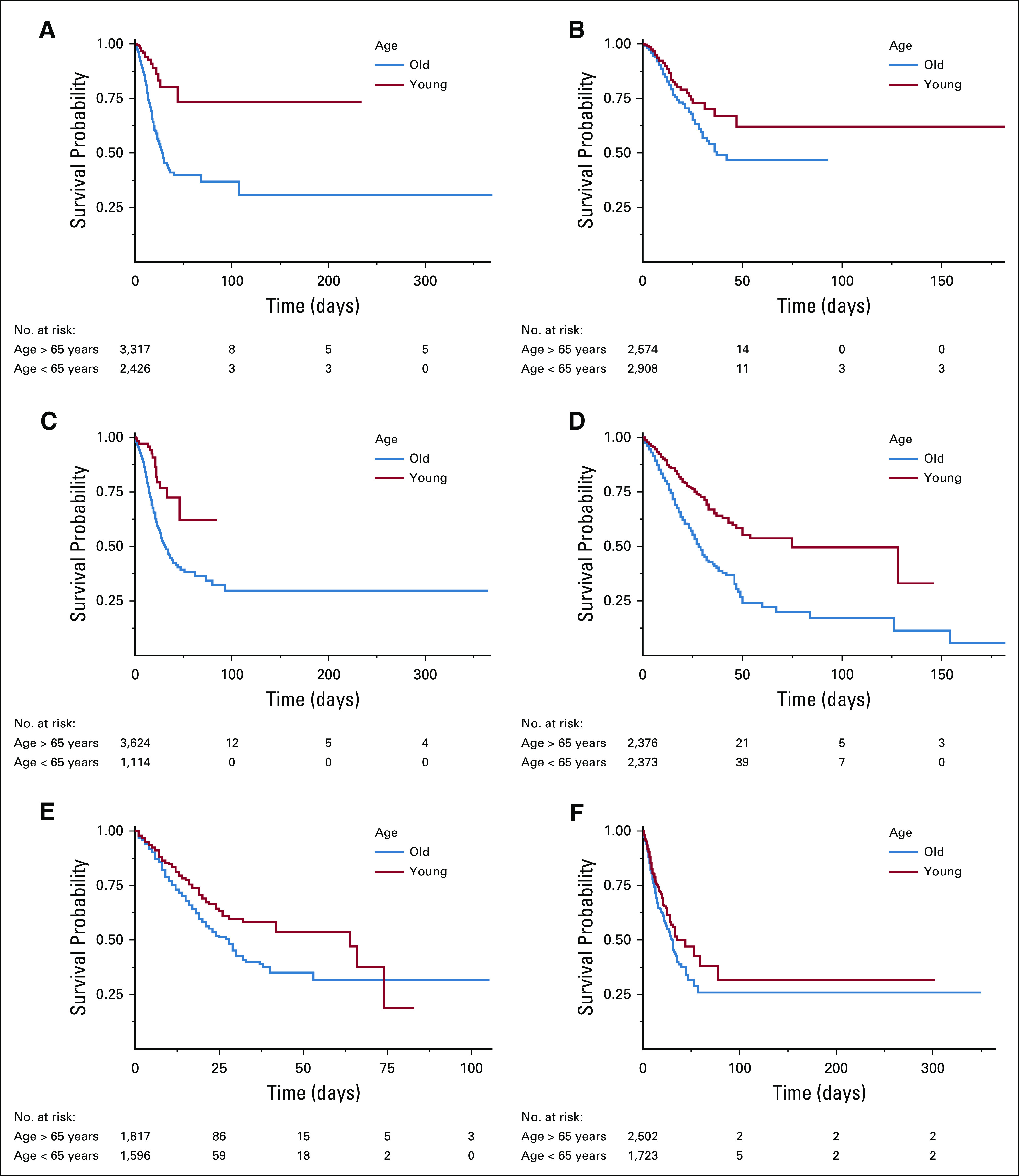
Survival probability curves by cancer type and age for COVID-19–positive patients. (A) Skin cancers, (B) breast cancer, (C) prostate cancer, (D) hematologic cancers, (E) GI cancers, and (F) multisite tumors.
TABLE A1.
Excluded Concepts From the Standard Malignant Neoplastic Disease Concept Set (SNOMED Concept Code 363346000)
TABLE A2.
List of Cancer Therapies Captured Within Each Therapy Category
TABLE A3.
Adjusted HRs for Association of Potential Risk Factors With All-Cause Mortality in Hematologic Malignancy Patients Compared With Solid Malignancy COVID-19–Positive Patients

TABLE A4.
Adjusted HRs for Association of Potential Risk Factors With All-Cause Mortality in Hematologic Malignancy COVID-19–Positive Patients

Benjamin Bates
Stock and Other Ownership Interests: Pfizer
Eileen Lee
Employment: Johnson & Johnson/Janssen
Yu Raymond Shao
Employment: GlaxoSmithKline, Suzhou Kintor Pharmaceuticals
Feifan Liu
Stock and Other Ownership Interests: Pfizer
Timothy Bergquist
Research Funding: Celgene
Justin Guinney
Consulting or Advisory Role: AstraZeneca
Research Funding: AstraZeneca, Bristol Meyers Squib, Roche/Genentech
Umit Topaloglu
Stock and Other Ownership Interests: CareDirections
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Authorship was determined using ICMJE recommendations.
PRIOR PRESENTATION
Presented in part at the ASCO 2021 Virtual Meeting, June 4-8, 2021.
SUPPORT
Supported by NCATS U24 TR002306 and the National Institute of General Medical Sciences, 5U54GM104942-04, and the analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave (https://covid.cd2h.org/enclave). This work was also partially financially supported by the Indiana University Precision Health Initiative funded to J.S. U.T. was partially supported by the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197). U.T. and Q.S. were supported in part by Bioinformatics Shared Resources under the NCI Cancer Center Support Grant to the Comprehensive Cancer Center of Wake Forest University Health Sciences (P30 CA012197). Additional support for Q.S. was provided by a Fellowship to Wei Zhang from the National Foundation for Cancer Research. N.S. was supported by the Leukemia and Lymphoma Society Career Development Award (LLS 3386-19). T.B. and J.G. were supported by the Bill and Melinda Gates Foundation (INV-018455).
AUTHOR CONTRIBUTIONS
Conception and design: Noha Sharafeldin, Benjamin Bates, Qianqian Song, Yu Raymond Shao, Feifan Liu, Jing Su, Umit Topaloglu
Financial support: Justin Guinney
Administrative support: Justin Guinney
Collection and assembly of data: Noha Sharafeldin, Benjamin Bates, Qianqian Song, Vithal Madhira, Yao Yan, Sharlene Dong, Eileen Lee, Nathaniel Kuhrt, Yu Raymond Shao, Timothy Bergquist, Justin Guinney, Jing Su, Umit Topaloglu
Data analysis and interpretation: Noha Sharafeldin, Benjamin Bates, Qianqian Song, Vithal Madhira, Yao Yan, Eileen Lee, Yu Raymond Shao, Feifan Liu, Timothy Bergquist, Jing Su, Umit Topaloglu
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Outcomes of COVID-19 in Patients With Cancer: Report From the National COVID Cohort Collaborative (N3C)
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Benjamin Bates
Stock and Other Ownership Interests: Pfizer
Eileen Lee
Employment: Johnson & Johnson/Janssen
Yu Raymond Shao
Employment: GlaxoSmithKline, Suzhou Kintor Pharmaceuticals
Feifan Liu
Stock and Other Ownership Interests: Pfizer
Timothy Bergquist
Research Funding: Celgene
Justin Guinney
Consulting or Advisory Role: AstraZeneca
Research Funding: AstraZeneca, Bristol Meyers Squib, Roche/Genentech
Umit Topaloglu
Stock and Other Ownership Interests: CareDirections
No other potential conflicts of interest were reported.
REFERENCES
- 1.Lima NT, Buss PM, Paes-Sousa R. COVID-19 pandemic: A health and humanitarian crisis. Cad Saude Publica. 2020;36:e00177020. doi: 10.1590/0102-311x00177020. [DOI] [PubMed] [Google Scholar]
- 2.Nicola M, Alsafi Z, Sohrabi C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): A review Int J Surg 78185–1932020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giannakoulis VG, Papoutsi E, Siempos II.Effect of cancer on clinical outcomes of patients with COVID-19: A meta-analysis of patient data JCO Glob Oncol 6799–8082020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunski MJ, Burton J, Tawagi K, et al. Multivariate mortality analyses in COVID-19: Comparing patients with cancer and patients without cancer in Louisiana Cancer 127266–2742021 [DOI] [PubMed] [Google Scholar]
- 5.Pathania AS, Prathipati P, Abdul BA, et al. COVID-19 and cancer comorbidity: Therapeutic opportunities and challenges Theranostics 11731–7532021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saini KS, Tagliamento M, Lambertini M, et al. Mortality in patients with cancer and coronavirus disease 2019: A systematic review and pooled analysis of 52 studies Eur J Cancer 13943–502020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hachem RY, Datoguia T, Siddiqui B, et al. 372. Comparing the outcome of COVID-19 in cancer and non-cancer patients: An international multicenter study. Open Forum Infect Dis. 2020;7:S256. [Google Scholar]
- 8.Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China Lancet Oncol 21335–3372020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Ouyang W, Chua MLK, et al. SARS-CoV-2 transmission in patients with cancer at a Tertiary Care Hospital in Wuhan, China JAMA Oncol 61108–11102020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker TJA.Risk of COVID-19 in patients with cancer JAMA Oncol 61470–14712020 [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Zhang J, Tu Y, et al. Cancer patients in SARS-CoV-2 infection: A single-center experience from Wuhan J Cancer 116243–62472020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai M, Liu D, Liu M, et al. Patients with cancer appear more vulnerable to SARS-CoV-2: A multicenter study during the COVID-19 outbreak Cancer Discov 10783–7912020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyashita H, Mikami T, Chopra N, et al. Do patients with cancer have a poorer prognosis of COVID-19? An experience in New York city Ann Oncol 311088–10892020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rüthrich MM, Giessen-Jung C, Borgmann S, et al. COVID-19 in cancer patients: Clinical characteristics and outcome—An analysis of the LEOSS registry Ann Hematol 100383–3932021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grivas P, Khaki AR, Wise-Draper TM, et al. Association of clinical factors and recent anti-cancer therapy with COVID-19 severity among patients with cancer: A report from the COVID-19 and Cancer Consortium Ann Oncol 32787–8002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee LY, Cazier J-B, Angelis V, et al. COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: A prospective cohort study Lancet 3951919–19262020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albiges L, Foulon S, Bayle A, et al. Determinants of the outcomes of patients with cancer infected with SARS-CoV-2: Results from the Gustave Roussy cohort Nat Cancer 1965–9752020 [DOI] [PubMed] [Google Scholar]
- 18.Haendel MA, Chute CG, Bennett TD, et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment J Am Med Inform Assoc 28427–4432021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett TD, Moffitt RA, Hajagos JG, et al. The National COVID Cohort Collaborative: Clinical Characterization and Early Severity Prediction. medRxiv; 2021. [Google Scholar]
- 20.Hripcsak G, Shang N, Peissig PL, et al. Facilitating phenotype transfer using a common data model. J Biomed Inform. 2019;96:103253. doi: 10.1016/j.jbi.2019.103253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation J Chronic Dis 40373–3831987 [DOI] [PubMed] [Google Scholar]
- 22.Golestaneh L, Neugarten J, Fisher M, et al. The association of race and COVID-19 mortality. EClinicalMedicine. 2020;25:100455. doi: 10.1016/j.eclinm.2020.100455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Berger NA, Xu R.Analyses of risk, racial disparity, and outcomes among US patients with cancer and COVID-19 infection JAMA Oncol 7220–2272021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price-Haywood EG, Burton J, Fort D, et al. Hospitalization and mortality among black patients and white patients with Covid-19 N Engl J Med 3822534–25432020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yehia BR, Winegar A, Fogel R, et al. Association of race with mortality among patients hospitalized with coronavirus disease 2019 (COVID-19) at 92 US hospitals. JAMA Netw Open. 2020;3:e2018039. doi: 10.1001/jamanetworkopen.2020.18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rentsch CT, Kidwai-Khan F, Tate JP, et al. Patterns of COVID-19 testing and mortality by race and ethnicity among United States veterans: A nationwide cohort study. PLoS Med. 2020;17:e1003379. doi: 10.1371/journal.pmed.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millett GA, Jones AT, Benkeser D, et al. Assessing differential impacts of COVID-19 on black communities Ann Epidemiol 4737–442020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones J, Sullivan PS, Sanchez TH, et al. Similarities and differences in COVID-19 awareness, concern, and symptoms by race and ethnicity in the United States: Cross-sectional Survey. J Med Internet Res. 2020;22:e20001. doi: 10.2196/20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brar G, Pinheiro LC, Shusterman M, et al. COVID-19 severity and outcomes in patients with cancer: A matched cohort study J Clin Oncol 383914–39242020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinato DJ, Scotti L, Gennari A, et al. Determinants of enhanced vulnerability to coronavirus disease 2019 in UK patients with cancer: A European study Eur J Cancer 150190–2022021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuderer NM, Choueiri TK, Shah DP, et al. Clinical impact of COVID-19 on patients with cancer (CCC19): A cohort study Lancet 3951907–19182020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar D, Dey T. Treatment delays in oncology patients during COVID-19 pandemic: A perspective. J Glob Health. 2020;10:010367. doi: 10.7189/jogh.10.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patt D, Gordan L, Diaz M, et al. Impact of COVID-19 on cancer care: How the pandemic is delaying cancer diagnosis and treatment for American seniors JCO Clin Cancer Inform 41059–10712020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for observational researchers Stud Health Technol Inform 216574–5782015 [PMC free article] [PubMed] [Google Scholar]