Abstract
Background: in current clinical practice, the standard evaluation for axillary lymph node (ALN) status in breast cancer has a low efficiency and is based on an invasive procedure that causes operative-associated complications in many patients. Therefore, we aimed to use machine learning techniques to develop an efficient preoperative magnetic resonance imaging (MRI) radiomics evaluation approach of ALN status and explore the association between radiomics and the tumor microenvironment in patients with early-stage invasive breast cancer.
Methods: in this retrospective multicenter study, three independent cohorts of patients with breast cancer (n = 1,088) were used to develop and validate signatures predictive of ALN status. After applying the machine learning random forest algorithm to select the key preoperative MRI radiomic features, we used ALN and tumor radiomic features to develop the ALN-tumor radiomic signature for ALN status prediction by the support vector machine algorithm in 803 patients with breast cancer from Sun Yat-sen Memorial Hospital and Sun Yat-sen University Cancer Center (training cohort). By combining ALN and tumor radiomic features with corresponding clinicopathologic information, the multiomic signature was constructed in the training cohort. Next, the external validation cohort (n = 179) of patients from Shunde Hospital of Southern Medical University and Tungwah Hospital of Sun Yat-Sen University, and the prospective-retrospective validation cohort (n = 106) of patients treated with neoadjuvant chemotherapy in prospective phase 3 trials [NCT01503905], were included to evaluate the predictive value of the two signatures, and their predictive performance was assessed by the area under operating characteristic curve (AUC). This study was registered with ClinicalTrials.gov, number NCT04003558.
Findings: the ALN-tumor radiomic signature for ALN status prediction comprising ALN and tumor radiomic features showed a high prediction quality with AUC of 0·88 in the training cohort, 0·87 in the external validation cohort, and 0·87 in the prospective-retrospective validation cohort. The multiomic signature incorporating tumor and lymph node MRI radiomics, clinical and pathologic characteristics, and molecular subtypes achieved better performance for ALN status prediction with AUCs of 0·90, 0·91, and 0·93 in the training cohort, the external validation cohort, and the prospective-retrospective validation cohort, respectively. Among patients who underwent neoadjuvant chemotherapy in the prospective-retrospective validation cohort, there were significant differences in the key radiomic features before and after neoadjuvant chemotherapy, especially in the gray-level dependence matrix features. Furthermore, there was an association between MRI radiomics and tumor microenvironment features including immune cells, long non-coding RNAs, and types of methylated sites.
Interpretation this study presented a multiomic signature that could be preoperatively and conveniently used for identifying patients with ALN metastasis in early-stage invasive breast cancer. The multiomic signature exhibited powerful predictive ability and showed the prospect of extended application to tailor surgical management. Besides, significant changes in key radiomic features after neoadjuvant chemotherapy may be explained by changes in the tumor microenvironment, and the association between MRI radiomic features and tumor microenvironment features may reveal the potential biological underpinning of MRI radiomics.
Keywords: Breast cancer, Axillary lymph node metastasis, Radiomics, Machine learning, Tumor microenvironment
Research in context.
Evidence before this study
We searched PubMed for publications without language restrictions, with the terms (“radiomics” OR “MRI-radiomics” OR “artificial intelligence” OR “AI”) AND (“breast cancer” OR “axillary lymph node” OR “ALN”). There have been several radiomics feature-based models to predict ALN metastasis in previous studies. However, the efficacy of their prediction models was not convincing enough, due to the small sample size, lack of independent validation cohorts and focus just on a single region. Thus, the clinical use of these models was greatly limited.
Added value of this study
Thinking of the association between ALN and the primary tumor site, we developed the ALN-tumor radiomic signature using both tumor and ALN radiomic features, and a higher AUC was observed compared with using ALN or tumor radiomic features alone. More importantly, the combination of ALN and tumor radiomic features with clinical and pathologic characteristics, as well as with molecular subtype could improve the predictive ability. With independent validation cohorts tested in our model, the competence of the constructed signatures was well verified. Furthermore, our findings revealed significant differences in MRI radiomics before and after neoadjuvant chemotherapy, and associations between MRI radiomic features and tumor microenvironment features, including immune cells, long non-coding RNAs, and types of methylated sites.
Implications of all the available evidence
This study indicated that the multiomic signature with a high practicability and universality has potential to serve as a preoperative tool to predict ALN metastasis more accurately in patients with early-stage invasive breast cancer. In particular, the precise prediction in patients can prevent unnecessary SLNB or ALND surgery and help to guide clinical decision-making. Besides, significant changes in MRI radiomics after neoadjuvant chemotherapy may be explained by changes in the tumor microenvironment, and the association between MRI radiomic features and tumor microenvironment features may reveal the potential biological underpinnings of MRI radiomics.
Alt-text: Unlabelled box
1. Introduction
The importance of axillary lymph node (ALN) status in early-stage invasive breast cancer was highlighted in a 10-year follow up of the ACOSOG Z0011 and IBCSG 23-01 randomized clinical trials [1–2]. The inclusion of the sentinel lymph node biopsy (SLNB) in standard diagnostic procedures of breast cancer since 2005 has greatly improved the accuracy of ALN status judgment and has allowed women to avoid axillary lymph node dissection (ALND) if they have fewer than three positive sentinel lymph nodes [3–4]. However, SLNB is still an invasive procedure that may cause complications such as lymphedema and upper limb numbness. Its accuracy is also limited by radiopharmaceuticals and the modalities of radiocolloid injection [5–6]. Moreover, it takes a long time for the frozen section of SLNB, which extends the surgery period. As a consequence, it is urgent and indispensable to develop a reliable and efficient preoperative evaluation approach of the ALN status in patients with early-stage invasive breast cancer.
The diagnostic performance of current non-invasive imaging modalities to assess ALN status, including magnetic resonance imaging (MRI) and axillary ultrasound, are limited by their high false-negative rates [7]. Nowadays, the development of radiomics in the aspects of tumor diagnosis, treatment decisions, and prognosis prediction is encouraging, and breast cancer is one of the focal areas in the exploration [[8], [9], [10]]. MRI examination has been recommended for preoperative estimation of breast cancer patients, and our previous study showed that MRI radiomic features of ALN region were helpful for ALN metastasis (ALNM) prediction [11]. In this retrospective multicenter study, we aimed to establish a new preoperative approach based on MRI radiomics focusing on both ALN and tumor regions for ALNM prediction in patients with early-stage invasive breast cancer; the final intention is to help tailor the surgical management and reduce unnecessary SLNB or ALND, thereby decreasing the incidence of associated postoperative complications and improving the life quality of patients. Moreover, we explored the association between MRI radiomics and breast tumor microenvironment.
2. Methods
2.1. Patients and study design
This multicenter study was conducted following the Declaration of Helsinki and the CLAIM guideline. The study protocol was approved by the ethics committee of each participating institution (Sun Yat-sen Memorial Hospital of Sun Yat-sen University, SYSEC-KY-KS-2019-054-001; Sun Yat-sen University Cancer Center, B2020-114-01; Shunde Hospital of Southern Medical University, KYLS-20190579; and Tungwah Hospital of Sun Yat-sen University, 2020DHLL018). The requirement for informed consent in retrospective cohorts was waived. Participants from the prospective phase III clinical trials [NCT01503905] [12] provided their informed consent, and the trial was approved by the ethics committee with the number of 2011-KY-012-001.
A total of 1,161 patients with early-stage invasive breast cancer were recruited from four institutions in China, of which 1,088 patients passed quality control. The inclusion criteria were as follows: (a) female patients aged at least 18 years with histologically confirmed staged I–III invasive breast cancer [13]; (b) patients who had been treated with surgery and SLNB or ALND, and had pathologically confirmed ALN status; (c) available preoperative MRI scan of breast tumor and/or ALN, including contrast-enhanced T1-weighted imaging (T1+C), T2-weighted imaging (T2WI), or diffusion-weighted imaging quantitatively measured apparent diffusion coefficients (DWI-ADC). The exclusion criteria were as follows: (a) biopsy conducted at an external institution and unavailable pathologic results; (b) lack of preoperative T1+C, T2WI, and DWI-ADC, simultaneously (c) previous or simultaneous presence of other tumors. The clinical T and clinical N stages were evaluated by imaging (MRI, ultrasonography, or positron emission tomography) or clinical examination according to the NCCN guideline [14]. The primary outcome was the ALN status, which was pathologically determined based on SLNB or ALND.
The patients were divided into three cohorts: (1) the training cohort (n = 803 from Sun Yat-sen Memorial Hospital of Sun Yat-sen University, a national hospital [Guangzhou, China], and Sun Yat-sen University Cancer Center, a national hospital [Guangzhou, China]), (2) the prospective-retrospective validation cohort (n = 106 from prospective phase III clinical trials [NCT01503905] collected from Sun Yat-sen Memorial Hospital of Sun Yat-sen University, a national hospital [Guangzhou, China]), (3) and the external validation cohort (n = 179 from the Shunde Hospital of Southern Medical University [Foshan, China] and Tungwah Hospital of Sun Yat-sen University [Dongguan, China]). All of the patients in the prospective-retrospective validation cohort underwent neoadjuvant chemotherapy before surgery.
Breast tumors were classified into four molecular subtypes according to St. Gallen Consensus Conference 2013 [15]. Patients who were estrogen receptor (ER) and progesterone receptor (PR)-positive, human epidermal growth factor receptor 2 (HER2)-negative, and had a Ki67 level of <14% in breast tumors were defined as Luminal A subtype patients. Luminal B subtype patients included ER-positive and HER2 overexpressed or amplified patients, or ER-positive and HER2-negative patients with at least one of the following situations: Ki67 >14%, PR negative or low PR. If ER and PR were absent, HER2-positive subtype patients were distinguished by HER2 overexpression or overamplification, whereas those with HER2-negative breast tumors were classified as the triple-negative subtype. These biomarkers were measured with immunohistochemical methods or in situ hybridization.
2.2. Radiomic features extraction
The protocol of multi-sequence MRI acquisition across four institutions and the parameters of MR scanners for patients are shown in appendix pp 9, 10. Multi-sequence MRI images from all of the cohorts were retrieved from the Picture Archiving and Communication System, and the N4ITK Bias Field Correction module (https://www.slicer.org/wiki/Documentation/Nightly/Modules/N4ITKBiasFieldCorrection) in the 3D Slicer software (https://www.slicer.org/, version 4.10.2) [16] was applied to obtain the standard normal distribution of image intensities and reduce the possible heterogeneity before feature extraction. Initially, the regions of interest (ROI) of tumors and lymph nodes were semi-automatically delineated on each slice obtained via T1+C sequence by 3D Slicer software. Then, the ROIs of the breast tumor area (ROI-1) and ALN area (ROI-2) comprising all lymph nodes that can be seen were applied to T2WI and DWI-ADC (delineated with b value of 800 or 1000 s/mm2 and then copied to the corresponding ADC maps) sequences, in which elaborated matching was done by the “transform section” in 3D Slicer software before radiomic feature extraction. In the process of image segmentation for identifying the ALN and tumor regions, MRI images were randomly assigned to and delineated by two radiologists (N Lu and XH Li) who were blinded to the patients’ clinical outcomes from different institutions. All delineated ROIs and ROIs for which consensus had not been reached were reassessed under the guidance of two senior radiologists (CM Xie and Z Wu) with more than 20 years of experience in MRI interpretation. Next, radiomic features corresponding to the quantitative data obtained after computational translation of images of ROIs were extracted using the SlicerRadiomics extension in 3D Slicer software, the in-house texture extraction platform developed based on the python package “PyRadiomics”, which has shown to conform to the Image Biomarker Standardization Initiative (IBSI) guidelines. A total of 863 radiomic features were extracted from each ROI in each sequence. The features included 12 diagnostic features, 107 original features (consisting of seven categories features: first-order statistics, shape-based, gray level co-occurrence matrix [GLCM], gray level run length matrix [GLRLM], gray level size zone matrix [GLSZM], neighbouring gray tone difference matrix [NGTDM], and gray level dependence matrix [GLDM]), and 744 wavelet features (also composed of seven categories of features). Therefore, a total of 5178 radiomic features were extracted from ROI-1s and ROI-2s in T1+C, T2WI, and DWI-ADC sequences. The extracted radiomic features from ROIs of each MRI sequence were normalized separately using the Z-score to make the dynamic ranges comparable before radiomic feature selection.
2.3. Tumor and lymph node radiomic signature construction and validation
The signatures were built based on the training cohort with MRI radiomic features that were extracted from the breast primary tumor and the ALN regions of T1+C, T2WI, and DWI-ADC sequences. First, a t- test was used to detect the associations between each feature and the patients’ ALN status; MRI radiomic features achieving significance at P < 0·05 during the t- test were entered into further selection. Next, the random forest algorithm [17] was used to select the top 30 radiomic features based on the mean decrease Gini index from the T1+C, T2WI, and DWI-ADC sequences of ROI-1 and ROI-2, respectively, in the training cohort. Then, a total of 180 top features identified by the random forest algorithm were applied for the construction of the signature via the support vector machine (SVM) algorithm [18], with the model type of c-classification and the kernel type of radial basis. The predictive accuracy of the ALN-tumor radiomic signature for predicting ALNM was initially assessed in the training cohort and then validated in the prospective-retrospective validation and external validation cohorts by receiver operating characteristic (ROC) curve analysis.
2.4. Multiomic signature construction and validation
The independent sample t- test was used to assess the association between clinical characteristics, pathologic characteristics, molecular subtype, and ALN status in the training cohort. To provide clinicians with a quantitative tool to predict individual probability of ALNM, all of these characteristics with P < 0·05 and key MRI radiomic features were used for the construction of the multiomic signature via the SVM algorithm. ROC curve analysis was also performed to evaluate the performance of the multiomic signature in the training cohort and was validated in the prospective-retrospective validation and external validation cohorts.
2.5. Radiomic features varied after neoadjuvant chemotherapy
A total of 45 (42%) of 106 patients from the prospective-retrospective validation cohort both received MRI examination before and after the neoadjuvant chemotherapy. The clustering heatmaps of the key radiomic features extracted from ALN and tumor regions were performed with the R package pheatmap. The differentiated key radiomic features after neoadjuvant chemotherapy were analysed using the paired-samples t- test.
2.6. MRI radiomics features associated with tumor microenvironment
The t- test was utilized to identify differentially expressed genes [19] associated with the radiomic score in 91 patients with T1+C and T2WI sequences MRI from The Cancer Genome Atlas (TCGA) and The Cancer Imaging Archive (TCIA). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using the clusterProfiler R package [20]. The GO terms and KEGG pathways were considered statistically significant if their P values and false-discovery rates were lower than 0.05.
The CIBERSORT algorithm [21] and the LM22 gene signature were used for highly sensitive and specific discrimination of 22 human immune cell phenotypes. CIBERSORT is a deconvolution algorithm that uses a set of reference gene expression values (a signature with 547 genes) that is considered a minimal representation for each cell type. Based on those values, CIBERSORT infers the cell type proportions in the data from bulk tumor samples with mixed cell types using support vector regression. The gene expression profiles were prepared using standard annotation files, the data were uploaded to the CIBERSORT web portal (http://cibersort.stanford.edu/), and the algorithm was run using the LM22 signature at 1,000 permutations.
Next, the data of 12,578 long non-coding RNAs of transcriptome RNA sequencing based on the Illumina platform and 23,381 types of methylated site data based on both the Illumina Human Methylation 27 and the Illumina Human Methylation 450 platforms were used for analysis. The top 30 ALNM-associated long non-coding RNAs and types of methylated sites were selected using the random forest algorithm. Correlation analysis was used to estimate the strength of the correlations with Spearman ρ.
3. Statistical analysis
The χ2 test was performed to examine the differences in the categorical variables, and the independent-samples t- test was applied to evaluate the differences in the continuous variables between two groups. The predictive accuracy of the signatures was assessed by ROC analysis. The area under ROC curve (AUC) was used to evaluate sensitivity and specificity in each signature. Additionally, decision curve analysis (DCA) was performed to assess the clinical utility of the prediction model by quantifying the net benefits when different threshold probabilities were considered [22]. The patients were categorized into high- and low-radiomic score groups, with the optimal cutoff values defined by the R package cutpointr. Heatmaps were generated to show the distribution and expression levels of radiomic features, immune cells, long non-coding RNAs, and types of methylated sites. Survival was calculated using the Kaplan-Meier method and the log-rank test, and hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated by Cox regression analysis. For all the analyses, two-sided P-values less than 0·05 were considered statistically significant. Statistical analyses were performed using R software (version 4.0.2). This study was registered with ClinicalTrials.gov, number NCT04003558 (Sun Yat-sen University Cancer Center are included in the training cohort now).
3.1. Role of the funding source
The funding sources had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data and the final responsibility, with the agreement of all authors, for the decision to submit for publication.
4. Results
4.1. Patients’ characteristics
A total of 1,161 early-stage invasive breast cancer patients were retrospectively recruited; of them, 1,088 patients were eligible for this study, while 73 patients did not pass the quality control (55 patients did not have histologically confirmed staged I–III invasive breast cancer, and 18 patients lacked MRI before surgery). The study design and the study workflow are shown in Fig. 1 and appendix p 1, respectively. Table 1 shows the clinicopathologic characteristics of the patients in the training cohort (n = 803), the prospective-retrospective validation cohort (n = 106), and the external validation cohort (n = 179). Of the 1,088 patients, 389 (35·75%) initially received the diagnosis of positive ALN status by radiologists through MRI, but 106 (27·25%) of them did not have ALNM and were confirmed as negative ALN status with pathologic examination. In contrast, 190 (28·27%) of 672 patients with clinical negative ALN status were found to have pathologic positive ALN status.
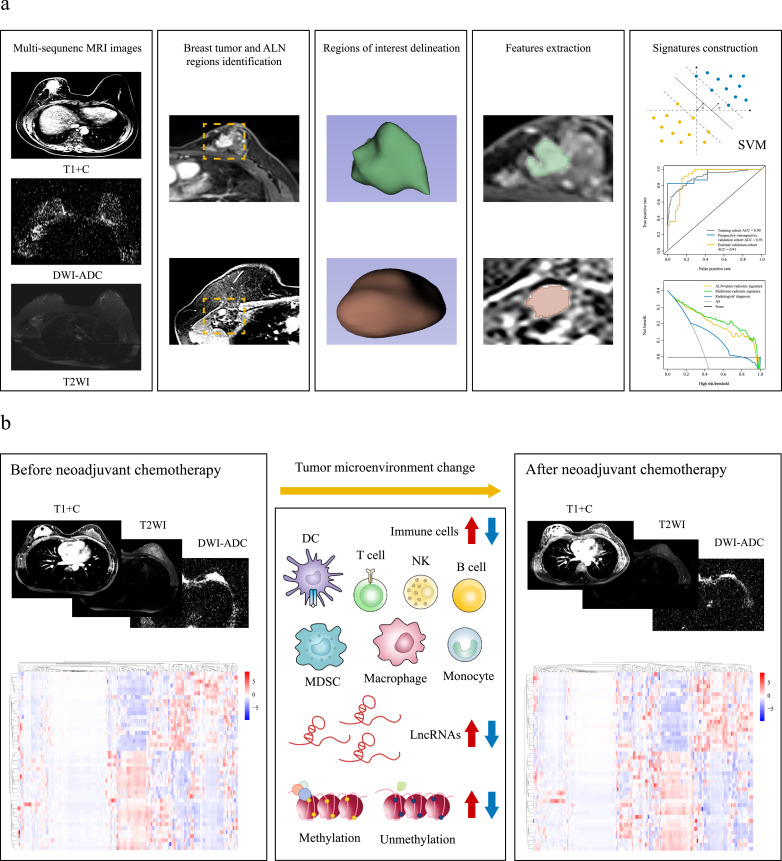
Fig. 1.
Study workflow
(a) Radiomic workflow. Multi-sequence MRI images were used for breast tumor and ALN region identification and the delineation of regions of interest, then features were extracted using 3D Slicer software for the signatures construction. (b) The association between MRI radiomics and tumor microenvironment. MRI=magnetic resonance imaging. ALN = axillary lymph node. T1+C=contrast-enhanced T1-weighted imaging. T2WI=T2-weighted imaging. DWI-ADC=diffusion-weighted imaging quantitatively measured the apparent diffusion coefficient. SVM=support vector machine. DC= dendritic cell. MDSC=myeloid-derived suppressor cell. NK=natural killer cell. LncRNA=long non-coding RNA.
Table 1.
Clinicopathologic characteristics between axillary lymph node positive and negative groups in the training, prospective-retrospective validation and external validation cohorts.
| Training cohort | Prospective-retrospective validation cohort | External validation cohort | ||||
|---|---|---|---|---|---|---|
| (No. of patients[n] = 803) | (n =106) | (n =179) | ||||
| Axillary lymph node status | Axillary lymph node status | Axillary lymph node status | ||||
| Characteristic | Negative | Positive | Negative | Positive | Negative | Positive |
| (n = 475) | (n = 328) | (n = 28) | (n = 78) | (n = 94) | (n = 85) | |
| Age, years (median [IQR]) | 48 [43,56] | 48 [41,56] | 42 [39,46] | 41 [38,45] | 49 [43,56] | 46 [42,56] |
| Number of tumors (%) | ||||||
| 1 | 421 (88·6) | 284 (86·9) | 21 (75·0) | 62 (79·5) | 76 (80·9) | 72 (84·7) |
| >1 | 54 (11·4) | 43 (13·1) | 7 (25·0) | 16 (20·5) | 18 (19·1) | 13 (15·3) |
| Tumor size, cm (median [IQR]) | 2·0 [1·6, 2·7] | 2·5 [2·0, 3·2] | 3·6 [2·6, 5·0] | 3·1 [2·5, 5·1] | 2·3 [1·7, 2·8] | 2·8 [2·1, 3·7] |
| Clinical T stage (%) | ||||||
| T1 | 241 (50·7) | 97 (29·6) | 0 (0·0) | 12 (15·4) | 39 (41·5) | 18 (21·2) |
| T2 | 213 (44·8) | 209 (63·7) | 22 (78·6) | 44 (56·4) | 50 (53·2) | 58 (68·2) |
| T3 | 16 (3·4) | 15 (4·6) | 4 (14·3) | 18 (23·1) | 3 (3·2) | 7 (8·2) |
| T4 | 5 (1·1) | 7 (2·1) | 2 (7·1) | 4 (5·1) | 2 (2·1) | 2 (2·4) |
| Clinical N stage (%) | ||||||
| N0 | 382 (80·4) | 132 (40·2) | 15 (60·0) | 21 (29·6) | 85 (96·6) | 37 (50·0) |
| N1 | 90 (18·9) | 178 (54·3) | 10 (40·0) | 45 (63·4) | 3 (3·4) | 20 (27·0) |
| N2 | 2 (0·4) | 18 (5·5) | 0 (0·0) | 4 (5·6) | 0 (0·0) | 16 (21·6) |
| N3 | 1 (0·2) | 0 (0·0) | 0 (0·0) | 1 (1·4) | 0 (0·0) | 1 (1·4) |
| Clinical TNM stage (%) | ||||||
| I | 207 (43·6) | 51 (15·5) | 0 (0·0) | 4 (5·6) | 37 (42·0) | 12 (16·2) |
| II | 253 (53·3) | 241 (73·5) | 22 (88·0) | 51 (71·8) | 48 (54·5) | 42 (56·8) |
| III | 15 (3·2) | 36 (11·0) | 3 (12·0) | 16 (22·5) | 3 (3·4) | 20 (27·0) |
| Histological grade (%) | ||||||
| Grade 1 (low) | 22 (5·0) | 2 (0·6) | 0 (0·0) | 0 (0·0) | 4 (4·9) | 5 (6·7) |
| Grade 2 (intermediate) | 235 (53·0) | 149 (46·1) | 10 (50·0) | 43 (72·9) | 46 (56·1) | 33 (44·0) |
| Grade 3 (high) | 186 (42·0) | 172 (53·3) | 10 (50·0) | 16 (27·1) | 32 (39·0) | 37 (49·3) |
| Pathologic T stage (%) | ||||||
| T1 | 272 (57·3) | 140 (42·7) | 19 (67·9) | 30 (40·0) | 45 (47·9) | 23 (27·1) |
| T2 | 188 (39·6) | 171 (52·1) | 7 (25·0) | 32 (42·7) | 45 (47·9) | 55 (64·7) |
| T3 | 13 (2·7) | 16 (4·9) | 1 (3·6) | 7 (9·3) | 2 (2·1) | 6 (7·1) |
| T4 | 2 (0·4) | 1 (0·3) | 1 (3·6) | 6 (8·0) | 2 (2·1) | 1 (1·2) |
| Pathologic TNM stage (%) | ||||||
| I | 271 (57·1) | 0 (0·0) | 19 (67·9) | 0 (0·0) | 45 (47·9) | 0 (0·0) |
| II | 202 (42·5) | 206 (62·8) | 8 (28·6) | 28 (37·3) | 47 (50·0) | 44 (51·8) |
| III | 2 (0·4) | 122 (37·2) | 1 (3·6) | 47 (62·7) | 2 (2·1) | 41 (48·2) |
| ER status (%) | ||||||
| Negative | 64 (13·6) | 51 (15·5) | 5 (17·9) | 8 (10·3) | 20 (21·3) | 25 (29·8) |
| Positive | 407 (86·4) | 277 (84·5) | 23 (82·1) | 70 (89·7) | 74 (78·7) | 59 (70·2) |
| PR status (%) | ||||||
| Negative | 128 (27·2) | 91 (27·7) | 14 (50·0) | 23 (29·5) | 31 (33·0) | 29 (34·5) |
| Positive | 343 (72·8) | 237 (72·3) | 14 (50·0) | 55 (70·5) | 63 (67·0) | 55 (65·5) |
| HER2 status (%) | ||||||
| Negative | 326 (72·0) | 208 (65·4) | 15 (68·2) | 43 (65·2) | 59 (71·1) | 44 (62·9) |
| Positive | 127 (28·0) | 110 (34·6) | 7 (31·8) | 23 (34·8) | 24 (28·9) | 26 (37·1) |
| Ki67 status (%) | ||||||
| <30 | 259 (55·0) | 150 (45·7) | 12 (42·9) | 63 (80·8) | 52 (57·1) | 49 (59·8) |
| ≥30 | 212 (45·0) | 178 (54·3) | 16 (57·1) | 15 (19·2) | 39 (42·9) | 33 (40·2) |
| Molecular subtypes (%) | ||||||
| Luminal A | 92 (20·0) | 30 (9·2) | 4 (16·0) | 21 (30·0) | 20 (23·0) | 15 (19·7) |
| Luminal B | 306 (66·7) | 249 (76·6) | 16 (64·0) | 42 (60·0) | 50 (57·5) | 40 (52·6) |
| HER2-positive | 31 (6·8) | 28 (8·6) | 1 (4·0) | 3 (4·3) | 6 (6·9) | 12 (15·8) |
| Triple negative | 30 (6·5) | 18 (5·5) | 4 (16·0) | 4 (5·7) | 11 (12·6) | 9 (11·8) |
| Type of surgery (%) | ||||||
| Breast-conserving surgery | 250 (52·7) | 127 (38·7) | 14 (50·0) | 28 (35·9) | 22 (23·4) | 18 (21·2) |
| Others | 224 (47·3) | 201 (61·3) | 14 (50·0) | 50 (64·1) | 72 (76·6) | 67 (78·8) |
| Follow-up time, months (median [IQR]) | 23·4 [15·8, 35·6] | 21·7 [15·3, 35·1] | 45·5 [21·5, 65·2] | 44·6 [25·5, 57·1] | 22·6 [10·6, 41·5] | 24·3 [9·4, 55·1] |
IQR=interquartile range. TNM=tumor–node–metastasis. ER=estrogen receptor. PR=progesterone receptor. HER2=human epidermal growth factor receptor 2. Ki67=proliferation marker protein Ki67.
4.2. Tumor and lymph node radiomic signature for ALN metastasis discrimination
From a total of 5178 quantitative features from ALN and tumor regions of three MRI sequences by the random forest algorithm, 180 key radiomic features were selected. The classification of these features and the process to determine the optimal number of feature are shown in appendix pp 11, 12. Fig. 2a demonstrates the overall distribution of key radiomic features from T1+C, T2WI, and DWI-ADC sequences among patients with and without ALNM in the training cohort, which shows the obvious difference between the two groups. Incorporating these key features of the ALN region to predict ALNM yielded AUC values of 0·85, 0·61, and 0·81 in the training cohort, the prospective-retrospective validation cohort, and the external validation cohort, respectively (appendix p 2). Simultaneously, incorporating the three-sequence key features of the tumor region for ALNM prediction achieved an AUC of 0·78, 0·59, and 0·63 in the training cohort, the prospective-retrospective validation cohort, and the external validation cohort, respectively (appendix p 3). The AUC values for ALNM prediction of combining the multi-sequence features were higher than those when incorporating single-sequence features in the training and the validation cohorts (appendix p 13). We found that the groups of patients classified correctly by ALN or tumor radiomic signature were disjoint, which indicated that the radiomic signature reflecting phenotypes of the tumor and ALN regions may achieve better performance. The cases’ classification results were shown in the Sankey diagram (appendix p 4). We also compared the clinicopathologic characteristics between intersecting and disjoint groups in the validation cohorts. Except for the case origin, there were no differences for other characteristics between intersecting and disjoint groups (appendix pp 14, 15).
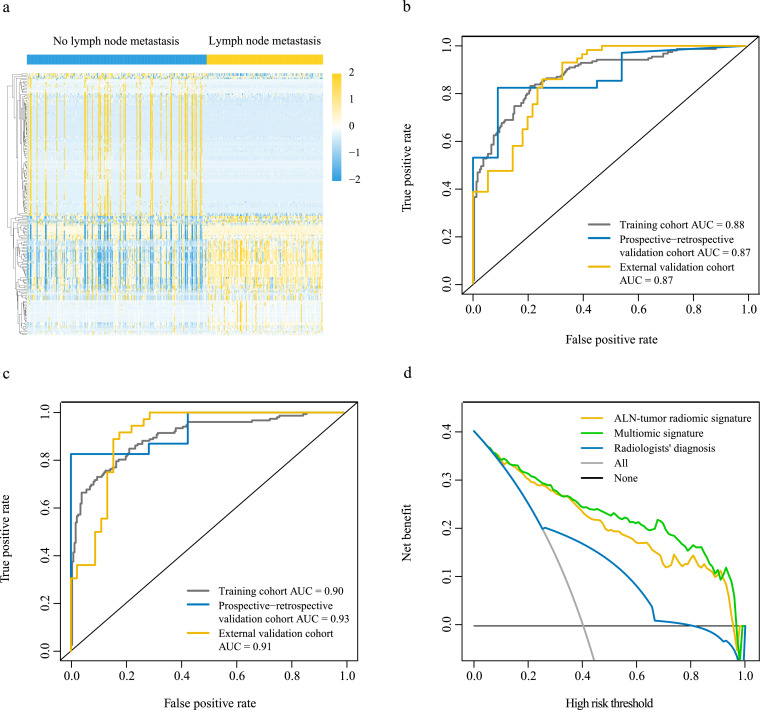
Fig. 2.
Performance and clinical value of magnetic resonance imaging radiomic signatures
(a) Overall distribution of key radiomic features from T1+C, T2WI, and DWI-ADC sequences among patients with and without ALN metastasis in the training cohort (n = 392). (b) Performance of the ALN-tumor radiomic signature for predicting ALN metastasis in the training (n = 392), the prospective-retrospective validation (n = 45), and the external validation (n = 112) cohorts. (c) Performance of the multiomic signature for predicting ALN metastasis in the training (n = 381), the prospective-retrospective validation (n = 30), and the external validation (n = 81) cohorts. (d) Decision curve analysis for the ALN-tumor radiomic signature and the multiomic signature (n = 381). ALN=axillary lymph node. AUC=area under the receiver operating characteristics curve. T1+C=contrast-enhanced T1-weighted imaging. T2WI=T2-weighted imaging. DWI-ADC=diffusion-weighted imaging quantitatively measured the apparent diffusion coefficient.
When combined both ALN and tumor regions features, the ALN-tumor radiomic signature was constructed and illustrated good performance in detecting ALNM in the training cohort (AUC, 0·88), the prospective-retrospective validation cohort (AUC, 0·87), and the external validation cohort (AUC, 0·87), which outperformed the ALN or tumor radiomic signature alone (Fig. 2b). The detailed evaluation indicators for model performance including sensitivity and specificity are summarized in appendix p 16.
When comparing the performance of the ALNM prediction between the ALN-tumor radiomic signature and radiologists, we found that the ALN-tumor radiomic signature achieved higher accuracy, AUC, and sensitivity than the diagnosis of radiologists on the same MRI in the training, the prospective-retrospective validation and the external validation cohorts (appendix p 17). Besides, we collected the information on radiologists’ evaluations of both MRI and ultrasound images of 348 patients from Sun Yat-sen Memorial Hospital; then, we compared the performance between the ALN-tumor radiomic signature and radiologists’ evaluations based on both MRI and ultrasound images. The results indicated that the ALN-tumor radiomic signature achieved higher sensitivity (0.83 vs. 0.60 and 0.49), and the clinical utility of the ALN-tumor radiomic signature was not worse than the standard clinical practice (appendix p 18).
4.3. Multiomic signature for ALN metastasis discrimination
In the univariate analysis, which is presented in appendix p 19, five differentially expressed clinical characteristics, including age (P = 0·012, independent-samples t- test), clinical T stage (P < 0·001, Chi-squared test), clinical N stage (P < 0·001, Chi-squared test), Ki67 expression (P = 0·010, Chi-squared test), and molecular subtype (P = 0·033, Chi-squared test), were found to be associated with ALN status in the training cohort. To develop a more precise and clinically applicable method that can predict an individual’s ALN status, we built a multiomic signature incorporating all key radiomic features of ALN and tumor regions, with clinical characteristics, pathologic characteristics, and molecular subtypes that were significantly associated with ALNM. The multiomic signature showed better performance in ALNM prediction and achieved a higher AUC (0·90) in the training cohort, the prospective-retrospective validation cohort (AUC, 0·93), and the external validation cohort (AUC, 0·91) (Fig. 2c). The sensitivity and specificity of the multiomic signature are listed in appendix p 16. As for the combination of these clinicopathologic characteristics and molecular subtype, it achieved an AUC of 0·74, 0·68, and 0·72 for predicting ALNM in the training cohort, the prospective-retrospective validation cohort, and the external validation cohort, respectively (appendix p 5).
The multiomic signature also exhibited the ability to discriminate ALNM patients with 1, 2, and 3 positive nodes (AUC of 0·88, 0·89, and 0·92, respectively, in the training cohort; AUC of 0·79, 1·00, and 0·93, respectively, in the prospective-retrospective validation cohort; and AUC of 0·97, 0·93, and 0·87, respectively, in the external validation cohort; appendix p 20). Moreover, to further assess the added value of the multiomic signature to the ALN status, we conducted subgroup analysis within patients with different molecular subtypes. Encouragingly, the multiomic signature could identify ALNM patients in the subgroups of Luminal A (AUC, 0·91, 0·91, respectively), Luminal B (AUC, 0·89, 0·92, respectively), human epidermal growth factor receptor 2 (Her-2) positive (AUC, 0·90, 0·76, respectively), and triple-negative breast cancer (TNBC) (AUC, 1·00, 1·00, respectively) patients in the training and external validation cohorts (appendix p 20). In addition, when stratified by other factors such as age, Ki67 expression level, and clinical T stage, the AUC value of the multiomic signature remained at 0·86–1·00 in these subgroups (appendix p 20).
According to the radiomic score of the multiomic signature, an optimal cutoff value of 0·334 was generated to classify the patients into high- and low-score groups in the training cohort. The high-score patients, with a high risk of ALNM, had significantly shorter disease-free survival compared with the low-score group (HR 0·43, 95% CI 0·21–0·86, P = 0·014, log-rank test; appendix p 6).
4.4. Multiomic signature for clinical decision-making
Decision curve analysis was conducted to determine the clinical usefulness of the multiomic signature by quantifying the net benefits at different threshold probabilities. The decision curve showed that if the threshold probability was >5%, the use of the multiomic signature to predict ALNM added more benefits than either the ALN-tumor radiomic signature or the radiologists’ diagnosis (Fig. 2d).
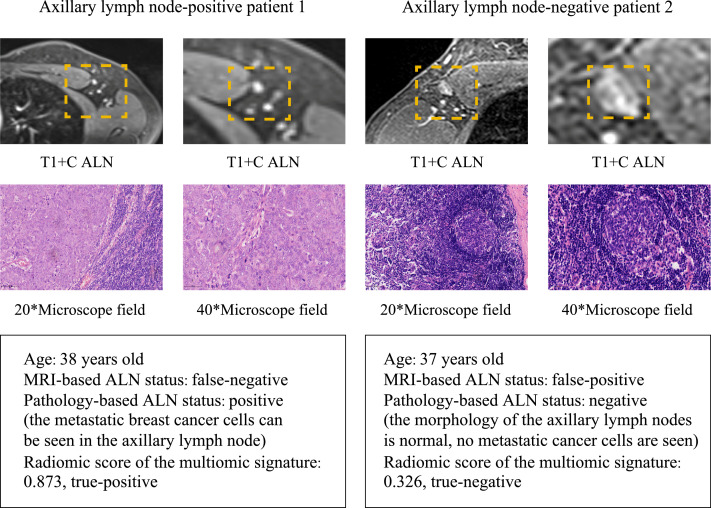
During clinical practice, patients’ clinical ALN status judged by the preoperative MRI is commonly inconsistent with the pathologic ALN status, and even senior radiologists may make mistakes. The multiomic signature could precisely recognize ALNM among patients with different clinical node stages in the training and validation cohorts (appendix p 20). As shown in Fig. 3, patient 1 had a pathologic positive ALN status but was considered as a non-ALNM patient by radiologists based on the MRI before surgery. On the contrary, patient 2 had initial diagnosis of ALNM by radiologists but was later found to be a non-ALNM patient by pathologic examination. The ALN status of the two patients was accurately assessed through the multiomic signature by the cutoff value of 0·334.
Fig. 3.
Patients with inconsistent MRI-based ALN status and pathology-based ALN status
Patient 1 had a pathologic positive ALN status, but the MRI-based status was considered negative by radiologists before surgery. In contrast, the MRI-based status of patient 2 had initially been marked as positive by radiologists, but it was later found to be negative on pathologic examination. The ALN status of two patients was accurately assessed through the multiomic radiomic signature by the cutoff value of 0·334. MRI=magnetic resonance imaging. ALN=axillary lymph node. T1+C=contrast-enhanced T1-weighted imaging.
4.5. Radiomics changed after neoadjuvant chemotherapy and was associated with the tumor microenvironment
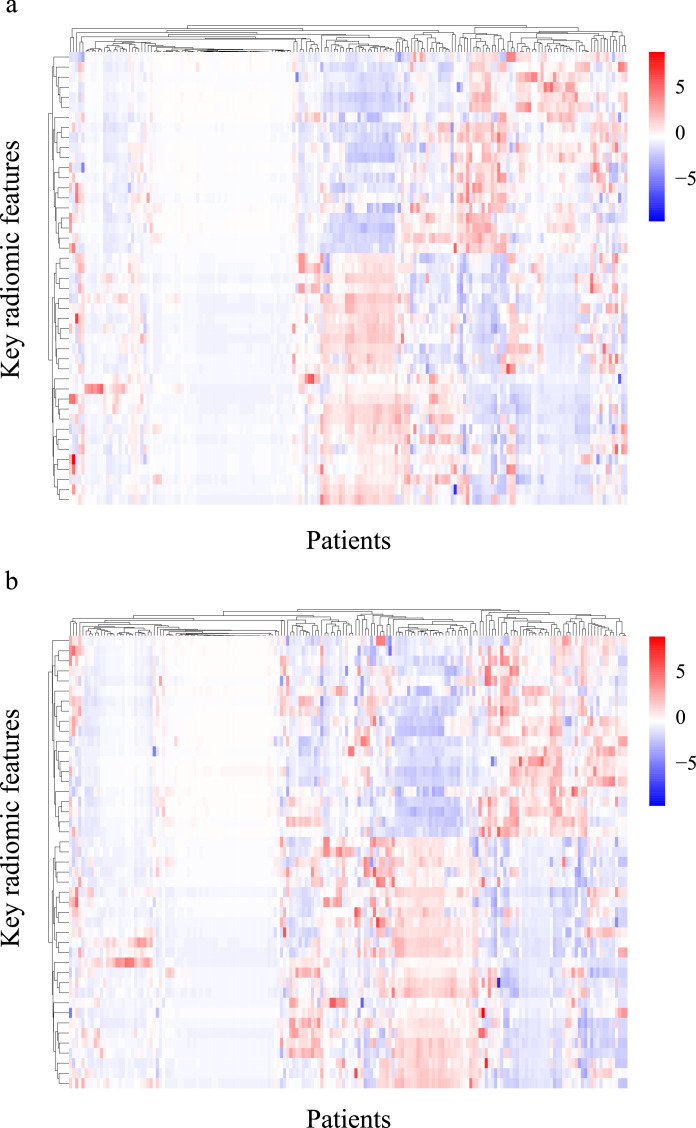
Among the 106 patients who underwent neoadjuvant chemotherapy before surgery in the prospective-retrospective validation cohort, 45 patients received MRI examination before the neoadjuvant chemotherapy. There were significant differences in the key radiomic features before and after neoadjuvant chemotherapy, especially in the GLDM-type features (Table 2). Moreover, the expression of 70 features was up-regulated, while the expression of 68 features was down-regulated. Comparison of the key radiomic features between neoadjuvant therapy response and non-response groups revealed two radiomic features (T1+C-wavelet-LLL-GLCM-Inverse difference moment normalized from tumor region and DWI-ADC-wavelet-HHH-glszm-Large area low gray level emphasis from ALN region) that were significantly associated with the response to the chemotherapy. Fig. 4 shows that the clustering of key radiomic features changed greatly after neoadjuvant chemotherapy, which may be explained by changes of the tumor microenvironment.
Table 2.
The differentiation of radiomics before and after neoadjuvant therapy.
| Radiomic features class | Tumor region |
ALN region |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1+C |
T2WI |
DWI-ADC |
T1+C |
T2WI |
DWI-ADC |
||||||||
| Up-regulation features | Down-regulation features | Up-regulation features | Down-regulation features | Up-regulation features | Down-regulation features | Up-regulation features | Down-regulation features | Up-regulation features | Down-regulation features | Up-regulation features | Down-regulation features | ||
| Shape | 1 | 2 | 1 | 1 | 2 | 0 | 0 | 2 | 0 | 2 | 4 | 0 | |
| First-order | 0 | 0 | 1 | 2 | 2 | 1 | 3 | 3 | 0 | 1 | 1 | 0 | |
| GLCM | 1 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| GLSZM | 2 | 1 | 2 | 4 | 7 | 2 | 3 | 0 | 0 | 0 | 2 | 0 | |
| GLDM | 5 | 2 | 3 | 2 | 6 | 1 | 4 | 5 | 2 | 6 | 3 | 2 | |
| GLRLM | 1 | 2 | 2 | 2 | 3 | 3 | 2 | 4 | 3 | 8 | 3 | 3 | |
| NGTDM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Number of key radiomic features under each class that differentiated (P < 0·05) before and after neoadjuvant therapy in 45 patients from the prospective-retrospective validation cohort. ALN=axillary lymph node. T1+C=contrast-enhanced T1-weighted imaging. T2WI=T2-weighted imaging. DWI-ADC=diffusion-weighted imaging quantitatively measured the apparent diffusion coefficient. GLCM=gray-level co-occurrence matrix. GLSZM=gray-level size zone matrix. GLDM=gray-level dependence matrix. GLRLM=gray-level run length matrix. NGTDM=neighboring gray-tone difference matrix.
Fig. 4.
Clustering of key radiomic features before and after neoadjuvant chemotherapy
The clustering of 180 key radiomic features (a) before neoadjuvant chemotherapy and (b) after neoadjuvant chemotherapy in 45 patients from the prospective-retrospective validation cohort.
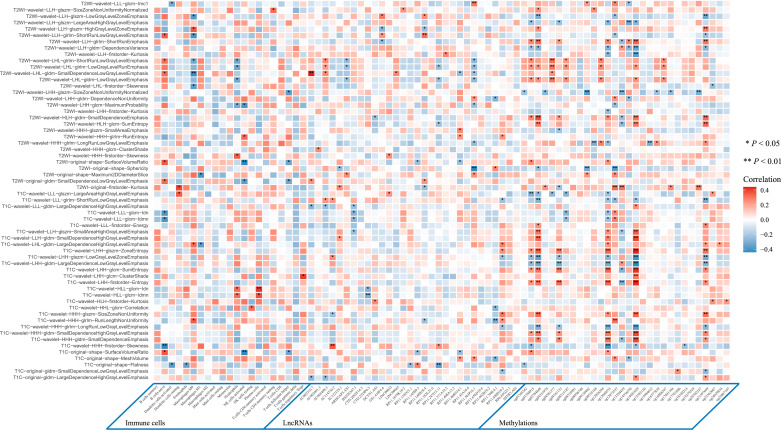
To determine whether radiomics can reflect the tumor microenvironment, we further explored the association between MRI radiomic features and the tumor microenvironment including 22 immune cells, long non-coding RNAs, and types of methylated sites in 90 patients with breast cancer from TCGA and TCIA with T1+C and T2WI sequences MRI. The expression level of 395 long non-coding RNAs and the enrichment level of 1,784 types of methylated sites were found to be significantly different between the ALNM and non-ALNM patients. Moreover, the top 30 ones were selected via the random forest algorithm for further analysis. The overall distribution of key tumor microenvironment features among patients with and without ALNM is shown in appendix p 7. Correlation analysis showed that the key radiomic features of ALN and tumor regions remarkably and linearly correlated with immune cells, such as M0 macrophages, B naïve cells, and neutrophils; long non-coding RNAs, such as P11.563P16.1 and RP11.888D10.3; methylated sites, such as cg14681629 and cg02784848 (Fig. 5).
Fig. 5.
The association between MRI radiomics and tumor microenvironment
The correlation between key MRI radiomic features and immune cells, lncRNAs and types of methylated sites in 90 breast cancer patients from The Cancer Genome Atlas and The Cancer Imaging Archive. Correlation analysis was used to estimate the strength of the correlations with Spearman ρ. T1+C=contrast-enhanced T1-weighted imaging. T2WI=T2-weighted imaging. LncRNA=long non-coding RNA. MRI= magnetic resonance imaging.
According to the cutoff value of the radiomic score from the T1+C and T2WI sequence signatures of the tumor region in the training cohort, the patients were classified into two groups. In total, 1,381 T1+C and T2WI sequence-based differentially expressed genes (DEGs) were obtained among the low-score and high-score patients. COX6B2 was most up-regulated and PPP2R2C was most down-regulated in high-score patients. Next, the GO and KEGG analyses were performed to further examine the biological functions of the identified radiomic-based genes. The GO enrichment analysis showed that the radiomic-based genes were enriched in various physiological metabolic processes, such as affection of transmembrane transporter activity, NADP binding, and ATPase complex (appendix pp 8, 21). The KEGG pathway enrichment analysis found that these genes were involved in the oxidative phosphorylation signaling pathway (appendix p 21).
5. Discussion
Unlike previous studies focusing exclusively on the tumor region or ALN region for predicting ALNM [11,23–24], in this study, the ALN-tumor radiomic signature combining multi-sequence key radiomic features of ALN and tumor regions showed a high predictive ability and could be applied to predict the ALN status precisely. The multiomic signature integrating the radiomic features above, ALNM-associated clinicopathologic characteristics, and molecular subtype demonstrated better performance in predicting ALNM. In addition, the multiomic signature improved the accuracy of ALN status evaluation in patients with inconsistent clinical and pathologic ALN status; moreover, it accurately recognized ALNM in patients with a different number of positive lymph nodes, different clinical T stages, different age, different Ki67 expression levels, and different molecule subtypes. Also, the key radiomic features were found to be associated with the tumor microenvironment. The findings indicated that the multiomic signature is an effective tool for ALNM prediction, which provides useful messages for diagnosis and treatment decision preoperatively. It can also prevent unnecessary ALND and even SLNB in patients with early-stage invasive breast cancer.
Breast cancer patients have distinct ALN status when they are newly diagnosed, and the treatment plans are also different depending on their ALN status. It has been demonstrated that patients with positive ALN status have poorer outcomes than those with negative ALN status [25]. It is important for clinician’s decision-making that patients with a positive ALN status are considered as high-risk patients and need to undergo adjuvant chemotherapy according to NCCN guidelines [14]. Some TNBC patients with a larger tumor burden and clinical positive ALN status are also considered for neoadjuvant therapy. It should be noted that the misjudgement of the ALN status is a cause of patients’ overtreatment and a waste in health care. As for the patients with negative ALN status and without other risk factors, they can be treated with endocrine therapy alone, which translates to lower costs and more comfortable treatment experience. Thus, it is very important to accurately and clearly distinguish the ALN status for patients with early-stage breast cancer.
During clinical practice, patients’ clinical ALN status judged by the preoperative MRI is commonly inconsistent with the pathologic ALN status, and even senior radiologists may make mistakes. To reduce the misjudgment, tumor size, grade and other tumor-related factors have been used to predict lymph node metastasis. Some researchers have shown that the radiomic features derived from tumors containing a lot of biological information could also predict tumor metastasis, therefore the combination of radiomic features derived from tumor and lymph node regions for ALNM prediction was explored in this study. According to the ‘seed and soil’ theory, ALNM initiation depends on the synergies of the tumor cells (seed) and the ALN microenvironment (soil), which may explain why including the MRI profiles of the tumor and ALN regions can improve the performance of the lymph node diagnostic model.
Besides MRI examination, ultrasound is also favored by clinicians in clinical practice because of its wide application and easily accessible data for study. Previous research demonstrated that an approach based on ultrasound radiomics also showed acceptable performance for ALN status prediction in early-stage breast cancer patients [8,26]. However, compared with MRI examination, operators’ experience and heterogeneity from the ultrasound beam’s angle of incidence often affect the standardization of ultrasound image data, which results in the lack of reproducibility of the model. As MRI examination possesses the advantage of showing the three-dimensional spatial position of each organ and tissue with high resolution, it can provide more comprehensive information for diagnosis and TNM staging; therefore, MRI has been recommended for lesions that are difficult to be visualized by ultrasound and mammography, and for preoperative estimation of breast cancer [27].
Additionally, the ALN-tumor signature based on MR radiomic in this study illustrated its superiority over general radiologists in detecting ALNM. Hence, it may be beneficial to combine MR radiomics with ultrasound radiomics, which may achieve better performance for ALNM prediction; this is especially useful for improving the diagnosis and treatment level in hospitals in rural areas as a compensation for the disadvantages in these areas with insufficient medical resources. Some ALNs that are not visible or not captured by breast MRIs would not be missed if combined with ultrasound radiomics.
Previous studies have indicated that the tumor microenvironment harbours a variety of immune cells, blood vessels, and extracellular matrix, and changes in the distribution of immune cells and blood vessel formation may facilitate the development and metastasis of tumors [28]. It has also been shown that the tumor microenvironment changes greatly after neoadjuvant chemotherapy. Therefore, many tumor-microenvironment-related factors, such as cancer-associated fibroblasts, myeloid-derived suppressor cells, and tumor-associated macrophages, have been used to predict tumor metastasis and therapeutic effects [[29], [30], [31]]. It has been reported that radiomic features could reflect the molecular and gene features of tumor and even provide a large volume of information on the tumor microenvironment [[32], [33], [34]]. In this study, the key radiomic features of ALN and tumor regions remarkably and linearly correlated with immune cells, long non-coding RNAs, and methylated sites. Besides, there were significant differences in the key radiomic features before and after neoadjuvant chemotherapy, which may be explained by changes of the tumor microenvironment. Moreover, the up- and down-regulation of genes in the patients with a high radiomic risk score and the function of these genes to develop metastases may reveal the potential biological underpinning of MRI radiomics [35–36]. Based on these results, we think radiomic profiles could help identify potential targets for molecular-based therapy of breast cancer, which shows implications for personalized medicine. In the future, we plan to conduct targeted regulation of these immune cells or epigenetic molecules in the breast cancer metastasis mouse model; the mice will receive an MRI examination during the treatment so that the dynamic changes of the radiomic features during the treatment can be monitored. In that way, we can verify the scientific hypothesis that targeted regulation of immune cells or epigenetic molecules can reverse the radiomic changes and improve the prognosis of breast cancer patients. This hypothesis will provide a new theoretical and experimental basis for the precision medicine of breast cancer. Based on the finding that the ALN-tumor radiomic signature with the integration of radiomic features from the tumor and the ALN regions could effectively predict ALNM, the combination of radiomics and tumor microenvironment characteristics is promising to improve the predictive ability for ALNM.
Several limitations still need to be addressed in our study. First, the clinical information and MRI sequences of some patients were missing because of the study’s retrospective nature, which led to a decrease in the sample size. Second, although the normalization was applied during data pre-processing, the heterogeneity of the MRI scans from multiple centers was inevitable. Third, the image segmentation method used in this study was based on manual delineation, which could be replaced by automatic delineation of ROIs using deep learning methods to improve the reproducibility of our model in the future. Fourth, because our finding that radiomic features may reflect the change of tumor microenvironment was based on indirect evidence, validation and experimental proof are needed in the future. Fifth, for the current study design cannot address the uncertainty and variability in sentinel lymph node diagnoses, we will construct a model for sentinel lymph node metastasis and extra-sentinel lymph node metastasis prediction respectively in the future to solve this problem. Besides, since the TCIA database only contains MRI images of the T1+C and T2WI sequences, without the DWI-ADC sequence, TCIA data were not used to verify our signature performance or in any other analyses. We also failed to construct an ALNM prediction model with a combination of tumor microenvironment features, MR radiomics, and ultrasound radiomics, but further exploration could be conducted in the future to realize more precise prediction for ALNM.
In conclusion, this study revealed the clinical value of machine learning algorithms and presented the multiomic signature incorporating MRI multi-sequence key radiomic features of ALN and tumor regions with clinicopathologic characteristics and molecular subtype. It can be conveniently used to identify ALNM patients among different molecular subtypes in early-stage invasive breast cancer and may eventually result in a preoperative approach to guide future clinical practice. Furthermore, we also found significant changes in MRI radiomics after neoadjuvant chemotherapy and the association between MRI radiomic features and tumor microenvironment features including immune cells, long non-coding RNAs, and types of methylated sites, which may reveal the potential biological underpinnings of MRI radiomics.
Contributors
All authors conceived and designed the study. HR Yao, YF Yu, ZF He, J Ouyang, YJ Tan, CM Xie analysed and interpreted the data, and did the statistical analysis. All authors drafted and revised the manuscript. HR Yao, YF Yu, ZF He, J Ouyang, YJ Tan, CM Xie provided administrative, technical, and material support. HR Yao, YF Yu, ZF He, J Ouyang, YJ Tan, CM Xie verified the underlying data. HR Yao provided study supervision. All authors reviewed the manuscript and approved the final version.
Data sharing
The codes, tabulated feature values and expected output involved in the body text are available in a publicly accessible repository (https://github.com/ZifanHe/Ebiomedicine.git). To protect the privacy of the patients, other data related to patients cannot be made available for public access but MRI datasets and all the codes used for analyses following feature extraction from this manuscript are saved at the Ethics Committee of Sun Yat-sen Memorial Hospital and are available from the corresponding author (yaoherui@mail.sysu.edu.cn) upon reasonable request approved by the Ethics Committee. TCGA and TCIA data are available online. All experiments and implementation details are described thoroughly in the Methods section and appendix.
Declaration of Competing Interest
We declare no conflicts of interest.
Acknowledgments
This study was supported by Grant 2020ZX09201021 from the National Science and Technology Major Project, Grant YXRGZN201902 from the Medical Artificial Intelligence Project of Sun Yat-Sen Memorial Hospital, Grants 81572596 and 81972471 from the National Natural Science Foundation of China, Grant 2017A030313828 from the Natural Science Foundation of Guangdong Province, Grant 201704020131 from the Guangzhou Science and Technology Major Program, Grant 2017B030314026 from the Guangdong Science and Technology Department, Grant 2018007 from the Sun Yat-Sen University Clinical Research 5010 Program, Grant SYS-C-201801 from the Sun Yat-Sen Clinical Research Cultivating Program, Grant A2020558 from the Guangdong Medical Science and Technology Program, Grant SYSU-81000-20200311-0001 and SYSU-05160-20200506-0001 from the Tencent Charity Foundation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103460.
Contributor Information
Chuanmiao Xie, Email: xiechm@sysucc.org.cn.
Herui Yao, Email: yaoherui@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Armando E., Karla V., Linda M.C. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: the ACOSOG Z0011 (alliance) randomized clinical trial. JAMA. 2017;318(10):918–926. doi: 10.1001/jama.2017.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galimberti V., Bernard F., Viale G. Axillary dissection versus no axillary dissection in patients with breast cancer and sentinel-node micrometastases (IBCSG 23-01): 10-year follow-up of a randomized, controlled phase 3 trial. Lancet Oncol. 2018;19(10):1385–1393. doi: 10.1016/S1470-2045(18)30380-2. [DOI] [PubMed] [Google Scholar]
- 3.Lyman G.H., Giuliano A.E., Somerfield M.R. American society of clinical oncology guideline recommendations for sentinel lymph node biopsy in early-stage breast cancer. J Clin Oncol. 2005;23(30):7703–7720. doi: 10.1200/JCO.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Gary H., Mark R., Linda D., Cheryl L., Donald L., Armando E. Sentinel lymph node biopsy for patients with early-stage breast cancer: American society of clinical oncology clinical practice guideline update. J Clin Oncol. 2017;35(5):561–564. doi: 10.1200/JCO.2016.71.0947. [DOI] [PubMed] [Google Scholar]
- 5.Giammarile F., Alazraki N., Aarsvold J.N. The EANM and SNMMI practice guideline for lymphoscintigraphy and sentinel node localization in breast cancer. Eur J Nucl Med Mol Imaging. 2013;40(12):1932–1947. doi: 10.1007/s00259-013-2544-2. [DOI] [PubMed] [Google Scholar]
- 6.George E., Roberts Sacha, Cheryl L. Quantifying the impact of axillary surgery and nodal irradiation on breast cancer-related lymphedema and local tumor control: long-term results from a prospective screening trial. J Clin Oncol. 2020;38(29):3430–3438. doi: 10.1200/JCO.20.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samiei S., Cornelis M., Marc B., Kristien B., Thiemo J., Marjolein LS. Diagnostic performance of noninvasive imaging for assessment of axillary response after neoadjuvant systemic therapy in clinically node-positive breast cancer: a systematic review and meta-analysis. Ann Surg. 2021;273(4):694–700. doi: 10.1097/SLA.0000000000004356. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X.Y., Zhao Y., Huang Y.N. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun. 2020;11(1):1236. doi: 10.1038/s41467-020-15027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z.Y., Li Z.L., Qu J.R. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 10.Park H., Lim Y.J., Eun S.K. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2018;24(19):4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y.F., Tan Y.J., Xie C.M. Development and validation of a preoperative magnetic resonance imaging radiomics-based signature to predict axillary lymph node metastasis and disease-free survival in patients with early-stage breast cancer. JAMA Netw Open. 2020;3(12) doi: 10.1001/jamanetworkopen.2020.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fengxi Su. Neoadjuvant chemotherapy for operable premenopausal breast cancer patients. Updated May 24, 2106. https://www.clinicaltrials.gov/ct2/show/NCT01503905?cond=NCT01503905&draw=2&rank=1. Accessed November 4, 2020.
- 13.Armando E., Stephen B., Gabriel N. Eighth edition of the AJCC cancer staging manual: breast cancer. Ann Surg Oncol. 2018;25(7):1783–1785. doi: 10.1245/s10434-018-6486-6. [DOI] [PubMed] [Google Scholar]
- 14.NCCN clinical practice guidelines in oncology breast cancer version 3.2021—March 29, 2021.
- 15.Goldhirsch A., Winer E., Coates A. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedorov A., Beichel R., Jayashree K.C. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 18.Noble W.S. What is a support vector machine? Nat Biotechnol. 2006;24(12):1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 19.Ritchie M., Phipson B., Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G., Wang L.G., Han Y., He QY. Cluster profiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman A.M., Liu C.L., Green M.R. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr K.F., Brown M.D., Zhu K., Janes H. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol. 2016;34(21):2534–2540. doi: 10.1200/JCO.2015.65.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Sun D., Chen L.L. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol. 2019;9:980. doi: 10.3389/fonc.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L., Zhu Y.B., Liu Z.Y. Radiomic nomogram for prediction of axillary lymph node metastasis in breast cancer. Eur Radiol. 2019;29(7):3820–3829. doi: 10.1007/s00330-018-5981-2. [DOI] [PubMed] [Google Scholar]
- 25.Tausch C., Taucher S., Dubsky P. Prognostic value of number of removed lymph nodes, number of involved lymph nodes, and lymph node ratio in 7502 breast cancer patients enrolled onto trials of the Austrian Breast and colorectal cancer study group (ABCSG) Ann Surg Oncol. 2012;19(6):1808–1817. doi: 10.1245/s10434-011-2189-y. [DOI] [PubMed] [Google Scholar]
- 26.Ohuchi N., Suzuki A., Sobue T. JSTART investigator groups. Sensitivity and specificity of mammography and adjunctive ultrasonography to screen for breast cancer in the Japan strategic anti-cancer randomized trial (J-START): a randomized controlled trial. Lancet. 2016;387(10016):341–348. doi: 10.1016/S0140-6736(15)00774-6. [DOI] [PubMed] [Google Scholar]
- 27.Bae M.S., Lee S.H., Chu A.J., Shin S.U., Ryu H.S., Moon WK. Preoperative MR imaging in women with breast cancer detected at screening US. Radiology. 2017;282(3):681–689. doi: 10.1148/radiol.2016160706. [DOI] [PubMed] [Google Scholar]
- 28.Arneth B. Tumor microenvironment. Medicina. 2019;56(1):15. doi: 10.3390/medicina56010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houthuijzen J.M., Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev. 2018;37(4):577–597. doi: 10.1007/s10555-018-9768-3. [DOI] [PubMed] [Google Scholar]
- 30.Safarzadeh E., Orangi M., Mohammadi H., Babaie F., Baradaran B. Myeloid-derived suppressor cells: important contributors to tumor progression and metastasis. J Cell Physiol. 2018;233(4):3024–3036. doi: 10.1002/jcp.26075. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y.X., Xu J.X., Lan HY. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. doi: 10.1186/s13045-019-0760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun R., Limkin E.J., Vakalopoulou M. A radiomics approach to assess tumor-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto S., Han W., Kim Y. Breast Cancer: Radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275(2):384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 34.Braman N., Prasanna P., Whitney J. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw Open. 2019;2(4) doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie K., Li J., He X.J. COX6B2 drives metabolic reprogramming toward oxidative phosphorylation to promote metastasis in pancreatic ductal cancer cells. Oncogenesis. 2020;9(5):51. doi: 10.1038/s41389-020-0231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluemn E.G., Spencer E.S., Mecham B. PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality. Mol Cancer Res. 2013;11(6):568–578. doi: 10.1158/1541-7786.MCR-12-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.