Abstract
Hypertrophic growth of cardiomyocytes is one of the major compensatory responses in the heart after physiological or pathological stimulation. Protein synthesis enhancement, which is mediated by the translation of messenger RNAs, is one of the main features of cardiomyocyte hypertrophy. Although the transcriptome shift caused by cardiac hypertrophy induced by different stimuli has been extensively investigated, translatome dynamics in this cellular process has been less studied. Here, we generated a nucleotide-resolution translatome as well as transcriptome data from isolated primary cardiomyocytes undergoing hypertrophy. More than 10,000 open reading frames (ORFs) were detected from the deep sequencing of ribosome-protected fragments (Ribo-seq), which orchestrated the shift of the translatome in hypertrophied cardiomyocytes. Our data suggest that rather than increase the translational rate of ribosomes, the increased efficiency of protein synthesis in cardiomyocyte hypertrophy was attributable to an increased quantity of ribosomes. In addition, more than 100 uncharacterized short ORFs (sORFs) were detected in long noncoding RNA genes from Ribo-seq with potential of micropeptide coding. In a random test of 15 candidates, the coding potential of 11 sORFs was experimentally supported. Three micropeptides were identified to regulate cardiomyocyte hypertrophy by modulating the activities of oxidative phosphorylation, the calcium signaling pathway, and the mitogen-activated protein kinase (MAPK) pathway. Our study provides a genome-wide overview of the translational controls behind cardiomyocyte hypertrophy and demonstrates an unrecognized role of micropeptides in cardiomyocyte biology.
Keywords: cardiac hypertrophy, translational landscape, Ribo-seq, micropeptide, short open reading frame, noncoding RNA
Graphical abstract
In the current study, authors generated a nucleotide-resolution translatome and detected more than 100 uncharacterized short open reading frames in lncRNA genes with micropeptide coding potential from hypertrophied cardiomyocytes. It provides a genome-wide overview of the translational controls behind cardiomyocyte hypertrophy and demonstrates an unrecognized role of micropeptides in cardiomyocyte biology.
Introduction
The heart undergoes remodeling in response to various physiological and pathological stimuli.1 Hypertrophic growth of cardiomyocytes, which account for 70% of the volume of the heart, is one of the major cellular responses during cardiac remodeling.2 Increased protein synthesis and sarcomere assembly occur in this process. Both phosphatidylinositol 3-kinase (PI3K)/AKT and mitogen-activated protein kinase (MAPK) signaling pathways have been widely reported as one of the main signaling pathways transducing external stimuli to a shift in the transcriptome in cardiac hypertrophy. For example, phenylephrine (PE) activates the PI3K/AKT and MAPK signal pathways through the α1-receptor, which induces hypertrophic growth of cardiomyocytes.3,4 Studies have demonstrated that PE induces protein synthesis in cardiomyocytes through the downstream targets of mammalian target of rapamycin (mTOR), S6K1, and 4EBP1;5 however, the mechanism of this induction at the translation level has yet to be determined. Whether the increased efficiency of protein synthesis results from an increase in the quantity or the translational rate of ribosomes is unknown.
The development of new technologies for massively parallel sequencing provides new tools for genome-wide investigations of regulation by various DNA and RNA modifications. The shift of the cardiac transcriptome under different conditions has been intensively investigated by our group6 and others7,8 in the past decade; yet, transcriptome data deviate from mass spectrum data, which reflect the protein levels, to some extent. Therefore, deep sequencing of actively translated transcripts might provide a more accurate and complete measure of gene expression in addition to conventional RNA sequencing (RNA-seq) analyses. Translating ribosome affinity purification sequencing (TRAP-seq) was previously applied to study the actively translating transcripts associated with polyribosomes in the heart.9 Recently, the cardiac translatome has been analyzed with a combination of deep sequencing of ribosome-protected fragments (Ribo-seq) and RNA-seq.10,11 Similar to the cardiac transcriptome analysis, extension of the analysis of the translational landscape in cardiac remodeling under different stimuli may help to obtain a full picture of how the gene program is shifted in cardiac disease.
Micropeptides refer to those translational products containing 100 amino acids (aa) or less derived from short open reading frames (sORFs) in the transcripts.12 Since the sORFs are too short, most of them are considered as false-positive hits during ORF scanning for genomic annotation.13 Therefore, genes producing, for example, sORF-containing transcripts, are usually mis-annotated as noncoding RNA genes. Accumulating evidence has demonstrated that a large number of micropeptides are present in mammalian cells with unknown function in most cases.10,14,15 The biological function of these “tiny” protein molecules needs to be explored because it could help distinguish whether the regulatory function of the DNA locus comes from the “noncoding” transcript or if they are encoded micropeptides. Recently, micropeptides have been shown to regulate striated muscle development and function.16 Myomixer, a micropeptide enriched in developing and regenerating skeletal muscle, was reported to control the critical step in myofiber formation during muscle development.17 Furthermore, dwarf open reading frame (DWORF), a sarcoplasmic reticulum-located micropeptide, activates sarcoplasmic endoreticulum calcium ATPase (SERCA), which is a target candidate for gene therapy of heart failure,18,19 and prevents pathological remodeling and Ca2+ dysregulation in a model of heart failure.20
In this study, we performed Ribo-seq and RNA-seq with hypertrophic cardiomyocytes induced by PE. Our transcriptional and translational profiling data, respectively, showed that the PI3K/AKT and MAPK signaling pathways were upregulated translationally and transcriptionally. Our data demonstrated that the increased efficiency of protein synthesis in PE-induced cardiomyocyte hypertrophy was mainly attributable to the increased quantity of ribosomes, as evidenced by the increased translation of ribosomal protein genes. In addition, hundreds of sORF candidates were identified in this genome-wide translational analysis with a portion of the micropeptides coded by transcripts from the complementary strand of protein-coding genes. Three micropeptides were further demonstrated to regulate the hypertrophic growth of cardiomyocytes through modulating various signaling pathways as assessed by a functional assay.
Results
Genome-wide translatomic and transcriptomic profiling in hypertrophic cardiomyocytes
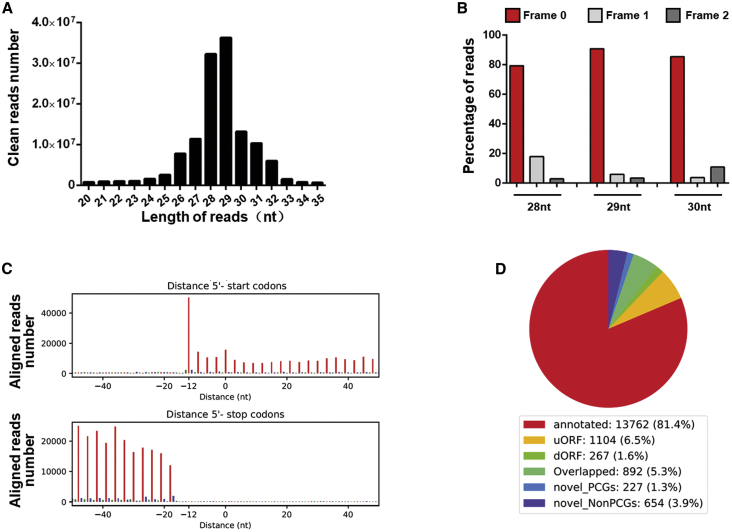
Enhanced protein synthesis is one of the main features of hypertrophic growth of cardiomyocytes under stimuli.21,22 To study how translation is regulated at a genome-wide scale during the process of hypertrophy, primary neonatal cardiomyocytes were isolated and stimulated with PE for 24 h. Hypertrophic growth was confirmed by measuring the size of cardiomyocytes (Figures S1A and S1B) and detecting the gene expression of markers, such as natriuretic peptide A (NPPA), NPPB, and ACTA1 (Figure S1C). Cells were then harvested and prepared for Ribo-seq, which detects ribosome-protected fragments for measuring ribosome occupancy of protein-coding genes, and RNA-seq, which allows the study of changes in the transcriptome during cardiomyocyte hypertrophy. In total, we obtained ∼185 M clean reads from Ribo-seq (∼90 M from control and ∼95 M from PE-treated cells; Table S1) and ∼820 M clean reads from RNA-seq (∼400 M from control and ∼420 M from PE-treated cells; Table S1). The majority of the detected ribosome-protected fragments showed a length distribution from 26 nt to 32 nt (Figure 1A). Uniquely mapped reads from Ribo-seq were analyzed with Ribocode,23 and we found that over 80% of the main ribosome-protected fragments (from 28 nt to 30 nt) were qualified reads (Figure 1B), which displayed the 3-nt codon movement characteristic of actively translating ribosomes (Figure 1C). Further analyses with Ribocode found that the majority of qualified reads (81.4%) mapped to ORFs of 13,762 annotated protein-coding genes. 6.5% qualified reads mapped to uORFs, a sORF localizing upstream of the main ORF of protein-coding genes. Interestingly, 3.9% of qualified reads mapped to long noncoding RNA (lncRNA) genes (Figure 1D; Table S2), indicating that these genes carried coding potential, which was studied below.
Figure 1.
Detection of the active translating RNA fragments from hypertrophic cardiomyocytes
(A) The distribution of length of reads in ribosome-protected fragments sequencing (Ribo-seq) in our study. (B) The percentage of Ribo-seq reads with different lengths matching the primary open reading frames (ORFs) versus other possible frames. (C) Representative bar plots showing the peptidyl-site (P-site) positions derived from Ribo-seq reads across the first 50 nt and last 50 nt of ORFs. (D) The proportion of different categories of ORFs detected in our study. Annotated, annotated ORFs for proteins; uORF, upstream ORF; dORF, downstream ORF; overlapped, uORF/dORF overlapped with main ORF; novel_PCGs, novel ORFs from protein-coding genes; novel_NonPCGs, novel ORFs from nonprotein-coding genes.
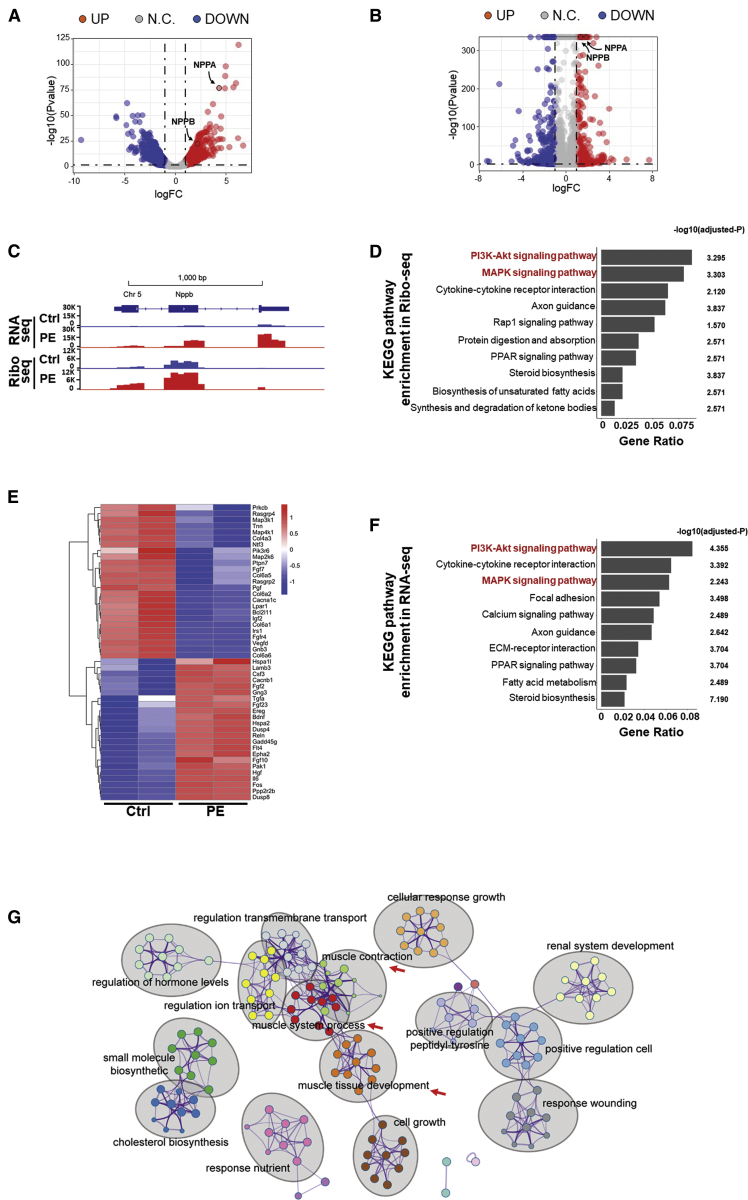
To identify the differentially expressed (DE) genes in transcription and translation, we analyzed RNA-seq and Ribo-seq data from PE-treated and control cardiomyocytes. As a result, we detected 1,201 transcriptionally DE genes (545 upregulated and 656 downregulated with |Log2(fold change)| > 1 and false discovery rate [FDR] < 0.05) (Figure 2A; Table S3) and 836 translationally DE genes (347 upregulated and 489 downregulated with |Log2(fold change)| > 1 and FDR < 0.05) (Figures 2B and 2C; Table S4). The increase of expressed and translating marker mRNAs, such as NPPA, NPPB, and ACTA1, were further confirmed in RNA-seq and Ribo-seq analyses (Figures 2A−2C; Tables S3 and S4. Next, we asked which signaling pathways were regulated during PE-induced cardiomyocyte hypertrophic growth. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that translationally dysregulated genes were enriched in several pathways known to regulate cardiac hypertrophy, such as “MAPK signaling pathway” and “PI3K/AKT signaling pathway” (Figures 2D and 2E). We performed a similar KEGG analysis for transcriptionally dysregulated genes. Similar KEGG terms were enriched with MAPK signaling pathway and PI3K/AKT signaling pathway at the top of the list (Figure 2F). Furthermore, enriched ontology clusters analysis of the transcriptionally dysregulated genes showed that terms related to muscle system process, contraction, and development were among the most highly enriched, which were expected in the process of cardiomyocyte hypertrophic growth (Figure 2G). The similarity of top KEGG lists of the dysregulated gene enrichment between Ribo-seq and RNA-seq provided us a hint that the rate of utilization of the ribosome for translation of the majority of mRNAs did not significantly change in PE-induced cardiomyocyte hypertrophy.
Figure 2.
Integrative genome-wide analyses of Ribo-seq and RNA-seq from PE-treated cardiomyocytes
(A and B) Volcano maps of dys-regulated genes in RNA-seq (A) and Ribo-seq (B). Red and blue dots represent the upregulated and downregulated genes, respectively (FDR < 0.05; |Log2(fold change)| > 1). (C) The genomic view of reads in RNA-seq and Ribo-seq for hypertrophic marker gene Nppb in the IGV browser. y axis shows the mapped reads number. (D) KEGG pathway analysis of significantly dys-regulated genes in translation (Ribo-seq). Enriched pathways are ranked by gene ratio. (E) Heatmap of dys-regulated genes in the “PI3K/AKT signaling pathway” and “MAPK signaling pathway” from Ribo-seq. (F) KEGG pathway analysis of significantly dys-regulated genes in transcription (RNA-seq). Enriched pathways are ranked by gene ratio. (G) Metascape analysis of PE-treated cardiomyocyte transcriptome. Enriched ontology clusters were colored by cluster ID and shown by gray ovals. Clusters described in the main text are indicated by red arrows.
Increased protein synthesis is mainly attributable to an enhanced amount of ribosomal proteins during PE-induced cardiomyocyte hypertrophy
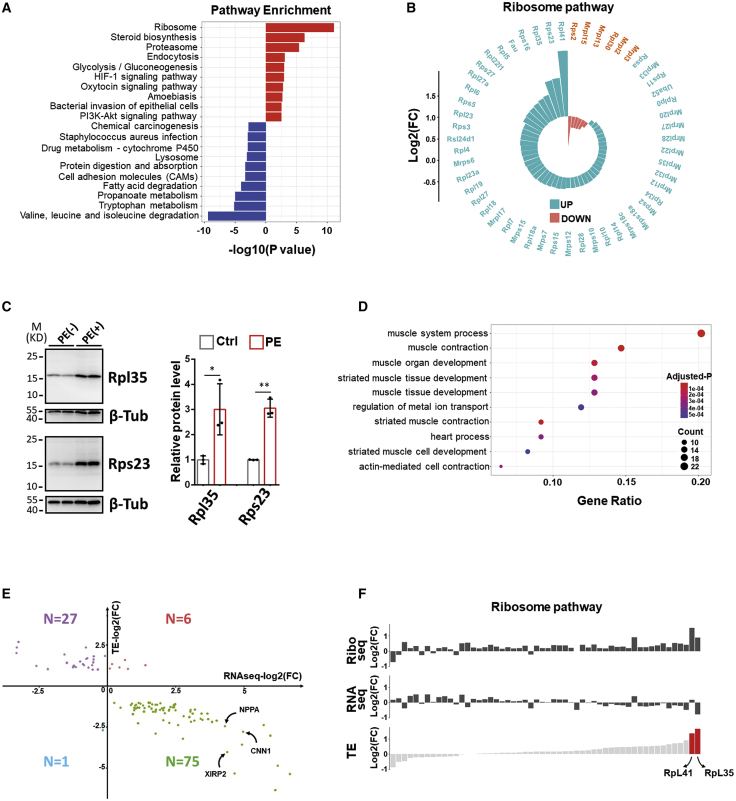
Increased protein synthesis during cardiomyocyte hypertrophy may result from an increased number of ribosomes (with stable translational rate) and/or an increased translational rate from these ribosomes. Analysis with a less stringent criteria (FDR < 0.05) provided us a broad view of the translatome shift during this cellular process. We found that “ribosome” is the first hit of the upregulated KEGG terms (Figure 3A), indicating that the synthesis of ribosomal proteins was enhanced in hypertrophic cardiomyocytes. In fact, 47 out of 53 significantly altered (FDR < 0.05) ribosomal protein genes were upregulated when we looked at the gene list of Ribo-seq (Figure 3B), indicating an increased ribosomal number and enhanced translational capacity. Increases in the levels of two ribosomal proteins, Rpl35 and Rps23, in PE-treated cardiomyocytes were also confirmed by western blotting (Figure 3C). Next, we asked whether increased translational rates play a role in PE-induced cardiomyocyte growth. Ribo-seq data were incorporated together with RNA-seq data for analysis of gene translational efficiency (TE) using Ribodiff.24 It turned out that only 109 protein-coding genes were significantly altered in TE (FDR < 0.05; |Log2(fold change)| > 1) (Table S5). Among these, most were genes related to muscle contraction (Figure 3D); however, most TE-altered genes showed a buffered effect in transcription versus translation (the tendency of dysregulation is opposite between transcript expression and TE) (Figure 3E). Then, we aligned the changes of translation, transcription, and TE for dysregulated genes in the MAPK signaling pathway, PI3K/AKT signaling pathway, and ribosome. No clear pattern of gene TE in these pathways was observed (Figures 3F and S2), although dysregulated ribosome genes have a tendency to shift upon TE enhancement. All of these results suggest an increased number of ribosomes but not an increased translational rate, contributed to the enhanced protein synthesis in PE-induced cardiomyocyte hypertrophy.
Figure 3.
The alteration of translatome in hypertrophic cardiomyocytes
(A) Enrichment of upregulated (red bars) and downregulated (blue bars) KEGG pathways from Ribo-seq analysis (genes with FDR < 0.05 only) in PE-treated cardiomyocytes. Enriched pathways are ranked by p value. (B) A wind rose map displaying the expression change of ribosome pathway-related genes with FDR < 0.05. (C) Detection of the expression of ribosomal proteins Rpl35 and Rps23 by western blotting. The expression of β-tubulin serves as the control. Quantification is provided on the right. n = 3 for each group. M, molecular weight. ∗p < 0.05; ∗∗p < 0.01. (D) Gene Ontology (GO) analysis of genes with significant change of translational efficiency (TE). GO terms are ranked by gene ratio. (E) A plot of TE-altered genes with ratio of fold change in transcription (RNA-seq) versus fold change in TE. Gene number in each sector is indicated. (F) An alignment of gene TE to expression level in translation and transcription for ribosome pathway-related genes with FDR < 0.05. Genes are indicated by bars in each column. Genes are sorted based on their TE. Bars in red indicate that TE for these genes increases significantly.
The translational activity of uORFs does not affect the TE of the main ORF in PE-induced cardiomyocyte hypertrophy
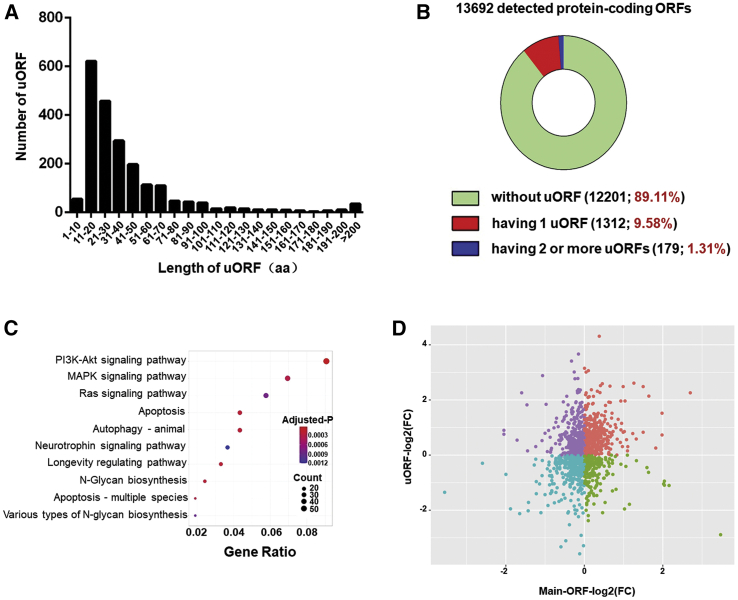
The uORF was reported to regulate the translational activity of the main ORF of protein-coding genes.25 In our study, we identified 1,834 uORFs in 1,491 annotated protein-coding genes (Table S6). The length of these uORFs is around 20−50 aa (60−150 nucleotides) (Figure 4A). Among 13,692 detectable ORFs for proteins identified in this study, 12,201 of them (89.11%) did not have an uORF; 1,312 of them (9.58%) possessed one uORF, whereas 179 of them (1.31%) possessed two or more uORFs (Figure 4B). Those uORF-containing protein-coding genes were further subjected to KEGG analysis. Interestingly, terms of PI3K/AKT signaling pathway and MAPK signaling pathway were shown at the top of the list (Figure 4C). Next, we tried to address whether uORFs regulate the translational activity of the main ORFs by checking the correlation of TE between the main protein-coding ORF and its preceding uORF (uORF-ORF pair). Multiple uORF-ORF pairs were applied if multiple preceding uORFs for one main protein-coding ORF were identified. No clear distribution pattern was found when we plotted TE ratios of all detected uORF-ORF pairs in Ribo-seq (Figure 4D). Similar negative results were obtained when we carefully checked uORF-ORF pairs associated with PI3K/AKT signaling pathway or MAPK signaling pathway (Figure S3). Therefore, the TE of primary ORFs was largely independent from the frequency of uORF translation in PE-induced cardiomyocyte hypertrophy.
Figure 4.
uORF and its regulation on main ORF translation
(A) The length distribution (in amino acids) of uORF detected in NRVCs. (B) The proportion of transcripts of annotated ORFs for proteins with or without uORF. (C) KEGG pathway analysis of uORF-containing protein-coding genes/ORFs. (D) A plot of the ratio of TE change of uORFs and ORFs for all detected uORF-ORF pairs in PE-treated cardiomyocytes.
Identification of small ORF-coded micropeptides from primary cardiomyocytes
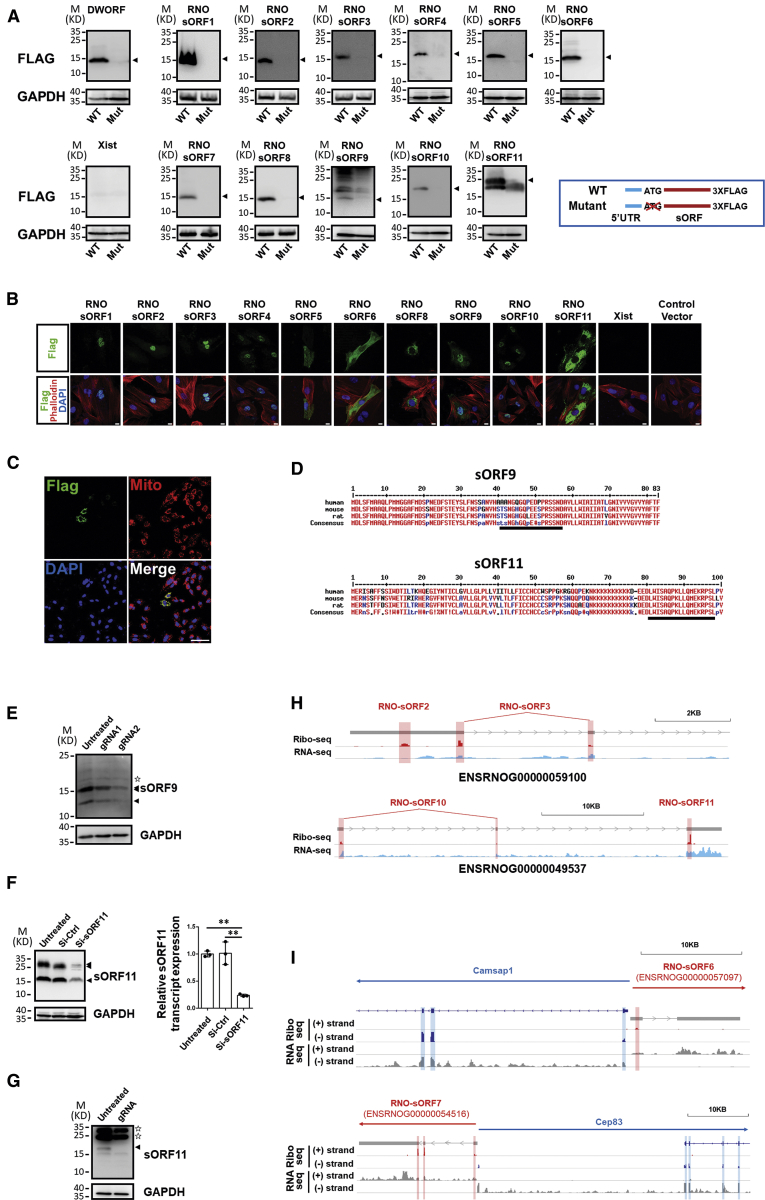
Emerging evidence shows that many micropeptides, which are 100 aa or less, are encoded by sORFs in transcripts originally annotated as noncoding RNAs. In order to identify novel sORFs on a genome-wide basis, we carefully examined reads from Ribo-seq, which displayed the 3-nt codon movement pattern, and matched these to noncoding transcripts, which indicated that actively translating ORFs were present in these noncoding transcripts. As a result, 265 novel ORF candidates were detected. Among them, 126 sORF candidates with 100 aa or less were identified (Table S7). These sORF candidates were matched to 103 annotated lncRNAs (8.67%, 103 out of 1,188). We found that 27 sORF candidates overlapped with sORFs detected in adult rat heart previously (Figure S4).10 To further evaluate the coding capability, 15 sORF candidates were randomly selected for further testing (Table 1). We built expression constructs containing a 3XFLAG epitope tag in-frame with the C terminus of sORFs. A previously reported sORF, DWORF,16 was used as a positive control. As expected, a 3XFLAG-tagged micropeptide was detected as ectopically expressed in transfected HEK293 cells. It turned out that 11 out of 15 constructs yielded 3XFLAG-tagged fusion micropeptides ranging from 10 to 20 kDa in HEK293 cells, as demonstrated by western blotting (Table 1; Figure 5A), and the length of 5′ UTR seemed to have a limited effect on the translation of sORF (Figure S5). More importantly, the expression of these fusion micropeptides was abolished when the ATG codon was deleted (Figure 5A) or a single nucleotide was inserted immediately after ATG to cause the frameshift of sORFs in mutant constructs (Figure S6). Many micropeptides were reported to be localized within mitochrondia.10 To further explore subcellular localizations of these identified micropeptides, we detected the cellular distribution of these ectopic-expressing 3XFLAG-tagged fusion micropeptides in H9C2 cells, a cardiomyocyte-like cell line, with immunostaining. As shown in Figures 5B and S7, most micropeptides showed a predominant nuclear or cytoplasmic localization. Only RNO-sORF7, which is encoded by transcript ENSRNOG00000054516, showed a mitochondrial-specific localization (Figure 5C). Analyses combined with bioinformatics searches and comparisons to recent data from humans10 found that most of the validated micropeptides were not conserved in human and mouse, except sORF9 and sORF11 (Table 1; Figure 5D).
Table 1.
Information of micropeptides tested in this study
| Designated_name | Gene_ID | Transcript_ID | sORF_tstart (nt in transcript) | sORF_tstop (nt in transcript) | Length (aa) | WB | IF | Peptide_location | Conservation |
|---|---|---|---|---|---|---|---|---|---|
| RNO-sORF1 | ENSRNOG00000057352 | ENSRNOT00000078986 | 250 | 408 | 52 | + | + | Nu | N |
| RNO-sORF2 | ENSRNOG00000059100 | ENSRNOT00000090348 | 198 | 392 | 64 | + | + | Nu | N |
| RNO-sORF3 | ENSRNOG00000059100 | ENSRNOT00000090348 | 850 | 1107 | 85 | + | + | Nu | N |
| RNO-sORF4 | ENSRNOG00000058926 | ENSRNOT00000081933 | 955 | 1101 | 48 | + | + | Nu | N |
| RNO-sORF5 | ENSRNOG00000057834 | ENSRNOT00000090803 | 269 | 571 | 100 | + | + | Cyto | N |
| RNO-sORF6 | ENSRNOG00000057097 | ENSRNOT00000081712 | 592 | 816 | 74 | + | + | Cyto | N |
| RNO-sORF7 | ENSRNOG00000054516 | ENSRNOT00000078509 | 202 | 486 | 94 | + | + | Mito | N |
| RNO-sORF8 | ENSRNOG00000051251 | ENSRNOT00000076492 | 29 | 289 | 86 | + | + | Cyto | N |
| RNO-sORF9 | ENSRNOG00000048986 | ENSRNOT00000075676 | 466 | 717 | 83 | + | + | Nu/Cyto | Y |
| RNO-sORF10 | ENSRNOG00000049537 | ENSRNOT00000076888 | 81 | 365 | 94 | + | + | Nu | N |
| RNO-sORF11 | ENSRNOG00000049537 | ENSRNOT00000076888 | 517 | 819 | 100 | + | + | Cyto | Y |
| ENSRNOG00000060475 | ENSRNOT00000082248 | 97 | 360 | 87 | − | + | |||
| ENSRNOG00000052518 | ENSRNOT00000086642 | 267 | 380 | 37 | − | − | |||
| ENSRNOG00000052394 | ENSRNOT00000083923 | 187 | 450 | 87 | − | − | |||
| ENSRNOG00000061921 | ENSRNOT00000089924 | 313 | 459 | 48 | − | − |
nt in transcript, the nucleotide number from the beginning of the transcript; WB, whether detected in western blot; IF, whether detected in immunofluorescence; Nu, a dominant nuclear location of the micropeptide was detected in immunofluorescence; Cyto, a dominant cytoplasmic location of the micropeptide was detected in immunofluorescence; Mito, a dominant mitochondrial location of the micropeptide was detected in immunofluorescence; Nu/Cyto, both strong nuclear and cytoplasmic signals were detected in immunofluorescence for the micropeptide. Y, the micropeptide is conserved in both human and mouse; N, the micropeptide is not conserved in human or mouse.
Figure 5.
Identification of short ORF (sORF)-coding micropeptide from hypertrophic cardiomyocytes
(A) Wild-type (WT) and mutant (Mut; ATG codon deletion) sORFs were cloned into a 3XFLAG-tagged expressing vector (inlet). Ectopic expression of flag-tagged micropeptides was detected by western blotting with anti-FLAG antibodies. The detected GAPDH expression serves as loading control. (B) The localization of micropeptides is determined by immunostaining for FLAG-tagged micropeptides in H9C2 cardiomyocytes transfected with micropeptide-overexpressing vectors. Cardiomyocytes were co-stained with phalloidin. Nuclei are indicated by DAPI. Scale bar, 10 μm. (C) The expression of FLAG-tagged RNO-sORF7 was detected in mitochondria in H9C2 cardiomyocytes transfected with overexpressing vector. Mitochondria are indicated with MitoTracker; nuclei are indicated by DAPI. Scale bar, 75 μm. (D) Amino acid sequence alignment of sORF9 and sORF11 from humans, mice, and rats. The peptide fragments used for antibody development are underlined. (E) Detection of sORF9 expression in untreated H9C2 cardiomyocytes and cells edited with the CRISPR-Cas9 system for frameshift mutation in sORF9. (F) Detection of sORF11 expression by western blotting and qRT-PCR in sORF11 knockdown H9C2 cardiomyocytes. n = 3 for each group. ∗∗p < 0.01. (G) Detection of sORF11 expression in untreated NIH 3T3 cells and cells edited with the CRISPR-Cas9 system for frameshift mutation in sORF11. The expression of GAPDH serves as the control. Arrowheads indicate specific bands, and stars indicate unspecific bands. (H and I) Examples for two micropeptides, which are coded by one transcript (H), and examples for micropeptides, which are coded by lncRNAs derived from the complementary DNA strand (I), in the IGV genome browser. Reads in Ribo-seq and RNA-seq are shown. sORF are shadowed in red, and annotated ORFs are shadowed in blue. Gene structures are shown by bars and lines with directions of transcription indicated (arrows).
To further demonstrate the existence of sORFs in vivo, two antibodies against sORF9 and sORF11 were developed to detect the endogenous expression of these micropeptides, respectively. As shown in Figure 5E, anti-sORF9 antibodies detected endogenous sORF9 micropeptides with molecular weights around 12 kDa and 15 kDa from H9C2 cardiomyocytes in a western blotting analysis. The detected signals were significantly decreased in two mixed H9C2 cell populations in which sORF9 DNA was edited with the CRISPR-Cas9 system using two independent guide RNAs (gRNAs), respectively (Figure S8). Similarly, anti-sORF11 antibodies detected endogenous sORF11 micropeptides with molecular weights around 17 kDa and 28 kDa from H9C2 cardiomyocytes (Figure 5F). A small interfering RNA (siRNA) targeting the transcripts of sORF11 was applied to knock down the expression of sORF11. As expected, both the expression of RNA and protein of sORF11 were significantly decreased (Figure 5F). To further test the conservation of sORF11, we edited sORF11 DNA with the CRISPR-Cas9 system in mouse NIH 3T3 cells. Since the editing induced frameshift mutations in most of the cells (11 out of 12 clones showed a frameshift mutation in a clonal analysis of DNA; Figure S9), a band with a molecular weight around 17 kDa detected in wild-type NIH 3T3 cells was undetected in CRISPR-Cas9-edited cells in a western blotting analysis (Figure 5G). We noticed that smeared bands larger than predicted size were detected in western blotting with anti-sORF9 and anti-sORF11 antibodies (15 kDa bands for sORF9 and 28 kDa bands for sORF11), which could possibly be caused by protein modifications of these micropeptides.
Interestingly, some transcripts, such as ENSRNOG00000059100 and ENSRNOG00000049537, were found to possess two sORFs (Figure 5H). In our analyses, we also noticed that ∼20% (21 out of 103) of micropeptide-coding lncRNAs were derived from the complementary strand and likely shared a bi-directional promotor driving expression of a protein-coding gene (Figure 5I), similar to Hand2-UPPERHAND gene pairs.26,27 Consistent with a previous report,10 sORFs within UPPERHAND were detected in our Ribo-seq from rat cardiomyocytes but were negative upon further validation (Table 1; ENSRNOG00000052518). However, our successful validation of the coding capability of some sORFs from these lncRNAs (Figure 5I; Table 1) suggests that partial of these complementary strand-derived lncRNAs could have micropeptide-coding capability.
Micropeptides modulate PE-induced cardiomyocyte hypertrophy through key signaling pathways
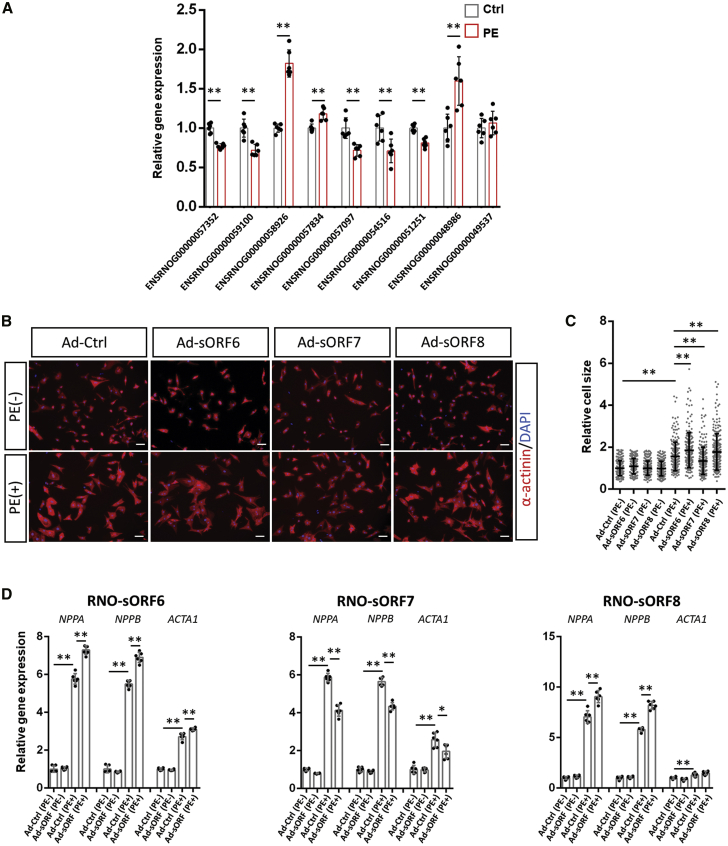
The micropeptide DWORF has been reported to benefit the failing heart by regulating Ca2+ dynamics.20 However, whether micropeptides participate in the regulation of cardiac hypertrophy has not been studied. To address this question, we first examined the expression of transcripts harboring sORFs validated above in PE-induced cardiomyocyte hypertrophy. The expression of these sORF-harboring transcripts was analyzed from RNA-seq data (Table S8). A more careful examination with quantitative reverse-transcriptase PCR (qRT-PCR) was applied to determine their dysregulation. The result showed that the expression of 5 out of 9 sORF-harbored transcripts was repressed by PE treatment, with transcripts ENSRNOG00000058926, ENSRNOG00000057834, and ENSRNOG00000048986 being significantly upregulated (Figure 6A). As shown above (Table S5), only ∼100 ORFs were significantly altered in TE genome-wide during PE treatment. All of these 11 sORFs were found being translated in a stable rate, indicating that their dysregulations in translation are similar to their dysregulations in transcription.
Figure 6.
Regulation of cardiomyocyte hypertrophy by micropeptides
(A) Determination of PE-induced dys-regulation of transcripts harboring sORFs for micropeptides by qRT-PCR. Data were obtained from biological replicates from six independent experiments (n = 6) for each group. (B) Representative images of NRVCs infected with Ad-sORF or control virus, with or without treatment of PE. α-actinin-labeled cardiomyocytes (red). DAPI marks nuclei. Scale bars, 50 μm. (C) Quantitative analysis of cardiomyocyte cell size. At least 200 cardiomyocytes from 8 images were measured for each group. (D) Determination of PE-induced expression of hypertrophy markers NPPA, NPPB, and ACTA1 when micropeptides were overexpressed in NRVCs by qRT-PCR. Data were obtained from biological replicates from six independent experiments (n = 6) for each group. ∗p < 0.05, ∗∗p < 0.01.
To further test whether some of these micropeptides modulate the hypertrophic growth of cardiomyocytes, adenovirus was applied to overexpress all validated micropeptides from Figure 5A in primary cardiomyocytes (Figure S10). As a result, 3 micropeptides—RNO-sORF6, RNO-sORF7, and RNO-sORF8—were found to alter hypertrophic growth of cardiomyocytes induced by PE, but not in basal conditions, as determined by the measurement of cell-surface area of α-actinin-positive cardiomyocytes. Cytoplasmic RNO-sORF6 and RNO-sORF8 promoted cardiac hypertrophy. On the contrary, the mitochondrial RNO-sORF7 exerted an inhibitive effect during this process (Figures 6B and 6C). The expression of hypertrophic molecular markers, such as NPPA, NPPB, and ACTA1, further demonstrated the regulatory functions of these micropeptides in cardiac hypertrophy (Figure 6D), consistent with the size measurement of cardiomyocytes described above. Furthermore, the regulatory function of these micropeptides were abolished when sORFs with ATG start codon deletion were overexpressed (Figure S11), indicating that micropeptides, but not RNA transcripts, regulate PE-induced cardiomyocyte hypertrophy.
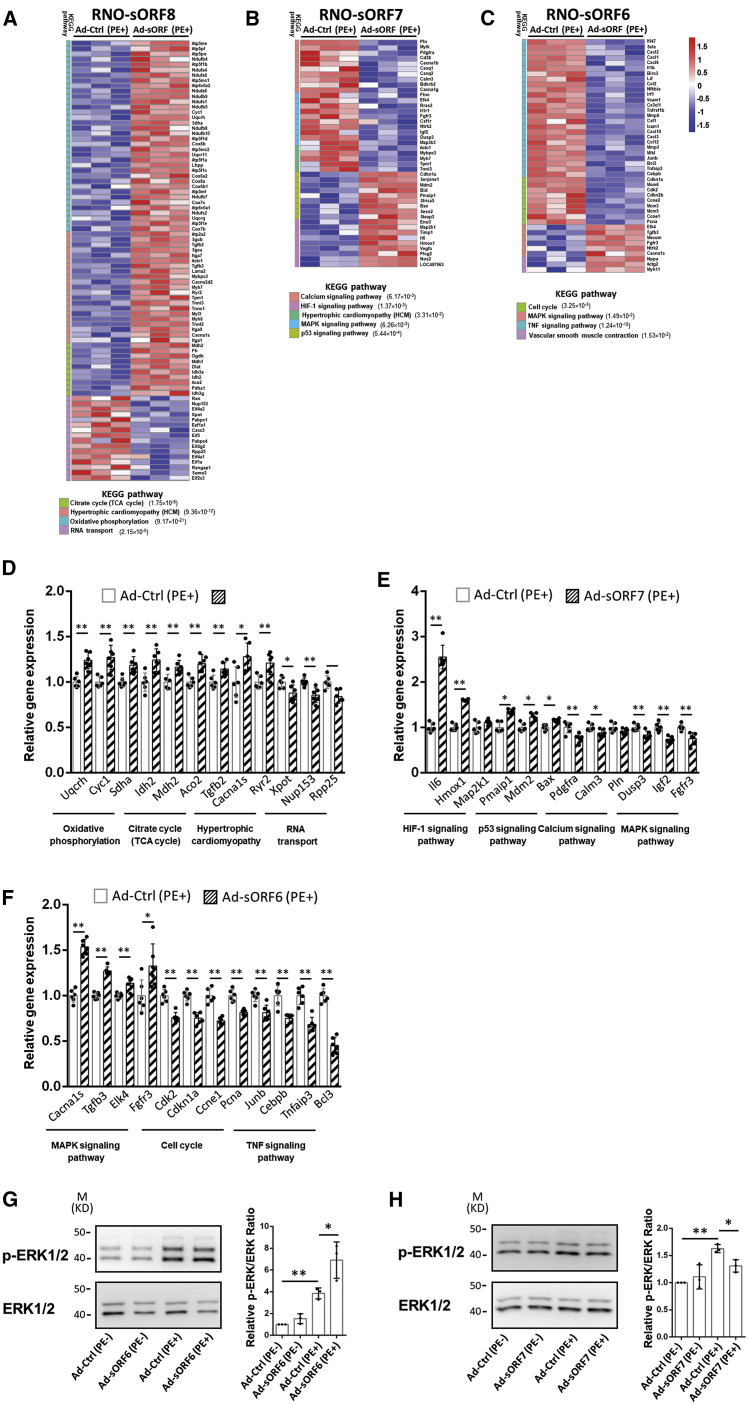
Finally, RNA-seq experiments were performed to explore how these micropeptides drive the shift of the transcriptome during the process of hypertrophy. In RNO-sORF8-overexpressing cardiomyocytes, the KEGG terms of “oxidative phosphorylation” and “citrate cycle (tricarboxylic acid [TCA] cycle)” were enriched in upregulated genes, along with the increase of “hypertrophic cardiomyopathy”-related genes, indicating that the micropeptide RNO-sORF8 induced hypertrophic growth of cardiomyocytes by regulating mitochondrial function (Figure 7A). Interestingly, mitochondrial RNO-sORF7 downregulated genes in the “calcium signaling pathway” and MAPK signaling pathway, which are central in the gene regulatory network for cardiac remodeling and repressed in PE-induced cardiomyocyte hypertrophy (Figure 7B). In addition, genes related to the MAPK signaling pathway were upregulated, whereas those related to “TNF signaling pathway” and “cell cycle” were downregulated during the promotion of cardiomyocyte hypertrophic growth of micropeptide RNO-sORF6, indicating that multiple signaling pathways were coordinated by RNO-sORF6 (Figure 7C). Dysregulated genes, induced by overexpression of sORFs during hypertrophy in each Gene Ontology (GO) term, were further confirmed by qPCR (Figures 7D−7F). Since the activity of extracellular signal-regulated kinase (ERK) in the MAPK signaling pathway, which was shown to be dysregulated in hypertrophic phenotypes modulated by sORF6 and sORF7, is critical to cardiac hypertrophy, the phosphorylation of ERK in sORF-overexpressing cardiomyocytes was examined by western blotting. Consistently, phosphorylation of ERK was increased in sORF6-overexpressing cardiomyocytes (Figure 7G) but decreased in sORF7-overexpressing cardiomyocytes during PE-induced hypertrophic growth (Figure 7H). The phosphorylation level of ERK remained unchanged in sORF8-overexpressing cardiomyocytes during PE treatment (Figure S12). Together, our data suggest that the micropeptides sORF6 and sORF7 modulate cardiac hypertrophy, at least in part, through the ERK signaling pathway.
Figure 7.
Analyses of dysregulated signaling pathways in cardiomyocyte hypertrophy regulated by micropeptides
(A–C) Heatmaps showing the expression of dys-regulated genes in significantly altered KEGG pathways in Ad-sORF8 (A)-, Ad-sORF7 (B)-, and Ad-sORF6 (C)-overexpressed NRVCs under the treatment of PE. (D–F) Selected gene expression was confirmed with qRT-PCR in Ad-sORF8 (D)-, Ad-sORF7 (E)-, and Ad-sORF6 (F)-overexpressed NRVCs under the treatment of PE. (G and H) Detection of the expression of phosphorylated and total ERK in Ad-sORF6 (G)- and Ad-sORF7 (H)-overexpressed NRVCs with or without the treatment of PE by western blotting. The ratio of phospho-ERK to total ERK was quantified. Data were obtained from biological replicates from six independent experiments (n = 6) for each group. ∗p < 0.05, ∗∗p < 0.01.
Discussion
Most biological processes depend on dynamic protein expression. The translation from mRNAs to proteins often undergoes post-transcriptional modifications. Therefore, the dynamics of the transcriptome reflect this process in a less accurate manner. The development of Ribo-seq provided us a chance to study the translatome in a genome-wide and nucleotide-resolution fashion. Combined with RNA-seq and Ribo-seq data, we found that the translational rate from ribosomes was altered for only a small portion of protein-coding genes (∼100 genes) during PE-induced cardiomyocyte hypertrophy. Among these TE-altered genes, most of them are contraction-related genes, and their decreased translational rate buffered the increase in mRNA transcription. In addition, PI3K/AKT and MAPK signaling pathways, which play critical roles in cardiac hypertrophy, were shown transcriptionally controlled with stable translational rates in PE-treated cardiomyocytes. Furthermore, the increase in translation of ribosomal protein genes provided indirect evidence supporting the idea that increased ribosomal numbers (instead of increased translational rates from ribosomes) were responsible for the enhanced protein synthesis during PE-induced cardiomyocyte hypertrophy. More direct evidence with a combination of RNA-seq data and genome-wide proteinomic data detecting newly generated peptides in hypertrophic cardiomyocyte should further prove this point in the future.
Recently, several groups have reported a shift in the translatome during cardiac remodeling.11,28 In the present study, we provided an integrative analysis using both transcriptome and translatome data of cardiomyocyte hypertrophy induced by PE. In recent studies, the translation landscape has been assessed in the failing human heart.10 The TE of “extracellular matrix” genes was found to be significantly upregulated, which reflects the activation of cardiac fibroblasts and the manifestation of fibrosis. Combined with the strategy of Ribo-tag, labeling ribosomes in vivo using a genetic approach, another recent report showed that transcripts related to translation, protein quality control, and metabolism are translationally controlled in cardiomyocytes when the heart undergoes pressure overload.11 Together with these recent studies, the translational landscape of PE-induced cardiomyocyte hypertrophy presented here shows a comprehensive picture of how translation controls cardiac remodeling in different situations.
More and more studies have demonstrated that large numbers of micropeptides encoded by sORFs, a group of previously overlooked molecules, are presented in eukaryotic cells.29,30 The function of most of these micropeptides has yet to be explored. Interestingly, we found that a portion of micropeptides encoded by transcripts derived from the complementary strand of protein-coding genes might share a bi-directional promotor. Several studies suggest that most of these complementary stranded transcripts are noncoding RNAs.31,32 Some of these noncoding transcripts, such as UPPERHAND26,27 and NEXN-AS1,33 have been shown to play important roles in cardiovascular development and pathology. The potential micropeptides derived from sORFs in these functional lncRNAs need to be carefully determined. If it is confirmative of micropeptides coding, whether the molecular functions of these transcripts are mediated by the micropeptides or RNA transcripts should be addressed.
Although the functions of some micropeptides have begun to be investigated,17,20,34,35 the majority of them remained uncharacterized. The current study identified three micropeptides—RNO-sORF6, RNO-sORF7, and RNO-sORF8—that possess a function of modulating cardiac hypertrophy. The regulatory functions of sORF6 and sORF7 were shown to be mediated, at least in part, by the ERK signaling pathway. Our data showed that sORF7 dominantly locates in mitochondria and mediates the downregulation of the calcium signaling pathway and MAPK signaling pathway in PE-induced cardiomyocyte hypertrophy, which is consistent with a previous report that repressing calcium signaling by chelating intracellular Ca2+ abolished Ang II-induced activation of MAPKs, including ERK.36 Recently, a mitochondrial micropeptide, mitoregulin, has been shown to regulate intracellular Ca2+ dynamics.34 Given that many micropeptides were found to locate in mitochondria in the heart,10 it is interesting to investigate whether a majority of these mitochondrial micropeptides share a similar signaling cascade in affecting cardiac physiology and pathology. Although detailed mechanisms warrant investigation in detail in the future, our study indicates that micropeptides are a group of new players in regulating cardiac remodeling. Unlike ORFs of conventional protein-coding genes, the majority of which are evolutionarily conserved, a large portion of sORFs identified in this study are not conserved among humans, mice, and rats (except sORF9 and sORF11). Similar observations were also found in the human heart.10 Although our study of rat-specific sORF-coding micropeptides may not directly contribute to the identification of novel regulators of human cardiac hypertrophy and failure, it suggests that it is promising to search for functional sORF-coding micropeptides in the human heart, as this may reveal novel therapeutic targets.
Materials and methods
Cardiomyocyte isolation, culture, and transfection
Neonatal rat ventricular cardiomyocytes (NRVCs) were prepared as previously described.37 Briefly, 2-day-old neonatal Sprague-Dawley rats were decapitated using sterile scissors, and the chest was opened to extract the heart. NRVCs were isolated by repeated enzymatic dissociation. All isolated cells were pre-plated for 1 h to remove noncardiomyocytes. Nonadherent cells were then plated on 0.5% gelatin-coated plates and cultured in DMEM high-glucose medium (Gibco), supplemented with sodium pyruvate (1×), GlutaMAX (1×), and 10% fetal bovine serum with antibiotics. Cardiomyocytes were transduced with adenovirus at a multiplicity of infection (MOI) of 20 for 24 h prior to treatment with hypertrophic agent PE (20 μM). Cells were harvested 24 h after PE treatment for RNA isolation, 48 h for detection of ribosomal proteins or immunochemistry, or 1 h for detection of phosphorylated protein. All animal procedures were approved by the Medical Ethics Committee of The First Affiliated Hospital, Sun Yet-sen University.
The H9C2 cardiomyocyte-like cells, HEK293 cells, and NIH 3T3 cells were cultured in DMEM, supplemented with 10% fetal bovine serum. The cells were maintained in a 5% CO2 atmosphere at 37°C. H9C2 and HEK293 cells were transfected with the indicated plasmids using Lipofectamine 3000 (Invitrogen; L3000-015) and PEI (1 μg/μL), respectively.
Ribo-seq
Ribo-seq was performed according to a previous report38 with minor modifications. A total of 5 × 106 NRVCs were prepared for cell lysis. The cell medium was aspirated, and cells were washed twice with ice-cold PBS containing 100 μg/mL cycloheximide (MP Biomedicals). After thorough removal of the PBS, the plates were immersed in liquid nitrogen for 10 s and transferred to dry ice. We then dripped 400 μL of mammalian polysome buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 5 mM MgCl2 with 1 mM DTT and 100 μg/mL cycloheximide [added freshly]), supplemented with 1% (v/v) Triton X-100, 0.1% NP-40, 500 U/mL RNase inhibitor (Promega; N2515), 10 U/mL DNase I (Thermo Scientific; EN0521), and protease inhibitor cocktail (Roche; 05892970001), onto the plates and then placed the plates on ice. Cells were scraped and lysed with a 26-gauge needle, and then cleared by centrifugation at 20,000 × g for 5 min. The resulting supernatant was used for library preparation. The prepared supernatant was digested with RNase I (Life Technologies; AM2294) for 45 min at room temperature. SUPERase⋅In RNase Inhibitor (Life Technologies; AM2694) was then added to stop the reaction. Monosomes generated from the digestion were purified with MicroSpin S-400 HR columns (GE Healthcare; 27-5140-01). The RNAs were then isolated from the flowthrough with RNAzol. To enrich the RFPs (ribosome footprints), the RNAs were subjected to the Ribo-Zero Gold rRNA Removal Kit (Illumina; MRZG12324) and then separated with a 17% 7.5 M urea-PAGE gel. RNAs, with sizes ranging from 26 nt to 30 nt, were isolated and purified by PAGE. Strand-specific ribosome-profiling libraries were prepared as described previously.39 Libraries were sequenced on the HiSeq X Ten platform (Illumina).
Micropeptide expression vector and adenoviral construction
The sORF sequences with an extra 15-nt sequence upstream of the ATG or with predicted full-length 5′ UTR sequence was cloned from NRVC cDNA and inserted into pCFH vector 3. Frameshift mutant vectors, in which an adenine (A) was inserted behind the ATG translation start codon of the sORF, or ATG translation start codon deletion mutant vectors were constructed according to mutagenesis, a protocol previously described.40
To yield adenoviral constructs, 3 × FLAG-tagged sORF cDNAs or sORF cDNAs with deletion of the ATG translation start codon were inserted into the adenoviral empty vector harboring the murine cytomegalovirus (mCMV) promoter. The prepared adenovirus vectors were then transfected in 293AD cells with PEI (1 μg/μL) to produce packed adenovirus. 293AD cells were subsequently transduced with adenovirus for viral amplification. Adenovirus was collected from 293AD cells and purified with the ViraTrap Adenovirus Purification Miniprep Kit, according to the manufacturer’s instructions (Biomiga; V1160).
Knockdown of micropeptide expression in cells with siRNA and gRNA
To knock down the expression of micropeptide in H9C2 cardiomyocyte with siRNA, 50 nM targeting siRNA or control siRNA was transfected into cardiomyocytes by using the Lipofectamine RNAiMAX transfection reagent. 6 h later, medium with transfection reagent was removed. Cells were then cultured with full medium for 48 h before harvest for protein extraction. To knock down the expression of micropeptides in cells with CRISPR-Cas 9 system, NIH 3T3 cells and H9C2 cardiomyocytes were transfected with the PX459 vector containing the targeting gRNA to create a frameshift mutation in sORF for 1 day, followed by another 3-day selection with puromycin (3.2 μg/mL). The mixed cell population was then cultured in full medium for protein extraction.
Generation of sORF9- and sORF11-specific antibodies
Antibodies specific to sORF9 and sORF11 were raised in a rabbit against the following synthetic peptides: NH2-TSNGHGQPEESPRSSND-COOH (for sORF9) and NH2-LWISAQPKLLQMEKRPS-COOH (for sORF11), respectively (GL Biochem, Shanghai).
Quantitative real-time PCR and western blotting analysis
Total RNA was isolated from cells, and 2 μg RNA was used for cDNA synthesis. 0.1 μL cDNA was used as a template for quantitative real-time PCR. Primers for qRT-PCR were listed in Table S9. Target gene expression levels were normalized against the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or 18S rRNA.
For western blotting analyses, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 1 mM PMSF and then denatured at 98°C for 10 min. Cell lysates were resolved by electrophoresis in 4%–20% precast SDS-PAGE gel (Beyotime; P0468S) and then transferred to 0.2 μm polyvinylidene fluoride (PVDF) membranes. Blocking and blotting with primary antibodies were performed in Tris-buffered saline with Tween 20 (TBST), supplemented with 5% and 3% skimmed milk, respectively. The antibodies used in this study included the following: anti-FLAG M2 (Sigma-Aldrich; F1804), anti-GAPDH (Proteintech; 60004-1-Ig), anti-phospho-Erk1/2 (Cell Signaling Technology; cat #4370), anti-total-Erk1/2 (Cell Signaling Technology; cat #4695), anti-Rpl35 (Signalway Antibody; cat #34357), anti-Rps23 (Signalway Antibody; cat #34336), anti-sORF9 (GL Biochem, Shanghai), and anti-sORF11(GL Biochem, Shanghai). The membrane was incubated overnight at 4°C with the primary antibodies and washed 3 times with TBST buffer before adding horseradish peroxidase (HRP)-conjugated secondary antibodies. Specific protein bands were visualized using the Immobilon Western chemiluminescent HRP substrate (Millipore; WBKLS0500). Chemiluminescent signal was captured with ImageQuant LAS4000 Mini (GE Healthcare).
Immunofluorescence staining
Cardiomyocytes were fixed with 4% paraformaldehyde for 15 min and then permeabilized with PBS, supplemented with 0.1% Triton X-100 in PBS for 10 min at room temperature. Cells were then blocked with 3% bovine serum albumin in PBS for 1 h, followed by incubation with anti-FLAG (1:200; Sigma-Aldrich; F1804) or anti-sarcomeric α-actinin (1:200; Abcam; ab9465) in blocking buffer overnight at 4°C. After 3 washes with PBS, cells were incubated with anti-mouse secondary antibodies conjugated with Alexa 488 (1:1,000; Thermo Fisher Scientific; A-11029) or Alexa 594 (1:1,000; Thermo Fisher Scientific; A-11032), together with the nuclear stain DAPI (0.1 mg/mL; Sigma; D9542) in PBS for 1 h. Images ware captured using confocal microscopy (Zeiss; LSM880). Phalloidin (Phalloidin-iFluor 647; Abcam; ab176759) was used to stain F-actin in H9C2 cells. For mitochondrial staining, MitoTracker probes were used according to the manufacturer’s instruction (Thermo Scientific; M7512). The surface area of cardiomyocytes was quantified with ImageJ software.
RNA-seq and Ribo-seq analyses
To generate clean reads, which are reads after quality trimming and contaminant filtering, from RNA-seq, adaptor sequences, low-quality and un-paired reads from raw RNA-seq data were removed to obtain clean data for further data analysis by using Cutadapt (version [v.]2.0.dev0) with the following parameters: –pair-filter = any–minimum-length = 50–maximum-length = 150 -q 20–match-read-wildcards–trim-n–max-n 0.25 -a 5′-AGATCGGAAGAGCACACGTC-3′ 5′-AACACGACGCTCTTCCGATCT-3′. Clean reads were then aligned to the rat genome (Rattus_norvegicus.Rnor_6.0) using STAR (v.2.4.2a)41 with ENCODE standard options for long RNA-seq pipeline and the Ensembl transcriptome (Rattus_norvegicus.Rnor_6.0.94) as a reference. We generated read counts using featureCounts (v.1.6.4)42 with default options. Significantly DE genes were analyzed using DESeq2 and edgeR. To view the distribution of reads, the bam files were converted to bigwig format using deepTools (v.3.3.1)43 and visualized with the Integrative Genomics Viewer (IGV).
To generate clean reads from Ribo-seq, Cutadapt (v.2.0.dev0) was used to trim the relevant adaptor sequence and remove the low-quality reads as well as filter reads out of the range of 24−35 nt with the following parameters:–minimum-length = 24–maximum-length = 35 -q 20–match-read-wildcards–max-n 0.25 -u 4 -a 5′-TGGAATTCTCGGGTGCCAAGG-3′. Clean reads that then aligned to rRNA were removed using bowtie (v.1.1.2)44 with default parameters. The remaining reads were then mapped to the rat genome (Rattus_norvegicus.Rnor_6.0) using STAR (v.2.4.2a) with default options, except for the following: –outFilterMismatchNmax 2, –outFilterMultimapNmax 5, –outFilterMatchNmin 16, –alignEndsType EndToEnd. ORF identification and quantification were performed according to the manual of RiboCode (v.1.2.10)23 with the following parameters: -l no -m 10 5′-A CTG,GTG,TTG-3′ -g -b. The TE of ORFs was then analyzed with Ribodiff (v.0.2.2).24 The workflow of Ribo-seq analysis was illustrated in Figure S13. The correlation between replicates of RNAs-seq and Ribo-seq was analyzed (Figure S14).
The raw data and processed data of RNA-seq and Ribo-seq in this study were deposited in Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) (series GEO: GSE155914).
Conservation analysis of micropeptide
To analyze the conservation of micropeptide, we first used the BLAT tool (with default parameters) in the University of California, Santa Cruz (UCSC), genome database to search the aa sequences of micropeptides against mouse and human genomes. Hits with over 80% length of an aa sequence matched, and over 80% identical were considered conserved micropeptides. In addition, we also used the BLASTP tool (with default parameters) in the NCBI database to search the aa sequences of micropeptides against nonredundant protein sequence databases. Similarly, hits from genomes other than the rat with over 80% length of aa sequence matched, and over 80% identical were considered conserved micropeptides.
Statistics
Values are reported as mean ± SD unless indicated otherwise. An analysis of variance (ANOVA) followed by Dunnett’s post hoc testing was used to evaluate the statistical significance for multiple-group comparisons. In addition, the 2-tailed Mann-Whitney U test was used for 2-group comparisons. Values of p <0.05 were considered statistically significant.
Acknowledgments
We thank members of the Huang laboratory for advice and technical support. We also thank Dr. Douglas Cowan for editing the manuscript. This work is supported by grants from the National Natural Science Foundation of China (81873463 to Z.-P.H. and 31900903 to B.L.); Guangdong Basic and Applied Basic Research Foundation (2019B151502003 to Z.-P.H.); Guangdong Science and Technology Department (2018A050506026 to Z.-P.H.); and Fundamental Research Funds for the Central Universities (20ykzd06 to Z.-P.H. and 20lgpy112 to B.L.).
Author contributions
Y.Y. and R.T. performed most of the experiments. B.L. and L.C. performed bioinformatic analyses. S.Y. and T.Y. built micropeptide expression vectors used in this study. Z.-P.H. and J.-H.Y. supervised the study and wrote the manuscript. Y.-C.H., C.L., K.O.L., Y.D., and L.-H.Q. performed quality control of all data and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.03.004.
Contributor Information
Jian-Hua Yang, Email: yangjh7@mail.sysu.edu.cn.
Zhan-Peng Huang, Email: huangzhp27@mail.sysu.edu.cn.
Supplemental information
References
- 1.Hill J.A., Olson E.N. Cardiac plasticity. N. Engl. J. Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 3.Schlüter K.D., Goldberg Y., Taimor G., Schäfer M., Piper H.M. Role of phosphatidylinositol 3-kinase activation in the hypertrophic growth of adult ventricular cardiomyocytes. Cardiovasc. Res. 1998;40:174–181. doi: 10.1016/s0008-6363(98)00171-0. [DOI] [PubMed] [Google Scholar]
- 4.Rolfe M., McLeod L.E., Pratt P.F., Proud C.G. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2) Biochem. J. 2005;388:973–984. doi: 10.1042/BJ20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rybkin I.I., Cross M.E., McReynolds E.M., Lin R.Z., Ballou L.M. alpha(1A) adrenergic receptor induces eukaryotic initiation factor 4E-binding protein 1 phosphorylation via a Ca(2+)-dependent pathway independent of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 2000;275:5460–5465. doi: 10.1074/jbc.275.8.5460. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z.P., Ding Y., Chen J., Wu G., Kataoka M., Hu Y., Yang J.H., Liu J., Drakos S.G., Selzman C.H. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc. Res. 2016;112:543–554. doi: 10.1093/cvr/cvw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y., Matkovich S.J., Hecker P.A., Zhang Y., Edwards J.R., Dorn G.W., 2nd Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc. Natl. Acad. Sci. USA. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amirak E., Fuller S.J., Sugden P.H., Clerk A. p90 ribosomal S6 kinases play a significant role in early gene regulation in the cardiomyocyte response to G(q)-protein-coupled receptor stimuli, endothelin-1 and α(1)-adrenergic receptor agonists. Biochem. J. 2013;450:351–363. doi: 10.1042/BJ20121371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Zhang Y., Ma Q., Gu F., Day D.S., He A., Zhou B., Li J., Stevens S.M., Romo D., Pu W.T. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc. Natl. Acad. Sci. USA. 2013;110:15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L. The Translational Landscape of the Human Heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Doroudgar S., Hofmann C., Boileau E., Malone B., Riechert E., Gorska A.A., Jakobi T., Sandmann C., Jürgensen L., Kmietczyk V. Monitoring Cell-Type-Specific Gene Expression Using Ribosome Profiling In Vivo During Cardiac Hemodynamic Stress. Circ. Res. 2019;125:431–448. doi: 10.1161/CIRCRESAHA.119.314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarewich C.A., Olson E.N. Mining for Micropeptides. Trends Cell Biol. 2017;27:685–696. doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H., Chan W.S., Li Z., Wang D., Liu S., Zhou Y. Small open reading frames: current prediction techniques and future prospect. Curr. Protein Pept. Sci. 2011;12:503–507. doi: 10.2174/138920311796957667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sousa M.E., Farkas M.H. Micropeptide. PLoS Genet. 2018;14:e1007764. doi: 10.1371/journal.pgen.1007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.-R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J.R., Shelton J.M., Sánchez-Ortiz E., Bassel-Duby R., Olson E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Periasamy M., Kalyanasundaram A. SERCA2a gene therapy for heart failure: ready for primetime? Mol. Ther. 2008;16:1002–1004. doi: 10.1038/mt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasala N.B., Yue Y., Lostal W., Wasala L.P., Niranjan N., Hajjar R.J., Babu G.J., Duan D. Single SERCA2a Therapy Ameliorated Dilated Cardiomyopathy for 18 Months in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. 2020;28:845–854. doi: 10.1016/j.ymthe.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarewich C.A., Munir A.Z., Schiattarella G.G., Bezprozvannaya S., Raguimova O.N., Cho E.E., Vidal A.H., Robia S.L., Bassel-Duby R., Olson E.N. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife. 2018;7:e38319. doi: 10.7554/eLife.38319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanford C.F., Griffin E.E., Wildenthal K. Synthesis and degradation of myocardial protein during the development and regression of thyroxine-induced cardiac hypertrophy in rats. Circ. Res. 1978;43:688–694. doi: 10.1161/01.res.43.5.688. [DOI] [PubMed] [Google Scholar]
- 22.Bachner L., Raymondjean M., Bogdanovsky D., Kneip B., Schapira G. Increase of protein synthesis in cell-free system prepared from hypertrophied rat heart during L-triiodothyronine treatment. Biochimie. 1977;59:863–868. doi: 10.1016/s0300-9084(78)80700-7. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Z., Huang R., Xing X., Chen Y., Deng H., Yang X. De novo annotation and characterization of the translatome with ribosome profiling data. Nucleic Acids Res. 2018;46:e61. doi: 10.1093/nar/gky179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Y., Karaletsos T., Drewe P., Sreedharan V.T., Kuo D., Singh K., Wendel H.G., Rätsch G. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics. 2017;33:139–141. doi: 10.1093/bioinformatics/btw585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson K.M., Anderson D.M., McAnally J.R., Shelton J.M., Bassel-Duby R., Olson E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X., Zhang J., Liu Y., Fan X., Ai S., Luo Y., Li X., Jin H., Luo S., Zheng H. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development. 2019;146:dev176198. doi: 10.1242/dev.176198. [DOI] [PubMed] [Google Scholar]
- 28.Chothani S., Schäfer S., Adami E., Viswanathan S., Widjaja A.A., Langley S.R., Tan J., Wang M., Quaife N.M., Jian Pua C. Widespread Translational Control of Fibrosis in the Human Heart by RNA-Binding Proteins. Circulation. 2019;140:937–951. doi: 10.1161/CIRCULATIONAHA.119.039596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galindo M.I., Pueyo J.I., Fouix S., Bishop S.A., Couso J.P. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepoivre C., Belhocine M., Bergon A., Griffon A., Yammine M., Vanhille L., Zacarias-Cabeza J., Garibal M.A., Koch F., Maqbool M.A. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:914. doi: 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei W., Pelechano V., Järvelin A.I., Steinmetz L.M. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y.W., Guo F.X., Xu Y.J., Li P., Lu Z.F., McVey D.G., Zheng L., Wang Q., Ye J.H., Kang C.M. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J. Clin. Invest. 2019;129:1115–1128. doi: 10.1172/JCI98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein C.S., Jadiya P., Zhang X., McLendon J.M., Abouassaly G.M., Witmer N.H., Anderson E.J., Elrod J.W., Boudreau R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018;23:3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarewich C.A., Baskin K.K., Munir A.Z., Bezprozvannaya S., Sharma G., Khemtong C., Shah A.M., McAnally J.R., Malloy C.R., Szweda L.I. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadoshima J., Qiu Z., Morgan J.P., Izumo S. Angiotensin II and other hypertrophic stimuli mediated by G protein-coupled receptors activate tyrosine kinase, mitogen-activated protein kinase, and 90-kD S6 kinase in cardiac myocytes. The critical role of Ca(2+)-dependent signaling. Circ. Res. 1995;76:1–15. doi: 10.1161/01.res.76.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z.P., Young Seok H., Zhou B., Chen J., Chen J.F., Tao Y., Pu W.T., Wang D.Z. CIP, a cardiac Isl1-interacting protein, represses cardiomyocyte hypertrophy. Circ. Res. 2012;110:818–830. doi: 10.1161/CIRCRESAHA.111.259663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 39.Giraldez M.D., Spengler R.M., Etheridge A., Godoy P.M., Barczak A.J., Srinivasan S., De Hoff P.L., Tanriverdi K., Courtright A., Lu S. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018;36:746–757. doi: 10.1038/nbt.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 43.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.