Abstract
Objectives
Serological studies have been critical in tracking the evolution of the COVID-19 pandemic. Data on anti-SARS-CoV-2 antibodies persistence remain sparse, especially from infected individuals with few to no symptoms. The objective of the study was to quantify the sensitivity for detecting historic SARS-CoV-2 infections as a function of time since infection for three commercially available SARS-CoV-2 immunoassays and to explore the implications of decaying immunoassay sensitivity in estimating seroprevalence.
Methods
We followed a cohort of mostly mild/asymptomatic SARS-CoV-2-infected individuals (n = 354) at least 8 months after their presumed infection date and tested their serum for anti-SARS-CoV-2 antibodies with three commercially available assays: Roche-N, Roche-RBD and EuroImmun-S1. We developed a latent class statistical model to infer the specificity and time-varying sensitivity of each assay and show through simulations how inappropriately accounting for test performance can lead to biased serosurvey estimates.
Results
Antibodies were detected at follow-up in 74–100% of participants, depending on immunoassays. Both Roche assays maintain high sensitivity, with the EuroImmun assay missing 40% of infections after 9 months. Simulations reveal that without appropriate adjustment for time-varying assay sensitivity, seroprevalence surveys may underestimate infection rates.
Discussion
Antibodies persist for at least 8 months after infection in a cohort of mildly infected individuals with detection depending on assay choice. Appropriate assay performance adjustment is important for the interpretation of serological studies in the case of diminishing sensitivity after infection.
Keywords: Latent class model, SARS-CoV-2, Seroprevalence, Serosurveillance, Seroepidemiology
Introduction
Serosurveys have played an important role during the COVID-19 pandemic by helping track the true extent of transmission in different populations [[1], [2], [3], [4]] and estimating key epidemiological indicators such as the infection fatality ratio [[5], [6], [7]]. Serological studies have employed dozens of different immunoassays designed to detect antibodies targeting primarily all or part of the spike (S) or nucleocapsid (N) proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [8]. The accuracy of serology-based estimates depends on the immunoassay antibody targets and their performance in detecting both recent and historic infections. Successive epidemic waves in different parts of the world have created a heterogeneous immune landscape, which poses challenges for SARS-CoV-2 serosurveillance.
Anti-SARS-CoV-2 antibody levels tend to decay after the convalescent period, leading to increasing chances of a negative immunoassay result [9,10]. False negatives can lead to underestimates of the true infection attack rate unless appropriately understood and accounted for in analyses [12]. Furthermore, post-infection antibody kinetics appear to depend on infection severity, with severe infections leading to larger increases in antibodies than in mild or asymptomatic infections [11]. However, few studies have characterized antibody kinetics past 6 months after infection [13,14], or described these kinetics in mild and asymptomatic infections [[15], [16], [17]], which comprise the vast majority of infections in the community [18], thus limiting the public health use of antibody testing for SARS-CoV-2 [19].
Here we aim at quantifying changes in anti-SARS-CoV-2 antibody levels across the spectrum of severity and age using three commercially available immunoassays. We do this through a longitudinal study up to 9 months from plausible infection dates of a cohort of seropositive individuals recruited through serosurveys in Geneva, Switzerland.
Materials and methods
Recruitment
Participants of the SEROCoV-POP [2] and SEROCoV-WORK+ [20] serosurveys (please see supplementary material) who were seropositive on the Euroimmun anti-S1 test (referred to as EI, OD ratio ≥1.1, EI-positive cohort) at their first study visit (referred to as baseline, between April and July 2020) were invited to return for another serological test in November 2020 (referred to as follow-up). To estimate the infection risk in the community over the period between the two visits, we also randomly selected participants initially Euroimmun anti-S1 negative (EI-negative) with a similar sex ratio and age range who returned for a second visit. All participants gave written informed consent, completed a questionnaire and provided a venous blood sample. This study was approved by the Geneva Cantonal Commission for Research Ethics (CCER project number 2020-00881).
Immunoassays
SARS-CoV-2 antibodies were measured using three commercially available tests: a semiquantitative anti-S1 ELISA detecting IgG (Euroimmun, Lübeck, Germany #EI 2606-9601 G), and the quantitative Elecsys anti-RBD (#09 289 275 190, Roche-RBD) and semiquantitative Elecsys anti-N (#09 203 079 190, Roche-N), both measuring total antibodies (IgG/A/M, Roche Diagnostics, Rotkreuz, Switzerland). Seropositivity was defined using the cut-off provided by the manufacturers: ≥1.1 for Euroimmun anti-S1 ELISA; ≥0.8 U/mL for Roche anti-RBD; and ≥1.0 for Roche anti-N. The Euroimmun immunoassay on samples from the baseline and follow-up visits was performed as samples came into the laboratory, using several different reagent lots. All Roche tests (anti-RBD and anti-N) on baseline and follow-up samples were performed using the same reagent lot in each case. Samples from the baseline visit were frozen after their first analysis and thawed several months later for retesting with the Roche immunoassays (please see supplementary material).
Statistical analyses
We determined the proportion of participants who seroconverted (negative to positive) or seroreverted (positive to negative), and tested the significance in proportions between sex and age class using a two-sample Wilson score interval test for equality of proportions with continuity correction [21]. We also compared test readout values at each visit and classified each participant's response as decreasing, increasing or stable for Roche-RBD test's quantitative readouts (Table S1). We assessed significance of changes taking into account the intra-lot variance of our internal positive control serum (Fig. S5). Significance of response changes was based on the z-score of the difference between follow-up and baseline results at a significance level of 5%.
We developed a statistical model to jointly infer each test's specificity and sensitivity accounting both for changes in sensitivity with time post-infection due to antibody decay as well as possible unknown SARS-CoV-2 infection times including the possibility of infection between visits (Fig. S8). We assumed that each test result is independent of the other test results conditional on its true status as positive or negative. Inference is drawn in a Bayesian framework incorporating multiple sources of test validation data and RT-PCR test results, when available (please see supplementary material).
We then used simulations to illustrate how seroprevalence estimates could be biased if only correcting for sensitivity and specificity using the Gland-Rogen estimator [22] with data from typical validation studies in the literature and package inserts (single time-invariant sensitivity, short follow-up times and more representative of severe infections). We considered three hypothetical scenarios of serosurveys using Geneva's epidemic curves: the first one occurring one month after the first wave peak, the second one as if it had occurred five months after a single wave, and the third one month after the peak of the second wave (Fig. 1 A). We then used the associated distributions of time after infection to simulate test results based on our estimates of specificity and time-varying test sensitivity. We finally estimated seroprevalence correcting for test performance but using the conventional approach with a single value for sensitivity [23]. For each scenario we simulated 2000 samples with 10–90% seroprevalence and compared these simulated data to the seroprevalence estimates that ignore changes in sensitivity (please see supplementary material).
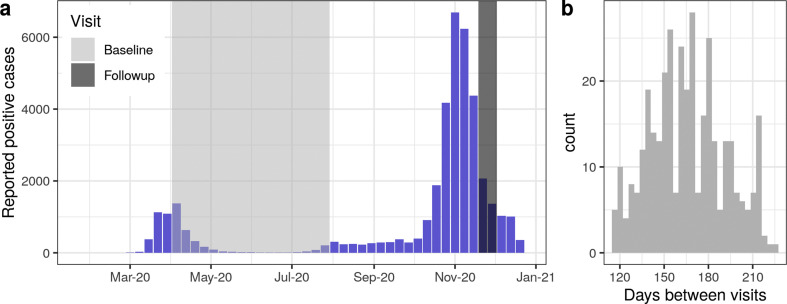
Fig. 1.
Study recruitment with respect to the SARS-CoV-2 epidemic curve in Geneva, Switzerland. (A) Weekly reported number of virologically-confirmed SARS-CoV-2 infections in the canton of Geneva (blue bars) and study timing for both the baseline (light grey) and follow-up (dark grey) visits. (B) Histogram of days between study visits for the EI-positive cohort (N = 354).
Results
Cohort characteristics
A total of 354 participants from previous serosurveys having a positive Euroimmun anti-S1 IgG test result at baseline constituted the EI-positive cohort (Fig. 1A, Fig. S1). Participants in this cohort were 18–84 years old, and 52% (183/354) were women (Table 1 ). Less than half reported having had an RT-PCR test prior to the baseline visit (148, 42%), 58 of whom reported a positive result (90 negative, positivity rate of 39%). Ten per cent (37/354) of participants reported having had no COVID-19-compatible symptoms before the baseline visit, while 69% reported four symptoms or more (Fig. S2). The majority of these participants did not require hospitalization (336/354, 95%). RT-PCR testing before the baseline visit did not necessarily imply having self-reported symptoms and vice versa. The median period between baseline and follow-up visits was 165 days (range 115–224 days) (Fig. 1B). Twenty per cent (71/354) of participants reported having a SARS-CoV-2 virologic (RT-PCR or rapid antigen) test between visits including four (6%) positive results. No participant reported being hospitalized between their baseline and follow-up visits.
Table 1.
Characteristics of baseline Euroimmun anti-S1 IgG (EI) positive and negative cohorts
| Characteristic | EI-positive cohort, n = 354 | EI-negative cohort, n = 187 |
|---|---|---|
| Sex | ||
| Female | 183 (52) | 93 (50) |
| Male | 171 (48) | 94 (50) |
| Age group | ||
| 18–65 | 313 (88) | 183 (98) |
| 65+ | 41 (12) | 4 (2.1) |
| RT-PCR test before baseline visit | ||
| No test | 206 (58) | 169 (90) |
| Positive | 58 (16) | 3 (1.6) |
| Negative | 90 (25) | 15 (8.0) |
| Hospitalization before baseline visit | ||
| Did not require hospitalization | 336 (94.9) | 187 (100) |
| Required hospitalization | 18 (5.1) | 0 (0) |
| Required ICUa | 2 (0.6) | 0 (0) |
| Number of COVID-19-compatible symptoms reported before baseline visitb | ||
| 0 | 37 (10) | 71 (38) |
| 1 | 16 (4.5) | 23 (12) |
| 2 | 28 (7.9) | 29 (16) |
| 3 | 31 (8.8) | 16 (8.6) |
| 4 | 242 (68) | 47 (25) |
| Symptoms onset before baseline visitc | ||
| 1 month before baseline visit | 19 (6.0) | 12 (9) |
| >1 month before baseline visit | 298 (94) | 103 (90) |
| RT-PCR/Rapid antigen test between visits | ||
| No test | 283 (80) | 126 (67) |
| Positive | 4 (1.1) | 15 (8.0) |
| Negative | 67 (19) | 46 (25) |
Data are presented as n (%).
ICU: intensive care unit. Five positive cohort participants with no responses to the ICU question.
Symptoms (self-reported): fever, cough, cold, throat pain, panting, headache, muscular and/or articular pain, fatigue, loss of appetite, nausea, diarrhoea, stomach pain, loss of taste and/or smell, other. One participant from the negative cohort did not reply to this question.
Percentages computed over the number of participants presenting at least 1 COVID-19 compatible symptom (EI-positive cohort n = 317, EI-negative cohort n = 115).
We also followed a cohort of 187 participants who had a negative EI test at baseline (EI-negative cohort) selected from previous participants to have a similar sex ratio and age range as the EI-positive cohort (Table 1).
Antibody detection and decay with three immunoassays
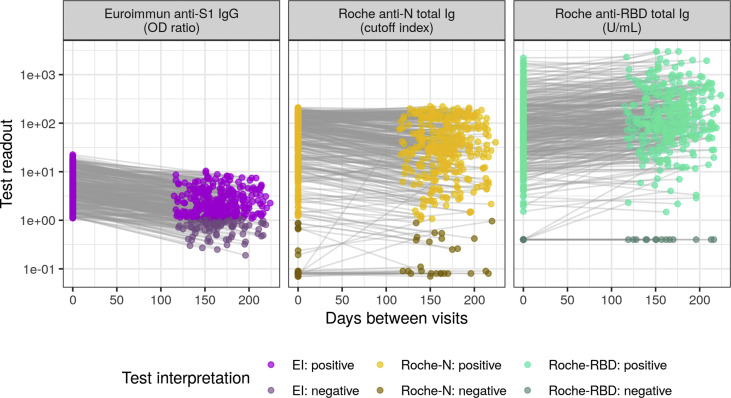
Within the EI-positive cohort, 93.2% (330/354) and 95.2% (337/354) were positive at baseline for the Roche-N and Roche-RBD assays, respectively (Fig. 2 and Fig. S3). At the individual level, 26% (91/354) of the EI-positive cohort was seronegative with the EI test at follow-up (i.e. seroreverted, Table 2 ). Seroreversions were less frequent with the Roche-N assay (1.2%, 6/330) and none were detected with the Roche-RBD assay. We identified no significant differences in seroreversions across age groups and sex (Table S1).
Fig. 2.
Test readout trajectories between baseline and follow-up visits. The cohort was composed of 354 participants with positive Euroimmun anti-S1 (EI) test at baseline. Test readout units and thresholds for positivity are assay-specific, Roche-RBD values below the limit of quantitation (0.4 U/mL) were set to the limit of quantitation for plotting and analysis. The dynamic range of both the EI and Roche-N tests are limited compared to the Roche-RBD thus leading to censoring of extremely high and low values. Baseline and follow-up samples were tested with different reagent lots of the EI immunoassay whereas the same Roche-N and Roche-RBD reagent lots were used for all samples (supplementary material). Trajectories for the EI-negative cohort are given in Fig. S4.
Table 2.
Serostatus and test readout changes between visits
| Serostatus change | EI-positive cohort (n = 354) |
EI-negative cohort (n = 187)a |
|||
|---|---|---|---|---|---|
| EI | Roche-N | Roche-RBD | Roche-N | Roche-RBD | |
| Reversion | 91/354 | 6/330 | 0/337 | 3/21 | 1/23 |
| Conversion |
— |
4/24 |
3/17 |
25/166 |
29/164 |
| No change | 263/354 | 344/354 | 351/354 | 159/187 | 157/187 |
Serostatus changes are given with respect to the baseline number of positives (negatives) for reversion (conversion) for each test. Statistics by sex and age group given in Table S1.
EI test results at follow-up were not available for the EI-negative cohort.
We quantified the change in test readout between visits for the Roche-RBD immunoassay, as the other two tests are considered qualitative or semiquantitative by the manufacturers. We found that 17% (61/354) of participants had a significant decrease in their test readout and 66% (235/354) had a significant increase. When subdivided by sex and age class, women had a significantly lower proportion of decaying as well as a higher proportion of increasing Roche-RBD responses, and no differences were found between age classes (Table S1).
In the EI-negative cohort, 11% of the participants were classified as seropositive at baseline with Roche-N (21/187) and 12% with Roche-RBD (23/187, Fig. S3). A total of 25 out of 166 (15%) Roche-N negatives, and 29/164 (17.5%) Roche-RBD negatives seroconverted at follow-up (Table 2). We identified no significant differences by age or sex in seroconversion rates (Table S1).
Estimation of time-varying test sensitivity and impact on serosurveillance
The trajectories of antibody detection in both cohorts, combined with data from assay validation studies (Table S2, Fig. S9) enabled us to produce model-based estimates of test specificity and changes in clinical sensitivity with time after infection.
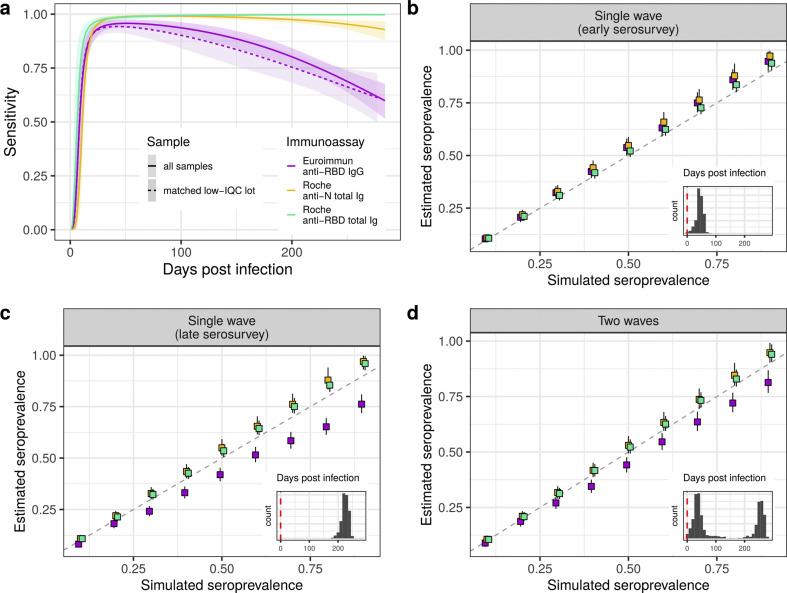
We estimated large differences in test sensitivity, which depended on the delay between infection and serologic assessment (Fig. 3 A). EI sensitivity decreased gradually reaching 61.2% (95% CI 53.4–68.5) at 284 days after infection, the longest time modelled. The decrease in sensitivity for Roche-N was smaller (mean at 284 days of 93.3%; 95% CI 88.7–96.7). Sensitivity estimates for Roche-RBD remained close to the peak value up to the maximal modelled time. As baseline and follow-up samples were tested with different EI test reagent lots, we also estimated performance where we restricted to 127 samples with baseline and follow-up reagent lots with similar test readout values for our internal positive control serum and found similar results for sensitivity and proportion of seroreversions, though with larger uncertainty (Fig. 3A, Fig. S7 and supplementary material).
Fig. 3.
Model test performance estimates and simulation. (A) Model estimates of sensitivity changes with time post infection. Due to EI reagent inter-lot variability (please see supplementary material) results are shown for the whole sample (N = 354), as well as for a subsample for which assay internal positive quality control (IQC) readout values were similar for baseline and follow-up reagent lots (N = 127, matched low IQC lot). (B–D) Simulation scenarios of seroprevalence estimation if the decay in sensitivity is not accounted for. Scenario in (B) is assumed to occur one month after the first epidemic wave peak in Geneva, with corresponding distribution of days between infection and the serosurvey; scenario in (C) the serosurvey occurs after a single wave and 180 days after the epidemic peak; and scenario in (D) assumes the serosurvey occurred one month after the peak of the second epidemic wave, yielding a bimodal distribution of days post infection (insets, vertical dashed line at x = 0 indicates infections that occurred on the serosurvey date).
When serosurveys were simulated after a single epidemic wave (times after infection 0–115 days, Fig. 3B), seroprevalence estimates for all three tests using conventional adjustment for sensitivity have 95% CI covering the true values of simulated seroprevalence between 10 and 30%, after which estimates had a slight tendency towards overestimation. Instead, when simulating serosurveys longer after a single wave (180–250 days after infection), as the true underlying seroprevalence increased, estimates based on the EI assay grew increasingly biased, with an under-estimation of up to 15% when the simulated seroprevalence was 90% (Fig. 3C). In contrast, estimates based on the two Roche assays tended to overestimate seroprevalence by 5% in this scenario. Finally, the results obtained for the two epidemic waves scenario stood in between, with a less severe seroprevalence underestimation for EI test (10% underestimation at 90% seroprevalence). The two Roche tests remained in closer agreement with the true seroprevalence throughout the simulated range with the 95% CrI covering the true value at seroprevalence below 30%, and slightly overestimating it by around 3% for higher seroprevalence (>60%).
Discussion
In a cohort of mostly mild/asymptomatic SARS-CoV-2-infected individuals, antibodies targeting either the nucleocapsid (N) or the spike (S) proteins of the virus generally persisted for at least 8 months after infection. The initial measurements taken within 4.5 months of participants' infections were consistent across all three assays. However, results diverged between assays upon re-evaluation 4–8 months later, with one-in-four participants seroreverting according to the EI IgG assay, with few to no seroreversions with the Roche anti-N and anti-RBD total Ig tests. We find that without appropriate quantification and adjustment for time-varying assay sensitivity, seroprevalence surveys may underestimate the true number of cumulative infections.
Seroprevalence underestimation due to decaying sensitivity [4] will depend on the timing of serosurveillance with respect to the number and amplitude of preceding epidemic waves. Larger and more distal waves being more severely underestimated due to the increasing relative frequency of false negatives compared with false positives. Even for mild infections, which may elicit less robust immune responses [25], the sensitivity of both Roche tests remained close to 100% after more than 8 months after infection. These results suggest that these Roche immunoassays are suitable for seroprevalence estimation at longer times post-exposure.
While previous studies suggest that anti-RBD antibody measurement correlates with neutralizing antibody titres at least up to 4–6 months after exposure [11,26], it remains unclear whether the persistence of antibodies we report here is a proxy of continued immune protection. We also highlight the increase of anti-RBD total Ig measured by the Roche-RBD assay, observed in other studies as well, however correlating poorly with neutralizing antibody measurements [16]. This increase may be explained by aspects of the assay design that lead to preferential binding of higher affinity antibodies (which increase after infection) or due to the fact that it measures total immunoglobulins versus a single isotype [27]. Differences in readout trajectories between sexes for the Roche tests are consistent with results from other studies [28].
Our results come with a number of limitations. While our estimates of seroreversion with the EI assay were in line with previously published data [10,29,30], we tested baseline and follow-up samples with the EI assay at different times and with different reagent lots (inter-lot coefficient of variation of 30%, Fig. S5). Sensitivity analyses accounting for these differences showed similar seroreversion patterns and sensitivity decay as the main analyses (Fig. 3A, Fig. S7 and supplementary material). Secondly, statistics on changes in sero-status and test response may have been influenced by (re-)exposures during the period between baseline and follow-up visits, which was accounted for in the latent class model. Our analyses are conditioned on antibody response to infection and therefore do not account for the small proportion of infected individuals that do not do so. This implies sensitivity estimates may be overestimated in general. In using time-varying sensitivity to estimate seroprevalence we assumed a constant case-to-infection ratio over the course of the epidemic, an assumption which may not hold in many settings. Finally, our sample included mostly working age adults, thus potentially limiting the generalizability of our results to other subpopulations such as children, elderly or immunosuppressed individuals.
Through quantifying anti-SARS-CoV-2 antibody persistence in a cohort of mostly mildly symptomatic and asymptomatic infections, we confirm that antibodies remain detectable after at least 8 months after infection. Using multiple immunoassays, we illustrate that test choice matters and can greatly affect the interpretation of results from population-level serologic studies, especially as the immune landscape becomes a more complex mix of recent and old infections. Continued multi-assay, multi-epitope characterization of post-infection kinetics over longer periods is important for appropriate analysis and interpretation of data from serosurveillance efforts aimed at tracking the evolution of this pandemic.
Author contributions
J.P.S.: Methodology, Software, Formal Analysis, Visualization, Writing – Original Draft, Writing – Review & Editing. M.E.Z.: Conceptualization, Methodology, Validation, Investigation, Data curation, Project Administration, Writing – Original Draft, Writing – Review & Editing. S.Y.: Investigation, Writing – Review & Editing. D.A.: Resources, Writing – Review & Editing. M.E.: Writing – Review & Editing. I.E.: Writing – Review & Editing. J.F.B.: Writing – Review & Editing. F.C.: Writing – Review & Editing. D.P.: Writing – Review & Editing. D.T.: Writing – Review & Editing. O.K.: Writing – Review & Editing. NV: Resources, Writing – Review & Editing. LK: Resources, Writing – Review & Editing. I.G.: Supervision, Funding acquisition, Writing – Review & Editing. S.S.: Conceptualization, Methodology, Validation, Supervision, Project Administration, Funding acquisition, Writing – Original Draft, Writing – Review & Editing. A.S.A.: Methodology, Formal Analysis, Visualization, Supervision, Writing – Original Draft, Writing – Review & Editing.
Transparency declaration
This work was supported by the Private Foundation of the Geneva University Hospitals, the General Directorate of Health of the Department of Safety, Employment and Health of the canton of Geneva, the Swiss Federal Office of Public Health, the Fondation des Grangettes, the Center for Emerging Viral Diseases, and the Corona Immunitas research network, coordinated by the Swiss School of Public Health (SSPH+), Switzerland. N.V. received restricted research grants from Roche SA with payment made to the Geneva University Hospitals in the past 36 months that were unrelated to the present work. The other authors declare that they have no conflicts of interest.
Acknowledgements
We thank all the participants, without whom this study would not have been possible.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.06.040.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., et al. ENE-COVID Study Group Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stringhini S., Wisniak A., Piumatti G., AAzman A.S., Lauer S.A., Baysson H., et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckerle I., Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396:514–515. doi: 10.1016/S0140-6736(20)31482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buss L.F., Prete C.A., Jr., Abrahim M.M., Mendrone A., Jr., Salomon T., de Almeida-Neto C., et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Saez J., Lauer S.A., Kaiser L., Regard S., Delaporte E., Guessous I., et al. Serocov-POP Study Group Serology-informed estimates of SARS-CoV-2 infection fatality risk in Geneva, Switzerland. Lancet Infect Dis. 2020;21:69–70. doi: 10.1016/S1473-3099(20)30584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen O.B., Nissen J., Dinh K.M., Schwinn M., Kaspersen K.A., Boldsen J.K., et al. SARS-CoV-2 infection fatality rate among elderly retired Danish blood donors – a cross-sectional study. Clin Infect Dis. 2020;ciaa1627 doi: 10.1093/cid/ciaa1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin A.T., Hanage W.P., Owusu-Boaitey N., Cochran K.B., Walsh S.P., Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol. 2020;35:1123–1138. doi: 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Chen Z., Azman A.S., Deng X., Sun R., Zhao Z., et al. Serological evidence of human infection with SARS-CoV-2: a systematic review and meta-analysis. Lancet Glob Health. 2021;6:e598–e609. doi: 10.1016/S2214-109X(21)00026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen S.C.A., Yang F., Hoh R.A., Jackson K.J.L., Roeltgen K., Lee J.-Y., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host & Microbe. 2020;28:516–525. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi S., Greenhouse B., Rodríguez-Barraquer I. Are seroprevalence estimates for severe acute respiratory syndrome coronavirus 2 biased? J Infect Dis. 2020;222:1772–1775. doi: 10.1093/infdis/jiaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan A., Najeeb J., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W.-H., Chiu S.S., et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12:63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection. bioRxiv. 2020 doi: 10.1101/2020.11.15.383323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L’Huillier A.G., Meyer B., Andrey D.O., Arm-Vernez I., Baggio S., Didierlaurent A., et al. Geneva Centre for Emerging Viral Diseases. Antibody persistence in the first six months following SARS-CoV-2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021;27 doi: 10.1016/j.cmi.2021.01.005. 784.e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duysburgh E., Mortgat L., Barbezange C., Dierick K., Fischer N., Heyndrickx L., et al. Persistence of IgG response to SARS-CoV-2. Lancet Infect Dis. 2021;21:163–164. doi: 10.1016/S1473-3099(20)30943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theel E.S., Slev P., Wheeler S., Couturier M.R., Wong S.J., Kadkhoda K. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00797-20. e00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stringhini S., Zaballa M.E., Pullen N., de Mestral C., Perez-Saez J., Dumont R., et al. Large variation in anti-SARS-CoV-2 antibody prevalence among essential workers in Geneva, Switzerland. Nat Commun. 2021;12:3455. doi: 10.1038/s41467-021-23796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson E.B. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 1927;22:209–212. [Google Scholar]
- 22.Rogan W.J., Gladen B. Estimating prevalence from the results of a screening test. Am J Epidemiol. 1978;107:71–76. doi: 10.1093/oxfordjournals.aje.a112510. [DOI] [PubMed] [Google Scholar]
- 23.Gelman A., Carpenter B. Bayesian analysis of tests with unknown specificity and sensitivity. J R Stat Soc Ser C Appl Stat. 2020;69:1269–1283. doi: 10.1111/rssc.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vetter P., Cordey S., Schibler M., Vieux L., Despres L., Laubscher F., et al. Geneva Center for Emerging Viral Diseases. Clinical, virological and immunological features of a mild case of SARS-CoV-2 re-infection. Clin Microbiol Infect. 2021;27 doi: 10.1016/j.cmi.2021.02.010. 791e.1–e.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzelak L., Velay A., Madec Y., Gallais F., Staropoli I., Schmidt-Mutter C., et al. Sex differences in the evolution of neutralizing antibodies to SARS-CoV-2. J Infect Dis. 2021;jiab127 doi: 10.1093/infdis/jiab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choe P.G., Kang C.K., Suh H.J., Jung J., Song K.-H., Bang J.H., et al. Waning antibody responses in asymptomatic and symptomatic SARS-CoV-2 Infection. Emerg Infect Dis. 2021;27 doi: 10.3201/eid2701.203515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korte W., Buljan M., Rösslein M., Wick P., Golubov V., Jentsch J., et al. SARS-CoV-2 IgG and IgA antibody response is gender dependent; and IgG antibodies rapidly decline early on. J Infect. 2021;82:e11–e14. doi: 10.1016/j.jinf.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.