Abstract
Compared to normal cells, cancer cells exhibit specific metabolic characteristics that facilitate the growth and metastasis of cancer. It is now widely appreciated that long non-coding RNAs (lncRNAs) exert extensive regulatory effects on a spectrum of biological processes through diverse mechanisms. In this review, we focus on the rapidly advancing field of lncRNAs and summarize the relationship between the dysregulation of lncRNAs and cancer metabolism, with a particular emphasis on the specific roles of lncRNAs in glycolysis, mitochondrial function, glutamine, and lipid metabolism. These investigations reveal that lncRNAs are a key factor in the complexity of malignant cancer metabolism. Only through understanding the relevance between lncRNAs and cancer metabolic reprogramming can we open a new chapter in the history of carcinogenesis, one that promises to alter the methods of cancer diagnosis and treatment.
Keywords: cancer metabolism, lncRNA, reprogramming, regulatory mechanism
Graphical abstract
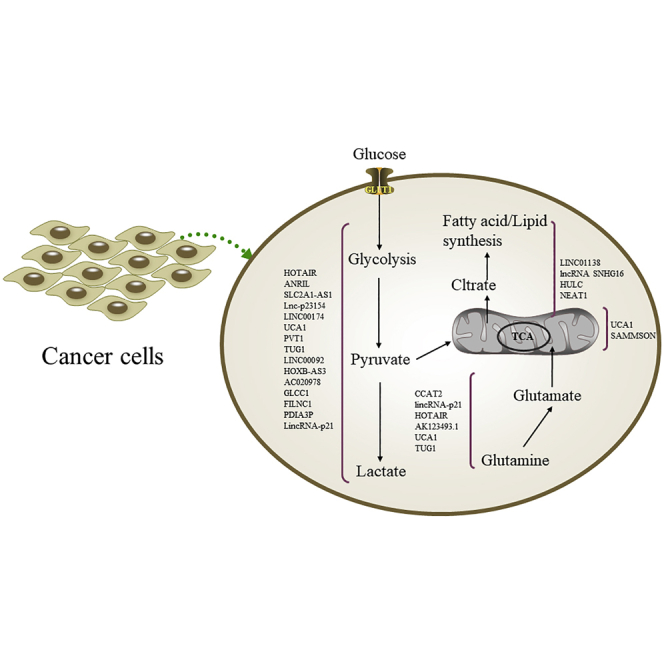
The reprogramming of the cellular metabolism is a well-established hallmark of cancer. In this review, Xu and colleagues highlighted the pivotal roles of lncRNAs in the regulation of cancer metabolism. It is expected that lncRNA-based diagnostics and therapeutics involved in cancer metabolism will one day be beneficial for cancer patients.
Introduction
Energy metabolism refers to the set of substances and chemical transformations via enzyme-catalyzed reactions within the cells of organisms.1 The process of metabolism can be divided into anabolism and catabolism, which focus either on using energy to make basic molecules or on harvesting energy from the breakdown of molecules. Compared with normal cells, the metabolic ecology of cancer cells is more complex. To satisfy the requirements of tumor growth and metastatic dissemination, cancer cells can reprogram metabolism, which is regarded as a hallmark of cancer.2,3 There are some cancer-associated metabolic changes including excessive glucose and/or glutamine uptake, alterations in lipid metabolism, and increased dependence on aerobic glycolysis.4 Accumulating evidence implies that cancer metabolism can be impacted by signaling pathways and eventually lead to the inactivation of tumor suppressor genes or activation of oncogenes, such as TP53, Myc, and hypoxia inducible factor-1 (HIF-1).5,6 However, we are still far from a comprehensive understanding of cancer-associated metabolic reprogramming, and thus, elucidation of the molecular mechanisms underlying metabolic reprogramming is of great importance.
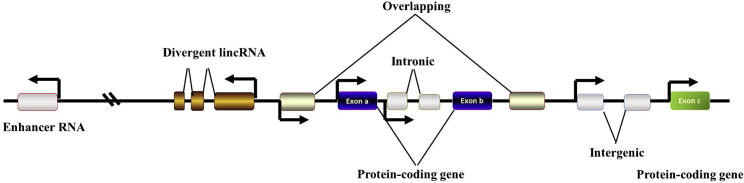
With rapid advancements in transcriptome sequencing technology and bioinformatics, an increasing number of non-coding RNAs (ncRNAs) have been annotated and investigated. According to size, these ncRNAs can be broadly grouped into two major classes. Long ncRNAs (lncRNAs) contain RNA transcripts longer than 200 nucleotides (nt) with no or limited protein coding potential.3,7 It is important to note that lncRNAs are broadly defined and can be subclassified into several classes, including intergenic transcripts, enhancer RNAs, and exonic or intronic transcripts in either the antisense or sense orientation (Figure 1).8 Analogous to mRNAs, many identified lncRNAs are transcribed by RNA polymerase II (RNA pol II) and undergo 5′ end capping and 3′ end polyadenylation.9,10
Figure 1.
Classification of lncRNAs
lncRNAs can be further subclassified into divergent, overlapping, intronic, and intergenic lncRNAs on the grounds of their intersection with protein-coding genes.
lncRNAs are diverse and numerous. In many instances, lncRNAs are located within cytosolic or nuclear fractions.11 Compared with mRNA, the half-lives of lncRNAs appear to be more important during biological processes because of their limited protein coding potential. In 2012, Marcel E. Dinger and John S. Mattick12 performed a genome-wide analysis of lncRNA stability by custom microarrays. They revealed that lncRNA half-lives vary over a wide range. cis-antisense or intergenic lncRNAs are more stable than those transcribed from introns. According to some reports, factors such as exosomes and microRNAs (miRNAs) degrade lncRNAs.13,14 Recently, studies have reported that RNA or ribosome-binding patterns can also affect lncRNA stability.15,16 For example, lncRNA-highly expressed in GBC (HGBC) specifically binds to the RNA-binding protein HuR and is stabilized by HuR. In addition, RNA methylation has also been revealed to be associated with RNA metabolic processes. Chen and colleagues17 found that ALKBH5-mediated m6A modification contributes to the stability of PVT1. Consistently, another study identified that NSUN2-mediated m5C modification of H19 lncRNA can increase its stability.18 However, the underlying regulatory pathways affecting lncRNA stability remain largely undefined.
Over the past several years, multiple studies have begun to advance the idea that lncRNAs are not just “junk products” of transcription but crucial supplements to proteins and other effectors in complex regulatory networks.19 Given that lncRNAs show strict biological regulation and participate in a spectrum of cellular processes, misregulated lncRNA expression can cause various human diseases and cancers.20,21 lncRNAs exhibit complex modulatory roles via various mechanisms, including functioning as scaffolds for chromatin-protein interactions and regulating mRNA splicing, protein translation, miRNA sequestration, and so on.22 Notably, although miRNA “sponges” have been a hot area of research for several years,23 the competing endogenous RNA (ceRNA) hypothesis remains controversial. The in vivo data of Denzler et al.24 indicated that modulation of miRNA target abundance is unlikely to lead to significant effects on gene expression through a ceRNA effect.25 Recent studies have discovered a novel regulatory connection between miRNAs and their target genes, named as target-directed miRNA degradation (TDMD). Extended base pairing with a target can expose the 3′ end of the miRNA from the argonaute (AGO) PAZ domain or AGO proteolysis through the ubiquitin-proteasome pathway, resulting in miRNA degradation.26, 27, 28, 29 lncRNA CYRANO mediates miRNA (miR)-7 degradation through TDMD.30 Owing to the major transcriptional and post-transcriptional regulatory ability of lncRNAs in cell proliferation, metastasis, and survival, it is now well recognized that lncRNAs are emerging stars in development and progression.31 In this review, we will discuss in detail the pivotal roles of lncRNAs in the regulation of cancer metabolism.
The overview of cancer metabolism and lncRNAs
Carcinogenesis is a multifactor and multistage process. During this process, cancer cells must overcome the biosynthetic demands for proliferation and metastasis. As a result, the absence of sufficient metabolic resources might have disastrous consequences for the cell if there is an attempt to grow rapidly. To avoid this, the aberrant loss of tumor suppressors and/or activated oncogenes has an impact on signaling pathways, which result in metabolic reprogramming, particularly in the hypoxic microenvironment.32,33 Metabolic reprogramming allows cancer cells to maintain unlimited proliferation and metastasis potential by supplying materials and corrupting the surrounding microenvironment.4 Emerging evidence favors the hypothesis that some cancer-related lncRNAs are key players in enabling cancer cells to overcome metabolic stress and complete metabolic reprogramming.34 In Table 1 and Figure 2, we provide an overview of how lncRNAs control the cancer metabolism.
Table 1.
lncRNAs associated with cancer metabolism
| Metabolic process | lncRNA | Tumor | Molecular mechanism | The level of evidence | Ref. |
|---|---|---|---|---|---|
| Glucose metabolism | HOTAIR | hepatocellular carcinoma | increases GLUT1 expression by activating mTOR signaling | in vitro | 35 |
| ANRIL | nasopharyngeal carcinoma | increases GLUT1 expression by activating mTOR signaling | in vitro | 36 | |
| SLC2A1-AS1 | hepatocellular carcinoma | represses GLUT1expression by inhibiting the STAT3/FOXM1 pathway | in vivo | 37 | |
| lnc-p23154 | oral squamous cell carcinoma | increases GLUT1 expression by interacting with miR-378a-3p | in vitro and in vivo | 38 | |
| LINC00174 | glioma carcinogenesis | increases GLUT1 expression by interacting with miR-152-3p | in vitro | 39 | |
| UCA1 | cervical cancer and pediatric AML | regulates hexokinase 2 (HK2) | in vitro | 40,41,42 | |
| PVT1 | osteosarcoma and gallbladder cancer | regulates HK2 by regulating miR-497 and miR-143 | in vitro | 43,44 | |
| UCA1 | esophageal cancer | increases HK2 expression by interacting with miR-203 | in vitro | 45 | |
| TUG1 | hepatocellular carcinoma | regulates HK2 by miR-455-3p | in vitro | 46 | |
| LINC00092 | ovarian cancer | interacts with PFKFB2 | in vitro | 47 | |
| HOXB-AS3 | colon cancer | binds to hnRNP A1 and regulates PKM splicing | in vitro | 48 | |
| AC020978 | non-small cell lung cancer | directly interacts with PKM2 and enhances its protein stability | in vitro | 49 | |
| GLCC1 | colorectal cancer | binds to HSP90 and stabilizes c-Myc protein | in vitro | 50 | |
| FILNC1 | renal cell carcinoma | represses c-Myc protein level through sequestering AUF1 | in vitro | 51 | |
| PDIA3P | multiple myeloma | interacts with c-Myc directly and binds to the promoter of G6PD | in vitro | 52 | |
| lincRNA-p21 | decreases HIF-1α ubiquitination and accumulation of HIF-1α | in vitro | 53 | ||
| Glutamine metabolism | CCAT2 | colorectal cancer | secondary structure | in vitro and in vivo | 54 |
| lincRNA-p21 | bladder cancer | regulates GLS level | in vitro | 55 | |
| HOTAIR | glioma | regulates GLS level by interacting with miR-126-5p | in vitro | 56 | |
| AK123493.1 | pancreatic cancer | regulates GLS level via ADAR/Dicer-dependent RNA interference | in vitro | 57 | |
| UCA1 | bladder cancer | regulates GLS2 level by miR-16 | in vitro | 58 | |
| TUG1 | intrahepatic cholangiocarcinoma | stabilizes Sirt3 mRNA by binding to miR-145 | in vitro | 59 | |
| Lipid metabolism | LINC01138 | ccRCC | stabilizes SREBP-1 protein by interacting with PRMT5 | in vitro | 60 |
| lncRNA SNHG16 | pancreatic cancer | modulates SREBP-2 expression by directly targeting miR-195 | in vitro | 61 | |
| lncRNA SNHG16 | colorectal cancer | Wnt signaling | in vitro | 62 | |
| HULC | hepatocellular carcinoma | upregulates PPARA and activates ACSL1 | in vitro | 63 | |
| NEAT1 | hepatocellular carcinoma | binds to miR-124-3p | in vitro | 64 | |
| Mitochondrial oxidative metabolism | UCA1 | bladder cancer | binds to miR-195 and induces ARL2 | in vitro | 65 |
| SAMMSON | melanomas | sequesters XRN2 | in vitro | 66,67 |
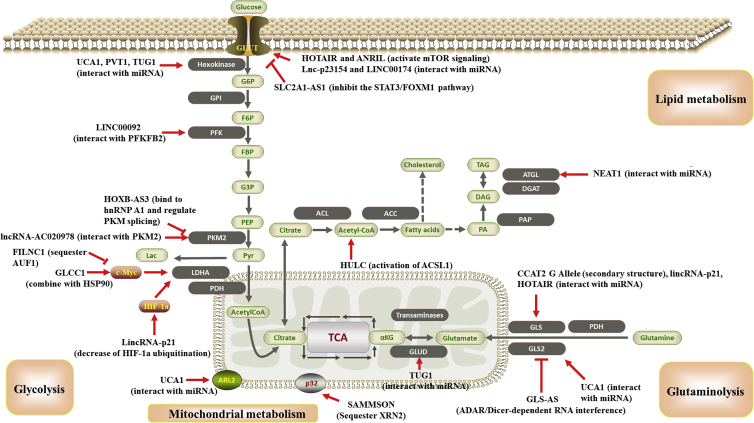
Figure 2.
Schematic illustration of lncRNAs involved in the metabolic rearrangement of cancer metabolism by targeting metabolism-related molecules or pathways
Detailed mechanisms of these lncRNAs are described in the main text.
lncRNAs regulate glucose metabolism in cancer
The preference for glycolysis over oxidative phosphorylation to generate energy regardless of oxygen availability, which is a unique metabolic phenotype that characterizes cancer cells, has been defined as “aerobic glycolysis” or the “Warburg effect.”68,69 Compared with mitochondrial oxidative phosphorylation, glycolysis produces low levels of ROS (reactive oxygen species) that can cause apoptosis in cancer cells.70,71 Additionally, glycolysis generates substrates and intermediates fulfilling the biosynthetic demands needed for rapid cell proliferation, such as acetyl-CoA.72 Glucose metabolic alterations often lead to enhanced lactate excretion and low pH values in the microenvironment, which drives malignant progression and is related to poor prognosis in human cancer.73 Hence, glucose metabolic reprogramming is an optimized approach in which cancer cells deal with cellular stress. Glucose transporters (GLUTs), multiple enzymes, and several signaling pathways may be involved in the glucose metabolism.74 To date, it has been reported that numerous lncRNAs can affect genes and pathways forming complex regulatory networks of glucose metabolism regulation.70
Regulation of glucose uptake
Malignant cells are famous for the avid uptake of glucose to meet energy and substance demands. What drives cancer cells to internalize more glucose than normal cells? Normally, GLUTs and Na+-glucose-linked transporters (SGLTs) are two important transmembrane proteins that facilitate glucose uptake into the eukaryotic cytoplasm.75 Hox transcript antisense intergenic RNA (HOTAIR) is one of the most studied lncRNAs involved in genome modification.76 It is transcribed from the opposite direction of the HOXC gene and represses transcription from the HOXD gene by recruiting PRC2 in fibroblasts.77 Further study showed that HOTAIR promotes hepatocarcinogenesis partially through increasing GLUT1 expression by activating mammalian target of rapamycin (mTOR) signaling or by binding GLUT1 directly in hepatocellular carcinoma (HCC) cells.35 Analogously, CDKN2B antisense RNA1 (lncRNA ANRIL) also upregulated GLUT1 via activation of the mTOR signaling pathway, resulting in nasopharyngeal carcinoma (NPC) cell glucose metabolism reprogramming.36 In addition to the mTOR signaling pathway, SLC2A1-AS1 suppresses HCC metastasis and aerobic glycolysis in vivo by repressing GLUT1 expression by inhibiting the STAT3/FOXM1 pathway.37 Moreover, lncRNAs can antagonize miRNA regulation of GLUT1 expression. For instance, by interacting with the miR-378a-3p promoter and repressing its transcription, lnc-p23154 increases GLUT1 expression, thus increasing glucose uptake and participating in metastasis of oral squamous cell carcinoma (OSCC) cells in vitro and in vivo.38 In addition, LINC00174 promotes glioma carcinogenesis, which was verified by a nude mouse-transplanted tumor model and further facilitates glycolysis by regulating miR-152-3p, which leads to GLUT1 (SLC2A1) expression upregulation.39 LINC00346 regulates cell proliferation and glycolysis in breast cancer by interacting with miR-148a/b and increasing GLUT1 expression.78 These findings indicate that lncRNAs can affect glucose uptake in cancer cells by regulating GLUT1 expression through a wide range of mechanisms.
Regulation of glycolytic enzymes
In addition to regulating glucose uptake, lncRNAs have been identified to impact glycolysis by activating the transcription of key enzymes directly or indirectly. During the glycolysis process, four regulatory enzymes play an important role in the Warburg effect, namely, hexokinase (HK), glucokinase, pyruvate kinase (PK), and phosphofructokinase.79 lncRNA urothelial carcinoma-associated 1 (UCA1) plays an oncogenic role, and its abundance is associated with therapeutic resistance in several malignancies. In cervical cancer and pediatric acute myeloid leukemia (AML), lncRNA UCA1 promotes glycolysis and radioresistance (cervical cancer) or chemoresistance (pediatric AML) by upregulating HK2, which is a crucial determining enzyme in the first irreversible step of the glycolysis process.40,41,42 Moreover, several lncRNAs have been found to regulate HK2 expression via affecting miRNAs of target genes. For instance, PVT1 can influence miR-497 and miR-143 and thereby upregulate HK2 expression in osteosarcoma and gallbladder cancer,43,44 whereas UCA1 regulates miR-203 to increase HK2 expression in esophageal cancer.45 In addition, a subsequent study demonstrated that HK2 can be impacted by TUG1 in HCC by miR-455-3p binding46. Fructose-2,6-bisphosphatase (PFKFB2) and PKM2 are also key enzymes in the process of glycolysis and can also be regulated by lncRNAs. Rather than interacting with miRNAs, LINC00092 can directly interact with PFKFB2 to induce the glycolytic phenotype and metastasis in vivo cancer, which mediates the features of cancer-associated fibroblasts (CAFs) in ovarian cancer.47 Under normal conditions, PKM2 is mainly expressed in stem or embryonic cells, whereas PKM1 is expressed in most differentiated tissues. HOXB-AS3 (HOXB cluster antisense RNA 3) can encode a conserved 53-amino acid small peptide that can competitively bind to hnRNP A1 and act as a switch in the conversion of PKM1 to PKM2, thereby suppressing the formation of PKM2 and subsequent glucose metabolism reprogramming in colon cancer cells.48 AC020978 can directly interact with PKM2, enhance its stability, and promote the nuclear translocation of PKM2 in non-small cell lung cancer.49 In addition to the molecular mechanisms we mentioned above, lncRNAs can also influence these key enzymes in other ways, such as impeding catalytic activity.
Regulation of related genes or signaling pathways
Several other lncRNAs have been reported to regulate glucose metabolism and cancer progression through some cancer-related genes or signaling pathways. Notably, c-Myc is a critical transcription factor that is increased in numerous human cancers and regulates genes either directly or indirectly involved in glycolysis.80 Accumulating evidence has revealed that lncRNAs can regulate c-Myc expression or interact with c-Myc to affect the underlying pathway via multiple mechanisms. For example, glycolysis-associated lncRNA of colorectal cancer (GLCC1) binds to heat shock protein (HSP)90 (HSP90AA1) directly and further stabilizes c-Myc protein from ubiquitination degradation, thus increasing its target gene expression level, especially lactate dehydrogenase (LDHA), in colorectal cancer cells.50 In addition, FoxO-induced lncRNA 1 (FILNC1) is specifically expressed in the kidney and can repress c-Myc protein levels by sequestering AUF1 from interacting with c-Myc mRNA under glucose starvation conditions. Thus, low FILNC1 levels promote glycolysis and are associated with poor patient survival outcomes in renal cell carcinoma (RCC).51 Moreover, in multiple myeloma, lncRNA protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P) upregulates G6PD expression and pentose phosphate pathway flux by interacting with c-Myc directly and recruiting it to the promoter of G6P.52
Generally, it is widely accepted that the microenvironment of cancer cells has a shortage of glucose and oxygen supply owing to delayed tumor angiogenesis during the rapid growth of solid tumors.72,81 Interestingly, several lncRNAs have been found to be involved in the HIF-1 pathway that is activated by hypoxic stress and have gained widespread attention, including lincRNA-p21 and AC020978. lincRNA-p21, a direct transcriptional target of HIF-1, is able to disrupt the von Hippel-Lindau disease (vHL)-HIF-1α complex interaction, leading to a decrease in HIF-1α ubiquitination and accumulation of HIF-1α.53 As a consequence, a positive-feedback loop between lincRNA-p21 and HIF-1α is established and promotes glycolysis under conditions of hypoxia. Unlike lincRNA-p21, lncRNA-AC020978 can bind to PKM2 directly, thereby stabilizing PKM2 protein from ubiquitination degradation, and translocation to the nucleus, resulting in enhanced transcription of HIF-α.49 Collectively, these findings suggest that lncRNAs are vital players in Warburg effect regulation and highlight therapeutic targets for glucose metabolic reprogramming.
lncRNAs in glutamine metabolism
Glutamine, another kind of principal growth-supporting substrate, provides not only carbon but also a reduced nitrogen source required for the biosynthesis of various nitrogen-containing compounds.82 Once taken up into the cytoplasm, glutamine can be converted to glutamate by glutaminase (GLS) and then catalyzed into alpha-ketoglutarate (α-KG) via transaminases or glutamate dehydrogenase (GLUD/GDH). Since α-KG is an important intermediate product of the tricarboxylic acid (TCA) cycle, to sustain rapid proliferation, cancer cells demand α-KG.83,84 In addition, glutamine has also been reported to facilitate the import of essential amino acids.4 Hence, glutamine internalization and metabolism are critical for a spectrum of biological processes in cancer cells, including the maintenance of redox balance and ROS levels, energy production, and macromolecular synthesis85.
GLS is the rate-limiting enzyme in the glutamine metabolism and has different functions depending on the isoform.86 In mammals, GLS is encoded by two genes known as liver-type GLS (GLS2) and kidney-type GLS.87,88 Of these two GLS enzymes, GLS can be further classified into GLS kidney isoform (KGA) and GLS isoform C (GAC) due to alternative splicing.89 Interestingly, the GLS2 and two GLS isoforms also show differences in their regulation and activity by lncRNAs and other factors, such as colon cancer-associated transcript 2 (CCAT2). CCAT2 was originally identified in colorectal cancer, and its upregulation was related to high risks for multifarious malignancies.90,91 Notably, Redis et al.54 reported that CCAT2 regulates glutamine metabolism in an allele-specific manner. Through hyperpolarized magnetic resonance imaging, metabolic changes were also detected in nude mice after injection with CCAT2-overexpressing HCT116 cells. Mechanistic studies showed that the rs6983267 SNP (G/T) altered the secondary structure of CCAT2, resulting in a transcript with the G allele preferentially interacting with CFIm25 instead of CFIm68. Through the complex and allele-specific regulatory mechanism, CCAT2 affects the alternative splicing of GLS and contributes to the preferential expression of the more aggressive splice isoform.
In addition to CCAT2, research to date has revealed other lncRNAs involved in the glutamine metabolism by regulating GLS. As discussed previously, lincRNA-p21 is well known to be involved in the glucose metabolism; interestingly, lincRNA-p21 also contributes to the glutamine catabolism. Zhou et al.55 found that the decrease in lincRNA-p21 in bladder cancer enhances glutamine catabolism and accelerates the growth of cancer cells by upregulating GLS levels. Nonetheless, the accurate molecular regulatory mechanisms between lincRNA-p21 and GLS remain unknown. In contrast, by miRNA binding, HOTAIR can modulate GLS expression by interacting with miR-126-5p.56 In addition, AK123493.1, a nuclear-enriched antisense lncRNA of GLS (GLS-AS), can form double-stranded RNA with GLS premessenger RNA (premRNA) via adenosine deaminase RNA-specific (ADAR)/Dicer-dependent RNA interference (RNAi) in pancreatic cancer.57 Further investigation showed that deprivation of glutamine and glucose can induce the downregulation of GLS-AS at the transcriptional level by Myc, thereby reducing the interaction of GLS-AS and GLS premRNA and upregulating GLS expression. Remarkably, the stability and level of Myc protein can also be decreased by GLS-AS, which implies that there is a reciprocal feedback loop between GLS-AS and Myc when pancreatic cancer cells respond to nutrient stress.57
In addition, GLS2 is also pivotal for the glutamine metabolism reprogramming in cancer cells, especially in redox balance maintenance, and the elimination of excessive ROS levels.92 A recent study conducted by Li et al.58 revealed a positive relationship between the expression levels in UCA1 and GLS2 mRNA or protein in bladder cancer. Mechanistically, it was proposed that UCA1 binds with miR-16, promoting the expression of GLS2 to inhibit ROS formation and protect cells from oxidative toxicity in bladder cancer. In addition to UCA1, TUG1 could also interact with miR-145 and protect Sirt3 mRNA from degradation, allowing Sirt3 to positively regulate GDH levels and α-KG production in intrahepatic cholangiocarcinoma.59
Overall, the multiple lines of evidence favor the viewpoint that the glutamine metabolism is crucial for tumorigenesis. lncRNAs related to the metabolism of glutamine might be an attractive therapeutic target against cancer. Therefore, further studies are required to explore the potential therapeutic roles of lncRNAs, especially in specific in vivo models.
lncRNAs and lipid metabolism in cancer
In addition to the established role of lncRNAs in the glucose and glutamine metabolism, participation in the lipid metabolism is another essential role of lncRNAs in energy metabolism. In particular, lncRNAs have been proposed to be crucial regulators of fatty acids, adipogenesis, phospholipid metabolism, and transport.93 Multiple studies have verified that aberrant lncRNA expression can cause various metabolism-related diseases and disorders, including dyslipidemia, obesity, and atherosclerosis.94 Abnormal alterations in lipid metabolism have also been observed in cancerous tissue. The distinctive changes in lipid metabolism in cancer cells include de novo lipid synthesis, storing lipids, and converting cholesterol esters to free cholesterol. Herein, we focus on the molecular mechanisms mediated by lncRNAs leading to lipid abnormalities in human cancer.
Sterol regulatory element-binding proteins (SREBPs) are considered the most important regulatory factors in the processes of lipid homeostasis. There are three isoforms of SREBPs, named SREBP-1a, SREBP-1c, and SREBP-2. Among them, SREBP-1a and SREBP-1c mainly promote fatty acid synthesis, whereas SREBP-2 seems to be essential for de novo lipogenesis and cholesterol synthesis, and therefore, they might be potential drug targets for cancer treatment.95 Recently, Dong et al.96 revealed crucial roles for SREBP-1 in lipid desaturation through regulation of nuclear factor κB (NF-κB) signaling in clear cell RCC (ccRCC). Subsequently, research has shown that LINC01138 is located at chromosome 1q21.2 and correlated with poor ccRCC patient survival. In vitro assays showed that the ectopic expression of LINC01138 strongly increased the ratios of unsaturated/saturated lipids in ccRCC cells and promoted ccRCC cell proliferation. This effect was attributed to the increase in SREBP-1 protein stability by interacting with PRMT5.60 Moreover, SNHG16 has been proven to modulate the SREBP-2 expression level by directly targeting miR-195 in pancreatic cancer.61 Surprisingly, another study showed that SNHG16 is positively regulated by Wnt signaling and implicated in lipid metabolism of colorectal cancer depending on stearoyl coenzyme A (CoA) desaturas.62 With the use of AGO-cross-linking and immunoprecipitation (CLIP) analysis, Claus L. Andersen62 found that one-half of the unique miRNA families with high-confidence targets on SNHG16 also target the 3′ UTR of stearoyl CoA desaturase, which implies that SNHG16 alters lipid metabolism through diverse mechanisms in different malignant tumors.
Another lncRNA, a 500-nt lncRNA, involved in abnormal lipid metabolism is highly upregulated in liver cancer (HULC).97 A recent study showed that HULC also contributes to the accumulation of intracellular triglycerides and cholesterol in hepatoma cells.63 Acyl-CoA synthetase long-chain family members (ACSLs) are enzymes that catalyze the conversion of long-chain fatty acids to fatty acid-CoA in mammals.98 ACSL1 is a member of the family and is transactivated by peroxisome proliferator-activated receptor (PPAR)α. In hepatoma cells, HULC inhibits miR-9 transcription by eliciting the methylation of CpG islands in its promoter, resulting in the upregulation of PPARα and activation of ACSL1. Of particular note, the cholesterol product of ACSL1 promotes HULC expression, in turn, by activating the transcription factor retinoid X receptor A (RXRA). Similar to HULC, nuclear paraspeckle assembly transcript 1 (NEAT1) can also modulate abnormal lipolysis to drive HCC proliferation but through a different mechanism. By directly binding to miR-124-3p, NEAT1 regulates adipose triglyceride lipase (ATGL), diacylglycerol (DAG), and free fatty acid (FFA) levels, which finally activate the PPARα signaling pathway and induce HCC cell growth.64 Taken together, lipid metabolism is obviously supervised by complex lncRNA regulation networks, and studying these networks could allow us to develop novel strategies or therapies for human cancers.
The role of lncRNAs in cancer mitochondrial oxidative metabolism
Mitochondria are the metabolic factories and primary powerhouses in human cells. Mitochondria are responsible for multiple biological processes, such as oxidative phosphorylation and cytosolic biosynthetic precursor synthesis.99,100 Warburg originally hypothesized that mitochondrial function is impaired in tumor cells.72 However, subsequent evidence indicates that tumor cells possess functional mitochondria and can carry out oxidative phosphorylation.4 Notably, some tumor cells are more reliant on mitochondrial metabolism. Accordingly, the exploration of the roles that lncRNAs play in mitochondrial metabolism is pivotal for understanding cancer progression, particularly cancer cell metabolism.
As mentioned above, UCA1 not only is involved in glutamine and glucose metabolism but can also execute regulatory roles in mitochondria. In bladder cancer, UCA1 interacts with miR-195 and induces ADP-ribosylation factor-like 2 (ARL2) expression, thereby elevating mitochondrial function.65 Previous data showed that ARL2 is located on the mitochondrial membrane and functions as an activator of ATP/ADP transporters.101 Thus, studies on the roles and underlying mechanisms of UCA1 in metabolic rearrangement demonstrate its potential applications in novel antitumor therapies. In addition to UCA1, another lncRNA, designated survival-associated mitochondrial melanoma-specific oncogenic noncoding RNA (SAMMSON), that is located in the cytoplasm and mitochondria, prevents the exoribonuclease XRN2 from binding to the RNA-binding protein CARF by forming a complex containing the CARF and p32 proteins.66 This favors the nuclear localization of XRN2 and mitochondrial localization of p32. As a result, SAMMSON alters ribosomal RNA (rRNA) maturation and protein synthesis in the cytosol and mitochondria to promote cell growth. Given that SAMMSON is highly, selectively expressed in melanomas, these data identify SAMMSON as an attractive biomarker and therapeutic target of melanoma.67 In summary, lncRNAs function as essential regulators of mitochondrial metabolism and function through different mechanisms. Further studies are needed to identify other mitochondria-related lncRNAs and to study their roles in cancer mitochondrial oxidative metabolism.
The functions of lncRNAs in normal cell metabolism
Currently, intensive research efforts are underway to better understand the moderating effect of lncRNAs on cancer metabolism. Of note, recent studies report that lncRNAs are also involved in the control of normal cell metabolism, such as adipogenesis and adipose tissue differentiation. Sun et al.102 demonstrated 175 lncRNAs that are significantly and specifically abnormally expressed during adipogenesis in 2013. Among them, lncRAPs (lncRNAs regulated in adipogenesis) were enriched within adipose tissues, and the functional roles of lncRAPs were further explored through RNAi. Additionally, lncRNA steroid receptor RNA activator (SRA), a well-known lncRNA that fulfills its activation function (AF) through the AF-1 domain of nuclear receptors, is implicated in the differentiation of adipose tissues and regulation of insulin sensitivity.103 Thus, there is strong interest in the regulation of cell metabolism under physiological and pathological conditions.
Conclusions and prospects
Altered cellular metabolism is a well-established hallmark of cancer cells.4 Cumulative evidence shows that many factors are involved in this process. In this review, we have highlighted lncRNAs as an important class of regulators in cancer metabolism. The encouraging results from functional studies demonstrate the potential applications of lncRNAs in tumor diagnosis and therapies. However, compared with miRNA and protein-coding genes, our knowledge of lncRNAs in cancer metabolism is still limited. Notably, the majority of insights concerning the metabolic functions and regulatory mechanism of lncRNAs was inferred from in vitro studies. To convincingly assess the role of lncRNAs in the control of metabolism, further in vivo models through lncRNA knockdown or overexpression are needed. Fortunately, thanks to the success of oligo-based and RNAi-based drugs, some clinical trials with ncRNAs have begun.104,105 lncRNAs were inhibited via approaches such as antisense oligonucleotide (ASO) technology in vivo, and the stability of lncRNA-targeting ASOs can be increased by specific chemical modifications, such as locked nucleic acid (LNA).106 To date, some lncRNA-targeting drugs are currently undergoing preclinical studies. For example, lncRNA MALAT1 depletion by ASO can impact the growth and metastasis of breast and lung cancer cells in murine models.107,108 It is expected that lncRNA-based diagnostics and therapeutics involved in cancer metabolism will one day be beneficial for cancer patients, even though the clinical applications of lncRNAs are still at an early stage.
Acknowledgments
We apologize to all researchers whose relevant contributions were not cited due to space limitations. The work was supported by grants from the National Natural Science Foundation of China (no. 81802309 to Y.X.); Medical and Health Research Project of Zhejiang Province (no. 2019RC284 to Y.X.); Young Talents Project of Huzhou Central Hospital (nos. 2020YC10 to Y.X. and 2020YC05 to X.S.); and Project of Zhejiang Basic Public Benefit Research of Zhejiang Province (no. LGF18H160005 to G.Y.).
Author contributions
Y.X., M.Q., M. Shen, S.D., and G.Y. retrieved the related literature and drafted the manuscript. X.S. and M. Shu participated in the design of the review and drafted the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xuefei Shi, Email: shixuefei1223@aliyun.com.
Ming Sun, Email: summing348@hotmail.com.
References
- 1.Sun H., Huang Z., Sheng W., Xu M.D. Emerging roles of long non-coding RNAs in tumor metabolism. J. Hematol. Oncol. 2018;11:106. doi: 10.1186/s13045-018-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Wang J., Zhang X., Chen W., Hu X., Li J., Liu C. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int. J. Cancer. 2020;146:906–916. doi: 10.1002/ijc.32277. [DOI] [PubMed] [Google Scholar]
- 4.Pavlova N.N., Thompson C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 6.Yeung S.J., Pan J., Lee M.H. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spizzo R., Almeida M.I., Colombatti A., Calin G.A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuck A.C., Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Clark M.B., Johnston R.L., Inostroza-Ponta M., Fox A.H., Fortini E., Moscato P., Dinger M.E., Mattick J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dijk E.L., Chen C.L., d’Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., Bertrand C., Silvain M., Legoix-Né P., Loeillet S. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 14.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlevaro-Fita J., Rahim A., Guigó R., Vardy L.A., Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melé M., Mattioli K., Mallard W., Shechner D.M., Gerhardinger C., Rinn J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S., Zhou L., Wang Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z., Xue S., Zhang M., Xu H., Hu X., Chen S., Liu Y., Guo M., Cui H. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906–6919. doi: 10.1038/s41388-020-01475-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 20.Hu Q., Egranov S.D., Lin C., Yang L. Long noncoding RNA loss in immune suppression in cancer. Pharmacol. Ther. 2020;213:107591. doi: 10.1016/j.pharmthera.2020.107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Z., Zhou J.K., Peng Y., He W., Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer. 2020;19:77. doi: 10.1186/s12943-020-01188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broderick J.A., Zamore P.D. Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol. Cell. 2014;54:711–713. doi: 10.1016/j.molcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Han J., LaVigne C.A., Jones B.T., Zhang H., Gillett F., Mendell J.T. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. 2020;370:eabc9546. doi: 10.1126/science.abc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheu-Gruttadauria J., Pawlica P., Klum S.M., Wang S., Yario T.A., Schirle Oakdale N.T., Steitz J.A., MacRae I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell. 2019;75:1243–1255.e7. doi: 10.1016/j.molcel.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi C.Y., Kingston E.R., Kleaveland B., Lin D.H., Stubna M.W., Bartel D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science. 2020;370:eabc9359. doi: 10.1126/science.abc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang A., Shao T.J., Bofill-De Ros X., Lian C., Villanueva P., Dai L., Gu S. AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2. Nat. Commun. 2020;11:2765. doi: 10.1038/s41467-020-16533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 32.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 34.Liu H., Luo J., Luan S., He C., Li Z. Long non-coding RNAs involved in cancer metabolic reprogramming. Cell. Mol. Life Sci. 2019;76:495–504. doi: 10.1007/s00018-018-2946-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei S., Fan Q., Yang L., Zhang X., Ma Y., Zong Z., Hua X., Su D., Sun H., Li H., Liu Z. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol. Rep. 2017;38:1902–1908. doi: 10.3892/or.2017.5840. [DOI] [PubMed] [Google Scholar]
- 36.Zou Z.W., Ma C., Medoro L., Chen L., Wang B., Gupta R., Liu T., Yang X.Z., Chen T.T., Wang R.Z. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7:61741–61754. doi: 10.18632/oncotarget.11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang R., Wang M., Dai B., Du J., Wang J., Liu Z., Qu S., Yang X., Liu J., Xia C. Long noncoding RNA SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol. Oncol. 2020;14:1381–1396. doi: 10.1002/1878-0261.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Zhang X., Wang Z., Hu Q., Wu J., Li Y., Ren X., Wu T., Tao X., Chen X. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi: 10.1016/j.canlet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Shi J., Zhang Y., Qin B., Wang Y., Zhu X. Long non-coding RNA LINC00174 promotes glycolysis and tumor progression by regulating miR-152-3p/SLC2A1 axis in glioma. J. Exp. Clin. Cancer Res. 2019;38:395. doi: 10.1186/s13046-019-1390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan L., Huang C., Li J., Gao T., Lin Z., Yao T. Long non-coding RNA urothelial cancer associated 1 regulates radioresistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int. J. Mol. Med. 2018;42:2247–2259. doi: 10.3892/ijmm.2018.3778. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Liu Y., Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J. Cell. Biochem. 2018;119:6296–6308. doi: 10.1002/jcb.26899. [DOI] [PubMed] [Google Scholar]
- 42.Robey R.B., Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Yu Y., Li H., Hu Q., Chen X., He Y., Xue C., Ren F., Ren Z., Li J. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer. 2019;18:33. doi: 10.1186/s12943-019-0947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song J., Wu X., Liu F., Li M., Sun Y., Wang Y., Wang C., Zhu K., Jia X., Wang B., Ma X. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 2017;490:217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Liu H.E., Shi H.H., Luo X.J. Upregulated Long Noncoding RNA UCA1 Enhances Warburg Effect via miR-203/HK2 Axis in Esophagal Cancer. J. Oncol. 2020;2020:8847687. doi: 10.1155/2020/8847687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y.H., Wu M.H., Huang Y.H., Yeh C.T., Cheng M.L., Chi H.C., Tsai C.Y., Chung I.H., Chen C.Y., Lin K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. 2018;67:188–203. doi: 10.1002/hep.29462. [DOI] [PubMed] [Google Scholar]
- 47.Zhao L., Ji G., Le X., Wang C., Xu L., Feng M., Zhang Y., Yang H., Xuan Y., Yang Y. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]
- 48.Huang J.-Z., Chen M., Chen D., Gao X.-C., Zhu S., Huang H., Hu M., Zhu H., Yan G.-R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 49.Hua Q., Mi B., Xu F., Wen J., Zhao L., Liu J., Huang G. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics. 2020;10:4762–4778. doi: 10.7150/thno.43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang J., Yan T., Bao Y., Shen C., Yu C., Zhu X., Tian X., Guo F., Liang Q., Liu Q. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019;10:3499. doi: 10.1038/s41467-019-11447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao Z.D., Han L., Lee H., Zhuang L., Zhang Y., Baddour J., Nagrath D., Wood C.G., Gu J., Wu X. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 2017;8:783. doi: 10.1038/s41467-017-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X., Ye H., He M., Zhou X., Sun N., Guo W., Lin X., Huang H., Lin Y., Yao R., Wang H. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem. Biophys. Res. Commun. 2018;498:207–213. doi: 10.1016/j.bbrc.2018.02.211. [DOI] [PubMed] [Google Scholar]
- 53.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Redis R.S., Vela L.E., Lu W., Ferreira de Oliveira J., Ivan C., Rodriguez-Aguayo C., Adamoski D., Pasculli B., Taguchi A., Chen Y. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell. 2016;61:640. doi: 10.1016/j.molcel.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Q., Zhan H., Lin F., Liu Y., Yang K., Gao Q., Ding M., Liu Y., Huang W., Cai Z. LincRNA-p21 suppresses glutamine catabolism and bladder cancer cell growth through inhibiting glutaminase expression. Biosci. Rep. 2019;39 doi: 10.1042/BSR20182372. BSR20182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L., Cui S., Wan T., Li X., Tian W., Zhang R., Luo L., Shi Y. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J. Cell. Physiol. 2018;233:6822–6831. doi: 10.1002/jcp.26432. [DOI] [PubMed] [Google Scholar]
- 57.Deng S.J., Chen H.Y., Zeng Z., Deng S., Zhu S., Ye Z., He C., Liu M.L., Huang K., Zhong J.X. Nutrient Stress-Dysregulated Antisense lncRNA GLS-AS Impairs GLS-Mediated Metabolism and Represses Pancreatic Cancer Progression. Cancer Res. 2019;79:1398–1412. doi: 10.1158/0008-5472.CAN-18-0419. [DOI] [PubMed] [Google Scholar]
- 58.Li H.J., Li X., Pang H., Pan J.J., Xie X.J., Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 2015;45:1055–1063. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]
- 59.Zeng B., Ye H., Chen J., Cheng D., Cai C., Chen G., Chen X., Xin H., Tang C., Zeng J. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8:113650–113661. doi: 10.18632/oncotarget.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X., Wu J., Wu C., Chen W., Lin R., Zhou Y., Huang X. The LINC01138 interacts with PRMT5 to promote SREBP1-mediated lipid desaturation and cell growth in clear cell renal cell carcinoma. Biochem. Biophys. Res. Commun. 2018;507:337–342. doi: 10.1016/j.bbrc.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 61.Yu Y., Dong J.T., He B., Zou Y.F., Li X.S., Xi C.H., Yu Y. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15:3831–3844. doi: 10.2217/fon-2019-0321. [DOI] [PubMed] [Google Scholar]
- 62.Christensen L.L., True K., Hamilton M.P., Nielsen M.M., Damas N.D., Damgaard C.K., Ongen H., Dermitzakis E., Bramsen J.B., Pedersen J.S. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol. Oncol. 2016;10:1266–1282. doi: 10.1016/j.molonc.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui M., Xiao Z., Wang Y., Zheng M., Song T., Cai X., Sun B., Ye L., Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 64.Liu X., Liang Y., Song R., Yang G., Han J., Lan Y., Pan S., Zhu M., Liu Y., Wang Y. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer. 2018;17:90. doi: 10.1186/s12943-018-0838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H.J., Sun X.M., Li Z.K., Yin Q.W., Pang H., Pan J.J., Li X., Chen W. LncRNA UCA1 Promotes Mitochondrial Function of Bladder Cancer via the MiR-195/ARL2 Signaling Pathway. Cell. Physiol. Biochem. 2017;43:2548–2561. doi: 10.1159/000484507. [DOI] [PubMed] [Google Scholar]
- 66.Vendramin R., Verheyden Y., Ishikawa H., Goedert L., Nicolas E., Saraf K., Armaos A., Delli Ponti R., Izumikawa K., Mestdagh P. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018;25:1035–1046. doi: 10.1038/s41594-018-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leucci E., Vendramin R., Spinazzi M., Laurette P., Fiers M., Wouters J., Radaelli E., Eyckerman S., Leonelli C., Vanderheyden K. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 68.Warburg O. The Chemical Constitution of Respiration Ferment. Science. 1928;68:437–443. doi: 10.1126/science.68.1767.437. [DOI] [PubMed] [Google Scholar]
- 69.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 70.Fan C., Tang Y., Wang J., Xiong F., Guo C., Wang Y., Zhang S., Gong Z., Wei F., Yang L. Role of long non-coding RNAs in glucose metabolism in cancer. Mol. Cancer. 2017;16:130. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Wu Y., Wang Y., Fu A., Gong L., Li W., Li Y. Bacillus amyloliquefaciens SC06 alleviates the oxidative stress of IPEC-1 via modulating Nrf2/Keap1 signaling pathway and decreasing ROS production. Appl. Microbiol. Biotechnol. 2017;101:3015–3026. doi: 10.1007/s00253-016-8032-4. [DOI] [PubMed] [Google Scholar]
- 72.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helmlinger G., Sckell A., Dellian M., Forbes N.S., Jain R.K. Acid production in glycolysis-impaired tumors provides new insights into tumor metabolism. Clin. Cancer Res. 2002;8:1284–1291. [PubMed] [Google Scholar]
- 74.Li Z., Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balon T.W. SGLT and GLUT: are they teammates? Focus on “Mouse SGLT3a generates proton-activated currents but does not transport sugar”. Am. J. Physiol. Cell Physiol. 2012;302:C1071–C1072. doi: 10.1152/ajpcell.00054.2012. [DOI] [PubMed] [Google Scholar]
- 76.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 77.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Li H., Wang W., Yu X., Xu Q. LINC00346 regulates glycolysis by modulation of glucose transporter 1 in breast cancer cells. Mol. Cell. Probes. 2020;54:101667. doi: 10.1016/j.mcp.2020.101667. [DOI] [PubMed] [Google Scholar]
- 79.Akram M. Mini-review on glycolysis and cancer. J. Cancer Educ. 2013;28:454–457. doi: 10.1007/s13187-013-0486-9. [DOI] [PubMed] [Google Scholar]
- 80.Dang C.V., Le A., Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subarsky P., Hill R.P. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis. 2003;20:237–250. doi: 10.1023/a:1022939318102. [DOI] [PubMed] [Google Scholar]
- 82.DeBerardinis R.J., Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 84.Jin L., Li D., Alesi G.N., Fan J., Kang H.B., Lu Z., Boggon T.J., Jin P., Yi H., Wright E.R. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang L., Venneti S., Nagrath D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 86.Ortiz-Pedraza Y., Muñoz-Bello J.O., Olmedo-Nieva L., Contreras-Paredes A., Martínez-Ramírez I., Langley E., Lizano M. Non-Coding RNAs as Key Regulators of Glutaminolysis in Cancer. Int. J. Mol. Sci. 2020;21:2872. doi: 10.3390/ijms21082872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Curthoys N.P., Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 88.Krebs H.A. Metabolism of amino-acids: The synthesis of glutamine from glutamic acid and ammonia, and the enzymic hydrolysis of glutamine in animal tissues. Biochem. J. 1935;29:1951–1969. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ling H., Spizzo R., Atlasi Y., Nicoloso M., Shimizu M., Redis R.S., Nishida N., Gafà R., Song J., Guo Z. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xin Y., Li Z., Zheng H., Chan M.T.V., Ka Kei Wu W. CCAT2: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:e12342. doi: 10.1111/cpr.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.D’Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 93.Zeng Y., Ren K., Zhu X., Zheng Z., Yi G. Long Noncoding RNAs: Advances in Lipid Metabolism. Adv. Clin. Chem. 2018;87:1–36. doi: 10.1016/bs.acc.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Mazidi M., Penson P., Gluba-Brzozka A., Rysz J., Banach M. Relationship between long noncoding RNAs and physiological risk factors of cardiovascular disease. J. Clin. Lipidol. 2017;11:617–623. doi: 10.1016/j.jacl.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 95.Rawson R.B. Control of lipid metabolism by regulated intramembrane proteolysis of sterol regulatory element binding proteins (SREBPs) Biochem. Soc. Symp. 2003:221–231. doi: 10.1042/bss0700221. [DOI] [PubMed] [Google Scholar]
- 96.Yang H., Zhang X., Liu F., Fan J., Wang B., Dong C. SREBP1-driven lipid desaturation supports clear cell renal cell carcinoma growth through regulation of NF-κB signaling. Biochem. Biophys. Res. Commun. 2018;495:1383–1388. doi: 10.1016/j.bbrc.2017.11.163. [DOI] [PubMed] [Google Scholar]
- 97.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mashek D.G., Bornfeldt K.E., Coleman R.A., Berger J., Bernlohr D.A., Black P., DiRusso C.C., Farber S.A., Guo W., Hashimoto N. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J. Lipid Res. 2004;45:1958–1961. doi: 10.1194/jlr.E400002-JLR200. [DOI] [PubMed] [Google Scholar]
- 99.Mishra P., Chan D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wallace D.C. Mitochondria and cancer. Nat. Rev. Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishi H., Ono K., Iwanaga Y., Horie T., Nagao K., Takemura G., Kinoshita M., Kuwabara Y., Mori R.T., Hasegawa K. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J. Biol. Chem. 2010;285:4920–4930. doi: 10.1074/jbc.M109.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D.R., Hendrickson D.G. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu B., Gerin I., Miao H., Vu-Phan D., Johnson C.N., Xu R., Chen X.W., Cawthorn W.P., MacDougald O.A., Koenig R.J. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Levin A.A. Treating Disease at the RNA Level with Oligonucleotides. N. Engl. J. Med. 2019;380:57–70. doi: 10.1056/NEJMra1705346. [DOI] [PubMed] [Google Scholar]
- 105.Slack F.J., Chinnaiyan A.M. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arun G., Diermeier S.D., Spector D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]