Abstract
Rapid and efficient biological sample preparation and pretreatment are crucial for highly sensitive, reliable and reproducible molecular detection of infectious diseases. Herein, we report a self-powered, integrated sample concentrator (SPISC) for rapid plasma separation, pathogen lysis, nucleic acid trapping and enrichment at the point of care. The proposed sample concentrator uses a combination of gravitational sedimentation of blood cells and capillary force for rapid, self-powered plasma separation. The pathogens (e.g., HIV virus) in separated plasma were directly lysed and pathogen nucleic acid was enriched by an integrated, flow-through FTA® membrane in the concentrator, enabling highly efficient nucleic acid preparation. The FTA® membrane of the SPISC is easy to store and transport at room temperature without need for uninterrupted cold chain, which is crucial for point of care sampling in resource-limited settings. The platform has been successfully applied to detect HIV virus in blood samples. Our experiments show that the sample concentrator can achieve a plasma separation efficiency as high as 95% and a detection sensitivity as low as 10 copies per 200 μL blood (~100 copies/mL plasma) with variability less than 7%. The sample concentrator described is fully compatible with downstream nucleic acid detection and has great potential for early diagnostics, monitoring and management of infectious diseases at the point of care.
INTRODUCTION
Recent advances in molecular detection technologies have offered great promise in rapid and accurate diagnostics and treatment of human diseases, such as infectious diseases and cancer. One of the emerging areas of such molecular detection is early diagnostics of human immunodeficiency virus (HIV), timely monitoring and management of antiretroviral therapy (ART). HIV/ Acquired immunodeficiency syndrome (AIDS) has become a global health threat and public economic burden. Early diagnosis followed by antiretroviral therapy along with continuous viral load monitoring post treatment is crucial to guide clinicians for appropriate treatment to allow better survival rates and control disease progression.1,2 As of 2019, about 38 million people have HIV infection with more than 95% of them in developing countries and 25.4 million having access to ART.3 Standard HIV viral load testing for ART monitoring is currently challenging in low- and middle-income countries because of lack of funds, need for expensive equipment, cold chain, testing facilities, and trained personnel.4 Therefore, there is an unmet need to develop a simple, low-cost, point of care, molecular diagnostic technology for HIV viral load testing in resource-constrained settings.
A rapid and efficient blood sample preparation method is one of the crucial steps for highly sensitive molecular detection of HIV virus in blood at the point of care. Due to variations in transportation and local environmental conditions in resource-constrained settings, on-site blood sampling and preparation have been widely used for rapid HIV diagnostic tests.5,6,7 For example, dried blood spots (DBS) sampling uses spotting of whole blood onto paper substrates like Whatman® FTA® (Flinders Technology Associates filter paper cards)8 or Whatman® 903 protein saver cards,9,10 for blood sampling and transportation, which is an attractive alternative to conventional blood sampling due to its simplicity, convenience and low-cost. However, the DBS sampling for HIV viral load testing has important limitations: i) it can result in higher HIV viral load measurement than plasma-based detection because cell-associated proviral DNA and intracellular RNA can be detected, which may misclassify HIV patients during antiretroviral therapy,11 and ii) it suffers from the blood hematocrit effect (or hematocrit bias), variability of hematocrit values.12 Variable hematocrit values are considered a critical drawback since it influences the sampling spot size and hence is not ideal for analyte quantitation.13
Recently, various plasma separation devices have been developed for rapid plasma separation at the point of care, such as membrane-based plasma separation,14,15,16 microfluidics based cross flow filtration,17,18 and continuous flow plasma separation.19,20 However, all of these devices require manual operation and a cold chain to conserve/ transport the liquid plasma for downstream molecular detection in a centralized laboratory. To this end, several paper-based plasma storage devices have been developed for plasma sample storage and transportation, such as dried plasma spots (DPS),21,22 Noviplex™ Plasma Prep Cards23 and HemaSpot-SE.24 However, these devices are standard prep cards and does not have capability to allow pre-concentration from higher volumes of blood to enrich pathogen nucleic acids, which is necessary for highly sensitive molecular diagnostics to meet stringent limit-of-detection specifications.
In this study, we develop a simple, self-powered integrated sample concentrator (SPISC) (Figure 1) that integrates cell-free plasma separation, pathogen lysis, nucleic acid capturing and enrichment, enabling point of care sampling and nucleic acid preparation. The device uses a combination of capillary force and cell gravitational sedimentation to rapidly separate plasma from blood cells, enabling self-powered sampling without manual interference. Self-powered platforms relieve the necessity of peripheral pumps and power source to operate crucial for applications addressed for remote settings. Incorporating capillary force to process samples allows for less complex platforms and have previously attracted many researchers integrating such technologies into novel point-of-care diagnostics.25,26 The flow-through FTA® membrane integrated in the device can lyse pathogens (e.g., HIV virus), trap and enrich nucleic acids in the paper matrix from separated plasma with volumes ranging from few microliters to milliliters, enabling highly efficient nucleic acid enrichment and highly sensitive molecular detection. The developed platform was evaluated based on detection of HIV virus in blood samples. The integrated sample concentrator described herein can be used as a stand-alone sample preparation module for plasma separation, nucleic acid enrichment and transportation. Accordingly, the platform is suitable for onsite blood sampling at home, in the clinic, as well as in resource-limited settings, allowing downstream analysis either onsite or by an external centralized laboratory through shipment (Figure 1).
Figure 1.
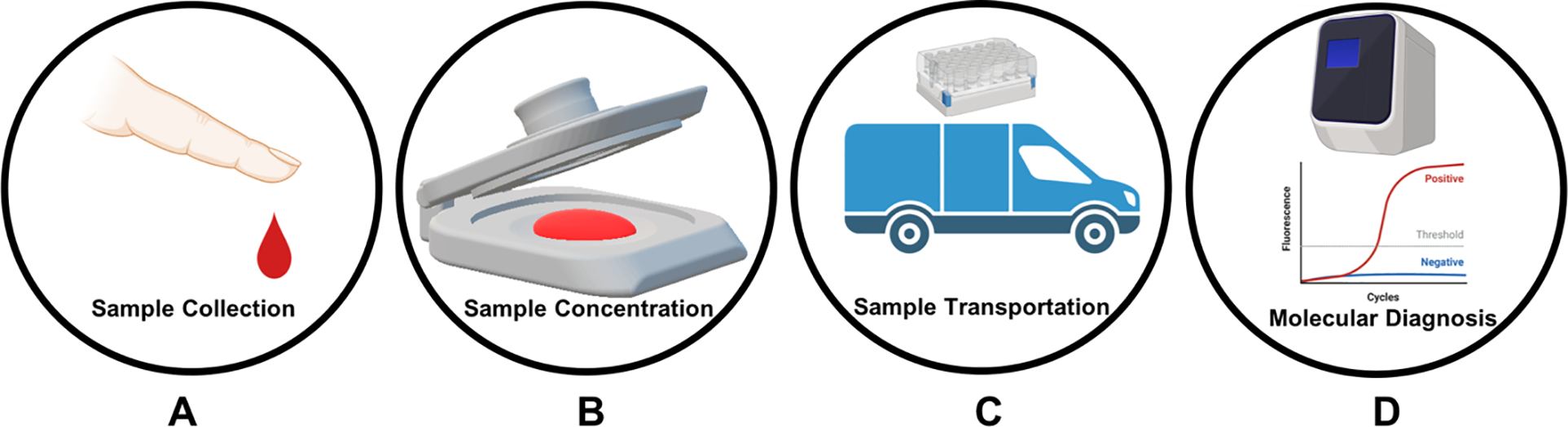
Schematic illustration of the workflow of point of sampling with our self-powered, integrated sample concentrator (SPISC) and testing. (A) Blood sample collection with a sterilized finger prick. (B) Plasma separation, pathogen lysis, and nucleic acid enrichment on the SPISC platform. (C) Drying, archiving (long term storage), point of care testing or transportation to centralized lab (if needed). (D) Analysis of isolated nucleic acids/analytes on centralized laboratories for accurate diagnosis
EXPERIMENTAL SECTION
Reagents, Primers & Targets: Whatman® FTA® micro card membrane, Whatman’s FTA purification reagents were purchased from Sigma Aldrich, Low EDTA TE buffer pH 8.0 was purchased from Thermo Fisher Scientific, Proteinase K from Qiagen, RT-PCR grade water from Ambion, RT-PCR amplification reagents from QuantiNova SYBR Green RT-PCR kit, Qiagen. RT-PCR for HIV-1 RNA amplification used previously published27 forward primer: 5’-TTTGGAAAGGACCAGCAAA-3’, reverse primer: 5’- CCTGCCATCTGTTTTCCA-3’ from IDT. Inactivated HIV-1 positive plasma samples for analytical performance testing were from AcroMetrix™. Whole blood was obtained commercially from Innovative Research from single donor and for control experiments from multiple donors. All experiments were performed in accordance with the Guidelines of University of Pennsylvania, and approved by the Institutional Review Board at University of Pennsylvania (IRB protocol #: 829539).
Fabrication of the integrated sample concentrator: Individual parts of the described sample concentrator were 3D printed using a high definition, low-cost Stereolithographic (SLA) laser-based Form 2 3D printer (Form labs) with Form labs methacrylate based clear resin (FLGPCL02). All the prototype designs as computer aided design (CAD) files were created using Autodesk 123D design and transformed to printable (.stl) format to be uploaded for printing at 50 μm print resolution. Average time to print 6 functional platforms is ~ 3 hours and design of the prototyped platform is suitable for mass production using a large-scale commercial 3D printer or injection molding. The printed parts are submerged in isopropanol (IPA) and subjected to ultra-sonication for 10 min post printing. The printed parts are washed and dried at 60°C for 30 mins prior to use. To improve the transparency of printed parts, a top coat of acrylic spray (Rust-Oleum Clear Gloss Enamel spray, 257884) was applied. The substrate surfaces that are exposed to whole blood are sprayed with Rust-Oleum® NeverWet® coating to form a superhydrophobic coating that enhances the plasma separation by forming a high contact angle of whole blood, minimizing the biomolecular adhesion, and reducing the hemolysis.16
RT-PCR assay: For downstream processing in diagnostics, the FTA® membrane was transferred to PCR amplification vials or 96 well PCR plates for high throughput analysis. 300 μL of Whatman® FTA® purification reagent and 20 μL of Proteinase K was added to the FTA® membranes, heated to ~55°C for 3 mins followed washing 1X with purification reagent further to remove trace amounts of PCR inhibitors, cell debris and weakly bound non-specific plasma protein. Finally, the FTA® membranes were washed 2X with low EDTA TE buffer pH 8.0 and one-step PCR amplification reaction buffer was added to amplify nucleic acid. 35 μL of one-step RT-PCR amplification reaction buffer was pre-pared from QuantiNova SYBR® Green one-step RT-PCR kit. Reaction mix composition contained 20 μL of 2X QuantiNova SYBR Green RT-PCR master mix, 0.4 μL of 100X QuantiNova SYBR Green RT mix, 1 μL of forward primer, 1 μL of reverse primer and 17.6 μL of RNase-Free water. Real-time cycler conditions for rapid amplification of pathogen nucleic acid include initial reverse-transcription stage for 10 min at 50°C via a HotStaRTScript Reverse Transcriptase, 2 min PCR initial activation step to activate QuantiNova DNA polymerase at 95°C and a two-step cycling of 5 sec denaturation phases at 95°C, combined annealing/extension phase for 25 sec at 60°C for 100 cycles.
RESULTS AND DISCUSSION
Self-Powered Integrated Sample Concentrator
Figure 2A presents an exploded schematic representation of our integrated blood sample concentrator. The sample concentrator consists of: i) one plasma absorption module filled with cotton adsorbent (Figure 2B), ii) one Whatman® FTA® membrane disc holder containing one FTA® membrane for pathogen lysis, nucleic acid capture and enrichment (Figure 2B), and iii) one plasma separation module with two hinged superhydrophobic substrates (Figure 2C and D). The absorption module has one adsorbent chamber (6.6 mm in diameter and 5.5 mm in depth) to pack the adsorbent material and two projections (1 mm in diameter and 0.8 mm in height) to fit the plasma separation module by simple twist & lock (Figure D-G). The absorption module and FTA® disc holder is connected through a patterned adhesive tape prior to locking into the plasma separation module via the twist-lock design (Figure 2E). In the plasma separation module, its top superhydrophobic substrate (36.6 (L) × 30.0 (W) × 5.0 (H) mm) equipped with a Vivid™ plasma separation mem-brane (Figure 2C) has a lock and twist port to lock the plasma absorption module/ FTA® disc. The FTA® disc is 9.0 mm in diameter and 1.6 mm in height which acts as casing to hold the flow-through FTA® membrane. To lyse pathogens and enrich their nucleic acids from separated plasma, the disc holder with the flow-through FTA® membrane was sandwiched between the plasma separation membrane and the absorption module. To reduce non-specific absorption and improve plasma yield, the surface of plasma separation module was coated with superhydrophobic layer using the NeverWet® spray kit. All individual modules of our sample concentrator were fabricated by simple 3D printing technology, allowing us to rapidly develop low-cost point of care diagnostic devices. Since the maximum volume of finger or heel-prick blood is 250–500 μL,28 our concentrator is designed to work with a blood volume of ~200 μL. If needed, our SPISC platform can be customized for higher sample volume for nucleic acid enrichment to detect lower abundant nucleic acids by adjusting adsorbent volume and FTA® membrane size.
Figure 2.
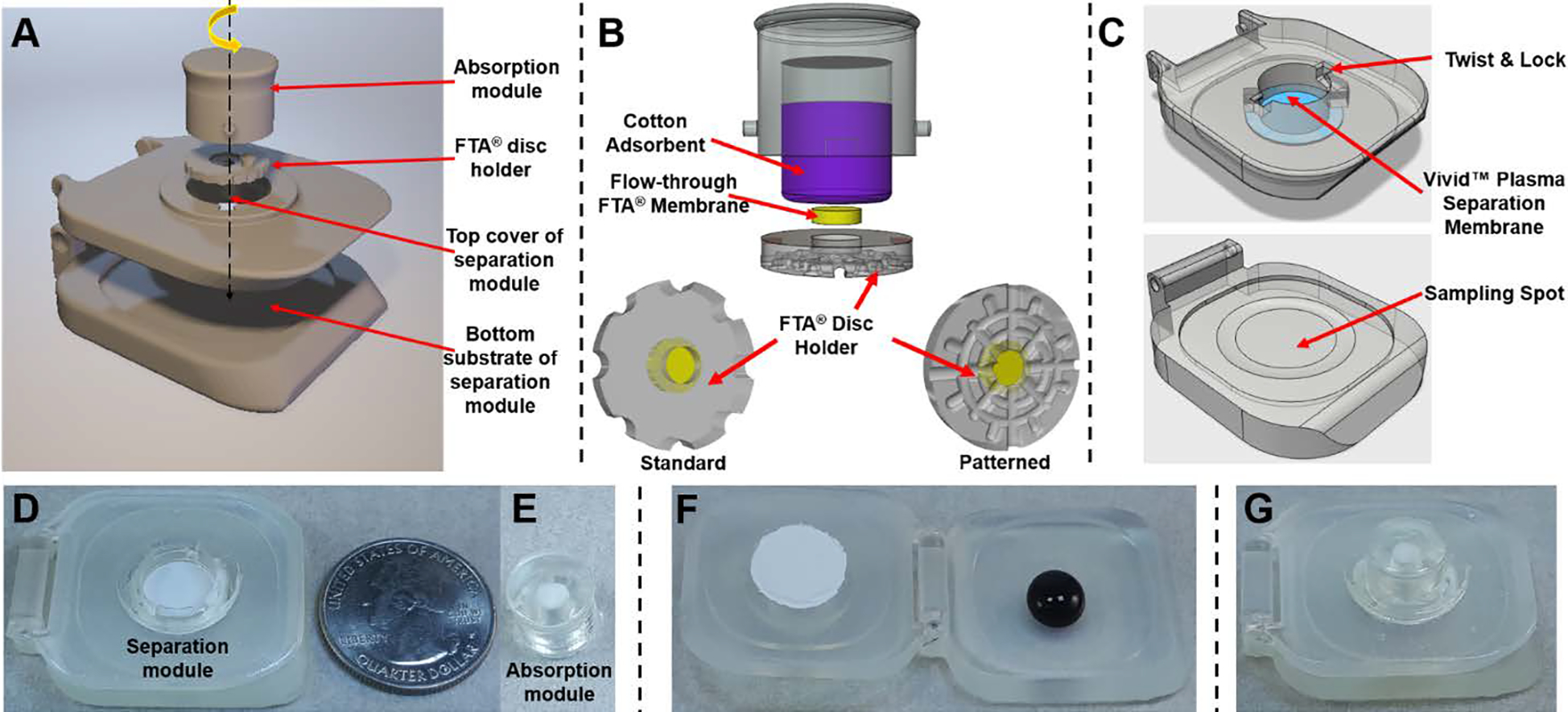
A self-powered integrated sample concentrator (SPISC). (A) Exploded view schematic representation of components of the blood sample concentrator with axis arrow showing position of each component, turn arrow on the top in yellow represents easy insertion and removing of the absorption module /FTA® disc holder via twist lock mechanism. (B) Representation of the plasma adsorbent chamber with cotton adsorbent and FTA® disc holder. (C) 3D CAD models of top cover and bottom sampling spots of sample concentrator along with representation of position of Vivid™ plasma separation membrane and twist-lock design. (D&E) Photographs of the plasma separation module with separation membrane and absorption module/FTA® disc holder. (F) Add whole blood on the bottom substrate of the plasma separation module. (G) Self-powered plasma separation, virus lysis and nucleic acid capturing on our sample concentrator.
To enable self-powered plasma separation and nucleic acid enrichment, whole blood sample was first loaded into the blood sampling spot of the bottom superhydrophobic substrate (Figure 2F). Next, the top substrate with plasma separation membrane was closed and blood sample was sandwiched between two superhydrophobic substrates, which allowed for gravitational sedimentation of the blood cells away from the top-positioned plasma separation membrane and increased the membrane separation capacity. Simultaneously, the absorption module/FTA® disc inserted into the top substrate via the twist & locking mechanism-initiated plasma separation by pulling plasma through the plasma separation membrane by capillary force. Since the Whatman® FTA® membrane possess chemicals to lyse the pathogen membranes and denature proteins, the pathogens were lysed and the released nucleic acid was captured and enriched by the cellulose fiber matrix of the flow-through FTA® membrane. Next, the FTA® membrane was allowed to sit inside the sample concentrator for ~5 min to enable efficient lysis of virus and trapping of viral nucleic acids. The entrapped nucleic acids were then ready for either onsite molecular detection or storage/transportation to a centralized lab at room temperature. Video S1 in ESI illustrates the operation of the self-powered 3D printed sample concentrator.
Next, the surface features of FTA® disc holder were optimized to enable efficient plasma separation. As shown in Figure 2B, bottom left, the FTA® disc holder equipped with the FTA® membrane with planar surface resulted in 67% separation efficiency with variability of 10% between similar separations. 55% of whole blood is comprised of plasma. The separation efficiency (percentage recovery) was calculated at 55% plasma recovery29 on the plasma separator from 200 μL whole blood. This lower efficiency is a result of loss of plasma that is trapped at the junction of Vivid™ plasma separation membrane and FTA® disc holder. To minimize the plasma trapping and improve efficiency, we designed the FTA® disc with a patterned surface that simulated fine capillaries/channels pointing towards flow path of FTA® membrane and adsorbent (Figure 2B, bottom right), thus minimizing plasma loss. The capillary force driven SPISC platform showed reproducible extraction of about 103 ± 4.4 μL within 5 min, which is ~1.6 times better separation efficiency than that of our previous generation.25 Therefore, our blood sample concentrator can achieve about 94% separation efficiency at variability of 4% within 5 min of 200 μL of fresh whole blood sampling.
Nucleic Acid Enrichment on the Integrated Flow-Through FTA® Membrane
To demonstrate the nucleic acid enrichment ability of the flow-through FTA® membrane in our sample concentrator, we first determined enrichment efficiency of HIV-1 RNA in a PBS solution. HIV viral RNA was extracted from inactivated HIV-1 positive plasma samples (AcroMetrix™, Thermo Fisher Scientific). The virus particles were lysed and purified using Qiagen QIAamp® Viral RNA mini extraction kit. Purified HIV-1 RNA was then spiked into PBS and added to the sampling spot of the sample concentrator, and the platform was closed to initiate automated nucleic acid enrichment. Viral RNA extracted by FTA® membranes was then added to PCR amplification vials and amplified using a one-step RT-PCR reaction mix. Figure 3A, demonstrates real-time amplification curves from RNA spiked samples ranging from 10–5000 copies/sample. Real-time amplification curves generated by plotting fluorescent intensity vs amplification cycle, indicate an increase in fluorescent intensity with time that eventually saturates. The higher the target HIV RNA concentration was, the earlier the intensity curve increased above the baseline. Time taken to reach the 50% saturation value was represented as cycle threshold (Ct), which was used to quantify the sample (Figure 3B). Reproducibility and reliability were established as variability between three independent experiments. No signal was seen in the absence of the target RNA. Relative standard deviations (RSD’s) ranging from 1.5 to 7.0 % demonstrated excellent reproducibility in nucleic acid enrichment.
Figure 3.
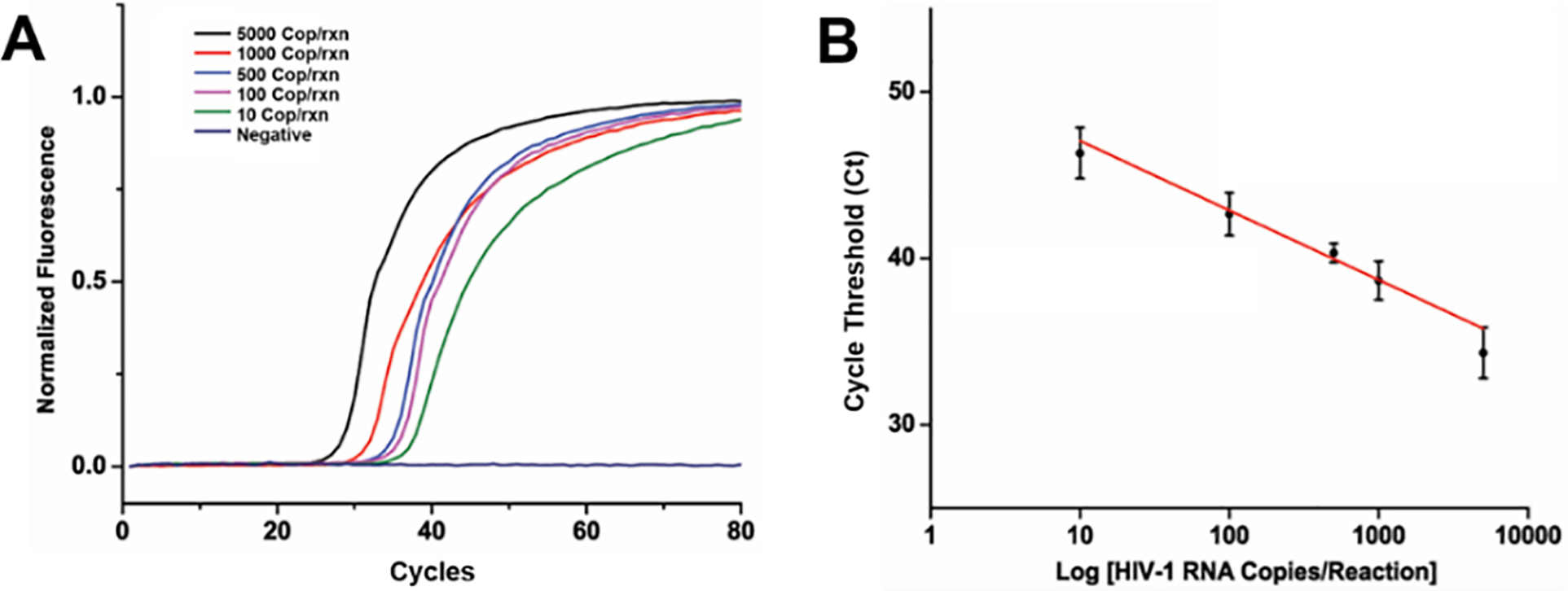
Nucleic acid extraction and enrichment of the flow-through FTA® membrane in the self-powered integrated sample concentrator. (A) Real-time RT-PCR amplification curves of HIV-1 RNA trapped from PBS buffer onto the flow-through FTA® membrane with concentrations ranging from 10 to 5,000 copies per sample. (B) Cycle threshold (Ct) used to develop calibration curve showing linear response with increase in HIV-1 RNA concentration, standard deviation with responses collected as n=3.
To demonstrate superior nucleic acid enrichment ability of our platform, we compared the performance of traditional dried plasma spot (DPS) and our sample concentrator by quantitatively detecting HIV virus in plasma. The 3 mm FTA® membrane has a sample volume capacity of 10 μL, if direct sampling is performed, we intend to demonstrate the effect of sampling higher volume as a pre-concentration step. Briefly, inactivated HIV-1 virus 10–500 copies was initially spiked into 100 μL human plasma. Next, 10 μL of sample was directly added to FTA® membrane (3 mm in diameter), simulating the traditional DPS platforms. This was compared to 90 μL of sample collected on the flow-through FTA® membrane (3 mm in diameter) via our sample concentrator. Then, FTA® membranes from both the experiments were further purified as described earlier and subjected to one step RT-PCR. As shown in Figure 4, the amplification times (e.g., Cycle threshold (Ct) value) significantly shorten for the same concentration samples due to nucleic acid enrichment of the flow-through FTA® membranes compared to the conventional DPS platforms. In our RT-PCR assay with FTA® membrane, the increased Ct values may be attributed to the following reasons: i) the existence of the FTA® membrane in the RT-PCR reaction potentially reduces amplification efficiency due to the absorption of polymerase and primers to the membrane as reported in previous literature.30 ii) Nucleic acid extracted by QIAamp® Viral RNA mini extraction kit has relatively higher quality with an absorbance ratio (A260/A280) of 1.96 than that of the FTA® membrane (1.82), which is consistent with the observed increase in the Ct values due to potential inhibition from plasma samples. The similar results have been reported in previous literature.31 iii) Rapid two-step cycling method with shorter extension phase was used in our RT-PCR assay. In addition, these sampled FTA® membranes are much easier to handle and less subject to contamination since no punching post sampling is required, and FTA® membrane encased in the FTA® disc holder is easy to store and transport. Therefore, our platform has the potential to replace conventional dried plasma spot sampling due to its simplicity, automation, high enriching ability and detection sensitivity.
Figure 4.
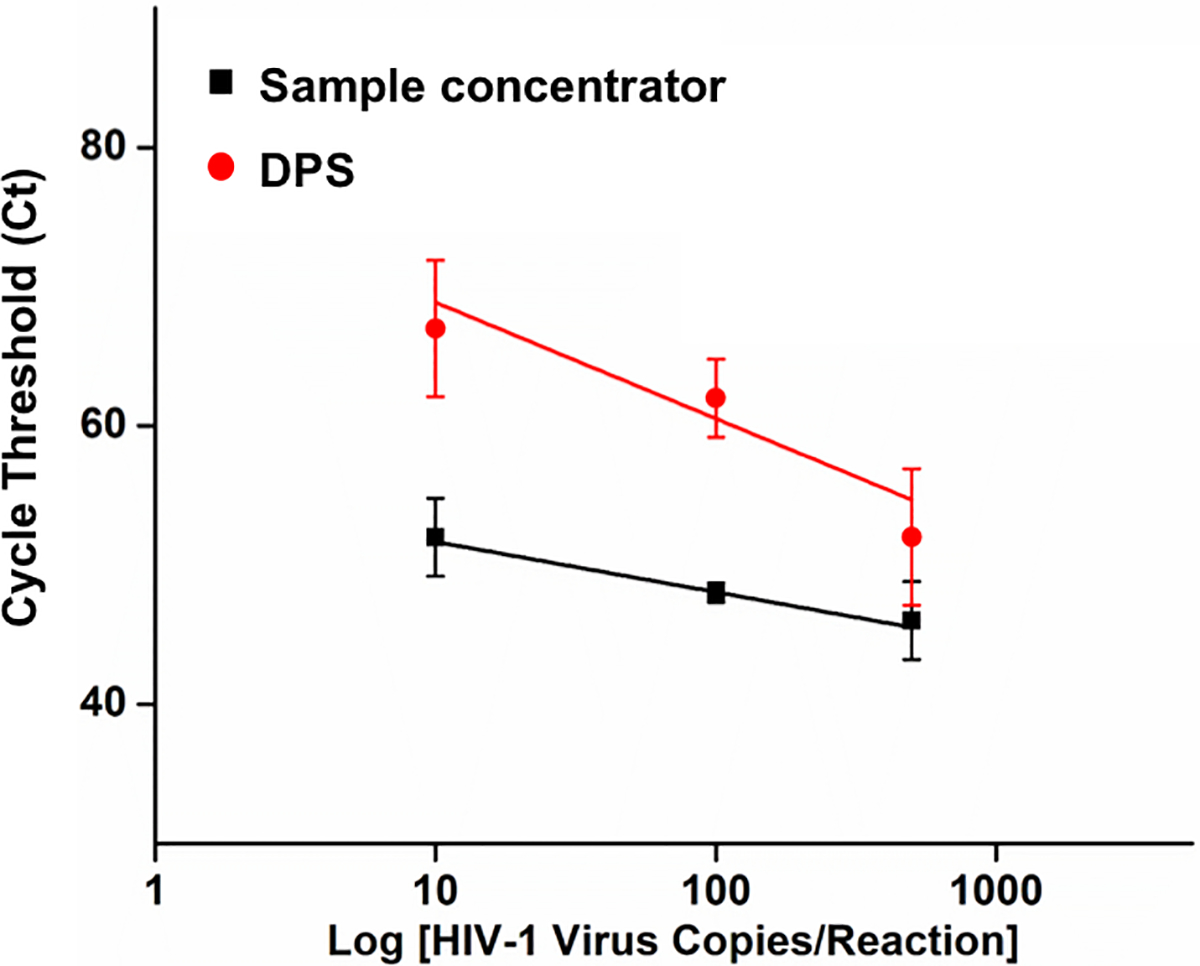
Performance comparison of HIV-1 virus detection by RT-PCR assay on traditional dried plasma spot (DPS) and the flow-through FTA® membrane of our sample concentrator. Standard deviation with responses collected as n=3.
Detection Sensitivity
We performed plasma separation from 200 μL of blood spiked with different concentration HIV-1 virus (10 to 5000 copies/sample), enriching nucleic acids on the flow-through FTA® membranes with 3-mm and 4-mm diameters. To enable highly sensitive detection of HIV viral RNA, we optimized washing steps after nucleic acid enrichment to minimize/eliminate the effect of inhibitors from blood plasma. Our optimized washing protocol includes: i) 2X washing of the FTA® membranes with FTA purification buffer (300 μL) with 3 min incubation to remove RT-PCR inhibitors, and ii) 2X washing with TE-buffer (pH 8.0) (300 μL) to stabilize the FTA® membrane for downstream amplification. In addition, treatment of the FTA® membrane with Proteinase K during the first wash step at ~55°C for 3 min yielded better reproducibility. This might be attributed to efficient removal of non-specifically adsorbed proteins from blood plasma that may inhibit subsequent enzymatic amplification. As shown in Figure 5, use of both 3-mm and 4-mm FTA® membranes enables detection of as low as 10 copies of HIV RNA. The 4-mm FTA® membranes showed faster detection time, while the 3-mm FTA® membrane has higher slope, hence better sensitivities for quantitative detection. Considering that the smaller FTA® membrane fits better with our PCR vials during real-time PCR amplification, we proceeded with 3-mm FTA® membranes for HIV virus detection. These results therefore demonstrate high efficiency recovery and a limit of detection of 10 viral copies per 200 μL of whole blood, approximately equal to 100 copies per mL plasma, which is well below the clinically relevant threshold value of 1,000 copies/mL that was recommended by the World Health Organization (WHO) and on par with FDA approved high end detection strategy.32 Thus, our sample concentrator enables highly sensitive detection of HIV virus in blood.
Figure 5.
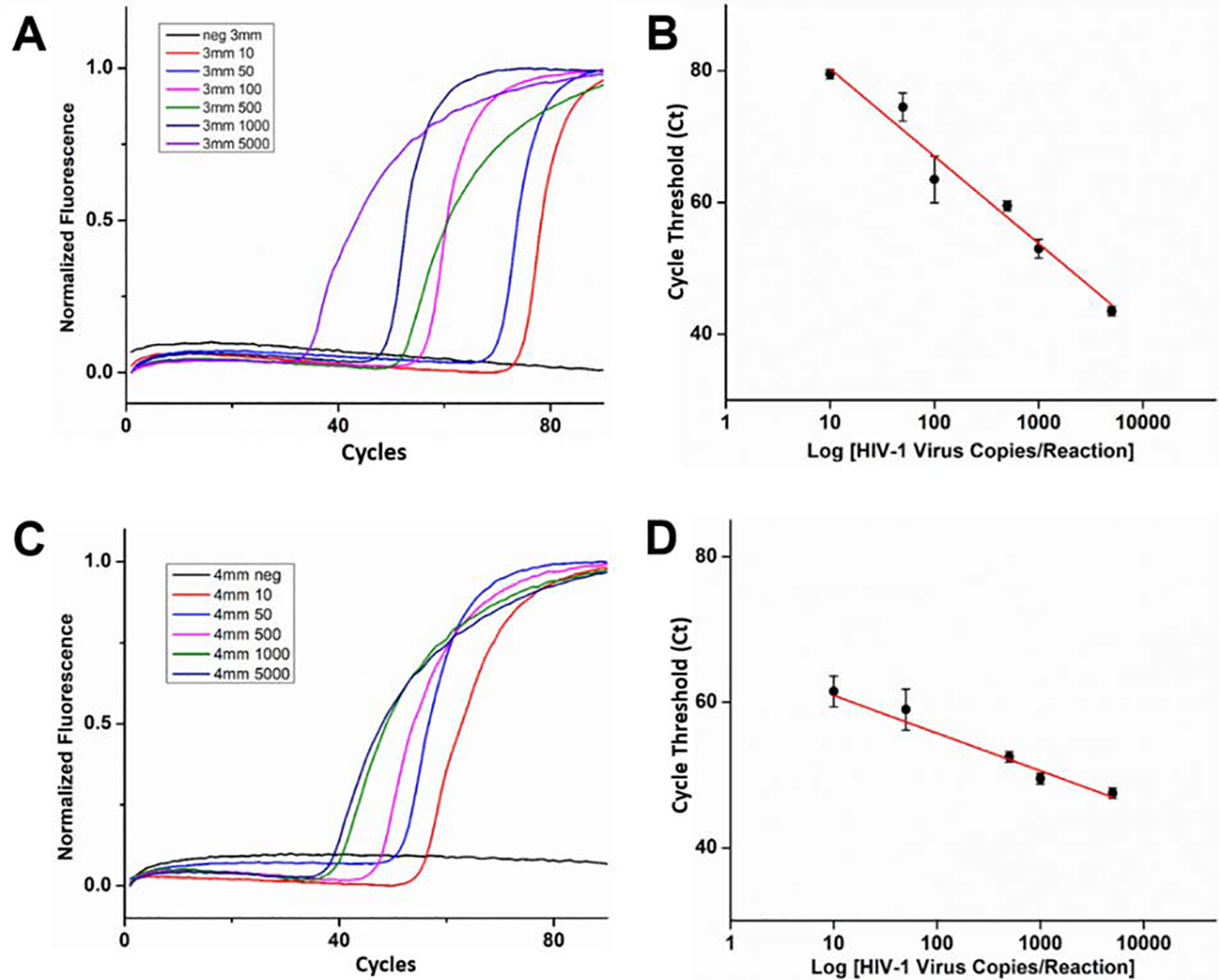
HIV virus enrichment and detection in blood samples by using different size flow-through FTA® membranes on our sample concentrator: A) Real time RT-PCR amplification curves of HIV-1 virus trapped from 200 μL blood (10 to 5000 copies) in the 3-mm FTA® membrane. (B) Cycle threshold (Ct) used to develop calibration curve showing linear response with in-crease in HIV-1 RNA concentration in the 3-mm FTA® membrane. C) Real time RT-PCR amplification curves of HIV-1 virus trapped from 200 μL blood (10 to 5000 copies) in the 4-mm FTA® membrane. (D) Cycle threshold (Ct) used to develop calibration curve showing linear response with increase in HIV-1 RNA concentration in the 4-mm FTA® membrane, standard deviation with responses collected as n=3.
Long-term Storage Evaluation of Nucleic Acid in Flow-Through FTA® Membrane
Efficient nucleic acid sample transportation and storage at room temperature allowed precise downstream molecular detection at remote centralized laboratories, also reducing costs of tests without a need for a cold chain. Whatman® FTA® card technology is known for its ability to lyse cells/pathogens and capture nucleic acids in addition to protecting nucleic acids from nucleases and atmospheric oxidation. To demonstrate the feasibility of our sample concentrator for remote transportation, we evaluated potential degradation of HIV viral RNA on the flow-through FTA® membranes after sampling at room temperature and −20°C over 4 weeks’ time, a reasonable period for establishing transportation challenges. Spiked human blood samples with 1,000 copies were used in the long-term stability studies. Post blood plasma separation, the FTA® disc holder (inset of Figure 6A) with the flow-through FTA® membrane were placed in double barrier foil pouches (Figure 6A) with an external desiccant pack and stored at different temperatures as needed. Cycle threshold of real time RT-PCR was used to evaluate the quality of HIV viral RNA of the flow-through FTA® membranes. As shown in Figure 6B, there was no significant degradation of HIV viral RNA over a 4-week period both at room temperature and −20°C. Therefore, the experimental results indicate that the flow-through FTA® membrane of our device can be easily transported at room temperature and have the potential applicability for point of care molecular diagnostics in resource limited settings.
Figure 6.
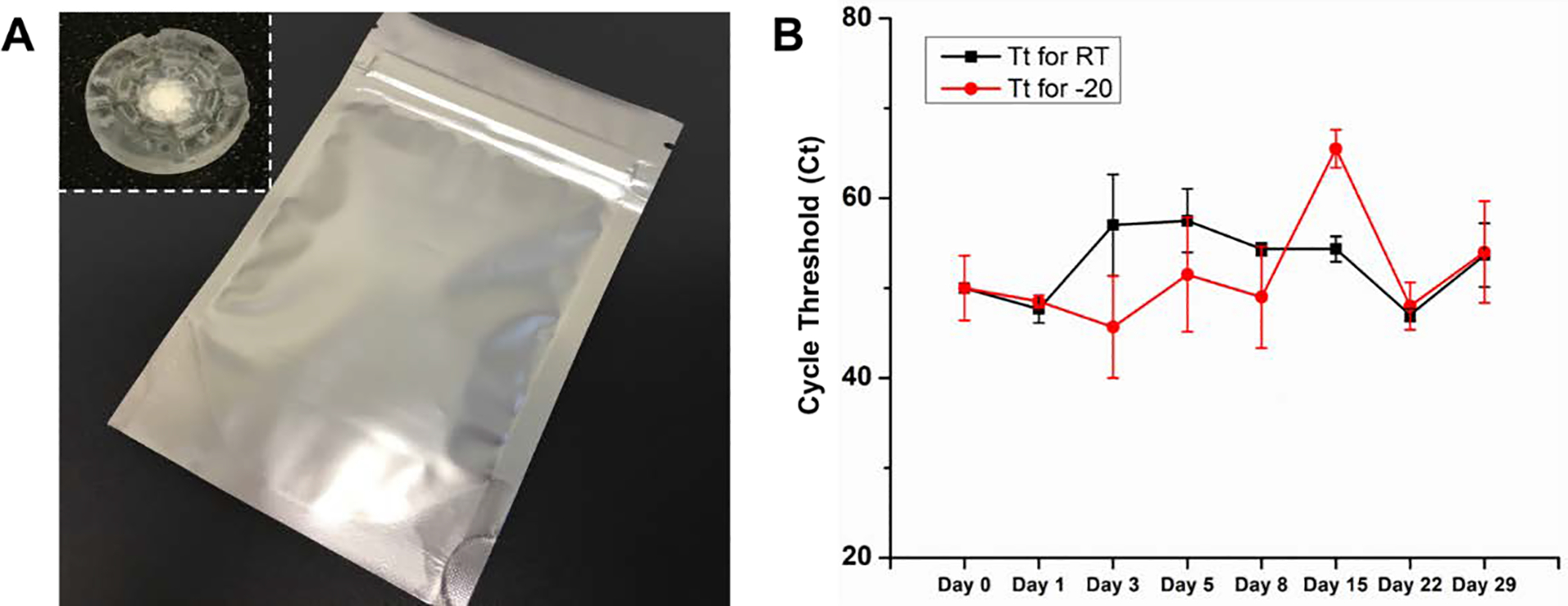
(A) A photo of the double barrier foil pouch with an external desiccant pack for long-term storage of the FTA® disc holder with one FTA® membrane. Inset is the photo of the flow-through FTA® disc holder. (B) Stability comparison of dried HIV plasma on the flow-through FTA® discs stored at room temperature and −20° C freezer. n=3.
CONCLUSION
We have successfully developed a simple, “all-in-one”, multi-functional blood sample concentrator that is able to perform high efficiency, power-free plasma separation, pathogen lysis and nucleic acid isolation/enrichment for molecular detection. On one hand, it combines cell gravitational sedimentation and capillary force to rapidly separate plasma from cellular components without need for an external pump. One the other hand, it takes advantage of the flow-through FTA® membrane for virus lysis and nucleic acid isolation/enrichment from the separated plasma. Our platform is suitable for rapid, automated, nucleic acid sample preparation and enrichment for highly sensitive molecular detection without need for complex equipment.
Our platform has several innovative features. i) Super high plasma separation yield. Due to the combination of cell gravitational sedimentation, membrane filtration and capillary force driving, our blood sample concentrator can automatically separate/extract 94% of plasma from whole blood within 5 minutes. ii) “All-in-one” integrated nucleic acid sample preparation. The concentrator integrates multiple sample preparation functions, including plasma separation, pathogen lysis, and nucleic acid capturing, eliminating the need of manual sample preparation operation. iii) Superior nucleic acid enrichment. Unlike conventional DBS or DPS sampling, the flow-through FTA® membrane of our platform showed highly efficient pathogen isolation and enrichment with recoveries as low as 10 copies from a relatively large blood sample volume (e.g., 200 μL), enabling point of care sampling and highly sensitive molecular detection. (iv) Self-powered fluid transport and waste collection. By taking advantage of capillary force, the plasma can be automatically separated, transported, and collected by the absorption module without need for any external pump. (v) Affordability. Our current device was fabricated by 3D printing technology with a low-cost desktop printer, providing ability of print to need for onsite applications. vi) Easy transportation and storage. The enriched pathogen nucleic acids by the flow-through FTA® membrane are stable at room temperature for at least four weeks, which facilitates to transport the pre-concentrated sample to centralized laboratories for molecular detection in resource-limited settings. We have demonstrated that our sample concentrator is able to separate plasma and enrich nucleic acid from whole blood within 5 minutes and transferred to perform RT-PCR to detect HIV virus with a high detection sensitivity (~100 copies/mL plasma). Therefore, the proposed sample concentrator has a great potential for broad applications in early diagnostics, monitoring and management of infectious diseases.
Supplementary Material
ACKNOWLEDGMENT
This work was supported, in part, by R01EB023607, R61AI154642 and R01CA214072. We acknowledge support and assistance of the Penn Center for AIDS Research P30AI045008.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
References
- 1.Stevens W, Gous N, Ford N and Scott L, BMC Medicine, 2014, 12, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.https://www.usaid.gov/global-health/health-areas/hiv-and-aids/technical-areas/antiretroviral-therapy (accessed March 2021)
- 3.https://www.unaids.org/en/resources/fact-sheet (accessed March 2021)
- 4.Magnani R, Sabin K, Saidel T and Heckathorn D, AIDS, 2005, 19, S67–S72. [DOI] [PubMed] [Google Scholar]
- 5.Kagulire S, Opendi P, Stamper P, Nakavuma J, Mills L, Makumbi F, Gray R, Shott J, Serwadda D and Reynolds S, International Journal of STD & AIDS, 2011, 22, 308–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plate D, AIDS Research and Human Retroviruses, 2007, 23, 1491–1498. [DOI] [PubMed] [Google Scholar]
- 7.Laksanasopin T, Guo T, Nayak S, Sridhara A, Xie S, Olowookere O, Cadinu P, Meng F, Chee N, Kim J, Chin C, Munyazesa E, Mugwaneza P, Rai A, Mugisha V, Castro A, Steinmiller D, Linder V, Justman J, Nsanzimana S and Sia S, Science Translational Medicine, 2015, 7, 273re1–273re1. [DOI] [PubMed] [Google Scholar]
- 8.Jefferies R, Ryan U and Irwin P, Veterinary Parasitology, 2007, 144, 20–27. [DOI] [PubMed] [Google Scholar]
- 9.Grivard P, Le Roux K, Laurent P, Fianu A, Perrau J, Gigan J, Hoarau G, Grondin N, Staikowsky F, Favier F and Michault A, Pathologie Biologie, 2007, 55, 490–494. [DOI] [PubMed] [Google Scholar]
- 10.Stresman G, Kamanga A, Moono P, Hamapumbu H, Mharakurwa S, Kobayashi T, Moss W and Shiff C, Malaria Journal, 2010, 9, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zida S, Tuaillon E, Barro M, Kwimatouo Lekpa Franchard A, Kagoné T, Nacro B, Ouedraogo A, Bolloré K, Sanosyan A, Plantier J, Meda N, Sangaré L, Rouzioux C, Rouet F and Kania D, Journal of Clinical Microbiology, 2016, 54, 1641–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denniff P and Spooner N, Bioanalysis, 2010, 2, 1385–1395. [DOI] [PubMed] [Google Scholar]
- 13.de Vries R, Barfield M, van de Merbel N, Schmid B, Siethoff C, Ortiz J, Verheij E, van Baar B, Cobb Z, White S and Timmerman P, Bioanalysis, 2013, 5, 2147–2160. [DOI] [PubMed] [Google Scholar]
- 14.Nabatiyan A, Parpia Z, Elghanian R and Kelso D, Journal of Virological Methods, 2011, 173, 37–42. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Mauk M, Gross R, Bushman F, Edelstein P, Collman R and Bau H, Analytical Chemistry, 2013, 85, 10463–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Liao S, Song J, Mauk M, Li X, Wu G, Ge D, Greenberg R, Yang S and Bau H, Lab on a Chip, 2016, 16, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CHEN X, CUI D, LIU C and LI H, Sensors and Actuators B: Chemical, 2008, 130, 216–221. [Google Scholar]
- 18.Kersaudy-Kerhoas M, Dhariwal R, Desmulliez M and Jouvet L, Microfluidics and Nanofluidics, 2009, 8, 105–114. [Google Scholar]
- 19.Yang S, Ündar A and Zahn J, Lab Chip, 2006, 6, 871–880. [DOI] [PubMed] [Google Scholar]
- 20.Dimov I, Basabe-Desmonts L, Garcia-Cordero J, Ross B, Ricco A and Lee L, Lab Chip, 2011, 11, 845–850. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Henion J, Abbott R and Wang P, Rapid Communications in Mass Spectrometry, 2012, 26, 1208–1212. [DOI] [PubMed] [Google Scholar]
- 22.Ryona I and Henion J, Analytical Chemistry, 2016, 88, 11229–11237. [DOI] [PubMed] [Google Scholar]
- 23.https://www.ssi.shimadzu.com/products/dried-plasma-spots/noviplex-cards.html (accessed March 2021)
- 24.https://www.spotonsciences.com/hemaspot-se (accessed March 2021)
- 25.Yeh EC, Fu C-C, Hu L, Thakur R, Feng J and Lee LP, Science Advances, 2017, 3, e1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin L, Vermesh O, Shi Q and Heath JR, Lab on a Chip, 2009, 9, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardi N, Secreto A, Shan X, Debonera F, Glover J, Yi Y, Muramatsu H, Ni H, Mui B, Tam Y, Shaheen F, Collman R, Karikó K, Danet-Desnoyers G, Madden T, Hope M and Weissman D, Nature Communications, 2017, 8,14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kattenberg J, Tahita C, Versteeg I, Tinto H, Traoré-Coulibaly M, Schallig H and Mens P, Tropical Medicine & International Health, 2012, 17, 550–557. [DOI] [PubMed] [Google Scholar]
- 29.https://www.pptaglobal.org/plasma (accessed March 2021)
- 30.Qiu X, and Mauk MG, Microsystem Technologies. 2015, 21, 841–50. [Google Scholar]
- 31.Melanson VR, Jochim R, Yarnell M, Ferlez KB, Shashikumar S, and Richardson J, Journal of Vector Borne Diseases. 2018, 54, 301–310. [DOI] [PubMed] [Google Scholar]
- 32.https://apps.who.int/iris/bitstream/handle/10665/255891/WHO-HIV-2017.22-eng.pdf?sequence=1 (accessed March 2021)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


