Abstract
Background
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver, and becoming the third-leading cause of cancer-related mortality worldwide. Despite the immune checkpoint inhibitors and molecular targeted therapies have shown preferable efficacy in HCC, large number of HCC patients do not respond effectively to anti-PD-1 reagents. Besides, the accumulation of genetic mutations in cancer cells may lead to the therapy resistant. Hence, there are clinical gaps between genetic and transcriptomic biomarkers for the HCC treatment.
Methods
To investigate the genetic mapping of liver cancer, targeted deep sequencing (TDS) and bioinformatics analysis were performed on hepatocellular carcinoma (HCC) tumor tissues and matched blood samples. Furthermore, copy number variants (CNVs) and Tumor mutation burden (TMB) were calculated. Immunohistochemistry was applied to determine the PD-L1 expression in HCC tumor tissues. Clinical characteristic, PD-L1 expression, and the TMB were analyzed in 32 HCC patients.
Results
This study indicated that the PD-L1 positive patients exhibited a lower TMB compared to the PD-L1 negative group, and PD-L1 positive patients were more likely to suffer from aggressive clinicopathologic features than PD-L1 negative patients. We also verified the top 30 mutated genes, including TP53, CTNNB1, KMT2D, AXIN1, ALK, and NOTCH1, in our dataset. Our results indicated that PD-L1 positive patients possessed more tumors with vascular invasion and advanced CCLC stage. Moreover, PD-L1 positive patients exhibited a lower TMB compared to the PD-L1 negative group.
Conclusions
These findings could improve our understanding of the effects of immune checkpoint therapies on prognosis, and could facilitate the monitoring of somatic mutations in HCC.
Keywords: PD-L1, targeted gene sequencing, hepatocellular carcinoma, tumor mutational burden (TMB), mutation landscape
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver, the fifth most common cancer, and the third-leading cause of cancer-related mortality worldwide (1,2). The poor prognosis of HCC is mainly due diagnosis in later stages, for which there are few effective treatment options. Given that chronic inflammation gives rise to a stromal environment that favors hepatocyte transformation and creates an immunosuppressive milieu that further leads to liver cancer progression, HCCs can be considered a paradigm for inflammation-induced cancers. For this tumor modality, immunotherapy is quite promising (3). Although, there are similar study illustrated the association between genetic and immunological background of HCC and expression of programmed cell death-1 and point out that CD8+ cells were densely infiltrated in PD-L1 positive tumors (4). Thus, there is a pressing need to explore the immune response signaling pathways and the disease microenvironment for HCC initiation and progression.
Immune checkpoint proteins, including PD-1, PD-L1 and CTLA-4, initiate signaling pathways that suppress T-cell function (5). PD-L1, a 40 kDa type I transmembrane protein, is composed of two side-by-side domains and extracellular IgV and IgC domains (6). Despite the approval of targeted inhibitors of sorafenib as first-line treatments for advanced HCC, which could improve survival by several months (7), small molecules of pathway inhibitors may induce drug resistance (8). At present, immune checkpoint inhibitors (ICIs) have shown preferable efficacy in several types of cancer, including lung, breast, bladder, and non-small-cell lung cancers (9-11). In 2017, the US Food and Drug Administration (FDA) approved nivolumab as the first anti-PD-1 antibody for the treatment of HCC. Thus, ICIs might play an indispensable role in HCC treatment, which brings new hope to cancer patients.
PD-L1 is universally expressed both inside and outside of cells (including cancer cells), which can lead to the exhaustion of T cells (12). Some studies have reported that patients with a higher expression of PD-L1 in cancer cells are correlated with preferential outcome in lung cancer (13). However, others have found that a high PD-L1 expression level is a poor prognostic factor in esophageal and cervical cancers (14,15). Although recent studies have been carried out (16-18), the association between the clinical response to anti-PD-1 antibodies and PD–L1 expression in HCC tumor cells remains unclear. Obviously, there is a pressing need to study the features of PD-L1 expression in HCC.
Cancer cells originate from a single cell with accumulated germ-line or somatic mutations and epigenetic alterations, which lead to the transformation of a normal cell into a malignant cell. Existing studies have demonstrated the diversity frequency rates of mutations, such as TP53, CTNNB1, AXIN1, KEAP1, and RB1, in the HCC population (19). The tumor mutational burden (TMB), coupled with PD-L1 expression, has been shown to be a powerful biomarker for ICB selection among different cancer types. The TMB has also been reported to be strongly correlated with the clinical response to immunotherapy using checkpoint inhibitors. Traditionally, the TMB was detected by whole-exome sequencing. However, more recently, targeted gene sequencing (TGS) has been extensively applied in clinical TMB evaluation. Moreover, taking advantage of whole-exome and TGS could provide valuable insight into tumor heterogeneity at the genetic and genomic levels (20).
In this study, we aimed to explore the correlation between PD-L1 expression and the TMB, mutation signature, and driver-gene mutations in 30 Chinese patients with HCC. We also analyzed patient outcomes based on the mutational landscape and the immune microenvironment. Our results highlight the roles of the immune microenvironment and the mutational landscape in patient prognosis, which will help to guide personalized immune-based therapy for Chinese patients with HCC. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-251).
Methods
Patients and clinical information collection
Thirty-two patients with primary HCC who were admitted to our hospital between May 2019 and November 2020 were included in this study. Written informed consent was obtained from all patients. This study was performed in accordance with the Helsinki Declaration (as revised in 2013) and was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University. Clinical information of the 30 HCC patients were in Table S1.
Hematoxylin-eosin staining and immunohistochemistry
For consensus judgement, all hematoxylin-eosin (HE) images were identified together independently by one otolaryngologist and two experienced pathologists, and the histological subtype classification was assessed according to the World Health Organization’s Classification of Tumors [2015]. Formalin-fixed, paraffin-embedded (FFPE) tissue specimens obtained from surgeries were examined in triplicate. Each FFPE tissue was selected from a centrally located area of the tumor that had been confirmed to contain tumor cells by HE staining. FFPE tissue were serial sectioned at a thickness of 4µm and deparaffinized, and then subjected to immunohistochemical staining using a previously described method with a previously validated rabbit monoclonal PD-L1 antibody (E1L3N, 1:800, Cell Signaling Technology, Danvers, MA). For visualization of the antigen, a peroxidase-labeled secondary antibody (DAKO, 22C3) was applied.
DNA extraction
Tumor tissues and matched blood DNA were extracted using the GeneJET FFPE DNA Purification Kit (#K0881, Thermo Scientific, Shanghai, China) according to the manufacturer’s instructions. The DNA samples were then identified using the Applied Biological Materials Inc. (ABM), and DNA quality was assessed on Thermo NanoDrop 2000 (Thermo Scientific, Shanghai, China).
Library preparation and sequencing
Genomic DNA were exacted from the tumor tissue sections, and then sonicated into 200 bp fragments in ultrasonic. To prepare the DNA library, we used Roche SeqCap EZ Exome V3 and TruePrep DNA Library Prep Kit V2 for Illumina (#TD501, Vazyme, Nanjing, China) to capture the target DNA, and Illumina HiSeq machines were used to acquire sequencing data. Agilent’s SureSelect Human All Exon V5 Kit (Agilent Technologies, Inc. Beijing, China) was applied to capture the whole exome, after amplified modified gDNA fragments in six cycles of PCR. In total, 50 Mb of DNA sequences, containing 33,4378 exons from 20,965 genes, were obtained. The average sequencing depth and coverage of the target region are summarized.
Copy number determination using exome-sequencing data
We aligned bisulfite reads to the reference genome at the candidate locations, and then applied GATA 4.1.4.0 to classify and eliminate PCR duplicates as previously described (21). Copy number variants (CNVs) from the next generation sequencing (NGS) data were calculated by CNVKIT (22). Finally, somatic mutations were converted to MAF and visualized using R package maftools.
Statistical analysis
All clinical and statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). Clinicopathologic variables of the high and low PD-L1 groups were compared using two-sample t-test or Fisher’s exact test for nominal variables. Given that this study was limited to a small sample size, all clinical-related statistical analysis were considered to be significant when p<0.05 without multiple correction test.
Results
Patient characteristics
Tumor tissue and blood were collected from 30 patients with HCC at the time of diagnosis. The HCC patients, including 26 males and four females, had an average age of 58 years (range, 34–69 years). Nineteen patients were former smokers, and the remaining 11 were non-smokers. Additionally, 24 male patients and one female patient were never-drinkers, while four male patients were ever-drinkers. Twenty-four patients had hepatitis, while the remaining six patients did not. The number of patients with CCLC stages Ia, Ib, IIa, IIb, and IIIa were 10, seven, seven, one, and four respectively, and the number of those with BCLC stages A and B were 12 and 17, respectively. The exclusion criteria were as follows: (I) patients with prior chemotherapy and radiotherapy, (II) patients with prior non-infectious pneumonitis; and (III) patients with HIV, autoimmune disease, or other conditions that could interfere with their participation in the trial. None of the included patients received radiation therapy previously. The baseline patient characteristics and detailed clinical information are depicted in Table 1 and Table S1.
Table 1. Baseline patient characteristics.
| Characteristic | Value |
|---|---|
| Median age in years [range] | 58 [34–69] |
| Gender (male/female) | 26/4 |
| Status (alive/dead) | 24/6 |
| PD-L1 expression: +/−/NA | 10/13/7 |
| Targeted deep sequencing | 30 |
| Smoker or ex-smoker/non-smoker | 7/23 |
| Never-drinker/ever drinker | 18/12 |
| Hepatitis (positive/never) | 24/6 |
| CCLC stage (Ia/Ib/IIa/IIb/IIIa) | 10/7/7/1/4 |
| BCLC stage (A/B) | 12/17 |
| Tumor number (1/2–3/over 3) | 19/5/4 |
| Tumor location (SF/C/DC/HF/R) | 10/4/5/8/3 |
| Median (range) tumor size, cm | 5.5 (1.7–13) |
| Microvascular invasion (yes/no) | 12/18 |
| α-fetoprotein (AFP) >20 μg/L | 17 |
NA, not available; SF, sigmoid flexure; C, cecum; DC, descending colon; HF, hepatic flexure; R, rectum.
Correlation between PD-L1 and clinicopathologic features
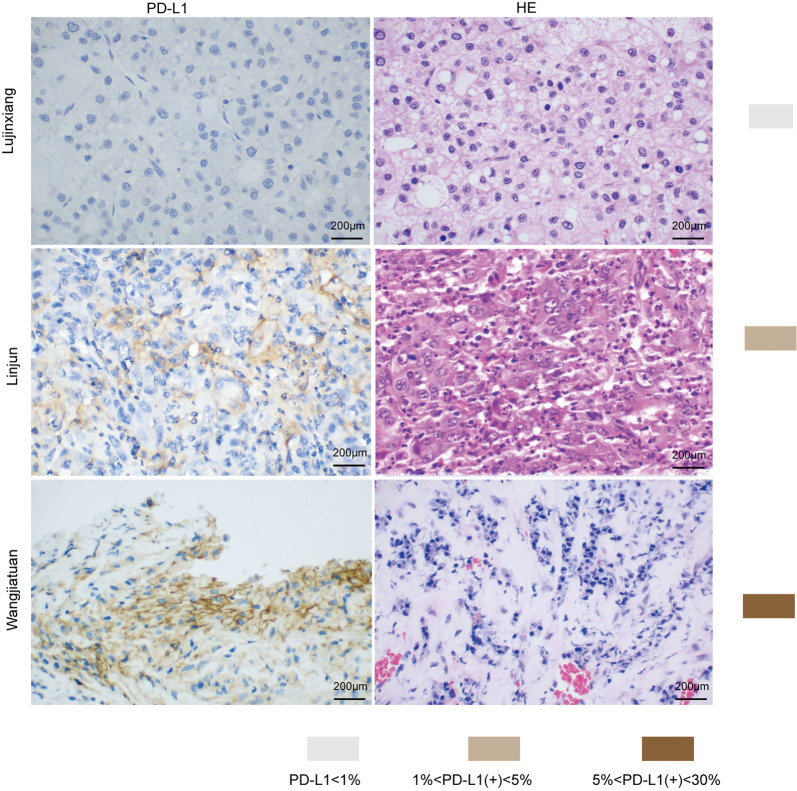
To evaluate the relationship between PD-L1 and tumor pathological features, we compared the clinicopathologic features with PD-L1 in each tissue sample. Patients were separated into PD-L1 positive and PD-L1 negative groups. We found that the PD-L1 positive patients were more likely to suffer from aggressive clinicopathologic features. These results demonstrate that PD-L1 positive patients possessed more tumors with vascular invasion and advanced CCLC stage (Figure 1 A,B,C,D,E,F and Table S1).
Figure 1.
PD-L1 expression in HCC tissue samples. (A) PD-L1 immunohistochemistry of Liujinxiang HCC biopsy. (B) HE staining of Liujinxiang HCC biopsy. (C) PD-L1 immunohistochemistry of Linjun HCC biopsy. (D) HE staining of Linjun HCC biopsy. (E) PD-L1 immunohistochemistry of Wangjiatuan HCC biopsy. (F) HE staining of a representative HCC biopsy. HCC, hepatocellular carcinoma. Scale bars =200 µm.
Relationship between PD-L1 and the TMB
The TMB is specified using whole genome sequencing (WGS), whole exome sequencing (WES), or targeted gene panels sequencing. In our study cohort, the PD-L1 positive patients exhibited a lower TMB compared to the PD-L1 negative group. Thus, we may infer that high PD-L1 patients in the Chinese HCC population have a low TMB (Figure 2).
Figure 2.
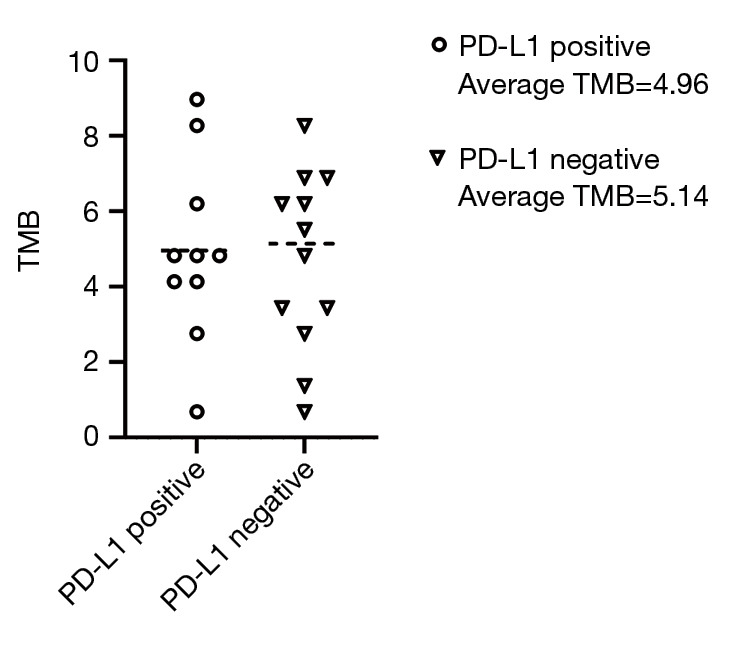
Correlation between PD-1 expression and the TMB. TMB, tumor mutational burden.
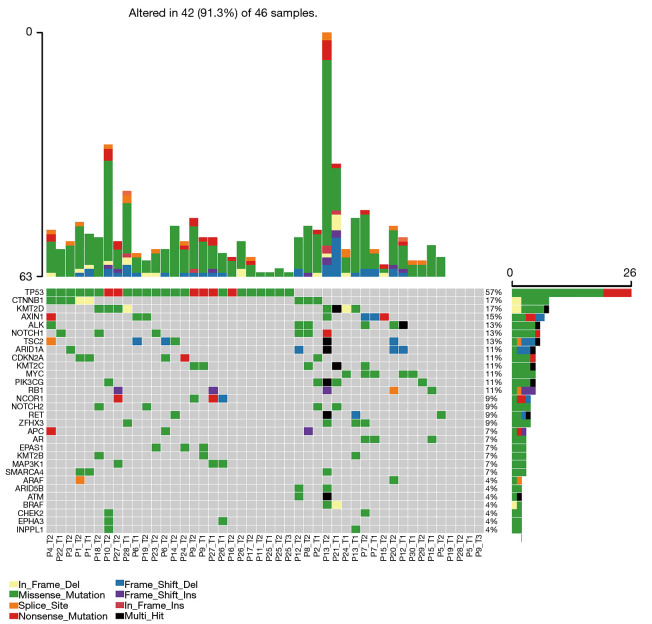
Landscape of somatic mutations in HCC patients
To assess somatic alternation, TGS was performed on tissue samples from the 30 HCC patients. The mean coverage depth for tumor and blood samples was 194×. There were 459 mutation events in 556 genes; the top 30 mutated genes are shown in Figure 3. In HCC, the most frequent driver gene mutations were found to be in TP53, CTNNB1, KMT2D, AXIN1, ALK, and NOTCH1 (Figure 3). The TMB (mutations/Mb) and CNV alteration (CNA) are shown in Table S2.
Figure 3.
The Oncoprint of the top 30 driver genes in 46 HCC samples. HCC, hepatocellular carcinoma.
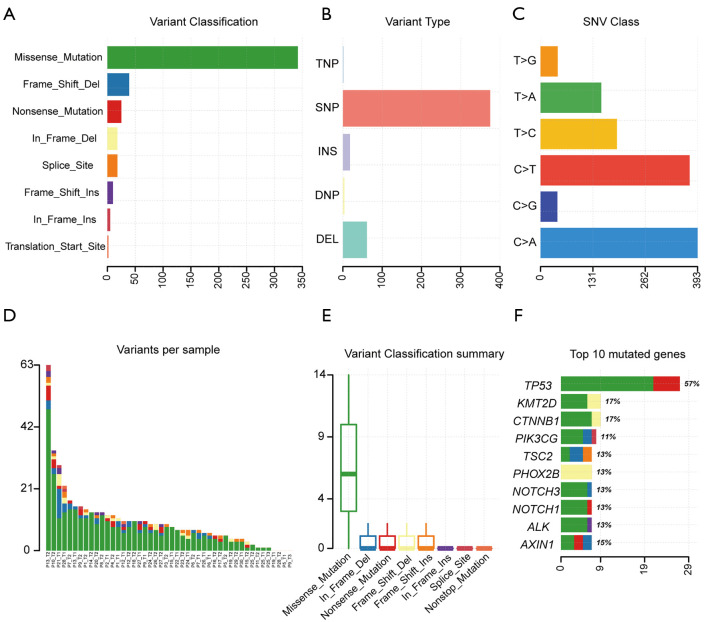
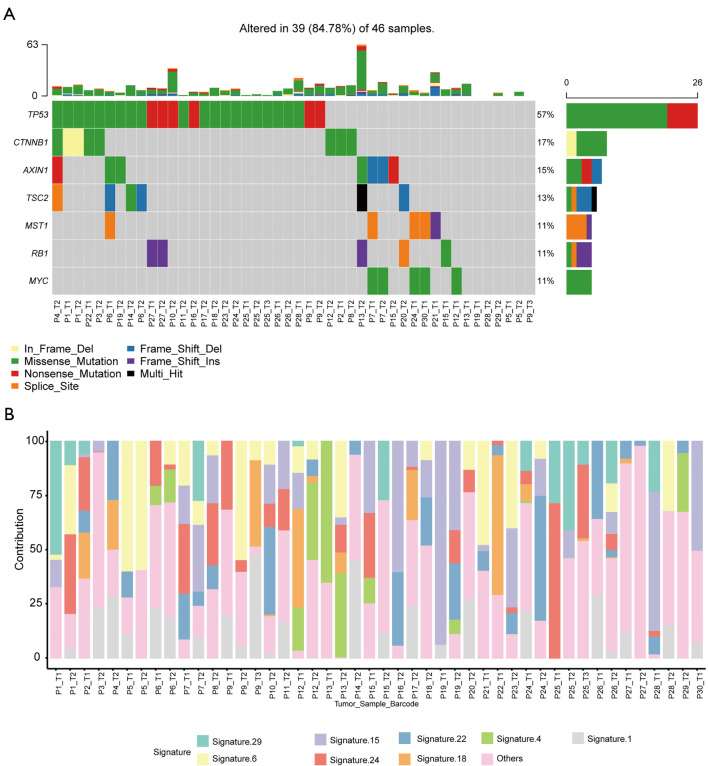
Mutation spectrum and signatures
Our results revealed that missense mutations were the most common mutation variant cases (Figure 4A); and single nucleotide polymorphisms (SNPs) were the most common variant types (Figure 4B). C > A and C > T were the most common base substitutions (Figure 4C); The mean variant of each sample was 9.97, and P13_T2 had the highest mutation number (Figure 4D). Variant classification summary also suggested that missense mutations and frame shift deletion was the primarily mutation variant cases (Figure 4E). To identify the HCC mutation spectrum and signatures, the top 10 mutated genes in Figure 4F were consistent with SMGs in Figure 5A. Moreover, we identified the significantly mutated genes (SMGs) with p<0.01 in the HCC mutation cohort, which included TP53, CTNNB1, AXIN1, TSC2, MST1, RB1, and MYC etc. (Figure 5A). We further analyzed mutational signatures 1 and 6 in 46 HCC samples, and found that they were correlated with age of cancer diagnosis and defective DNA mismatch repair, respectively (Figure 5B).
Figure 4.
Landscape of somatic mutations of driver genes in HCC. (A) Classification of variant; (B) number of variants; (C) SNV class; (D) number of variants; (D) variants per sample; (E) variant classification summary; (F) top 10 mutated genes. HCC, hepatocellular carcinoma.
Figure 5.
Significant mutated gene and signature distribution. (A) Significant mutated gene of HCC samples; (B) catalogue of somatic mutations of 46 HCC samples according to COSMIC. HCC, hepatocellular carcinoma; COSMIC, Catalogue of Somatic Mutations in Cancer.
Discussion
To demonstrate the genetic mapping of liver cancer, we enrolled 30 Chinese HCC patients (involving 46 HCC tissue samples) in this study. TGS was performed on HCC tumor tissues and matched normal tissues, and their clonal evolution relationships were analyzed. The prevalence of somatic mutations, including nucleotide substitutions, CNA, TMB, and small insertions/deletions, was evaluated by bioinformatics analysis using the R packages, Mutect2 (23,24) and VarDict (25). We also determined the correlation between PD-L1 expression and clinicopathologic features, and TMB. Finally, the landscape of somatic mutations, as well as the mutation spectrum and signatures were calculated as previously described.
Our results suggested that tumors with positive PD-L1 were more likely to suffer from aggressive clinicopathologic features, which is consistent with the results of a previous study (26). Specifically, PD-L1 positive patients possessed more tumors with vascular invasion, and advanced CCLC stage. Moreover, these patients exhibited a lower TMB compared to the PD-L1 negative group. We might speculate that high PD-L1 patients in the Chinese HCC population have a low TMB.
Furthermore, the TMB and overall neoantigen load analysis, as predictors of response to immune checkpoint inhibitors and anti–PD-1 therapy, can alter neoantigen-specific T cell reactivity (27-29). Tumors with high microsatellite instability (MSI-H) accumulate substantial numbers of somatic mutations secondary to deficits in DNA mismatch repair (MMR) (30). The top 30 mutated genes in our dataset indicated that the most frequent driver gene mutations, including TP53, CTNNB1, KMT2D, AXIN1, ALK, and NOTCH1, were consistent with the SMGs, including TP53, CTNNB1, AXIN1, TSC2, MST1, RB1, and MYC etc. However, these results were inconsistent with the findings of Guichard et al. (19) and David et al. (31), who reported that the most common driver mutations were in TP53 and CTNNB1.
Missense mutations were the most common mutation variant cases, while C>A and C>T were the most common base substitutions. This is consistent with the findings of Julián et al. (31), who described the base substitution differential frequency distribution in HDV+ patients relative to HDV- patients in the Mongolian HCC population. Our results also indicated that SNPs were the most common variant types. The mean variant of each sample was 9.97, and P13_T2 had highest mutation number (63). We further analyzed mutational signatures 1 and 6 in 46 HCC samples, and found that they were correlated with age of cancer diagnosis and defective DNA mismatch repair, respectively.
In this study, we found that the PD-L1 positive HCC patients have a lower TMB than the PD-L1 negative group. It was recommended for PD-L1 positive HCC patients taking anti-PD-1 antibody administration. We also identified the most frequent driver gene mutations in our cohort, including TP53, CTNNB1, KMT2D, AXIN1, ALK, and NOTCH1. This study promotes the potential for PD-L1 inhibitor treatment development for HCC and provides a deep understanding of the HCC mutational landscape, thereby encouraging personalized immune-based therapy for Chinese HCC patients.
Acknowledgments
The authors thank Shanghai Tongshu Biotechnology Co., Ltd. for their technical support.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed in accordance with the Helsinki Declaration (as revised in 2013) and was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-251
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-251
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-251). The authors report that this work was sponsored by the Shanghai Tongshu Biotechnology Co., Ltd. The authors have no other conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- 2.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog 2017;16:1. 10.4103/jcar.JCar_9_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich B, Czauderna C, Marquardt JU. Immunotherapy of Hepatocellular Carcinoma. Oncol Res Treat 2018;41:292-7. 10.1159/000488916 [DOI] [PubMed] [Google Scholar]
- 4.Nishida N, Sakai K, Morita M, et al. Association between Genetic and Immunological Background of Hepatocellular Carcinoma and Expression of Programmed Cell Death-1. Liver Cancer 2020;9:426-39. 10.1159/000506352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang K, Li J, Sun Z, et al. Retreatment with immune checkpoint inhibitors in solid tumors: a systematic review. Ther Adv Med Oncol 2020;12:1758835920975353. 10.1177/1758835920975353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Zhu G, Tamada K, et al. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999;5:1365-9. 10.1038/70932 [DOI] [PubMed] [Google Scholar]
- 7.Zongyi Y, Xiaowu L. Immunotherapy for hepatocellular carcinoma. Cancer Lett 2020;470:8-17. 10.1016/j.canlet.2019.12.002 [DOI] [PubMed] [Google Scholar]
- 8.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526-31. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunger A, Kiessler M, Wehner R, et al. Immune Monitoring of Cancer Patients Prior to and During CTLA-4 or PD-1/PD-L1 Inhibitor Treatment. Biomedicines 2018;6:26. 10.3390/biomedicines6010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Z, Liang Y, Yang X, et al. A meta-analysis of the efficacy and safety of PD-1/PD-L1 immune checkpoint inhibitors as treatments for metastatic bladder cancer. Onco Targets Ther 2019;12:1791-801. 10.2147/OTT.S186271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jing W, Li M, Zhang Y, et al. PD-1/PD-L1 blockades in non-small-cell lung cancer therapy. Onco Targets Ther 2016;9:489-502. 10.2147/OTT.S94993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Chen W, Xu ZP, et al. PD-L1 Distribution and Perspective for Cancer Immunotherapy-Blockade, Knockdown, or Inhibition. Front Immunol 2019;10:2022. 10.3389/fimmu.2019.02022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okita R, Maeda A, Shimizu K, et al. PD-L1 overexpression is partially regulated by EGFR/HER2 signaling and associated with poor prognosis in patients with non-small-cell lung cancer. Cancer Immunol Immunother 2017;66:865-76. 10.1007/s00262-017-1986-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X, Dong M, Liu Z, et al. Elevated PD-L1 expression predicts poor survival outcomes in patients with cervical cancer. Cancer Cell Int 2019;19:146. 10.1186/s12935-019-0861-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito M, Yajima S, Suzuki T, et al. High serum PD-L1 level is a poor prognostic biomarker in surgically treated esophageal cancer. Cancer Med 2020;9:1321-7. 10.1002/cam4.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai X, Pi G, Yang SL, et al. Association of PD-L1 and HIF-1alpha Coexpression with Poor Prognosis in Hepatocellular Carcinoma. Transl Oncol 2018;11:559-66. 10.1016/j.tranon.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CY, Wang Y, Luo GY, et al. Relationship Between PD-L1 Expression and CD8+ T-cell Immune Responses in Hepatocellular Carcinoma. J Immunother 2017;40:323-33. 10.1097/CJI.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 18.Liu CQ, Xu J, Zhou ZG, et al. Expression patterns of programmed death ligand 1 correlate with different microenvironments and patient prognosis in hepatocellular carcinoma. Br J Cancer 2018;119:80-8. 10.1038/s41416-018-0144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. 10.1038/ng.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallou A, Jennings J, Kabla AJ. Tumour heterogeneity promotes collective invasion and cancer metastatic dissemination. R Soc Open Sci 2017;4:161007. 10.1098/rsos.161007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A, Chia WK, Toh HC. Immunotherapy for nasopharyngeal cancer-a review. Chin Clin Oncol 2016;5:22. 10.21037/cco.2016.03.08 [DOI] [PubMed] [Google Scholar]
- 22.Cao C, Wei Q, Tang X, et al. PD-1 and PD-L1 in locoregionally advanced nasopharyngeal carcinoma: Substudy of a randomized phase III trial. Head Neck 2019;41:1427-33. 10.1002/hed.25601 [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Conroy JM, Morrison CD, et al. Structural variation discovery in the cancer genome using next generation sequencing: computational solutions and perspectives. Oncotarget 2015;6:5477-89. 10.18632/oncotarget.3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. 10.1093/nar/gkw227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Q, Wang XY, Qiu SJ, et al. Overexpression of PD-L1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res 2009;15:971-9. 10.1158/1078-0432.CCR-08-1608 [DOI] [PubMed] [Google Scholar]
- 27.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S nyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [DOI] [PMC free article] [PubMed]
- 29.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 2015;521:81-4. 10.1038/nature14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network . Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41 e23. 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]