Abstract
Background
The immune microenvironment is a critical regulator of clear cell renal cell carcinoma (ccRCC) progression. However, the underlying mechanisms the regulatory role of immune-related long non-coding RNAs (irlncRNAs) in the ccRCC tumor microenvironment (TME) are still obscure. Herein, we investigated prognostics role of irlncRNAs for ccRCC.
Methods
The raw data of patients with ccRCC were downloaded from The Cancer Genome Atlas (TCGA) database, and immune-related genes were obtained from the ImmPort database. First, we investigated the correlation between the immune-related genes and irlncRNAs. Then, we identified the differentially expressed irlncRNA pairs (ILRPs) between normal and cancer tissue samples, and prognostic model was constructed with the differentially expressed ILRPs. We further explored whether the signature risk scores of ILRPs had a considerable impact on immune cell infiltration. Finally, we performed a drug sensitivity analysis based on risk score.
Results
There were 13 upregulated and 40 downregulated irlncRNAs between the ccRCC and normal tissue samples. We further selected the irlncRNAs that significantly affect the prognosis of patients with ccRCC via univariate Cox, lasso regression, and multivariate regression analyses. Twelve ILRPs were used to construct a prognostic signature. The model showed the ILRPs model could be used to assess the prognosis of ccRCC patients. Study of the influence of risk score and clinical characteristics on the prognosis of patients with ccRCC showed risk score to be an independent factor affecting the outcome of ccRCC. We further performed the difference analysis of immune cell abundance between ccRCC and normal tissue samples. The results showed that patients with higher abundance of M0 macrophages, plasma cells, follicular helper T cells, and regulatory T cells (Tregs) had a poor outcome. Finally, we performed a drug sensitivity analysis based on risk score. The results showed that high-risk score patients are sensitive to orafenib, sunitinib, temsirolimus, cisplatin, and gemcitabine.
Conclusions
Our study has developed a novel and reasonable ILPRs model for prognostic prediction, which does not require transcriptional levels to be detected.
Keywords: Immune-related long non-coding RNAs (irlncRNAs), prognostic biomarkers, clear cell renal cell carcinoma (ccRCC)
Introduction
Kidney cancer ranks 6th and 8th out of all male and female cancers for new diagnoses in the United States, and in 2020, approximately 73,750 new cases were recorded (1). Among the types of kidney cancer, clear cell renal cell carcinoma (ccRCC) is the commonest and most aggressive subtype that originates from the renal epithelium (2). Despite the advances in diagnostic and therapeutic methods, the 5-year survival rate of patients with advanced ccRCC is only 12%. The reason responsible for this high mortality is about one-third will suffer localized or metastatic disease, and 30% patients may suffer relapsing after undergoing surgical resection, and this make is big challenge for treating the advances patients (3). However, early diagnosis of patients whose ccRCC is still localized or has only spread locally may significantly improve the survival rate at 5 years (67% to 92%). The main reason for this situation is that there still are standard therapeutic regimens for patients with advanced ccRCC (1). Therefore, a novel diagnostic biomarker which combines sensitivity and specificity is urgently needed for ccRCC, as are potential therapeutic targets.
Risk factors for ccRCC including smoking, obesity, and hypertension. Aside from these risk factors, aberration in the genome is an important event for ccRCC tumorigenesis, and loss of the short arm of chromosome 3 is the most common genomic change leading to ccRCC. The second most common genetic change in ccRCC is gain of chromosome 5q (4). A significant correlation also exists between ccRCC and aberration in the von Hippel-Lindau (VHL) gene, which is located on chromosome 3p. Inhibition or change of VHL results in transformation of the cellular oxygen-sensing pathway (5). Other genetic and epigenetic events also contribute to the development of ccRCC, such as mutations in polybromo 1 (PBRM1), SET domain containing 2, histone lysine methyltransferase (SETD2), BRCA1-associated protein 1 (BAP1), lysine demethylase 5C (KDM5C), and mechanistic target of rapamycin (MTOR) (6).
Over the years, long non-coding RNAs (lncRNAs) have been shown to play a key role in multiple cancers (7-9). Qu et al.’s research suggested that lncRNAs can serve as diagnostic biomarkers for ccRCC (10). Ye et al.’s study showed that lncRNA-LET can inhibit ccRCC cell growth by regulating microRNA-373-3p (11). Further, lncRNA MRCCAT1 can promote metastasis of ccRCC by inhibiting PR3 and activating the p38-mitogen-activated protein kinase (MAPK) signaling pathway (12). Further study showed that lncRNAs can also effect the therapeutic response of ccRCC. Jiang et al. showed that lncRNA 01094 induces radioresistance via regulation of the miR-577/CHEK2/FOXM1 axis (13). Recently, the results of several studies have shown that lncRNAs also have a significant impact on the immune system in cancers such as triple-negative breast cancer, hepatocellular carcinoma (HCC), and glioblastoma multiforme (14-16), and these lncRNAs have been termed immune-related lncRNAs (irlncRNAs). Liu et al.’s study revealed that LINC00973 acts as an immune suppressor in ccRCC via its positive regulation of Siglec-15 (17). Jiang et al.’s research also indicated that irlncRNAs can serve as diagnostic and prognostic biomarkers of ccRCC (18). Previous studies have investigated the role of irlncRNAs in ccRCC, but only in relation to immune cell infiltration and prognosis, and the mechanisms underlying the regulatory role of irlncRNAs in the drug sensitivity of ccRCC have yet to be investigated. The present study aimed to construct a novel model for predicting the prognostics role of ILRPs for ccRCC patients.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/tau-21-445).
Methods
Raw data
Transcriptome data and clinical data of 539 kidney renal clear cell carcinoma (CCRCC) tissue samples and 72 normal tissue samples were extracted from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/). An immune-related gene list was collected from the Immport website (https://www.immport.org/).
Obtaining irlncRNAs
We extracted the lncRNAs of the TCGA-CCRCC cohort from the RNA-seq expression matrix and analyzed their correlations with immune-related genes. The threshold values for the correlation analysis were set as coefficient R>0.4 and P<0.001 (Spearman’s correlation).
Screening of differentially expressed irlncRNAs
Differences in irlncRNA expression between tumor and normal tissue samples were analyzed with the limma package in R software (https://www.r-project.org/). The filtering criteria were log two-fold change (|logFC|) >1 and false discovery rate (FDR) <0.05. A heatmap and volcano plot were created using the ggplot2 package.
Pairing of irlncRNAs
The differentially expressed irlncRNAs were paired one by one. In each irlncRNA pair (ILRP), the expression of the 1st lncRNA was higher than that of the 2nd lncRNA. The expression value of this ILRP was recorded as 1; otherwise, it was recorded as 0.
Construction of a prognostic ILRP signature
First, we performed univariate Cox regression analysis of all the ILRPs (screening criterion P<0.01). Then, we used lasso regression to obtain a more refined signature by constructing a penalty function. Finally, stepwise multivariate Cox regression was used to construct a prognostic signature from the genes screened by lasso regression. The Kaplan-Meier method was adopted to perform survival analysis. The best cutoff value was identified according to the maximum Youden index (sensitivity + specificity – 1) of the 1-year receiver operating characteristic (ROC) curve, using the R packages survivalROC and survminer. Only patients with complete survival data were included to construct the signature. P<0.05 was considered to be statistically significant.
Tumor immune infiltration analysis
The CIBERSORT deconvolution algorithm can estimate the relative abundance of 22 immune cell types by characterizing the cell composition of complex tissues based on gene expression profiles (19). We used the CIBERSORT algorithm to calculate the abundance of 22 immune cells in all samples in the TCGA-CCRCC cohort. The correlation results of risk scores of the prognostic signature and immune cells were visualized using ggplot2. Kaplan-Meier survival analysis was used to analyze the correlation between the relative abundance of immune cells and overall survival (OS) in patients with CCRCC. Log-rank P<0.05 indicated statistical significance.
Analysis of risk scores and drug sensitivity
The R package pRRophetic, which is based on gene expression and drug sensitivity data from a large number of cancer cell lines (20). The relationship between the risk scores of the prognostic signature and drug sensitivity predicted using the pRRophetic package was analyzed. P<0.05 was considered to be statistically significant.
Statistical analysis
We used limma package to screened differential irlncRNAs, and the thresholds were set as |logFC| >1 and FDR <0.05. Univariate cox regression, lasso regression and multivariate (stepwise) cox regression were used to construct the prognostic signature. Wilcoxon test and Spearman correlation were used for difference analysis and correlation analysis, respectively. P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
ILRPs
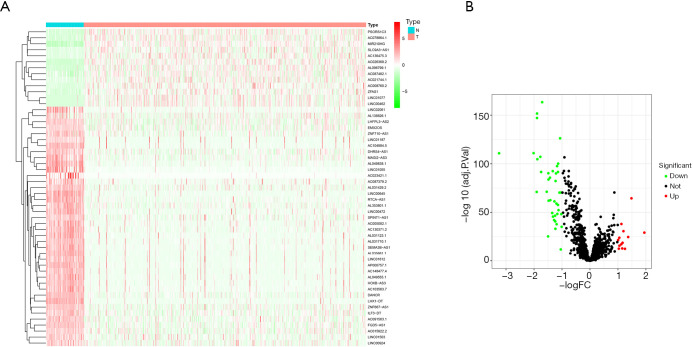
A total of 13,413 lncRNAs were extracted from the TCGA-BRCA cohort. Their correlations with immune-related genes were analyzed, and 1,397 irlncRNAs were obtained. The top 50 upregulated and downregulated genes are shown in the heatmap plot in Figure 1A. Differential expression analysis indicated that 13 and 40 irlncRNAs were upregulated and downregulated, respectively (Figure 1B). Subsequently, we paired up the 53 differentially expressed irlncRNAs, obtaining a total of 619 ILRPs.
Figure 1.
Differential analysis of irlncRNAs. Heatmap of the top 50 upregulated and downregulated lncRNAs; blue and red represent low and high expression, respectively (A). Volcano plot of differential lncRNAs; green and red represent downregulated and upregulated lncRNAs, respectively (B). IrlncRNAs, immune-related long non-coding RNAs.
Prognostic ILRP signature
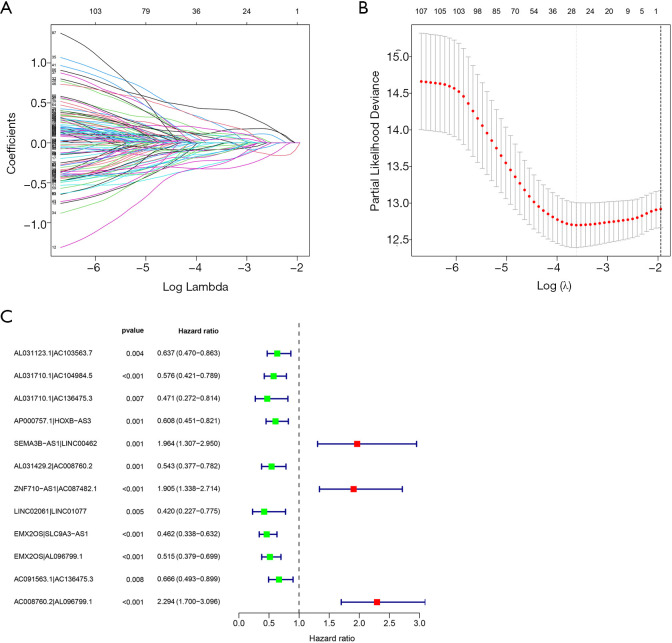
The univariate Cox regression results showed that there were 111 ILRPs which significantly impact the OS of patients with CCRCC. The results of lasso regression indicated λ=–3.6 to be the optimal value, and 27 ILRPs were obtained for further analysis (Figure 2A,B). Through multivariate regression analysis, 12 lncRNA pairs were obtained and incorporated into the ILRP prognostic signature (Figure 2C).
Figure 2.
Prognostic signature of ILRPs. (A,B) Lasso regression analysis results indicate that λ=–3.6 is the optimal value based on the prognostic ILRPs. (C) Forest map showing the 12 ILRPs included in the prognostic signature; green and red respectively represent favorable and unfavorable lncRNA pairs for the OS of patients with CCRCC. ILRPs, irlncRNA pairs; irlncRNAs, immune-related long non-coding RNAs; OS, overall survival; CCRCC, kidney renal clear cell carcinoma.
Optimal cutoff value determination and Kaplan-Meier survival analysis of the ILRP signature
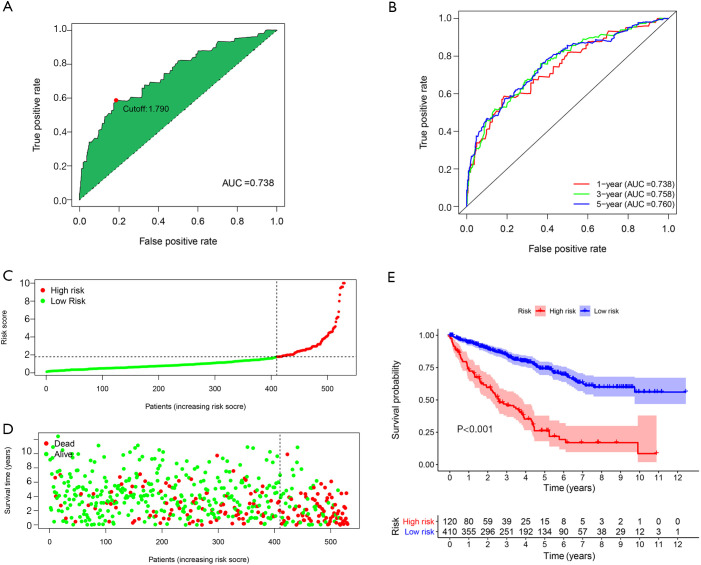
The 1-year ROC curve of the ILRP signature showed an area under the curve (AUC) value of 0.738, with the optimal cutoff value for the risk score reaching 1.790 (Figure 3A). The AUC values of the 1-, 3- and 5-year ROC curves of the signature were 0.738, 0.758 and 0.760, respectively (Figure 3B). As shown by the risk score curve and survival status map, 530 patients with CCRCC were divided into the high-risk group (n=120) and low-risk group (n=410) according to the optimal cutoff value (Figure 3C,D). Kaplan-Meier survival analysis indicated that patients in the low-risk group had better OS than those in the high-risk group (log-rank P<0.001) (Figure 3E).
Figure 3.
Optimal cutoff value determination and Kaplan-Meier survival analysis of the ILRP signature. The 1-year ROC curve and the determination of the optimal cutoff value (A). The 1-, 3-, and 5-year ROC curves (B): the X- and Y-axes represent the false and true positive rate, respectively. The risk score curve: the X-axis represents increasing risk score of patients, and Y-axis represents risk score (C). The survival status map: the X-axis represents increasing risk score of patients; the Y-axis represents survival time; and the green and red represent alive and dead, respectively (D). Kaplan-Meier survival analysis of patients with high and low risk scores based on the ILRP signature (E). ILRPs, irlncRNA pairs; irlncRNAs, immune-related long non-coding RNAs; ROC, receiver operating characteristic; AUC, area under the curve.
Relationship between signature risk score and clinical features
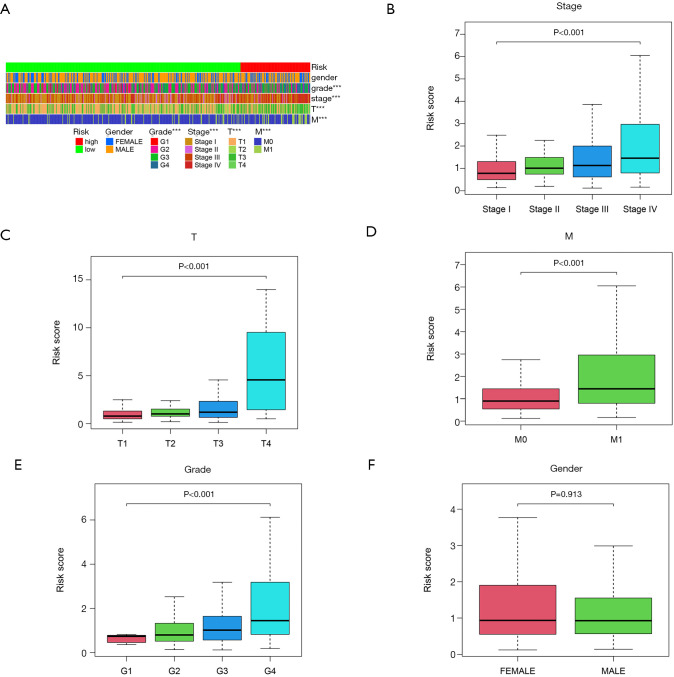
An overview heatmap of the correlation between risk score and clinical characters (Figure 4A). The results of correlation analysis with clinical features revealed that the higher the risk score in patients with ccRCC, the higher the tumor stage, the larger the tumor size, the more likely distant metastasis was, and the higher the histological grade (P<0.001, Figure 4B,C,D,E). However, risk score showed no significant sex difference in patients with ccRCC (Figure 4F).
Figure 4.
Relationship between signature risk score and clinical features. Heatmap of difference analysis of clinical features and risk scores (***, P<0.001) (A); correlation analysis results of risk score with stage (B), tumor size (C), distant metastasis (D), histological grade (E), and sex (F) in patients with CCRCC. CCRCC, kidney renal clear cell carcinoma.
Prognostic signature risk score and clinical features
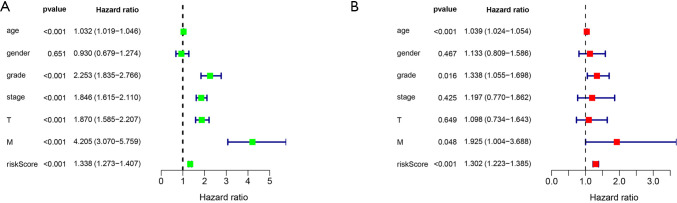
Univariate and multivariate Cox regression analyses of risk score and clinical features indicated that risk score was an independent prognostic factor in patients with ccRCC (P<0.001, Figure 5A,B).
Figure 5.
Prognostic analysis of signature risk score and clinical features. Forest map of univariate Cox regression analysis of risk score and clinical features (A); forest map of multivariate Cox regression analysis of risk score and clinical features (B).
Relationship between risk score and immune cells
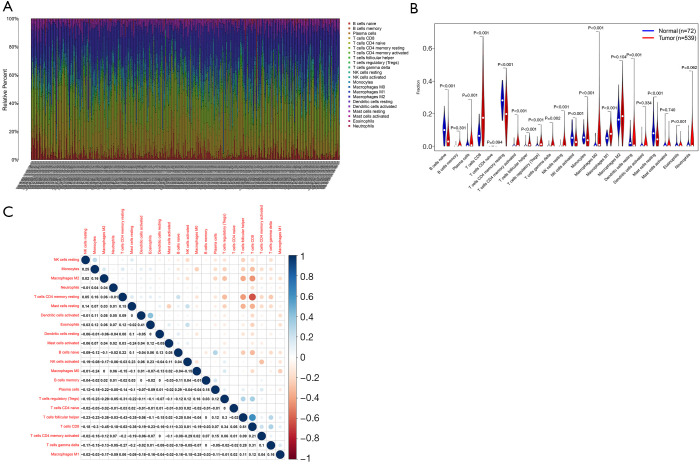
We used the CIBERSORT algorithm to calculate the relative abundance of 22 types of immune cells in 539 tumor samples and 72 normal samples in the TCGA-ccRCC cohort. The histogram in Figure 6A shows the relative abundance of the 22 immune cell types in all samples. Differential analysis indicated that resting dendritic cells, resting mast cells, and resting memory CD4 T cells showed low infiltration in ccRCC tumor tissue, while M0 macrophages, plasma cells, follicular helper T cells, and regulatory T cells (Tregs) exhibited high infiltration in ccRCC tumor tissues (Figure 6B). Correlation analysis results of the 22 types of immune cells revealed that CD8 T cells had the strongest negative correlation with resting memory CD4 T cells (R=–6.3) and the strongest positive correlation with follicular helper T cells (R=0.61) (Figure 6C).
Figure 6.
Relationship between risk score and immune cells. The relative abundance of 22 types of immune cells in the TCGA-CCRCC cohort: the X-axis represents individual samples, and the Y-axis represents the relative abundance of immune cells; individual colors represent individual immune cells (A). Violin plot of the expression of the 22 types of immune cells in tumor tissue and normal tissue in the TCGA-CCRCC cohort: X-axis represents 22 types of immune cells, and the Y-axis represents the relative abundance of immune cells (B). Heatmap of the correlation between 22 types of immune cells: blue and red represent positive and negative correlations, respectively (C). TCGA, The Cancer Genome Atlas; CCRCC, kidney renal clear cell carcinoma.
Correlation between the relative abundance of immune cells and OS in patients with ccRCC
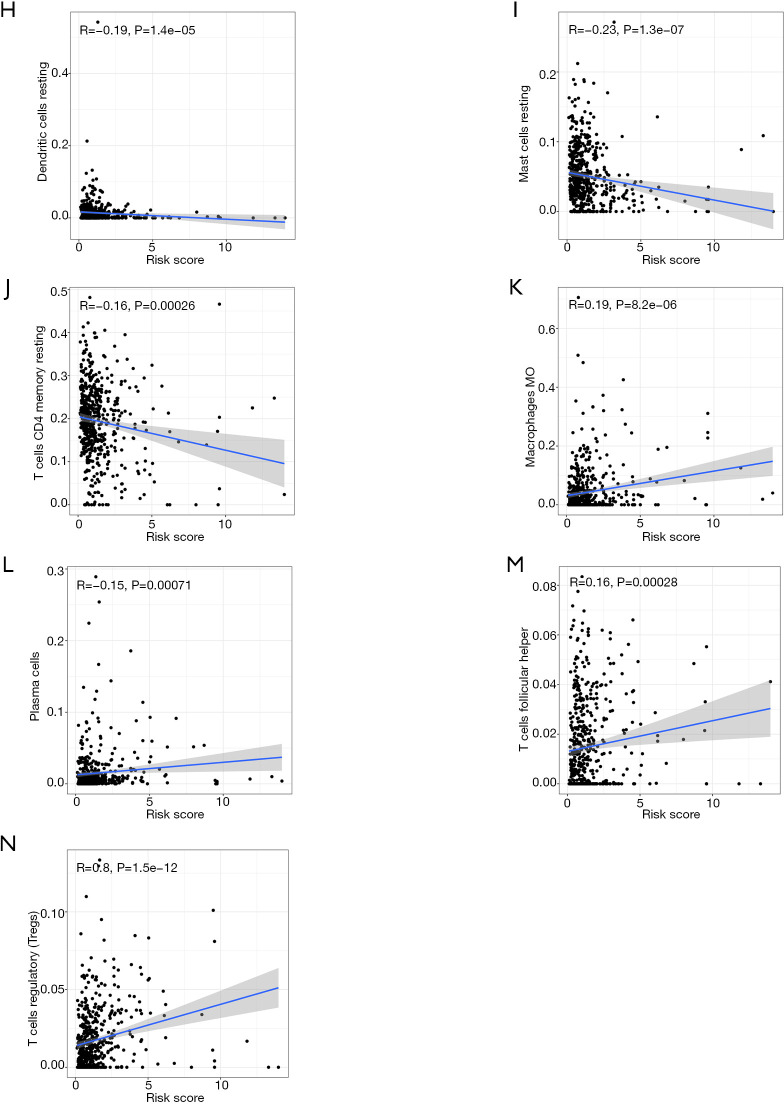
The results of Kaplan-Meier survival analysis demonstrated that patients with a high abundance of resting dendritic cells, resting mast cells, and resting memory CD4 T cells had better OS than those with low abundances of these cells (Figure 7A,B,C). Meanwhile, a high abundance of M0 macrophages, plasma cells, follicular helper T cells, and Tregs were associated with reduced OS in patients with CCRCC (Figure 7D,E,F,G). Correlation analysis results indicated that risk score was negatively correlated with resting dendritic cells, resting mast cells, and resting memory CD4 T cells (Figure 7H,I,J), but was positively correlated with M0 macrophages, plasma cells, follicular helper T cells, and Tregs (Figure 7K,L,M,N).
Figure 7.
Immune cells that affect the OS of patients with CCRCC, and their correlations with risk score. Kaplan-Meier survival analysis of resting dendritic cells (A), resting mast cells (B) and resting memory CD4 T cells (C), M0 macrophages (D), plasma cells (E), follicular helper T cells (F), and Tregs (G). Correlation analysis of risk score and resting dendritic cells (H), resting mast cells (I), resting memory CD4 T cells (J), M0 macrophages (K), plasma cells (L), follicular helper T cells (M), and Tregs (N). OS, overall survival; CCRCC, kidney renal clear cell carcinoma; Tregs, regulatory T cells.
Results of risk score and drug sensitivity analysis
Finally, we analyzed the half maximal inhibitory concentration (IC50) of some chemotherapeutic drugs commonly used to treat ccRCC, using the R package pRRophetic. The results revealed that sorafenib (P=0.0086, Figure 8A), sunitinib (P=9.2e–08, Figure 8B), temsirolimus (P=0.00017, Figure 8C), cisplatin (P=0.00089, Figure 8D), and gemcitabine (P=0.026, Figure 8E) had lower IC50 values in the high-risk ccRCC population, which suggested that these drugs have better efficacy in high-risk patients.
Figure 8.
Relationship between risk scores and drug sensitivity. Sorafenib (A), sunitinib (B), temsirolimus (C), cisplatin (D), and gemcitabine (E).
Discussion
With the advances in sequencing methods, coding RNA and non-coding RNA can now be detected in normal and cancer tissue samples, and numerous differentially expressed RNAs have been discovered to date. Studies have proved that coding RNAs and non-coding RNAs can serve as diagnostic and prognostics biomarkers for multiple cancer types, including liver, kidney, pancreatic, cervical, and esophageal cancer (21-24). In almost all existing studies, the researchers screened the biomarkers based on transcriptional expression. It has been proved that immune-related gene pairing can serve as a reasonable model for the diagnosis and prognostic prediction of cancer. Herein, we investigated whether a novel model that incorporated pairs of irlncRNAs, and for which quantification of irlncRNA transcriptional expression was not necessary, could act as a reasonable prognostic and diagnostic model for patients with ccRCC.
First, the raw data of patients with ccRCC were downloaded from TCGA, and confirmed immune-related genes were obtained from the ImmPort database. Next, we investigated the correlation between immune-related genes and irlncRNAs, and identified the differentially expressed irlncRNAs between normal and cancer tissue samples. Then, a novel model was constructed with the differentially expressed ILRPs. Since the lncRNAs were irlncRNAs, which may play a key role in immune cell infiltration of the tumor microenvironment (TME), we further explored whether the ILRP signature risk score had a significant influence on immune cell infiltration. Finally, we performed a drug sensitivity analysis based on risk score.
In recent decades, as more knowledge of the TME has been acquired, analysis of the TME has mainly been focused on tumor cells, immune cells, and stromal cells. Among of these contents of the TME, immune cells act as the main significant regulators of progression in multiple cancers (25). Immune cell infiltration of the TME can be influenced by several factors, including immune-related genes, irlncRNAs, tumor cells, and stromal ingredients. Among these factors, immune-related genes may be the most significant regulators, and they are regulated by a number of irlncRNAs. Herein, we supposed that irlncRNAs may influence immune cell infiltration via the regulation of immune-related genes. A recent study revealed that irlncRNAs can act as diagnostic or prognostic biomarkers for multiple cancer types; however, that study was based on the transcriptional expression levels of irlncRNAs (26). According to our study, we identified 12 ILRPs that significantly influence the prognosis of patients with ccRCC. The 12 ILRPs included AL031123.1|AC103563.7, AL031710.1|AC104984.5, AL031710.1|AC136475.3, AP000757.1|HOXB-AS3, SEMA3B-AS1|LINC00462, AL031429.2|AC008760.2, ZNF710-AS1|AC087482.1, LINC02061|LINC01077, EMX2OS|SLC9A3-AS1, EMX2OS|AL096799.1, AC091563.1|AC136475.3, and AC008760.2|AL096799.1. The ILRPs that served as biomarkers of a favorable prognosis were AL031123.1|AC103563.7, AL031710.1|AC104984.5, AL031710.1|AC136475.3, AP000757.1|HOXB-AS3, LINC02061|LINC01077, EMX2OS|SLC9A3-AS1, EMX2OS|AL096799.1, AC091563.1|AC136475.3, AL031429.2|AC008760.2, and AC008760.2|AL096799.1. Meanwhile, SEMA3B-AS1|LINC00462 and AC008760.2|AL096799.1 acted as biomarkers of an adverse prognosis of ccRCC.
Among these selected ILRPs, the roles of several irlncRNAs have previously been investigated in some cancers. Ye et al. indicated that AC136475.3 can serve as a prognostic biomarker for patients with HCC who have cirrhosis (27). HOXB-AS3 has a dual role in various cancers; in colon cancer, it acts as a tumor suppressor, but in lung cancer, it acts as a tumor promoter (28,29). Zhong et al. found that downregulation of lnc-EMA3B-AS1 may lead to a poor outcome in patients with HCC (30). LINC00462 enhanced the invasion ability of pancreatic cancer cells by regulating the miR-665/TGFBR1-TGFBR2/SMAD2/3 signaling pathway (31), which indicated that LINC00462 serves as a tumor promoter in pancreatic cancer. Jiang et al. revealed that the downexpression of lnc-EMX2OS led to a poor outcome in patients with ccRCC (32). On the basis of these findings, we can conclude that these ILRPs have a significant role in tumorigenesis and therapeutic response of ccRCC. Regrettably, only the role of lnc-EMX2OS has been explored in ccRCC so far. Except for exploring the prognostic role of these ILRPs in ccRCC, we estimated the risk score and explored whether it could be reasonably and independently used to predict the prognosis of ccRCC. The results showed that the risk score based on the ILRPs was an independent factor for predicting the prognoses of patients with ccRCC. Also, the risk score had a significant association with tumor grade, primary lesion, and metastasis, with a high-risk score indicating a poor prognosis for patients with ccRCC.
We further explored the relationship between the 12 ILRPs and immune cell infiltration in ccRCC samples. The results indicated that ccRCC samples exhibited significantly lower infiltration of resting dendritic cells, resting mast cells, and resting memory CD4 T cells, but significantly higher infiltration of M0 macrophages, plasma cells, follicular helper T cells, and Tregs, than normal samples. Furthermore, a high abundance of resting dendritic cells, resting mast cells, and resting memory CD4 T cells indicated a better prognosis in patients with ccRCC. However, a high abundance of M0 macrophages, plasma cells, follicular helper T cells, and Tregs shortened the OS of patients with ccRCC. Interestingly, Sammarco et al. showed that mast cells performed a protumorigenic role in gastric cancer by enhancing angiogenesis and lymphangiogenesis (33). Also, Derakhshani et al.’s study indicated that mast cells have a dual role (promoter vs. suppressor) in different cancers (34). Xiong et al. revealed that mast cells inhibit anticancer immunity via secreting interleukin-10 and transforming growth factor-β (35). Tregs have also been explored in multiple cancer types (36,37). These findings indicate that irlncRNAs can influence the prognosis of patients with ccRCC by regulating immune cell infiltration of the TME.
Despite the advance of therapeutic regimens for ccRCC, it is still incurable. In this study, we investigated the drug sensitivity of ccRCC based on patient risk score. The results showed that ccRCC patients with a high-risk score are sensitive to sorafenib, sunitinib, temsirolimus, cisplatin, and gemcitabine. Sorafenib and sunitinib are effective therapeutic regimens that have been put into practice in the treatment of kidney cancer for many years (38-40). However, in some cases of kidney cancer, resistance to sunitinib or sorafenib can develop (41,42). Temsirolimus has also been used as kidney cancer therapy. Payton’s study showed that temsirolimus did not achieve a good therapeutic response in metastatic renal cell carcinoma (43), whereas in another study, Hudes et al. showed that it could significantly improve the prognosis of patients with metastatic renal cell carcinoma (44). These conflicting results suggest that further research is needed to explore the effects of temsirolimus on metastatic renal cell carcinoma. Gemcitabine plus cisplatin has been researched in advanced urothelial cancer (45); however, no study has explored the efficacy and safety of this regimen in ccRCC. One study showed that the responses of kidney cancer cells to gemcitabine differed (46). By selecting therapeutic regimens based on patient risk score, we can overcome drug resistance and ensure efficacy.
Conclusions
Our study has constructed a novel and reasonable ILRP model that can be used to diagnose and predict the prognosis of patients with ccRCC without the need to perform transcriptional level quantification. This model may aid clinicians in the selection of more effective therapeutic regimens for patients with ccRCC based on the risk score.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-445
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-445). The authors have no conflicts of interest to declare.
(English Language Editor: J. Reynolds)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005;353:2477-90. 10.1056/NEJMra043172 [DOI] [PubMed] [Google Scholar]
- 3.Lam JS, Belldegrun AS, Pantuck AJ. Long-term outcomes of the surgical management of renal cell carcinoma. World J Urol 2006;24:255-66. 10.1007/s00345-006-0055-5 [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225-33. 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelin WG, Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 2008;8:865-73. 10.1038/nrc2502 [DOI] [PubMed] [Google Scholar]
- 6.D'Avella C, Abbosh P, Pal SK, et al. Mutations in renal cell carcinoma. Urol Oncol 2020;38:763-73. 10.1016/j.urolonc.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 7.Li J, Li Z, Zheng W, et al. LncRNA-ATB: An indispensable cancer-related long noncoding RNA. Cell Prolif 2017;50:e12381. 10.1111/cpr.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta 2018;485:229-33. 10.1016/j.cca.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 10.Qu L, Wang ZL, Chen Q, et al. Prognostic value of a long non-coding RNA signature in localized clear cell renal cell carcinoma. Eur Urol 2018;74:756-63. 10.1016/j.eururo.2018.07.032 [DOI] [PubMed] [Google Scholar]
- 11.Ye Z, Duan J, Wang L, et al. LncRNA-LET inhibits cell growth of clear cell renal cell carcinoma by regulating miR-373-3p. Cancer Cell Int 2019;19:311. 10.1186/s12935-019-1008-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer 2017;16:111. 10.1186/s12943-017-0681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Li W, Yan Y, et al. LINC01094 triggers radio-resistance in clear cell renal cell carcinoma via miR-577/CHEK2/FOXM1 axis. Cancer Cell Int 2020;20:274. 10.1186/s12935-020-01306-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchatskiĭ LP, Litvinov GS, Podrezova LM, et al. Effect of physical and chemical factors on the iridovirus of the mosquito Aedes cantans. Mikrobiol Zh 1986;48:55-8. [PubMed] [Google Scholar]
- 15.Hong W, Liang L, Gu Y, et al. Immune-related lncRNA to construct novel signature and predict the immune landscape of human hepatocellular carcinoma. Mol Ther Nucleic Acids 2020;22:937-47. 10.1016/j.omtn.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Zhang Z, Zhao H, et al. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol 2018;55:3684-97. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Li X, Zhang C, et al. LINC00973 is involved in cancer immune suppression through positive regulation of Siglec-15 in clear-cell renal cell carcinoma. Cancer Sci 2020;111:3693-704. 10.1111/cas.14611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Gou X, Wei Z, et al. Bioinformatics profiling integrating a three immune-related long non-coding RNA signature as a prognostic model for clear cell renal cell carcinoma. Cancer Cell Int 2020;20:166. 10.1186/s12935-020-01242-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. 10.1371/journal.pone.0107468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Xu T, Li S, et al. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY) 2019;11:12165-76. 10.18632/aging.102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Z, Jiao Y, Li Y, et al. TYMS presents a novel biomarker for diagnosis and prognosis in patients with pancreatic cancer. Medicine (Baltimore) 2019;98:e18487. 10.1097/MD.0000000000018487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao Y, Li Y, Fu Z, et al. OGDHL Expression as a prognostic biomarker for liver cancer patients. Dis Markers 2019;2019:9037131. 10.1155/2019/9037131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Zhang Q, Lou Q, et al. Differential analysis of lncRNA, miRNA and mRNA expression profiles and the prognostic value of lncRNA in esophageal cancer. Pathol Oncol Res 2020;26:1029-39. 10.1007/s12253-019-00655-8 [DOI] [PubMed] [Google Scholar]
- 25.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014-22. 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun 2020;11:1000. 10.1038/s41467-020-14802-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J, Li H, Wei J, et al. Risk scoring system based on lncRNA expression for predicting survival in hepatocellular carcinoma with cirrhosis. Asian Pac J Cancer Prev 2020;21:1787-95. 10.31557/APJCP.2020.21.6.1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang JZ, Chen M, Chen, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 2017;68:171-84.e6. 10.1016/j.molcel.2017.09.015 [DOI] [PubMed] [Google Scholar]
- 29.Jiang W, Kai J, Li D, et al. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J Cell Physiol 2020;235:7194-203. 10.1002/jcp.29618 [DOI] [PubMed] [Google Scholar]
- 30.Zhong Y, Li Y, Song T, et al. MiR-718 mediates the indirect interaction between lncRNA SEMA3B-AS1 and PTEN to regulate the proliferation of hepatocellular carcinoma cells. Physiol Genomics 2019;51:500-5. 10.1152/physiolgenomics.00019.2019 [DOI] [PubMed] [Google Scholar]
- 31.Zhou B, Guo W, Sun C, et al. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis 2018;9:706. 10.1038/s41419-018-0724-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang H, Chen H, Wan P, et al. Downregulation of enhancer RNA EMX2OS is associated with poor prognosis in kidney renal clear cell carcinoma. Aging (Albany NY) 2020;12:25865-77. 10.18632/aging.202151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sammarco G, Varricchi G, Ferraro V, et al. Mast cells, angiogenesis and lymphangiogenesis in human gastric cancer. Int J Mol Sci 2019;20:2106. 10.3390/ijms20092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derakhshani A, Vahidian F, Alihasanzadeh M, et al. Mast cells: a double-edged sword in cancer. Immunol Lett 2019;209:28-35. 10.1016/j.imlet.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y, Liu L, Xia Y, et al. Tumor infiltrating mast cells determine oncogenic HIF-2alpha-conferred immune evasion in clear cell renal cell carcinoma. Cancer Immunol Immunother 2019;68:731-41. 10.1007/s00262-019-02314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol 2014;27:1-7. 10.1016/j.coi.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 37.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res 2017;27:109-18. 10.1038/cr.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nassif E, Thibault C, Vano Y, et al. Sunitinib in kidney cancer: 10 years of experience and development. Expert Rev Anticancer Ther 2017;17:129-42. 10.1080/14737140.2017.1272415 [DOI] [PubMed] [Google Scholar]
- 39.Stone L. Kidney cancer: No advantage of adjuvant sunitinib or sorafenib. Nat Rev Urol 2016;13:240-1. 10.1038/nrurol.2016.63 [DOI] [PubMed] [Google Scholar]
- 40.Tatsugami K, Oya M, Kabu K, et al. Evaluation of efficacy and safety of sorafenib in kidney cancer patients aged 75 years and older: a propensity score-matched analysis. Br J Cancer 2018;119:241-7. 10.1038/s41416-018-0129-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishibashi K, Koguchi T, Matsuoka K, et al. Interleukin-6 induces drug resistance in renal cell carcinoma. Fukushima J Med Sci 2018;64:103-10. 10.5387/fms.2018-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016;29:653-68. 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 43.Payton S. Kidney cancer: Temsirolimus fails to expand its role in patients with mRCC. Nat Rev Urol 2014;11:2. 10.1038/nrurol.2013.308 [DOI] [PubMed] [Google Scholar]
- 44.Hudes G, Carducci M, Tomczak P, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 2007;356:2271-81. 10.1056/NEJMoa066838 [DOI] [PubMed] [Google Scholar]
- 45.Izumi K, Iwamoto H, Yaegashi H, et al. Gemcitabine plus cisplatin split versus gemcitabine plus carboplatin for advanced urothelial cancer with cisplatin-unfit renal function. In Vivo 2019;33:167-72. 10.21873/invivo.11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papadopoulos EI, Yousef GM, Scorilas A. Gemcitabine impacts differentially on bladder and kidney cancer cells: distinct modulations in the expression patterns of apoptosis-related microRNAs and BCL2 family genes. Tumour Biol 2015;36:3197-207. 10.1007/s13277-014-2190-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as