SUMMARY
The phenotypic effect of perturbing a gene’s activity depends on the activity level of other genes, reflecting the notion that phenotypes are emergent properties of a network of functionally interacting genes. In the context of cancer, contemporary investigations have primarily focused on just one type of functional relationship between two genes—synthetic lethality (SL). Here, we define the more general concept of “survival-associated pairwise gene expression states” (SPAGEs) as gene pairs whose joint expression levels are associated with survival. We describe a data-driven approach called SPAGE-finder that when applied to The Cancer Genome Atlas (TCGA) data identified 71,946 SPAGEs spanning 12 distinct types, only a minority of which are SLs. The detected SPAGEs explain cancer driver genes’ tissue specificity and differences in patients’ response to drugs and stratify breast cancer tumors into refined subtypes. These results expand the scope of cancer SPAGEs and lay a conceptual basis for future studies of SPAGEs and their translational applications.
INTRODUCTION
Cellular functions are mediated by functionally interacting networks of genes. Functional relationships between pairs of genes (x, y) in which the phenotypic effects of gene x’s activity are modified by the activity of gene y, are thus a key to understanding complex diseases, including cancer, which involves an interplay among a myriad of genes (Ashworth et al., 2011; Jerby-Arnon et al., 2014; Kelley and Ideker, 2005; Lu et al., 2013; Wong et al., 2004; Zhong and Sternberg, 2006). For instance, synthetic lethality (SL) is the most studied type of functional interaction between a pair of genes where joint inactivity, or low expression, of two genes, but not inactivity of individual genes, is associated with lower tumor fitness and, consequently, better cancer patient survival (O’Neil et al., 2017). Flere, we generalize this concept as “survival-associated pairwise gene expression states” (SPAGEs), whereby specific joint expression levels of a pair of genes as one entity is associated with cancer survival. SPAGEs are of particular interest in cancer because the dependence of one gene’s phenotypic effect on the activity of another gene provides opportunities for selective killing of cancer cells (Kaelin, 2005) and the functional partners of a drug’s target gene can buffer its effect leading to resistance (Fong et al., 2015; Miyamoto et al., 2015). In the cancer genomics literature, such inferred functional interactions between genes have been referred to as genetic interactions (GIs) (Boucher and Jenna, 2013; Lee et al., 2010; Wong et al., 2004) or epistasis (Matlak and Szczurek, 2017). However, the terms genetic interaction and epistasis have a specific meaning in the genetics literature, and here we avoid them.
In cancer genomics, three types of SPAGEs have been studied so far showing major roles in disease progression and patient survival and suggesting novel therapeutic avenues. The vast majority of functional gene pair studies in cancer to date have focused on SL gene pairs, describing the relationship between two genes whose individual inactivation results in a viable phenotype, while their combined inactivation is lethal to the cell (Ashworth et al., 2011; Miyamoto et al., 2015; Sajesh et al., 2013; Stuhlmiller et al., 2015). They provide selective treatment opportunities by drugs that inhibit an SL partner of a gene that is specifically inactivated or lost in a given tumor, thus selectively killing the tumor cells (Ashworth et al., 2011; Jerby-Arnon et al., 2014; Kroll et al., 1996). Another related class of SPAGEs are synthetic dosage lethal (SDL) interactions, where the underactivity of one gene together with the overactivity of another gene is lethal, but neither event alone is lethal (Megchelenbrink et al., 2015; Stuhlmiller et al., 2015; Szappanos et al., 2011). SDLs are promising for oncogenes, many of which are difficult to target directly, by targeting their SDL partners (Weinstein et al., 2013; Luo et al., 2009a; Rathert et al., 2015). A third class of SPAGEs are synthetic rescues (SRs), where a change in the activity of one gene is lethal to the cell but an alteration in its SR partner “rescues” cell viability. SRs may play a key role in tumor relapse and emergence of resistance to therapy (Brough et al., 2011; Hartwell et al., 1997; McLornan et al., 2014; Sahu et al., 2019). Indeed, previous investigations have shown that the overall numbers of functionally active SLs and SDLs in a given tumor sample are highly predictive of patient survival (Megchelenbrink et al., 2015). These three interaction types, however, represent just the “tip of the iceberg,” as there are many additional types of SPAGEs that can be defined at a conceptual level and whose systematic exploration may have important functional ramifications for cancer therapy.
In our previous works, we have established the efficacy of unbiased data-driven computational approaches to predict SL and SR gene pairs (Lee et al., 2018; Sahu et al., 2019). Instead of defining functional gene interactions based on gene expression levels, one could use mutation data to identify functionally interacting gene pairs, as was done previously in CoMEt (Leiserson et al., 2015) and SurvLRT (Matlak and Szczurek, 2017). CoMEt identifies functionally interacting gene pairs based on mutual exclusivity of their mutation status, while SurvLRT identifies gene pairs based on associations between mutation status and survival.
Here we present a data-driven computational pipeline called SPAGE-finder. We applied SPAGE-finder to analyze 5,157 The Cancer Genome Atlas (TCGA) samples (STAR Methods) of 18 different cancer types, identifying SPAGEs of 12 distinct types that are significantly associated with clinical outcome. Using drug response data from TCGA and molecular drug target information, we show that the detected SPAGEs are associated with response to therapy by specific drugs. Their activation patterns can account for the tissue specificity of known driver genes and stratify breast cancer into clinically relevant subtypes. In sum, SPAGE-finder substantively expands the current knowledge of functionally related gene pairs in cancer, laying a strong conceptual and computational foundation for future studies of additional types of SPAGEs.
RESULTS
Overview of the SPAGE-Finder Pipeline
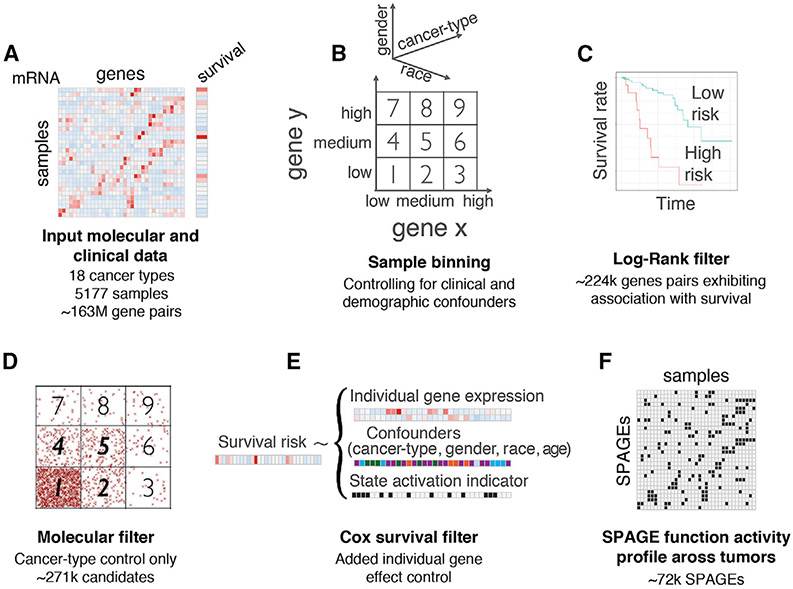
The overall SPAGE-finder pipeline is summarized in Figure 1, and the details are provided in STAR Methods. Given a large set of tumor transcriptomes (Figure 1A), following previous data-driven approaches to identify SLs (Lee et al., 2018) and SRs (Sahu et al., 2019), we used a quantile-based partition approach to bin gene expression into discrete states relative to all tumor samples. However, extending previous methods, we divided gene expression into three states (low, medium, and high), instead of only low and high states, allowing us to explore dosage-sensitive relationships between genes. Notably, we accounted for differences in expression patterns across clinical and demographic cohorts by performing the binning in a stratified manner (STAR Methods).
Figure 1. Overview of the SPAGE-Finder Pipeline.
(A and B) Given a large set of tumor transcriptomes (A), we first partition the expression level of each gene into low, medium, and high activity state, resulting in 9 joint activity state bins for any two genes (B). Each combination of a gene pair and bin b induces a bipartition of the set of tumor samples based on whether the co-activity levels of the gene pair in a specific tumor is in bin b.
(C) The first step of SPAGE-finder screens for the gene pairs that show distinct survival trends in the two sets of tumors in any of the bins, based on log rank test.
(D) Next, for a gene pair and a bin identified in (C), we test whether the putative gene interaction in bin b has a differential effect on tumor fitness, by testing for depletion or enrichment of samples in the bin b relative to expectation based on individual genes.
(E) Finally, for each retained gene pair, in each of the 9 bins separately, we fit a Cox proportional hazards model to assess whether being in a particular bin is associated with a distinct (positive or negative) pattern of patient survival, followed by correction for multiple hypotheses testing.
(F) The output of SPAGE-finder is (1) a list of SPAGEs of each of the 12 types studied and (2) SPAGE profile in each of the individual tumor samples, defined as activity state of each SPAGE in the tumor sample.
Thus, for a pair of genes, there are 9 = 3 × 3 combinations, or bins, of possible co-activity states for the two genes (Figure 1B). For a given ordered pair of genes, each tumor sample maps to exactly one of the 9 bins. Our goal is to identify SPAGE pairs of the form (x, y, b, ± α) such that for the specific gene pair (x, y), the tumors in which the joint activity of (x, y) maps to bin b have a significant fitness advantage (+) or disadvantage (−) with effect size α, relative to all other tumors whose activity of (x, y) maps to a different bin. The effect size α is estimated by measuring the difference in the survival curves between those patients where the activity of (x, y) is in bin b in their tumors and those where it is not, as depicted in Figure 1C; note that for most gene pairs, there may not be any bin exhibiting a significant fitness differential. A significant SPAGE (x, y, b, ± α) is termed “functionally active” in a particular tumor if the co-activity states of (x, y) in that tumor fall in bin b. We hypothesized that the patients whose tumors have a larger number of functionally active interactions with negative tumor fitness effects will have a better prognosis and, conversely, the patients whose tumors have a larger number of functionally active interactions with positive tumor fitness effects will have a poorer prognosis.
We analyzed 5,157 TCGA samples for 18 cancer types (see STAR Methods). First, as an initial screening, we performed a log rank survival test (depicted in Figure 1C) for each gene pair in each of the 9 bins. To make this computationally feasible and to limit the burden of multiple testing correction in the subsequent steps, we used an extremely stringent cutoff for the log rank test leading to the retention of about 1/1,000 gene pairs surveyed (STAR Methods), resulting in 223,946 gene pairs that exhibit a significant association with survival in one of the 9 bins. Second, if a potential SPAGE in bin b has a differential effect on tumor fitness, we expect the number of tumors that map to bin b to be relatively enriched (for a “+” relationship positively affecting tumor survival) or depleted (for a “−” relationship negatively affecting tumor survival). Thus, we applied an additional filter (Figure 1D) to retain the SPAGEs exhibiting a consistent patient survival and tumor fitness enrichment or depletion statistic (STAR Methods), yielding 179,444 gene pairs. Third, for each retained gene pair, in each of the 9 bins, we implemented a Cox proportional hazards model, specifically controlling for age, cancer type, sex, and race, to assess whether a tumor being in a particular bin is associated with patient survival, either positively or negatively (Figure 1E; STAR Methods). Finally, we applied an empirical false discovery rate (FDR) correction based on the significance of the Cox interaction term of the 179,444 gene pairs relative to those obtained for randomly shuffled gene pairing as the null control. At FDR < 1 %, this resulted in 71,946 predicted SPAGEs across the 9 bins, of the form(x, y, b, ± α), which form the final set of TCGA inferred SPAGEs (Figure 1E). Considering the symmetry among bins (bin 2–bin 4 corresponding to low-medium expression interaction; bin 3–bin 7 corresponding to low-high expression interaction; bin 6–bin 8 corresponding to medium-high expression interaction), there are 6 unique types of interaction bins, and considering the two directions of the effect size yields a total of 12 basic types of SPAGEs. We ascertained the robustness of the pipeline to changes in the quantile boundaries for the 3 × 3 bins and to changes in the log rank and FDR thresholds (STAR Methods). By perturbing the binning thresholds and inducing substantial changes to the bin size and composition, we were able to demonstrate that the vast majority (57%–96%) of the gene pairs are recapitulated across all log rank threshold perturbations (STAR Methods). Additionally, we quantified the extent to which the binning strategy may partition multimodal expression distributions unreasonably, depending on where the modes are relative to the two partition values. The dip test for multimodality (a.k.a. Hartigan’s dip statistic [HDS]) (Freeman and Dale, 2013) applied on each gene in each stratum indicated that the percentage of genes with significant evidence for multimodality in the majority of strata was a reassuringly low 7.6% (dip p value < 0.05).
The Landscape of Different SPAGE Types
SPAGE-finder identified 71,946 SPAGEs of 12 different types that are significantly associated with clinical outcome, accounting for ~0.02% of all the possible candidate gene pairs and SPAGE types tested (Table S1). Considering the expectation that neighboring genes in the protein interaction network (PIN) are more likely to be involved in a SPAGE (Schaefer et al., 2012), to obtain a smaller but more biologically grounded PIN-supported SPAGE network, we retained only the gene pairs that are separated by two or fewer edges in the human PIN. This PIN-supported SPAGE network was composed of 1,704 SPAGEs involving 1,786 genes (Table S1), which included 133 known cancer genes (Cosmic dataset; Futreal et al., 2004) associated with various cancer types (enrichment p = 2.5 × 10−22) and 50 breast-cancer-specific (Intogen dataset; Gonzalez-Perez et al., 2013) driver genes (enrichment p = 7.7 × 10−13).
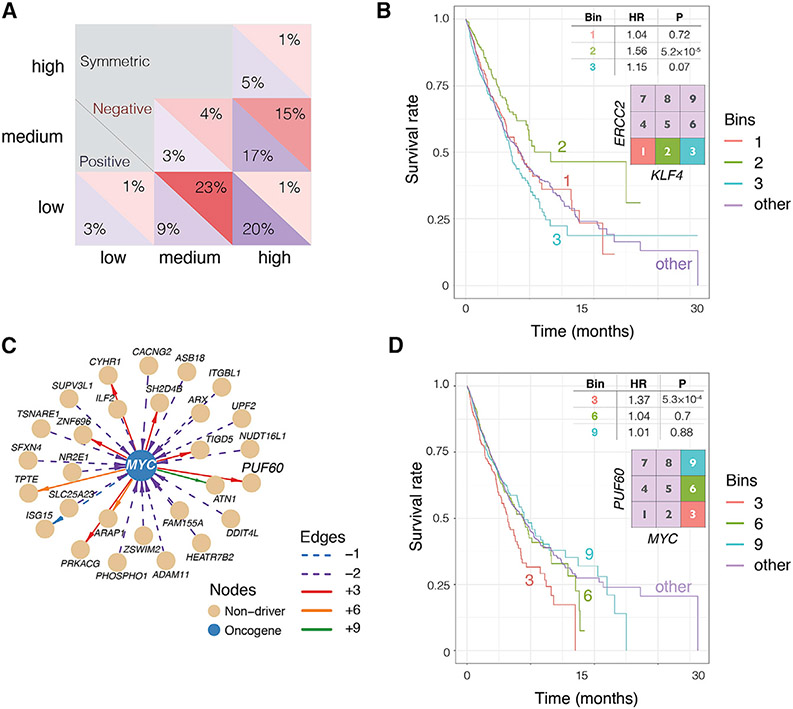
The distribution of the detected 1,704 PIN-supported SPAGEs across the 12 SPAGE types reveals that previously characterized interactions may represent only a small fraction of the overall interaction landscape (Figure 2A). SL interactions are surprisingly one of the least abundant types of identified SPAGEs, and so are SDLs (1% of all SPAGEs). Remarkably, the positive “anti-symmetric” type of SLs, in which the joint low activity of the two interacting genes is associated with a higher tumor fitness, is three times more abundant than SLs. The interaction between the Cosmic Cancer Census genes GNAQ and JAK2 is one example of such a positive interaction in bin 1 (Figure S1A). GNAQ, encoding Gq, and JAK2 are both downstream targets in a signaling pathway with several functions pertinent to cancer, including endothelial cell maintenance and vascular remodeling (Kawai et al., 2017). Interestingly, the two most abundant types of pan-cancer SPAGEs correspond to bin 2 and bin 6, where one of the genes has medium level of activity and only the extreme activity of its partner gene reveals a phenotypic effect. For most SPAGE bins, we see a higher proportion of SPAGEs exerting a positive effect on tumor fitness, consistent with the hypothesis that the SPAGEs uncovered during the evolution of cancer are under positive selection.
Figure 2. Broad Distribution and Characteristics of the Detected SPAGEs and Context-Specific Effect of Cancer Driver Genes on Survival.
(A) Distribution of the 1,704 significant PIN-supported SPAGEs across 9 joint activity bins. The fractions of SPAGEs in each bin are shown for SPAGEs with positive (blue) and negative (red) effect on tumor fitness. Only the data in the lower triangle of the matrix are shown as the SPAGEs are symmetric relative to the genes in a pair.
(B) The Kaplan-Meier (KM) survival curve of SPAGE involving ERCC2, a transcription-coupled DNA excision repair gene known to be a breast cancer tumor suppressor, and KLF4, a zinc-finger transcription factor known to be oncogenic in breast cancer, reveals increasingly poor survival by overactivation of the oncogene and underactivation of the tumor suppressor (bin 3). Strikingly, the effect of ERCC2 inactivity on survival is reversed when KLF4 has medium activity level (bin 2).
(C) The predicted SPAGEs involving the oncogene MYC.
(D) KM survival curve of SPAGE involving MYC and its regulator PUF60. High expression of MYC is associated with poor prognosis, specifically at low activation of PUF60.
We find that the distributions of various SPAGE types among the PIN-supported SPAGEs in Figure 2A are similar to those for the full 71,946 SPAGE network (Figure S1B), suggesting that the PIN-supported SPAGEs are not skewed either by the PIN curation process or by biological network structures (Kelley and Ideker, 2005). Additionally, we ascertained that the inferred SPAGEs are not dominated by correlated gene expression patterns (STAR Methods; Figure S1C).
Cancer genes that encode transcription factors, such as MYC and KLF4, have proven difficult to target directly (Lambert et al., 2018; Li et al., 2018). One important application of SPAGE-finder is to identify candidate interaction partners of the difficult-to-target cancer genes for indirect interventions. To assess this capability, we identified the SPAGE partners of several cancer genes using target-specific FDR (STAR Methods). Figure 2B shows survival patterns for different activity state combinations of breast tumor suppressor ERCC2, a transcription-coupled DNA excision repair gene (Benhamou and Sarasin, 2002; Bernard-Gallon et al., 2008), and a breast cancer oncogene KLF4, a zinc-finger transcription factor (Akaogi et al., 2009). It reveals two interesting trends: as expected, the overactivation of the oncogene and underactivation of the tumor suppressor (bin 3) results in poorer patient survival than expected from the individual gene effects (bins, 1, 2, 6, and 9). However, surprisingly, the survival curve reveals a reversal of the effect of the tumor suppressor ERCC2 inactivity on survival when the oncogene KLF4 has medium activity (bin 2), whose individual activity is associated with better survival; the (ERCC2, KLF4) interaction exemplifies the relevance of medium expression bins in this study. This and several other examples of SPAGEs involving a cancer driver gene demonstrate that the context-specific effects of driver genes may show very different trends than their previously established effects as individual genes. Figure S1D shows the pairs within the extended SPAGE network (71,946 SPAGEs prior to PIN filtering) that involve at least one of the Cosmic or Intogen driver genes. In addition, and consistent with MYC’s role as an oncogene, the SPAGEs occurring when MYC has low activity mostly have a negative effect on tumor fitness. However, when MYC is activated, our results suggest that the effect on tumor fitness may be significantly dependent on the activity level of another gene. For example, we find that in the high MYC state, the low expression of PUF60, one of the known regulators of MYC (Matsushita et al., 2014; Rahmutulla et al., 2014), is associated with higher tumor fitness (type +3 SPAGE, hazard ratio [HR] = 1.37, p = 5.0 × 10−4) (Figure 2D, bin 3). In contrast, we find that high expression of MYC does not significantly contribute to poorer prognosis when PUF60 is expressed at medium or high levels (p = 0.9; Figure 2D, bins 6 and 9). This example underscores the importance of molecular context in designing trials to test possible anti-MYC treatments; expression levels of other genes such as PUF60 can determine the extent to which patients with overexpressed MYC will respond to downregulation of MYC.
Exemplifying the putative role of medium bins in dosage-sensitive gene functions, such as targets of morphogens (Rogers and Schier, 2011) or stoichiometric relationships among the genes in a regulatory complex (Birchler et al., 2001; Veitia and Potier, 2015), we identified evidence for dosage-sensitive effects of multiple genes involved in SPAGEs. We curated 15 dosage-sensitive genes with literature evidence for both haploinsufficiency and triplosensitivity, suggesting that either deletion or addition of a copy would have a phenotypic effect. We find that 10 of these 15 genes are involved in SPAGEs with medium expression level. One of these genes, EFNB1 (coding Ephrin B1) is associated with increased migration and invasion capacity in cancer by interacting with RhoGDI1 and CNK1 (Cho et al., 2014, 2018). Consistent with the expectation for dosage-specific effects and association of EFNB1high expression with increased survival risk, we find 5 negative interactions involving the EFNB1medium expression and 17 positive interactions involving EFNB1high expression. This evidence supports the idea that deviations in EFNB1 expression levels are linked to distinct phenotypic effects.
We validated SPAGE-finder by comparing its SL predictions to previously reported SLs identified via large in vitro screens (Bommi-Reddy et al., 2008; Lord et al., 2008; Luo et al., 2009b; Martin et al., 2009; Steckel et al., 2012; Turner et al., 2008). Each of the three filtering steps of SPAGE-finder (Figures 1C-1E) could discriminate the experimentally determined SLs from the non-SLs, with an area under receiver operating characteristic curve (ROC-AUC) of 0.63 (p = 0.0005), 0.62 (p = 0.001), and 0.59 (p = 0.012), respectively. These results are significant, albeit of modest accuracy (reflecting the widely known discrepancy between in vitro and in vivo data; Williams et al., 2000), and support the contribution of each of the individual steps in SPAGE-finder. In addition, we find the PIN-supported SPAGEs to be predictive of patient survival both in a cross-validation setting in TCGA (Weinstein et al., 2013) as well as in an independent breast cancer Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset (Curtis et al., 2012; Figure S2A). The prediction accuracy quantified via the concordance index (CI) shows that SPAGE-based prediction compares favorably with the gene-wise approach (STAR Methods). A bigger improvement is observed in the independent METABRIC dataset (concordance = 0.64), testifying that the SPAGE-related approach is generalizable, while the individual gene-based approach fails to generalize (concordance ≈ 0.51). Figures S2B and S2D depict the survival prediction accuracy of each SPAGE type by multiple metrics and the full compendium of 12 (positive and negative) SPAGE types (STAR Methods). Interactions involving both genes in their wild-type mid-activity levels (i.e., bin 5) have negligible predictive power on survival, testifying that more extreme levels of expression of at least one of the two genes tend to be involved in functional SPAGEs affecting survival.
Differential Activity of Drug Target SPAGEs between Responders and Non-responders
Aiming to test the ability of our detected SPAGEs to predict drug response, we applied SPAGE-finder to identify SPAGEs based only on TCGA samples that do not have drug response information and tested the predicted SPAGEs’ ability to discriminate responders from non-responders in the “unseen” TCGA samples where the drug response information is available (STAR Methods). Notably, because the considered drugs are inhibitory, it suffices to focus on SPAGE bins 1, 2, and 3, where one of the genes (the drug’s target) has low activity. For a given drug and cancer type having data on responders and non-responders, we analyzed the SPAGEs involving each of the drug targets (identified via target-specific FDR; STAR Methods). We then tested whether the frequencies of SPAGE activation in responders and the non-responders are significantly different using a Fisher’s exact test (Fisher, 1922; STAR Methods). For positive SPAGEs, we expect a lower SPAGE activation frequency among responders and the opposite for negative SPAGEs (e.g., as in the case of SL-type SPAGEs). However, owing to very small and unbalanced numbers of responders and non-responders (5 to 35 samples per response group per drug), the Fisher’s test is underpowered, and we therefore compared the Fisher’s test p values of the SPAGEs (equivalently, ratio of SPAGE frequency in responders and non-responders), segregated over all SPAGEs of a specific type, with those obtained using randomly shuffled drug-response labels, using paired Wilcoxon tests (Wilcoxon, 1945; STAR Methods) performed separately for each drug-cancer type pair.
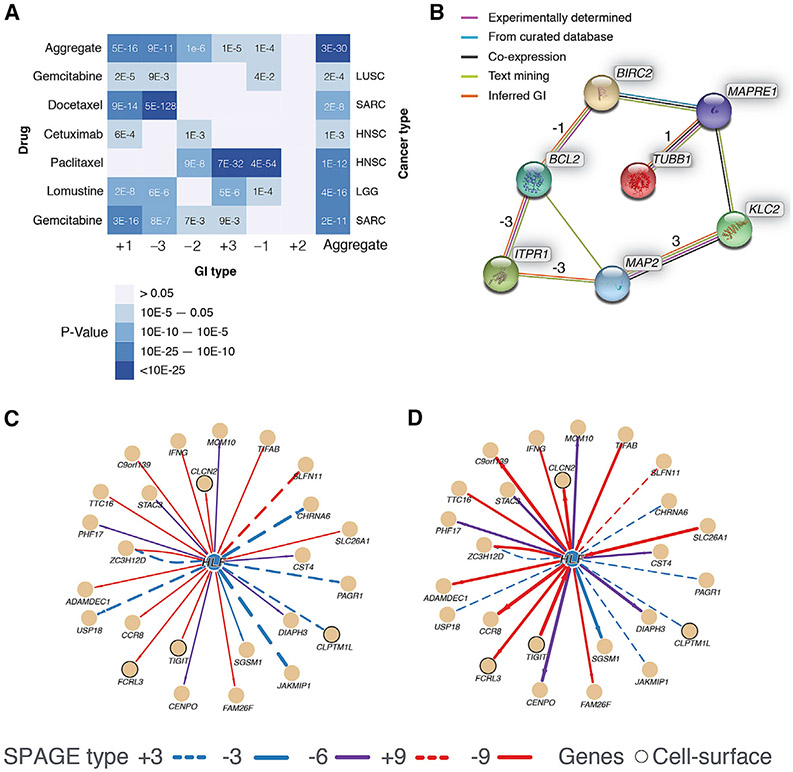
We considered the 12 drug-cancer type pairs that have RECIST (response evaluation criteria in solid tumors) (Eisenhauer et al., 2009) drug response following treatment for at least 10 patients (at least 5 responders and 5 non-responders) in TCGA. Each of the 6 basic SPAGE types was tested for the 12 drug-cancer type pairs. Overall, in 18 of the 72 tests (5-fold enrichment for p ≤ 0.05) of drug-cancer type combinations, SPAGEs of a particular type exhibit statistically significant differential activation frequencies between responders and non-responders consistent with the expected effects of the SPAGEs (Figure 3A). Reassuringly, several of those significant drug target SPAGEs are in bin 1, which contains the SLs, consistent with previous reports showing the role of SLs in mediating drug response (Jerby-Arnon et al., 2014). Among the drugs, gemcitabine, lomustine, and paclitaxel exhibit differential SPAGE activation for most SPAGE types (aggregate p values ranging from 4 × 10−16 to 2 × 10−11). We also explored the most differentially activated individual SPAGEs. We imposed an empirical FDR threshold of 0.01 on the Fisher’s test p value and that yielded 521 SPAGEs for the 12 drug-cancer type combinations (STAR Methods; Table S2). Individual genes comprising the 521 SPAGEs are closer to each other in the protein-protein interaction (PPI) network relative to shuffled pairs (Wilcoxon p = 0.001; STAR Methods) and have a significantly increased number of direct PPIs between them (Fisher’s p = 0.02; STAR Methods).
Figure 3. Differential SPAGE Activation Patterns across Drug Response Groups and Tissues.
(A and B) Differential SPAGE activation between drug response groups. (A) For each drug (left row labels) and each cancer type (right row labels) combination, and for each SPAGE type (columns), the heat plot shows the significance of differential activation of SPAGEs in responders and non-responders consistent with expectation. The last column shows the significance when all SPAGE types are aggregated. (B) The network shows the inferred functional interactions (based on STRING; Szklarczyk et al., 2015) among the genes interacting with paclitaxel targets, as well as inferred SPAGE types.
(C and D) Differential SPAGE activation between tissues in gene-specific SPAGE network. For the HLF-specific SPAGE network, the figure shows the activity states of SPAGEs in breast and lung cancers (C, foreground tissues) and in other cancer types (D, background tissues). The edge weight (thickness) represents the fraction of samples in which the SPAGE was functionally active. Several SPAGEs can be seen as differentially active in the two sets of cancer types. The figure also depicts cell surface proteins among the HLF’s SPAGE partners. The SPAGE-network-based sample-specific risk score is significantly higher (q value < 1.07 × 10−12) in breast and lung cancer relative to other cancer types, potentially mediated by a selective activation of positive SPAGEs in the foreground tissues and negative SPAGEs in the background tissues.
As an illustrative test case, we explored the SPAGEs associated with the response to paclitaxel in the TCGA head and neck squamous cell carcinoma (HNSC) cohort. Paclitaxel inhibits the proteins encoded by BCL2, TUBB1, and MAP based on DrugBank (Law et al., 2014). We identified a SPAGE involving the inactivation of BCL2 (known to suppress apoptosis, indirectly inhibited through phosphorylation; Ruvolo et al., 2001) and the overactivation of ITPR1 (Inositol 1,4,5-trisphosphate receptor type 1, also known as IP3 receptor type 1), negatively affecting tumor fitness (SPAGE type −3). Interestingly, our analysis shows that this SPAGE is functionally active among the responders at a significantly higher ratio than among non-responders (odds ratio ≈ 11.1). Upon treatment with paclitaxel, the activity level of BCL2 is expected to decrease, regardless of its pre-treatment level. Therefore, we reasoned that variations in drug response should depend only on the level of genes (ITPR1 in this case) in SPAGEs for which the other gene is BCL2. Therefore, post-treatment, the interaction discussed here is active in ITPR1high and inactive in ITPR1low or ITPR1medium tumors. Thus, the comparison of responders versus non-responders is divided based on ITPR1 expression levels alone. In general, the analysis of gene pairs and drug response is asymmetric because one gene (x, BCL2 in the example) is the target of a drug, while the other gene (y, ITPR1 in the example) is found via a target-specific search for gene partners of x.
The PPI between ITPR1 and BCL2 is well characterized (Chen et al., 2004; Oakes et al., 2005; Rong et al., 2009); one of these studies suggests that BCL2 also interacts with the two other human paralogs ITPR2 and ITPR3, but these interactions are not represented in the PIN used in this study and were therefore not detected. BCL2 exerts its oncogenic effect by inhibiting ITPR3-mediated channel opening and Ca2+ release from the endoplasmic reticulum and thus preventing cancer cell apoptosis. Our analysis strongly suggests that BCL2 inhibition by paclitaxel is especially effective when the ITPR1 expression is abundant, enabling effective Ca2+ release. Additional paclitaxel targets TUBB1 and MAP2 are also linked with ITPR1/BCL2 through SPAGEs with literature evidence for experimentally validated or putative interactions (in STRING database; Szklarczyk et al., 2015; Figure 3B), suggesting promising avenues for additional studies.
SPAGEs Can Explain Tissue-Specific Oncogenic Effects of Some Cancer Driver Genes
Many of the known cancer driver genes affect tumor initiation and development in a tissue-specific manner, despite the cancer gene being expressed in other tissues as well. Next, we explored whether the SPAGEs can explain the tissue specificity of cancer driver genes. Toward this, we identified 15 oncogenes and 20 tumor suppressors whose effects are likely to be restricted to specific cancer types, based on preferentially high mutation rates in those cancer types, including breast, bladder, and gastric cancer (STAR Methods; Table S3). For each cancer driver, we assigned a risk score to each patient by aggregating functionally active SPAGEs involving the driver gene defined using target-specific FDR (STAR Methods); for oncogenes, only the bins with high oncogene activity were considered, and for tumor suppressors, only the bins with low tumor suppressor activity were considered (STAR Methods). We hypothesized that for a cancer gene, the risk score will be greater in tissues where the cancer gene is implicated relative to other tissues. Indeed, for 15 out of 35 (~43%; 5 oncogenes and 10 tumor suppressors) driver genes, the observations are consistent with our hypothesis (Wilcoxon rank-sum test, FDR < 0.1; Table S3).
For instance, HLF, a bZIP transcription factor, has been linked to lung and breast cancer based on its significantly greater missense mutation frequency in those cancer types (Gonzalez-Perez et al., 2013). We observed a significant difference (FDR < 1.07 × 10−12) in the SPAGE activation risk scores for breast and lung cancer relative to the other tissues. Specifically, we found that positive SPAGEs are preferentially activated in these two tissues, while negative SPAGEs are preferentially activated in the other tissues, consistent with the increased tumor fitness in these two foreground tissues (Figures 3C and 3D). Overall, these results suggest that cancer-type-specific effects of many driver genes may be explained by their tissue-specific SPAGE network activity.
Stratifying Breast Cancer Tumors into Distinct Subtypes Based on their SPAGE Profiles
Next, we investigated whether functionally active pan-cancer SPAGEs in a tumor may provide an alternative methodology to tumor stratification into subtypes. We focus on breast cancer because it has a large number of samples in TCGA and because a second independent dataset, METABRIC, is publicly available. The proportion of breast cancer samples in TCGA is ~17%, indicating that any circularity in using all cancer samples to determine the SPAGEs rather than only the ~83% of samples of other types is minor. We represent each sample by the functional activity (a binary indicator) of each PIN-supported SPAGE detected in TCGA, rather than generating a BRCA-specific network, thus avoiding potential circularity of inference and prediction within samples sharing similar characteristics. Based on this 1,704-dimensional binary vector representation of each tumor sample, we clustered the 1,981 breast cancer samples in the independent METABRIC dataset (Curtis et al., 2012) using a conventional non-negative matrix factorization (NMF) (STAR Methods). Optimal clustering was achieved (maximum value of the Dunn index; STAR Methods) for 10 clusters (Figure S3A). Upon closer inspection of the distributions of known breast cancer subtypes in these clusters (Figure S3B), we merged 2 of the clusters, thus yielding 9 clusters for further analyses.
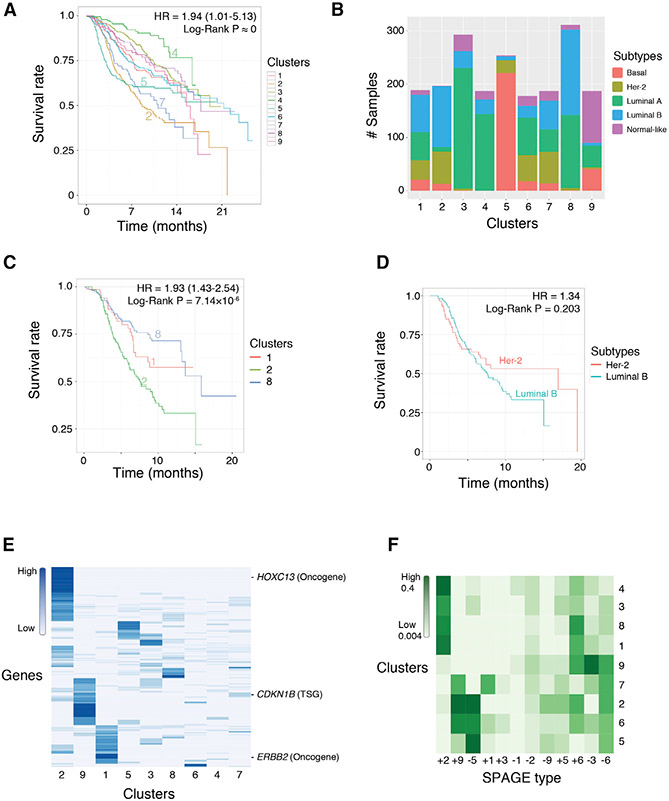
Kaplan-Meier curves (Figure 4A) and statistical analysis show that the 9 clusters have distinct survival characteristics with an overall mean HR difference of 1.94 (p value below the lowest reportable threshold and shown as 0). The distinct survival characteristics are consistent with the analysis performed using the full 71,946 SPAGE network (Figure S3C). Figure S3D shows the survival characteristics obtained for the previously published clustering of the METABRIC samples (Curtis et al., 2012). As evident, both approaches obtain similar survival separation levels but exhibit differences in their histopathological composition (Figure S3E). Currently, breast cancer has 5 well-established clinically distinct subtypes based on the tumors’ histopathological attributes. Figure 4B shows the fractions of each known subtypes among the 9 SPAGE-based clusters. Several clusters are highly associated with specific subtypes such as basal (triple-negative) (cluster 5), luminal A (clusters 3 and 4), etc. Others show association with several subtypes, e.g., luminal A and B both have high fractions in cluster 8. Interestingly, the basal subtype, which are largely triple-negative and have poor prognosis, correspond to a distinct cluster in our analysis (cluster 5), consistent with their distinct clinical status. In the original METABRIC publication (Curtis et al., 2012), 50% of the samples were left unassigned to any of their 10 clusters, while our SPAGE-based clustering covers all samples.
Figure 4. Breast Cancer Stratification by SPAGE Activation Patterns.
(A) Mean survival curves of the individuals in the 9 inferred SPAGE-based breast cancer subtypes.
(B) Cluster subtype composition based on PAM50 breast cancer sub typing (Parker et al., 2009).
(C and D) Survival trends of tumors of known histopathological cancer subtypes within and across SPAGE-based clusters. Luminal B samples that are split across SPAGE-based clusters 1, 2, and 8 show significant survival differences C). Her-2- and luminal-B-type tumors that are included within the SPAGE-based cluster 2 exhibit similar survival trends, as expected (D).
(E) Mutational profile of SPAGE-based breast cancer subtypes. Mutational profiles of 196 genes (rows) across the 9 SPAGE-based clusters (columns). For each gene and each cluster, the figure depicts the fraction of samples in the cluster in which the gene is mutated.
(F) SPAGE-type composition of the SPAGE-based breast cancer subtypes in the METABRIC dataset. The x axes represent the 12 SPAGE types (6 activity bins and 2 directional effects on survival), and the y axes represent the clusters. The colors represent the fraction of cluster-assigned SPAGEs per SPAGE type.
We find the SPAGE-based approach to clustering is able to separate subsets of patients that have different survival characteristics even though they belong to the same histopathological class (Figures 4C and 4D). More specifically, there are two situations in which the SPAGE-based clustering leads to a different classification of patients for survival analysis: (1) cases where known histopathological breast cancer subtypes are split across multiple SPAGE-based clusters (e.g., luminal B across clusters 1, 2, and 8) and, conversely, (2) cases where one SPAGE-based cluster harbors multiple known histopathological subtypes (e.g., cluster 2 contains Her-2 and luminal B subtypes). In the former case, we find that the 1,989 luminal B tumors that are split across different SPAGE-based clusters exhibit statistically significant (p = 7.14 × 10−6) distinct survival trends (Figure 4C), supporting the SPAGE approach in separating the luminal B tumors. Likewise, in the latter case, we find that the survival trends of Her-2 and luminal B histopathological subtype samples that are assigned to the same SPAGE-based cluster 8 do not show a significant difference (p = 0.203) in their survival trends (Figure 4D), suggesting that the SPAGE-based stratification may in some instances provide a better survival prognosis relative to histopathology-based stratification. We systematically identified 6 additional instances of the above two scenarios where (1) a known tumor histopathological subtype was split across multiple SPAGE-based clusters or (2) multiple known histopathological subtypes were assigned to the same SPAGE-based cluster (and each cluster has at least 30 samples). In each instance of the first kind, we tested for statistically significant differences in survival, and in each instance of the second kind, we tested for lack thereof. As shown in Figure S4A, in 4 out of 6 instances we found that the SPAGE-based clusters provided a more accurate survival prognosis. In contrast, we identified 4 cases of the second kind in the original METABRIC clusters (Curtis et al., 2012) and found that none of their survival trends were significantly different (Figure S4B). Thus, these results provide proof of principle that incorporating combined expression state information may provide an improved phenotypic characterization of tumors relative to histopathological subtypes or METABRIC clustering, which is based on gene expression profiles alone.
To explore the potential mutational basis of the SPAGE-based clusters in another way, we assessed whether the samples in SPAGE-based clusters harbor distinct mutations patterns; no mutation data were used by SPAGE-finder. We identified 196 genes with significantly greater mutation frequency in one or more of the clusters, relative to their overall mutation frequency in breast cancer (STAR Methods). Figure 4E shows the mutational frequency profiles of these genes across the 9 clusters. Overall, the differentially mutated genes across the SPAGE-based clusters include 10 cancer drivers: CDK12, CDKN1B, DNAJB1, ERBB2, EXT2, FCGR2B, FNBP1, HOXC13, PDGFRB, and SEC24D.
The mutation analysis results, while consistent with the literature, provide a more refined view. For instance, the Her-2 subtype is characterized by overexpression of ERBB2 and, in rare cases, by activating mutations of ERBB2 (Sun et al., 2015). We find an enrichment of ERBB2 mutations in cluster 1, which includes only a small subset of annotated Her-2 subtype tumors. Most of the ERBB2 missense mutations (11/13) are predicted to be functional (SIFT and PolyPhen measures; refer to STAR Methods), suggesting a distinct oncogenic mechanism underlying this subclass. Likewise, FCGR2B mutations, known to modulate Her-2 tumor’s response to trastuzumab (Norton et al., 2014), are enriched in cluster 6, which includes several other breast cancer subtypes besides Her-2, suggesting common mechanisms spanning multiple subtypes.
Finally, we quantified the fraction of each of the 12 types of functionally active SPAGEs among the samples in each cluster (see STAR Methods). Figure 4F shows the active SPAGE profiles of each cluster and reveals two broad subgroups: one including clusters 4, 3, 8, and 1 and another including clusters 2, 5, 6, and 7. Interestingly, the two subgroups clearly segregate in terms of their survival, testifying that the classification into SPAGE types captures a simplified yet robust characterization of the clinical prognosis. The first broad subgroup of tumors (clusters 4, 3, 8, and 1) are characterized by high fractions of type +2 and +6 SPAGEs, both of which involve a medium-expression and lowexpression bin. Therefore, this analysis demonstrates the relevance of considering medium expression states in molecular stratification. Figure S4C compares the SPAGE profiles of clusters revealed in the TCGA and the METABRIC breast cancer data and shows a high degree of consistency. A global comparison of the SPAGE profiles of the 9 clusters in the two datasets shows a Spearman correlation of 0.67 (p = 2.4 × 10−14) between the SPAGE types composition of these clusters, implying that SPAGE profiles are a robust characteristic of breast cancer tumors across different tumor collections. Thus, the SPAGE-based clustering demonstrates a proof of principle for improved stratification of breast cancer tumors into classes with distinct survival prognosis and mutational profiles.
DISCUSSION
Analyzing molecular and clinical data across thousands of tumors of dozens of types, we comprehensively map the landscape of 12 basic SPAGE types in cancer. Our work extends previous investigations of gene interactions in cancer, which have been almost entirely focused on SL (corresponding to positive effect on survival in bin 1), with a few studies of SDL (corresponding to bins 3 and 7), to a total of 12 types of interactions. The identified SPAGEs are predictive of patient survival and drug response, explain tissue specificity of cancer driver genes, and reveal functionally and clinically relevant breast cancer subtypes. The set of functionally active SPAGEs thus provides a complementary molecular characterization of tumors to those obtained by histology and contemporary individual gene-centric transcriptomic and sequence-based profiles. Distinct survival trends between the SPAGE-based breast cancer subtypes may be partly because SPAGEs were inferred based on their impact on survival. However, interestingly, the detected subtypes are additionally marked by distinct mutational profiles, which were not used in inferring the SPAGEs. An alternative approach would be to use mutation data and other genomics data to perform multidimensional molecular subtyping. Overall, these results underscore the importance of molecular context represented by functionally active SPAGEs.
Multiple factors are worth considering when deciding on the strategy to bin tumors into bins based on gene activity levels. For instance, a previous study of SL in glioblastoma (Szczurek et al., 2013) defined the high (low) state as mRNA expression higher than the 80th quantile (lower than the 20th quantile, respectively). However, the continuous nature of expression data allowed us to partition gene activity into 3 quantiles of low, medium, and high activity levels within cancer samples. This binning strategy allows to expand the scope of the analysis and identify dosage-sensitive fitness effects, depending on the states of other genes (e.g., bins 2 and 6). While in some instances of non-unimodal gene expression, the location of peaks near the 1/3 or 2/3 thresholds may render the medium bin compositions not biologically meaningful, we have demonstrated above that this scenario is limited to a modest (~7.6%) fraction of genes. This limitation is outweighed by the advantages of a simple, non-parametric and uniform binning construction across all genes that provides for robust statistical inference as described above. Furthermore, we have shown the robustness of the detected SPAGEs (STAR Methods), which could be further optimized by parameter adjustment in future studies of specific genes of interest.
Identifying pairwise gene interaction is only a first step toward capturing the true complexity of cellular networks. Future work can go beyond the 12 basic SPAGE types studied here to investigate more complex SPAGE types that involve different compositions of these basic types; for instance, a given interacting gene pair can confer tumor fitness benefit in multiple co-activity bins and reduce tumor fitness in others. Thus, while the results presented here go markedly beyond previous definitions and analyses of SPAGEs, they only begin to explore the full scope and clinical potential of gene-pair-based analyses of cancer, awaiting future investigations.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Sridhar Hannenhalli (sridhar@umiacs.umd.edu).
METHOD DETAILS
Cancer datasets
We downloaded The Cancer Genome Atlas (TCGA) (Weinstein et al., 2013) molecular profiles and clinical covariates via the Broad Firehose (https://gdac.broadinstitute.org, downloaded on Jan 28, 2016). This covers RSEM-normalized RNaseq data, mutation, and clinical information such as age, sex, race, tumor types, and overall survival of the 8,749 patients. Drug response information was downloaded from TCGA data portal available in the form of RECIST criteria (Eisenhauer et al., 2009) and mapped using DrugBank (Law et al., 2014) database V4.0. For the drug response analysis, to consider only gene the inactivation mechanism, we excluded those drugs whose DrugBank mechanism of action label is either potentiator, inducer, positive allosteric modulator, intercalation, stimulator, positive modulator, activator, partial agonist, or agonist.
We performed gene expression binning specifically for each combination of cancer type, race, and sex. Furthermore, we control for various clinical and demographic group specific effects. Because using small groups of samples may result in insufficiently robust models, we filtered rare combinations of clinical and demographic groups, resulting in 5,157 mRNA samples derived from patients spanning 18 cancer types, 3 races and 2 sexes. The data were not stratified by stage and grade for three reasons: 1) the grade is missing for most cancer types 2) the stage/grade system varies across tumor types 3) further stratification would result in further loss of data due to small group sizes. The METABRIC breast cancer dataset (Curtis et al., 2012) (as described in reference Jerby-Arnon et al., 2014) consists of 1,989 microarray samples and was used for independent validation.
Protein interaction Network (PIN)
The PIN was obtained from a previously published resource called HIPPIE (version 2.0, http://cbdm-01.zdv.uni-mainz.de/~mschaefer/hippie/), which aggregates physical protein interaction data from 10 source databases and 11 studies (Schaefer et al., 2012). This network consists of 15,673 human proteins and 203,159 interacting pairs.
Identification of SPAGEs associated with cancer patient survival
As shown in Figure 1, we divided the range of each gene’s expression across tumor samples into 3 equal-sized bins that correspond to the 3 activity states: low, medium, and high expression. Given a gene pair, each tumor sample is thus mapped into one of the 9 joint activity states of the two genes. The choice of dividing a gene’s activity into three classes, while somewhat arbitrary, was made in consideration of interpretability of functional states and robustness of inference. However, to account for differences in expression distributions across clinical and demographic confounders, we apply a subpopulation-specific binning approach. We considered the following categorical confounders: cancer type, race, and sex, as provided in TCGA. We considered the combination of confounder states for which there were at least 100 tumor samples. Our binning is thus not confounded by various clinical and demographic variables.
The SPAGE-finder pipeline consists of three steps that successively refine the predictions to arrive at high-confidence set of predicted SPAGEs. As such, the number of all pairwise combinations of genes is excessively large to apply a comprehensive Cox regression model. For the principal analysis shown in the manuscript, specific parameter thresholds were chosen to make the subsequent analysis tractable, but users of SPAGE-finder may choose other thresholds. To inform such decisions, we performed a robustness analysis of the parameter settings with a smaller input set of gene pairs (see the subsection entitled “Robustness analysis” later in STAR Methods).
Step 1: Log-Rank
In the first step, for each of the ~163 million gene pairs, say (x,y), we compute the Log-rank statistics (Harrington and Fleming, 1982) estimating the survival difference between the samples that map to one of the 9 activity bins and the other 8 bins. We implemented the log-rank test in C++ for computational speed. To control for gene-wise effects, we compare the Log-rank statistics of the gene pair (x,y) (in a bin) with those for (x,U) and separately with those for (V,y), where U and V represent all other genes (refer to Quantification and Statistical Analysis for additional information). We retain a gene pair if it is deemed significant in any of the 9 bins. This procedure retained 223,946 gene pairs of the total of ~163M.
Step 2: Molecular enrichment and depletion
For a gene interaction having positive (respectively, negative) effect on survival, we expect the tumor having that interaction to be under negative (respectively, positive) selection, and therefore we expect the fraction of such tumors (i.e., those mapped to the corresponding activity bin relative to the interacting gene pair to be depleted (respectively, enriched). We only retained the potentially interacting gene pairs for which the fraction of samples in a particular bin, suggested by the log-rank test, was lower (bottom 45 percent) or respectively, greater (top 45 percent among all gene pairs) than expectation, reducing the number of SPAGEs to 271,096 across 179,444 gene pairs. Recall that a gene pair can participate in multiple SPAGEs corresponding to multiple bins and effect on survival.
Step 3: Cox proportional hazard test
The Cox proportional hazards model is the most widely accepted approach for modeling survival while accounting for censored data as well as confounding factors. For each gene pair passing the filter at step 2, we modeled its effect on survival in each of the 9 bins using the interaction status λ (active if the sample mapped to the bin and inactive otherwise), along with the confounders. Specifically, we introduced the expression levels of the two individual genes σ1, σ2 to model each gene’s independent effect on survival, and additionally, clinical and demographic confounders, namely, cancer type, race, sex, and age, as provided in TCGA. Conceptually, λ represents an interaction term similar to: bij × xi, × yj in which the coefficient bij can be different for each pair of activation states for the genes x and y.
The model is stratified based on the discrete confounders, to account for differences in the baseline hazard (risk) characteristics. We did not control for tumor stage and grade as these classifications reflect the very same tumor characteristics our model aims to capture, and such control would prevent us from learning an important element of the disease. Control for genomic-stability and tumor purity as a potential confounder is described in STAR Methods.
Cox modeling provides a p value representing the significance of the effect of joint gene pair activity on survival.
To obtain a null distribution for the p values, we performed random shuffling analysis (referto Quantification and Statistical Analysis for additional information).
Optional Step 4: Filtering by protein interactions
To gain additional confidence in the predicted SPAGEs, given the greater tendency (and expectation) for neighbors in the Protein Interaction Network (PIN) to exhibit functional interactions (see Results), we further refined the SPAGE set by retaining the pairs that are found within distance of 2 in the HIPPIE PIN. Overall, we obtain a set of 1704 SPAGEs that exhibit molecular and clinical evidence in cancer as well as evidence from the PIN network.
The pipeline is implemented in R and C++ in a distributed computing environment using SLURM (Yoo et al., 2003) to run many jobs in parallel on a computer cluster.
Survival risk score computation
We applied a semi-supervised approach to assign a risk score to each patient according to the functionally active SPAGEs in the sample. Consider a SPAGE involving genes x and y conferring a positive effect on the tumor fitness in a particular bin B (Figure 1). If in a sample, genes x and y fall in bin b, then the SPAGE is said to be ‘functionally active’ in the sample, and a score of +1 is contributed to the overall tumor ‘fitness’. Likewise, if the SPAGE has negative effect on the tumor fitness, then a score of −1 is contributed. The overall risk score given a set of SPAGEs is the sum of the individual SPAGE +1 or −1 scores.
Assessment of potential confounding factors
We assessed genomic-instability and tumor purity as potential confounding factors and found that the vast majority of the discovered SPAGEs remain highly significant. The genomic instability index measures the relative amplification or deletion of genes in a tumor based on the SCNA (Bilal et al., 2013) and the tumor purity is an estimate of the proportion of cancer cells in the sample (Aran et al., 2015). We recomputed the controlled Cox survival step (Figure 1E) for all the candidates and shuffled candidates obtained from the previous step of the pipeline, with the addition of the two covariates - genomic instability and tumor purity, to calculate the empirical FDR threshold (0.01 quantile). We obtain 77630 confounder-corrected SPAGEs where (1) 60302 of the 71946 original SPAGEs and (2) 1405 of the 1704 original PPI SPAGEs were detected. Modifying the FDR threshold to 0.1 yields a list of 154450 confounder-corrected SPAGEs which contain almost all of the 72k original SPAGEs with the exception of 48 SPAGEs that are not included (these 48 SPAGEs remain the exception when increasing the FDR quantile threshold to 0.2, reflecting the set of confounded SPAGEs). 45 of the confounded SPAGEs involve DEFB21, a member of the beta subfamily of defensins (antimicrobial peptides), suggesting that the majority of the confounded effects are limited to one gene as oppose to a widespread phenomenon. None of the SPAGEs is discussed in the main text are affected by this additional confounding control.
Robustness analysis
We assessed the robustness of threshold selection throughout the SPAGE-finder pipeline, including:
Modifications of bin quantile boundaries: we kept the 3 × 3 structure and either increased or decreased the corner bins by 10%, corresponding to ~170 added or removed samples per bin
Log-rank p value based most significant quantile: scanning between top 5% – 30%
Cox regression FDR threshold: scanning between 0.01 – 0.1
Notably, the relatively small changes in binning thresholds described in (a) result in moving hundreds of samples in or out of the bins (170-340 samples), thus it is a significant change of the bin size and composition. For computational tractability of exploring a large parameter space, we limited the set of genes to 557 Cosmic Cancer Census genes (Tier 1 and 2) corresponding to 154,846 possible gene pair combinations. As shown in Table S4, for the alternate binning, across all log-rank thresholds, 57%–96% of the gene pairs are recapitulated. Likewise, setting the log-rank quantile threshold to 0.8 (top 20% retained), across various Cox FDR thresholds, 45%–56% of the SPAGE detected using the default setting are captured. For instance, at log-rank threshold = 0.8 and Cox regression FDR = 0.1 yields ~33,000 SPAGEs (regardless of the binning threshold), and these include 53%~55% of the SPAGEs defined using the original binning method. While the SPAGE-finder pipeline offers users to set the above parameters, to demonstrate the utility of SPAGE-finder in the main results we have used a more stringent Log-rank p value based quantile and Cox FDR in the genome-wide analysis to make the execution time of the pipeline tractable.
Gene correlation analysis
To assess whether the full SPAGEs network is dominated by correlated gene expression patterns, we compared between correlations found between SPAGE pairs to shuffled SPAGEs. For a pair of SPAGEs A = (x1,y1) and B = (x2,y2), we defined the Pearson correlation statistic (PCS) between A and B as Min(ρ(x1,x2),ρ(y1,y2)), quantifying whether the two SPAGEs are independently inferred. Then, we calculated the PCS for SPAGE pairs within each SPAGE type and compared them to PCS calculated for the shuffled SPAGE pairs from the same SPAGE type. Given the high number of SPAGEs and the infeasible number of pairs to test, we selected up to 1000 SPAGEs randomly from each SPAGE type and calculated the corresponding shuffled SPAGE list of the same length. The PCS comparisons indicated similar distributions of the actual and shuffled SPAGEs across all SPAGE types (Figure S2B). Although SPAGE type +9 exhibited higher PCS relative to shuffled, the effect size was marginal. Thus, we conclude that the full SPAGE network is not dominated by correlated gene expression patterns.
Patient survival risk prediction
Recall that an object we call “Survival-associated Pairwise Gene Expression states” (SPAGE) is represented by a quadruple comprising of a gene pair (x,y), a symmetric bin b (1, 2, 3, 5, 6, 9; Figure 1), and its effect on tumor fitness (positive or negative), and a SPAGE is deemed functionally active in a specific tumor sample if the joint expression levels of the genes x, y, in the tumor fall in bin b. We turned to assess the extent to which the aggregate effect of functionally active SPAGEs in a tumor predicts patient survival (the individual SPAGEs are indeed inferred while considering survival but here, we are interested in the predictive power of their combined effects). For each sample, we computed the overall score conferred by functionally active SPAGEs (either in a binspecific and effect direction-specific fashion, or overall) in the sample. The higher the tumor ‘fitness’ score the lower the survival potential. However, to make our approach comparable to gene-wise approaches (Yuan et al., 2014), we assigned each gene the sum of the contributions by all active SPAGEs involving that gene, with multiplicity for gene pairs involved in multiple SPAGEs. The estimated gene-wise SPAGE score is used as a predictor variable in a Cox model along with the confounding factors discussed above to predict patient’s survival.
For cross validation, this model was trained on the same data used for the SPAGEs training and then validated on its cross-validation counterpart. For independent validation, the model was trained on the full TCGA dataset, and tested on the independent METABRIC breast cancer data with 1989 samples (Curtis et al., 2012). The prediction accuracy is estimated in terms of the C-index (Harrell et al., 2005). Several previous publications have assessed survival risk prediction accuracy based on dichotomized analysis where samples are separated into distinct low- and high-survival groups and their survival curves then compared (Harrington and Fleming, 1982), which is prone to overestimating prediction accuracy. For comparison, we also performed accuracy estimation following the dichotomized comparison of survival risks between the extreme cases of predicted survival risk groups, for variable thresholds to define the extreme (such as top versus bottom 10% or top versus bottom 20% and so on; Figure S2C).
To compare the SPAGE-based survival prediction with the individual gene approach, we implemented an analogous scheme for individual genes where the gene expression values were discretized into 3 expression levels (low, medium, and high), and the discretized representation was used as a predictor variable in a controlled Cox regression model to obtain the significance (p value) of each gene with respect to survival prediction. The most significant predictors (top 5%) were chosen and precisely used as described for the SPAGEs survival prediction procedure. An analogous procedure was used to estimate the prediction accuracy based on both individual gene effects and SPAGEs. Figure S2A shows the survival prediction accuracy using the widely used C-index (CI) metric, both via cross-validation within TCGA (Weinstein et al., 2013) and on independent METABRIC breast cancer dataset (Curtis et al., 2012). As shown, pan-cancer risk prediction based on the predicted SPAGEs compares favorably with the comparable gene-level approach both in cross-validation, and more so on the METABRIC independent dataset - suggesting good generalizability. Applying the fully supervised individual gene-level approach (Huang et al., 2016) (individual gene feature selection based on association with survival, further dimensionality reduction with LASSO Cox and a final Cox model to obtain genes’ coefficients for survival risk prediction) yields a comparable accuracy in cross validation (CI ≈ 0.63) but a lower accuracy over the independent dataset (CI ≈ 0.55). Finally, a previous study has reported a CI of 0.71 using a supervised approach specifically on Kidney Renal Clear Cell Carcinoma (KIRC). We ensured that a lower gene-wise accuracy that we observe is not simply due to our filtering and implementation; applying the fully supervised individual gene-level approach on KIRC subset of the filtered dataset yields CI ≈ 0.71, recapitulating the accuracy reported in the original publication (Huang et al., 2016).
In Figure S2B, we present the performance of the SPAGE-based approach using each of the 6 SPAGE types. As evident, interactions involving both genes in their wild-type mid-activity levels have negligible predictive power on survival, testifying that more extreme levels of gene expression tend to be involved in functional SPAGEs affecting survival. Figure S2C shows the performance based on an alternative metric where we dichotomized the extreme (at varying thresholds from 10% to 50%) predicted low- and high-risk groups and quantified the difference in their area under their KM survival curves (refer to the next section Quantification and Statistical Analysis for additional information). The survival prediction performance of the full compendium of 12 (positive and negative) SPAGE types is shown in Figure S2D.
Identifying gene target(s)-specific SPAGEs
To investigate the SPAGEs involving specific genes of interest (e.g., one or more target genes inhibited by a drug), we used a modified gene set-specific FDR approach. For a set of one or more genes X, we compare the SPAGE significance (Cox regression p value) of SPAGEs involving any gene in X across the quadruples derived from step 2 (Molecular enrichment/depletion) (refer to Quantification and Statistical Analysis for additional information).
Characterization of differential SPAGE activation between drug-response groups
We retrieved the drug response data as explained in the first subsection of STAR Methods; some patients have response information, and some do not. We inferred the SPAGEs involving each drug’s known target gene based on the TCGA samples that do not have that drug’s response information to avoid circularity and filtered based on FDR restricted to the target-specific SPAGEs (using target-specific FDR above). For each of the drug-specific SPAGEs, we compared its activation frequency (whether the SPAGE was functionally active or inactive) among the responders (stable disease, partial response, and complete response categories) and non-responders group (clinical progressive disease categories), using one-sided Fisher’s exact test (Fisher, 1922), where the alternative hypothesis was that negative (respectively, positive) SPAGEs are more frequently active among responders (respectively, non-responders). However, given the extremely small and imbalanced sample sizes, and the conservative nature of Fisher’s exact test (Berkson, 1978), we tested whether the overall distribution of the obtained odds-ratios are lower than those obtained using randomly shuffled drug-response labels (refer to Quantification and Statistical Analysis for additional information).
Characterization of tissue-specific effect of cancer driver genes
A study (Rubio-Perez et al., 2015) of genes’ somatic mutation profile across cancer types has identified and characterized the tissue specificity of 459 candidate drivers. For each such candidate, we matched the driver role annotation (oncogene or tumor-suppressor) obtained from the Cosmic Census (Futreal et al., 2004) cancer genes dataset, to obtain a set of 33 tumor suppressors and 25 oncogenes matching the tissue/tumor type annotations. For each of these 58 genes, we calculated the significant SPAGEs involving this gene (target-specific FDR). We further excluded genes with 5 or fewer interactions or with 300 or fewer samples where they are expressed, reducing the set of genes of interest to 20 tumor suppressors and 15 oncogenes spanning 10 cancer types (Table S3). For a gene, a sample-specific risk score was calculated based on the functionally active SPAGE partners of the gene (as above for the drug response analysis above), but only considering high activity bin for oncogenes and low-activity bins for tumor suppressors. For each gene, the cancer types are partitioned into affected types (cancer types affected by the driver) and the other unaffected cancer types. Using a one-sided Wilcoxon rank-sum test, we tested for higher risk score in the samples in the affected cancer type in comparison to those in unaffected types. After correcting for multiple hypotheses testing, 15 out of the 35 (~43%) driver genes were found to have significant tissue-specific SPAGE-based risk score (FDR q-value < 0.1; Table S3).
Breast cancer tumor stratification
We represent a tumor sample as a vector indicating the functional activity status of each predicted SPAGE. This provides a survivalcognizant alternative to the widely used gene expression profile representation of a sample. We used this representation to partition the METABRIC (as well as independently for TCGA) breast cancer patients into clusters using Non-Negative Matrix Factorization (NMF using the brunet algorithm and assigning each sample to the cluster with the highest weight) (Lee and Seung, 2000; Paatero and Tapper, 1994), which has suitable statistical properties and has been shown to be effective in a variety of contexts (Lee and Seung, 1999). Since NMF requires a predetermined number of clusters, we performed the analysis for 2-15 clusters, and assessed their fitness using Dunn’s index (Dunn, 1974), which quantifies compactness within and separation across clusters (refer to Quantification and Statistical Analysis for additional information). For comparison purposes, to match our estimated clusters’ sizes to previously published METABRIC cluster sizes (~900 samples), we constrained the number of samples in each cluster to the 1000 samples that were found to be most highly associated with the cluster. Each cluster’s SPAGE profiles were constructed as follows. Our clustering approach – NMF, assigned each SPAGE to a single cluster. For each cluster, and for each of the 12 SPAGE-types (6 bins in Figure 1 and the two directional effects on survival), we obtain the frequency of SPAGEs of that type, relative to all SPAGE assigned to the clusters.
Mutation frequency analysis was performed on the TCGA clusters. We defined the gene-wise mutation frequency as the fraction of samples in the cluster in which the gene has a mutation predicted to be deleterious as explained in the next paragraph. Then, we tested whether the mutation frequency distribution of each gene differs significantly across clusters using Chi-square tests. The genes with significant Chi-square statistic (FDR q-value < 0.1) were then used to illustrate the mutational profiles of the clusters.
Each mutation’s predicted effect on the protein function was obtained from the cBioPortal repository (Cerami et al., 2012; Gao et al., 2013). Out of the 196 differentially mutated genes, 138 genes had matching extended mutation information indicating their SIFT (Ng and Henikoff, 2003) (sorts intolerant from tolerant amino acid substitutions) and PolyPhen (Adzhubei et al., 2010) (polymorphism phenotyping) predictions. We calculated a gene-wise fraction of mutations predicted to have a significant effect on the protein, separately for SIFT and PolyPhen.
QUANTIFICATION AND STATISTICAL ANALYSIS
Datasets
The Cancer Genome Atlas (TCGA) (Weinstein et al., 2013) dataset consists of 8,749 patients which were filtered to 5,157 mRNA samples. We applied quantile normalization within each sample of the expression data. The METABRIC breast cancer dataset (Curtis et al., 2012) consists of 1,989 microarray samples. Similarly, quantile normalization was applied in each sample.
Log-Rank tests
For a candidate gene pair (x,y), we consider Log-rank(x,y) to be significant if it is among the top 0.1% relative to all (x,U) and top 0.1% among all (V,y) gene pairs. This threshold of 0.1% (1/1000) can be controlled by the user.
Molecular enrichment and depletion
We retained the potentially interacting gene pairs for which the fraction of samples in a particular bin, suggested by the log-rank test, were lower (bottom 45 percent) or respectively, greater (top 45 percent among all gene pairs) than expectation. The threshold of 45% can be controlled by the user. Our choice of threshold ascertained that molecular enrichment/depletion is consistent with log-rank test without being overly punitive.
Cox proportional hazard test
Cox modeling provides a p value representing the significance of the effect of joint gene pair activity on survival. To obtain a null distribution for the p values, we performed the following random shuffling. Among all the pairs passing the molecular enrichment and depletion filter (step 2), we randomly permuted the second genes (y) that get paired with the first genes (x) and computed the p values for the shuffled pairs. Among the real pairs, we retained only the ~71K gene pairs whose observed p value is lower than the lowest 1% of p values among the permuted, random gene pairs.
Patient survival risk prediction
Survival prediction accuracy was measured using the widely used C-index (CI) metric, both via cross-validation within TCGA (Weinstein et al., 2013) and on the independent METABRIC breast cancer dataset (Curtis et al., 2012). In addition, we measured performance based on an alternative metric in which we dichotomized the extreme (at varying thresholds from 10% to 50%) predicted low- and high-risk groups and quantified the difference in their area under their KM survival curves.
Identifying gene target(s)-specific SPAGEs
We defined significant SPAGEs as those where the significance is more extreme than (lower p value, higher quantile) the 90th quantile of shuffled SPAGEs involving any member of X.
Characterization of differential SPAGE activation between drug-response groups
We tested whether the overall distribution of the obtained odds-ratios are lower than those obtained using randomly shuffled drug-response labels, using one-sided Wilcoxon tests (Wilcoxon, 1945). Then, for each gene pair in the inferred SPAGEs list and, as a control, in a shuffled list of size 10x the original SPAGEs list size, we computed the distance between the genes in the PPI network (Schaefer et al., 2012). We thus obtained a p value for each drug-cancer type pair, segregated by SPAGE type. We then used one-sided Wilcoxon tests (Wilcoxon, 1945) to assess whether SPAGE gene pairs are closer to each other than random expectation. Alternatively, we also compare the number of directly SPAGE gene pairs having direct interaction using one-sided Fisher’s exact test (Fisher, 1922).
Characterization of tissue-specific effect of cancer driver genes
We used a one-sided Wilcoxon rank-sum test; we tested for higher risk score in the samples in the affected cancer type in comparison to those in unaffected cancer types.
Breast cancer tumor stratification
Hazard-ratio significance values were computed for each pair of clusters, while the p values were generated using multi-class log-rank test.
Mutation frequency analysis significance was quantified by the Chi-square statistic (FDR q-value < 0.1).
Extended mutation information was determined based on SIFT (Ng and Henikoff, 2003) (sorts intolerant from tolerant amino acid substitutions) and PolyPhen (Adzhubei et al., 2010) (polymorphism phenotyping) predictions. We calculated a gene-wise fraction of mutations predicted to have a significant effect on the protein, separately for SIFT and PolyPhen.
The breast cancer subtypes were derived using the widely accepted PAM50 algorithm (Parker et al., 2009). The METABRIC PAM50 subtypes were annotated in the original publication (Curtis et al., 2012), while the TCGA breast cancer subtypes were calculated using the original published class centroids (Parker et al., 2009).
DATA AND CODE AVAILABILITY
The SPAGE-finder software is available on GitHub: https://github.com/asmagen/SPAGEfinder. The code is archived by Zenodo and can be cited via https://doi.org/10.5281/zenodo.3239265. The pipeline requires access to a SLURM high-performance computing core for efficient simulation analyses.
The 72k SPAGE network is accessible online via a web portal: https://amagen.shinyapps.io/spage.
The web portal codes are available on: https://github.com/asmagen/SPAGEPortal. The code is archived by Zenodo and can be cited via https://doi.org/10.5281/zenodo.3239267.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Publicly Available Data | ||
| The Cancer Genome Atlas (TCGA) | Weinstein et al., 2013 | N/A |
| METABRIC | Curtis et al., 2012 | N/A |
| Software and Algorithms | ||
| SPAGE-finder | This paper | https://github.com/asmagen/SPAGEfinder |
| SPAGEPortal | This paper | https://amagen.shinyapps.io/spage |
| SLURM (We used primarily the installation at U. Maryland and tested on a second installation at the National Institutes of Health) | Yoo et al., 2003 | https://slurm.schedmd.com |
| Individual gene-wise survival risk prediction | Yuan et al., 2014 | https://doi.org/10.7303/syn1710282 |
| Other | ||
| TCGA Data Acquisition - Broad Firehose | N/A | https://gdac.broadinstitute.org |
| Drug response information - RECIST criteria | Eisenhauer et al., 2009 | N/A |
| DrugBank V4.0 | Law et al., 2014 | https://www.drugbank.ca |
| HIPPIE V2.0 | Schaefer et al., 2012 | http://cbdm-01.zdv.uni-mainz.de/~mschaefer/hippie |
| Cosmic Census | Futreal et al., 2004 | https://cancer.sanger.ac.uk/census |
| cBioPortal | Cerami et al., 2012; Gao et al., 2013 | https://www.cbioportal.org |
| SIFT | Ng and Henikoff, 2003 | https://sift.bii.a-star.edu.sg |
| PolyPhen2 (evaluation of predicted pathogenicity of amino acid substitutions) | Adzhubei et al., 2010 | http://genetics.bwh.harvard.edu/pph2 |
ACKNOWLEDGMENTS
This research is supported in part by the Intramural Research Program of the NIH, National Cancer Institute. S.H. is funded in part by NSF award 1564785.
Footnotes
DECLARATION OF INTERESTS
E.R. is a co-founder (divested) and a non-paid scientific adviser of Pangea Therapeutics, a startup company that aims to harness genetic interactions to advance cancer treatments.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.Org/10.1016/j.celrep.2019.06.067.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, and Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaogi K, Nakajima Y, lto I, Kawasaki S, Oie SH, Murayama A, Kimura K, and Yanagisawa J (2009). KLF4 suppresses estrogen-dependent breast cancer growth by inhibiting the transcriptional activity of ERalpha. Oncogene 28, 2894–2902. [DOI] [PubMed] [Google Scholar]
- Aran D, Sirota M, and Butte AJ (2015). Systematic pan-cancer analysis of tumour purity. Nat. Commun 6, 8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A, Lord CJ, and Reis-Filho JS (2011). Genetic interactions in cancer progression and treatment. Cell 145, 30–38. [DOI] [PubMed] [Google Scholar]
- Benhamou S, and Sarasin A (2002). ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis 17, 463–469. [DOI] [PubMed] [Google Scholar]
- Berkson J (1978). In dispraise of the exact test. Do the marginal totals of the 2X2 table contain relevant information respecting the table proportions? J. Stat. Plan. Inference 2, 27–42. [Google Scholar]
- Bernard-Gallon D, Bosviel R, Delort L, Fontana L, Chamoux A, Rabiau N, Kwiatkowski F, Chalabi N, Satih S, and Bignon YJ (2008). DNA repair gene ERCC2 polymorphisms and associations with breast and ovarian cancer risk. Mol. Cancer 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal E, Dutkowski J, Guinney J, Jang IS, Logsdon BA, Pandey G, Sauerwine BA, Shimoni Y, Moen Vollan HK, Mecham BH, et al. (2013). Improving breast cancer survival analysis through competition-based multidimensional modeling. PLoS Comput. Biol 9, e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, and Auger DL (2001). Dosagedependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol 234, 275–288. [DOI] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, Kaelin WG Jr., and Grueneberg DA (2008). Kinase requirements in human cells: III. Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proc. Natl. Acad. Sci. USA 105, 16484–16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher B, and Jenna S (2013). Genetic interaction networks: better understand to better predict. Front. Genet 4, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R, Frankum JR, Costa-Cabral S, Lord CJ, and Ashworth A (2011). Searching for synthetic lethality in cancer. Curr. Opin. Genet. Dev 27, 34–41. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Valencia I, Zhong F, McColl KS, Roderick HL, Bootman MD, Berridge MJ, Conway SJ, Holmes AB, Mignery GA, et al. (2004). Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J. Cell Biol 766, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Hwang YS, Mood K, Ji YJ, Lim J, Morrison DK, and Daar IO (2014). EphrinB1 interacts with CNK1 and promotes cell migration through c-Jun N-terminal kinase (JNK) activation. J. Biol. Chem 289, 18556–18568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Hwang YS, Yoon J, Lee M, Lee HG, and Daar IO (2018). EphrinB1 promotes cancer cell migration and invasion through the interaction with RhoGDI1. Oncogene 37, 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. ; METABRIC Group (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JC (1974). Well-Separated Clusters and Optimal Fuzzy Partitions. J. Cybern 4, 95–104. [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. (2009). New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247. [DOI] [PubMed] [Google Scholar]
- Fisher RA (1922). On the Interpretation of χ 2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc 85, 87–94. [Google Scholar]
- Fong CY, Gilan O, Lam EYN, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. (2015). BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JB, and Dale R (2013). Assessing bimodality to detect the presence of a dual cognitive process. Behav. Res. Methods 45, 83–97. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, and Stratton MR (2004).Acensusofhuman cancer genes. Nat. Rev. Cancer 4, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A, and Lopez-Bigas N (2013). IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 70, 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE, Lee KL, and Mark DB (2005). Prognostic/Clinical Prediction Models: Multivariable Prognostic Models: Issues in Developing Models, Evaluating Assumptions and Adequacy, and Measuring and Reducing Errors. In Tutorials in Biostatistics, Statistical Methods in Clinical Studies, pp. 223–249. [DOI] [PubMed] [Google Scholar]
- Harrington DP, and Fleming TR (1982). A class of rank test procedures for censored survival data. Biometrika 69, 553–566. [Google Scholar]
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, and Friend SH (1997). Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Huang X, Stern DF, and Zhao H (2016). Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival–Evidence from TCGA Pan-Cancer Data. Sci. Rep 6, 20567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, Seashore-Ludlow B, Weinstock A, Geiger T, Clemons PA, et al. (2014). Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 158, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. (2005). The concept of synthetic lethality in thecontext of anticancer therapy. Nat. Rev. Cancer 5, 689–698. [DOI] [PubMed] [Google Scholar]
- Kawai T, Forrester SJ, O’Brien S, Baggett A, Rizzo V, and Eguchi S (2017). AT1 receptor signaling pathways in the cardiovascular system. Pharmacol. Res 725 (Pt A), 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, and Ideker T (2005). Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol 23, 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, and Li JJ (1996). Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M, Jambon S, Depauw S, and David-Cordonnier MH (2018). Targeting transcription factors for cancer treatment. Molecules 23, 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. (2014). DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, and Seung HS (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. [DOI] [PubMed] [Google Scholar]
- Lee DD, and Seung HS (2000). Algorithmsfor Non-negative Matrix Factorization. https://papers.nips.cc/paper/1861-algorithms-for-non-negative-matrix-factorization.pdf. [DOI] [PubMed] [Google Scholar]
- Lee AY, Perreault R, Harel S, Boulier EL, Suderman M, Hallett M, and Jenna S (2010). Searching for signaling balance through the identification of genetic interactors of the Rab guanine-nucleotide dissociation inhibitor gdi-1. PLoS ONE 5,e10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Das A, Jerby-Arnon L, Arafeh R, Auslander N, Davidson M, McGarry L, James D, Amzallag A, Park SG, et al. (2018). Harnessing synthetic lethality to predict the response to cancer treatment. Nat. Commun 9, 2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MDM, Wu HT, Vandin F, and Raphael BJ (2015). CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 76, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fang Y, Niu C, Cao H, Mi T, Zhu H, Yuan J, and Zhu J (2018). Inhibition of cIAP1 as a strategy for targeting c-MYC-driven oncogenic activity. Proc. Natl. Acad. Sci. USA 115, E9317–E9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, McDonald S, Swift S, Turner NC, and Ashworth A (2008). A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst.) 7, 2010–2019. [DOI] [PubMed] [Google Scholar]
- Lu X, Kensche PR, Huynen MA, and Notebaart RA (2013). Genome evolution predicts genetic interactions in protein complexes and reveals cancer drug targets. Nat. Commun 4, 2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, and Elledge SJ (2009a). Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, and Elledge SJ (2009b). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, McCarthy A, Barber LJ, Burgess DJ, Parry S, Lord CJ, and Ashworth A (2009). Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Mol. Med 1, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlak D, and Szczurek E (2017). Epistasis in genomic and survival data of cancer patients. PLoS Comput. Biol 13, e1005626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Shimada H, Ueda Y, Inoue M, Hasegawa M, Tomonaga T, Matsubara H, and Nomura F (2014). Non-transmissible Sendai virus vector encoding c-myc suppressor FBP-interacting repressor for cancer therapy. World J. Gastroenterol 20, 4316–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLornan DP, List A, and Mufti GJ (2014). Applying synthetic lethality for the selective targeting of cancer. N. Engl. J. Med 371, 1725–1735. [DOI] [PubMed] [Google Scholar]
- Megchelenbrink W, Katzir R, Lu X, Ruppin E, and Notebaart RA (2015). Synthetic dosage lethality in the human metabolic network is highly predictive of tumor growth and cancer patient survival. Proc. Natl. Acad. Sci. USA 112, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, et al. (2015). RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, and Henikoff S (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Olson RM, Pegram M, Tenner K, Ballman KV, Clynes R, Knutson KL, and Perez EA (2014). Association studies of Feγ receptor polymorphisms with outcome in HER2+ breast cancer patients treated with trastuzumab in NCCTG (Alliance) Trial N9831. Cancer Immunol. Res 2, 962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil NJ, Bailey ML, and Hieter P (2017). Synthetic lethality and cancer. Nat. Rev. Genet 18, 613–623. [DOI] [PubMed] [Google Scholar]
- Oakes SA, Scorrano L, Opferman JT, Bassik MC, Nishino M, Pozzan T, and Korsmeyer SJ (2005). Proapoptotic BAX and BAK regulate the type 1 inositol trisphosphate receptor and calcium leak from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paatero P, and Tapper U (1994). Positive Matrix Factorization: A Non-negative Factor Model With Optimal Utilization of Error Estimates of Data Values. Environ metrics 5, 111–126. [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmutulla B, Matsushita K, Satoh M, Seimiya M, Tsuchida S, Kubo S, Shimada H, Ohtsuka M, Miyazaki M, and Nomura F (2014). Alternative splicing of FBP-interacting repressor coordinates c-Myc, P27Kip1/cyclinE and Ku86/XRCC5 expression as a molecular sensor for bleomycin-induced DNA damage pathway. Oncotarget 5, 2404–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, et al. (2015). Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, and Schier AF (2011). Morphogen gradients: from generation to interpretation. Annu. Rev. Cell Dev. Biol 27, 377–407. [DOI] [PubMed] [Google Scholar]
- Rong Y-P, Bultynck G, Aromolaran AS, Zhong F, Parys JB, De Smedt H, Mignery GA, Roderick HL, Bootman MD, and Distelhorst CW (2009). The BH4 domain of Bcl-2 inhibits ER calcium release and apoptosis by binding the regulatory and coupling domain of the IP3 receptor. Proc. Natl. Acad. Sci. USA 106, 14397–14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Perez C, Tamborero D, Schroeder MP, Antolin AA, Deu-Pons J, Perez-Llamas C, Mestres J, Gonzalez-Perez A, and Lopez-Bigas N (2015). In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396. [DOI] [PubMed] [Google Scholar]
- Ruvolo PP, Deng X, and May WS (2001). Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 15, 515–522. [DOI] [PubMed] [Google Scholar]
- Sahu AD, S Lee J, Wang Z, Zhang G, Iglesias-Bartolome R, Tian T, Wei Z, Miao B, Nair NU, Ponomarova O, et al. (2019). Genome-wide prediction of synthetic rescue mediators of resistance to targeted and immunotherapy. Mol. Syst. Biol 15, e8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajesh BV, Guppy BJ, and McManus KJ (2013). Synthetic genetic targeting of genome instability in cancer. Cancers (Basel) 5, 739–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, Fontaine JF, Vinayagam A, Porras P, Wanker EE, and Andrade-Navarro MA (2012). HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS ONE 7, e31826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel M, Molina-Arcas M, Weigelt B, Marani M, Warne PH, Kuznetsov H, Kelly G, Saunders B, Howell M, Downward J, and Hancock DC (2012). Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 22, 1227–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, Angus SP, Collins KAL, Granger DA, Reuther RA, et al. (2015). Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep. 11, 390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Shi Y, Shen Y, Cao L, Zhang W, and Guan X (2015). Analysis of different HER-2 mutations in breast cancer progression and drug resistance. J. Cell. Mol. Med 19, 2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos B, Kovács K, Szamecz B, Honti F, Costanzo M, Baryshnikova A, Gelius-Dietrich G, Lercher MJ, Jelasity M, Myers CL, et al. (2011). An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat. Genet 43, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurek E, Misra N, and Vingron M (2013). Synthetic sickness or lethality points at candidate combination therapy targets in glioblastoma. Int. J. Cancer 133, 2123–2132. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, lorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, and Ashworth A (2008). A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 27, 1368–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, and Potier MC (2015). Gene dosage imbalances: action, reaction, and models. Trends Biochem. Sci 40, 309–317. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Butterfield YSN, et al. ; Cancer Genome Atlas Research Network (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F (1945). Individual Comparisons by Ranking Methods. Biom. Bull 1, 80–83. [Google Scholar]
- Williams CS, Watson AJM, Sheng H, Helou R, Shao J, and DuBois RN (2000). Celecoxib prevents tumor growth in vivo without toxicity to normal gut: lack of correlation between in vitro and in vivo models. Cancer Res. 60, 6045–6051. [PubMed] [Google Scholar]
- Wong SL, Zhang LV, Tong AHY, Li Z, Goldberg DS, King OD, Lesage G, Vidal M, Andrews B, Bussey H, et al. (2004). Combining biological networks to predict genetic interactions. Proc. Natl. Acad. Sci. USA 101, 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AB, Jette MA, and Grondona M (2003). SLURM: Simple Linux Utility for Resource Management. In Job Scheduling Strategies for Parallel Processing, Feitelson D, Rudolph L, and Schwiegelshohn U, eds. (Springer Berlin Heidelberg; ), pp. 44–60. [Google Scholar]
- Yuan Y, Van Allen EM, Omberg L, Wagle N, Amin-Mansour A, Sokolov A, Byers LA, Xu Y, Hess KR, Diao L, et al. (2014). Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat. Biotechnol 32, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, and Sternberg PW (2006). Genome-wide prediction of C. elegans genetic interactions. Science 311, 1481–1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SPAGE-finder software is available on GitHub: https://github.com/asmagen/SPAGEfinder. The code is archived by Zenodo and can be cited via https://doi.org/10.5281/zenodo.3239265. The pipeline requires access to a SLURM high-performance computing core for efficient simulation analyses.
The 72k SPAGE network is accessible online via a web portal: https://amagen.shinyapps.io/spage.
The web portal codes are available on: https://github.com/asmagen/SPAGEPortal. The code is archived by Zenodo and can be cited via https://doi.org/10.5281/zenodo.3239267.