Abstract
Background
We report here for the first time, a comprehensive characterization of biological and clinical features of early-stage triple negative Invasive Lobular Carcinomas(TN-ILCs)
Methods
We analyzed all consecutive patients with early-stage TN-ILC operated at two reference cancer-centers between 1994 and 2012.
Primary objective was to assess the invasive disease-free survival(iDFS).
Co-primary objective was to assess biological features of TN-ILCs, including molecular intrinsic subtypes based on PAM-50 assay, expression of androgen receptor (AR) and mutational status of ERBB2-gene.
Additionally, DNA mutational status of an independent cohort of 45 TN-ILCs from three databases were analyzed, to confirm mutations in ERBB2-gene and to identify other recurrently mutated genes.
Results
Among 4152 ILCs, 74(1.8%) were TN and were analyzed.
The iDFS at 5 and 10 years of FUP were 50.4%(95%CI,38.0–61.6) and 37.2%(95%CI,25.5–48.8), respectively.
The molecular subtype was defined through PAM50-classifier for 31 out of 74 TN-ILCs: 48% were Luminal-A(15/31), 3% luminal-B(1/31), 32% HER2-enriched (10/31), and only 16% basal-like(5/31).
Luminal tumors expressed AR more frequently than non-luminal tumors (AR≥1% in 94% of luminal tumors versus 53% in non-luminal tumors; p-value = 0.001).
20% of TN-ILCs analyzed(7/35), harbored a pathogenetic and actionable mutation in the ERBB2-gene. Analysis of the independent cohort of 45 TN-ILCs from three different databases, confirmed similar percentage of pathogenetic and actionable mutations in ERBB2-gene(20%; 9/45).
Among the top 10 molecular pathways significantly enriched for recurrently mutated genes in TN-ILCs(FDR<0.05), there were ErbB-signaling and DNA-damage-response pathways.
Conclusions
TN-ILCs are rare tumors with poor prognosis. Their specific biological features require newly defined targeted therapeutic strategies
Keywords: Triple negative invasive lobular carcinoma, ERBB2 gene mutations, HER2/PI3K/AKT pathway, Androgen receptor, Luminal androgen receptor subtype
Highlights
-
•
Triple-negative ILCs are distinct from both TN-IDCs and endocrine-responsive ILC.
-
•
TN-ILCs are enriched for actionable mutations in ErbB2 gene and DNA Damage Response genes.
-
•
The Luminal Androgen Receptor (LAR) is the most prevalent subtype in TN-ILCs.
1. Introduction
Hormone receptors negative and HER2 negative (i.e. triple negative, TN) Invasive Lobular Carcinoma (ILC) of the breast, is very rare tumor [[1], [2], [3], [4]]. Two recent analyses based on SEER and NCDB databases, reported that among 19,828 and 38,913 TN breast cancers respectively, only 201 (1.0%) and 540 (1.4%) were ILCs [5,6]. To the best of our knowledge, there are no studies specifically focused on the biology and clinical features of such rare tumors. Limited evidences suggest that HER2 and AR pathways may have a role in biology of TN-ILCs. In a seminal work that characterized molecular alterations of ILCs, it has been reported a non-significant trend for an enrichment of ERBB2 gene mutations in a small group of estrogen receptor negative ILCs analyzed, as compared with ER + ILCs.1 In another work in which TNBCs were assessed according to Lehmann's classification, half of cases of a small cohort of TN-ILCs were classified as luminal androgen receptor (LAR) subtype [7]. Here we reported for a first time a comprehensive characterization of TN-ILCs, including both clinical features as well as molecular alterations, particularly focused on HER2 and Androgen Receptor pathways, in the light of both, the available evidences suggesting a potential role in TN-ILC biology, and their actionability.
2. Methods
We extracted information from prospectively collected institutional databases on all consecutive patients with ILCs operated at two reference cancer centers, the European Institute of Oncology (EIO) and Jules Bordet Institute (JBI), between June 1994 and December 2012. We included in our analysis only triple negative ILCs (TN-ILCs), defined as ER and PgR status <1% in immunoreactive tumor cells and HER2 negative according with 2018 ASCO/CAP guidelines. Patients with a previous primary tumor, with a mixed lobular/ductal histotype, were excluded. Histological types were classified according to WHO criteria and the Armed Forces Institute of Pathology criteria. Tumor grade, peritumoral vascular invasion, ER and PgR status, Ki-67 labelling index, HER2 overexpression and/or amplification were evaluated as previously reported [8,9].The original pathology reports were used.
We also extracted sub-cohorts from the hormone-receptors positive and HER2 negative ILCs (HR+/HER2- ILCs) as well as from the triple negative invasive ductal carcinomas (TN-IDCs) treated during the same time period.
Patients in the sub-cohorts were 1:1:1 matched to patients in the TN-ILCs cohort according to patients age group (<40, 40–49, 50–59 and ≥ 60 years old), nodal status, T stage and year of surgery (before 2003, 2003–2006, 2007–2010, after 2010).
TN-ILCs treated at EIO and JBI, for which adequate FFPE material was available, were evaluated to determine: a) the intrinsic molecular subtype; b) expression of AR protein through IHC as previously described [3]; and c) ERBB2 gene mutational status through targeted NGS.
The molecular subtype classification was carried out by using the PAM50 signature (NanoString Technologies, USA) on the nCounter FLEX Analysis System, following the manufacturer's instruction [10].Subtype classification was performed via NanoString service.
ERBB2 gene mutational status was evaluated by next generation sequencing using an in-house designed GeneRead® QIAact Custom Panels (Qiagen, Germany), following the manufacturer's instruction. Data were analyzed by QIAGEN Clinical Insight (QCI™) Analyze software to evaluate QC and to call sequence variants.
Finally, we analyzed data from three large databases - that are, METABRIC (Molecular Taxonomy of Breast Cancer International Consortium), MSKCC-IMPAKT and Desmedt et al. databases - on gene mutational status of TN-ILCs as well as of HR+/HER2- ILCs and TN-IDCs [11,12].
2.1. Statistical analysis
The primary objective of this study was to assess the outcome of patients affected by early stage TN-ILCs in terms of disease-free survival (DFS) and cumulative incidence of distant metastases (CI-DM). Active follow-up was conducted to determine patient status as of October 2020. Surviving patients were censored at the date of last follow-up. Patients were followed up with physical examination every six months, annual mammography and breast ultrasound, blood tests every 6–12 months and further evaluations only in case of symptoms.
Events considered in the iDFS computations were invasive relapse (categorized as loco-regional events, including ipsilateral invasive breast recurrence, and distant metastases), appearance of a second primary cancer (including contralateral breast cancer), or death, whichever occurred first. The DFS function were estimated using the Kaplan–Meier method. The log-rank test was used to assess differences between groups. The CI-DM curve function was estimated according to methods described by Kalbfleisch and Prentice, taking into account the competing causes of recurrence. The Gray's test was used to assess cumulative incidence differences between groups [13,14].
Co-primary endpoints were: a)to compare outcome of patients with TN-ILCs with that of the matched cohorts of patients with HR+/HER2- ILCs and TN-IDCs; b)to describe the distribution and outcome of patients with TN-ILCs according to tumor intrinsic molecular subtype classification (i.e. luminal A or B tumors versus HER2-enriched or basal-like tumors); c) to assess the expression of androgen receptor (AR) protein in the whole cohort of TN-ILCs as well as in the different intrinsic molecular subtypes; d) to assess the rate of pathogenetic mutations in ERBB2 gene e) to describe genes recurrently mutated in TN-ILCs as compared with HR+/HER2- ILCs and TN-IDCs, and to identify molecular pathways significantly enriched for mutated genes in TN-ILCs.
All analyses were performed with SAS software v.9.4 (SAS Institute, NC). The validated Enrichr tool was used to perform enrichment analyses [15]. All tests were two-sided and p-values<0.05 were considered statistically significant; the false discovery rate (FDR) was used to correct for multiple comparisons.
3. Results
Between June 1994 and December 2012, 2952 and 1200 patients with ILC of the breast were operated respectively at the EIO and JBI. Forty-four (1.5%) patients treated at EIO and 30 (2.5%) at JBI, had TN-ILCs without a history of other primary tumors, and therefore fulfilled the inclusion criteria for the analysis.
The baseline characteristics of the whole cohort of patients with TN-ILCs are shown in Table 1. The majority of patients were treated with mastectomy (55%), and received an adjuvant chemotherapy (71%). Twenty-two percent of patients received anthracycline-containing adjuvant chemotherapies (i.e. AC, FEC or CEF), 28% classic CMF, 25% sequential chemotherapy with the combination of an anthracycline and cyclophosphamide followed by either CMF or a taxane, and 25% different other types of chemotherapy including taxane-containing regimens, oral metronomic combination of cyclophosphamide with methotrexate, or high-dose chemotherapy in clinical trials. The median follow-up was 5.8 (IQR 3.3–10.1)years. Sixty-two percent of patients (46 out of 74) had disease recurrence, of which 46% (21/46) were distant metastases (Table S1). The iDFS at 5 and 10 years of FUP were 50.4% (95% CI, 38.0–61.6) and 37.2% (95% CI, 25.5–48.8), respectively(Fig. 1A). The CI-DM at 5 and 10 years of FUP were 23.2% (95% CI, 14.0–33.9) and 28.2% (95% CI, 17.9–39.4), respectively(Fig. 1B). The DFS and CI-DM assessed separately for the EIO and JBI cohorts of patients were not significantly different (data not shown).
Table 1.
Distribution of patient baseline characteristics.
| Variable | Level | EIO (N = 44) | JBI (N = 30) | P-value | Overall (N = 74) |
|---|---|---|---|---|---|
| Year of surgery, N (%) | Before 2003 | 15 (34) | 9 (30) | 0.49 | 24 (32) |
| 2003–2006 | 12 (27) | 8 (27) | 20 (27) | ||
| 2007–2010 | 10 (23) | 4 (13) | 14 (19) | ||
| After 2010 | 7 (16) | 9 (30) | 16 (22) | ||
| Age (years), N (%) | <40 | 2 (5) | 1 (3) | 0.97 | 3 (4) |
| 40–49 | 6 (14) | 4 (13) | 10 (14) | ||
| 50–59 | 8 (18) | 7 (23) | 15 (20) | ||
| 60+ | 28 (64) | 18 (60) | 46 (62) | ||
| Median (IQR) | 66 (53–71) | 64 (53–71) | 65 (53–71) | ||
| Menopausal status, N (%) | Premenopausal | 7 (16) | 7 (23) | 0.55 | 14 (19) |
| Postmenopausal | 37 (84) | 23 (77) | 60 (81) | ||
| pN, N (%) | pN0 | 16 (36) | 20 (67) | 0.09 | 36 (49) |
| pN1 | 9 (20) | 5 (17) | 14 (19) | ||
| pN2 | 5 (11) | 1 (3) | 6 (8) | ||
| pN3 | 10 (23) | 4 (13) | 14 (19) | ||
| pNX | 4 (9) | 0 | 4 (5) | ||
| pT, N (%) | pT0 | 0 | 3 (10) | 0.14 | 3 (4) |
| pT1 | 21 (48) | 11 (37) | 32 (43) | ||
| pT2 | 15 (34) | 8 (27) | 23 (31) | ||
| pT3/4 | 8 (18) | 8 (27) | 16 (22) | ||
| Tumor grade, N (%) | G1 | 1 (2) | 1 (3) | <0.001 | 2 (3) |
| G2 | 13 (30) | 25 (83) | 38 (51) | ||
| G3 | 22 (50) | 4 (13) | 26 (35) | ||
| Unknown | 8 (18) | 0 | 8 (11) | ||
| Peritumoral vascular invasion, N (%) | No | 32 (73) | 26 (87) | 0.25 | 58 (78) |
| Yes | 12 (27) | 4 (13) | 16 (22) | ||
| Neo-adjuvant treatment, N (%) | No therapy | 38 (86) | 23 (77) | 0.01 | 61 (82) |
| ET | 0 | 1 (3) | 1 (1) | ||
| CT | 1 (2) | 6 (20) | 7 (9) | ||
| CT + ET | 1 (2) | 0 | 1 (1) | ||
| Unknown | 4 (9) | 0 | 4 (5) | ||
| Local treatment, N (%) | Mastectomy w/o RT | 14 (32) | 9 (30) | 0.16 | 23 (31) |
| Mastectomy w RT | 7 (16) | 11 (37) | 18 (24) | ||
| Quadrantectomy w/o RT | 4 (9) | 3 (10) | 7 (9) | ||
| Quadrantectomy w RT | 19 (43) | 7 (23) | 26 (35) | ||
| Adjuvant treatment, N (%) | No therapy | 5 (11) | 10 (33) | 0.007 | 15 (20) |
| ET | 2 (5) | 3 (10) | 5 (7) | ||
| CT | 34 (77) | 12 (40) | 46 (62) | ||
| CT + ET | 2 (5) | 5 (17) | 7 (9) | ||
| Unknown | 1 (2) | 0 | 1 (1) | ||
| Ki67 (%) | Median (IQR) | 22 (13–34) | 10 (10–15) | 0.001 | 15 (10–30) |
Fig. 1.
Title: Invasive disease-free survival of patients with TN-ILCs, overall (panel A) and compared with HR+/HER2- ILCs and TN-IDCs (panel C); cumulative incidence of distant metastases of patients with TN-ILCs (panel B). Legend: Panel A) invasive disease-free survival in TN-ILCs; Panel B) cumulative incidence of distant metastases in TN-ILCs; Panel C) invasive disease-free survival in TN-ILCs compared with HR+/HER2- ILCs and TN-IDCs.
As compared with the two matched sub-cohorts of patients with HR+/HER2- ILCs and TN-IDCs, patients with TN-ILCs had the worst prognosis (TableS2): the iDFS at 10 years of FUP were 39.3% (95%CI, 26.6–51.8) for TN-ILCs, versus 54.1% (95%CI, 39.7–66.4) for TN-IDCs and 68.6% (95%CI, 51.1–80.9) for HR+/HER2- ILCs (log-rank test p-value<0.001; Fig.1C and Fig. S1).
Since a previous work suggested that TN-ILCs could be enriched for the luminal molecular subtype driven by AR, we assessed distribution of molecular intrinsic subtype and AR expression in our cohort of TN-ILCs, and their association with patients’ prognosis.
The molecular intrinsic subtype was defined through PAM50 classifier for 31 out of 74 TN-ILCs (Table S3): 48% were classified as luminal-A (15 out of 31), 3% luminal-B (1/31), 32% HER2-enriched (10/31), and only 16% basal-like (5/31).
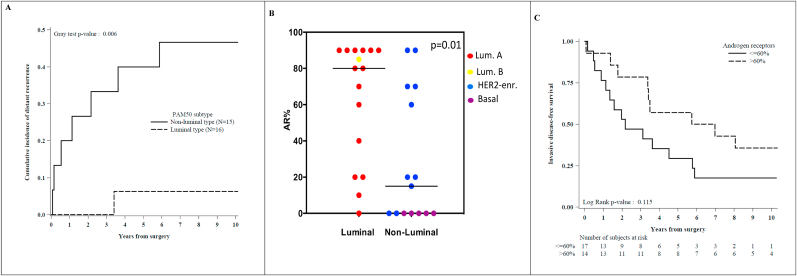
The iDFS was not significantly different between luminal (i.e. luminal A and B tumors) and non-luminal tumors (i.e. HER2-enriched and basal-like tumor; p-value = 0.53, fig. S2). The CI-DM was significantly worse in non-luminal tumors: the CI-DM at 10 years was 46.7% (95%CI, 19.6–70.0) in the group of HER2-enriched or basal-like tumors, while only one distant metastasis was observed in the group of luminal A or B tumors (p-value = 0.006; Fig. 2A). Twenty three out of 31 (74%) TN-ILCs analyzed expressed the androgen receptor (AR) in at least 1% of immunoreactive tumor cells (Table S3).
Fig. 2.
Title: Patients outcome according to molecular intrinsic subtype and AR expression levels. Legend: Panel A) cumulative incidence of distant metastases of TN-ILCs classified as luminal (i.e. Luminal A or B) versus non-luminal (i.e. HER2-enriched or Basal-like) subtypes; Panel B) percentage of AR-positive cells in luminal versus non-luminal tumors; Panel C) invasive disease-free survival of TN-ILCs grouped according to AR expression levels (i.e. above or below the overall AR median value).
Luminal tumors expressed AR more frequently than non-luminal tumors (AR≥1% in 94% of luminal tumors versus 53% in non-luminal tumors; p-value = 0.01; Table S3 and Fig. 2B), and at significantly higher levels (median percentage of AR-positive cells was 80% [IQR, 30–90] in luminal tumors tumors vs. 40% [IQR, 15–70] in HER2-enriched while all the basal-like tumors were AR-negative; p-value = 0.01; Table S3 and Fig. 2B). Tumors expressing higher AR levels (i.e. above the median value of 60%) had a trend for better iDFS (p-value = 0.11; Fig. 2C), and better CI-DM (p-value = 0.21; Fig.S3), as compared with tumors expressing lower AR levels.
Frequency of ERBB2 gene mutation was assessed independently in our cohort of TN-ILCs and in those included in public datasets, to confirm the hypothesis that TN-ILCs harbored a high frequency of pathogenetic mutations in this gene. ERBB2 gene mutational status was evaluated in 35 out of 74 TN-ILCs of our cohort.
Twenty percent of cases analyzed (7 out of 35) harbored a pathogenetic mutation in the ERBB2 gene, and two further cases had a gene variant of unknown significance (VUS). The average Variant Allele Fraction (VAF) was 0.38 (range: 0.19 to 0.79; Table S4). Notably, all but one case of ERBB2-mutated TN-ILCs and assessed through PAM50 assay were classified as HER2-enriched subtype.
ERBB2 was mutated in 20% of 45 TN-ILCs included overall in METABRIC, MSKCC-IMPACT and Desmedt et al. databases (9 out of 45; Fig. 3A); the average Variant Allele Fraction (VAF) was 0.21 (range: 0.1 to 0.57).
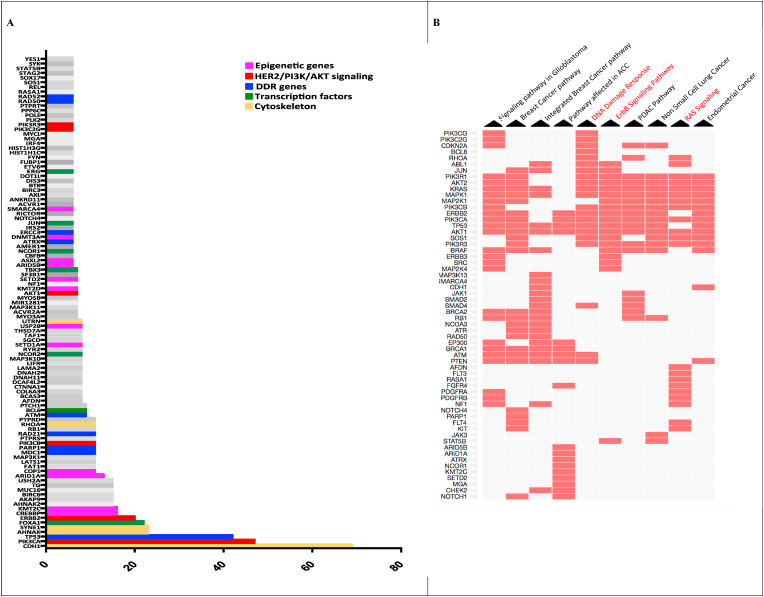
Fig. 3.
Title: Recurrently mutated genes and molecular pathways significantly enriched for mutated genes in TN-ILCs. Legend:Panel A) On y-axis, are represented genes recurrently mutated (i.e. ≥ 5% of cases) in TN-ILCs; on x-axis is reported the percentage of mutation of each gene. Panel B) The Clustergram shows the top 10 pathways significantly enriched for mutated genes in TN-ILCs, ranked according to p-value for enrichment. Red cells in the matrix, indicate for each pathway the belonging genes found mutated in TN-ILCs (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Totally, all but two ERBB2 mutations found in our cohort of TN-ILCs as well as in public datasets analyzed, were known to be pathogenetic in BCs, to activate the HER2 pathway and to predicte response to neratinib in BCs (Table S4).
Finally, we analyzed METABRIC, MSKCC-IMPACT and Desmedt et al. databases., to identify genes recurrently mutated in TN-ILCs as compared with HR+/HER2- ILCs and TN-IDCs, and to discover molecular pathways significantly enriched for mutated genes in TN-ILCs.
Data on genes mutational status of 45 TN-ILCs, 703 HR+/HER2- ILCs and 445 TN-IDCs, were analyzed overall.
Fig. 3A shows 105 genes found recurrently mutated in TN-ILCs in ≥5% of cases, while Table S5 reports all mutated genes. The 5 genes most frequently mutated were CDH1 (mutated in 69% of cases), PIK3CA (47%), TP53 (42%), SYNE1 and AHNAK (both 23%).
Fig. 3B shows the top 10 molecular pathways significantly enriched for mutated genes in TN-ILCs, ranked accordingly to the adjusted p-value for enrichment. Table S7 reports all molecular pathways significantly enriched for mutated genes. Among the top 10 significantly enriched pathways, there were ErbB signaling (namely WikiPathways [WP] number 673), RAS signaling (WP673) and DNA damage response (DDR; WP710) pathways (Fig. 3B).
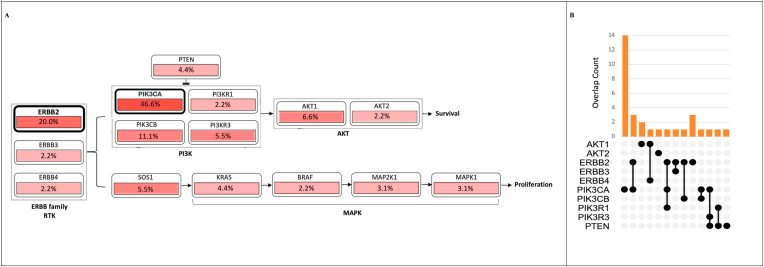
Recurrent mutations were found along the entire ErbB signaling pathway, from RTK (i.e ERBB2, ERBB3 and ERBB4) to regulatory and catalytic subunit of PI3K, PTEN, AKT1 and 2 isoforms, as well as KRAS and MAPK genes (Fig. 4A).
Fig. 4.
Title: Mutation rate in genes belonging to ErbB signaling pathway. Legend:Panel A) Mutation rate found in each of the genes belonging to ErbB signaling pathway (WikiPathways 673). Panel B) Distribution of mutations found in ERBB2/PTEN/PI3K/AKT genes. On x-axis are reported the genes analyzed. On y axis it is reported the number of sample harboring mutation in each single gene exclusively or in combination with the other genes analyzed.
The majority of mutations in HER2/PTEN/PI3K/AKT genes were mutually exclusive(Fig. 4B).
DDR-related genes found mutated in TN-ILCs included DNA damage sensors (i.e. TP53,ATM,ATR,MDC1 and CHECK2) and genes involved in DNA repair mechanisms through homologous recombination (i.e. PALB2,RAD21,RAD50, RAD52,BRCA1,BRCA2), non-homologous end joining (PARP1), mismatch repair (MLH1) and nucleotide excision repair (ERCC1, ERCC3; Fig. 3A and Table S5).
Enrichment analysis using “gene ontology biological process” and “gene ontology cellular components” terms, revealed respectively a significant enrichment of mutated genes coding for proteins involved in the androgen receptor signaling pathway (namely GO:0030521, including MED12,DAXX,RB1,NCOA3 and ARID1A genes; FDR<0.001) and in cell cytoskeleton (namely GO:0005856, including CDH1,AHNAK, NF2,UTRN, SYNE1,DDR2 and RHOA genes; FDR = 0.07).
Fifty-four genes had a significant higher frequency of mutation (FDR<0.05) in TN-ILCs as compared with HR+/HER2- ILCs (Fig. 5 and Table S5).
Fig. 5.
Title: Summary plot of differences in the mutation frequency of recurrently mutated genes in TN versus HR+/HER2- ILCS and TN-IDCs. Legend: Figure shows mutation frequency of each gene in TN-ILCs, TN-IDCs and HR+/HER2-tumor samples.
Among the genes with the highest difference in mutation rate between the two tumor groups, there were TP53 (mutated in 42% of TN-ILCs vs 8% of HR+/HER2- ILCs), ERBB2 (20% versus 7%) and CREBBP (15% versus 1%).
Molecular pathways significantly enriched for genes more frequently mutated in TN-ILCs as compared with HR+/HER2- ILCs, included: ErbB signaling pathway (namely WP673; FDR<0.001); DDR-pathways (namely WP710,WP4016, WP3959 and WP707; FDR<0.001); androgen receptor signaling pathway (namely WP138; FDR<0.001) and Focal adhesion signaling pathway (namely WP306; FDR<0.001; Fig. 5 and Table S5). No gene has been found significantly more frequently mutated in HR+/HER2- ILCs as compared with TN ILCs (Fig. 5 And Table S5).
Twenty-five genes had a significant higher frequency of mutation (FDR<0.05) in TN-ILCs as compared with TN-IDCs (Fig. 5 and Table S5). Among the genes with the highest difference in mutation rate between the two tumor groups, there were CDH1 (mutated in 68% of TN-ILCs versus 1% of TN-IDCs), ERBB2 (20% versus 3%), PI3KCA (46% versus 14%) and FOXA1 (22% versus 8%).
Molecular pathways significantly enriched for genes more frequently mutated in TN-ILCs as compared with TN-IDCs, included: ErbB signaling pathway (namely WP673; FDR = 0.006) and DDR-pathways (namely WP710, WP4016 and WP3959; FDR<0.01; Fig. 5 and Table S5).
TP53 was the only gene significantly more frequently mutated in TN-IDCs as compared with TN-ILCs (respectively 82% versus 42%, FDR<0.001; Fig. 5 and Table S5).
In line with the significantly higher frequency of mutations in a number of DDR genes observed in TN-ILCs, we found a significantly greater tumor mutational burden in such tumor group as compared with both TN-IDCs and HR+/HER2- ILCs (the median number of mutations for sample was respectively 9.9 [IQR 9–16] in TN-ILCs, 4.5 [IQR 3–6] in TN-IDCs and 4 [IQR 3–6] in HR+/HER2; p-value <0.01; fig.S4).
4. Discussion
According to our knowledge, data reported here, provided for the first time a portrait of biologic and clinical features of TN-ILCs.
From a clinical point of view, TN-ILCs were characterized by the worst prognosis when compared with TN-IDCs and HR+/HER2- ILCs, matched for stage of disease.
The majority of TN-ILCs tumors were in advanced pT and pN stage of disease at diagnosis, and only 23% of patients were disease free at 10 years from surgery. The risk of disease recurrence persisted throughout the follow up, with some late recurrence events (i.e. after the first five years from surgery).
From a biological point of view, TN-ILCs were characterized by dysregulation of AR and HER2 pathways.
In gene expression profiling studies, it has been reported that up to 70–80% of TN-IDCs are basal-like [16]. In our analysis only 15% of TN-ILCs were classified as basal-like, whereas the majority were luminal-A.
The prognosis of patients with TN-ILCs was significantly impacted by the intrinsic molecular subtypes classification: both luminal and non-luminal tumors had a substantial risk of locoregional recurrence, but luminal tumors seemed to have a very low risk of distant recurrence, although the small number of patients analyzed does not permit definitive conclusions.
We provided evidences suggesting that AR could be the driver of the luminal phenotype observed in TN-ILCs.
Indeed, almost all the TN-ILCs classified as luminal A or B expressed high levels of AR, and the tumor levels of AR seemed to be associated with the prognosis of patients.
Furthermore, we found a significant enrichment of mutated genes involved in AR signaling pathway, including transcriptional AR-coregulators such as DAXX and NCOA3, or genes associated with castration-resistance in prostate cancer, such us MED12 [17,18].
Such our results are in line with a previous work in which 550 TNBCs were classified according to Lehmann's classification, and half of TN-ILCs assessed (8 out of 17) were classified as LAR subtype, as compared with only 16% of TN-IDCs (67 out of 406) [19]. Notably, the luminal androgen receptor (LAR) subtype was characterized by a distinct mutational profile when compared with other subtypes, with significant enrichment of mutations in the same genes that we found recurrently and highly mutated in TN-ILCs, including PIK3CA, KMT2C, CDH1 and AKT1 genes [7,19].
Taken together all these data indicate that AR could be the driver of the luminal phenotype in TN-ILCs and may represent a potential therapeutic target. Preclinical evidences suggested that PIK3CA mutations in AR-positive TNBC confer meaningful sensitivity to the combination of PI3K and androgen receptor inhibitors. Such therapeutic strategy should be tested in TN-ILCs, considering the high frequency of both AR expression and PI3KCA mutations found in these tumors [20].
The second most frequent molecular subtype in TN-ILCs, was the HER2-enriched.
The analysis of the DNA mutational status of TN-ILCs of our cohort as well as of those from three large public databases, revealed that 20% of cases harbored a mutation in the ERBB2 gene.
The VAF of the ERBB2 mutations was considerably high, with an average value ranging from 0.21 to 0.38, indicating the high clonality of these alterations.
The vast majority of the ERBB2 gene mutations found, were missense point mutations, concentrated in the tyrosine kinase domain of the protein, and are known to be oncogenic and to activate the HER2 pathway [[21], [22], [23]]. All but one of the ERBB2 mutations found in TN-ILCs, are classified as Level 3A alterations for neratinib according to the OncoKB tool, indicating that compelling clinical evidence support these mutations as being predictive of response to the drug in BC.
Furthermore, the ErbB signaling pathway was one of the most enriched for mutated genes in TN-ILCs, with alterations affecting a number of genes along the entire pathway.
All these data indicate HER2/PI3K/AKT pathway as a driver in TN-ILCs, and emphasize the actionable nature of gene mutations found, that should be studied as potential target for therapy [24].
Finally, we identified a number of genes more frequently mutated in TN-ILCs as compared with both TN-IDCs and HR+/HR- ILCs, including transcription factor, epigenetic genes, and genes involved in the DNA repair mechanisms. DNA damage response was one of the pathways most enriched for mutated genes, and some of the DDR genes mutated in TN-ILCs have been demonstrated (i.e. BRCA1 and 2) or are under evaluation(i.e. ATM, ATR, CHECK2, PALB2) as potential predictive factors for response to PARP inhibitors and/or platinum-based chemotherapy, in BCs or in other tumors [25,26].Consistently with this finding we show that TN-ILCs are characterized by a tumor mutational burden significantly higher than both TN-IDCs and HR+/HR- ILCs, that may predict for sensitivity to anticancer immunotherapy.
Strength points of this work were the comprehensive biological characterization of such rare tumors, providing new data at genomic, transcriptomic and immunohistochemistry levels as well as the complete and rigorously curated collection of all the relevant clinical and pathological data on the cohort of patients analyzed and treated in two referral centers for the disease.
The weakness of our work was the retrospective nature of the analysis as well as the limited number of patients and tumor samples analyzed, that however is related to the extreme rarity of the disease.
In conclusion, our data show that TN-ILCs are rare tumors that warrant further investigation in light of the poor prognosis of patients and evidence of specific biological features potentially needing targeted therapeutic strategies.
Contributors
All authors contributed equally.
Declaration of interests
The authors declared no conflicts of interest.
Funding
Beretta Foundation for cancer research and treatment.
Role of funding source
No funding sources had a role in the preparation of this paper.
Acknowledgments
We thank Giorgia Santomauro for data management assistance and Shari Gelber for editorial assistance.
We thank Beretta Foundation for cancer research and treatment, for the the financial support.
Footnotes
Dedicated to our loved friend and mentor, Professor Aron Goldhirsch, with endless gratitude. Fabio and Laura.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.breast.2021.06.011.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Desmedt C., Zoppoli G., Gundem G. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34:1872–1881. doi: 10.1200/JCO.2015.64.0334. [DOI] [PubMed] [Google Scholar]
- 2.Ciriello G., Gatza M.L., Beck A.H. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colleoni M., Rotmensz N., Maisonneuve P P. Outcome of special types of luminal breast cancer. Ann Oncol. 2012;23:1428–1436. doi: 10.1093/annonc/mdr461. [DOI] [PubMed] [Google Scholar]
- 4.Pestalozzi B.C., Zahrieh D. Mallon E Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- 5.Plasilova M.L., Hayse B., Killelea B.K. Features of triple-negative breast cancer.Analysis of 38,813 cases from the national cancer database. Medicine. 2016;95:35. doi: 10.1097/MD.0000000000004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao S., Ma D., Xiao Yi. Clinicopathologic features and prognoses of different histologic types of triple-negative breast cancer: a large population-based analysis. Eur J Surg Oncol. 2018;44:420e428. doi: 10.1016/j.ejso.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Bereche Y., Venet D., Ignatiadis M. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol. 2018;29:895–902. doi: 10.1093/annonc/mdy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavassoli F.A., Devilee P. IARC Press; Lyon, France: 2003. Pathology and genetics of tumours of the breast and female genital organs: WHO classification of tumours series; pp. 13–48. [Google Scholar]
- 9.Rosen P.P., Oberman H.A. Armed Forces Institute of Pathology; Washington, DC: 1992. Atlas of tumor pathology: tumors of the mammary gland. [Google Scholar]
- 10.Parker J.S., Mullins M., Cheang M.C. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis C. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razavi P., Chang M.T., Xu G. The genomic landscape of endocrine-resistant advanced breast cancers. Canc Cell. 2018;34:427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalbfleisch J.D., Prentice R.L. Wiley & Sons Ltd; Hoboken, NJ: 1980. The statistical analysis of failure time data. [Google Scholar]
- 14.Gray R.J. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. [Google Scholar]
- 15.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016:gkw377. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheang M., Martin M., Nielsen T.O. Defining breast cancer intrinsic subtypes by quantitative receptor expression. Oncol. 2013;20:474–482. doi: 10.1634/theoncologist.2014-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbieri E., Baca S., LawrenceM Exome sequencing identifies recurrent SPOP, FOXA1and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin D.Y., Fang H.I., Ma A.H. Negative modulation of androgen receptor transcriptional activity by Daxx. Mol Cell Biol. 2004;24:10529–10541. doi: 10.1128/MCB.24.24.10529-10541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D Lehmann B., Bauer J.M., Chen X. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann B.D., Bauer J.A., Schafer J.M. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors Breast Cancer. Res. 2014;16:406. doi: 10.1186/s13058-014-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bose R., Kavury S., Searleman A. Activating HER2 mutations in HER2 gene amplification negative breast cancer cancer discov. 2013;2:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyman D.M., Piha-Paul S.A., Won H. HER Kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554:189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B.T., Shen R., Buonocore D. Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: results from a phase II basket trial. J Clin Oncol. 2019;37:362–2019. doi: 10.1200/JCO.2018.77.9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakravarty D., Gao J., Phillips S. OncoKB: a precision Oncology knowledge base. JCO Prec Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo J., Porta N., Bianchini D. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2019 Dec 2;(19):30684–30689. doi: 10.1016/S1470-2045(19)30684-9. pii: S1470-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tung N., Robson M., Ventz S. TBCRC 048: a phase II study of olaparib monotherapy in metastatic breast cancer patients with germline or somatic mutations in DNA damage response (DDR) pathway genes (Olaparib Expanded) J Clin Orthod. 2020;38(15_suppl) 1002-1002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.