Summary
Turnover of the primary cilium (PC) is critical for proliferation and tissue homeostasis. Each key component of the PC resorption machinery, the HEF1/Aurora kinase A (AurA)/HDAC6 pathway harbors cis-elements potentially targeted by the transcriptional co-activator myocardin-related transcription factor (MRTF) and/or its partner serum response factor (SRF). Thus we investigated if MRTF and/or SRF regulate PC turnover. Here we show that (1) both MRTF and SRF are indispensable for serum-induced PC resorption, and (2) they act via both transcriptional and local mechanisms. Intriguingly, MRTF and SRF are present in the basal body and/or the PC, and serum facilitates ciliary MRTF recruitment. MRTF promotes the stability and ciliary accumulation of AurA and facilitates SRF phosphorylation. Ciliary SRF interacts with AurA and HDAC6. MRTF also inhibits ciliogenesis. It interacts with and is required for the correct localization of the ciliogenesis modulator CEP290. Thus, MRTF and SRF are critical regulators of PC assembly and/or disassembly, acting both as transcription factors and as PC constituents.
Subject areas: cell biology, organizational aspects of cell biology, functional aspects of cell biology
Graphical abstract

Highlights
-
•
MRTF and SRF are required for primary cilium (PC) resorption
-
•
MRTF and SRF localize to the basal body/PC and interact with ciliary proteins
-
•
MRTF/SRF impact cilium turnover also by non-transcriptional, local mechanisms
-
•
MRTF interacts with CEP290 and inhibits ciliogenesis
Cell biology; Organizational aspects of cell biology; Functional aspects of cell biology
Introduction
The primary cilium (PC) is a microtubule-based, mechano- and chemosensory organelle, which integrates extracellular stimuli (e.g. flow, growth factors, hedgehog ligands) with intracellular signals emanating from the cell cycle, the cytoskeleton and metabolic pathways (Izawa et al., 2015; Malicki and Johnson, 2017; Nachury and Mick, 2019; Song et al., 2018; Spasic and Jacobs, 2017; Wang and Dynlacht, 2018). Using these inputs, the PC orchestrates complex responses affecting proliferation, differentiation, and morphogenesis (Anvarian et al., 2019; Marra et al., 2016; McNeill, 2009; Youn and Han, 2018). Accordingly, genetic defects of the PC cause a wide spectrum of “ciliopathies” (Braun and Hildebrandt, 2017; Reiter and Leroux, 2017; Wheway and Mitchison, 2019) (with clinical manifestations encompassing neurological and skeletal anomalies, polycystic diseases and blindness), while acquired ciliary defects contribute to the pathogenesis of fibrosis (Egorova et al., 2011; Rozycki et al., 2014; Teves et al., 2019) and cancer (Higgins et al., 2019; Liu et al., 2018). Nearly all quiescent animal cells possess a PC, which develops from the basal body (BB) that arises from the membrane-anchored mother centriole. Upon cell cycle reentry the PC gets resorbed, thereby liberating the mother centriole to function as a mitosis organizing center (Sanchez and Dynlacht, 2016; Wang and Dynlacht, 2018). Despite the key roles of PC assembly and disassembly in cell cycle regulation and differentiation, the molecular mechanisms underlying and coordinating these processes are incompletely understood.
Previously, we have described a cell cycle-independent form of deciliation, which was associated with epithelial-mesenchymal/myofibroblast transition (EMT/EMyT) of kidney tubular cells (Rozycki et al., 2014). Similar EMT-related loss of the PC was reported during neurodevelopment (Das and Storey, 2014). Such phenotype shift-induced PC loss likely occurs via ciliary scission (severing) (Das and Storey, 2014). Both studies indicated that this process requires myosin IIA-dependent contractility. We have also shown that the transcriptional co-activator myocardin-related transcription factor (MRTF) (a driver of myosin expression) is indispensable for the EMT-induced PC loss (Rozycki et al., 2014).
MRTF is a central, Rho- and actin-dependent regulator of gene expression. Under resting conditions, the majority of MRTF molecules are bound to monomeric actin; upon actin polymerization, G-actin dissociates from MRTF, thereby unmasking its nuclear localization signal and allowing its nuclear entry. In the nucleus, MRTF associates with serum response factor (SRF), and the complex drives gene expression via the CC(A/T-rich)6GG cis-element, the so-called CArG box (Esnault et al., 2014; Miano et al., 2007; Olson and Nordheim, 2010). Since EMT requires actin polymerization/reorganization (Fan et al., 2007; Masszi et al., 2010), and MRTF drives myosin IIA expression (Esnault et al., 2014; Rozycki et al., 2014), we attributed the MRTF-dependence of EMT-induced deciliation primarily to the impact of MRTF on myosin. Nonetheless, several observations prompted us to ask whether MRTF might play a more general role in PC homeostasis, which goes beyond its impact on myosin-dependent regulation of PC scission. Namely, we found that the PC was longer in MRTF-depleted cells (Rozycki et al., 2014). We have also noted that key components of the cilium resorption apparatus harbor CArG box(es) in their promoter (see below). Furthermore, a recent high-throughput BioID analysis (Gupta et al., 2015) (and see Gingras lab, ProHits database at https://prohits-web.lunenfeld.ca/) suggests that MRTF may be in the vicinity of (i.e. in potential physical contact with) ciliary/centrosomal/BB proteins. Based on this scenario, we asked whether (1) MRTF might be necessary for “classic”, serum-induced (cell cycle-dependent) cilium resorption; (2) MRTF might exert both transcriptional and direct (local) effects on PC homeostasis; and (3) MRTF might also impact ciliogenesis, the other key determinant of cilium turnover.
The best characterized mechanism for serum-induced PC resorption is mediated by the human enhancer of filamentation-1 (HEF1) — Aurora A kinase (AurA) — Histone deacetylase 6 (HDAC6) pathway (Plotnikova et al., 2012; Pugacheva et al., 2007; Ran et al., 2015). Briefly, upon serum addition the adaptor HEF1 (also known as NEDD9) binds to and activates AurA, which in turn phosphorylates and activates HDAC6. This enzyme catalyzes tubulin deacetylation, which is believed to destabilize (ciliary) microtubules (Matsuyama et al., 2002; Pugacheva et al., 2007). Because we noticed that each of the three pathway components has CArG box(es) in their promoter, and high-throughput ChiPseq analyses suggested the presence of functional CArGs in HEF1 and AurA promoters (Bruna et al., 2012; Esnault et al., 2014), we started our inquiry by investigating the impact of MRTF and SRF on this deciliation pathway.
Here we show that MRTF and SRF are major regulators of PC homeostasis, as they are required for PC resorption. Moreover, they exert their effects not only as transcription factors. Namely, both MRTF and SRF interact with components of the major deciliation pathway, and MRTF is critical for maintaining AurA protein stability. Intriguingly, both MRTF and SRF localize to various parts of the PC, and MRTF is involved in the proper recruitment of components of the deciliation machinery. Finally, MRTF is also an inhibitor of ciliogenesis; it interacts with and impacts the localization of CEP290, a protein involved in cilium assembly. Together these findings define SRF and MRTF as key PC regulators, unraveling new, direct mechanisms whereby these factors exert their biological effects, beyond mere transcriptional control.
Results
MRTF and SRF are required for serum-induced resorption of the PC
Having shown that MRTF is indispensable for EMT-provoked loss of the PC (Rozycki et al., 2014), we sought to investigate if MRTF also impacts serum-induced PC resorption. Upon serum deprivation the majority of LLC-PK1 kidney tubular cells develop a long PC (Figure 1A), making this cell type ideal to follow PC turnover. We transfected these cells with non-related siRNA (siNR) or a mix of MRTF-A and MRTF-B-specific siRNAs (together referred to as siMRTF) (Figure 1B) as in our previous studies (Masszi et al., 2010; Rozycki et al., 2014; Speight et al., 2016), and then exposed them to 10% serum-containing medium for the indicated times (Figures 1A and 1C). Serum induced robust resorption of the PC in siNR-transfected cells, as revealed by acetylated tubulin (AcTub) staining (Figures 1A and 1C). MRTF downregulation slightly increased the degree of ciliation under serum-free conditions, and, more importantly, drastically inhibited serum-induced PC resorption (Figures 1A and 1C)l. Namely, after a 24-hr serum treatment the percentage of ciliated cells was six-fold higher in the MRTF-silenced group, as ciliation dropped from ≈70% to ≈10% in the siNR-transfected control, while it decreased from ≈80% to ≈60% in the siMRTF-transfected cultures (Figure 1C). Moreover, inhibition of serum-induced PC loss by MRTF silencing was not restricted to LLC-PK1 cells but could also be observed in retinal epithelial cells (RPE), a cell type frequently used for ciliary studies (Figures 1D–1F). As MRTF (at least as a transcriptional co-activator) acts in conjunction with SRF, we next tested the impact of SRF on serum-induced PC resorption. Elimination of SRF (Figure 1B) repressed PC resorption (Figure 1A) to almost the same extent as MRTF silencing (Figure 1C). (Extended Western blots for antibodies used in this study are shown in Figure S1.)
Figure 1.
MRTF and SRF are necessary for serum-induced primary cilium resorption
(A) LLC-PK1 cells were transfected with control (non-related, NR) or specific siRNAs against pig MRTF A/B (siMRTF) or SRF (siSRF) (50 nM for each gene) for 24 hr under serum-free conditions. The cells were then exposed to fresh serum-free or 10% serum-containing medium (serum) for 24 hr. Primary cilia were visualized by acetylated tubulin staining (red) and nuclei by Dapi (blue). Bar: 20 μm.
(B) Effective silencing of MRTF and SRF in LLC-PK1 cells was verified by immunoblotting.
(C) Time course of serum-induced primary cilium resorption in LLC-PK1 cells transfected with NR, siMRTF or siSRF. The percentage of ciliated cells was determined using acetylated tubulin staining for each time point (0, 4, 8, 12, 24 hr); n = 5–6 for each time point, with ≈150 cells counted/condition. The plot shows each individual point, and the mean ± SEM. The percentage of ciliated NR-transfected cells was compared to that of siMRTF- or siSRF-transfected cells at each corresponding time point using unpaired t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(D) Human retinal pigment epithelial cells were transfected with siNR and human siMRTF and treated and stained as in A.
(E) MRTF knockdown was verified by western blotting.
(F) Time course of ciliation percentage, quantified as in C, n = 6.
(G and I) Verification of the effect of MRTF silencing on serum-induced cilium resorption using an alternative cilium marker, Arlb13 in LLC-PK1 (G) and RPE (H) cells. Conditions were as in A.
(H and J) Quantitation of the results shown in G and I, respectively; n = 4, groups were compared by ANOVA.
Since, in principle, MRTF might affect tubulin acetylation (Fernandez-Barrera et al., 2018), we wished to verify these results using an alternative PC membrane marker, Arl13b (Caspary et al., 2007). Downregulation of MRTF efficiently preserved the PC in serum-treated cells as visualized by Arl13b staining as well, both in LLC-PK1 (Figures 1G and 1H) and in RPE cells (Figures 1I and 1J). Together, these results revealed that MRTF and SRF are essential contributors to the process of serum-induced deciliation in epithelial cells.
Finally, we wished to substantiate that serum-induced PC resorption and the EMT-associated PC loss, albeit both MRTF-dependent, represent two fundamentally different types of deciliation. To this end, we visualized morphological changes in the PC using time-lapse confocal microscopy in Arl13b-mCherry-tranfected live cells. As expected, serum induced a gradual shortening of the PC, eventually leading to its complete resorption (Figure S2A, upper panels). This process was prevented by MRTF silencing; the PC remained long, although some beading of the Arl13b labeling was noted along the axoneme (Figure S2A. lower panels). To visualize PC loss during EMT, this process was induced by the combined stimuli of cell contact disassembly (using low Ca2+ medium) and TGFβ as in our earlier studies (Masszi et al., 2010; Rozycki et al., 2014). Since in EMT PC alterations occur after an induction phase, the process was monitored after 12 hr of stimulation. In siNR-transfected cells the PC exhibited dramatic morphological changes (Figure S2B): after a lag phase, a bulge developed at the ciliary tip, which continued to grow, eventually forming a large vesicle. Initially the enlarged tip remained connected to the plasma membrane via a shortening stalk. Later the vesicle separated from the membrane, presumably via fission. Thus EMT leads to PC loss by shedding of a large ciliary vesicle. These gross morphological changes and the subsequent shedding were prevented by MRTF depletion (Figure S2C); only a slight bulging of the tip and occasional beading of Arl13b distribution along the axoneme were observed. Taken together, these findings imply that MRTF is indispensable for various types of PC loss, presumably through distinct mechanisms. In this study, we chose to further explore the role of MRTF in PC resorption.
Mechanisms underlying the requirement for MRTF and SRF for PC resorption
To gain insight into the mechanism whereby MRTF and SRF contribute to PC resorption, we initially considered the most plausible possibility, namely that they might regulate, as transcription factors, the expression of key components of the cilium resorption machinery. This rationale was strengthened by in silico findings that each component of the HEF1-AurA-HDAC6 pathway contains CArG box(es) in their promoters (Figure 2A). Therefore, we determined the impact of MRTF or SRF silencing on both mRNA (Figures 2B, 2E, and 2G) and protein (Figures 2C, 2D, 2F, and 2H) expression of each component of the pathway under resting (serum-deprived) conditions and upon serum treatment (2 hr). Serum addition increased mRNA expression for each pathway component (Figures 2B, 2E, and 2G). Strikingly, however, MRTF downregulation substantially elevated HEF1 mRNA expression both with and without serum treatment (Figure 2B), and this effect was matched by a robust rise in HEF1 protein expression (Figures 2C and 2D for quantitation). In contrast, SRF silencing reduced the basal HEF1 mRNA level and eliminated the serum-induced increase in HEF1 mRNA (Figure 2B) and protein expression (Figures 2C and 2D). Together, these findings indicate that HEF1 expression is SRF-dependent, but it does not require (in fact it is suppressed by) MRTF. This may be due to a competition between MRTF and other potent SRF-binding transcriptional co-activators (ternary complex factors, TCFs (Cen et al., 2003; Gualdrini et al., 2016; Miano, 2003; Wang et al., 2004) (see Discussion)). Collectively these results imply that reduced HEF1 expression may explain the inhibitory effect of SRF silencing on PC resorption but cannot account for the similar effect of MRTF silencing.
Figure 2.
The effect of MRTF or SRF downregulation on basal and serum-induced mRNA and protein expression of the components of the HEF1-AurA-HDAC6 cilium resorption pathway
(A) The promoters of HEF1, AuroraA, and HDAC6 all contain CArG boxes.
(B, E, and G) Cells were transfected with siNR, siMRTF, or siSRF as in Figure 1A, serum-deprived for 48 hr and then left untreated or exposed to 10% serum-containing medium for 3 hr. mRNA levels were determined by qPCR. Values are expressed as fold changes compared to the siNR-transfected, serum-deprived controls (n = 3 for each gene and each condition).
(C) Cells were transfected and treated as in B, and then processed for immunoblotting for the indicated proteins. A representative blot is shown.
(D, F, and H) Densitometric quantitation of protein expression for the same conditions as shown in C. Each protein was normalized to the corresponding GAPDH level and expressed as fold change compared to siNR-transfected, serum-deprived controls (n = 3). ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001, determined by ANOVA while & and $ p < 0.05 where the fold change in corresponding two groups were determined by ratio t test (in D NR/serum-free vs. NR/serum ($) and NR/serum vs. siSRF/serum (&); in NR/serum vs. siSRF/serum ($)).
In this regard, changes in AurA expression provided further interesting insights. While the mRNA for this critical enzyme was not substantially affected by either MRTF or SRF downregulation (less than 1.4 fold) (Figure 2E), elimination of MRTF significantly reduced the level of AurA protein expression, both in the absence and presence of serum (Figures 2C and 2F). The robust decrease in AurA protein but not in mRNA expression suggested a non-transcriptional mechanism, which we subsequently analyzed further (see next section). Finally, HDAC6 mRNA expression was modestly but significantly reduced by MRTF or SRF silencing (Figure 2G), and siMRTF slightly decreased HDAC6 protein expression, as well as in the investigated time frame (Figures 2C and 2H). While these modest effects on HDAC6 might contribute, they are likely insufficient to account for the major impact of MRTF elimination on PC resorption.
To assess the generality of the observed effects, we checked the entire MRTF profile in RPE cells as well (Figure S3). The findings recapitulated those obtained in LLC-PK1 cells, showing that MRTF silencing resulted in a robust increase in HEF1 expression, no profound change in AurA mRNA levels yet strong decrease in AurA protein expression and essentially no effect on HDAC6. Taken together, SRF and MRTF had significant (and opposite) impact on HEF1 expression, while MRTF reduced AurA expression via a non-transcriptional mechanism.
MRTF exerts non-transcriptional effects: it interacts with AurA and is required to maintain AurA stability
AurA levels are known to be regulated by protein degradation, a process controlled via the interaction of AurA with various adaptor proteins (Jia et al., 2014; Kiat et al., 2002; Lim and Gopalan, 2007a, b; Lindon et al., 2015). Having seen no impact of MRTF downregulation on AurA mRNA we wondered if it might alter AurA protein stability. To approach this question, we initially treated cells with the proteasome inhibitor MG-132, and tested if this compound affects the MRTF silencing-induced reduction in AurA protein. As a positive control, we silenced HEF1, which has been shown to bind to and thereby stabilize AurA (Pugacheva et al., 2007). Downregulation of either HEF1 or MRTF caused a strong decrease in AurA protein and these effects were entirely prevented by MG-132 (Figures 3A and 3B).
Figure 3.
MRTF interacts with AurA and inhibits its degradation; non-transcriptional effects of MRTF
(A) LLC-PK1 cells were transfected with the indicated siRNAs, and 24 hr later placed in serum-free medium supplemented with vehicle (DMSO) or the proteasome inhibitor MG-132 (20 μM) as indicated. After an additional 24 hr, cell lysates were prepared for Western blotting and probed for the indicated proteins.
(B) Experiments as in A were quantified by densitometry, and results were normalized to the level of NR-transfected DMSO-treated control is each experiment (n = 3). ∗p < 0.05, ∗∗p < 0.01, the corresponding groups were compared by t test.
(C) LLC-PK1 cells were transiently transfected with plasmids encoding HA-tagged MRTF or Myc-tagged AurA or the combinations of these and 24 hr later HA-MRTF was immunoprecipitated with anti-HA. Protein levels in the input and the precipitates were detected by immunoblot using anti-HA or anti-Myc antibodies.
(D) Endogenous MRTF was immunoprecipitated from LLC-PK1 cells. Total cell lysates and immunoprecipitates were probed with the indicated antibodies.
(E) LLC-PK1 cells were transfected with siNR or siMRTF, and 24 hr later, the level of indicated proteins was examined in the total cell lysates (left) and in immunoprecipitates after pull-down with anti-AurA (right).
(F) Quantification of the various protein-protein interactions. The amount of coprecipitating proteins were normalized to the corresponding amount of immunoprecipitated AurA (n = 3-4, mean ± SD). Results were compared to interactions determined in NR-transfected cells, using ratio t test, ∗p < 0.05, ∗∗p < 0.01.
(G) Differential capacity of wild-type (WT) and TADless (TL) MRTF to exert transcriptional effects. LLC-PK1 cells were co-transfected with the smooth muscle actin (SMA) promoter firefly luciferase construct and renilla TK reporter (for normalization) along with either an empty vector (pcDNA, control) or plasmids encoding HA-tagged WT or TL MRTF. (n = 3, mean ± SD). The results verify that TL MRTF has lost its transactivation capacity.
(H) LLC-PK1 cells were serum-deprived and transfected with the porcine-specific MRTF (psMRTF) siRNA, then re-transfected with siRNA-resistant (murine) HA-tagged wild type (WT) or transcriptionally inactive (TADless, TL) MRTF. Cells were then re-exposed to serum and after 24 hr fixed and co-stained with anti-HA (to visualize MRTF-retransfection) and acetylated tubulin (to visualize the PC). Successful downregulation of MRTF-A and B by psMRTF is shown under the images.
(I) Quantification of cells with cilia in non-transfected (NT) neighbors and in WT- or TL-expressing cells. In each experiment, 25–35 cells were counted; ∗∗p < 0.01. Note that both WT and TL support cilium resorption. Bar: 20 μm.
Having observed that MRTF protects AurA from proteolytic degradation, we asked if MRTF could form a complex with AurA. First, we tested this possibility using heterologous expression. Cells were transfected with Hemagglutinin (HA)-tagged MRTF and/or Myc-tagged AurA, and MRTF was immunoprecipitated by an anti-HA antibody. The HA-MRTF precipitates contained Myc-AurA, indicating that complex formation can indeed occur between these proteins (Figure 3C). Next, we tested whether this can be demonstrated for the endogenous proteins as well. Endogenous MRTF pulled down AurA, both in the presence and absence of serum (Figure 3D).
In light of these findings, we investigated the mechanism whereby MRTF interferes with AurA degradation. Various protein-protein interactions have been reported to regulate this process. A major one of these involves formation of a complex between AurA, AurA interacting protein (AIP), and antizyme 1 (AZ1), which ultimately targets AurA for a non-conventional, proteasome-dependent but ubiquitin-independent degradation (Kiat et al., 2002; Lim and Gopalan, 2007a, b). An alternative pathway is mediated by Smad4 binding to AurA, which promotes ubiquitin-dependent proteasomal degradation (Jia et al., 2014). To test the potential involvement of these interactions, we immunoprecipitated AurA from control and MRTF-silenced cells and probed the precipitates for the degradation-regulating proteins. In control cells, MRTF was readily detectable in the AurA precipitates, verifying our previous findings (Figure 3E) by reverse immunoprecipitation. In addition, AurA also pulled down some Smad4, AIP, and AZ1. Importantly downregulation of MRTF increased the association of each of these proteins with AurA (Figure 3E), despite reduced total AurA levels (input) upon MRTF elimination. Normalization to precipitated AurA revealed significantly higher association of AurA with Smad4, AIP, and AZ1 (Figure 3F). These results suggest that MRTF inhibits the association of AurA with key components of both ubiquitin-dependent and -independent degradation systems, overall enhancing AurA stability.
To substantiate that MRTF contributes to deciliation, at least in part, independent of its transcriptional activity, we performed complementation experiments. We silenced endogenous MRTF in LLC-PK1 cells and re-expressed siMRTF-resistant HA-tagged MRTF constructs. One group of cells were transfected with HA-tagged wild-type (WT) MRTF, while the other with an HA-tagged truncation mutant, lacking the C-terminal transactivation domain (TAD) (TADless (TL) MRTF). Transcriptional activity of WT HA-MRTF and inactivity of TL HA-MRTF was independently verified by luciferase assays using the SMA promoter-Luc construct, a CArG box-driven reporter (Speight et al., 2016) (Figure 3G). Cells were first serum-deprived then incubated with serum for 24 hr and the percentage of cells with cilia was quantified. As expected, retransfection with WT-MRTF significantly reduced ciliation (Figure 3H); remarkably TL-MRTF exerted a similar effect (Figure 3H). These results further suggest that the transcriptional activity of MRTF is not a prerequisite for its capacity to promote serum-induced deciliation.
MRTF and SRF are present in BB/centrosome or the PC
Since AurA is found in the PC/BB, we asked if MRTF might also localize to these structures. This assumption was supported by recent BioID data available in the ProHits repository, showing proximity of MRTF to various centrosomal/ciliary baits (Gupta et al., 2015). To address this question, we initially used double immunostaining for MRTF and γ-tubulin, a centrosome/BB marker. Confocal microscopy images revealed that MRTF clustered in punctate aggregates below the apical membrane in LLC-PK1 cells (Figure 4A, left side). These MRTF punctae showed extensive colocalization with γ-tubulin clusters (Figure 4A, left side, bottom two rows). The overlap between the two labels was often partial in that the MRTF clusters were in close association with but extended beyond the boundaries of the γ-tubulin dots (see enlarged image, bottom row). While the MRTF signal was strongly enriched in the BB/centrosome areas, occasionally it also penetrated the ciliary shaft. Similar observations were made in RPE cells as well (Figure 4A, right side, Figures S4D and S4E). To substantiate the specificity of these findings, we tested 5 different MRTF antibodies (3 against MRTF-A and two against MRTF-B) in 3 different epithelial cell types (LLC-PK1, RPE, and IMCD3 cells). A table summarizing the results and representative images are shown in (Figures S4A and S4B). Although the efficiency of the different MRTF antibodies was, in some cases, cell-type dependent (see table), each of them visualized similar clusters, which consistently overlapped with γ-tubulin staining. These experiments verified that both MRTF-A and B could localize to the BB/centrosome (and to a much lesser extent to the axoneme) in various kidney and retinal epithelial cells from 3 different species (pig, human, mouse).
Figure 4.
MRTF and SRF localize to the basal body and the primary cilium
(A) LLC-PK1 (left panels) and RPE cells (right panels) were serum-deprived and co-stained for MRTF and the basal body marker γ-tubulin. Staining was visualized by confocal microscopy. Scale bar 20 μm and 5 μm for full and boxed images, respectively. The last panels (Box) show the magnified regions demarcated by the dashed rectangles in the corresponding merged images. Note that distinct MRTF patches colocalize or partially overlap with γ-tubulin (BB), but MRTF staining occasionally penetrates the cilium as well.
(B) LLC-PK1 cells (left panels) and RPE cells (right panels) were co-stained for SRF and the cilium marker acetylated tubulin. The last panels (Box) show the magnified regions demarcated by the dashed rectangles in the corresponding merged images. Note that SRF, as expected, resides primarily in the nucleus. In addition, SRF also shows clear localization to the primary cilium in a subset of cells.
Next, we checked if SRF also exhibited some ciliary/BB distribution. As expected SRF staining showed predominantly nuclear localization, both in LLC-PK and in RPE cells (Figure 4B left and right side, respectively), and we did not observe colocalization with γ-tubulin (not shown). However, over this large nuclear “background”, a faint but definitive SRF labeling was clearly detectable in a subset of primary cilia, as visualized by AcTub staining, in both cell types (Figures 4B and S4C). SRF was distributed along the entire axoneme, often showing bead-like accumulations in the ciliary shaft. The presence of SRF in the PC is consistent with a single previous report, showing SRF staining in motile cilia of airway epithelial cells (Nordgren et al., 2014). Taken together, our results indicate that both MRTF and SRF can localize to ciliary structures, and they reside predominantly in the BB region and along the axoneme, respectively. These findings further strengthen the notion that MRTF and SRF could regulate PC homeostasis not only as transcriptional (co)activators but also as distinct PC constituents.
MRTF is recruited to the PC and impacts ciliary protein-protein interactions
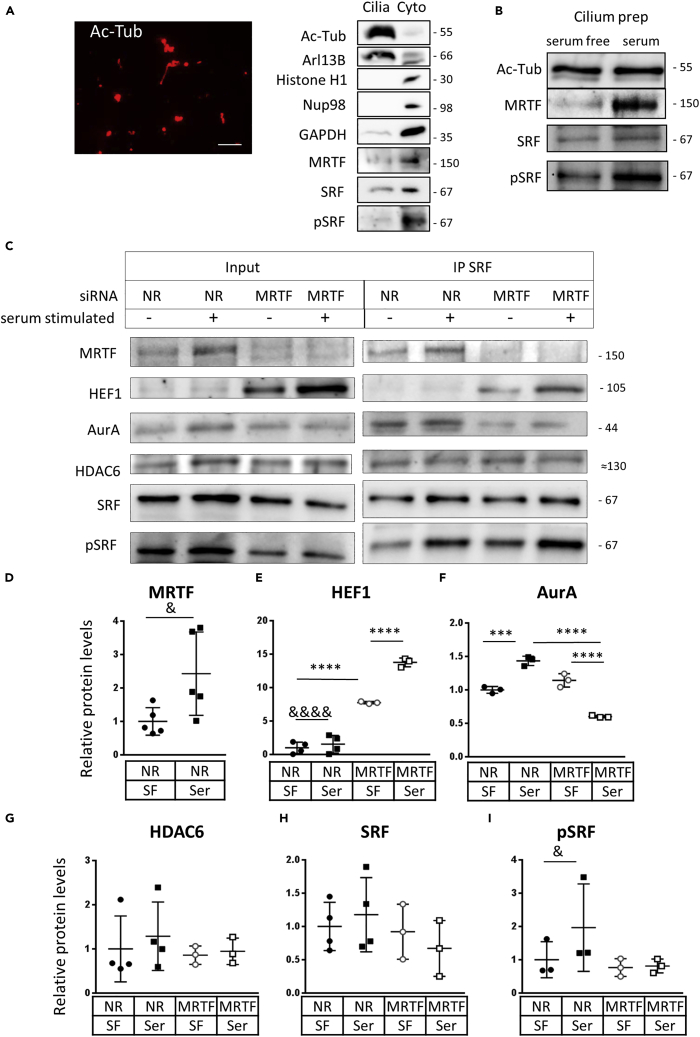
To substantiate these morphological findings by biochemical means and to assess if serum treatment induces changes in ciliary content of MRTF and SRF and/or their interactions with other cilium components, we isolated primary cilia from LLC-PK1 cells (Mitchell, 2013; Rozycki et al., 2014) (see STAR Methods). The cells were first serum-deprived and then either left untreated or re-exposed to serum for a brief time period (2 hr) wherein cilium resorption was still marginal in the majority of cells. Microscopic analysis of the obtained ciliary prep showed the presence of AcTub-positive tubular and vesicular structures, corresponding to PC-derived material (Figure 5A, left panel). Successful isolation of the cilium fraction was also verified biochemically using ciliary (Ac-Tub, Arlb13), nuclear (histone), nuclear/cytosolic (Nup98) and predominantly cytosolic (GAPDH) markers (Figure 5A, right panel). Western blot analysis of these preps indicated similar AcTub content without or with short serum exposure (Figure 5B). Importantly, MRTF was present in the cilium preparations at low levels (Figures 5A and 5B), and serum substantially increased the ciliary MRTF content (Figure 5B). SRF was readily detectable under basal conditions and did not change upon serum treatment (Figures 5A and 5B). However, serum addition increased the phosphorylation of ciliary SRF (Figures 5B and 5C and see also 5H and I for quantification, as detailed below). In the following experiments, we wished to determine if serum induces changes in the ciliary content of the main components of the PC resorption machinery and to assess if MRTF played a role in their presence or serum-induced recruitment to the PC. To this end, cells were transfected with NR or MRTF siRNAs, serum-deprived to induce PC assembly and then left untreated or re-exposed to serum prior to PC isolation. As observed before (Figure 5B), serum treatment caused a significant increase in the ciliary MRTF content (Figures 5C and 5D), and the specific siRNAs efficiently depleted MRTF from the PC as well (Figure 5C). Resting ciliary HEF1 levels were very low, and serum induced only weak (albeit significant) HEF1 recruitment to the PC. However, MRTF depletion caused a dramatic increase in ciliary HEF1 content, which was further elevated by serum treatment (Figures 5C and 5E). These results were in full agreement with the strong inhibitory effect of MRTF on HEF1 expression and also indicated that HEF1 recruitment to the PC is independent of MRTF.
Figure 5.
MRTF and SRF are present in isolated cilia and interact with the components of the deciliation machinery
(A) Primary cilia were isolated from LLC-PK1 cells as described in STAR Methods. Left panel: An aliquot of the preparation was layered onto a coverslip, fixed, stained with acetylated tubulin, and visualized by immunofluorescence microscopy. Note the compact tubular and vesicular acetylated tubulin-positive structures, indicating large enrichment of PC-derived material. Right panel: Quality control of the cilium preparation. Equal amount of protein (5 μg) from cilium preparations and the non-ciliary fraction (residual whole cell lysates) of serum-deprived cells were analyzed by Western blotting using ciliary (Ac-Tub, Arl13b), nuclear (histone), nuclear/cytosolic (Nup98) and cytosolic (GAPDH) markers. Note the robust enrichment of ciliary markers and the absence of the nuclear marker in the cilium preparation.
(B) The effect of serum stimulation on the level of indicated proteins in PC preparations as determined by Western blotting. LLC-PK1 cells were serum-deprived for 48 hr, and then left untreated or exposed to 10% serum-containing medium for 2 hr. Note that serum induces substantial recruitment of MRTF and increases SRF phosphorylation in the PC.
(C) LLC-PK cells were transfected with siNR or siMRTF for 24 hr, serum-deprived for 48 hr, and then incubated for 3h in the absence or presence of serum. Primary cilia were then isolated from each cell population. The ciliary content of the indicated proteins (Input) is shown in the left panel. These preparations were then subjected to immunoprecipitation with an anti-SRF antibody (IP SRF), and the precipitates were probed for SRF and the indicated proteins, shown in the right panel.
(D–I) Quantification of the indicated proteins in cilium fractions isolated from control (NR) or MRTF-silenced (MRTF) cells, under serum-free (SF) or serum-stimulated (Ser) conditions. The average density obtained from the control fractions (siNR-transfected cells, SF condition) was set to 1 for each protein, and means ± SD are shown. ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001, determined by ANOVA; & p < 0.05, &&&& p < 0.0001 when fold changes were compared only in the corresponding two groups by ratio t test.
See also Figure S5 for changes in each individual experiment.
In cells transfected with control (NR) siRNA, serum triggered AurA recruitment to the PC, as expected (Figures 5C and 5F). MRTF depletion in itself did not significantly change basal ciliary AurA levels. Given that MRTF elimination can reduce total cellular AurA content, this finding was somewhat surprising and may be related to the fact that in serum-deprived cells ciliary MRTF level is very low to begin with. Remarkably, however, MRTF downregulation entirely prevented the serum-induced rise in ciliary AurA content. In fact, serum induced a net reduction in ciliary AurA protein in the absence of MRTF (Figures 5C and 5F). Thus, efficient serum-induced ciliary accumulation/recruitment of AurA depends on MRTF. Moreover, these findings (i.e. that basal ciliary AurA levels were not substantially affected by siMRTF, while the serum-induced response was abolished/reversed) suggest that MRTF regulates ciliary AurA localization beyond its effect on AurA expression.
We did not find significant changes upon serum treatment or MRTF downregulation in ciliary HDAC6 or SRF content (Figure 5C, and for quantitation G and H). However, serum-induced SRF phosphorylation was reduced in MRTF-downregulated cells (Figures 5C and 5I) (See also the individual datasets for the same experiments in Figure S5).
Finally, we immunoprecipitated SRF from the abovementioned isolated cilium samples and analyzed co-precipitating proteins, using Western blots (Figure 5C, right panel). In preparations from cells transfected with control siRNA, ciliary SRF pulled down MRTF, AurA and HDAC6 but not HEF1. Upon MRTF depletion, HEF1 was detectable in the SRF precipitates and showed an increase upon serum stimulation, whereas AurA exhibited a major reduction (uncoupling from SRF) without or with serum stimulation. No obvious change was observed in HDAC6. Taken together, these results imply that ciliary SRF can form a complex with the major regulators of deciliation, and its interaction with HEF1 and AurA is controlled by MRTF, which inhibits and facilitates their association, respectively. These changes, at least in part, might be due to MRTF's differential effect on the expression of these proteins.
MRTF is an inhibitor of ciliogenesis, and this action does not require the transactivation domain
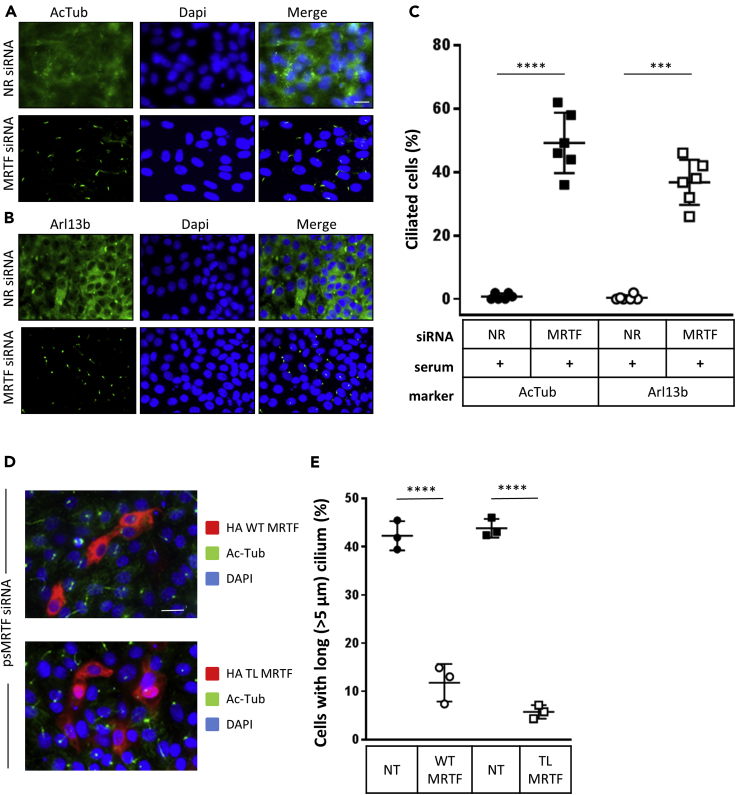
So far our data have implicated MRTF as an important regulator of PC resorption and suggested that a substantial part of its effects is mediated via non-transcriptional mechanisms. As PC homeostasis is determined by the relative rates of PC resorption and ciliogenesis, it was conceivable that MRTF might affect the latter process as well. In support of this possibility, we have shown previously that MRTF downregulation increases average PC length under serum-free conditions (Rozycki et al., 2014). The following experiments were designed to discern whether MRTF might impact (presumably constitutively inhibit) ciliogenesis and if so whether this effect could be, at least partially, related to non-transcriptional actions of MRTF. To address these questions, LLC-PK1 cells were constantly kept in a serum-containing medium, a non-permissive condition for ciliogenesis. Indeed, under these conditions hardly any cell developed a PC, as determined by AcTub or Arl13b staining (Figures 6A and 6B, and quantitation in C); instead consistent with earlier observations, the ciliary markers showed some diffuse cytosolic distribution. Remarkably, despite the incessant presence of serum, downregulation of MRTF resulted in the development of a PC in approximately half of the cells as detected by either marker (Figures 6A–6C). To address whether non-transcriptional mechanisms could contribute to the ciliogenesis-inhibiting capacity of MRTF, we applied the same strategy as in Figure 3H; we silenced MRTF in LLC-PK1 cells and re-expressed HA-tagged WT or TL MRTF constructs (Figure 6D). Experiments were carried out in the constant presence of serum. Forty hours after transfection, cells were stained for the HA-epitope and AcTub (Figure 6D), and efficient ciliogenesis was quantified by determining the percentage of cells that developed a >5μm PC (Figure 6E). MRTF depletion resulted in the formation of such cilia in approximately 40% of the cells that did not express WT HA-MRTF. As expected, re-introduction of WT HA-MRTF profoundly inhibited PC formation, inducing a four-fold reduction in the frequency of ciliated cells. Remarkably, TL HA-MRTF had a similar capacity to prevent ciliation. Taken together, these results suggest that MRTF can exert constitutive inhibition on ciliogenesis, and this effect does not require the transcriptional activity of MRTF.
Figure 6.
MRTF silencing promotes ciliogenesis even in the presence of serum, partially via a transcription-independent mechanism
(A and B) LLC-PK1 cells were transfected with siNR or siMRTF and continuously kept in serum-containing medium for 48 hr before processing for immunofluorescence. Cells were stained with primary cilia markers Acetylated tubulin (A) or Arl13b (B). Scale bar: 20 μm.
(C) The percentage of ciliated cells was determined using both cilium markers (mean ± SD, n = 6, ≈200 cells/condition). ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001 using t test.
(D) LLC-PK1 cells were first transfected with the porcine-specific MRTF (psMRTF) siRNA, then re-transfected with siRNA-resistant (murine) HA-tagged wild-type (WT) or transcriptionally inactive (TL) truncation mutant of MRTF. Cells were then co-stained with anti-HA (to visualize MRTF-retransfection) and acetylated tubulin (to visualize the PC). Scale bar 20 μm.
(E) Cells possessing a long (>5μm) PC and expressing either HA WT or HA TL MRTF were counted and compared to the percentage of cells with a long PC without expressing re-transfected MRTF construct on the same coverslip (i.e. percentage of the neighboring control cells); mean ± SD, n = 3, ≈100 cells/condition in each experiment).∗∗∗∗p < 0.0001, t test.
MRTF interacts with CEP290 and is necessary for the correct localization of CEP290 at the ciliary base
CEP290, a protein localized to the centrosome and the ciliary transition zone (TZ), is a key regulator of ciliogenesis (Garcia-Gonzalo et al., 2011; Kim et al., 2008; Tsang et al., 2008), whose mutations were associated with a variety of ciliopathy syndromes (Coppieters et al., 2010). Since BioID data suggested that CEP290 could be in complex with MRTF, as well as with several other potentially MRTF-interacting ciliary proteins (see Discussion), we initially sought to assess if CEP290 and MRTF can co-localize in renal tubular cells. As expected, CEP290 staining was dot-like and showed strong overlap with γ-tubulin staining, indicating centriolar/BB localization (Figure 7A). Co-staining with AcTub confirmed that CEP290 resided at the base of the cilium (Figure 7B). Remarkably, CEP290 also exhibited robust co- or closely apposed localization with MRTF punctae (Figures 7C and 7C’), indicating that these proteins reside in partly overlapping, adjacent structures. To substantiate these findings by biochemical means, we immunoprecipitated endogenous MRTF from LLC-PK1 cells and probed the precipitates with an anti-CEP290 antibody. MRTF but not control IgG pulled down CEP290. Two CEP290-immunoreactive bands (255 and 270 kDa) were visualized both in the input fraction and in the MRTF precipitates (Figure 7D). To gain some insight into the structural requirements of the MRTF-B/CEP290 interaction, we transfected cells with Myc-tagged WT MRTF, an N-terminal MRTF construct (NT, amino acids 1–544) and a C-terminal MRTF construct (CT, amino acids 545–1080) and precipitated these MRTF proteins through the Myc-tag (Figure 7E). WT and NT MRTF readily pulled down endogenous CEP290 (mostly the 275 kDa band), while CT MRTF, which includes the transactivation domain, failed to do so (Figure 7E). These results imply that CEP290 and MRTF can interact via N-terminal (transcriptionally inactive) sequence(s) in MRTF. Next, we tested if MRTF downregulation alters CEP290 localization. In control (siNR-transfected) cells, distinct, dot-like CEP290 staining localized almost exclusively to the ciliary base. Upon MRTF downregulation, however, CEP290 was mislocalized in approximately half of the cells, such that CEP290 either entirely resided at the ciliary tip, or two distinct CEP290 dots were visible, one at the base and one at the tip (Figure 7F and quantified in G). Together these results indicate that MRTF has a critical role in the correct localization of CEP290 at the ciliary base, or conversely, the absence of MRTF leads to striking redistribution of CEP290, an effect that might contribute to dysregulated ciliogenesis and PC turnover.
Figure 7.
MRTF is necessary for the correct localization of CEP290 at the ciliary base
(A and B) To assess the localization of CEP290 in relation to PC-related structures, serum-starved RPE cells were fixed and co-stained with antibodies against CEP290 and either the BB marker γ-tubulin (A) or the PC marker (shaft) Ac-Tubulin (B). Nuclei were visualized by Dapi, and cells were viewed by confocal microscopy. Bar 10 μm.
(C) Co-staining of CEP290 and MRTF in a monolayer of RPE cells. The merged image (last panel) shows strong colocalization between distinct MRTF-positive patches and CEP290. Bar: 20 μm. (C’) Magnified view of the two areas demarcated by the dashed rectangles in C. Note that MRTF- and CEP290-containing clusters strongly co-localize but do not completely overlap.
(D and E) MRTF and CEP290 are in a complex. (D) RPE cells were serum-deprived, followed by lysis and immunoprecipitation of endogenous MRTF. Whole cell lysates (input) and precipitates were probed with anti-MRTF and anti-CEP290 antibodies. (E) RPE cells were transfected with Myc-tagged WT (full-length) MRTF or the Myc-tagged N- or C-terminal half of the MRTF protein. Immunoprecipitation was performed with anti-Myc antibody. Note that the full-length and the N-terminal MRTF exhibited strong association with CEP290, while the C-terminal segment showed marginal if any binding.
(F) To determine the impact of MRTF on CEP290 localization, RPE cells were transfected with siNR or siMRTF and allowed to ciliate for 48 hr under serum-free conditions. Cells were then co-stained for CEP290 and acetylated tubulin to determine the localization of CEP290 vis-à-vis the PC. In cells transfected with siNR CEP290 unequivocally localized to the base of the PC. In cells transfected with siMRTF CEP290 exhibited varied localization, in three different subsets; CEP290 resided 1) at the base of PC or 2) at the tip of the PC or 3) at both locations. Representative images for these categories are shown. Bar 10 μm. The insets depict further enlarged images of the corresponding cilium tips to document ectopic CEP290 staining.
(G) Quantification of CEP290 localization, as percentage of cells in each category (localization at the base, tip, or both). Only those cells were counted where the localization could be unambiguously determined (mean ± SD, n = 3 independent experiments, ≈100 cells/experiment/condition); ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001, determined by ANOVA.
MRTF affects cilium turnover beyond its impact on the cell cycle
The regulation of cell cycle and deciliation has been shown to be related. On one hand cell cycle entry is associated with cilium resorption, while on the other, the persistence of the cilium was proposed to act as a break/decelerator on cell cycle entry (Kim and Tsiokas, 2011; Li et al., 2011; Phua et al., 2017; Sanchez and Dynlacht, 2016). MRTF has been shown to exert subtle effects on the cell cycle, although this modest impact was observed only in serum-starved cells (Shaposhnikov et al., 2013). Nonetheless, we wished to address whether an impact on the cell cycle might be a key determinant in MRTF's capacity to promote serum-induced resorption and/or prevent ciliogenesis. We found that downregulation of MRTF modestly (and non-significantly) increased the percentage of cells in G0/G1 and reduced those in S/G2 (Figure S6A). In agreement with this finding, nuclear staining of the proliferation marker Ki67 was modestly but significantly reduced upon MRTF silencing (Figure S6B). Retransfection of WT vs. TL MRTF however had differential effect on the cycle. While WT MRTF restored the cycle, TL MRTF strongly reduced the proportion of Ki67 + cells (Figure S6C). This effect was observed both with and without downregulation of MRTF, suggesting that TL, as opposed to WT inhibits cell cycle progression, possibly by exerting a dominant negative effect. Importantly, however, while having differential effect on the cycle, both WT and TL similarly suppressed serum-induced cilium resorption (Figure 3H) and inhibited ciliogenesis (Figure 6D) in the presence of serum. Thus, MRTF has the capacity to directly influence cilium turnover, independent of (or in addition to) its impact on the cell cycle. Nonetheless, further studies are needed to discern whether MRTF downregulation might slow the cell cycle partly by inhibiting deciliation.
Discussion
Our studies identify SRF and MRTF as two novel regulators of PC turnover. Both proteins are necessary for efficient serum-induced PC resorption and MRTF also acts as a constitutive inhibitor of ciliogenesis. Importantly, these proteins exert their effects at multiple levels, which include conceptually new and hitherto unrecognized modes of action. Namely, they impact PC homeostasis not only as transcription factors/cofactors but also via non-transcriptional mechanisms (protein-protein interactions).
Direct and indirect transcriptional effects
The transcriptional effects can be direct (affecting the expression of components of the PC turnover machinery) or indirect (affecting other genes, particularly those encoding cytoskeletal proteins, see below). Considering the direct effects, we show that SRF is necessary for the expression of HEF1, which in turn is indispensable for deciliation (Pugacheva et al., 2007). However, while the expression of the HEF1 gene is fully SRF-dependent, it is not MRTF driven. In fact, MRTF acts as an inhibitor of HEF1 expression. Several possible mechanisms may underlie this effect. SRF can bind TCFs or MRTF, in a mutually exclusive manner (Wang et al., 2004; Zaromytidou et al., 2006), and certain promoter regions preferentially bind TCF/SRF or MRTF/SRF complexes (Chang et al., 2001, Miralles et al., 2003, Sotiropoulos et al., 1999). TCFs have been documented to suppress MRTF/SRF target genes (Gualdrini et al., 2016; Lee et al., 2010; Wang et al., 2004), and mutual inhibition has also been proposed (Gualdrini et al., 2016). Thus, elimination of MRTF might lead to SRF redistribution to preferential TCF/SRF sites (Cen et al., 2003; Chang et al., 2001; Esnault et al., 2014) or TCF/SRF complexes might exert stronger stimulation than SRF/MRTF. It is noteworthy that the HEF1 promoter harbors a direct TCF binding sequence adjacent to the CArG box. Finally, MRTF might also drive suppressors of HEF1 expression or might itself act as a suppressor. Future experiments should address these possibilities.
Our results also show that MRTF positively contributes to HDAC6 expression. Interestingly a recent study has suggested that the expression of α-tubulin acetyltransferase (ATAT1) may be driven by SRF/MRTF in a cell type-specific manner (Fernandez-Barrera et al., 2018). According to this finding, downregulation of SRF or MRTF would be expected to inhibit PC growth or stability, which is the opposite of what we observed, using either AcTub or Arlb13, an alternative PC marker. Moreover, we did not detect a significant change in ATAT1 protein mRNA levels upon MRTF silencing in our cells (Figure S7). Several possibilities could account for these differences. It is conceivable that ATAT1 expression is maintained by other transcriptional inputs or prolonged mRNA stability in various cells. More importantly, our results imply that any potential positive (PC-building) effect of SRF or MRTF is outweighed by their negative effects on the PC, both at the transcriptional and non-transcriptional levels.
Considering indirect transcriptional roles, MRTF or the SRF/MRTF complex may also impact the PC by regulating the expression of cytoskeletal proteins. Indeed, the state of the cytoskeleton has been recognized as a major determinant of PC assembly and disassembly, in that subtly regulated actin dynamics is central for both ciliogenesis and PC resorption (Copeland, 2020; Copeland et al., 2018). Excessive actin polymerization (Drummond et al., 2018; Hernandez-Hernandez et al., 2013; Pitaval et al., 2010; Smith et al., 2020), myosin contractility (Pitaval et al., 2010; Rozycki et al., 2014) and activation of RhoA or Rho kinase (Hernandez-Hernandez et al., 2013; Rangel et al., 2019; Stewart et al., 2016) suppress ciliogenesis or restrict PC growth. Moreover, evidence is accumulating that ciliation is controlled by local Rho activity and cytoskeleton organization. Notably, a fraction of p190 RhoGAP (ARHGAP35) resides at the ciliary base and its loss-of-function mutation, which results in enhanced Rho activation, abrogates PC growth (Stewart et al., 2016). These data, along with our findings, suggest that actin remodeling impacts PC homeostasis by a double mechanism. First cytoskeletal changes may structurally alter the PC, e.g. by acting on the ciliary gate, intraflagellar transport, or BB docking (Copeland, 2020). Second, actin polymerization promotes MRTF-dependent expression of many cytoskeletal proteins (Esnault et al., 2014), several of which (e.g. myosin heavy chain, Arp3) have been shown to counter cilium formation (Drummond et al., 2018; Pitaval et al., 2010; Rozycki et al., 2014). It is also noteworthy in this regard that two cytoskeleton- and Hippo pathway-regulated transcriptional co-activators TAZ and YAP were recently proposed to inhibit PC formation (Kim et al., 2015a). Our earlier studies have shown that MRTF is a key transcriptional regulator of TAZ expression (Miranda et al., 2017; Speight et al., 2016). This effect may thus be a significant (synergistic) contributor to the inhibitory action of MRTF on the cilium. Since actin polymerization enhances MRTF activity, which in turn promotes the expression of cytoskeletal proteins, leading to further actin polymerization and contractility, this self-augmenting mechanism may render MRTF an efficient link between actin remodeling and PC turnover.
MRTF and SRF in PC homeostasis: beyond transcriptional effects
The present work provides several lines of evidence that the role of MRTF in PC homeostasis must go well beyond transcriptional control, namely (1) MRTF is localized to the BB/centriolar region under resting conditions; and (2) it is further recruited to ciliary structures upon serum stimulation, preceding PC resorption; (3) MRTF binds to and stabilizes AurA, presumably by preventing its association with proteins (Smad4, AIP, AZ1 (Jia et al., 2014; Kiat et al., 2002; Lim and Gopalan, 2007a, b)) that promote its degradation; (4) a transcriptionally inactive MRTF fragment remains capable of promoting cilium resorption and counteracting the formation of long cilia; (5) MRTF interacts with CEP290, a centrosomal/BB protein/TZ protein, and is necessary for its localization at the ciliary base; and (6) a recent BioID analysis indicates that MRTF forms complexes with several ciliary proteins, including, RGRIP1, TMEM67, NPHP1, LC5A, and others (Gupta et al., 2015), the mutations of which have been associated with various ciliopathies (Braun and Hildebrandt, 2017; Reiter and Leroux, 2017). Considering SRF, we show that a pool of this protein is localized to the PC, where it undergoes phosphorylation upon serum addition and interacts with key PC resorption proteins. These findings are consistent with a single previous report, which showed the presence of SRF in motile cilia, albeit the functional significance of this phenomenon remained open (Nordgren et al., 2014).
As is the case with explorative studies, while our work has uncovered and entirely new layer of PC regulation via SRF and MRTF, it has also generated a large number of new questions, which define the direction of future research. Here we would like to focus on two major ones of these: the mechanism of ciliary localization/recruitment of MRTF/SRF and their potential mechanisms of action, both in terms of cilium resorption and ciliogenesis.
Centrosomal or ciliary localization of proteins may be guided by two mechanisms: a direct one, when a protein harbors cilium localization signal(s) (CLS) (Berbari et al., 2008; Malicki and Avidor-Reiss, 2014; Mazelova et al., 2009; Nachury et al., 2010) or centrosomal targeting sequence(s) (CTS) (Matsumoto and Maller, 2004; Pascreau et al., 2010) and/or by an indirect one, via binding to centrosomal/ciliary proteins. Interestingly, a potential CLS sequence AX [S/A]XQ is present in SRF, and MRTF contains CTS-like motifs ((D/S)WL), although the latter ones are less well-defined and conserved. Future mutagenesis studies should address if these candidate sequences contribute to the BB/PC localization and recruitment. Our present work also reveals a variety of new protein-protein interactions. These include complex formation between MRTF and AurA, as well as between SRF and HEF1, AurA, and HDAC6, as indicated by coimmunoprecipitation studies from PC preparations. It remains to be established if some of these interactions contribute to ciliary retention or recruitment of these partners. Interestingly, most of the BioID-predicted interactors of MRTF are localized in the BB and TZ area at the ciliary base, and our data are consistent with MRTF accumulation in these regions. Super-resolution microscopy studies will be useful to determine the exact molecular anatomy of ciliary MRTF interactions, and the potential role of MRTF and its partners in the targeting and retention of each other (see also next section regarding CEP290).
From a functional standpoint two important questions emerge: (1) how do local SRF and MRTF contribute to cilium resorption; and (2) what is the mechanism underlying the impact of MRTF on ciliogenesis? Since serum induces SRF phosphorylation in the PC, and SRF interacts with members of the HEF1-AurA-HDAC6 pathway, it is conceivable that SRF phosphorylation triggers or augments the activation of components of this deciliation route. MRTF, which is also present in ciliary SRF immunoprecipitates, and which can bind AurA, might act as a local scaffold to support PC disassembly. SRF might also contribute to the serum-induced ciliary recruitment of MRTF, a new phenomenon described in our work. In the absence of MRTF, there is a profound loss of AurA from the PC and from the ciliary SRF precipitates. These findings suggest that the local or global AurA-stabilizing effect of MRTF, described herein, is a central component of MRTF's permissive role in cilium resorption. Furthermore, serum is known to induce MRTF phosphorylation as well; 23 phosphorylatable Ser/Thr residues, targeted by a variety of kinases, were found in MRTF-A (Panayiotou et al., 2016). Accordingly, it remains to be established whether posttranslational modifications of MRTF contribute to its recruitment and/or local actions. In addition, alternative PC resorption mechanisms (e.g. the Polo kinase (Lee et al., 2012; Wang et al., 2013a) and the NEK kinase (Kim et al., 2015b) pathways) have also been reported, and the involvement of SRF/MRTF in these remains to be explored. It is also worth noting that in addition to local effects, cilium-derived phosphorylated SRF might also serve as a global signal. As the PC is being resorbed, phospho-SRF might get liberated and enter the nucleus. In fact, SRF was found to leave motile cilia of the bronchial epithelium upon induction of airway inflammation (Nordgren et al., 2014), a process which was suggested to promote injury repair. Since PC disassembly is a prerequisite for cell division (Sanchez and Dynlacht, 2016), and SRF is a key inducer of proliferation by driving early response genes (Chai and Tarnawski, 2002); growth factor-induced ciliary modification of SRF could be one of the coordinating links between PC removal and cell division. It will be interesting to test if SRF undergoes other, possibly PC-specific posttranslational modifications, and if ciliary SRF enters the nucleus.
Finally, our studies show that MRTF is a constitutive inhibitor of ciliogenesis, and this effect is conferred by the N-terminal (transcriptionally inactive) region of the molecule. Remarkably, we found that the N-terminus also interacts with CEP290, and MRTF is critical for the correct distribution of this protein at the ciliary base. CEP290 is a centriolar and TZ protein and a key integrator of PC assembly, primarily by its capacity to interact with Rab8 (Kim et al., 2008), a small GTPase necessary for ciliogenesis, and with CP110, a key inhibitor of axonemal growth, whose removal is a requisite for ciliogenesis (Tsang et al., 2008). It is therefore conceivable that the MRTF downregulation-induced mislocalization of CEP290 prevents the proper localization or action of CP110, thereby hindering its suppressive action on PC growth. Ectopic localization of CEP290 (at the ciliary tip) might also support ciliogenesis. A similar, and potentially related, phenomenon has been described for CEP162, another regulator of ciliogenesis. This protein resides in the centrosome distal end and the TZ, where it bridges TZ components to the axoneme (Wang et al., 2013b). Intriguingly, CEP162 can form a complex with CEP290 (Wang et al., 2013b), and was identified in the BioID screen as an MRTF partner (Gupta et al., 2015). Truncation mutants of CEP162, which cannot bind to the centrosome, were shown to relocate to the ciliary tip and recruit CEP290 at this site, which in turn results in ectopic TZ assembly, PC elongation, and the bulging of the cilium tip (Wang et al., 2013b). These morphological features are very similar to those seen upon MRTF knockdown. Taken together, MRTF may be essential for the correct spatial distribution of the TZ, via its interaction(s) with CEP290, CEP162, and other TZ constituents. In addition, PC lengthening may also indicate an imbalance between anterograde and retrograde intraflagellar transport. Finally, since MRTF is an actin binding protein, and recent studies indicate the presence of actin in the PC (Lee et al., 2018b), the question arises whether the molecular interactions of MRTF at the PC are modulated by local actin dynamics. Future studies should address these intriguing possibilities.
In aggregate, our results show that SRF and MRTF, through a variety of mechanisms, play central roles in the control of three distinct processes determining cilium homeostasis, namely PC resorption, ciliogenesis, and PC scission. Correct regulation of each of these is paramount to human health. In this regard, MRTF and SRF are central mediators in the pathogenesis of organ fibrosis ((Bialik et al., 2019; Fan et al., 2007; Luchsinger et al., 2011; Small et al., 2010; Tian et al., 2015) and reviewed in (Gasparics and Sebe, 2018; Small, 2012)) and dysregulated PC homeostasis has been associated with fibrotic conditions (Arrighi et al., 2017; Egorova et al., 2011; Han et al., 2018; Lee et al., 2018a; Li et al., 2020; Rozycki et al., 2014; Villalobos et al., 2019). While our understanding is rudimentary as yet, the PC alterations might play a biphasic role (Rozycki et al., 2014; Teves et al., 2019), wherein an initial PC growth or enhanced ciliation might contribute to repair or early fibrogenic signaling (Arrighi et al., 2017; Lee et al., 2018a; Rozycki et al., 2014), whereas subsequent loss of the cilium is associated with EMT, terminal myofibroblast differentiation and advanced fibrosis (Han et al., 2018; Li et al., 2020; Rozycki et al., 2014). Future research should address if functional or genetic alterations in SRF or MRTF, or their interactions with PC components, contribute to the pathogenesis of acquired ciliary disorders (e.g. in fibrosis and cancer) or inherited ciliopathies.
Limitations of the study
Our studies clearly imply SRF and MRTF as new regulators of cilium turnover that act both as transcription factors and as BB/cilium constituents. However, the exact mechanisms whereby MRTF and SRF locally support cilium resorption or MRTF suppresses ciliogenesis remain to be identified. Similarly, further studies are warranted to analyze the precise relationship between the effect of MRTF on cilium dynamics and the cell cycle. It is noteworthy in this regard that our work identifies MRTF as a novel regulator of AurA stability, and AurA is a central component of both the cilium resorption machinery and cell cycle regulation. New studies should follow up these findings “in vivo”. Since whole-body MRTF-A/B KO is lethal, and tissue-specific and inducible systems are needed to investigate their impact on ciliary homeostasis. Finally, as mentioned above, it remains an exciting question whether mutations in MRTF or SRF could underlie or contribute to the pathobiology of certain ciliopathies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MRTFA | Cell Signaling Technology | Cat# 14760; RRID:AB_2798598 |
| Rabbit anti-MRTFB | Santa Cruz Biotechnology | Cat# sc-98989; RRID:AB_2250525 |
| Mouse anti-MRTFA | Santa Cruz Biotechnology | Cat# sc-398675 |
| Rabbit anti-HA | Abcam | Cat# ab-13834; AB_443010 |
| Rabbit anti-SRF | Cell Signaling Technology | Cat# 5147, AB_10694554 |
| Mouse anti-c-Myc 9E10 | Santa Cruz Biotechnology | Cat# sc-40; RRID:AB_627268 |
| Mouse anti-GAPDH | Santa Cruz Biotechnology | Cat# sc-47724; RRID:AB_627678 |
| Rabbit anti-Smad4 | Cell Signaling Technology | Cat# 9515; RRID:AB_2193344 |
| Rabbit anti-HDAC6 | Cell Signaling Technology | Cat# 7612; RRID:AB_10889735 |
| Mouse anti-HEF1 | Cell Signaling Technology | Cat# 4044; RRID:AB_1904002 |
| Rabbit anti-CEP290 | Novus Biologicals | Cat# NB100-86991; RRID:AB_1201171 |
| Mouse anti-Aurora A | Thermo Fisher Scientific/Invitrogen | Cat# 45-8900; RRID:AB_2533839 |
| Rabbit anti-Tubulin | Cell Signaling Technology | Cat# 2128; RRID:AB_823664 |
| Mouse anti- β-actin | Sigma-Aldrich | Cat# A1978; AB_476692 |
| Rabbit anti-Antizyme1 | Enzo Lifesciences | Cat# BML-PW8885; RRID:AB_2156011 |
| Rabbit anti-AURKAIP1 | Mybiosource | Cat# MBS2516763 |
| Rabbit anti-C6orf134 | Abcam | Cat# ab58742; RRID:AB_940731 |
| Mouse anti-Acetylated Tubulin | Sigma-Aldrich | Cat# T7451; RRID:AB_609894 |
| Mouse anti-Acetylated Tubulin | Abcam | Cat# ab24610; RRID:AB_448182 |
| Rabbit anti- Arl13b | Proteintech | Cat# 17711-1-AP, RRID:AB_2060867 |
| Mouse anti-CEP290 | Santa Cruz Biotechnology | Cat# sc-390637 |
| Rabbit anti-gamma Tubulin | Abcam | Cat# ab16504; RRID:AB_443396 |
| Mouse anti-gamma Tubulin | Abcam | Cat# ab27074; RRID:AB_2211240 |
| Rabbit anti-SRF | Cell Signaling Technology | Cat# 5147; RRID:AB_10694554 |
| Goat anti-MRTFB | Santa Cruz Biotechnology | Cat# sc-47282; RRID:AB_2142502 |
| Rabbit anti-MRTFA | Santa Cruz Biotechnology | Cat# sc-32909; RRID:AB_2142495 |
| Rabbit-anti-Ki67 | Abcam | Cat# ab15580; RRID:AB_443209 |
| Normal Rabbit IgG | Santa Cruz Biotechnology | Cat# sc-2027; RRID:AB_737197 |
| Normal Mouse IgG | Santa Cruz Biotechnology | Cat# sc-2025; RRID:AB_737182 |
| Protein A/G UltraLink Resin | Thermofisher scientific | Cat# 53132 |
| Chemicals, peptides, and recombinant proteins | ||
| Human TGF-β1 | R&D | 240-B; GenPept: P01137 |
| MG-132 | Sigma-Aldrich | Cat# 474790, CAS 133407-82-6 |
| Lipofectamine 2000 | Thermofisher Scientific | Cat# 11668027 |
| Jetprime transfection reagent | Polyplus transfection | Cat# 114-07 |
| Lipofectamine RNAimax | Invitrogen | Cat# 13778075 |
| Critical commercial assays | ||
| iScript reverse transcription supermix | BioRad | Cat# 1708840 |
| Dual luciferase Reporter assay | Promega | Cat# E1910 |
| Experimental models: cell lines | ||
| LLCPK1 – Kidney tubular epithelial cell line | Gift - R.C. Harris, Vanderbilt University School of Medicine | |
| hTERT-RPE | ATCC | Cat# CRL-4000, RRID:CVCL_4388 |
| IMCD3 | ATCC | Cat# CRL-2123, RRID:CVCL_0429 |
| Oligonucleotides | ||
| siRNA MRTF A 5'-CCAAGGAGCUGAA GCCAAA-3' (and human sequences |
Thermo Scientific/Sigma-Aldrich | Sus scrofa |
| siRNA MRTF B 5'-CGACAAACACCGTAGCAAA-3' | Thermo Scientific/Sigma-Aldrich | Sus scrofa |
| siRNA SRF 5'-GGAACUGUGCUGAAGAGUA -3' | Thermo Scientific/Sigma-Aldrich | Sus scrofa |
| siRNA psMRTFA 5'-GGGAAUUGUAAACAAG GUU-3' (where ps denotes specificity to porcine only; no homology to mouse or human) |
Thermo Scientific/Sigma-Aldrich | Sus scrofa |
| siRNA psMRTFB 5'-GGACUCCAGUGUGAAG GAA-3' (where ps denotes specificity to porcine only; no homology to mouse or human) |
Thermo Scientific/Sigma-Aldrich | Sus scrofa |
| siRNA MRTF A 5'-CCAAGGAGCUGAAGCCAAA-3' | Thermo Scientific/Sigma-Aldrich | Homo sapiens |
| siRNA MRTF B 5'-UGACAAACACCGUAGCAAA -3' | Thermo Scientific/Sigma-Aldrich | Homo sapiens |
| Control Non-related (NR) siRNA | Applied Biosystems | AM4635 |
| See Table S1 for primer pairs used for quantitative PCR | ||
| Recombinant DNA | ||
| Arl13b-mCherry | Gift: Kristen Verhey | (Kee et al., 2012) |
| pcDNA5 FRT TO Myc ArA WT | Addgene | Plasmid# 59804, RRID:Addgene_59804 |
| MRTF-WT myc-tag |
Masszi et al., 2010, Speight et al., 2016 |
N/A |
| MRTF-N-terminal myc-tag | Speight et al., 2016 | N/A |
| MRTF-C-terminal myc-tag |
Masszi et al., 2010, Speight et al., 2016 |
N/A |
| MRTF-WT HA-tag |
Masszi et al., 2010, Speight et al., 2016 |
N/A |
| MRTF-TL (TADless mutant) HA-tag | This paper | NA |
| p765-SMA-Luc (WT) | Masszi et al., 2003 | NA |
| pRL-Renilla Luciferase-TK control reporter vector | Promega | E224A |
| Software and algorithms | ||
| Quantity one software | BioRad | https://www.bio-rad.com/en-ca/product/quantity-one-1-d-analysis-software?ID=1de9eb3a-1eb5-4edb-82d2-68b91bf360fb |
| Image lab software | BioRad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
| Metamorph premier Image analysis software | Molecular devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref |
| Fiji | ImageJ | https://imagej.net/Fiji |
| Quantstudio 7 pro software | Applied Biosystems | https://www.thermofisher.com/ca/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-systems/models/quantstudio-6-7-pro.html |
| Prism software version 8.2.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Andras Kapus (andras.kapus@unityhealth.to)
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze [datasets/code].
Experimental model and subject details
Cells
Tissue culture media and reagents were from Thermo Fisher/Life Technologies. Culture media was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and cells were grown in an atmosphere containing 5% CO2.
LLC-PK1, a kidney tubule epithelial cell line (male) was a gift from R.C. Harris, Vanderbilt University School of Medicine and was cultured in low-glucose DMEM as described in (Masszi et al., 2010). The following cells were from the American Type Culture Collection (ATCC): hTERT RPE cells, a Telomerase immortalized human retina pigmented epithelial cell line (female) (ATCC Cat# CRL-4000, RRID:CVCL_4388); IMCD3, mouse inner medullary collecting duct cell line (ATCC Cat# CRL-2123, RRID:CVCL_0429) . These cells were cultured in DMEM-F12.Where indicated, cells were incubated under serum-free conditions for varying times depending on the experimental procedure. Cell contact disassembly was achieved by thoroughly washing cultures in PBS and placing them in nominally calcium chloride-free DMEM (low calcium medium, LCM) containing TGFβ1.
Methods details
Reagents
TGFβ1 (R&D Systems) was used at 10 ng/ml. MG-132 (Sigma-Aldrich) was used at 20μM.
Antibodies
Commercial antibodies used were as follows: For western blot analysis; rabbit anti-MRTFA (1:1000, Cell Signaling Technology,14760), rabbit anti-MRTFB (1:1000, Santa Cruz Biotechnology, 98989), mouse anti-MRTFA (1:1000, Santa Cruz Biotechnology, 398675), rabbit anti-HA (1:1,000, Abcam, ab-13834), rabbit anti-SRF (1:1000, Cell Signaling Technology, 5147), mouse anti-c-Myc 9E10 (1:1,000, Santa Cruz Biotechnology, sc-40), mouse anti-GAPDH (1:20,000, Santa Cruz Biotechnology, sc-47724,), rabbit anti-Smad4 (1:1000, Cell Signaling Technology, 9515), rabbit anti-HDAC6 (1:1000, Cell Signaling Technology, 7612), mouse anti-HEF1 (1:1000, Cell Signaling Technology, 4044), rabbit anti-CEP290 (1:500, Novus Biologicals, NB100-86991), mouse anti-Aurora A (1:1000,Thermo Fisher Scientific/Invitrogen, 45-8900), rabbit anti-Tubulin (1:10,000, Cell Signaling Technology, 2128), mouse anti- β-actin ( 1:10,000, Sigma-Aldrich, A1978), rabbit anti-Antizyme1 (1:250, Enzo Life Sciences, BML-PW8885), rabbit anti-AURKAIP1 (1:500, MyBiosource, MBS2516763), rabbit anti-C6orf134 (1:500, Abcam, ab58742), mouse anti-Acetylated Tubulin (1:500, Sigma-Aldrich, T7451). For immunofluorescence; mouse anti-Acetylated Tubulin (1:200, Abcam, ab24610), rabbit anti- Arl13b (1:100, Proteintech, 17711-1-AP), rabbit anti-Nup98 (1:1000, Santa Cruza Biotechnology, sc-74578); rabbit anti-Histone H1 FL-219 (1:100, Santa Cruz, Biotechnology, sc-10806), mouse anti-CEP290 (1:50, Santa Cruz Biotechnology, sc-390637), rabbit anti-gamma Tubulin (1:100, Abcam, ab16504), mouse anti-gamma Tubulin (1:100, Abcam, ab27074), rabbit anti-SRF (1:1000, Cell Signaling Technology, 5147), rabbit anti-MRTFA (1:100, Cell Signaling Technology, 14760), rabbit anti-MRTFB (1:50, Santa Cruz Biotechnology, 98989), mouse anti-MRTFA (1:50, Santa Cruz Biotechnology, sc-398675), goat anti-MRTFB (1:50, Santa Cruz Biotechnology, sc-47282), rabbit anti-MRTFA (1:50, Santa Cruz Biotechnology,sc-32909) and rabbit-anti-Ki67 (1:100, Abcam, ab15580).
Normal rabbit and mouse IgG were from Santa Cruz Biotechnology (sc-2027 and sc-2025, respectively). Jackson ImmunoResearch Laboratories was the source for all horseradish peroxidase-conjugated secondary antibodies (1:5,000), and all fluorescent secondary antibodies were from Thermo Thermo Fisher Scientific/Invitrogen (1:1000).β
Plasmid and small-interfering RNA (siRNA) transfection
The Arl13b-mCherry was a kind gift from Kristen Verhey (Kee et al., 2012). The WT myc-tagged AurA was a gift from Stephen Taylor (plasmid# 59804; Addgene, Cambridge, MA). Standard PCR techniques were used to create the myc tagged WT, N- or C-terminal MRTF, the HA tagged WT MRTF and the MRTF transactivation domain deletion construct (TADless, TL) (Masszi et al., 2010; Speight et al., 2016). Transfection of DNA construct was performed with Lipofectamine 2000 (Invitrogen) or jetPrime (PolyPlus Transfection SA) according to the manufacturer's protocol.
The following siRNA duplexes used were directed against the indicated porcine sequences: MRTF A 5'-CCAAGGAGCUGAAGCCAAA-3' ; MRTF B 5'-CGACAAACACCGTAGCAAA-3' ; SRF 5'-GGAACUGUGCUGAAGAGUA -3'; psMRTFA 5'-GGGAAUUGUAAACAAGGUU-3'; psMRTFB 5'-GGACUCCAGUGUGAAGGAA-3' (where ps denotes specificity to porcine only; no homology to mouse or human); AurA 5' GAGAAGAGAAGUAGAAAUAUU 3'; and human sequences: MRTF A 5'-CCAAGGAGCUGAAGCCAAA-3'; and MRTF B 5'-UGACAAACACCGUAGCAAA -3'. Oligonucleotides were synthesized and purchased from Thermo Scientific or Sigma-Aldrich. NR control siRNA was obtained from Applied Biosystems and used under the same experimental conditions. Transfections with siRNA alone were performed using Lipofectamine RNAiMAX (Invitrogen) and co-transfections with cDNA and siRNA were carried out using jetPrime. siRNAs were applied at 50 nM.
Luciferase reporter assay
The p765-SMA-Luc (WT) reporter construct was previously described in detail (Masszi et al., 2003). The luciferase assay was performed as in (Masszi et al., 2010; Speight et al., 2016). Cells were transfected with reporter constructs together with the normalizing plasmid pRL-TK (Promega) and the indicated expression plasmids. Renilla luciferase and firefly luciferase activities in cell lysates were measured using a reporter assay system (Dual Luciferase; Promega) in a luminometer (Lumat 9507; Berthold). Transfections and measurements were performed in triplicates for each experiment and experiments were repeated three times. Results are expressed as fold changes compared with the mean firefly/renilla ratio of the control taken as a unit.
Immunoprecipitation and Western blotting
To detect the interaction between endogenous MRTF and AuroraA or the indicated, transiently transfected, tagged proteins, cells were collected from 10-cm dishes following any described treatment. The lysis buffer (30 mM HEPES (pH 7.4), 100 mM NaCl, 1 mM EGTA, 20 mM sodium fluoride and 1% Triton X-100) was supplemented with 1 mM PMSF, 1 mM sodium vanadate and Complete Mini Protease Inhibitor (Roche). Lysates were spun at 12,000 r.p.m. for 5 min and analyzed for protein content (BCA Protein Assay; Pierce Biotechnology). After reserving samples for analyzing input, supernatants were precleared by rotating with Protein A/G UltraLink Resin (Thermo Scientific) for 30 min. Precleared supernatants were incubated with the precipitating antibody or a control IgG and then with aliquots of Protein A/G Resin. Bound proteins were eluted from the washed beads and analyzed by western blot along with samples from input to monitor expression levels. Immunodetection was performed using the ECL method (ECL/ECL Plus reagents from GE Healthcare, Life Sciences) or Clarity Western/ Clarity Max Western ECL substrates from BioRad). The signal was either captured on film and analyzed by densitometry using a GS800 densitometer and Quantity One software (BioRad) or with the ChemiDocTM Touch Imaging System version 1.2.0.12. and Image Lab software (Bio-Rad). All experiments were performed a minimum of three times and representative immunoblots are shown.
Isolation of primary cilia
LLC-PK1 cells were grown in multiple 150-mm culture dishes, and where indicated, transfected with NR or MRTF siRNA. Following 48h serum depletion, cells were washed, placed in fresh PBS, and agitated at 350 rpm at room temperature for 4min. The cell solution was subsequently spun at 3000 rpm for 10min at 4°C. The clarified supernatant was subjected to a high speed spin (40,000rpm) for 30min at 4°C using a Type 42.1 rotor and an L8-80M ultracentrifuge (Beckman). A small fraction of the isolated primary cilia was placed on a glass coverslip, dried with a stream of nitrogen, fixed and stained for acetylated tubulin to monitor the purification procedure. Cell pellets were resuspended directly in Laemmli buffer for analysis by immunoblotting, or in RIPA buffer for immunoprecipitation as described above.
Immunofluorescence microscopy
Cells were cultured on sterile coverslips and fixed with either methanol cooled to -20°C, or 4% paraformaldehyde (room temperature), or a combination of the two.
Next, samples were blocked with bovine serum albumin and incubated with relevant primary antibodies. After washing, the coverslips were incubated with the corresponding fluorescently labeled secondary antibodies along with 4',6-diamidino-2-phenylindole (Dapi) (Thermo Fisher) to counterstain the nuclei. Coverslips were mounted on slides using fluorescent mounting medium (Dako). Images were captured using an Olympus IX81 microscope (Olympus, Tokyo, Japan) coupled to an Evolution QEi Monochrome camera (Media Cybernetics, Silver Spring, MD) or by confocal microscopy using a WaveFX spinning-disk confocal microscope system (Quorum Technologies, Guelph, Canada) equipped with an ORCA-Flash4.0. Images were processed and analyzed with MetaMorph Premiere software (Molecular Devices).
Live cell imaging
LLC-PK1 cells plated on 25-mm glass coverslips were co-transfected with Arl 13b-mCherry along with NR or MRTF-specific siRNA. Cells were placed in a TC-L-10 live incubation device, treated as indicated and time-lapse confocal microscopy was performed using the WaveFX spinning-disk microscopy system using the MetaMorph Premiere software.
Quantitative PCR
LLC-PK1 or RPE cells were transfected or treated as indicated and total RNA was extracted using the RNeasy Kit (Qiagen). Following cDNA synthesis using iScript reverse transcriptase (BioRad), SYBR green-based real time PCR was performed to evaluate gene expression of HEF1, HDAC6, and AuroraA using GAPDH as the reference standard. For LLC-PK1 cells, porcine specific primer pairs for GAPDH ( 5' -GCAAAGTGGAC ATGGTCGCCATCA-3' and 5' -AGCTTCCCATTCTCAGCCTTGACT-3') ; HEF1 (5'-AGGACCTGGTCAGCCAACTA-3' and 5'-CGCCATAAGATTCTTTGCCC-3'); HDAC6 (5'-CTAGAGTACATTGATCTGATG-3' and 5'-CACCAGCCTGAGGACGGAGCC-3') and AurA (5'- CAGTCCCACCTTAGGCATCC-3' and 5' CTCCGAGAGGTGCGTATTCC-3') were used. For analysis of RPE cells, human specific primer pairs for GAPDH (5'-GGAAGGTGAAGGTCGGAGTC-3’ and 5’-ACATGTAAACCATGTAGTTGAGGTC-3'); HDAC6 (5'-GGAGGGTCCTTATCGTAGATTG-3' and 5'-GTAGCGGTGGATGGAGAAATAG-3'); HEF1 (5'- GGAACTCATCCTCCACAACAA-3' and 5'-GGACCAGCTGCACTCATTTA-3') and AurA (5'-GTGTGCCTTAACCTCCCTATTC-3' and 5'-CGAACCTTGCCTCCAGATTAT-3') were employed. Each sample was analyzed in triplicate and experiments were performed three times. All qPCR experiments were performed using an IQ cycler (BioRad) or QuantStudio 7 (Applied Biosystems). mRNA expression was quantified using the ΔCT (Litvak) method (Bialik et al., 2019), and data are expressed as fold changes compared to the average of the control condition.
Cell cycle analysis by flow cytometry
Propidium iodide staining was done as previously described (Riccardi and Nicoletti, 2006). Briefly, 24 h post siRNA transfection, cells were collected by trypsinization, counted, washed with PBS and fixed with 80% methanol for 15 min on ice. After washing with PBS, the pellet was incubated with 50 μg/ml RNAse A (Sigma, R6513) for 15 min at 37°C. Cells were then stained for with 10 μg/mL propidium iodide (Sigma, 81845) (PI) in PBS. SP6800 Spectral Cell Analyzer (Sony Biotechnology) was used to measure fluorescence from 100,000 events per sample. Data was analyzed using FlowJo software v7 (TreeStar). DNA histograms were generated by gating on single cell events and cell cycle phases de-convoluted using the built-in Dean-Jet-Fox algorithm.
Quantification and statistical analysis
Data are presented as blots or images from at least three similar experiments or as the mean ± standard deviation (SD) or standard error of the mean (SEM) for the number of experiments indicated (n) in the figure legends. Statistical significance was determined by one-way analysis of variance (ANOVA, Tukey posthoc testing) for comparison of multiple groups or the appropriate version of Student’s t-test for comparison of two groups using GraphPad Prism software (version 8.2.0, GraphPad Software Inc.). p < 0.05 was accepted as significant; ∗, ∗∗, ∗∗∗, ∗∗∗∗∗ correspond to p < 0.05, <0.01, <0.001 and <0.0001, respectively.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR, PJT-162360, PJT-148608 and MOP- 13046), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-05222), and the Kidney Foundation of Canada to A.K. A.K. is the Thor Eaton professor in fibrosis research.
Author contributions
P.S., M.R., and S.V. performed the majority of the experiments and contributed to the generation of the figures and the STAR Methods section. K.S. performed experiments and both K.S. and M.K. contributed to the critical analysis of the data and the completion of the manuscript; A.K. conceived the project and the central ideas, developed the experimental approach, contributed to data analysis and figure making, and wrote the majority of the manuscript.
Declaration of interests
The authors declare no competing interest.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102739.
Supplemental information
References
- Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L.B., Christensen S.T. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi N., Lypovetska K., Moratal C., Giorgetti-Peraldi S., Dechesne C.A., Dani C., Peraldi P. The primary cilium is necessary for the differentiation and the maintenance of human adipose progenitors into myofibroblasts. Sci. Rep. 2017;7:15248. doi: 10.1038/s41598-017-15649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., Johnson A.D., Lewis J.S., Askwith C.C., Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik J.F., Ding M., Speight P., Dan Q., Miranda M.Z., Di Ciano-Oliveira C., Kofler M.M., Rotstein O.D., Pedersen S.F., Szaszi K. Profibrotic epithelial phenotype: a central role for MRTF and TAZ. Sci. Rep. 2019;9:4323. doi: 10.1038/s41598-019-40764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.A., Hildebrandt F. Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017;9:a028191. doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A., Greenwood W., Le Quesne J., Teschendorff A., Miranda-Saavedra D., Rueda O.M., Sandoval J.L., Vidakovic A.T., Saadi A., Pharoah P. TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat. Commun. 2012;3:1055. doi: 10.1038/ncomms2039. [DOI] [PubMed] [Google Scholar]
- Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cen B., Selvaraj A., Burgess R.C., Hitzler J.K., Ma Z., Morris S.W., Prywes R. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J., Tarnawski A.S. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J. Physiol. Pharmacol. 2002;53:147–157. [PubMed] [Google Scholar]
- Chang P.S., Li L., McAnally J., Olson E.N. Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 2001;276:17206–17212. doi: 10.1074/jbc.M010983200. [DOI] [PubMed] [Google Scholar]
- Copeland J. Actin-based regulation of ciliogenesis - the long and the short of it. Semin. Cell Dev. Biol. 2020;102:132–138. doi: 10.1016/j.semcdb.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Copeland S.J., McRae A., Guarguaglini G., Trinkle-Mulcahy L., Copeland J.W. Actin-dependent regulation of cilia length by the inverted formin FHDC1. Mol. Biol. Cell. 2018;29:1611–1627. doi: 10.1091/mbc.E18-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F., Lefever S., Leroy B.P., De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- Das R.M., Storey K.G. Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science. 2014;343:200–204. doi: 10.1126/science.1247521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M.L., Li M., Tarapore E., Nguyen T.T.L., Barouni B.J., Cruz S., Tan K.C., Oro A.E., Atwood S.X. Actin polymerization controls cilia-mediated signaling. J. Cell Biol. 2018;217:3255–3266. doi: 10.1083/jcb.201703196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova A.D., Khedoe P.P., Goumans M.J., Yoder B.K., Nauli S.M., ten Dijke P., Poelmann R.E., Hierck B.P. Lack of primary cilia primes shear-induced endothelial-to-mesenchymal transition. Circ. Res. 2011;108:1093–1101. doi: 10.1161/CIRCRESAHA.110.231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N., Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Sebe A., Peterfi Z., Masszi A., Thirone A.C., Rotstein O.D., Nakano H., McCulloch C.A., Szaszi K., Mucsi I. Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol. Biol. Cell. 2007;18:1083–1097. doi: 10.1091/mbc.E06-07-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Barrera J., Bernabe-Rubio M., Casares-Arias J., Rangel L., Fernandez-Martin L., Correas I., Alonso M.A. The actin-MRTF-SRF transcriptional circuit controls tubulin acetylation via alpha-TAT1 gene expression. J. Cell Biol. 2018;217:929–944. doi: 10.1083/jcb.201702157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparics A., Sebe A. MRTFs- master regulators of EMT. Dev. Dyn. 2018;247:396–404. doi: 10.1002/dvdy.24544. [DOI] [PubMed] [Google Scholar]
- Gualdrini F., Esnault C., Horswell S., Stewart A., Matthews N., Treisman R. SRF Co-factors control the balance between cell proliferation and contractility. Mol. Cell. 2016;64:1048–1061. doi: 10.1016/j.molcel.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud E., Goncalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.J., Jung J.K., Im S.S., Lee S.R., Jang B.C., Park K.M., Kim J.I. Deficiency of primary cilia in kidney epithelial cells induces epithelial to mesenchymal transition. Biochem. Biophys. Res. Commun. 2018;496:450–454. doi: 10.1016/j.bbrc.2018.01.079. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hernandez V., Pravincumar P., Diaz-Font A., May-Simera H., Jenkins D., Knight M., Beales P.L. Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Hum. Mol. Genet. 2013;22:3858–3868. doi: 10.1093/hmg/ddt241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M., Obaidi I., McMorrow T. Primary cilia and their role in cancer. Oncol. Lett. 2019;17:3041–3047. doi: 10.3892/ol.2019.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I., Goto H., Kasahara K., Inagaki M. Current topics of functional links between primary cilia and cell cycle. Cilia. 2015;4:12. doi: 10.1186/s13630-015-0021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Lee H.S., Wu C.F., Kundu J., Park S.G., Kim R.N., Wang L.H., Erkin O.C., Choi J.S., Chae S.W. SMAD4 suppresses AURKA-induced metastatic phenotypes via degradation of AURKA in a TGFbeta-independent manner. Mol. Cancer Res. 2014;12:1779–1795. doi: 10.1158/1541-7786.MCR-14-0191. [DOI] [PubMed] [Google Scholar]
- Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., Verhey K.J. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiat L.S., Hui K.M., Gopalan G. Aurora-A kinase interacting protein (AIP), a novel negative regulator of human Aurora-A kinase. J. Biol. Chem. 2002;277:45558–45565. doi: 10.1074/jbc.M206820200. [DOI] [PubMed] [Google Scholar]
- Kim J., Jo H., Hong H., Kim M.H., Kim J.M., Lee J.K., Heo W.D., Kim J. Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nat. Commun. 2015;6:6781. doi: 10.1038/ncomms7781. [DOI] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S.R., Gleeson J.G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee K., Choi J.H., Ringstad N., Dynlacht B.D. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat. Commun. 2015;6:8087. doi: 10.1038/ncomms9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Tsiokas L. Cilia and cell cycle re-entry: more than a coincidence. Cell Cycle. 2011;10:2683–2690. doi: 10.4161/cc.10.16.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Oh D.H., Park K.C., Choi J.E., Kwon J.B., Lee J., Park K., Sul H.J. Increased primary cilia in idiopathic pulmonary fibrosis. Mol. Cells. 2018;41:224–233. doi: 10.14348/molcells.2018.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Johmura Y., Yu L.R., Park J.E., Gao Y., Bang J.K., Zhou M., Veenstra T.D., Yeon Kim B., Lee K.S. Identification of a novel Wnt5a-CK1varepsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 2012;31:3104–3117. doi: 10.1038/emboj.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Tan H.Y., Geneva I.I., Kruglov A., Calvert P.D. Actin filaments partition primary cilia membranes into distinct fluid corrals. J. Cell Biol. 2018;217:2831–2849. doi: 10.1083/jcb.201711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Vasishtha M., Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. J. Biol. Chem. 2010;285:22036–22049. doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Saito M., Chuang J.Z., Tseng Y.Y., Dedesma C., Tomizawa K., Kaitsuka T., Sung C.H. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat. Cell Biol. 2011;13:402–411. doi: 10.1038/ncb2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wei Z., Li G., Zhang Q., Niu S., Xu D., Mao N., XChen S., Gao X., Cai W. Silica perturbs primary cilia and causes myofibroblast differentiation during silicosis by reduction of the KIF3A-repressor GLI3 complex. Theranostics. 2020;10:1719–1732. doi: 10.7150/thno.37049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.K., Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- Lim S.K., Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem. J. 2007;403:119–127. doi: 10.1042/BJ20061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon C., Grant R., Min M. Ubiquitin-mediated degradation of Aurora kinases. Front. Oncol. 2015;5:307. doi: 10.3389/fonc.2015.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kiseleva A.A., Golemis E.A. Ciliary signalling in cancer. Nat. Rev. Cancer. 2018;18:511–524. doi: 10.1038/s41568-018-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger L.L., Patenaude C.A., Smith B.D., Layne M.D. Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J. Biol. Chem. 2011;286:44116–44125. doi: 10.1074/jbc.M111.276931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J., Avidor-Reiss T. From the cytoplasm into the cilium: bon voyage. Organogenesis. 2014;10:138–157. doi: 10.4161/org.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J.J., Johnson C.A. The cilium: cellular antenna and central processing unit. Trends Cell Biol. 2017;27:126–140. doi: 10.1016/j.tcb.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra A.N., Li Y., Wingert R.A. Antennas of organ morphogenesis: the roles of cilia in vertebrate kidney development. Genesis. 2016;54:457–469. doi: 10.1002/dvg.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masszi A., Di Ciano C., Sirokmany G., Arthur W.T., Rotstein O.D., Wang J., McCulloch C.A., Rosivall L., Kapus A. Central role for Rho in TGF-beta1-induced alpha-smooth muscle actin expression during epithelial-mesenchymal transition. Am. J. Pysiol. Renal Physiol. 2003;284:F911–F924. doi: 10.1152/ajprenal.00183.2002. [DOI] [PubMed] [Google Scholar]
- Masszi A., Speight P., Charbonney E., Lodyga M., Nakano H., Szaszi K., Kapus A. Fate-determining mechanisms in epithelial-myofibroblast transition: major inhibitory role for Smad3. J. Cell Biol. 2010;188:383–399. doi: 10.1083/jcb.200906155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Maller J.L. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelova J., Astuto-Gribble L., Inoue H., Tam B.M., Schonteich E., Prekeris R., Moritz O.L., Randazzo P.A., Deretic D. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H. Planar cell polarity and the kidney. J. Am. Soc. Nephrol. 2009;20:2104–2111. doi: 10.1681/ASN.2008111173. [DOI] [PubMed] [Google Scholar]
- Miano J.M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Miano J.M., Long X., Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Miranda M.Z., Bialik J.F., Speight P., Dan Q., Yeung T., Szaszi K., Pedersen S.F., Kapus A. TGF-beta1 regulates the expression and transcriptional activity of TAZ via a Smad3-independent, myocardin-related transcription factor-mediated mechanism. J. Biol. Chem. 2017;292:14902–14920. doi: 10.1074/jbc.M117.780502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K.A. Isolation of primary cilia by shear force. Curr. Protoc. Cell Biol. 2013;3:3 42 41–49. doi: 10.1002/0471143030.cb0342s59. [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Mick D.U. Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 2019;20:389–405. doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Seeley E.S., Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren T.M., Wyatt T.A., Sweeter J., Bailey K.L., Poole J.A., Heires A.J., Sisson J.H., Romberger D.J. Motile cilia harbor serum response factor as a mechanism of environment sensing and injury response in the airway. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014;306:L829–L839. doi: 10.1152/ajplung.00364.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotou R., Miralles F., Pawlowski R., Diring J., Flynn H.R., Skehel M., Treisman R. Phosphorylation acts positively and negatively to regulate MRTF-A subcellular localisation and activity. Elife. 2016;5:e15460. doi: 10.7554/eLife.15460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascreau G., Eckerdt F., Churchill M.E., Maller J.L. Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding. Proc. Natl. Acad. Sci. U S A. 2010;107:2932–2937. doi: 10.1073/pnas.0914874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua S.C., Chiba S., Suzuki M., Su E., Roberson E.C., Pusapati G.V., Schurmans S., Setou M., Rohatgi R., Reiter J.F. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 2017;168:264–279 e215. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaval A., Tseng Q., Bornens M., Thery M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J. Cell Biol. 2010;191:303–312. doi: 10.1083/jcb.201004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova O.V., Nikonova A.S., Loskutov Y.V., Kozyulina P.Y., Pugacheva E.N., Golemis E.A. Calmodulin activation of Aurora-A kinase (AURKA) is required during ciliary disassembly and in mitosis. Mol. Biol. Cell. 2012;23:2658–2670. doi: 10.1091/mbc.E11-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., Golemis E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J., Yang Y., Li D., Liu M., Zhou J. Deacetylation of alpha-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci. Rep. 2015;5:12917. doi: 10.1038/srep12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel L., Bernabe-Rubio M., Fernandez-Barrera J., Casares-Arias J., Millan J., Alonso M.A., Correas I. Caveolin-1alpha regulates primary cilium length by controlling RhoA GTPase activity. Sci. Rep. 2019;9:1116. doi: 10.1038/s41598-018-38020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., Leroux M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi C., Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006;1:1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- Rozycki M., Lodyga M., Lam J., Miranda M.Z., Fatyol K., Speight P., Kapus A. The fate of the primary cilium during myofibroblast transition. Mol. Biol. Cell. 2014;25:643–657. doi: 10.1091/mbc.E13-07-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez I., Dynlacht B.D. Cilium assembly and disassembly. Nat. Cell Biol. 2016;18:711–717. doi: 10.1038/ncb3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaposhnikov D., Kuffer C., Storchova Z., Posern G. Myocardin related transcription factors are required for coordinated cell cycle progression. Cell Cycle. 2013;12:1762–1772. doi: 10.4161/cc.24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E.M. The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J. Cardiovasc. Transl. Res. 2012;5:794–804. doi: 10.1007/s12265-012-9397-0. [DOI] [PubMed] [Google Scholar]
- Small E.M., Thatcher J.E., Sutherland L.B., Kinoshita H., Gerard R.D., Richardson J.A., Dimaio J.M., Sadek H., Kuwahara K., Olson E.N. Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ. Res. 2010;107:294–304. doi: 10.1161/CIRCRESAHA.110.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.E.L., Lake A.V.R., Johnson C.A. Primary cilia, ciliogenesis and the actin cytoskeleton: a little less resorption, A little more actin please. Front. Cell Dev. Biol. 2020;8:622822. doi: 10.3389/fcell.2020.622822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.K., Choi J.H., Kim M.S. Primary cilia as a signaling platform for control of energy metabolism. Diabetes Metab. J. 2018;42:117–127. doi: 10.4093/dmj.2018.42.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Spasic M., Jacobs C.R. Primary cilia: cell and molecular mechanosensors directing whole tissue function. Semin. Cell Dev. Biol. 2017;71:42–52. doi: 10.1016/j.semcdb.2017.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speight P., Kofler M., Szaszi K., Kapus A. Context-dependent switch in chemo/mechanotransduction via multilevel crosstalk among cytoskeleton-regulated MRTF and TAZ and TGFbeta-regulated Smad3. Nat. Commun. 2016;7:11642. doi: 10.1038/ncomms11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart K., Gaitan Y., Shafer M.E., Aoudjit L., Hu D., Sharma R., Tremblay M., Ishii H., Marcotte M., Stanga D. A point mutation in p190A RhoGAP affects ciliogenesis and leads to glomerulocystic kidney defects. PLoS Genet. 2016;12:e1005785. doi: 10.1371/journal.pgen.1005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves M.E., Strauss J.F., 3rd, Sapao P., Shi B., Varga J. The primary cilium: emerging role as a key player in fibrosis. Curr. Rheumatol. Rep. 2019;21:29. doi: 10.1007/s11926-019-0822-0. [DOI] [PubMed] [Google Scholar]
- Tian W., Hao C., Fan Z., Weng X., Qin H., Wu X., Fang M., Chen Q., Shen A., Xu Y. Myocardin related transcription factor A programs epigenetic activation of hepatic stellate cells. J. Hepatol. 2015;62:165–174. doi: 10.1016/j.jhep.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Tsang W.Y., Bossard C., Khanna H., Peranen J., Swaroop A., Malhotra V., Dynlacht B.D. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos E., Criollo A., Schiattarella G.G., Altamirano F., French K.M., May H.I., Jiang N., Nguyen N.U.N., Romero D., Roa J.C. Fibroblast primary cilia are required for cardiac fibrosis. Circulation. 2019;139:2342–2357. doi: 10.1161/CIRCULATIONAHA.117.028752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen Q., Zhang X., Zhang B., Zhuo X., Liu J., Jiang Q., Zhang C. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J. Cell Sci. 2013;126:1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- Wang L., Dynlacht B.D. The regulation of cilium assembly and disassembly in development and disease. Development. 2018;145:dev151407. doi: 10.1242/dev.151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.J., Tay H.G., Soni R., Perumal G.S., Goll M.G., Macaluso F.P., Asara J.M., Amack J.D., Tsou M.F. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat. Cell Biol. 2013;15:591–601. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang D.Z., Hockemeyer D., McAnally J., Nordheim A., Olson E.N. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wheway G., Mitchison H.M. Opportunities and challenges for molecular understanding of ciliopathies-the 100,000 genomes project. Front. Genet. 2019;10:127. doi: 10.3389/fgene.2019.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn Y.H., Han Y.G. Primary cilia in brain development and diseases. Am. J. Pathol. 2018;188:11–22. doi: 10.1016/j.ajpath.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaromytidou A.I., Miralles F., Treisman R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol. Cell. Biol. 2006;26:4134–4148. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze [datasets/code].