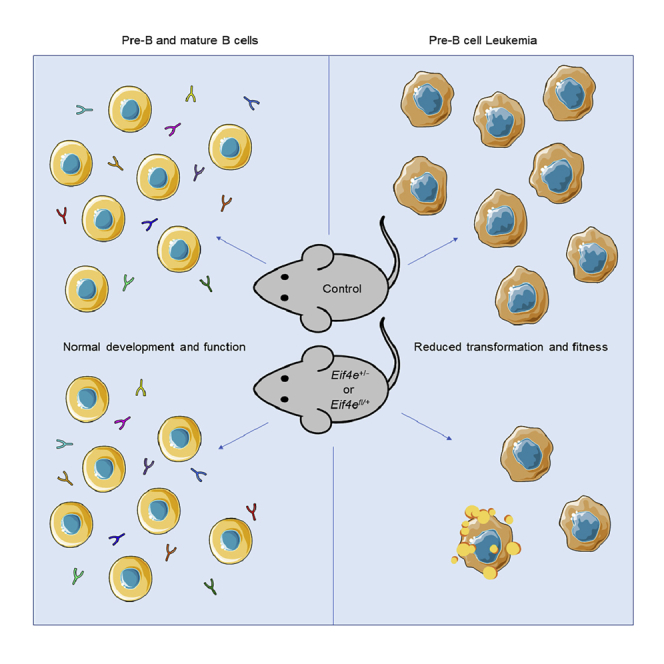
Summary
The cap-binding protein eukaryotic initiation factor 4E (eIF4E) promotes translation of mRNAs associated with proliferation and survival and is an attractive target for cancer therapeutics. Here, we used Eif4e germline and conditional knockout models to assess the impact of reduced Eif4e gene dosage on B-cell leukemogenesis compared to effects on normal pre-B and mature B-cell function. Using a BCR-ABL-driven pre-B-cell leukemia model, we find that loss of one allele of Eif4e impairs transformation and reduces fitness in competition assays in vitro and in vivo. In contrast, reduced Eif4e gene dosage had no significant effect on development of pre-B and mature B cells or on survival or proliferation of non-transformed B lineage cells. These results demonstrate that inhibition of eIF4E could be a new therapeutic tool for pre-B-cell leukemia while preserving development and function of normal B cells.
Subject areas: molecular biology, cell biology, cancer
Graphical abstract
Highlights
-
•
Loss of one allele of Eif4e impairs pre-B-cell leukemia transformation
-
•
Eif4e+/- leukemia cells had reduced competitive fitness both in vitro and in vivo
-
•
Reduced Eif4e dosage had no effect on B-cell development, survival, or proliferation
Molecular biology; Cell biology; Cancer
Introduction
The eukaryotic translation initiation factor 4E (eIF4E) protein binds to the 7-methylguanosine cap present in most mRNAs and recruits the scaffolding protein eIF4G, and the RNA helicase eIF4A to form the translation initiation complex known as eIF4F. In cancer, eIF4F contributes to progression of the disease by preferentially translating mRNAs involved in tumor hallmarks including sustained proliferative signaling, evasion of growth suppression, resistance to programmed cell death, replicative immortality, angiogenesis, invasion and metastasis (Pelletier et al., 2015; Malka-Mahieu et al., 2017). eIF4F activity and protein synthesis were found to be drivers in T cell lymphomas driven by active AKT (Hsieh et al., 2010), and in anti-BRAF and anti-MEK resistance in BRAF(V600)-mutant melanoma, colon, and thyroid cancer cell lines (Boussemart et al., 2014). This makes cap-dependent translation an attractive target for cancer therapy (Pelletier et al., 2015), and efforts are underway to develop small molecule inhibitors of eIF4E, eIF4G, and eIF4A for oncology (Moerke et al., 2007; Boussemart et al., 2014; Feng et al., 2015; Chu et al., 2016; Ernst et al., 2020).
Several cancers show increased expression of eIF4E and initial studies using siRNA knock-down inhibited cell growth in head and neck squamous cell carcinomas and in triple-negative breast cancer (Oridate et al., 2005; Soni et al., 2008). They also showed that eIF4E knock-down inhibited both rapamycin-insensitive and rapamycin-sensitive cell lines without activating the feedback loop to increase AKT phosphorylation (Soni et al., 2008). Importantly, reducing eIF4E dosage to heterozygosity is compatible with normal life and eIF4E+/- mice do not show any impairment in embryonic development and global protein synthesis. However, eIF4E haploinsufficiency significantly reduced cellular transformation and tumor development in a mouse model of Kras-driven lung cancer, due to selective changes in the translation of specific mRNA networks critical for cancer formation (Truitt et al., 2015). Conversely, in a gain-of-function model, eIF4E transgenic (Tg) mice displayed enhanced Myc lymphomagenesis (Ruggero et al., 2004). These findings establish that altering eIF4E dosage has significant yet selective biological consequences during tumor progression.
While several studies have investigated the role of eIF4E in cap-dependent translation of mRNAs in cancer cells, only recently has translational regulation been gaining attention in lymphocyte activation and differentiation (Bjur et al., 2013; Araki et al., 2017). In lymphocytes, the 4E-binding protein (4E-BP)/eIF4E axis coordinates cell growth and proliferation during lymphocyte activation (So et al., 2016). This suggests that the 4E-BP/eIF4E axis is critical for the initial translation of specific mRNAs needed during lymphocyte activation. Our lab also found that the mechanistic target of rapamycin (mTOR) complex-1 (mTORC1) is important in both normal B-cell function and tumorigenesis, specifically regulating B-cell differentiation and antibody class switching (Chiu et al., 2019) as well as pre-B cell leukemogenesis driven by the BCR-ABL oncogene (Kharas et al., 2008; Janes et al., 2010). The mTORC1 pathway controls the activity of eIF4E, through the phosphorylation of 4E-BPs, as well as the phosphorylation of additional proteins involved in many cellular processes. Notably, elevated phospho-4E-BP correlates with poor prognosis in pediatric acute lymphoblastic leukemia (ALL) (Nemes et al., 2013). This suggests that high eIF4E activity is an oncogenic driver in aggressive leukemia. Small molecules targeting eIF4F components have cytostatic and cytotoxic activity in blood cancer models, and some lead candidates have entered clinical trials. To better predict the potential therapeutic window of agents targeting eIF4F, genetic loss-of-function models are needed to assess eIF4F addiction in leukemia and lymphoma cells versus normal lymphocytes. Using Eif4e germline heterozygous mice (Truitt et al., 2015) and a novel conditional knockout model, we show that reduced eIF4E protein slows tumorigenesis in a mouse model of leukemia, while normal mouse B cells can maintain proliferation and antibody class switching. These data collectively strengthen the conclusion that cancer cells are selectively sensitive to reductions in eIF4E protein (Truitt et al., 2015) and provide a strong rationale for testing eIF4E antagonists in B-cell malignancies.
Results
Genetic deletion of one allele of Eif4e reduces eIF4E protein expression in B-ALL cells
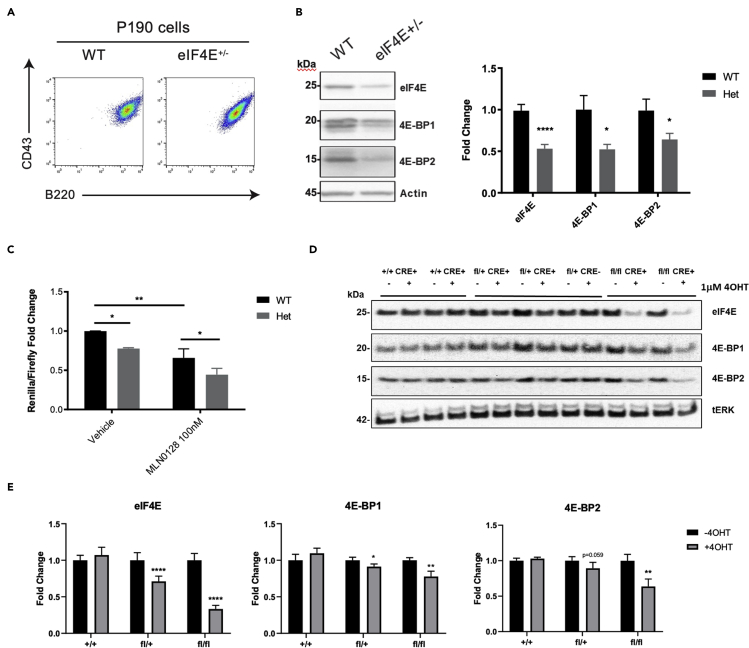
To study the role of eIF4E in leukemogenesis, we used a murine model of B-precursor acute lymphoblastic leukemia (B-ALL). Ph+ B-ALL is initiated by the BCR-ABL oncogene that results from the Philadelphia chromosome (Ph) translocation. When human p190 BCR-ABL (p190 is the most common isoform of BCR-ABL found in Ph+ B-ALL) is retrovirally expressed in infected mouse bone marrow cells, the transformed progenitor B cell lines (termed p190 cells) will initiate B-ALL when transferred to recipient mice. We generated p190 cells from wild-type mice (WT) and eIF4E heterozygous mice (eIF4E+/-) and confirmed the outgrowth of B-ALL cells with similar pro-B immunophenotype (that are B220+ CD43+) in both genotypes (Figure 1A).
Figure 1.
Genetic deletion of Eif4e reduces eIF4E protein expression in p190 cells
(A) Established p190 WT and eIF4E+/- leukemia cells have similar immunophenotype characterized by B220 and CD43 expression. Representative flow cytometry plots of p190 cells.
(B) Protein analyses of eIF4E, 4E-BP-1, 4E-BP-2 and actin in WT and Het p190 cells. Relative protein quantification performed by densitometric analysis using ImageJ64 software. Data are expressed as mean ± SEM. Fold change was calculated using actin as loading control and normalized to WT sample for each independent experiment. Significance was calculated using unpaired t test (∗p < 0.05; ∗∗∗∗p < 0.001, n = 7).
(C) p190s were generated from WT or Het mice and we used a bicistronic dual Renilla-Firefly luciferase reporter construct to measure cap-dependent translation (Renilla luciferase) relative to cap-independent, Coxsackie virus IRES mediated translation (firefly luciferase) as an internal control. MLN0128 (100 nM) treated cells were used as a control. Data are represented as mean ± SEM. Fold change was calculated using WT vehicle condition. Significance was calculated using a paired one tailed student's t test. (∗p < 0.05; ∗∗p < 0.01; n = 3 or 4 per group).
(D) Western blots of eIF4E, 4E-BP-1, 4E-BP-2, and total ERK (tERK) in eIF4E+/+, eIF4E fl/+ and eIF4E fl/fl p190 after 72hr of 4OHT (1 μM) treatment.
(E) Relative protein quantification performed by densitometric analysis using ImageJ64 software. Fold change calculated using tERK as loading control. Data are expressed as mean ± SEM. Significance was calculated using paired t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.001, n = 4–6 per group).
Measurement of eIF4E protein amounts showed that eIF4E was reduced by approximately 50% in the eIF4E+/- p190 cells compared to WT (Figure 1B). eIF4E binding proteins 4E-BP1 and 4E-BP2 were also reduced (Figure 1B), consistent with studies showing that 4E-BP stability is affected by changes in eIF4E expression (Yanagiya et al., 2012). We measured cap-dependent translation using a bicistronic dual Renilla-Firefly luciferase reporter construct. This construct measures cap-dependent translation by synthesis of the Renilla luciferase, as well as cap-independent, Coxsackie virus IRES-mediated translation by synthesis of firefly luciferase as an internal control for transfection efficiency and viable cell recovery. Dual luciferase reporter assays showed that cap-dependent translation was reduced approximately 25% in eIF4E+/- p190 cells (Figures 1C, S1A, and S1B). The mTOR kinase inhibitor MLN0128 was used as a positive control to reduce cap-dependent translation (Figure 1C), and in most samples also reduced cap-independent translation (Figures S1A and S1B).
As a distinct way to interfere with eIF4E expression, we established a conditional eIF4E loss of function model. We obtained mice with a conditional floxed allele of eIF4E from a knockout mouse repository and crossed them with Rosa26-Cre mice. We generated p190 cells from Cre-positive eIF4Efl/+, eIF4Efl/fl, and control eIF4E+/+ mice and assessed deletion efficiency by Western blot (Figure 1D). After 72hr of treatment with 4-hydroxytamoxifen (4OHT; 1 μM), eIF4E protein expression was significantly reduced by 30–35% in p190 cells of fl/+ genotype (p < 0.0001) and 70% in fl/fl (p < 0.0001) (Figure 1E). Expression of 4E-BP1 and 4E-BP2 were modestly reduced following Cre-mediated deletion in heterozygous fl/+ cells, with more significant reductions observed in fl/fl cells with homozygous deletion of Eif4e (Figure 1E). These results show that in both germline and conditional knockout systems the deletion of Eif4e alters the expressions of 4E-binding proteins in p190 cells.
In various cell systems, partial inhibition of cap-dependent translation selectively reduces translation efficiency of a subset of mRNAs without inhibiting global protein synthesis rates (Pelletier et al., 2015; Malka-Mahieu et al., 2017). In accord, puromycin incorporation assays showed that protein synthesis was not significantly reduced following conditional inactivation of one allele of Eif4e in p190 cells (Figure S1C).
eIF4e haploinsufficiency reduces leukemic transformation by BCR-ABL
To test the effects of reduced Eif4e gene dosage on leukemic transformation, we first conducted colony formation assays comparing WT and eIF4E+/- bone marrow cells infected with retroviruses expressing p190-BCR-ABL. The results showed significantly reduced leukemogenic potential in B-cell progenitors of eIF4E+/- mice (Figure S2A).
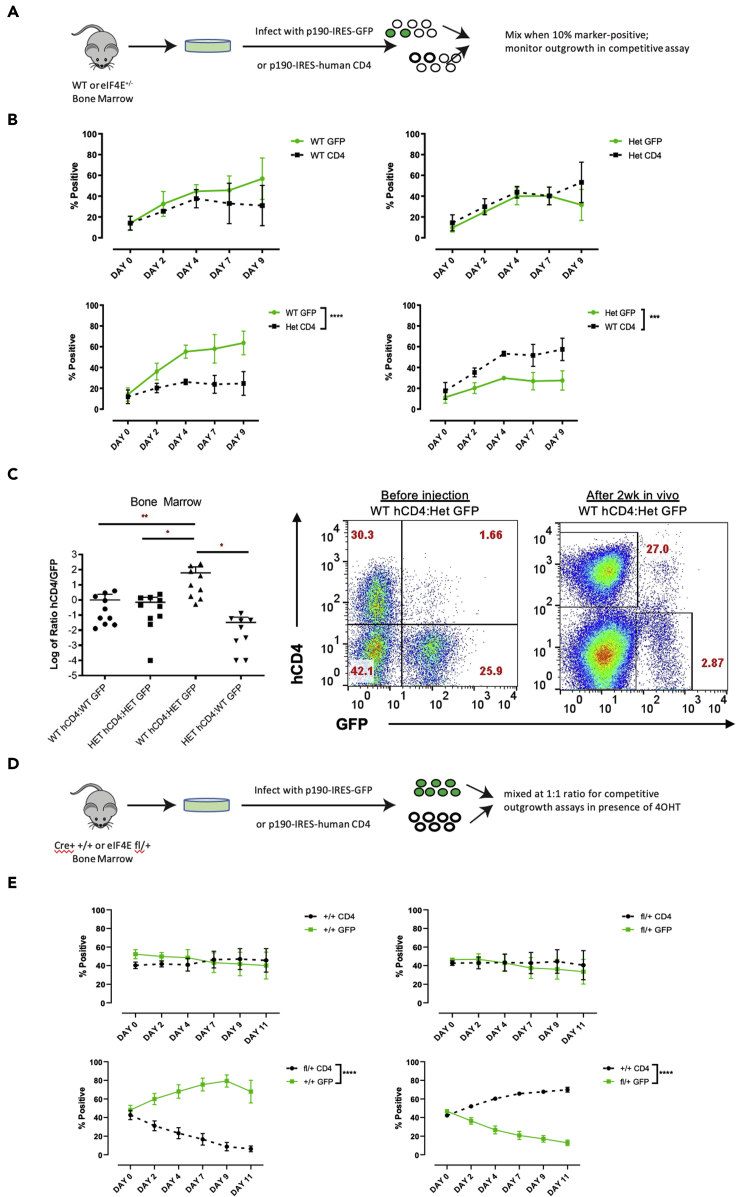
To further assess the fitness of leukemia cells in a stringent manner, we conducted competitive outgrowth assays. We co-cultured WT and germline eIF4E+/- (Het) B cells after viral infection when cells were 10–20% marker positive expressing either GFP (MSCV-IRES-GFP) or human CD4 (MSCV-IRES-hCD4) (experimental design outlined in Figure 2A). Cells were mixed at 1:1 ratios of WT:WT, WT:Het, Het:WT, and Het:Het, and their growth was monitored in vitro. We found that the WT p190 cells significantly outgrew the Het p190 cells during in vitro culture (Figure 2B). In the control mixtures (WT:WT and Het:Het), the two cell populations grew out at similar rates. The eIF4E+/- p190 cells also showed a dramatic disadvantage in vivo (Figure 2C). For this experiment, we injected WT syngeneic recipient mice with an equal ratio of p190 in all four combinations of hCD4 and GFP markers (Figure S2B). After 14 days, we collected bone marrow and analyzed for the presence of leukemia cells. The mean of Log10(ratio) of the WT:Het mixture was significantly different than the WT:WT, and the Het:WT mixture different than the Het:Het mixture (Figure 2C). These data indicate that reducing eIF4E expression levels can significantly delay leukemogenesis in vivo and that eIF4E gene dosage impacts BCR-ABL-mediated transformation.
Figure 2.
eIF4E+/- and eIF4Efl/+ p190 have a growth disadvantage
(A) Experimental scheme of competitive outgrowth assay of WT or eIF4E+/- p190 BCR-ABL.
(B) Growth of p190 cells was monitored in vitro over 9 days. Percentage of hCD4 and GFP marker positive cells were measured with flow cytometry. Data are expressed as mean ± SEM. (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 two-way ANOVA, n = 3 per genotype).
(C) Log of ratio of hCD4+/GFP+ was measured by flow cytometry. Data are expressed as mean ± SEM. (∗p < 0.05, ∗∗p < 0.01, one way ANOVA, n = 9 or 10 mice/group). Representative flow cytometry plots of bone marrow analyzed for frequency of WT and Het leukemia cells before and after injection of an equal ratio of hCD4 WT and GFP Het p190.
(D) Experimental scheme of maintenance assay of WT or eIF4E fl/+ p190 BCR-ABL with 100% marker positivity.
(E) Competitive growth assay of established hCD4 and GFP p190 eIF4E+/+ Cre+ and eIF4E fl/+ Cre+ treated with 1 μM 4OHT. Percentage of hCD4 and GFP marker positive cells were measured by flow cytometry. Data are expressed as mean ± SEM. (∗∗∗∗p < 0.0001 two-way ANOVA, n = 6–10/each combination).
We then conducted competition experiments using the conditional deletion (Eif4e-flox) system. Here, we induced deletion of the eIF4E gene after p190 leukemia cell pools were fully established, allowing us to test whether reduced gene dosage affected leukemia maintenance. We obtained bone marrow from 4 independent Cre+ mice from each genotype Eif4efl/+ and Eif4e+/+ (3–5-weeks-old) and infected with p190 viruses marked with GFP or hCD4. After 7 days (when cultures were ∼100% marker-positive), cells were mixed at a 1:1 ratio in different combinations (+/+CD4:+/+GFP, fl/+CD4:fl/+GFP, fl/+CD4:+/+GFP, +/+CD4:fl/+GFP) for competitive growth assays in presence of 4OHT (1 μM) (experimental design outlined in Figure 2D). Samples were withdrawn every 2 days for FACS analysis of the ratio of GFP vs. hCD4+ populations. In presence of 4OHT, the eIF4Efl/+ cells had a significant growth disadvantage vs. eIF4E+/+ p190 in both combinations, whereas combinations of the same genotype grew at similar rates (Figure 2E). Moreover, the addition of 4OHT to combinations of GFP and hCD4 fl/fl Cre-positive cells led to rapid cell death (Figure S2C). We confirmed reduced eIF4E expression in pure cultures of eIF4Efl/+ p190 cells treated with 4OHT (Figure S2D). Together, the germline and conditional deletion systems revealed that reduced eIF4E expression impairs BCR-ABL-dependent B-ALL leukemogenesis and maintenance.
eIF4e haploinsufficiency does not impair B-cell development
We were also interested in the role of eIF4E in normal B-cell function. We first assessed B-cell development using FACS analysis of splenic B cell subsets in WT and eIF4E+/- mice. There were no differences in the percentages of transitional (T1, T2, T3), marginal zone or follicular B cell subsets (Figures S3A and S3B). There were also no differences in the overall cell counts of total splenocytes (data not shown). We next analyzed pre-B cell subsets (B220+CD43+7AAD−) in the bone marrow of WT and eIF4E+/- mice cultured ex vivo. We isolated bone marrow cells and kept in culture for 48hr with or without IL-7; no differences were observed between the two genotypes (Figure S4A). Previous studies have shown that IL-7-dependent pre-B-cell outgrowth is dependent on mTOR-complex-1 (mTORC1) (Iwata et al., 2016). In agreement, both the mTORC1-selective inhibitor rapamycin and the dual mTORC1/mTORC2 inhibitor MLN0128 caused concentration-dependent reductions in pre-B-cell viability in the presence of IL-7. Notably, the mTOR inhibitors reduced viability to the same extent in both WT and eIF4E+/- pre-B cells (Figure S4B), indicating that the heterozygous cells have not rewired signaling to become mTORC1-independent. Similar results were obtained from the conditional knockout system (Figures S4C and S4D).
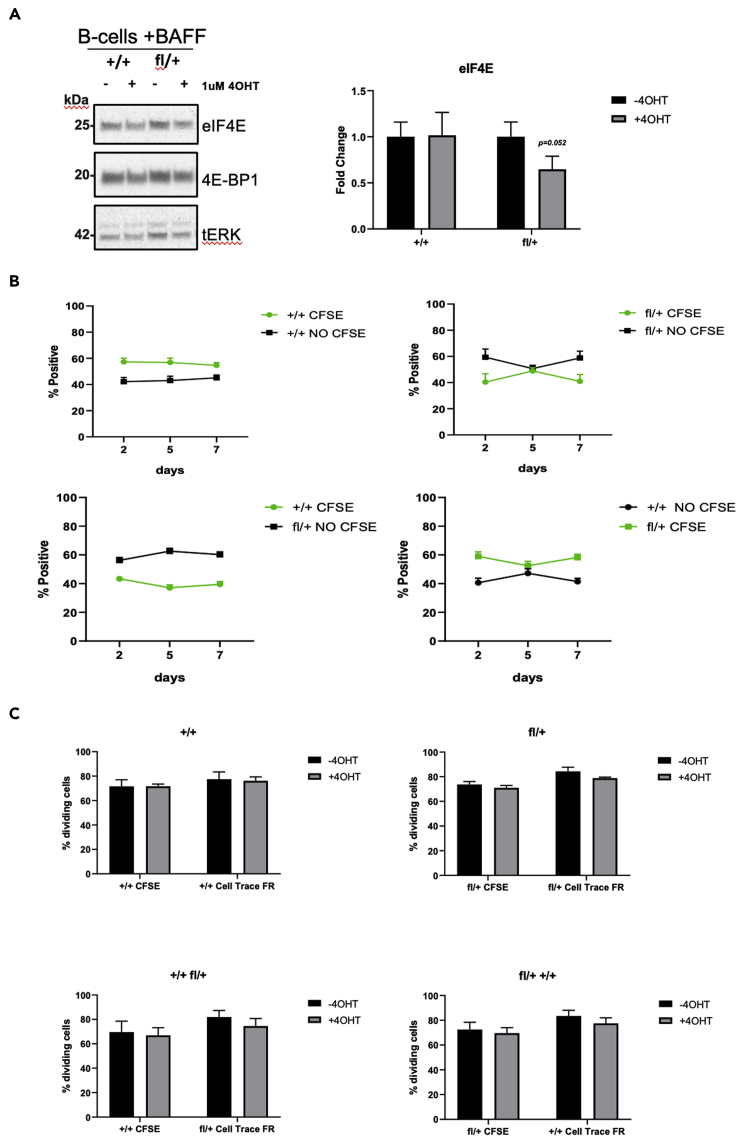
Figure 4.
B cell survival and proliferation is not affected by deletion of Eif4e
(A) Protein analyses of eIF4E, 4E-BP-1 and tERK in purified splenic B cells from eIF4E+/+, eIF4Efl/+ after 96hr of 4OHT treatment in the presence of BAFF survival factor. Fold change calculated using tERK as loading control. Data are expressed as mean ± SEM. (actual p value is shown in graph, paired one-tailed t test, n = 4 per group).
(B) Competitive survival assay of purified eIF4E+/+, eIF4Efl/+ splenic B cells labeled with or without CFSE in presence of BAFF 100ng/mL. Data are expressed as percentage of positive cells +/− SEM., n = 3–5 per group.
(C) Competitive proliferation assay of purified eIF4E+/+, eIF4Efl/+ B cells labeled with cell division tracking dyes CFSE or FAR RED (630nM). Following 48hr culture in presence or absence of 4OHT and with BAFF to maintain survival, cells were stimulated to proliferate with 10 μg/mL anti-IgM and 10 ng/mL IL-4. Data are expressed as percentage of dividing cells +/− SEM. (n = 3 for each combination).
Normal B-cell function is not altered by Eif4e haploinsufficiency
To evaluate B-cell function in vivo, we immunized WT and eIF4E+/- mice with sheep red blood cells (SRBC). SRBC-specific IgM and IgG1 antibodies were measured after 8 days using a FACS-based assay (McAllister et al., 2017). eIF4E+/- mice produced an equivalent titer of anti-SRBC antibodies of IgG1 isotype and a significant IgM response, although this was weaker than in WT mice (Figures S5A and S5B). Additionally, there were no differences in naturally occurring germinal center B cell populations (B220+/Fas+/GL7+) isolated from Peyer's patches from the eIF4E+/- mice compared to WT (data not shown).
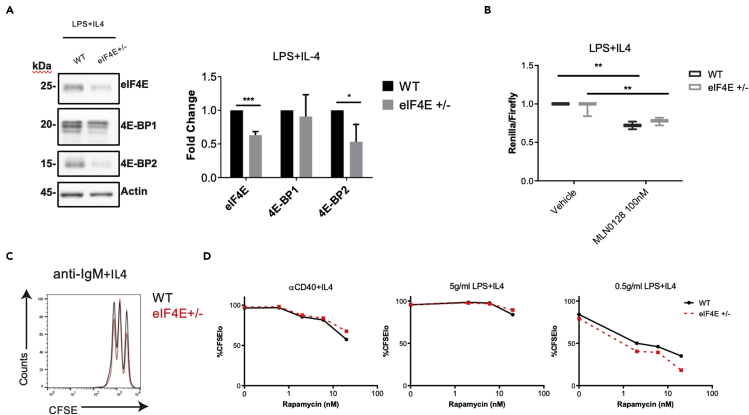
We purified splenic B cells from WT and eIF4E+/- mice and measured eIF4E, 4E-BP1, and 4E-BP2 protein levels before and after stimulation with LPS + IL-4 for 48 hr. As observed previously (Truitt et al., 2015), eIF4E expression was reduced in unstimulated B cells from eIF4E+/- mice (Figure S3C). Similarly, eIF4E protein expression was reduced approximately 50% in stimulated B cells from eIF4E+/- mice (Figure 3A). Interestingly, we observed that after stimulation, 4E-BP2 protein levels were significantly reduced in the eIF4E+/- cells (Figure 3A) despite no change in mRNA amounts (Figure S3D). Next, we assessed cap-dependent translation after B cell stimulation by employing the Renilla-Firefly dual luciferase assay. We observed that cap-dependent translation measured in activated eIF4E+/- B cells was just as efficient as in the WT control cells and remained sensitive to mTOR inhibition (Figure 3B). Considering that activated eIF4E+/- cells have reduced amounts of the inhibitory 4E-BP2, it is possible that, at least in part, the efficiency of cap-dependent mRNA translation might be preserved despite reduced eIF4E expression.
Figure 3.
Functional analyses of eIF4E+/- mouse splenic B cells
(A) Protein analyses of eIF4E, 4E-BP-2, 4E-BP-1 and actin in purified WT or eIF4E+/- B cells after 16 hr of LPS + IL-4 stimulation. Relative protein quantification performed by densitometric analysis using ImageJ64 software. Data are expressed as mean ± SEM. (∗p < 0.05, ∗∗∗p < 0.001, unpaired one-tailed student's t-test, n = 3 or 4 per group).
(B) Renilla-Firefly luciferase cap-dependent translation of WT and eIF4E+/- B cells stimulated with LPS + IL-4 for 48hr. Data are expressed as mean ratio of Renilla/Firefly +/− SEM. Significance was calculated using a paired one tailed student's t test. (∗∗p < 0.01).
(C) Flow cytometry histogram of CFSE staining of WT and eIF4E+/- B cells in presence of anti-IgM and IL-4. Similar results were obtained in a repeat experiment.
(D) Percentage of CFSE-low WT and eIF4E+/- B cells stimulated with αCD40+IL-4, 5 μg/mL LPS + IL-4, or 0.5 μg/mL LPS + IL-4 and treated with indicated concentrations of rapamycin was measured at 72 hr by flow cytometry. Similar results were obtained in a repeat experiment.
Next, we evaluated functional responses of splenic B cells ex vivo. We stimulated B cells with anti-IgM + IL-4 and measured proliferation by cell division tracking dye CFSE at 72 hr using flow cytometry. There were no differences in cell division of eIF4E+/- compared with WT cells under these conditions (Figure 3C) or upon stimulation with IL-4 together with either anti-CD40 or with two different concentrations of LPS (Figure 3D). Reduced Eif4e gene dosage also did not impair class switching to IgG1 (Figure S5C). We also tested if eIF4E+/- cells had altered sensitivity to the effects of rapamycin on switching to IgG1 or proliferation; again, there were no differences (Figures 3D and S5D).
Eif4e deletion does not affect normal B cell survival and proliferation
To evaluate if Eif4e deletion affects B-cell survival ex vivo, we purified B cells from Cre+ mice of different Eif4e-flox genotypes and treated with 4OHT in the presence of the cytokine BAFF, which maintains survival with minimal proliferation. We first confirmed reduced eIF4E protein expression after 4OHT treatment in isolated B cells by Western blot (Figure 4A). Next, we generated mixed cultures of B cells labeled with or without CFSE (+/+CFSE:+/+ NO CFSE, fl/+CFSE:fl/+ NO CFSE, +/+CFSE:fl/+ NO CFSE, fl/+ CFSE:+/+ NO CFSE). In this case, CFSE was used to distinguish cell populations rather than to track cell division. Survival of B cells with BAFF and 4OHT was monitored for 7 days four combinations and the initial ratios were not significantly changed in any of the four combinations (Figure 4B).
For proliferation assays, we generated mixed cultures of B cells labeled with cell division tracking dyes CFSE or Cell Trace Far Red (630nM) (+/+CFSE:+/+FAR RED, fl/+CFSE:fl/+FAR RED, +/+CFSE:fl/+FAR RED, fl/+ CFSE:+/+FAR RED ratio 1:1). After B cell mixtures were cultured for 48hr in presence of BAFF and 4OHT, we added anti-IgM and IL-4 to stimulate proliferation. After an additional 48hr of culture the CFSE and Far Red signals were measured by FACS (Figure S6). Again, we observed no difference in proliferation rate of B cells in all combinations (Figure 4C; proliferation was assessed by percent dividing cells based on tracker dye dilution). Together, these data indicate that deletion of one allele of Eif4e did not affect normal B cells in the same way as leukemia B cells. Reducing eIF4E protein slowed tumorigenesis of mouse p190 leukemia cell lines, whereas activated mouse splenic B cells were able to divide without impairment.
Discussion
The cap-binding protein eIF4E is a potential vulnerability of cancer cells (Truitt et al., 2015), and efforts are underway to develop eIF4E inhibitors for clinical testing (https://effector.com/pipeline/). eIF4E is a major effector of mTORC1 signaling, and cap-dependent translation is elevated in many B-cell leukemias and lymphomas (Prabhu et al., 2007; Pourdehnad et al., 2013; Schwarzer et al., 2015; Demosthenous et al., 2015). However, cap-dependent translation is important for both normal B-cell function and tumorigenesis (Ruggero et al., 2004; Prabhu et al., 2007; Pourdehnad et al., 2013; Demosthenous et al., 2015; So et al., 2016; Chiu et al., 2019); thus, it is important to understand the effects of targeting eIF4E directly in both systems. In this study we used both germline and conditional knockout models to reduce eIF4E protein expression and found that B cell development and function is preserved, whereas pre-B-cell transformation by BCR-ABL is impaired. In competitive growth assays with wild-type p190 cells, leukemia cells with reduced Eif4e gene dosage displayed reduced proliferation and/or survival. In contrast, constitutive or inducible loss of one Eif4e allele did not affect the survival or proliferation of mature splenic B cells.
One notable observation was that activated mature B cells from Eif4e+/- mice maintained normal levels of cap-dependent translation in a luciferase reporter assay (Figure 3B), whereas cap-dependent translation was reduced in p190 leukemia cells from Eif4e+/- mice (Figure 1C). This suggests that p190 cells require the full eIF4E dose for maintaining mRNA translation. Additionally, expression of the inhibitory 4E-BP2 protein was reduced in both the activated mature B cells and in p190 cells from Eif4e+/- mice and 4E-BP1 was also reduced in p190 cells. This suggests that p190 cells adapt to preserve eIF4E activity by further decreasing the inhibitory proteins as a compensatory mechanism and that mature B cells may partially do the same, consistent with previous findings (Yanagiya et al., 2012). The presence of constitutively active BCR-ABL in leukemia cells may drive other mechanisms of dependency on eIF4E dosage including altered total expression of eIF4E and/or eIF4G scaffolding proteins or altered function of other translational regulators. In order to gain insight into the mechanism for reduced leukemia cell fitness, an important future research direction will be to use polysome profiling to assess global changes in mRNA translation efficiency in eIF4E-deficient p190 cells versus non-transformed pre-B cells and mature B cells.
The conditional floxed system allowed the inducible deletion of Eif4e in established p190 leukemia cells and normal pre-B and mature B cells. This approach serves as a model of acute eIF4E inactivation and suggests that pharmacological targeting of this translation initiation factor might have therapeutic value in B-ALL. Experiments using the germline knockout (Figures 2B and S2A) suggested that reduced Eif4e dosage also impairs leukemia initiation. However, further experiments are needed to rule out the possibility of altered pre-B cell development and precursor B-cell function in mice with germline Eif4e deletion. It would also be interesting to determine whether conditional Eif4e deletion affects leukemia cell fitness in models driven by oncogenes other than BCR-ABL, such as those found in Ph-like B-ALL.
In a previous study, we established that mice with germline loss of one allele of Eif4e are viable and healthy, yet resistant to development of Kras-driven solid tumors (Truitt et al., 2015). The present study extends this paradigm to blood cancer and another classical oncogene, BCR-ABL. The data show that Eif4e haploinsufficiency is selectively detrimental to oncogene-driven leukemia cells yet does not impair survival of normal pre-B and mature B cells. In other words, cancer cells but not normal cells are addicted to full dosage of eIF4E. This result is somewhat surprising given the growing evidence that the 4E-BP/eIF4E axis controls key aspects of B-cell proliferation and differentiation (So et al., 2016; Chiu et al., 2019). As compounds that target eIF4E activity advance into clinic trials, future studies should address whether this dichotomy in responses between pre-B leukemia and normal B cells is observed when using chemical inhibitors or degraders of eIF4E. Conversely, crossing the Eif4e-flox model with different Cre driver strains may be more powerful than chemical tools to define the cell-intrinsic function of eIF4E in different lymphocyte subsets.
Limitations of the study
We would like to mention the following caveats to the conclusions of the study. First, the reduced outgrowth of p190 leukemia cells from germline Eif4e+/- mice might be due in part to altered frequencies or function of normal B-cell progenitors that are the targets for transformation. Further support for a defect in leukemic transformation would require more detailed comparison of B-cell progenitor frequencies, along with serial replating experiments. In contrast, we could make a more definitive conclusion about a defect in leukemia cell maintenance; in this case, we used the conditional knockout approach in which the target cells for initiation of transformation were equivalent and deletion was induced in established pools of p190 cells. A second caveat is that the transformation initiation and maintenance experiments were done with a single mouse model of B-ALL, driven by the human BCR-ABL oncogene. Additional models would be needed to generalize the conclusions. Finally, our study did not reveal the molecular mechanisms by which BCR-ABL-driven leukemia cells, compared to normal pre-B and mature B cells, have greater addiction to full Eif4e gene dosage.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human CD4 (RPA-T4) | Biolegend | 300505 (FITC); AB_314073 |
| Mouse B220 (RA3-6B2) | eBioscience ThermoFisher Scientific | 17-0452-82 (APC); AB_469395 |
| 53-0452-80 (AF488); AB_469907 | ||
| Mouse CD43 (eBIOR2/60) | eBioscience ThermoFisher Scientific | 11-0431-82; AB_465040 |
| Mouse IgG1 (A85-1) | BD Bioscience | BDB560089; AB_1645625 |
| Mouse AA4.1 | eBioscience ThermoFisher Scientific | 11-5892-81 (FITC); AB_465297 |
| 12-5892-81 (PE); AB_466017 | ||
| Mouse IgM | eBioscience ThermoFisher Scientific | 16-5092-85; AB_2573088 |
| Mouse IgM | Jackson Immunoresearch | 115-006-020; AB_2338469 |
| CD23 | BioLegend | 101607 (PE); AB_312832 |
| GL7 | EBioscience | 51-5902-82 (AF647); AB_1311298 |
| Fas | BD Pharmingen | 554258 (PE); AB_395330 |
| 7-Aminoactinomycin D | eBioscience ThermoFisher Scientific | A1310 |
| Anti-CD40 (HM40-3) | eBioscience ThermoFisher Scientific | 16-0402-86; AB_468947 |
| Anti-puromycin | EMD Millipore | MABE343-AF488 |
| b-actin | Cell Signaling Technologies | 3700S; AB_2242334 |
| Total ERK | Cell Signaling Technologies | 4696S; AB_390780 |
| eIF4E | Cell Signaling Technologies | 2067S; AB_2097675 |
| 4E-BP1 | Cell Signaling Technologies | 9644S; AB_2097841 |
| 4E-BP2 | Cell Signaling Technologies | 2845S; AB_10699019 |
| Goat anti mouse peroxidase conjugated IgG | Promega | W4021; AB_430834 |
| Goat anti rabbit peroxidase conjugated IgG | Promega | W4011; AB_430833 |
| Chemicals, peptides, and recombinant proteins | ||
| CFSE | ThermoFisher Scientific | C34554 |
| Cell Trace far red | ThermoFisher Scientific | C34564 |
| BAFF | R&D Systems | 8876-BF-010 |
| Cycloheximide | Sigma Aldrich | 01,810-1G |
| Puromycin | GIBCO Fisher Scientific | A1113803 |
| Hydroxytamoxifen | Sigma Aldrich | H7904 |
| IgM | Jackson Immuno Research Laboratories | 115-006-020 |
| LPS | Sigma Aldrich | L3012 |
| MLN0128 | Active Biochem | A-1023 |
| Rapamycin | LC Laboratories | R-5000 |
| Recombinant mouse IL-4 | R&D Systems | 404-ML-010 |
| Recombinant mouse IL-7 | R&D Systems | 407-ML-005 |
| Critical commercial assays | ||
| EasySep™ mouse B Cell Isolation Kit | Stemcell Technologies | 19854 |
| Bradford | Bio-Rad Laboratories | 5000006 |
| Dual luciferase assay kit | Promega | E1910 |
| Foxp3-staining kit fix/perm buffer | Biolegend | 421403 |
| Experimental models: organisms/strains | ||
| C57BL/6NJ mice | The Jackson Laboratory | 005304 |
| B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J mice | The Jackson Laboratory | 008463 |
| Oligonucleotides | ||
| Genotyping DNA primer for eIF4E set1 Forward: AGAGCAAATACGGAACCGACGTGTGC |
IDT | N/A |
| Genotyping DNA primer for eIF4E set1 Reverse: ATGCAGGGTTTGGGTGCTTACACAG |
IDT | N/A |
| Genotyping DNA primer for eIF4E set2 Forward: GAGCAAATACGGAACCGACGTGTGC | IDT | N/A |
| Genotyping DNA primer for eIF4E set2 Reverse: GAAGTTATCTCGACGAAGTTCC |
IDT | N/A |
| Genotyping DNA primer for eIF4E set3 Forward: GCGCAACGCAATTAATGATAAC |
IDT | N/A |
| Genotyping DNA primer for eIF4E set3 Reverse: TCTGCTAGCTTGTTCTCACGCACCC |
IDT | N/A |
| Genotyping DNA primer for Cre Forward: GCGGTCTGGCAGTAAAAACTATC |
IDT | N/A |
| Genotyping DNA primer for Cre Reverse: GTGAAACAGCATTGCTGTCACTT |
IDT | N/A |
| Software and algorithms | ||
| ImageJ1.52a | NIH | https://imagej.nih.gov/ij/ |
| FlowJo software (10.7.1) | BD | https://www.flowjo.com |
| GraphPad Prism8 | Graphpad | https://www.graphpad.com |
| Other | ||
| Novocyte Flow Cytometer | ||
| BD FACScalibur Flow Cytometer | ||
Resource availability
Lead contact
David Fruman (dfruman@uci.edu).
Materials availability
This study generated a conditional knockout model of the Eif4e gene. This mouse strain has been cryopreserved at UC Irvine and is available subject to a Material Transfer Agreement.
Data and code availability
This article includes all analyzed data.
Experimental model and subject details
Mice and reagents
C57Bl/6J (B6) mice were bred at the University of California, Irvine, and used at between 6 and 12 weeks of age. Eif4e+/- mice with a heterozygous loss-of-function allele were obtained from the Ruggero lab (UC San Francisco) (Truitt et al., 2015). All animals were studied in compliance with a protocol approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. Eif4e-flox mice were obtained from the NorCOMM2 mouse repository (C57BL/6N-Eif4etm1c(EUCOMM)Hmgu/Tcp) and were crossed with Rosa26-Cre mice (Rosa26-CreERT2). Primer sets used to genotype Eif4e-flox mice and Cre mice are shown in Table S1. MLN0128 and rapamycin were purchased from LC Laboratories (Woburn, MA, USA), and 4-hydroxytamoxifen was purchased from MilliporeSigma (St. Louis, MO). Inhibitors were included throughout the indicated cell treatment periods.
Generation of p190 BCR-ABL cell lines
Bone marrow (BM) cells were flushed from the tibias and femurs of 3-5 week-old mice; an equal number of male and female donors were used. Cells were incubated overnight with retroviral supernatants (p190 BCR-ABL-IRES-hCD4 or p190 BCR-ABL-IRES-GFP) in the presence of 5 μg/ml polybrene at 37°C, 5% CO2, with RPMI culture medium (GE Healthcare Life Science HyClone Laboratories South Logan, Utah) supplemented with 20% FCS, recombinant mouse IL-7 (10 ng/ml; R&D Systems Minneapolis, MN) to promote cell cycle entry. Following overnight incubation, cells were spun down to remove virus and cultured with fresh RPMI+20% FCS medium supplemented with recombinant mouse (rm) IL-7 for 1 week. Cells were then cultured without IL-7 for a second week and then gradually reduced to 10% FCS until a pure 100% marker-positive (human CD4+ or GFP+), cytokine- and stromal- independent culture was established. In vitro mixing experiments were initiated after removal of rm-IL-7 and cells were mixed at a 1 to 1 ratio for CD4+ and GFP+ cells. Cells were cultured in RPMI+20% FCS medium during the outgrowth period. For colony assays, 4h after the infection 50000 cells were plated in M3630 methocult (Stem Cell Technologies Kent, WA) with mIL-7 (10 ng/ml).
Method details
In vivo transplant experiments with mouse p190 leukemia
Mouse p190 transformed BM cells were used to initiate leukemia in irradiated (400 cGy) syngeneic female B6 recipients. In all in vivo experiments, p190 transformed BM were prepared fresh (after the removal of rmIL-7 step) to initiate leukemia. Leukemic engraftment was determined by retro-orbital bleeds and analyzed by flow cytometry using anti-human CD4 (Biolegend, San Diego, CA)
Mouse B cell culture
Equal numbers of male and female B6 mice were used for isolation of B cells. Splenic B cells were purified using The EasySep™ Mouse B Cell Isolation Kit (Stemcell Technologies Kent, WA). B-cell purity measured by FACS analysis (FACSCalibur and CellQuest software; BD Biosciences) using anti-B220 antibody (eBioscience, ThermoFisher Scientific, Waltham, MA) yielded >95% B220+. Purified B cells were seeded at a final concentration of 1.5 x 106 cells per milliliter. For class switching experiments, B cells were stimulated 5 μg/mL anti-CD40 (eBioscience) agonistic antibody, or 5 μg/mL LPS (MilliporeSigma), together with 5 ng/mL mIL-4 (R&D Systems) for 96 h to induce switching to IgG1. All cells were cultured with RPMI 1640 supplemented with 10% heat-inactivated FCS, 5 mM Hepes, 2 mM L-glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin, 50 μM 2-mercaptoethanol. To evaluate B-cell survival ex vivo, purified B cells from mice of different genotypes were treated with 4OHT in vitro in the presence of BAFF (100ng/ml, R&D Systems). For proliferation assays, B cells were stimulated with anti-IgM (10μg/ml, Jackson ImmunoResearch Laboratories, West Grove, PA) and IL4 (10ng/ml, R&D Systems). For pre-B cell culture, bone marrow cells were cultured for 5 days in the absence or presence of recombinant murine IL-7 (10ng/ml).
Western blotting analysis
Total cell lysates were prepared using RIPA buffer. Protein concentration was measured with Bradford assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein (20 μg) were heat-denatured in sample buffer (Bio-Rad Laboratories), resolved by SDS-PAGE using Novex 4–20% Tris-Glycine MiniProtein Gels (Thermo Fisher Scientific, Waltham, MA), and transferred to nitrocellulose membranes (Bio-Rad Laboratories). The filters were blocked in 5% BSA for 1h at room temperature and then incubated overnight at 4° with specific antibodies for β-actin (1:1000), total ERK (1:1000), BCR-ABL (1:1000), eIF4E (1:1000), total 4E-BP1 (1:1000), and total 4E-BP2 (1:1000) purchased from Cell Signaling Technologies (Danvers, MA). Goat anti-mouse or anti rabbit-peroxidase conjugated IgG (1:5000, Promega Madison, WI) was used as secondary antibody. Protein on western blots were analyzed using ImageJ64 software to quantitate signal of each band. Signal was normalized with actin or total ERK measurements and fold change was calculated using the stimulated/no drug control.
Flow cytometry, CFSE labeling, and antibodies
Cells were incubated with TruStain fcX in FACS buffer (0.5% BSA +0.02% NaN3 in 1× HBSS) for 10 min on ice. Staining with antibodies was performed with FACS buffer and on ice for 20 min. Flow cytometry antibodies and other reagents used: B220 (eBioscience), CD43 (eBioscience), IgG1 (BD Bioscience San Jose, CA) and 7-Aminoactinomycin D (Tonbo Bioscience San Diego, CA). CFSE (ThermoFisher Scientific) and Cell Trace Far Red labeling (ThermoFisher Scientific) of B cells was performed by resuspending cells to a concentration of 10 × 106 cells per mL with a concentration of 2.5 μM. Flow cytometric data were analyzed using FlowJo software (10.7.1).
Luciferase assay
Cap-dependent translation was measured using the luciferase reporter construct pRSTF-CVB3 containing the 5′ non-coding region of the Coxsackie B3 virus cloned between the firefly and renilla luciferase (Jang et al., 2004). The construct was electroporated in serum-free media and cells recovered in complete RPMI media for 2 hr followed by inhibitor treatment for 8 hr. Following treatment, cells were lysed and renilla and firefly luciferase expression were measured using the Dual luciferase assay kit (Promega) using a luminometer. Renilla luciferase expression was normalized to Firefly luciferase expression and results were expressed relative to vehicle treated.
Puromycin incorporation
10 μg/mL cycloheximide (CHX) (MilliporeSigma) was added to the control well 30 min before adding the puromycin (GIBCO, ThermoFisher Scientific). After 72hr of treatment with 4OHT, puromycin 10μg/mL was added to each well for 15 min. Cells were harvested, spun down and washed in PBS before fixation with Foxp3-staining kit fix/perm buffer (Biolegend) for 20 min RT. Cells were spun down 800g for 5min and washed twice with of Permeabilization buffer (Biolegend). Incubation with FITC-conjugated anti-puromycin Ab (1:100) (EMD Millipore Burlington, MA) for 45min RT in permeabilization buffer. After 2 washes, pellet was resuspended in PBS and analyzed using a Novocyte flow cytometer.
Quantification and statistical analysis
All samples represent biological replicates. Unless otherwise specified in figure legends, all center values shown in graphs refer to the mean. For statistical significance of the differences between the means of two groups, we used two-tailed Student's t-tests. Statistical significance of differences among multiple groups (≥3) was calculated by performing ANOVA multiple comparisons of the means for each group. No samples or animals were excluded from analysis, and power analyses to determine sample size were not used. Studies were not conducted blinded.
Additional resources
Please see the key resources table.
Acknowledgments
We thank Dr. Bert Semler for the pRSTF-CVB3 luciferase reporter plasmid. DAF was supported by National Institutes of Health grant R21-AI099656, American Society of Hematology Bridge grant ASH-5557789, and UC Irvine Chao Family Comprehensive Cancer Center pilot grant from the 2018 Anti-Cancer Challenge. The authors wish to acknowledge the support of the Chao Family Comprehensive Cancer Center Transgenic Mouse Facility Shared Resource, supported by the National Cancer Institute of the National Institutes of Health under award number P30CA062203 (PI: Van Etten). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. HC and LH were supported by National Institutes of Health grant T32 AI 060573. CSC was funded by the American Cancer Society (PF-14-212-01-RMC). DR was supported by NIH grants R35CA242986 and by the American Cancer Society RP-19-181-01-RMC (American Cancer Society Research Professor Award).
Author contributions
H.C., R.B., L.V.J., L.H., and S.M. designed experiments, performed research, analyzed data, and wrote the manuscript. C.S.C. and D.R. provided materials, designed experiments, analyzed data, and wrote the manuscript. D.F. designed experiments, supervised research staff, analyzed data, and wrote the manuscript.
Declaration of interests
D.R. is a shareholder of eFFECTOR Therapeutics, Inc. and a member of its scientific advisory board. The other authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102748.
Supplemental information
References
- Araki K., Morita M., Bederman A.G., Konieczny B.T., Kissick H.T., Sonenberg N., Ahmed R. Translation is actively regulated during the differentiation of CD8. Nat. Immunol. 2017;18:1046–1057. doi: 10.1038/ni.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjur E., Larsson O., Yurchenko E., Zheng L., Gandin V., Topisirovic I., Li S., Wagner C.R., Sonenberg N., Piccirillo C.A. Distinct translational control in CD4+ T cell subsets. PLoS Genet. 2013;9:e1003494. doi: 10.1371/journal.pgen.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemart L., Malka-Mahieu H., Girault I., Allard D., Hemmingsson O., Tomasic G., Thomas M., Basmadjian C., Ribeiro N., Thuaud F. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513:105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- Chiu H., Jackson L.V., Oh K.I., Mai A., Ronai Z.A., Ruggero D., Fruman D.A. The mTORC1/4E-BP/eIF4E Axis promotes antibody class switching in B lymphocytes. J. Immunol. 2019;202:579–590. doi: 10.4049/jimmunol.1800602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Galicia-Vázquez G., Cencic R., Mills J.R., Katigbak A., Porco J.A., Pelletier J. CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep. 2016;15:2340–2347. doi: 10.1016/j.celrep.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demosthenous C., Han J.J., Stenson M.J., Maurer M.J., Wellik L.E., Link B., Hege K., Dogan A., Sotomayor E., Witzig T., Gupta M. Translation initiation complex eIF4F is a therapeutic target for dual mTOR kinase inhibitors in non-Hodgkin lymphoma. Oncotarget. 2015;6:9488–9501. doi: 10.18632/oncotarget.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J.T., Thompson P.A., Nilewski C., Sprengeler P.A., Sperry S., Packard G., Michels T., Xiang A., Tran C., Wegerski C.J. Design of development candidate eFT226, a first in class inhibitor of eukaryotic initiation factor 4A RNA helicase. J. Med. Chem. 2020;63:5879–5955. doi: 10.1021/acs.jmedchem.0c00182. [DOI] [PubMed] [Google Scholar]
- Feng Y., Pinkerton A.B., Hulea L., Zhang T., Davies M.A., Grotegut S., Cheli Y., Yin H., Lau E., Kim H. Vol. 75. Cancer Res; 2015. SBI-0640756 attenuates the growth of clinically unresponsive melanomas by disrupting the eIF4F translation initiation complex; pp. 5211–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A.C., Costa M., Zollo O., Davis C., Feldman M.E., Testa J.R., Meyuhas O., Shokat K.M., Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T.N., Ramírez J.A., Tsang M., Park H., Margineantu D.H., Hockenbery D.M., Iritani B.M. Conditional disruption of raptor reveals an essential role for mTORC1 in B cell development, survival, and metabolism. J. Immunol. 2016;197:2250–2260. doi: 10.4049/jimmunol.1600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes M.R., Limon J.J., So L., Chen J., Lim R.J., Chavez M.A., Vu C., Lilly M.B., Mallya S., Ong S.T. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat. Med. 2010;16:205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G.M., Leong L.E., Hoang L.T., Wang P.H., Gutman G.A., Semler B.L. Structurally distinct elements mediate internal ribosome entry within the 5'-noncoding region of a voltage-gated potassium channel mRNA. J. Biol. Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- Kharas M.G., Janes M.R., Scarfone V.M., Lilly M.B., Knight Z.A., Shokat K.M., Fruman D.A. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J. Clin. Invest. 2008;118:3038–3050. doi: 10.1172/JCI33337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka-Mahieu H., Newman M., Désaubry L., Robert C., Vagner S. Molecular pathways: the eIF4F translation initiation complex-new opportunities for cancer treatment. Clin. Cancer Res. 2017;23:21–25. doi: 10.1158/1078-0432.CCR-14-2362. [DOI] [PubMed] [Google Scholar]
- McAllister E.J., Apgar J.R., Leung C.R., Rickert R.C., Jellusova J. New methods to analyze B cell immune responses to thymus-dependent antigen sheep red blood cells. J. Immunol. 2017;199:2998–3003. doi: 10.4049/jimmunol.1700454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke N.J., Aktas H., Chen H., Cantel S., Reibarkh M.Y., Fahmy A., Gross J.D., Degterev A., Yuan J., Chorev M. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Nemes K., Sebestyén A., Márk A., Hajdu M., Kenessey I., Sticz T., Nagy E., Barna G., Váradi Z., Kovács G. Mammalian target of rapamycin (mTOR) activity dependent phospho-protein expression in childhood acute lymphoblastic leukemia (ALL) PLoS One. 2013;8:e59335. doi: 10.1371/journal.pone.0059335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oridate N., Kim H.J., Xu X., Lotan R. Growth inhibition of head and neck squamous carcinoma cells by small interfering RNAs targeting eIF4E or cyclin D1 alone or combined with cisplatin. Cancer Biol. Ther. 2005;4:318–323. doi: 10.4161/cbt.4.3.1504. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Graff J., Ruggero D., Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourdehnad M., Truitt M.L., Siddiqi I.N., Ducker G.S., Shokat K.M., Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. U S A. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S., Saadat D., Zhang M., Halbur L., Fruehauf J.P., Ong S.T. A novel mechanism for Bcr-Abl action: bcr-Abl-mediated induction of the eIF4F translation initiation complex and mRNA translation. Oncogene. 2007;26:1188–1200. doi: 10.1038/sj.onc.1209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Schwarzer A., Holtmann H., Brugman M., Meyer J., Schauerte C., Zuber J., Steinemann D., Schlegelberger B., Li Z., Baum C. Hyperactivation of mTORC1 and mTORC2 by multiple oncogenic events causes addiction to eIF4E-dependent mRNA translation in T-cell leukemia. Oncogene. 2015;34:3593–3604. doi: 10.1038/onc.2014.290. [DOI] [PubMed] [Google Scholar]
- So L., Lee J., Palafox M., Mallya S., Woxland C.G., Arguello M., Truitt M.L., Sonenberg N., Ruggero D., Fruman D.A. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci. Signal. 2016;9:ra57. doi: 10.1126/scisignal.aad8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A., Akcakanat A., Singh G., Luyimbazi D., Zheng Y., Kim D., Gonzalez-Angulo A., Meric-Bernstam F. eIF4E knockdown decreases breast cancer cell growth without activating Akt signaling. Mol. Cancer Ther. 2008;7:1782–1788. doi: 10.1158/1535-7163.MCT-07-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt M.L., Conn C.S., Shi Z., Pang X., Tokuyasu T., Coady A.M., Seo Y., Barna M., Ruggero D. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A., Suyama E., Adachi H., Svitkin Y.V., Aza-Blanc P., Imataka H., Mikami S., Martineau Y., Ronai Z.A., Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article includes all analyzed data.