Abstract
Background
Traditional prognostic risk assessment in patients with coronary artery disease undergoing percutaneous coronary intervention (PCI) is based on a limited selection of clinical and imaging findings. Machine learning (ML) can consider a higher number and complexity of variables and may be useful for characterising cardiovascular risk, predicting outcomes, and identifying biomarkers in large population studies.
Methods
We prospectively enrolled 9,680 consecutive patients with coronary artery disease who underwent PCI at our institution between January 2013 and December 2013. Clinical features were selected and used to train 6 different ML models (support vector machine, decision tree, random forest, gradient boosting decision tree, neural network, and logistic regression) to predict cardiovascular outcomes, 10-fold cross-validation to evaluate the performance of models.
Results
During the 5-year follow-up, 467 (4.82%) patients died. Eighty-seven risk baseline measurements were used to train ML models. Compared with the other models, the random forest model (RF-PCI) exhibited the best performance on predicting all-cause mortality (area under the receiver operating characteristic curve: 0.71±0.04). Calibration plots demonstrated a slight overprediction for patients using the RF-PCI model (Hosmer-Lemeshow test: P>0.05). The top 15 features related to PCI candidates’ long-term prognosis, among which 11 were laboratory measures.
Conclusions
ML models improved the prediction of long-term all-cause mortality in patients with coronary artery disease before PCI. The performance of the RF model was better than that of the other models, providing a meaningful stratification.
Keywords: Machine learning (ML), coronary heart disease, percutaneous coronary intervention (PCI), personalised medicine, predictive model
Introduction
In the past few decades, percutaneous coronary intervention (PCI) has been a revolutionary innovation in the treatment of coronary artery disease. Risk stratification for prognosis is essential for the individualised management of patients undergoing PCI (1). However, an evaluation system for the long-term prognosis of PCI patients may need to integrate powerful multi-faceted factors. Traditional prognostic risk assessment has limited power to develop risk stratification under these conditions.
The Global Registry of Acute Coronary Events (GRACE) and the Thrombolysis in Myocardial Infarction (TIMI) scores commonly used in clinical practice mainly evaluate the short-term prognosis of patients with ST-segment elevation myocardial infarction (STEMI) (2,3). For non-elective coronary artery disease patients who need PCI, a more accurate scoring system is needed to assess long-term outcomes. It is necessary to integrate multiple clinical indicators to make the scoring system easier to apply in daily clinical practice.
Machine learning (ML) is a field of computer science that applies a variety of complex rules and multidimensional recognition pattern technologies to process large datasets (4). In the past few years, these types of algorithms and technologies have been effectively used in the clinical field for various applications, including diagnostic radiology, cardiac electrophysiology, dermatology, and accurate determination of phenotypes, which are effective in the diagnosis and prognosis of patients (5-8). To date, the benefit of utilising ML in predicting hard-prognostic endpoints has not been evaluated on a large scale.
Given its practicality, accessibility, and the impressive results achieved so far, we expect the application of ML in the healthcare sector to increase in the next few years. Despite the large number of cardiac patients requiring PCI, there is no accurate score based on ML applied to this target population.
The purpose of this study was to (I) develop and validate different ML-based models to predict five-year all-cause mortality in patients with coronary heart disease prior to PCI and (II) identify the important predictors for clinical outcomes in a large population.
We present the study in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/cdt-21-37).
Methods
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Fuwai Hospital Research Ethics Committee (No. 2013-449). The institutional review board approved the study protocol, and all patients provided written informed consent.
Study design
In this prospective study, we enrolled 10,724 consecutive patients with coronary artery disease who underwent PCI between January 2013 and December 2013 at Fuwai Hospital (Beijing, China). Individuals with prior coronary artery bypass grafting were excluded from the study.
Data collection
Based on previous models of patients with PCI, 87 risk factors were identified, as shown in Table 1. We mined the following groups of variables from the electronic health records: demographic and physical characteristics, comorbid conditions, medication, laboratory biomarkers, and electrophysiological and echocardiographic results. Hypertension was characterised by a history of systolic blood pressure of at least 140 mmHg. Diabetes mellitus was defined by a diagnosis made previously by a physician and/or the use of insulin or oral hypoglycaemic agents. Smoking history was defined as current smoking or cessation of smoking within the last 3 months. A family history of premature coronary artery disease was defined as myocardial infarction in a first-degree relative, 55 years (male) or 65 years (female). Dyslipidemia was defined as known but untreated dyslipidemia or current treatment with lipid-lowering medications. Laboratory data were based on fasting venous blood samples collected on the day before PCI. Echocardiographic parameters were collected by certified sonographers using two-dimensional echocardiography (Vivid 7 Dimension/Pro System, GE Healthcare, USA) to measure left ventricular end-diastolic diameter and left ventricular ejection fraction (LVEF) according to the modified Simpson’s rule on the day before device implantation.
Table 1. Features used in final model development.
| Domain | Individual features |
|---|---|
| Demographics | Gender, age, height, body mass index, systolic blood pressure, diastolic blood pressure, smoking, alcohol, family history of premature coronary artery disease |
| Comorbid conditions | Heart failure, hypertension, duration of hypertension, diabetes, duration of diabetes, atrial fibrillation, stroke, pulmonary artery hypertension, structure heart disease, hyperuricemia, hyperlipidemia, peripheral vascular diseases, cerebrovascular, previous myocardial infarction, previous PCI |
| Electrophysiology | Heart rate, PR duration, QRS duration, QT duration, P-axis, QRS axis, middle of ST segment amplitudes, end of ST segment amplitudes, left bundle branch block, ventricular tachycardia |
| Coronary angiography | Number of lesions, location of lesion, origin of coronary artery, distribution of coronary artery |
| PCI procedure | Stent placement, number of DES, number of BMS, preoperative Syntax score, postoperative Syntax score, intervention time, operative complications |
| Medications | ACE inhibitor or angiotensin-receptor blocker, beta-blocker, calcium channel blocker, spironolactone, digoxin, diuretics, statins, aspirin, clopidogrel, low molecular weight heparin, IIb/IIIa receptor antagonist |
| Laboratory test results | Hemoglobin, erythrocyte sedimentation rate, platelet count, platelet distribution width, white blood cell count, total bilirubin, direct bilirubin, albumin, alanine aminotransferase, aspartate aminotransferase, total triglycerides, total cholesterol, low density lipoprotein cholesterol, high density lipoprotein cholesterol, B-type natriuretic peptide, high-sensitivity C-reactive protein, hemoglobin A1c, serum sodium concentration, uric acid, glomerular filtration rate, big endothelin-1, serum creatinine, creatine phosphokinase-isoenzyme-MB, creatine phosphokinase-isoenzyme-BB, cardiac troponin I, cardiac troponin T, urine microalbumin |
| Echocardiogram | Left ventricular ejection fraction, left ventricular end diastolic diameter, left atrial diameter, right ventricular diameter |
PCI, percutaneous coronary intervention; DES, drug eluting stent; BMS, bare metal stent; ACE, angiotensin converting enzyme.
Follow-up and endpoints
All patients participated in follow-up outpatient clinic visits or telephone interviews. Primary endpoint events were defined as all-cause mortality.
Machine learning
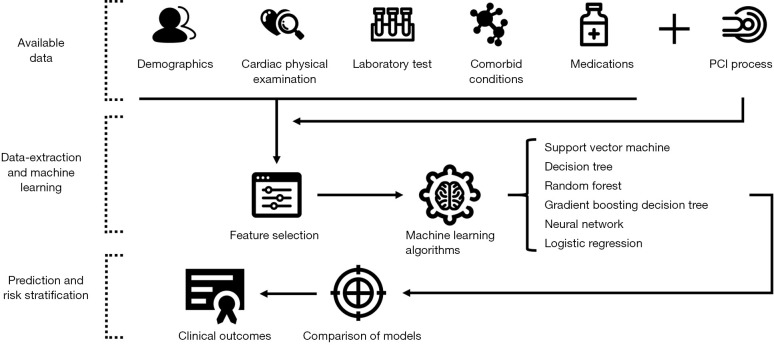
Feature selection: Information gain attribute ranking was used to select appropriate features (6,9). Information gain is a measure of the effectiveness of an attribute in classifying the training data. It is defined as the amount at which the entropy of a class decreases, which reflects additional information about the class provided by the feature (Figure 1).
Figure 1.
Workflow for machine learning. Information such as clinical examination and treatment of percutaneous coronary intervention (PCI) patients was collected, and different machine-learning methods were evaluated after feature selection to establish the best clinical outcomes prediction model.
ML algorithms: We compared the performance of six ML models (support vector machine, decision tree, random forest, gradient boosting decision tree, neural network, and logistic regression) in predicting all-cause death in coronary heart disease patients who underwent PCI (10,11).
Cross-validation: we used 10-fold cross-validation to evaluate the performance of all six models. The entire dataset was divided into ten mutually exclusive subsets, nine of which were used for training and one for evaluation. This process was repeated 10 times using 10 different but overlapping training sets and 10 unique testing sets. Model performance was assessed according to the area under the receiver operating characteristic curve (AUC). The calibration of scores was assessed using the Hosmer-Lemeshow test. Models with similar expected and observed event rates were defined as well calibrated (Hosmer-Lemeshow test: P>0.05) (12,13). Discrimination was also assessed graphically by plotting the cumulative incidence of death over 5 years for patients classified into ten risk groups, as published (14,15).
Statistical analysis
All analyses were performed using SPSS Statistics version 23 (IBM, Armonk, NY, USA) and Python 3.7.3 (Python Software Foundation, Python Language Reference, version 2.7; available at http://www.python.org). Data are presented as mean and standard deviation for continuous variables and as counts and percentages for categorical variables. For non-normally distributed data, the results are presented as medians (interquartile range). Two-sided P values <0.05 were considered to be significantly different.
Results
Study population and baseline characteristics
In total, 9,680 patients with coronary heart disease who underwent PCI were included. During a median follow-up period of 5.5 years, 467 (4.82%) patients died. Baseline characteristics of this cohort are shown in Table 2. The mean age of the cohort was 58.5±10.3 years; 76.92% (7,446/9,680) were male, 64.61% (6,254/9,680) had a history of hypertension, and 30.33% (2,936/9,680) had diabetes.
Table 2. Baseline characteristics for our cohort.
| Variable | Overall |
|---|---|
| Age, (years) | 58.5±10.3 |
| Male, n (%) | 7,446 (76.92) |
| Body mass index (kg/m2) | 24.53±4.46 |
| Systolic blood pressure (mmHg) | 127.1±17 |
| Diastolic blood pressure (mmHg) | 77.5±10.8 |
| Hypertension, n (%) | 6,254 (64.61) |
| Hyperlipidemia, n (%) | 6,502 (67.17) |
| Diabetes, n (%) | 2,936 (30.33) |
| Total cholesterol (mg/dL) | 4.2±1.1 |
| HDL-C (mg/dL) | 1±0.3 |
| BNP (pg/mL) | 756.9±584.4 |
| Serum creatinine (μmol/L) | 935±3,534.8 |
| Leukocyte (×109/L) | 6.8±1.7 |
| ESR (mm/h) | 10.9±11.6 |
| Big ET-1 (fmol/mL) | 0.3±0.2 |
| Hemoglobin (g/dL) | 143.1±15.4 |
| HbA1c (%) | 6.6±1.2 |
| hsCRP (pg/mL) | 3.1±3.7 |
| LVEF (%) | 62.8±7.3 |
| Smoking, n (%) | 5,531 (57.14) |
| Aspirin, n (%) | 9,556 (98.72) |
| Clopidogrel, n (%) | 9,529 (98.44) |
| Statin, n (%) | 9,287 (95.94) |
| All-cause death, n (%) | 467 (4.82) |
HDL-C, high-density lipoprotein cholesterol; BNP, B-type natriuretic peptide; ESR, erythrocyte sedimentation rate; Big ET-1, big endothelin-1; HbA1c, hemoglobin A1c; hsCRP, high sensitivity C-reactive protein; LVEF, left ventricular ejection fraction.
Summary measures of discrimination and calibration of six ML models for all-cause mortality in patients undergoing PCI
Figure 2 shows the performance of the six ML models for predicting the primary endpoint. When algorithms were trained with the basic input variables (without advanced laboratory biomarkers), the decision tree model showed the worst discrimination ability (AUC: 0.51±0.07), whereas the random forest model showed the best discrimination ability (AUC: 0.71±0.04). In addition, the result of the Hosmer-Lemeshow goodness-of-fit test suggested that the random forest model was well calibrated (P>0.05). A calibration plot confirmed close agreement between the random forest model (RF-PCI) scores and the observed 5-year risk of all-cause death (Figure 3).
Figure 2.
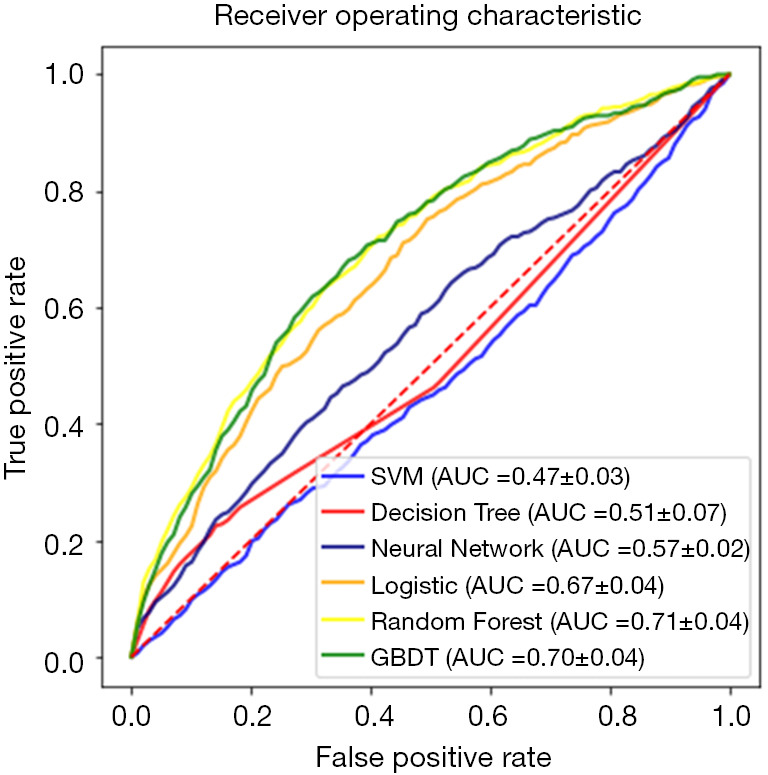
Receiver operating characteristic curves of the models. Six machine-learning algorithms [support vector machine (SVM), decision tree, random forest, gradient boosting decision tree (GBDT), neural network, and logistic regression] were evaluated by area under the curve (AUC) for all-cause death.
Figure 3.
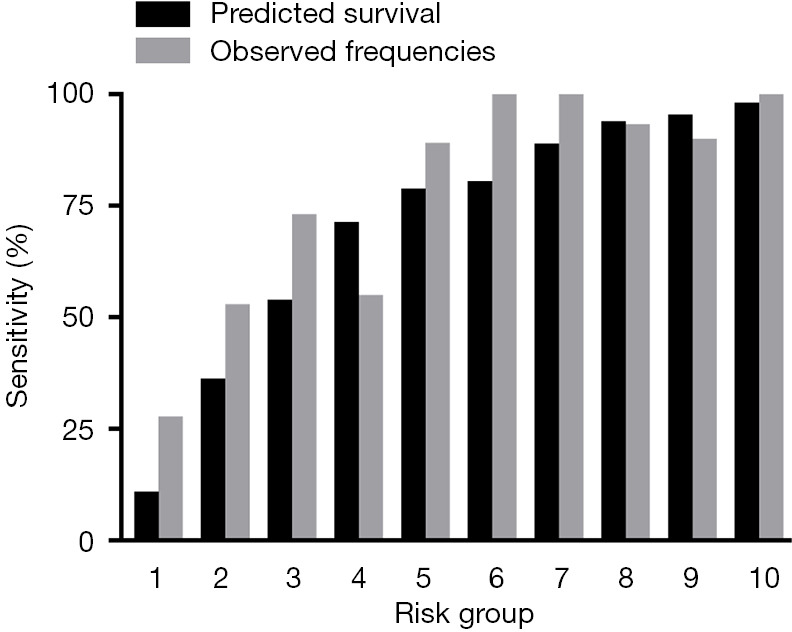
Observed frequencies vs. model-predicted 5-year survival in 10 risk groups. Risk groups 1–10 represent risk scores 0.7–26.4, 26.4–49.0, 49.0–64.5, 64.5–72.5, 72.5–79.7, 79.7–86.1, 86.1–90.9, 90.9–93.8, 93.8–95.9 and 95.9–99.6, respectively.
Feature selection
According to the information gain ranking criteria for this study, the 15 top-ranked features in RF-PCI are shown in Figure 4. As expected, 11 of the 15 were laboratory factors with top 15 features for 5-year all-cause mortality; diastolic blood pressure, age, and LVEF were important features in the RF-PCI model.
Figure 4.
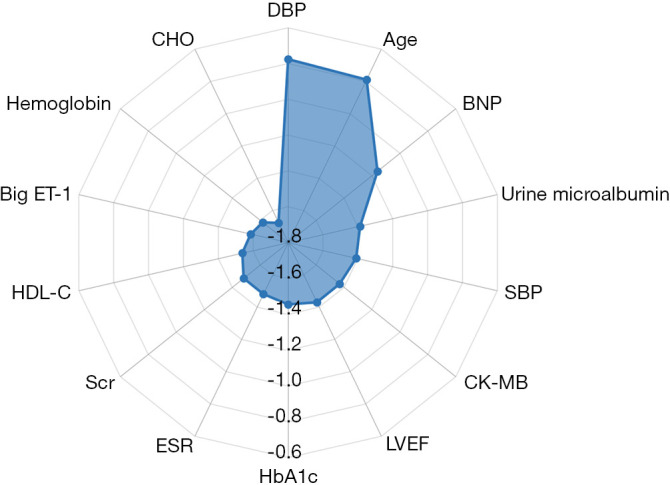
Feature selection. Information gain ranking was used to evaluate the worth of each variable by measuring the entropy gain with respect to the outcome. The importance of each feature was quantified by calculating the decrease in the model’s performance after permuting its values. The higher its value, the more important the feature is. As the feature importance values were spread over a wide range (multiple orders of magnitude), base-10 logarithmic transformation was performed to facilitate plotting. The top 15 variables in the random forest model were shown. DBP, diastolic blood pressure; BNP, B-type natriuretic peptide; SBP, systolic blood pressure; CK-MB, creatine phosphokinase-isoenzyme-MB; LVEF, left ventricular ejection fraction; HbA1c, haemoglobin A1c; ESR, erythrocyte sedimentation rate; Scr, serum creatinine; HDL-C, high-density lipoprotein cholesterol; Big ET-1, big endothelin-1; CHO, cholesterol.
Discussion
In the present study, among the evaluated ML classifiers, the RF model demonstrated the best performance. With an average AUC of over 0.71, the RF-PCI score can be used to predict the long-term prognosis of PCI patients. We also found the top 15 features that were related to PCI candidates’ long-term prognosis, among which 11 were laboratory measures.
Prognosis assessment is the key to coronary heart disease diagnosis and treatment. It can assist clinicians in choosing the best intervention methods according to the different risk stratification. In recent years, the predictive scores of coronary heart disease have been widely used for risk assessment before revascularisation (16). The GRACE score, one of the most commonly used scores, has been used to predict the short-term clinical prognosis of patients with acute coronary syndrome (2,17). The higher the score, the greater the risk of death of the patient, and the greater the need for earliest possible revascularisation to reduce the occurrence of adverse endpoints. Furthermore, it is crucial to conduct a risk assessment for patients after PCI treatment as it will help provide more targeted control and management of cardiovascular disease and other accompanying risk factors, thereby improving the long-term prognosis of patients.
Independent factors have limited predictive value for disease prognosis. Currently, traditional cardiovascular risk assessment models predict cardiovascular event endpoints based on traditional statistical models such as linear models. For diseases with multiple complex risk factors, traditional statistical models have limited power. For patients with coronary heart disease who are planning to receive PCI treatment, the prognostic factors are more complex (4,7,18). The current cardiovascular prediction tools cannot correctly perform effective risk stratification and guide effective intervention in the next step. It is urgent to develop a personalised risk stratification tool for patients with coronary heart disease undergoing PCI.
ML is a subset of artificial intelligence, which refers to the ability of a machine to learn independently and make accurate predictions. In recent years, artificial intelligence technology has developed rapidly and has begun to penetrate the fields of medicine and cardiovascular medicine. It has been used with large amount of medicine data in diagnosis, classification, and prediction (19-21). ML, as an effective means of information processing, may innovate prediction methods for cardiovascular diseases and improve the prediction accuracy, thus benificial to the diagnosis and treatment of clinical cardiovascular diseases (19).
Our study was based on patients with coronary heart disease who underwent PCI, without referring to specific subgroups of the type of coronary heart disease or the timing of PCI. Therefore, the results shows that ML-based predictive scores provide good practical prediction of the long-term prognostic risk stratification of patients after PCI and are easy to apply in clinical practice. This feature of ML methods is particularly important, given the ease with which the machine can seamlessly incorporate new data to continually update and optimise its algorithm and thus continually improve its predictive performance over time. Diastolic blood pressure, age, and brain natriuretic peptide are the top three risk factors for 5-year mortality in patients undergoing PCI. High-risk patients who were screened before PCI should be managed more strictly after PCI.
In addition to these clinical findings, our method has some methodological implications. First, it demonstrates the feasibility of a data mining approach for predicting the long-term prognosis of patients with PCI. Second, we found that non-parametric algorithms such as RF maintain high predictive performance, even in multidimensional scenarios with dozens of variables; this finding is consistent with previous studies using ML to predict clinical endpoints (22-24). With the emergence of electronic medical records, the completeness of disease-related information of PCI patients has been accumulated to an unprecedented degree, and the practical application of ML, a new type of disease prognosis prediction method, is highly feasible.
In the current study, to ensure the generalizability of our model, we used 10-fold cross-validation to evaluate the feasibility and performance of ML-based models in patients undergoing PCI. However, prospective practical implementation in the external population is scarce. As a prospective single-centre study, our ML-based models should be validated in external centres to confirm their generalizability. Another limitation is the lack of impact analyses to determine how the utilisation of the models improves patient care and outcomes. Further research is needed for targeted interventions for high-risk patients identified by this score. Although we evaluated 87 distinct variables with ML, along with variable-variable interactions, we did not consider the totality of features that may offer improved risk prediction due to the limitation of data collection. Because of the popularisation of electronic medical records, the evaluation model can be embedded in the electronic medical record system, and the risk prediction score can be continuously optimised to further improve the prediction ability.
Conclusions
In this study, we developed and validated an ML-based model to predict the clinical outcomes of coronary heart patients undergoing PCI. The performance of the RF model is better than that of the other models, providing a meaningful stratification of long-term outcomes in coronary patients following PCI. This study provides useful information for a predictive model for PCI recipients.
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (2017YFC1307800).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Fuwai Hospital Research Ethics Committee (No. 2013-449). The institutional review board approved the study protocol, and all patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/cdt-21-37
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-21-37
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (Available at http://dx.doi.org/10.21037/cdt-21-37). The authors have no conflicts of interest to declare.
References
- 1.Granger CB, Goldberg RJ, Dabbous O, et al. Predictors of hospital mortality in the global registry of acute coronary events. Arch Intern Med 2003;163:2345-53. 10.1001/archinte.163.19.2345 [DOI] [PubMed] [Google Scholar]
- 2.Fox KA, Dabbous OH, Goldberg RJ, et al. Prediction of risk of death and myocardial infarction in the six months after presentation with acute coronary syndrome: prospective multinational observational study (GRACE). BMJ 2006;333:1091. 10.1136/bmj.38985.646481.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antman EM, Cohen M, Bernink PJ, et al. The TIMI risk score for unstable angina/non-ST elevation MI: A method for prognostication and therapeutic decision making. JAMA 2000;284:835-42. 10.1001/jama.284.7.835 [DOI] [PubMed] [Google Scholar]
- 4.Al'Aref SJ, Anchouche K, Singh G, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975-86. 10.1093/eurheartj/ehy404 [DOI] [PubMed] [Google Scholar]
- 5.Ghorbani A, Ouyang D, Abid A, et al. Deep learning interpretation of echocardiograms. NPJ Digit Med 2020;3:10. 10.1038/s41746-019-0216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokodi M, Schwertner WR, Kovács A, et al. Machine learning-based mortality prediction of patients undergoing cardiac resynchronization therapy: the SEMMELWEIS-CRT score. Eur Heart J 2020;41:1747-56. 10.1093/eurheartj/ehz902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J 2017;38:500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litjens G, Ciompi F, Wolterink JM, et al. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc Imaging 2019;12:1549-65. 10.1016/j.jcmg.2019.06.009 [DOI] [PubMed] [Google Scholar]
- 9.Mandal I, Sairam N. Accurate prediction of coronary artery disease using reliable diagnosis system. J Med Syst 2012;36:3353-73. 10.1007/s10916-012-9828-0 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Zhao H. Variable importance-weighted Random Forests. Quant Biol 2017;5:338-51. 10.1007/s40484-017-0121-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deo RC. Machine Learning in Medicine. Circulation 2015;132:1920-30. 10.1161/CIRCULATIONAHA.115.001593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nashef SA, Roques F, Michel P, et al. European system for cardiac operative risk evaluation (EuroSCORE). Eur J Cardiothorac Surg 1999;16:9-13. 10.1016/S1010-7940(99)00134-7 [DOI] [PubMed] [Google Scholar]
- 13.Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem 2008;54:17-23. 10.1373/clinchem.2007.096529 [DOI] [PubMed] [Google Scholar]
- 14.Sartipy U, Dahlström U, Edner M, et al. Predicting survival in heart failure: validation of the MAGGIC heart failure risk score in 51,043 patients from the Swedish heart failure registry. Eur J Heart Fail 2014;16:173-9. 10.1111/ejhf.32 [DOI] [PubMed] [Google Scholar]
- 15.Rahman MS, Ambler G, Choodari-Oskooei B, et al. Review and evaluation of performance measures for survival prediction models in external validation settings. BMC Med Res Methodol 2017;17:60. 10.1186/s12874-017-0336-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houston M. The role of noninvasive cardiovascular testing, applied clinical nutrition and nutritional supplements in the prevention and treatment of coronary heart disease. Ther Adv Cardiovasc Dis 2018;12:85-108. 10.1177/1753944717743920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roffi M, Patrono C, Collet JP, et al. 2015 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: Task Force for the Management of Acute Coronary Syndromes in Patients Presenting without Persistent ST-Segment Elevation of the European Society of Cardiology (ESC). Eur Heart J 2016;37:267-315. 10.1093/eurheartj/ehv320 [DOI] [PubMed] [Google Scholar]
- 18.Poss AM, Maschek JA, Cox JE, et al. Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J Clin Invest 2020;130:1363-76. 10.1172/JCI131838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial Intelligence in Cardiology. J Am Coll Cardiol 2018;71:2668-79. 10.1016/j.jacc.2018.03.521 [DOI] [PubMed] [Google Scholar]
- 20.Johnson KW, Shameer K, Glicksberg BS, et al. Enabling Precision Cardiology Through Multiscale Biology and Systems Medicine. JACC Basic Transl Sci 2017;2:311-27. 10.1016/j.jacbts.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antman EM, Loscalzo J. Precision medicine in cardiology. Nat Rev Cardiol 2016;13:591-602. 10.1038/nrcardio.2016.101 [DOI] [PubMed] [Google Scholar]
- 22.Churpek MM, Yuen TC, Winslow C, et al. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit Care Med 2016;44:368-74. 10.1097/CCM.0000000000001571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handelman GS, Kok HK, Chandra RV, et al. eDoctor: machine learning and the future of medicine. J Intern Med 2018;284:603-19. 10.1111/joim.12822 [DOI] [PubMed] [Google Scholar]
- 24.Sengupta PP, Kulkarni H, Narula J. Prediction of Abnormal Myocardial Relaxation From Signal Processed Surface ECG. J Am Coll Cardiol 2018;71:1650-60. 10.1016/j.jacc.2018.02.024 [DOI] [PubMed] [Google Scholar]