Abstract
Filifactor alocis, a fastidious Gram-positive obligate anaerobic bacterium, is a newly appreciated member of the periodontal community that is now proposed to be a diagnostic indicator of periodontal disease. Its pathogenic characteristics are highlighted by its ability to survive in the oxidative stress–rich environment of the periodontal pocket and to significantly alter the microbial community dynamics by forming biofilms and interacting with several oral bacteria. Here, we describe the current understanding of F. alocis virulence attributes, such as its comparative resistance to oxidative stress, production of unique proteases and collagenases that can cause structural damage to host cells, and dysregulation of the immune system, which enable this bacterium to colonize, survive, and outcompete other traditional pathogens in the inflammatory environment of the periodontal pocket. Furthermore, we explore the recent advancements and future directions for F. alocis research, including the potential mechanisms for oxidative stress resistance and our evolving understanding of the interactions and mechanisms of bacterial survival inside neutrophils. We also discuss the current genetic tools and challenges involved in manipulating the F. alocis genome for the functional characterization of the putative virulence genes. Collectively, this information will expedite F. alocis research and should lead to the identification of prime targets for the development of novel therapeutics to aid in the control and prevention of periodontal disease.
Keywords: oxidative stress, bacterial virulence, biofilm, cytokine, host pathogen interactions, periodontal disease/periodontitis
Introduction
The ecologic environment of the human oral cavity, which is home to >700 microbial species, is one of the most complex and dynamic microbial communities in the human body (Deng et al. 2017). While less than half of these microbial species have been characterized and can be readily cultivated, several oral infectious diseases, including caries and periodontitis, have known etiologies by specific members of the microbial community (Dewhirst et al. 2010). Periodontitis, a multifaceted chronic inflammatory disease that affects the supporting structures of the teeth, is attributed to dysbiosis of the oral microbiota (Deng et al. 2017; Lamont et al. 2018). It is the sixth-most common infectious disease in the world and affects >65 million people in the United States (Eke et al. 2018). The severe form of periodontitis, which affects about 8.5% of adults in the United States, may not only cause tooth loss but can also affect systemic health by increasing patients’ susceptibility to cardiovascular disease, diabetes, rheumatoid arthritis, Alzheimer’s disease, adverse pregnancy outcomes, aspiration pneumonia, and cancer (Hajishengallis 2015; Dominy et al. 2019).
A triadic group of oral anaerobic bacteria designated the “red complex,” comprising Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia, has traditionally been categorized as the causative agents of periodontitis (Socransky et al. 1998). A paradigm shift based on the current model of polymicrobial synergy and dysbiosis suggests that the progression of periodontitis is induced by a more comprehensive dysbiotic microbial community rather than by select periopathogens (Hajishengallis and Lamont 2012). The development of novel culture-independent techniques, such as 16S pyrosequencing and next-generation sequencing, has identified several previously underappreciated bacteria—including the Gram-positives Filifactor alocis and Peptostreptococcus stomatis and other species from the genera Prevotella, Megasphaera, Selenomonas, and Desulfobulbus. These are as-yet-unculturable species but have nonetheless shed new light on pathogenic bacterial communities in the periodontal pocket (Dewhirst et al. 2010; Griffen et al. 2012; Abusleme et al. 2013). F. alocis is a fastidious asaccharolytic anaerobic rod with typical virulence features of a periodontopathogen, such as its unique ability to tolerate oxidative stress and cause substantial proinflammatory responses (Aruni et al. 2011; Moffatt et al. 2011). Consistent with Socransky’s modified Koch’s postulate, F. alocis shows a strong correlation with periodontal disease and is undetected in the healthy population (Kumar et al. 2006; Schulz et al. 2019). In this review, the current state of F. alocis pathogenesis is discussed, highlighting the recent advances on oxidative stress resistance and host-microbe interactions as well as the challenges for further characterization. For a detailed historical perspective, see Aruni, Chioma, and Fletcher (2014) and Aruni et al. (2015).
F. alocis: Prevalence and Characteristics
F. alocis was first identified in 1985 as Fusobacterium alocis from the gingival sulci of patients with periodontitis (Cato et al. 1985) and later reclassified as Filifactor alocis based on sequencing of 16S rRNA genes (Jalava and Eerola 1999). The natural habitat of F. alocis appears to be gingival sulcus (Gomes et al. 2006). Multiple studies have shown a high prevalence of F. alocis in patients with chronic and generalized aggressive periodontitis, apical periodontitis, peri-implantitis, and endodontic infections (reviewed by Aruni et al. 2015). Due to the initial observation of increased occurrence of F. alocis in diseased subgingival sites with its description as an etiologic agent of marginal periodontitis (Cato et al. 1985; Paster et al. 2001), it is now suggested to be used as a diagnostic indicator of periodontal disease (Wade 2011; Na et al. 2020).
F. alocis has been also linked to gingivitis and diabetes during pregnancy (Gogeneni et al. 2015) and oral squamous cell carcinoma (Yang et al. 2018). The ability of F. alocis to spread to extraoral sites, including the spleen, lungs, and kidneys, has been demonstrated in the mouse subcutaneous chamber model (Wang et al. 2014). Consistent with this study, 2 recent reports identified F. alocis at extraoral sites in association with a disease other than periodontitis. The first case report identified F. alocis in the pleural fluid of a patient with thoracic empyema, whereas in the second report, the bacterium was found in the fluid bronchoalveolar lavage of patients with lung cancer (Gray and Vidwans 2019; Wang et al. 2019). Additionally, F. alocis has been identified in brain abscesses (Hishiya et al. 2020) and shown to aid progression of periodontitis in patients with rheumatoid arthritis (Ayala Herrera et al. 2019). In these studies, the presence of F. alocis was identified via 16S rRNA sequencing. Future studies identifying the live organism from these extraoral sites are necessary to confirm any role of F. alocis in the etiology of these diseases. Unless otherwise stated, the studies discussed are associated with the type strain F. alocis ATCC 35896. Specific growth characteristics of F. alocis are discussed elsewhere (Aruni et al. 2011).
F. alocis: Virulence Attributes
To colonize and survive in the stress environment of the periodontal pocket, F. alocis shares virulence properties consistent with a typical periodontopathogen, as described below.
Interaction with Other Oral Organisms/Biofilms
A clinical study focusing on identifying oral pathogens in disease sites showed a F. alocis–centered co-occurrence group of 8 potential pathogens associated with periodontitis across different oral habitats: P. gingivalis, Porphyromonas endodontalis, T. forsythia, F. alocis, Eubacterium nodatum, Fretibacterium species, Lachnospiraceae species, and Peptostreptococcaceae species. This suggests possible synergistic interactions among those pathogens and a high diagnostic value for periodontitis (Chen et al. 2015).
In vitro studies have shown that F. alocis ATCC 35896 and D-62D interact with a variety of oral bacteria and participate in community development (Wang et al. 2013). Growth of F. alocis was reduced during interaction with Streptococcus gordonii. Interactions between Fusobacterium nucleatum and F. alocis were synergistic. Aggregatibacter actinomycetemcomitans stimulated the growth of F. alocis ATCC 35896 but had no effect on D-62D. P. gingivalis and F. alocis formed heterotypic communities; however, while the abundance of P. gingivalis was enhanced in the presence of F. alocis, accumulation of F. alocis was inhibited. The inhibitory effect of P. gingivalis on F. alocis was partially dependent on the minor fimbriae Mfa1. Sensing/communication between species required LuxS of P. gingivalis (autoinducer 2 dependent signaling) (Wang et al. 2013). In another clinical study, F. alocis formed in vivo biofilms in the apical and middle-third parts of the gingival pocket and colocalized with P. gingivalis, Prevotella intermedia, A. actinomycetemcomitans, T. denticola, T. forsythia, and F. nucleatum (Schlafer et al. 2010). Colocalization may suggest the involvement of Filifactor in coaggregation (a prerequisite step during the establishment and maturation of biofilms) and might indicate the necessary symbiotic relationship between F. alocis and other organisms. Consistent with this, in an in vitro study, P. gingivalis ATCC 33277 in coculture with F. alocis showed a significant increase in biofilm formation, which may indicate a commensal relationship between species (Aruni et al. 2011).
Invasion and Adhesion
A successful pathogen must adhere and invade epithelial cells. The ability of F. alocis ATCC 35896 and D-62D to adhere and invade epithelial cells was enhanced in coculture with P. gingivalis (Aruni et al. 2011). How P. gingivalis assists F. alocis invasion is unclear. While vesicle-mediated internalization was observed to occur, the specific components and their regulation that are vital for this process are unclear. Proteomic analysis of F. alocis during coinfection of epithelial cells with P. gingivalis revealed an increase in several membrane adhesion proteins and surface components recognizing adhesive matrix molecules, which may mediate adherence and colonization of host tissues during infection (Aruni, Zhang, et al. 2014).
Collagenases, Proteases, and Toxins
To cause infection, periodontopathogenic bacteria must penetrate the host’s structural barrier, such as the collagen-rich extracellular matrix. Collagen, the major component of the gingival connective tissue, is extremely resistant to degradation and can be cleaved only by collagenases. The F. alocis peptidase HMPREF0389_00504 (PrtFAC) has been demonstrated to bind and degrade type I collagen and gelatin in a calcium-dependent manner. Additionally, PrtFAC may induce caspase-dependent and caspase-independent apoptosis of normal oral keratinocytes (Chioma et al. 2017) and therefore may play a role in the pathogenesis of F. alocis by inducing host cell death; however, more studies are needed to outline a specific pathway and/or mechanism.
F. alocis can induce the release of neutrophil granule matrix content, including members of the matrix metalloproteinase (MMP) family, such as collagenase and gelatinase, which can contribute to periodontal tissue damage (Armstrong et al. 2016). An increase in the levels of MMP-1, a collagenase, has been demonstrated in gingival tissue and crevicular fluid from patients with periodontitis (Alfant et al. 2008; Popat et al. 2014). A recent study evaluated gingival biopsies from patients with periodontitis and healthy individuals for the presence ofF. alocis and MMP-1 (Nokhbehsaim, Nogueira, Damanaki, et al. 2020). F. alocis was highly prevalent in patients’ biopsies in contrast to the healthy gingiva. Similarly, higher levels of MMP-1 were observed in the inflamed gingiva as compared with healthy individuals. Moreover, F. alocis enhanced the production of MMP-1 by gingival fibroblastic and monocytic cells, suggesting that the bacterium may contribute to breakdown of the periodontal tissue matrix through MMP-1 (Nokhbehsaim, Nogueira, Damanaki, et al. 2020).
The genome of F. alocis ATCC 35896 also encodes several uncharacterized proteases (reviewed by Aruni et al. 2015), and elucidating their functions could be important in understanding the disease etiology, as they may be implicated in tissue destruction, the inactivation of key proteins in host defenses, or the processing of virulence factors of other bacteria.
Additionally, a recent report identified a novel RTX exotoxin FtxA in F. alocis ATCC 35896 and 9 clinical isolates (Oscarsson et al. 2020). At present, the particular role of FtxA in the pathogenicity of F. alocis is unknown and needs further characterization.
Oxidative Stress Resistance
Overcoming oxidative stress is one of the essential mechanisms to the survival of F. alocis in the periodontal community. Earlier studies showed that F. alocis is relatively more resistant to H2O2-induced oxidative stress as compared with P. gingivalis (Aruni et al. 2011). Additionally, under H2O2-induced stress conditions, the survival of P. gingivalis is increased >4-fold when grown in coculture with F. alocis (H.M. Fletcher, personal communication). These observations suggest that F. alocis may have an innate ability to detoxify the local inflammatory microenvironment of the periodontal pocket.
The characterization of the first antioxidant enzyme “superoxide reductase” (SOR) in F. alocis showed that it plays an important role in defense against superoxide radicals, air exposure, and H2O2-induced oxidative stress (Mishra et al. 2020). SORs are iron-containing enzymes that catalyze the reduction of superoxide radicals to H2O2. The SOR mutant was sensitive to air after 30 min of exposure and lost viability after 2 h. It also showed increased sensitivity to H2O2-induced stress. Furthermore, the mutant had reduced invasion capabilities to telomerase immortalized gingival keratinocyte cells and had a reduced ability to form in vitro biofilms. Taken together, these results have established a role of F. alocis SOR (HMPREF0389_00796) in oxidative stress resistance and the pathogenicity of F. alocis (Mishra et al. 2020).
The roles of other genes and/or mechanisms of oxidative stress resistance in F. alocis are unknown. In most anaerobic organisms, the H2O2 scavenging enzymes include catalases, peroxidases, bacterioferritin comigratory protein (Bcp), ruberythrin, and alkyl hydroperoxide reductase enzyme system (AhpC/AhpF) (Mishra and Imlay 2012). Although the genome of F. alocis ATCC 35896 lacks genes coding for catalases and ruberythrin (National Center for Biotechnology Information 2020), it does encode for a sole peroxidase: glutathione peroxidase (HMPREF0389_01233). Additionally, F. alocis encodes the alkyl hydroperoxide reductase subunit AhpC (HMPR EF0389_00768), though the partner AhpF homolog was not identified in the genome. However, a reductase annotated as thioredoxin-disulfide reductase (HMPREF0389_00608) exhibits 34% identity with P. gingivalis W83 AhpF (PG0619). Interestingly, F. alocis AhpC is 30% identical to P. gingivalis W83 Bcp (PG0880). Whether F. alocis utilizes these enzymes in oxidative stress resistance merits further study.
Another gene from F. alocis, HMPREF0389_00519 (FA519; encoding a hypothetical protein), was shown to be upregulated in coinfection of epithelial cells with F. alocis and P. gingivalis (Aruni, Zhang, et al. 2014). Our preliminary studies have shown that the ∆FA519 mutant is significantly more sensitive to H2O2 and nitric oxide–induced stress as compared with the wild type (data not provided). Yet another gene, HMPREF0389_01654 (FA1654), is currently being investigated in the laboratory for its ability to protect DNA from damage under H2O2-induced oxidative stress (data not provided). In summary, F. alocis has a plethora of yet-to-be-characterized proteins that could mediate its enhanced survival in the proinflammatory environment of the periodontal pocket (Fig. 1). Further studies of these proteins are needed to confirm their functional/mechanistic significance.
Figure 1.
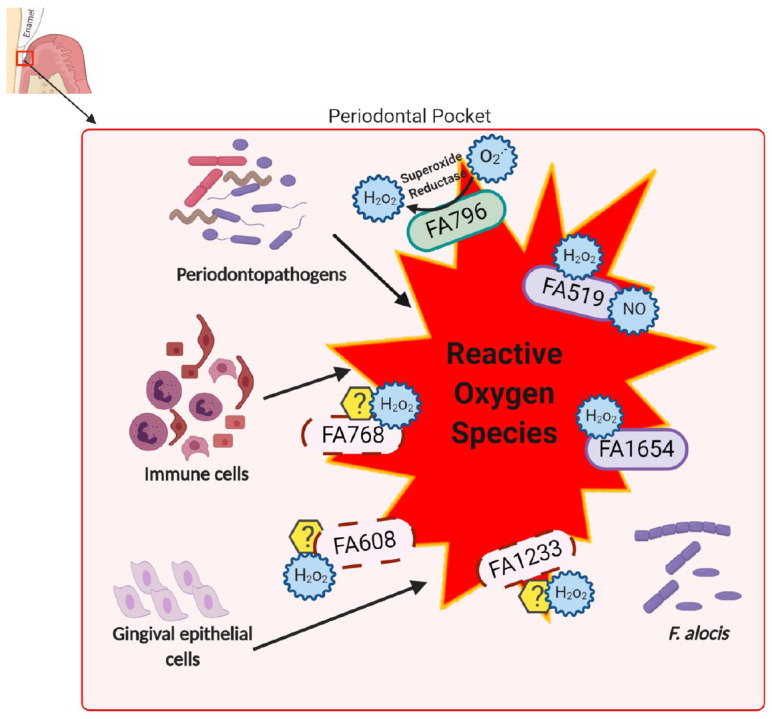
Proposed proteins involved in oxidative stress resistance mechanisms in Filifactor alocis. F. alocis is constantly exposed to oxidative stress, which may arise from other periodontopathogens and the host inflammatory response, with the concomitant release of reactive oxygen species such as superoxide radicals, hydrogen peroxide, and nitric oxide. The antioxidant enzyme superoxide reductase reduces superoxide radicals (O2 .−) to hydrogen peroxide (H2O2), which in turn could be detoxified by DPS protein FA1654 (DNA protection during starvation) and/or hypothetical protein FA519. Other proteins predicted to play a role in H2O2-induced stress resistant mechanisms are glutathione peroxidase (FA1233), thioredoxin-disulfide reductase (FA608), and alkyl hydroperoxide reductase subunit AhpC (FA768). FA519 may also be part of the nitric oxide detoxification system.
Host-Microbe Interactions
Host Cell Modulation
The epithelial cells in the gingival crevice are the first host cells encountered by periodontal bacteria (Moffatt et al. 2011). These gingival epithelial cells not only provide a mechanical barrier against invading microorganisms but also produce innate immunity effectors and act as sensors of infection to signal immune cells of the periodontal tissues. In gingival epithelial cells, F. alocis can induce the secretion of proinflammatory cytokines interleukin 1β (IL-1β), IL-6, and tumor necrosis factor α (TNF-α) (Moffatt et al. 2011). Extracellular vesicles purified from F. alocis can induce the expression of IL-1β, IL-1 receptor antagonist, IL-6, IL-8, and TNF-α in human monocytic cells and IL-6 and IL-8 in human oral keratinocyte cells (Kim et al. 2020). The induced cytokines IL-6 (Baker et al. 1999), IL-1β, and TNF-α (Graves and Cochran 2003) may upregulate pathways promoting osteoclast generation and increase alveolar bone loss leading to disease progression.
In the periodontal tissues, resident and infiltrating immune cells, including monocytes and fibroblasts, may also increase the level of another proinflammatory mediator, cyclooxygenase 2 (COX2), which is an inducible enzyme expressed only during the inflammatory process (Zidar et al. 2009). COX2 has been implicated in mediating periodontal tissue damage by converting arachidonic acid into prostaglandin E2, which in turn may alter connective tissue metabolism and increase osteoclastic bone resorption (Ricciotti and FitzGerald 2011). High levels of COX2 and prostaglandin E2 have been found in the saliva, gingiva, and gingival crevicular fluid of periodontal patients (Noguchi and Ishikawa 2007). Although there is no study describing the coidentification of F. alocis with prostaglandin E2, a recent report showed significantly upregulated COX2 expression by F. alocis in gingival fibroblastic and monocytic cells (Nokhbehsaim, Nogueira, Nietzsche, et al. 2020). These results may implicate F. alocis as a contributor to the elevated levels of COX2 in the gingival tissues of patients with periodontitis.
F. alocis Can Subvert Antimicrobial Mechanisms of Neutrophils
Neutrophils are the first innate immune cells to be recruited to the periodontal pocket to challenge the invading microbes and maintain homeostasis. They may utilize various strategies to detect, localize, and kill the invading pathogen, including induction of respiratory burst response, phagocytosis, degranulation, and formation of neutrophil extracellular traps (NETs) (Amulic et al. 2012). However, successful periodontal pathogens may use different mechanisms to manipulate neutrophil responses to prevent killing (Uriarte et al. 2016). The high occurrence of F. alocis in patients with periodontitis could imply that it disrupts the antimicrobial actions of neutrophils and effectively grows under inflammatory conditions (Miralda et al. 2019). F. alocis is likely to be in direct contact with neutrophils since it mostly inhabits areas of the biofilm that are in contact with soft tissue (Schlafer et al. 2010).
F. alocis is shown to modulate neutrophil effector functions, including chemotaxis and degranulation (Armstrong et al. 2016). A transcriptome analysis of the F. alocis–challenged neutrophils showed significant changes in the expression of genes involved in different neutrophil effector functions (Miralda et al. 2020). In its interaction with human neutrophils, F. alocis increased their random and directed migration toward IL-8 via recognition of TLR2, resulting in exocytosis of specific granules, gelatinase granules, and secretory vesicles.F. alocis–induced neutrophil migration and limited degranulation have contributed to dysregulated and sustained inflammation as well as tissue damage (Armstrong et al. 2016). Additionally, F. alocis can survive within human neutrophils by minimizing the respiratory burst response and preventing phagosome maturation. Collectively, this promotes the survival of the organism by reducing the release of lysozyme, lactoferrin, and other antimicrobial peptides inside the phagosome (Edmisson et al. 2018).
NETs are extracellular fibrous structures of decondensed nuclear chromatin from neutrophils, which can be formed in the gingival epithelium of the oral cavity with the ability of eliciting a first response to invading bacteria (Doke et al. 2017). F. alocis–challenged human neutrophils did not trigger NET formation independent of time of challenge, opsonization, and bacterial dose; however, it significantly reduced the ability of neutrophils to produce NETs in response to a known pharmacologic NET inducer, phorbol 12-myristate 13-acetate (Armstrong et al. 2018). This phenomenon could favor the survival of F. alocis itself as well as other members of the periodontal cavity by helping other bacteria go unnoticed, which would otherwise be killed by NETs (Armstrong et al. 2018). This would be important for a bacterium that cannot be engulfed by neutrophils; therefore, extracellular killing mechanisms would be beneficial to clear the infection.
Neutrophils have the ability to synthesize and store cytokines and chemokines in their granules, which enables a rapid release of these inflammatory mediators to infection sites (Scapini et al. 2005). Thus, to maintain periodontal health, a balance between neutrophil recruitment and activation in the periodontal pocket is important. F. alocis can likely limit the recruitment of neutrophils/monocytes and the amplification of the inflammatory reaction at the site of infection. A recent study showed that a challenge of human neutrophils with F. alocis induced mild release of proinflammatory cytokines IL-1β and TNF-α and chemokines CXCL1 and CXCL8 as compared with P. gingivalis and another emerging periodontal pathogen, P. stomatis. In contrast to F. alocis, P. stomatis can induce a more robust inflammatory response; however, their synergistic impact on neutrophil function still needs to be determined (Armstrong et al. 2018).
To summarize, F. alocis battles with host neutrophils in a very moderate way as compared with other pathogens such as P. stomatis. AlthoughF. alocis can enhance the chemotactic activity of neutrophils (when phagocytosed), the bacterium has evolved strategies to survive within neutrophils. Dysregulated and sustained inflammation as well as tissue damage may still take place through the limited exocytosis and mild production of cytokines and chemokines (Fig. 2).
Figure 2.
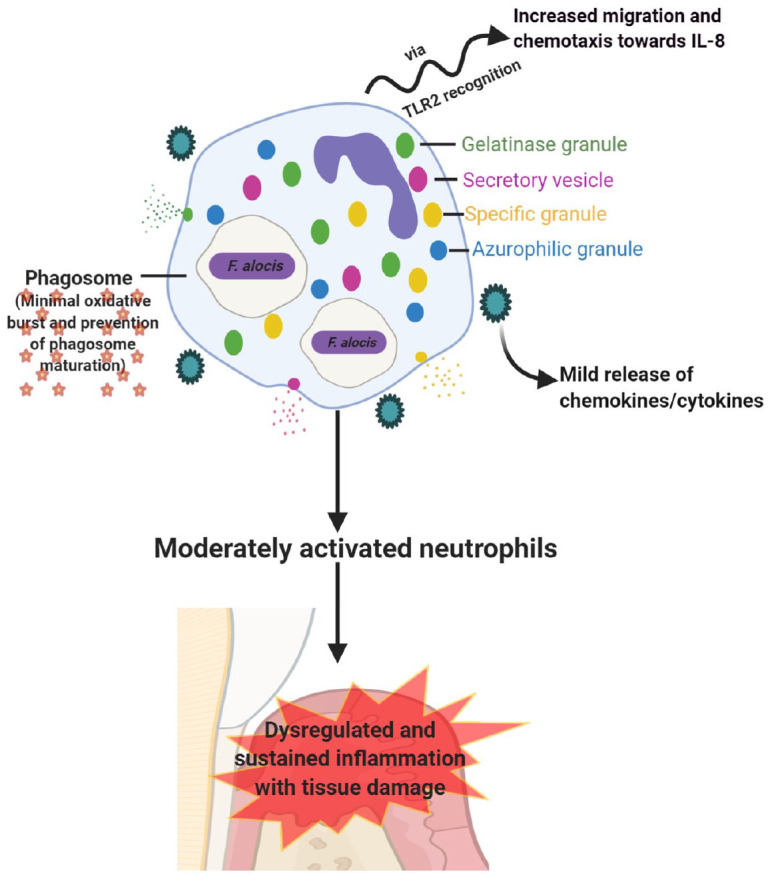
Interaction of Filifactor alocis with host neutrophils. Interaction between F. alocis and neutrophils is more of a moderate type. When phagocytosed, F. alocis enhances neutrophil migration and chemotaxis via TLR2 recognition. The bacterium also induces limited exocytosis of granules (specific and gelatinase) and secretory vesicles but survives within neutrophils by minimizing the oxidative burst and prevention of phagosome maturation. Due to the limited exocytosis, neutrophils are moderately activated, resulting in mild release of proinflammatory cytokines and chemokines, which causes sustained inflammation with tissue damage (image adapted from Miralda et al. 2019).
F. alocis Can Inhibit the Host Complement System
One of the hallmark features of periodontitis is the ability of periodontal bacteria to subvert the complement system, which leads to intensified inflammation and dysbiosis (Lamont et al. 2018). Complement proteins and their activation fragments are found in high concentrations in gingival cervical fluid; therefore, oral bacteria must subvert this system to perpetuate infection (Jusko et al. 2016).
Jusko et al. (2016) uncovered a novel F. alocis cytoplasmic protein, FACIN (F. alocis complement inhibitor), that once present on the bacterial surface can inhibit complement by binding to C3 and consequently suppressing the activity of all 3 complement pathways. FACIN has been classified as a rare complement inhibitor since very few molecules have been shown to act at the C3 level.
F. alocis: Challenges for Further Molecular Characterization
The functional characterization of putative virulence factors in F. alocis remains understudied due to the unavailability of an efficient genetic toolkit to allow genetic manipulations of its genome. The construction of the first isogenic gene deletion mutant (∆FA796) in F. alocis ATCC 35896 via a long polymerase chain reaction–based fusion method was demonstrated in the laboratory (Mishra et al. 2020). While the erythromycin resistance cassette (ermF) was used to replace the FA796 gene, this method is not as facile and efficient as in other oral bacteria such as P. gingivalis. It is likely that this may be due to low electroporation efficiency, degradation of foreign DNA, or low frequency of homologous recombination in F. alocis. The thick peptidoglycan cell wall of Gram-positive bacteria may restrict entry of exogenous DNA to the cell. To overcome this, pretreatment of bacterial cells with cell wall–weakening agents is recommended (Aune and Aachmann 2010). Conjugation could also be optimized and tested in Filifactor as a means to deliver plasmid/DNA (Wang and Jin 2014). F. alocis ATCC 35896 genome encodes 6 competence proteins, including the ComEC/Rec2 family competence protein. These proteins may play a role in the development of natural competence, which could be a major mechanism of DNA transfer in F. alocis. Moreover, the expression efficiency of the ermF cassette in F. alocis cannot be ruled out. When compared with P. gingivalis, F. alocis is more resistant to tetracycline (TetQ), which will make use of the tetQ cassette challenging. Given the limitations of available antibiotic resistance markers for use in F. alocis, a markerless system would be an advantage. A markerless gene deletion system could be used to create multiple gene inactivations in the same strain. Furthermore, for a markerless mutant, erythromycin (the only available marker for Filifactor) could be used for complementation studies.
In parallel to the development of more effective traditional tools, the implementation of modern methods should also be considered, including transposon mutagenesis coupled with high-throughput sequencing and a CRISPR-Cas9 system, which could be a breakthrough in understanding the prevalence of F. alocis in patients with periodontitis.
To verify that the phenotypic changes in any mutant are due to the loss of a particular gene, it would be desirable to complement the defective allele in the mutant in an attempt to restore the wild-type phenotype. Currently, there is no Escherichia coli/F. alocis shuttle vector available to do complementation in Filifactor. Preliminary assessment of several F. alocis strains in our laboratory has not identified any plasmids (unpublished data). Because Filifactor is distantly related to Clostridium (Aruni et al. 2011), plasmids belonging to the genus Clostridium, such as pJIR750 (chloramphenicol resistant) and pJIR751 (erythromycin resistant) (Bannam and Rood 1993), should be evaluated for use in Filifactor. Furthermore, the ermF cassette in the mutant could be replaced with a wild-type copy of the gene fused to a different antibiotic marker, which will facilitate counterselection.
Conclusions and Future Directions
F. alocis is emerging as a “novel” oral bacterium with distinct virulence attributes. The co-occurrence assembly of F. alocis with other important periodontopathic bacteria and its current putative status as a diagnostic indicator of periodontal disease are likely multifactorial and await full characterization.
The host response to F. alocis and its likely unique neutrophil survival strategy requires a comprehensive understanding of its molecular pathogenic mechanisms. The F. alocis–induced host damage is now beginning to emerge. “Omic” studies have begun to shed light on components vital for host-microbe and microbe-microbe interactions. While the majority of these components are identified as hypothetical or of unknown function, one of our challenges is their characterization, which should reveal their relative significance in the pathogenic process. Molecular genetics studies in F. alocis have been hampered due to the lack of an efficient genetic system. While there is limited success with the ermF cassette to replace target genes, this system is not as accomplished as in other periodontal bacteria. Moreover, knowing that ermF is the only currently available antibiotic resistance marker for use in F. alocis, developing a markerless genetic system would be promising. Based on the complex interactions of the microbial community and their likely effect on disease progression, it is vital to explore the functional significance of genes modulated in that environment.
What are some of the virulence factors of F. alocis that are induced by its interaction with other emerging pathogens?
How are they regulated under varying conditions in the periodontal pocket?
What are the vital F. alocis components that are critical for environmental stress adaptation/virulence of P. gingivalis and possibly other periodontal pathogens?
What are the molecular genetic requirements of F. alocis in the stabilization of the periodontal microbial community via its interaction with other periodontal pathogens, such as T. denticola, T. forsythia, andF. nucleatum, under environmental-induced stress?
The outcome of studies addressing these questions should advance our understanding of the multifactorial inflammatory disease. Furthermore, it should lead to the identification of novel targets that would facilitate the development of therapeutic interventions to aid in the control and prevention of periodontal disease.
Author Contributions
E. Aja, A. Mishra, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; M. Mangar, contributed to conception, design, and data acquisition, drafted the manuscript; H.M. Fletcher, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
Due to the limited number of references that could be included in the review, we apologize that several historical studies are not included, and readers are directed to other citations.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Public Health Service (grants R-56-DE13664, DE019730, DE022508, and DE022724) from the National Institute of Dental and Craniofacial Research to H.M.F.
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfant B, Shaddox LM, Tobler J, Magnusson I, Aukhil I, Walker C. 2008. Matrix metalloproteinase levels in children with aggressive periodontitis. J Periodontol. 79(5):819–826. [DOI] [PubMed] [Google Scholar]
- Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. 2012. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 30:459–489. [DOI] [PubMed] [Google Scholar]
- Armstrong CL, Klaes CK, Vashishta A, Lamont RJ, Uriarte SM. 2018. Filifactor alocis manipulates human neutrophils affecting their ability to release neutrophil extracellular traps induced by PMA. Innate Immun. 24(4):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CL, Miralda I, Neff AC, Tian S, Vashishta A, Perez L, Le J, Lamont RJ, Uriarte SM. 2016. Filifactor alocis promotes neutrophil degranulation and chemotactic activity. Infect Immun. 84(12):3423–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni AW, Mishra A, Dou Y, Chioma O, Hamilton BN, Fletcher HM. 2015. Filifactor alocis—a new emerging periodontal pathogen. Microbes Infect. 17(7):517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 79(10):3872–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni AW, Zhang K, Dou Y, Fletcher H. 2014. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect Immun. 82(8):3261–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni W, Chioma O, Fletcher HM. 2014. Filifactor alocis: the newly discovered kid on the block with special talents. J Dent Res. 93(8):725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune TE, Aachmann FL. 2010. Methodologies to increase the transformation efficiencies and the range of bacteria that can be transformed. Appl Microbiol Biotechnol. 85(5):1301–1313. [DOI] [PubMed] [Google Scholar]
- Ayala Herrera JL, Apreza Patron L, Martinez Martinez RE, Dominguez Perez RA, Abud Mendoza C, Hernandez Castro B. 2019. Filifactor alocis and Dialister pneumosintes in a mexican population affected by periodontitis and rheumatoid arthritis: an exploratory study. Microbiol Immunol. 63(9):392–395. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, Evans RT, Dufour L, Johnson E, Roopenian DC. 1999. Cd4(+) T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect Immun. 67(6):2804–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannam TL, Rood JI. 1993. Clostridium perfringens–Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid. 29(3):233–235. [DOI] [PubMed] [Google Scholar]
- Cato EP, Moore LVH, Moore WEC. 1985. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol. 35(4):475–477. [Google Scholar]
- Chen H, Liu Y, Zhang M, Wang G, Qi Z, Bridgewater L, Zhao L, Tang Z, Pang X. 2015. A Filifactor alocis–centered co-occurrence group associates with periodontitis across different oral habitats. Sci Rep. 5:9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioma O, Aruni AW, Milford TA, Fletcher HM. 2017. Filifactor alocis collagenase can modulate apoptosis of normal oral keratinocytes. Mol Oral Microbiol. 32(2):166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZL, Szafranski SP, Jarek M, Bhuju S, Wagner-Dobler I. 2017. Dysbiosis in chronic periodontitis: key microbial players and interactions with the human host. Sci Rep. 7(1):3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke M, Fukamachi H, Morisaki H, Arimoto T, Kataoka H, Kuwata H. 2017. Nucleases from Prevotella intermedia can degrade neutrophil extracellular traps. Mol Oral Microbiol. 32(4):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, et al. 2019. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 5(1):eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmisson JS, Tian S, Armstrong CL, Vashishta A, Klaes CK, Miralda I, Jimenez-Flores E, Le J, Wang Q, Lamont RJ, et al. 2018. Filifactor alocis modulates human neutrophil antimicrobial functional responses. Cell Microbiol. 20(6):e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Thornton-Evans GO, Wei L, Borgnakke WS, Dye BA, Genco RJ. 2018. Periodontitis in US adults: national health and nutrition examination survey 2009–2014. J Am Dent Assoc. 149(7):576–588, e576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogeneni H, Buduneli N, Ceyhan-Ozturk B, Gumus P, Akcali A, Zeller I, Renaud DE, Scott DA, Ozcaka O. 2015. Increased infection with key periodontal pathogens during gestational diabetes mellitus. J Clin Periodontol. 42(6):506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes BP, Jacinto RC, Pinheiro ET, Sousa EL, Zaia AA, Ferraz CC, Souza-Filho FJ. 2006. Molecular analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola associated with primary endodontic infections and failed endodontic treatment. J Endod. 32(10):937–940. [DOI] [PubMed] [Google Scholar]
- Graves DT, Cochran D. 2003. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 74(3):391–401. [DOI] [PubMed] [Google Scholar]
- Gray RM, Vidwans M. 2019. Mixed anaerobic thoracic empyema: the first report of Filifactor alocis causing extra-oral disease. New Microbes New Infect. 29:100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6(6):1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 27(6):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya N, Uno K, Amano M, Asada K, Masui K, Ishida Y, Suzuki Y, Hirai N, Nakano A, Nakano R, et al. 2020. Filifactor alocis brain abscess identified by 16S ribosomal RNA gene sequencing: a case report. J Infect Chemother. 26(2):305–307. [DOI] [PubMed] [Google Scholar]
- Jalava J, Eerola E. 1999. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol. 49(Pt 4):1375–1379. [DOI] [PubMed] [Google Scholar]
- Jusko M, Miedziak B, Ermert D, Magda M, King BC, Bielecka E, Riesbeck K, Eick S, Potempa J, Blom AM. 2016. Facin, a double-edged sword of the emerging periodontal pathogen Filifactor alocis: a metabolic enzyme moonlighting as a complement inhibitor. J Immunol. 197(8):3245–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Lim Y, An SJ, Choi BK. 2020. Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis. Mol Oral Microbiol. 35(1):1–9. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing.J Clin Microbiol. 44(10):3665–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, Hajishengallis G. 2018. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 16(12):745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralda I, Vashishta A, Rogers MN, Rouchka EC, Li X, Waigel S, Lamont RJ, Uriarte SM. 2020. Whole transcriptome analysis reveals that Filifactor alocis modulates TNFα-stimulated MAPK activation in human neutrophils. Front Immunol. 11:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralda I, Vashishta A, Uriarte SM. 2019. Neutrophil interaction with emerging oral pathogens: a novel view of the disease paradigm. Adv Exp Med Biol. 1197:165–178. [DOI] [PubMed] [Google Scholar]
- Mishra A, Aja E, Fletcher HM. 2020. Role of superoxide reductase FA796 in oxidative stress resistance in Filifactor alocis. Sci Rep. 10(1):9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Imlay J. 2012. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 525(2):145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. 2011. Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol. 26(6):365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na HS, Kim SY, Han H, Kim HJ, Lee JY, Lee JH, Chung J. 2020. Identification of potential oral microbial biomarkers for the diagnosis of periodontitis.J Clin Med. 9(5):1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. 2020. Filifactor alocis ATCC 35896, complete sequence. Bethesda (MD): National Center for Biotechnology Information, U.S. National Library of Medicine. https://www.ncbi.nlm.nih.gov/nuccore/NC_016630.1 [Google Scholar]
- Noguchi K, Ishikawa I. 2007. The roles of cyclooxygenase-2 and prostaglandin E2 in periodontal disease. Periodontol 2000. 43:85–101. [DOI] [PubMed] [Google Scholar]
- Nokhbehsaim M, Nogueira AVB, Damanaki A, Dalagiorgou G, Eick S, Adamopoulos C, Piperi C, Basdra EK, Papavassiliou AG, Deschner J. 2020. Regulation of matrix metalloproteinase-1 by Filifactor alocis in human gingival and monocytic cells. Clin Oral Investig. 24(6):1987-1995. [DOI] [PubMed] [Google Scholar]
- Nokhbehsaim M, Nogueira AVB, Nietzsche S, Eick S, Deschner J. 2020. Regulation of cyclooxygenase 2 by Filifactor alocis in fibroblastic and monocytic cells. Mediators Inflamm. 2020:4185273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscarsson J, Claesson R, Bao K, Brundin M, Belibasakis GN. 2020. Phylogenetic analysis of Filifactor alocis strains isolated from several oral infections identified a novel RTX toxin, FtxA. Toxins (Basel). 12(11):687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol. 183(12):3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat PR, Bhavsar NV, Popat PR. 2014. Gingival crevicular fluid levels of matrix metalloproteinase-1 (MMP-1) and tissue inhibitor of metalloproteinase-1 (TIMP-1) in periodontal health and disease. Singapore Dent J. 35:59–64. [DOI] [PubMed] [Google Scholar]
- Ricciotti E, FitzGerald GA. 2011. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 31(5):986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapini P, Carletto A, Nardelli B, Calzetti F, Roschke V, Merigo F, Tamassia N, Pieropan S, Biasi D, Sbarbati A, et al. 2005. Proinflammatory mediators elicit secretion of the intracellular β-lymphocyte stimulator pool (BLyS) that is stored in activated neutrophils: implications for inflammatory diseases. Blood. 105(2):830–837. [DOI] [PubMed] [Google Scholar]
- Schlafer S, Riep B, Griffen AL, Petrich A, Hubner J, Berning M, Friedmann A, Gobel UB, Moter A. 2010. Filifactor alocis . BMC Microbiol. 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, Porsch M, Grosse I, Hoffmann K, Schaller HG, Reichert S. 2019. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch Oral Biol. 99:169–176. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Uriarte SM, Edmisson JS, Jimenez-Flores E. 2016. Human neutrophils and oral microbiota: a constant tug-of-war between a harmonious and a discordant coexistence. Immunol Rev. 273(1):282–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG. 2011. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol. 38 Suppl 11:7–16. [DOI] [PubMed] [Google Scholar]
- Wang K, Huang Y, Zhang Z, Liao J, Ding Y, Fang X, Liu L, Luo J, Kong J. 2019. A preliminary study of microbiota diversity in saliva and bronchoalveolar lavage fluid from patients with primary bronchogenic carcinoma. Med Sci Monit. 25:2819–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jotwani R, Le J, Krauss JL, Potempa J, Coventry SC, Uriarte SM, Lamont RJ. 2014. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect Immun. 82(3):1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. 2013. Oral community interactions of Filifactor alocis in vitro. PLoS One. 8(10):e76271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XK, Jin JL. 2014. Crucial factor for increasing the conjugation frequency in Streptomyces netropsis SD-07 and other strains. FEMS Microbiol Lett. 357(1):99–103. [DOI] [PubMed] [Google Scholar]
- Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, Huang PJ, Hu SN, Liao CT, Chang KP, et al. 2018. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. 9:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidar N, Odar K, Glavac D, Jerse M, Zupanc T, Stajer D. 2009. Cyclooxygenase in normal human tissues—is COX-1 really a constitutive isoform, and COX-2 an inducible isoform? J Cell Mol Med. 13(9B):3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]


