Abstract
Purpose:
To generate dynamic, volumetric maps of hyperpolarized [1-13C]pyruvate and its metabolic products in vivo.
Methods:
Maps of chemical species were generated with iterative least squares (IDEAL) reconstruction from multiecho echo-planar imaging (EPI) of phantoms of thermally polarized 13C-labeled chemicals and mice injected with hyperpolarized [1-13C]pyruvate on a preclinical 3T scanner. The quality of the IDEAL decomposition of single-shot and multishot phantom images was evaluated using quantitative results from a simple pulse-and-acquire sequence as the gold standard. Time course and area-under-the-curve plots were created to analyze the distribution of metabolites in vivo.
Results:
Improved separation of chemical species by IDEAL, evaluated by the amount of residual signal measured for chemicals not present in the phantoms, was observed as the number of EPI shots was increased from one to four. Dynamic three-dimensional metabolite maps of [1-13C]pyruvate, [1-13C]pyruvatehydrate, [1-13C]lactate, [1-13C]bicarbonate, and [1-13C]alanine generated by IDEAL from interleaved multishot multiecho EPI of live mice were used to construct time course and area-under-the-curve graphs for the heart, kidneys, and liver, which showed good agreement with previously published results.
Conclusions:
IDEAL decomposition of multishot multiecho 13C EPI images is a simple, yet robust method for generating high-quality dynamic volumetric maps of hyperpolarized [1-13C]pyruvate and its products in vivo and has potential applications for the assessment of multiorgan metabolic phenomena.
Keywords: hyperpolarized 13C, IDEAL, metabolic imaging, multiecho echo-planar imaging
1 |. INTRODUCTION
Dynamic nuclear polarization (DNP) provides an increase of up to four orders of magnitude in signal-to-noise ratio for hyperpolarized (HP) MRI studies, giving an unprecedented opportunity for studying metabolism at high spatial and temporal resolution in vivo.1 This technique has been especially useful for studying [1-13C]pyruvate, a key intermediate in several metabolic pathways, which is converted in the cell to [1-13C]lactate by lactate dehydrogenase, to [1-13C]alanine by alanine aminotransferase, and to [1-13C]bicarbonate upon entry into the tricarboxylic cycle.2 Given the success of HP pyruvate imaging, multiple groups have pursued clinical translation with the hope of leveraging metabolic imaging to aid in disease characterization.3–8 Preclinically, HP MRI has been applied mostly to study the conversion of [1-13C] pyruvate to [1-13C]lactate in tumor xenograft models for both diagnosis and assessing treatment response,9–13 but also to the study of cardiac,14 kidney,9,15 and liver16,17 metabolism. Most of these studies typically required imaging a limited part of the anatomy, such as the tumor region or a single organ. However, for applications such as the study of cancer metastasis, drug delivery, and clearance, treatment bystander effects, and kinetic modeling, there is a need to develop imaging approaches to robustly cover multiple organs.
HP 13C metabolic imaging has constraints that make it challenging to acquire dynamic maps covering an extended field of view (FOV). The most generally used image acquisition approach, spectroscopic imaging, acquires data that can be categorized as spatially localized spectra (high spectral resolution, low spatial resolution) or images corresponding to a single metabolite (low spectral resolution, high spatial resolution). In chemical-shift imaging (CSI), spatial localization of spectra is achieved using phase-encoding for both in-plane dimensions and phase-encoding or slice selection to acquire slices. It is robust to motion artifacts and gradient errors because of the lack of a readout gradient and to off-resonance errors because of the acquisition of an entire spectrum.18 However, CSI requires radiofrequency (RF) excitations equal to the number of image voxels, making it an inefficient use of HP magnetization. To more efficiently use the available magnetization, sequences such as echo-planar spectroscopic imaging or spiral CSI sequence19,20 use readout gradients to encode k-space with fewer excitations. Although this results in a more efficient use of the available magnetization, these spectroscopic imaging sequences must make a compromise between spectral and spatial resolution.
Alternatively, imaging sequences can be used to acquire higher spatial resolution at a limited number of chemical shifts, corresponding to the metabolites of interest. An echo-planar imaging (EPI) readout gradient can be used to acquire the entirety of k-space in a single shot using a broadband or spectral-spatial (SPSP) RF excitation.14,21 When broadband excitation is used, data acquired at different echo times can be decomposed into metabolite maps using a model of the chemical spectra similar to the IDEAL formalism originally described for water–fat separation.22,23 SPSP excitation can be used to achieve even more efficient use of HP magnetization by exciting different metabolites with different flip angles (FAs). The injected substrate, which is available in a higher concentration, can be detected with a lower FA, preserving magnetization for acquisition of more dynamic time points. Despite its efficiency, there are a few disadvantages to this approach. Single-shot EPI may suffer from severe distortion and chemical-shift artifacts that require the acquisition of reference data, such as a field inhomogeneity map, for correction.24 The long duration of SPSP pulses results in signal loss from transverse relaxation, and their frequency selectivity requires accurate frequency calibration and may place limitations on slice thickness.
To efficiently use the HP magnetization, we propose acquiring images at multiple echo times following a broadband RF excitation, as in a fast-gradient-echo imaging sequence,25 using an interleaved multishot multiecho EPI sequence. This method has the advantage of enabling the use of higher FAs and/or the acquisition of more time points than is feasible with conventional spectroscopic imaging method, whereas compared with single-shot EPI, the shorter duration of each interleave of multishot EPI provides a shorter minimum ΔTE, which potentially improves the robustness of decomposition of the multiecho EPI images into metabolite maps by enabling a more optimal sampling rate. Additionally, the overall scan time may be shorter because the multishot EPI acquisition does not require additional reference scans for artifact correction.24 This robustness and efficiency makes interleaved multishot, multiecho EPI ideal for acquiring multiple slices over an extended FOV, to be reformatted into a time-resolved volumetric image of HP metabolites, which can facilitate the study of diseases such as cancer metastases or the measurement of an arterial input function from a blood vessel for the calculation of kinetic parameters in a distant organ.
2 |. METHODS
2.1 |. Data model
In a dynamic MRI experiment, the signal, S(t), in a voxel at time t caused by N chemical species each with relative intensity, ρn, and chemical shift frequency, Δfn, can be modeled by
| (1) |
where is the effective transverse relaxivity constant and φ is a phase shift caused by static field inhomogeneity.26 The pervoxel and φ maps can be estimated using a method such as direct phase estimation,27,28 and after applying these maps as corrections to the acquired images, Equation (1) can be written in the matrix form as
| (2) |
where x is the unknown vector of the intensities of the species (ρn), b is a vector of field-map demodulated, R2*-corrected MR images (Sj) at echo time tj, A is a matrix with the accrued phase for each species , and Δfn is the chemical-shift offset frequency.22,23 The least squares solution to this system is
| (3) |
where A† is the Moore-Penrose pseudo-inverse. Several approaches have been developed for obtaining this solution.29 For this study, we use the hierarchical IDEAL multiresolution framework,28 in which Equation (2) is solved for overlapping subdivisions of the initial image. The solutions for the subdivisions are interpolated and used to generate the final spatial map.
2.2 |. Optimal image acquisition parameters
The number and spacing (ΔTE) of gradient echoes used for image acquisition can affect the separation of chemical species. These variables determine the condition number of the matrix A in Equation 2, which quantifies the stability of its inversion in Equation (3).23 The condition number can be shown to decrease with an increasing number of echoes and decreasing ΔTE (assuming sufficient phase evolution).30 The optimal ΔTE for resolving two chemical species is the inverse of twice the chemical-shift frequency difference, that is, when their spins are in opposed phase.26 Therefore, a relatively long ΔTE will suffice for resolving two species that are close in chemical shift (for example, ΔTE = 5.2 ms is optimal for separating vitamin C (177 ppm) from its oxidized form dehydroascorbate (174 ppm) at 3T), whereas a shorter ΔTE is desired for spins separated by a larger chemical-shift difference (ΔTE = 1.3 ms is optimal for separating lactate [183 ppm] and pyruvate [171 ppm]).2 The ΔTE for the former case may be realized for the spatial resolutions typically used in HP imaging with a single-shot gradient-echo EPI sequence, but the latter will likely require the use of a multishot sequence on most scanners. Other factors to consider when selecting optimum imaging parameters are the of the chemical species, the desired image quality, and the required number of time points. As an example, our initial condition number simulation for five chemical species, pyruvate, acetate, pyruvate-hydrate, alanine, and urea in matrix A in Equation 2, showed insignificant improvement in the condition number beyond 10 echoes (Figure 1C). After initial experiments on our scanner showed it was possible to acquire multiecho EPI images of enriched carbon phantoms with signal in 10 echo images, a number of signal averages simulation using 10 echoes determined ΔTEs of 1.34, 2.49, and 3.46 ms for optimal performance of IDEAL (Figure 1D). These ΔTEs could be realized on our scanner using four-, two- and one-shot EPI protocols, respectively. Because our application required high-quality images, but few time points, we chose to use four-shot EPI, which has the benefit of reducing ghosting, chemical shift, and distortion artifacts by virtue of having a larger phase-encoding bandwidth, although this comes at the potential cost of increased sensitivity to motion artifacts31–34 and less efficient use of HP magnetization compared to two- or one-shot EPI.
FIGURE 1.
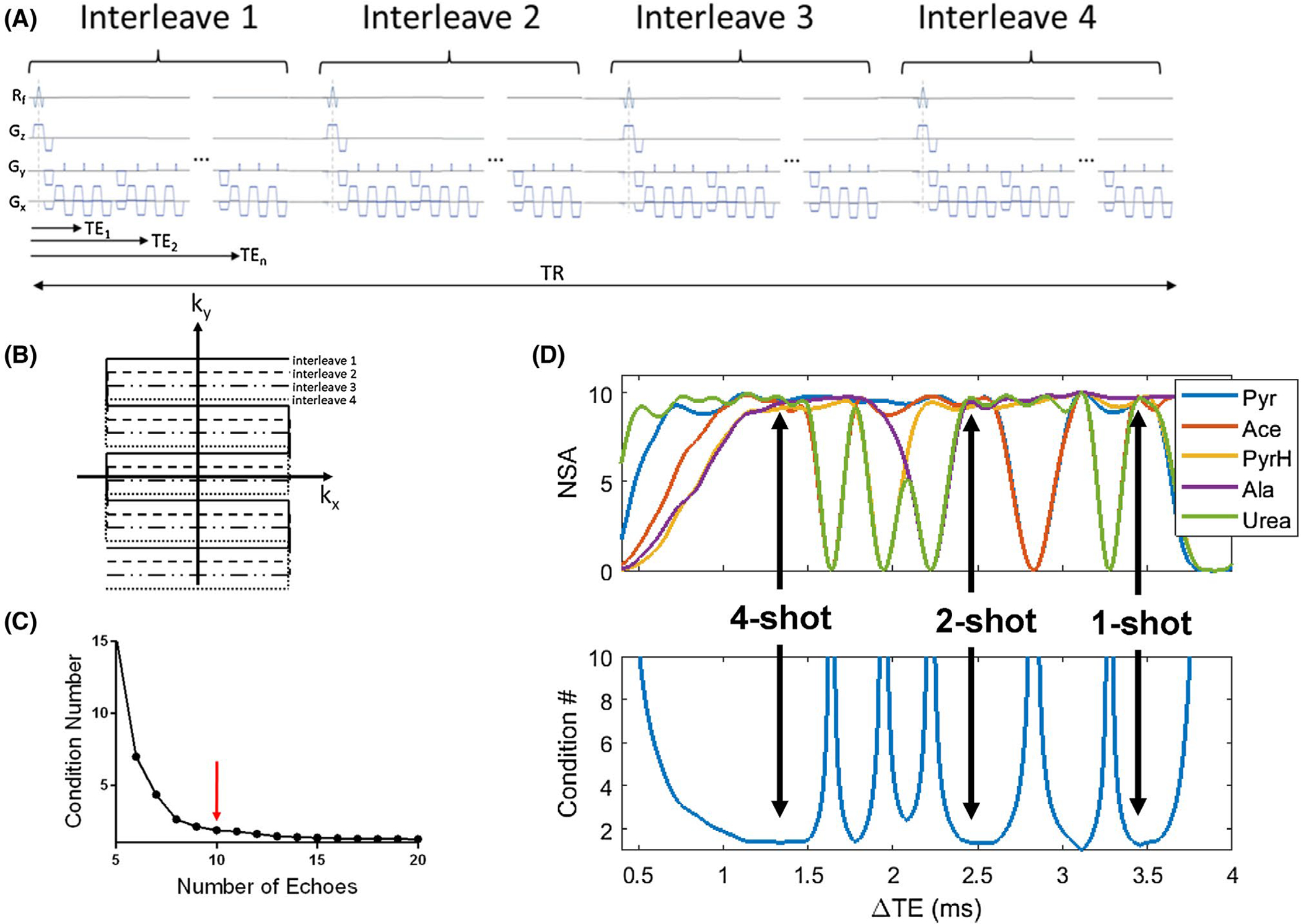
A, Pulse sequence and (B) k-space diagram for a four-shot interleaved 13C-EPI gradient-echo sequence. A slightly different phase-encoding is used for each interleave so that all lines in k-space are ultimately acquired. This interleaved acquisition of k-space shortens the time needed to reach the center of k-space, resulting in shorter echo times and better image quality. C, Plot of condition number against number of echoes for a simulation of the phase coefficient matrix for pyruvate, pyruvate hydrate, acetate, urea, and alanine. The improvement in the condition number is negligible after about 10 echoes (red arrow). D, Result of a number of signal averages (NSA) and condition number simulation for pyruvate, pyruvate hydrate, acetate, urea, and alanine to determine the optimal ΔTEs to be used for a phantom experiment. The black arrows identify three optimal ΔTEs (3.46 ms, 2.49 ms, and 1.34 ms), for acquisition high signal and low condition numbers for the coefficient matrix
2.3 |. Thermal phantom experiments
All experiments were performed on a preclinical Bruker BioSpec 3T scanner (Bruker, Kontich, Belgium; maximum gradient strength = 959 mT/m, maximum slew rate = 6393 T/m/s), using a dual-tuned transmit/receive 1H/13C 42-mm-diameter birdcage coil and a gradient-echo EPI sequence (Figure 1A). Thermal phantom experiments were performed to compare the performance of four-, two-, and one-shot multiecho EPI. The phantoms were two plastic syringes, filled with approximately 1 mL of 4 M [1-13C1]acetate and 0.8 mL of 4 M [1-13C1]urea, respectively (Figure 2A), selected to mimic [1-13C1]lactate and [1-13C1]bicarbonate in in vivo experiments because of their greater stability. Both syringes were doped with 1 mM gadolinium chloride to shorten T1 and eliminate saturation effects. The common acquisition parameters for the three EPI scans were pulse repetition time = 1 second, matrix size = 20 × 20, FOV = 60 × 60 × 50 mm, excitation = 90°, receiver bandwidth = 150 kHz, echo spacing = 0.132 ms, one slice, and 30 averages. The ΔTE values were determined from an number of signal averages simulation to be 1.34, 2.49, and 3.46 ms for four-, two-, and one-shot EPI, respectively (Figure 1C).
FIGURE 2.
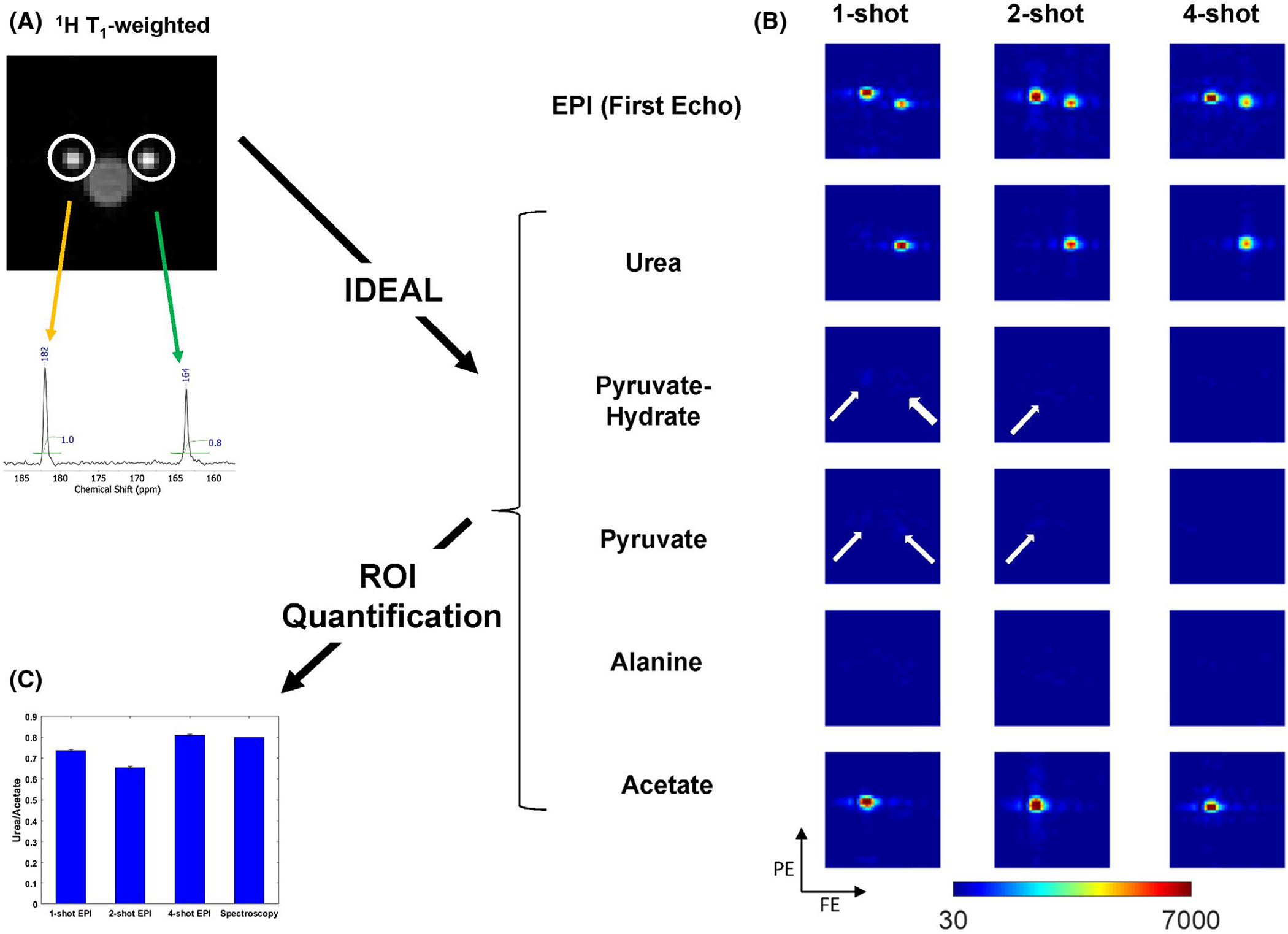
A, 1H T1-weighted images showing two Eppendorf tubes containing approximately 1 mL of 4 M [1-13C]acetate and 0.8 mL of 4 M [1-13C]urea. The spectrum from a nonlocalized spectroscopic scan is shown confirming the chemical shifts and relative quantity of the two chemicals. B, IDEAL was used to generate maps of acetate, pyruvate, pyruvate-hydrate, alanine, and urea from one-shot (ΔTE=3.46 ms), two-shot (ΔTE=2.49 ms), and four-shot (ΔTE=1.34 ms) 13C multiecho echo-planar (EPI) complex Digital Imaging and Communications in Medicine (DICOM) images. Images were resized twofold for display. The improvement in image quality from one- to two-shot EPI can be appreciated by decreasing width of the chemical-shift artifact. The chemical shift of urea and acetate images on the one-, two- and four-shot EPI images in the phase-encoding direction measured relative to the T1-weighted image were 2.5, 1, and 0.5 voxels. The chemical shift in the frequency-encoding was 0.5 voxels for all shots. White arrows placed on the maps for pyruvate, pyruvate-hydrate and alanine generated from one- and two-shot EPI highlight residual signal artifacts from incomplete separation of the chemical species by IDEAL. The fraction of total residual signal (pyruvate, pyruvate-hydrate, and alanine) on the one-, two- and four-shot images was 16%, 9%, and 6% for the urea ROI; and 11%, 7%, and 5% for the acetate region of interest (ROI), respectively. C, An estimate of the urea/acetate ratio obtained by measuring the mean voxel intensity within ROIs drawn on the urea and acetate maps. The urea/acetate ratio from the integration of the peaks in the spectrum shown in (B) is also plotted in the bar graph for comparison. The urea/acetate ratios obtained for one-shot, two-shot, four-shot EPI, and from spectroscopy were 0.74, 0.66, 0.81, and 0.8, respectively. The error bars are the propagated standard deviation of the ratio of voxel intensities within each ROI
2.4 |. Hyperpolarized imaging experiments
DNP was performed using a 5T SPINlab Hyperpolarizer (GE Healthcare) to generate HP pyruvate for infusion into nude mice (n = 3). All murine studies were conducted under an MSK Institutional Animal Care and Use Committee-approved protocol. Thirty-five μL of a mixture of 14.2 M [1-13C]pyruvic acid and 15 mM trityl radical (GE Healthcare) was polarized for 2 hours (5 T, 0.8 K, 139.960 GHz) to achieve spin polarization of approximately 30%. The mixture was dissolved with 10 mL of buffer (10 mM tris(hydroxymethyl)aminomethane in D2O at pH 7.4, 1 mM ethylenediamine tetraacetic acid) into a flask containing a stoichiometric amount of NaOH. Two-hundred μL of HP [1-13C]pyruvate was injected into the mouse tail vein via a catheter over a duration of 10 seconds starting approximately 20 seconds after dissolution. After field-map–based shimming of the FOV using the Bruker MAPSHIM protocol, EPI was initiated approximately 15 seconds before dissolution to capture the full dynamics of perfusion, metabolism, and washout. Acquisition parameters were TE1 = 3.98 ms, ΔTE = 1.5 ms, pulse repetition time = 3 seconds, matrix size = 20 × 20, FOV = 60 × 60 × 50 mm, excitation = 20°, receiver bandwidth = 150 kHz, one average, and 10 slices. The effective FA for a four-shot EPI with a nominal FA of 20° is 38.8°; that is, four 20° excitations consume the same amount of longitudinal magnetization as one 38.8° excitation. The optimal ΔTE for the in vivo experiment was determined from a number of signal averages simulation using the chemical shifts for pyruvate, lactate, pyruvate-hydrate, alanine, and bicarbonate. Anatomical images of the mouse were acquired with the same FOV and a matrix size of 96 × 96.
2.5 |. Data processing and analysis
Complex 13C Digital Imaging and Communications in Medicine (DICOM) EPI images were exported from the MRI scanner, and linearized by applying R2* and B0 inhomogeneity maps estimated by direct phase-fitting of all echoes,28 and processed into metabolite maps in MATLAB (R2017a; MathWorks, Natick, MA) using the IDEAL implementation described in the 2. Methods section using relative chemical shift values of 0, 6, 8, 12, and −10 ppm for [1-13C]pyruvate, [1-13C]alanine, [1-13C]pyruvatehydrate, [1-13C]lactate, and [13C]bicarbonate. The metabolite maps were interpolated fourfold and then overlaid on anatomical images using ITK-SNAP (http://itksnap.org) 35 for display. Regions-of-interest (ROIs) were prescribed on the liver, heart, and kidneys on the anatomical images (Figure 3), and then, copied to the interpolated metabolite maps to quantify the area-under-the-curve (AUC) metric, which is defined for the ith metabolite as
| (4) |
where M(j) is the mean value in the ROI for the jth metabolite at each time point, and δ(i − j) is 1 if i = j and 0 otherwise.
FIGURE 3.

A, The time courses for all metabolites summed over the entire field of view (FOV). B, The plot for each metabolite is baseline-corrected by subtracting the maximum value of the noise (time points before the injection). The red and blue rectangles mark the durations defined as early (9 to 15 seconds) and late (18 to 27 seconds) time points. C, Representative sum of metabolite maps reconstructed from 13C echo-planar imaging (EPI) acquired 9 to 15 seconds (early time points) and 18 to 27 seconds (late time points) after injection. Axial and coronal 1H T2-weighted images are shown for reference. The axial slice displays the heart, and the coronal slice displays slices for the abdominal aorta and liver. White lines show the plane of coronal slice on the axial image and the plane of the axial slice on the coronal image. The intensity of [1-13C]pyruvate in the heart is high at the early time points and sharply reduced at the late time points, but still maintained in the liver. The metabolic products are localized in the heart in early time points and are visible in other organs in the late time points. AUC, area under the curve
3 |. RESULTS
Figure 1A shows the pulse sequence for a four-shot interleaved EPI acquisition, which acquires all of k-space in four alternate lines. Because only a quarter of k-space is acquired per shot, this sequence has a higher effective bandwidth compared with a single-shot sequence, resulting in improved image quality. A comparison of the effect of using a different number of EPI shots is shown in Figure 2. This figure shows the maps’ IDEAL reconstructs from complex 13C-EPI DICOM images of [1-13C]urea and [1-13C]acetate phantom for chemical-shift inputs of 171 ppm([1-13C]pyruvate), 179 ppm ([1-13C]pyruvatehydrate), 177 ppm ([1-13C]alanine), 163 ppm ([1-13C]urea), and 182 ppm ([1-13C]acetate). The quality of separation of the chemical species by IDEAL for a different number of EPI shots can be assessed by the number of artifacts in the pyruvate, pyruvate-hydrate, and alanine maps, which decreases in the maps from one- to four-shot EPI (Figure 2B). The ROI-based quantification of the ratio of urea to acetate intensity shows an underestimation of the ratio by the one- and two-shot methods compared with the ratio calculated from the nonlocalized spectrum (Figure 2C).
Considering the results of the phantom study, the four-shot acquisition method was used in in vivo experiments to acquire HP data for generating metabolite maps of [1-13C] pyruvate, [1-13C]pyruvatehydrate, [1-13C]lactate, [1-13C] bicarbonate, and [1-13C]alanine with IDEAL. Time course and AUC metrics were determined for all metabolite maps. Figure 3B depicts a plot of the mean absolute value of the intensity of each metabolite map over the entire FOV shown in Figure 3A. The plot for each metabolite is baseline-corrected by subtracting the maximum value for the first time point from all other time points. The time-to-peak values for the entire FOV displayed were 12.4 ± 0.4 seconds, 12.4 ± 0.4 seconds, 14.4 ± 0.2 seconds, 12.8 ± 1.1 seconds, and 12.9 ± 1.04 seconds for [1-13C]pyruvate, [1-13C]pyruvatehydrate, [1-13C]lactate, [1-13C]bicarbonate, and [1-13C]alanine, respectively. Using these time courses, we defined two summations of the metabolite maps’ time series for qualitative assessment, which include early time points that summarize the pyruvate bolus (9 to 15 seconds), and late time points that are weighted to the metabolic products (18 to 27 seconds). In Figure 3C, the axial slice highlights the region of the heart, with accompanying coronal slice showing the liver in the same FOV, with window levels set separately for each metabolite to best demonstrate their spatial distribution. [1-13C]pyruvate has a high intensity in the heart and abdominal aorta at the early time points and is reduced and localized in the liver in the late time points. All the products of metabolism, [1-13C]lactate, [1-13C]bicarbonate, and [1-13C]alanine show increased intensity in the late time points than in the early time points.
The average values of metabolite time courses for all mice within ROIs prescribed in heart, liver, and kidneys (Figure 4A) are given in Table 1. Representative dynamics for each ROI reveal slight differences in organ time-to-peak as well as organ-dependent metabolic fluxes (Figure 4B). The ratio of the amount of each metabolite product to the total amount of metabolites is shown in Figure 4C. The ratios for [1-13C]lactate, [1-13C]bicarbonate, and [1-13C]alanine were 0.23 ± 0.15, 0.08 ± 0.05, 0.02 ± 0.02 in the kidneys, 0.18 ± 0.04, 0.13 ± 0.008, 0.02 ± 0.02 in the liver, and 0.06 ± 0.08, 0.5 ± 0.4, 0.02 ± 0.02 in the heart.
FIGURE 4.

4Average region-of-interest (ROI) measurements for metabolic biodistribution in selected organs. A, ROIs are prescribed on anatomical 1H T2-weighted images of slices containing the kidneys, liver, and heart and copied to metabolite maps for quantifying the average metabolite image intensity. B, The average for three mice of the mean organ intensity for each metabolite is shown plotted over time. C, The area-under-the-curve (AUC) ratio in each organ is shown for each metabolite
TABLE 1.
TTP measurements for [1-13C]lactate, [1-13C] bicarbonate, and [1-13C]alanine relative to the [1-13C]pyruvate peak within regions of interest prescribed in the heart, liver, and kidneys
| Metabolite | [1-13C] lactate |
[1-13C] bicarbonate |
[1-13C] alanine |
|---|---|---|---|
| TTP in heart (s) | 2 ± 1 | 5 ± 1 | 1 ± 1 |
| TTP in liver (s) | 4 ± 1.7 | 6 ± 5.2 | 3 ± 0.5 |
| TTP in kidneys (s) | 3 ± 2.5 | 3 ± 2.5 | 2 ± 1.4 |
Note: The data shown are the average and standard deviation for three mice and reported in seconds.
Abbreviation: TTP, Time-to-peak.
4 |. DISCUSSION
Our goal in this study was to describe a simple robust technique for the accurate quantification of HP metabolite spatiotemporal biodistribution to enable the study of metabolic processes that involve multiple organs. In phantom experiments with tubes of 13C-enriched chemicals, we showed that using IDEAL to reconstruct data acquired with a four-shot multiecho EPI scan is an effective way to accomplish this goal. We acquired multislice imaging data of a mouse over a FOV covering the major organs in the mouse body and calculated maps of the spatial distribution of [1-13C]pyruvate, [1-13C]pyruvatehydrate, [1-13C]lactate, [1-13C]bicarbonate, and [1-13C]alanine from the complex EPI DICOM images. From these maps, we calculated metabolite time courses for ROIs prescribed over the entire imaging FOV, kidneys, liver, and heart. The metabolite time courses showed similar dynamics in response to a bolus delivery for all ROIs, but with variations for each organ that correspond well to previously published animal data acquired using other approaches in the heart,14,36 liver,16,37 and kidneys,15 providing further confidence in the accuracy of the results.
Various methods have been described for HP 13C imaging, ranging from robust but inefficient spectroscopy-based techniques such as CSI to imaging methods such as SPSP EPI that are extremely efficient, but may require specialized RF pulses and a separate proton scan to correct geometric distortion and chemical-shift artifacts. Multishot multiecho EPI provides a method that is more suitable than the former for multislice dynamic imaging, without the requirement of specialized pulses or hardware that may be necessary for SPSP EPI. The number of shots used can be chosen as a trade-off between the required image quality and efficiency in the use of HP magnetization.
The spatial distribution of [1-13C]pyruvate and its metabolites in mice using multiecho EPI agree with maps generated using CSI38 and SPSP EPI.39 In addition, the ratios of the amount of each metabolite product to the total amount of carbon were comparable to previously published values.16 The AUC ratio in the each organ represents the enzyme catalyzed flux of the metabolite from [1-13C]pyruvate in the organ. For example, [1-13C]bicarbonate has a high AUC ratio in the heart, which is well-documented in the literature,14,40 primarily as the result of its flux from [1-13C]pyruvate through PDH.36 Its AUC ratios were much lower in the liver and kidneys, where [1-13C]bicarbonate may have been transported from the heart or created by additional mechanisms,37 including decarboxylation of [4-13C]oxaloacetate by phosphoenolpyruvatecarboxykinase (PEPCK).
The results obtained in this study suggest that the use of multishot, multiecho EPI may have a beneficial role in human studies through its improved image quality and HP metabolite quantification compared with single-shot EPI. The use of a broadband RF excitation also compensates for local B0 inhomogeneity, making it possible to obtain high-resolution, time-resolved, multislice imaging over a large region of interest that can be reformatted into volumetric images to facilitate the study diseases such as cancer metastasis using HP probes. In future studies, we aim to bring this approach into the clinic to better visualize in vivo metabolic dynamics.
ACKNOWLEDGMENTS
This work was sponsored by the National Institutes of Health (NIH R00 EB014328, R01 CA195476, R01 CA237466 and S10 OD016422; NIH/NCI Cancer Center Support grant P30 CA008748); Geoffrey Beene Cancer Research Center, Center for Molecular Imaging and Nanotechnology at MSKCC; The Thompson Family Foundation; The Center for Experimental Therapeutics at MSKCC; Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research; and the Peter Michael Foundation. R.E. is supported by Tow Foundation Postdoctoral Fellowship from Center for Molecular Imaging and Nanotechnology (CMINT) at Memorial Sloan Kettering.
Footnotes
CONFLICT OF INTEREST
Unrelated to this work, K.R.K. serves on the SAB of NVision Imaging Technologies.
REFERENCES
- 1.Golman K, in ‘t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci USA. 2006;103:11270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keshari KR, Wilson DM. Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization. Chem Soc Rev. 2014;43:1627–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Sci Transl Med. 2013;5:198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miloushev VZ, Granlund KL, Boltyanskiy R, et al. Metabolic imaging of the human brain with hyperpolarized. Cancer Res. 2018;78:3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granlund KL, Tee SS, Vargas HA, et al. Hyperpolarized MRI of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metabolism. 2020;31:105–114. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham CH, Lau JY, Chen AP, et al. Hyperpolarized 13C metabolic MRI of the human heart: Initial experience. Circ Res. 2016;119:1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon JW, Chen HY, Autry A, et al. Translation of carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magn Reson Med. 2019;81:2702–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, et al. Hyperpolarized. Neoplasia. 2019;21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupo JM, Chen AP, Zierhut ML, et al. Analysis of hyperpolarized dynamic 13C lactate imaging in a transgenic mouse model of prostate cancer. Magn Reson Imaging. 2010;28:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66:10855–10860. [DOI] [PubMed] [Google Scholar]
- 11.Park I, Hu S, Bok R, et al. Evaluation of heterogeneous metabolic profile in an orthotopic human glioblastoma xenograft model using compressed sensing hyperpolarized 3D 13C magnetic resonance spectroscopic imaging. Magn Reson Med. 2013;70:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Gialleonardo V, Aldeborgh HN, Miloushev V, et al. Multinuclear NMR and MRI reveal an early metabolic response to mTOR inhibition in sarcoma. Cancer Res. 2017;77:3113–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Y, Eskandari R, Ray C, et al. Hyperpolarized MRI visualizes Warburg effects and predicts treatment response to mTOR inhibitors in patient-derived ccRCC xenograft models. Cancer Res. 2019;79:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau AZ, Chen AP, Ghugre NR, et al. Rapid multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magn Reson Med. 2010;64:1323–1331. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Mayer D, Gu M, et al. Quantification of in vivo metabolic kinetics of hyperpolarized pyruvate in rat kidneys using dynamic 13C MRSI. NMR Biomed. 2011;24:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S, Chen AP, Zierhut ML, et al. In vivo carbon-13 dynamic MRS and MRSI of normal and fasted rat liver with hyperpolarized 13C-pyruvate. Mol Imaging Biol. 2009;11:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spielman DM, Mayer D, Yen YF, Tropp J, Hurd RE, Pfefferbaum A. In vivo measurement of ethanol metabolism in the rat liver using magnetic resonance spectroscopy of hyperpolarized [1-13C]pyruvate. Magn Reson Med. 2009;62:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durst M, Koellisch U, Frank A, et al. Comparison of acquisition schemes for hyperpolarised 13C imaging. NMR Biomed. 2015;28:715–725. [DOI] [PubMed] [Google Scholar]
- 19.Yen YF, Kohler SJ, Chen AP, et al. Imaging considerations for in vivo 13C metabolic mapping using hyperpolarized 13C-pyruvate. Magn Reson Med. 2009;62:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiesinger F, Weidl E, Menzel MI, et al. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate. Magn Reson Med. 2012;68:8–16. [DOI] [PubMed] [Google Scholar]
- 21.Lau JYC, Geraghty BJ, Chen AP, Cunningham CH. Improved tolerance to off-resonance in spectral-spatial EPI of hyperpolarized [1-. Magn Reson Med. 2018;80:925–934. [DOI] [PubMed] [Google Scholar]
- 22.Reeder SB, Wen Z, Yu H, et al. Multicoil Dixon chemical species separation with an iterative least-squares estimation method. Magn Reson Med. 2004;51:35–45. [DOI] [PubMed] [Google Scholar]
- 23.Reeder SB, Brittain JH, Grist TM, Yen YF. Least-squares chemical shift separation for (13)C metabolic imaging. J Magn Reson Imaging. 2007;26:1145–1152. [DOI] [PubMed] [Google Scholar]
- 24.Geraghty BJ, Lau JYC, Chen AP, Cunningham CH. Dual-Echo EPI sequence for integrated distortion correction in 3D time-resolved hyperpolarized. Magn Reson Med. 2018;79:643–653. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein MA, King KF, Zhou XJ. CHAPTER 14 - BASIC PULSE SEQUENCES. Handbook of MRI Pulse Sequences. Burlington: Academic Press; 2004:579–647. [Google Scholar]
- 26.Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. Hoboken, NJ: Wiley-Blackwell; 1999. [Google Scholar]
- 27.Xiang QS, An L. Water-fat imaging with direct phase encoding. J Magn Reson Imaging. 1997;7:1002–1015. [DOI] [PubMed] [Google Scholar]
- 28.Tsao J, Jiang Y. Hierarchical IDEAL: Fast, robust, and multiresolution separation of multiple chemical species from multiple echo times. Magn Reson Med. 2013;70:155–159. [DOI] [PubMed] [Google Scholar]
- 29.Hu HH, Börnert P, Hernando D, et al. ISMRM workshop on fat-water separation: Insights, applications and progress in MRI. Magn Reson Med. 2012;68:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of fast spiral chemical shift imaging using least squares reconstruction: application for hyperpolarized (13)C metabolic imaging. Magn Reson Med. 58;2007:245–252. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein MA, King KF, Zhou XJ. Echo Train Pulse Sequences. Handbook of MRI Pulse Sequences. Burlington: Academic Press; 2004:723. [Google Scholar]
- 32.Butts K, Riederer SJ, Ehman RL, Thompson RM, Jack CR. Interleaved echo planar imaging on a standard MRI system. Magn Reson Med. 1994;31:67–72. [DOI] [PubMed] [Google Scholar]
- 33.Buonocore MH, Gao L. Ghost artifact reduction for echo planar imaging using image phase correction. Magn Reson Med. 1997;38:89–100. [DOI] [PubMed] [Google Scholar]
- 34.McKinnon GC. Ultrafast interleaved gradient-echo-planar imaging on a standard scanner. Magn Reson Med. 1993;30:609–616. [DOI] [PubMed] [Google Scholar]
- 35.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. [DOI] [PubMed] [Google Scholar]
- 36.Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc Natl Acad Sci USA. 2007;104:19773–19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt ME, Harrison C, Sherry AD, Malloy CR, Burgess SC. Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc Natl Acad Sci USA. 2011;108:19084–19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler SJ, Yen Y, Wolber J, et al. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58:65–69. [DOI] [PubMed] [Google Scholar]
- 39.Gordon JW, Hansen RB, Shin PJ, Feng Y, Vigneron DB, Larson PEZ. 3D hyperpolarized C-13 EPI with calibrationless parallel imaging. J Magn Reson. 2018;289:92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golman K, Petersson JS, Magnusson P, et al. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn Reson Med. 2008;59:1005–1013. [DOI] [PubMed] [Google Scholar]


