Abstract
Advancement of tissue engineering and regenerative medicine (TERM) strategies to replicate tissue structure and function has led to the need for noninvasive assessment of key outcome measures of a construct’s state, biocompatibility, and function. Histology based approaches are traditionally used in preclinical animal experiments, but are not always feasible or practical if a TERM construct is going to be tested for human use. In order to transition these therapies from benchtop to bedside, rigorously validated imaging techniques must be utilized that are sensitive to key outcome measures that fulfill the FDA standards for TERM construct evaluation. This review discusses key outcome measures for TERM constructs and various clinical- and research-based imaging techniques that can be used to assess them. Potential applications and limitations of these techniques are discussed, as well as resources for the processing, analysis, and interpretation of biomedical images.
Background
Rapid innovation in tissue engineering and regenerative medicine (TERM) has driven development of novel approaches to fabricate constructs capable of replicating complex tissue structure and function.1 TERM constructs are designed to integrate with the local host environment in order to repair and/or regenerate, and restore function in damaged tissues.2 While the specific approach varies between techniques, generally TERM models can be separated into three categories: (1) acellular scaffolds, (2) cell-only, scaffold-free designs, and (3) hybrid cellularized scaffolds. Advancements in TERM have been driven by the development of new 3D bioprinting fabrication techniques, which allow for precision fabrication of tissue informed constructs consisting of various cell types,3–5 biomaterials,6,7 and growth factors.8 Many TERM applications are currently in the preclinical stage, utilizing in vivo animal experiments to demonstrate potential translation to humans and clinical viability. To advance these pre-clinical studies from benchtop to bedside, serial testing in the same animal rather than separate cohorts must be implemented, prior to even opening a phase I FDA-approved clinical trial. As human studies require fundamentally different approaches and techniques to validate and monitor function of TERM constructs, robust analysis techniques must be developed and utilized in parallel to pre-clinical studies.
Histology is currently the gold standard used to validate the biological performance and compatibility of TERM constructs. However, histology is highly invasive, semi-quantitative, destructive to the tissue, does not allow for longitudinal analysis, requires the use of multiple animals, can change the structure of a construct upon processing, is not representative of the entire volume of a tissue, and does not directly test function. This has driven the need for quantitative imaging technologies that are capable of accurately assessing TERM constructs non-destructively in vivo, over time, and in 3D, to monitor how they integrate with and affect tissue physiology. In order to support rapid advancement and translation of TERM constructs, a basic understanding of the potential medical imaging modalities and techniques that can be used is required to evaluate the effectiveness and function of constructs that may be implanted in vivo. In order to facilitate development of both TERM constructs and the noninvasive imaging techniques required to evaluate them in vivo, there needs to be continued collaboration between the tissue engineering, physiology, and radiology communities.
The purpose of this review is to: (1) identify specific outcome measurements of interest that are critical in the evaluation TERM constructs, (2) discuss imaging modalities and techniques that can be used to assess TERM constructs, and (3) review recent advancements in imaging TERM constructs. The aim of this review is to inform tissue engineers of specific imaging modalities and approaches that can be used for evaluating TERM constructs in vivo, for common outcome measures of interest, and facilitate communication with the imaging community. This review will highlight various tools and techniques available using the three most common and available clinical imaging modalities: ultrasound, X-ray based imaging, and magnetic resonance imaging (MRI).
Key considerations for quantitative imaging of TERM constructs
The initial considerations that must be made when choosing an imaging modality are dependent upon specific outcome measures of interest that fulfill the FDA standards for TERM construct evaluation. If outcome measures that are assessed in pre-clinical studies meet FDA requirements, the likelihood of the FDA accepting these measurements in a phase I trial increases. While specific outcome measures vary depending on the tissue of interest and the proposed application of the TERM construct, the main outcome measurements of interest can be broadly categorized as: (1) state of the construct (i.e. size, degradation, mechanical properties, presence of key cells or materials), (2) biocompatibility and biointegration of the construct (i.e. perfusion, inflammation, fibrous encapsulation, cell viability), and (3) function of the construct (i.e. stimulation of de novo tissue production, microstructural organization, mechanical functionality, biological functionality) (Fig. 1).
Fig. 1.
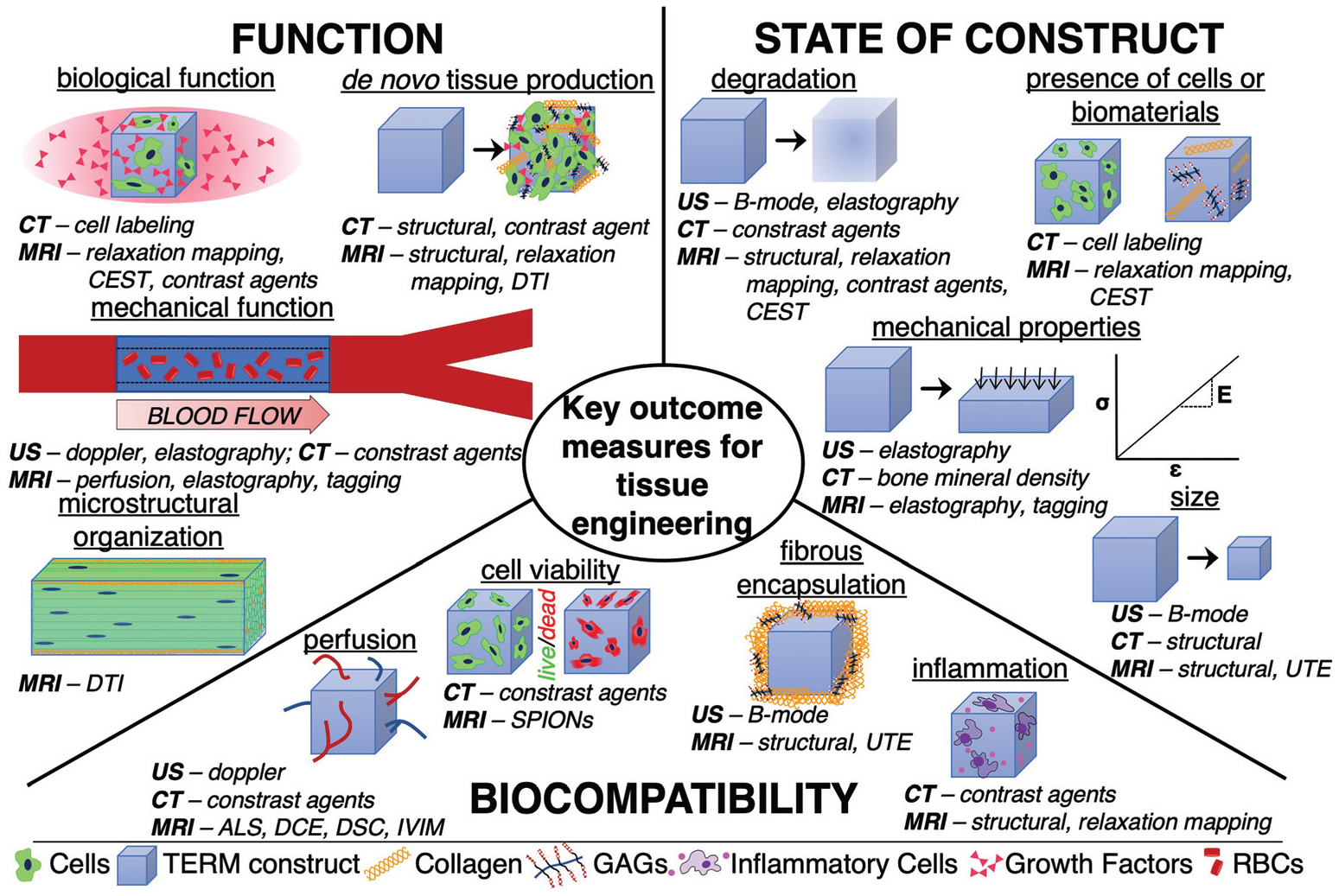
Schematic depicting key outcome measures for assessing tissue engineering and regenerative medicine constructs as well as the specific imaging techniques that can be used to assess them (italic). GAGs – glycosaminoglycans. RBCs – Red blood cells.
It is important to note that there is a need to serially assess these outcome measurements, as they change with time, and can influence which imaging modality is most suitable. For example, immediately after implantation, biocompatibility is a key biomarker of success, while at extended timepoints, stimulation of de novo tissue production is required. Furthermore, serial assessment minimizes the use of animals and humans in early stage work, as well as maximizes statistical power in a study. While most imaging modalities are multi-parametric (able to assess different outcome measures of tissue using the same equipment), no single imaging modality may be able to assess all features of interest.
Once outcome measures of interest have been identified, the strengths and weaknesses of various imaging approaches can be weighed. For each imaging modality, there are clinically oriented methods which can be easily and quickly utilized, typically yielding simple structural information about a TERM construct, from which basic volumetric or shape-based analyses can be performed. Research-oriented imaging techniques that are more sensitive to many of the outcome measures listed above are typically not standard on every machine or require acquisition optimization, depending on the application. This optimization process attempts to maximize signal at physiologically relevant voxel (volumetric pixel) size during a standard imaging session (minutes – 1 hour). A number of techniques for each imaging modality, their potential applications for detecting TERM outcome measures, their clinical-or research-oriented status, approximate scan time, and relative cost can be found in Table 1. In addition to the sensitivity of the imaging modality and technique to the key outcome measures at sufficient spatial and temporal resolution, several other factors are important in determining the practical applicability, such as the availability of the scanners, the ability to collect data in a reasonable amount of time and cost, the use of radiation or contrast agents, and the need for computational resources to analyze and visualize the data. In what follows we present an overview of the three advanced techniques that have demonstrated utility in tissue engineering applications and are most readily available.
Table 1.
Summary of the technical and functional capabilities of the 3 imaging modalities in this review. Approximate scan time and cost are listed as ranges depicting the approximate minimum and maximum values. Scan time generally increases with increased resolution (smaller in-plane resolution) and increasing the number of slices or acquisitions. The cost of scanning varies widely due to the institution, equipment used, use of a trained technician, and if the imaging time is designated as preclinical or clinical. Preclinical imaging time is traditionally much less expensive than time reserved for clinical imaging
| Modality | In-plane resolution | Depth of penetration | Approximate scan time | Cost ($ per hour) | Imaging techniques sensitive to TERM outcome measures | ||
|---|---|---|---|---|---|---|---|
| State | Biocompatibility | Function | |||||
| Ultrasound | ~0.1 mm | ~cm | Real-time | 50–300 | B-Modea | Dopplera | Dopplera, elastographya |
| X-Ray/CT | ~0.1 mm–~0.5 mm | Unlimited | Seconds – 10 minutes | 200–1000 | CTa, contrast agentsb | Cell labelingb | Contrast agentsb |
| MRI | ~mm | Unlimited | 1 minutes – 20 minutes | 400–2000 | Structural MRIa, relaxationa, UTEb | Perfusionb, UTEb, contrast agentsb | Relaxationa, CESTb, DTIb, elastographyb, taggingb, contrast agentsb |
Indicates clinically oriented imaging technique.
Indicates research-oriented imaging technique.
TERM – Tissue engineering and regenerative medicine. CT – computed tomography. MRI – magnetic resonance imaging. UTE – ultrashort echo time. CEST – chemical exchange saturation transfer. DTI – diffusion tensor imaging.
Ultrasound imaging
Overview
Ultrasound imaging is based on the generation and reception of sound waves as they penetrate a material and are partially reflected at interfaces between tissues of different density (i.e., acoustic impedance). Ultrasound imaging uses a probe, capable of transmitting and receiving sound waves, that must be held against tissue (normally skin), near a TERM construct and requires a trained operator to visualize underlying structures. Tissues with different impedance result in gray-scale contrast in the reconstructed image. Typical spatial resolution of ultrasound can be as small as 0.1 mm, and with higher ultrasound frequency, spatial resolution increases, albeit with more limited depth of penetration. Ultrasound additionally has great temporal resolution, capable of displaying images in near real time (24 Hz – 120 Hz), but has difficulty penetrating hard materials such as bone. Although ultrasound imaging is most often utilized to non-invasively assess constructs near the skin, alternate probe designs allow for more invasive assessment such as intravenous ultrasound, transesophageal echocardiography, and transvaginal ultrasound. Ultrasound imaging is the cheapest and most mobile of the imaging modalities covered in this review, with current models compatible with smartphones costing less than $2000.9 However, as the acoustic impedance of tissue is similar across tissue types, ultrasound generally has poor contrast and it is difficult to differentiate nearby structures, especially when compared to X-ray and MRI. For a comprehensive review on the fundamentals of ultrasound imaging, please see Ng et al.10
Structural imaging
One of the primary outcome measures assessed with standard 2D (e.g. B-mode) ultrasound imaging is the size and morphology of a TERM construct. For example, in vascular tissue engineering studies, lumen diameter and wall thickness of the implanted construct is an important measurement related to tissue formation, mechanical durability, and potential occulsion11,12 (Fig. 2). Additionally, ultrasound imaging may provide insight into integration or potential fibrous encapsulation of a TERM construct from increased extracellular matrix deposition by changes in signal attenuation around the region of interest.13–17
Fig. 2.
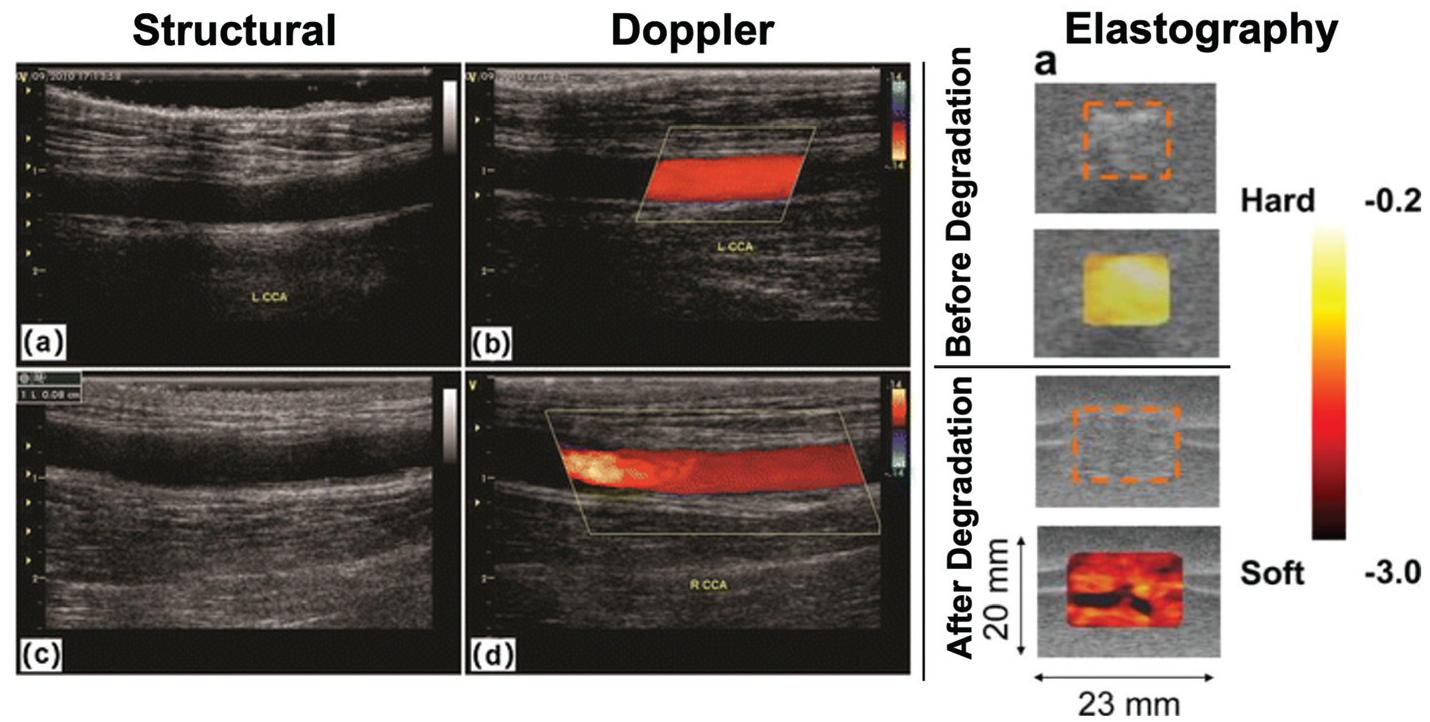
Ultrasound: B-mode structural imaging (left) of a tissue engineered vascular graft can be used to measure the size of the lumen and assess patency of the graft at 3 months (top) and 6 months (bottom) after implantation. Doppler shift imaging can be used to measure blood flow through the tissue engineered vascular graft (middle column). Figure reproduced with permission from Springer Nature (2017).22 Strain maps assessed with elastography (right) of a subcutaneously implanted tissue engineered scaffold before (top) and after (bottom) in vivo degradation, demonstrating that ultrasound elastography can be used to assess changes in mechanical properties of a scaffold associated with degradation. The dashed orange boxes represent the boundaries of the scaffold, the color overlay represents the in vivo strain map. Figure reproduced with permission from Elsevier (2008).29
Blood flow (Doppler) imaging
The Doppler shift occurs when a sound wave is reflected off of a moving object, causing a change in its frequency related to the object’s speed. Using the Doppler shift, ultrasound imaging can also provide measurements of macrovascular blood flow. This has driven interest in using ultrasound imaging for monitoring the function of tissue engineered vascular grafts.18,19 While standard ultrasound imaging can be used to identify the size of the lumen and wall of a graft, Doppler imaging can quantify the rate of blood flow (typically in cm s−1). Rapid, accurate, quantitative measurements of blood flow are useful for assessing patency of a graft,20–22 identifying collateral vasculature surrounding tissue engineered vascular grafts,11 identifying potential mechanical defects in grafts,23 and graft failure12,13 (Fig. 2). However, Doppler imaging is unable to directly measure blood flow in microvasculature (diameter less than ~100 μm) due to limitations of resolution. To increase sensitivity to blood flow in microvasculature, some studies include microbubbles as an exogenous contrast agent in order to enhance blood flow signal.24,25 While microbubble-enhanced US imaging does not provide blood flow of individual capillaries, it can help detect neovascularization and quantify perfusion in TERM constructs. Also, biomaterials can be directly incorporated into the microbubble construct in order to allow for therapeutic applications such as localized growth factor delivery.26
Mechanical property (elastography) imaging
Elastography is the process of extracting mechanical properties out of a tissue or TERM construct in response to an external mechanical force using ultrasound imaging. There are two main types of elastography typically found on commercial ultrasound machines: (1) strain-based elastography, which utilizes force from the application of probe pressure or through endogenous mechanical force and (2) shear wave ultrasound elastography, which uses the ultrasound probe to generate waves perpendicular to the ultrasound beam, causing transient displacements.27,28 As mechanical properties are a key outcome measure for TERM constructs, and mechanical properties of a construct are often closely tied to degradation, ultrasound elastography can be a valuable tool for quickly assessing the state of soft materials (i.e. hydrogels). Several groups have used either strain-based or shear-wave ultrasound elastography to monitor changes in the mechanical properties of biodegradable scaffolds due to degradation29–32 (Fig. 2). Elastography can be used to assess the functionality of a TERM construct in vivo and identify if it is at risk for failure.33 Furthermore, there is some evidence that elastography may be sensitive to TERM construct remodeling due to tissue ingrowth.33 However, like most ultrasound techniques, ultrasound elastography is operator dependent and is sensitive to subcutaneous fat, which attenuates signal and decreases measurement accuracy.34,35 For a comprehensive review on ultrasound elastography please see Sigrist et al.36
X-ray based imaging
Overview
X-ray imaging is based on the differential absorption of X-rays by tissues of different densities. By placing the subject between the X-ray source and an X-ray detector, a projection (or “shadow”) of these density variations is acquired. In computed tomography (CT), these projections are acquired at multiple angles, allowing the spatial distribution of these density variations (i.e., a tomographic 2D image) to be reconstructed. By doing this for multiple adjacent slices, a 3D (volumetric) image can be reconstructed. Current state-of-the-art human CT scanners are able to achieve high spatial resolution (150 microns). X-rays are able to penetrate the entire body, however, X-ray based imaging has poor soft tissue contrast and is better for resolving hard tissues, such as bone.37 To increase contrast in soft tissues and TERM constructs, exogenous contrast agents are often used either systemically or are directly incorporated into a construct. CT is inherently quantitative, where pixel intensity can be converted to Hounsfield units, which describes the attenuation coefficient of a tissue. While X-ray based imaging does provide excellent resolution, full body penetration, and is very fast, harmful ionizing radiation is utilized. Therefore, X-ray based imaging may lead to tissue damage or unacceptable radiation dosing when serial imaging is needed.
Micro computed tomography
A variation of CT called microcomputed tomography (μCT) utilizes the same principles as CT, but is capable of much higher resolution (down to ~1μm) and is often used in preclinical studies of TERM constructs. μCT machines are much smaller and cheaper than traditional human CT scanners, making them an easy tool to use for preclinical development of TERM constructs. Although most current TERM studies use μCT, many of the same techniques can be utilized in human CT scanning as well.
Typically, μCT is used for high resolution imaging of bone or TERM bone constructs as it is sensitive to many outcome variables of interest. For example, in many studies of TERM bone therapies, μCT measured ratio of bone volume to total volume, trabecular number, trabecular thickness, and trabecular spacing are key outcome measurements that describe size of the construct, stimulation of de novo tissue production, microstructural organization, and biological functionality38–41 (Fig. 3). Furthermore, since Hounsfield units are linearly related to bone mineral density and bone mineral density is directly related to the elastic modulus of bone tissue, 3D mechanical properties of bone TERM constructs can be assessed using μCT.42,43 In addition to assessing mechanical properties of bone, recent advances in X-ray elastography are being developed, which can potentially be used to assess the elastic modulus of softer tissues.44
Fig. 3.
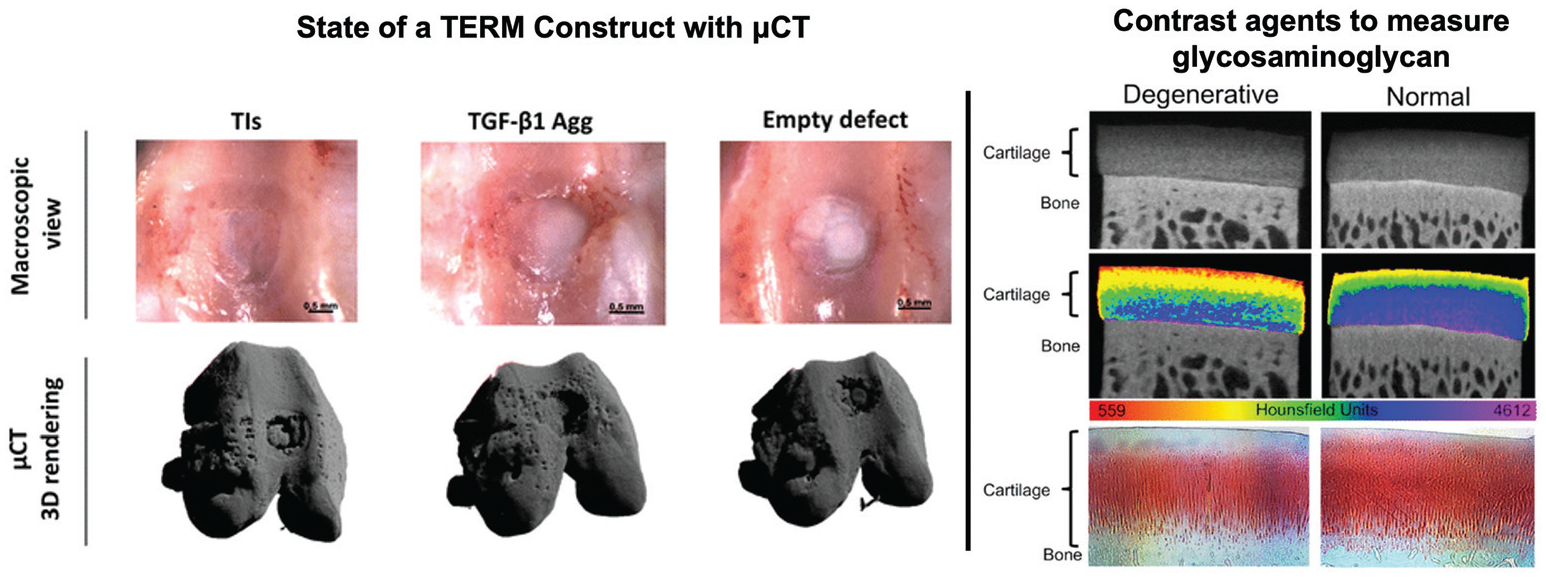
Computed tomography: Macroscopic view (top row) of TERM treated (left, middle) and untreated (right) osteochondral defects. μCT was used to render the osteochondral defect and subchondral bone to assess state of the TERM constructs and de novo tissue production. Figure reproduced with permission from Springer Nature (2018).39 (Right) A cationic contrast agent that is sensitive to glycosaminoglycan distribution in degenerated and normal cartilage. The contrast agent is attracted to the strong negative charge of glycosaminoglycans and increases radiopacity regions with high glycosaminoglycan concentration. This demonstrates how contrast agents can be used to assess the presence of biomaterials. Figure reproduced with permission from Elsevier (2018).53
While not an in vivo application, TERM scaffold characterization using μCT before implantation is common. Cell arrangement and function are directly impacted by its surrounding microenvironment. Therefore, it is important to confirm that a TERM construct is likely to be mechanically or biologically functional prior to in vivo implantation. Since μCT allows for high resolution assessment of 3D scaffold structure, scaffold alignment, size, and porosity are often quantified in addition to qualitative 3D visualization.45 Furthermore, comparing μCT data for a TERM construct prior to implantation to in vivo can be useful for understanding construct integration and function of the local tissue environment. For more information on the applications of μCT characterization of scaffolds, please see the review by Cengiz et al.45
Contrast agents.
The use of exogenous contrast agents with CT increases sensitivity to assessing the state, biocompatibility, and function of TERM constructs. For example, with the addition of radiopaque contrast agents, it is possible to map angiogenesis in and around a new tissue implant, increasing sensitivity to detecting perfusion in TERM constructs.46–48 Also, contrast agents can be utilized which have ionic interactions with specific biomaterials in vivo in order to identify de novo tissue production and mechanical functionality of a TERM construct. For instance, glycosaminoglycans (GAGs) are large negatively charged molecules in the extracellular matrix that attract cations and water molecules, leading to strong hydrostatic pressures. Both anionic and cationic contrast agents have been developed which either are repelled by, or are attracted to GAGs, in order to assess biological functionality and de novo tissue regeneration in cartilage49–54 (Fig. 3).
Rather than administering exogenous contrast agents to a tissue after implantation, another approach to increase the sensitivity of CT is to directly incorporate them into a TERM construct itself. Hydrogels – polymeric networks capable of retaining a large volume fraction of water – are one of the most common categories of TERM constructs. There are two main approaches that have been taken in order to incorporate contrast agents directly into a hydrogel: (1) physically mixing contrast agents with a hydrogel and (2) covalently bonding contrast agents into a hydrogel backbone. Although relatively easy to produce, simply mixing commercial contrast agents into a hydrogel can affect material properties of the hydrogel like increase viscosity and delay gelation rate.55 Chemically modifying a hydrogel’s structure provides more control over the amount of contrast to be imparted, as well as degradation of the hydrogel system. As the average Hounsfield unit signal will decrease as the scaffold degrades, it is easy to monitor scaffold degradation over time.56,57 Several different radiopaque compounds can be attached to polymers using common synthesis approaches, depending on the type of hydrogel that is being synthesized and the amount of contrast attenuation that is required for the desired application.56–59 For a comprehensive review of CT of hydrogels, please see Lei et al.60 For a comprehensive review on contrast agents for X-ray and CT imaging applications, please see the recent reviews by Hsu et al.61 and de Bournonville et al.62
Cell labeling.
The development of in vivo techniques to monitor TERM implants is not limited to scaffold design, but also applies to the direct labeling of cells. A simple approach to labeling cells for μCT tracking is to culture them in barium sulphate (a radiopaque contrast agent), which allows for cells to be tracked as they migrate within a scaffold.63 Another approach is to use gold nanoparticles, which are gold particles coated with various ligands that facilitate cell uptake.64,65 The amount of uptake by a cell is regulated by several factors including the size, shape, nanoparticle surface functionalization (chemical modification of the surface to modify cellular interaction), incubation time, and particle concentration.66,67 This allows for several different types of cells to be labeled including T cells,68 mesenchymal stem cells,65,69,70 and monocytes.71 In vivo experiments have demonstrated that gold nanoparticle labeling allows for the quantitative longitudinal cell tracking, which provides insight into de novo tissue production, microstructural organization, and biological functionality.69 For a review of cell labeling with CT, please see Kim et al.72
Dual energy/spectral CT.
First conceptualized in the 1970s,73–75 recent innovation in CT technology has led to the development and commercialization of dual energy or spectral CT scanners. Unlike conventional CT where a single projection is acquired at a single X-ray energy level, spectral CT simultaneously acquires two projections (90° to 95° apart76) at two different X-ray energy levels.77 This improves the ability to differentiate between tissues or cells that may have similar attenuation using traditional CT or even multiple contrast agents within the same tissue, by exploiting the energy-specific attenuation profiles of different materials.78–81 This can be used to differentially label and track cells within a bioengineered scaffold,80 which provides insight into the state of a construct, de novo tissue production, perfusion, and biological function. Furthermore, FDA approved contrast agents can be utilized with this method, reducing the need for extensive toxicity studies and the development of new contrast agents, which reduces the burden for clinical translation.
Magnetic resonance imaging
Overview
Magnetic resonance imaging (MRI) uses the intrinsic magnetic properties of hydrogen nuclei (protons) in tissue water in a large constant external magnetic field to create signal contrast. A signal is generated from the water in the tissue by the application of a smaller rapidly oscillating (radiofrequency, RF) magnetic field. Then the signal is spatially encoded with small amplitude, spatially varying magnetic fields (“gradients”). Unlike the aforementioned methods which rely on an external source reflecting off tissue boundaries, MRI generates signal directly from a tissue. The sensitivity of MRI to tissue water means it is particularly useful in soft tissues and provides multiple methods for generating image contrast. The sensitivity of MRI to a wide range of structural and physiological parameters make it by far the most versatile of the methods, and the most complicated. While hydrogen MRI is most common – as it is the most abundant element in the body – other elements such as sodium (23Na) and phosphate (31P) can be also imaged, though typically at much lower sensitivity.
This versatility comes at the expense of much more complex acquisition methods, higher cost, and a greater need for technical support. The sensitivity to different tissue parameters requires different acquisition types, or “pulse sequences”, which involve different patterns of RF and gradients pulses. The cost, complexity, and maintenance of MRI scanners results in large scan costs for users (from hundreds to thousands of dollars per hour), which can be excessive for certain applications. The complexity of MRI methods also means that high level technical support is often needed, particularly for the use of advanced research-oriented methods. Nevertheless, MRI provides the capability of acquiring quantitative data on a remarkably wide range of structural and physiological tissue characteristics.
Structural imaging
The most commonly employed MRI scans are T1- and T2-weighted structural imaging sequences. These are the “workhorse” pulse sequences, capable of relatively high resolution (hundreds of microns in-plane resolution) and are relatively quick (1–10 minutes) depending on the size of the volume scanned. The main difference in contrast between these pulse sequences are T1-weighted imaging creates contrast in tissues with different microstructural properties and has high adipose tissue signal, and T2-weighted imaging provides high contrast between free and “bound” water. Structural MRI scans are useful for visualizing the state of TERM constructs in vivo as well as biocompatibility and biointegration of the construct. If the TERM construct is being degraded by the body, serial MRI scans can monitor changes in the size and shape of the construct. If the TERM construct is being rejected by the body, at acute timepoints, increased signal around the construct on T2-weighted, but not T1-weighted MRI may suggest inflammation is present. At chronic timepoints, a lack of signal around the TERM construct may be indicative of fibrous connective tissue encapsulation (Fig. 4). In cases where the signal arising from the TERM construct is similar to that of the surrounding tissue, contrast agents can be either systemically applied or directly conjugated to the construct.
Fig. 4.
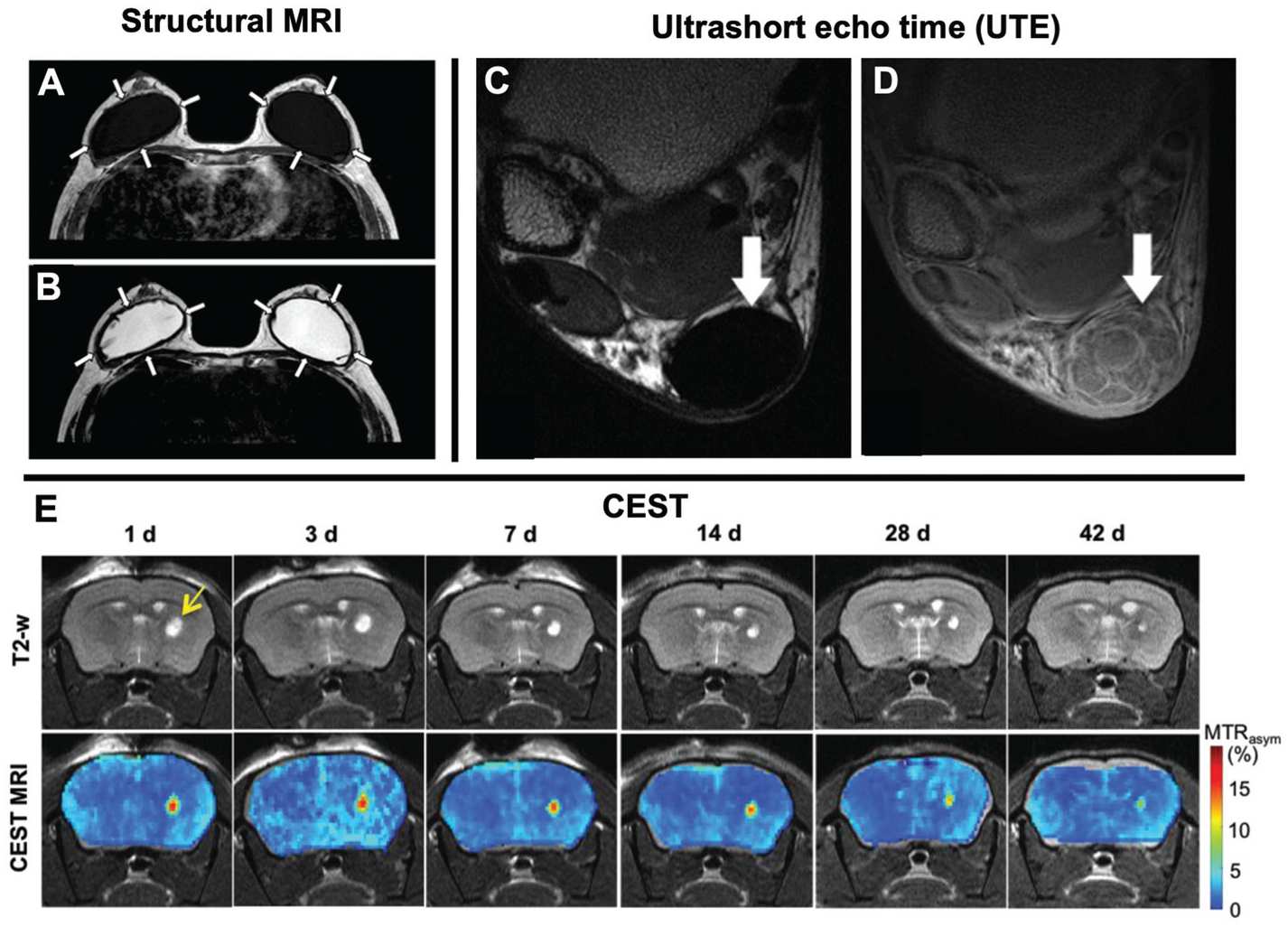
Structural MRI: T1-weighted (A) and T2-weighted (B) images of saline filled breast implants. Arrows highlight a thick, low signal fibrous capsule around the implant has formed. These images demonstrate how structural MRI can be used to visualize different features of TERM constructs (i.e. water rich regions) and its interaction with nearby tissues. Figure reproduced with permission from Springer Nature (2016).140 UTE: axial MRIs of a Achilles tendon repair. The internal structures of the Achilles tendon are not visible in a proton density weighted MRI (C; arrow), but are visible in a UTE pulse sequence (D; arrows). This demonstrates how UTE can be used to visualize fibrous structures with short echo times. Figure reproduced with permission from John Wiley and Sons (2015).106 CEST: Time course of a hydrogel injected into a mouse brain striatum over the course of 42 days (E). Serial T2-weighted imaging was used to identify the hydrogel (top row; arrow) which is easily identified due to its high water content. Relatively little change in the hydrogel size is observed over 42 days, even though the hydrogel is degrading, due to the large amount of unbound water in the hydrogel. Using CEST MRI, a continuous decrease in hydrogel signal was observed, consistent with hydrogel degeneration in vivo. This demonstrates how CEST can be used to monitor the presence of biomaterials and degradation of the construct better than routine structural imaging. Figure reproduced with permission from John Wiley and Sons (2019).93
Relaxation mapping
Structural imaging using T1- and T2-weighted imaging utilizes the variations in T1 and T2 but does not actually measure them. However, with modified pulse sequences it is possible to map the spatial distribution of T1 and T2 values, which is called relaxation mapping. Since T1 and T2 are intrinsic properties related to tissue microstructure, relaxation mapping can be a useful method for assessing biochemical properties associated with tissue composition. For example, T2-mapping provides quantitative spatial maps of tissue relaxation times – related to water content – which is a key component in hydrogel-based TERM constructs. This makes T2-mapping a valuable tool for assessing hydrogel-based TERM constructs, where decreased water content is associated with degeneration and diminished function.82,83 Furthermore, T2-relaxation can be used to quantitatively assess inflammation associated increases in water content around a TERM construct, associated with surgical implantation or incompatibility with the local environment.84,85 Another useful mapping technique, sensitive to proteoglycan concentration is T1ρ-mapping. Specifically, T1ρ is related to slow-motion interactions between water molecules that are hydrophilically attracted to their microenvironment.86 As GAG concentration is an important functional outcome measurement for articular cartilage and intervertebral disc TERM strategies, T1ρ-mapping can be used to quantify biological functionality and de novo tissue production for these tissues.87
Spectroscopy and CEST
Magnetic Resonance Spectroscopy (MRS) is the predecessor of modern day MRI. Different chemical species resonate at different frequencies, which can be used to distinguish them from one another. MRS can be used to assess metabolite concentration by imaging over a spectral range. Signal amplitudes at different frequencies can be mapped to specific metabolites, which can be used to provide quantitative information on the type and number of molecules in a TERM construct in vivo. This can be useful for measuring biological functionality of a TERM construct such as number of viable cells,88 as well as assessing products associated with degradation and regeneration.89,90 However MRS is usually only performed on a single voxel (~20mm3 on a clinical scanner), thus it does not provide detail about a constructs shape or size. Chemical exchange saturation transfer (CEST) MRI is a similar technique that allows for spatial quantification of interaction between water and molecules with exchangeable protons, such as amide and hydroxyl groups.91,92 CEST can be used to provide detailed quantitative information about the degeneration state of a construct, that can possibly be masked if that construct has similar magnetic properties to its adjacent tissues, such as hydrogels93 (Fig. 4). Furthermore, CEST has been used to provide functional assessment of the local microenvironment such as measuring pH94,95 or determining drug delivery kinetics from in vivo implanted TERM constructs.96
Perfusion
Vascularization of TERM constructs is a key outcome measure that is currently a major limiting factor in the development of large scale TERM implants. Therefore, there is a need to have accurate, quantitative tools to assess blood flow. Several MRI based tools exist including arterial spin labeling (ASL), dynamic susceptibility contrast (DSC), dynamic contrast enhanced (DCE), and intravoxel incoherent monition (IVIM) imaging, that are sensitive to changes in blood flow in a tissue. ASL is a perfusion technique that magnetically labels blood and measures the change in signal in a tissue of interest.97 DSC and DCE are perfusion techniques that rely on the use of exogenous contrast agents (normally gadolinium-based) that rely on changes in signal intensity in tissues of interest and have high spatial resolution. IVIM simultaneously assesses diffusion and pseudo-diffusion (collective movement of fluid due to blood flow in randomly oriented capillaries) properties of a tissue, but has the longest scan times and the lowest resolution.98,99 The decision to use a certain pulse sequence relies on the desired resolution, the use of contrast agents, scan time, and technical complications arising from labeling blood. Previously, neovascularization of TERM bone implants with different biomaterial compositions was evaluated with DCE perfusion MRI.100 DCE was able to detect perfusion changes associated with neovascularization associated with different biomaterial composition, which was confirmed by μCT and histological analysis. For detailed reviews of these perfusion MRI techniques, please see Jahng et al.,101 Essig et al.,102 and Le Bihan et al.103
Ultrashort echo time
While MRI is effective at assessing most soft tissues of the body, several tissues that have strong dipolar interactions which result in exceedingly short T2 values, their signal decays away too rapidly in the interval between the RF excitation and the data collection time (“echo time”, TE) to be “seen” by standard pulse sequences and thus making them essentially invisible in routine structural imaging. In order to make such tissues visible, it is necessary to shorten the TE in order to capture the signal before it decays away. Ultrashort echo time (UTE) pulse sequences are a specialized form of MRI that allows for increased contrast for tissues that are normally difficult to visualize including tendons, ligaments, meniscus, and cortical bone (Fig. 4).104–106 This makes UTE a potentially useful tool for assessing the state of TERM constructs like these and potentially increase sensitivity to detecting biointegration of TERM constructs due to its sensitivity to fibrotic, collagen rich tissues.107 UTE can be combined with other types of pulse sequences such as relaxation mapping in order to provide quantitative measurements of tissues with short T2 relaxation times.108,109 However, as UTE is a predominately research-focused pulse sequence, it may warrant designing TERM constructs to have a longer relaxation times in order to increase sensitivity.
Diffusion tensor imaging (DTI)
DTI is a form of MRI that measures the restricted diffusion of water in tissues, which is related to underlying microstructure. In particular, this technique is effective in evaluating tissues with anisotropic, organized microstructure, such as white matter, nerve, and muscle. The most reported outcome measurements from DTI are mean diffusivity (average overall diffusion) and fractional anisotropy (how anisotropic or restricted the diffusion profile is), which are sensitive to features of microstructure.110 In particular, mean diffusivity and fractional anisotropy are sensitive to cell size, inflammation, and cell permeability,111–114 making it a useful tool to assess organization and functionality of a TERM construct. Furthermore tractography – a post processing technique that allows for the assessment of macroscopic tissue properties such as fiber orientation and length (Fig. 5)115 – can be used to assess how well TERM constructs are integrating with the local microenvironment and alignment of tissue. By combining both the capacity for microstructural and macrostructural analysis of DTI, it is possible to assess how a TERM construct is integrating and functioning with the local tissues.116 For a review on techniques and applications of DTI, please see Oudeman et al.110
Fig. 5.
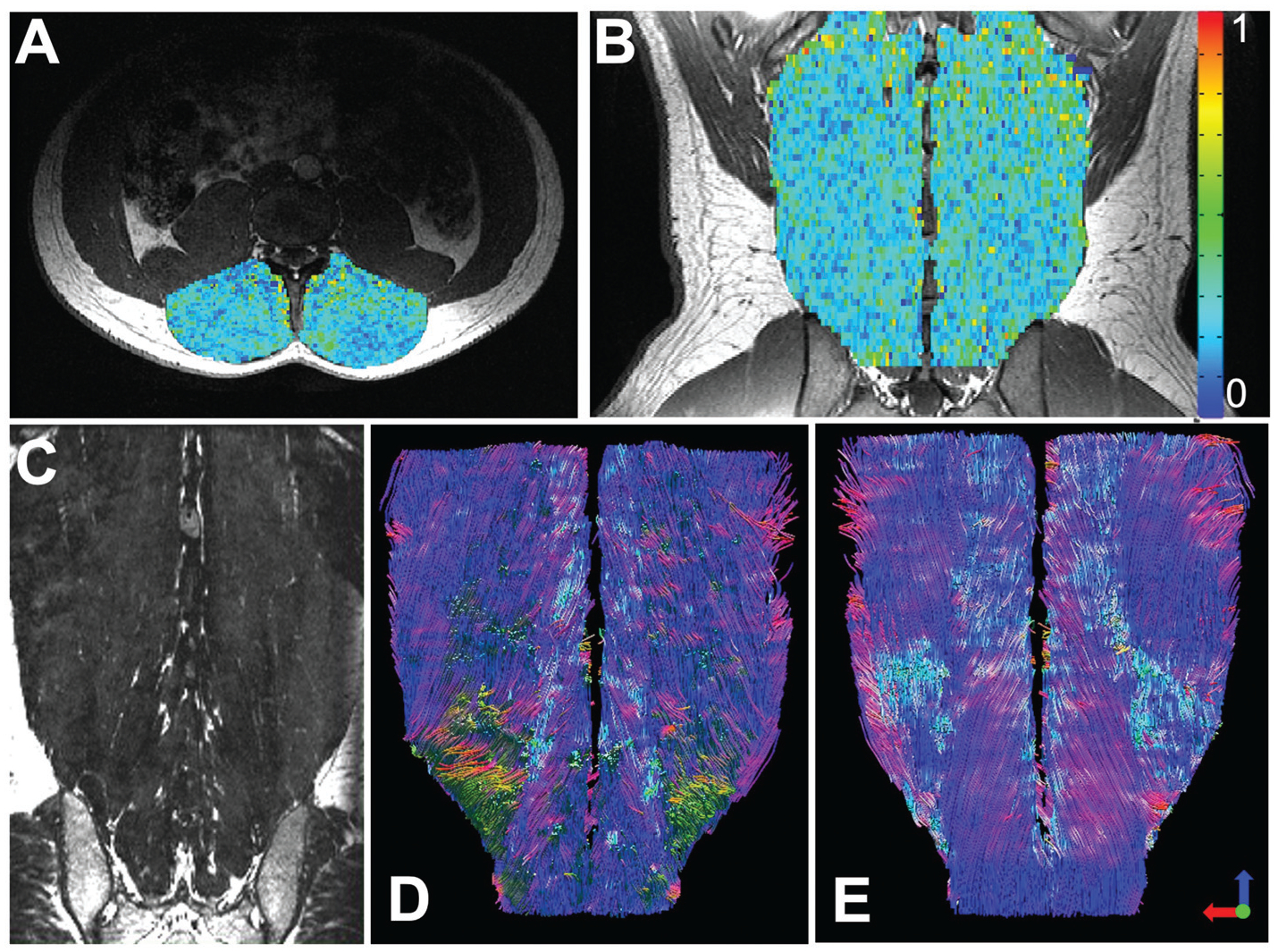
Diffusion tensor MRI: Axial (A) and coronal (B) fractional anisotropy maps overlaid on structural images of the lumbar paraspinal muscles, demonstrating the variance in tissue microstructural properties throughout a normal muscle. These maps can be used to assess microstructural organization of a TERM construct. In a coronal structural MRI scan (C) it is difficult to assess the 3D orientation of the paraspinal muscle fibers or assess fiber length. However, using tractography the orientation and length of the paraspinal muscles fibers can be measured. This technique can be used to assess how well a TERM construct is aligned and integrating with local tissue. Figure reproduced with permission from John Wiley and Sons (2020).115
Mechanical properties
Two main approaches can be taken in order to assess mechanical properties of a tissue in vivo with MRI: (1) magnetic resonance elastography (MRE) to assess shear modulus and (2) MRI tagging to assess tissue strain. Similar to ultrasound elastography, MRE utilizes small shear displacements on the surface of an object in order to map the shear modulus and viscosity of a tissue. MRE may be used to noninvasively estimate mechanical properties, which may be directly related to integrity and organization of a TERM construct. Generally, MRE is used in softer tissues, as the mechanical displacement for MRE of stiff tissues requires frequencies in the kHz range, which results in the dampening of shear waves. Furthermore, as sensitivity of MRE decreases with distance away from the mechanical source, MRE may not be an effective tool for measuring TERM construct properties that are deep in the body. MRE is predominantly a research-focused tool, but it provides a larger field of view and is more robust than ultrasound elastography. For more details on MRE, please see references.117,118
MRI tagging is the process of magnetically labeling grids or stripes on a tissue. As the tissue deforms due to active or passive motion, the lines will deform, allowing for the assessment of strain and strain rate of a tissue. This technique may be used to assess the mechanical function of TERM constructs such as cardiac patches or volumetric muscle loss scaffolds. MRI tagging is largely a research-focused tool, that requires post processing in order to calculate strain properties. Various forms of spatial tagging exist on MRI scanners such as SPAMM,119 DANTE,120 DENSE,121 and SENC.122 For a review on MRI tagging, please see the review by Chitiboi et al.123
Contrast agents
While most forms of 1H MRI use water as an endogenous contrast agent, several contrast agents for MRI have been developed in order to increase sensitivity to TERM construct state, biocompatibility, and function. Superparamagnetic iron oxide (SPIO) nanoparticles124 are a contrast agent that has been used to assess scaffold size,125 cell tracking,126,127 inflammation,128 and drug delivery126 in TERM constructs. SPIO nanoparticles decrease the overall signal in MR images. Therefore, if one were to serially image SPIO-containing scaffolds over time, an increase of signal intensity would be indicative of degeneration of the TERM construct. Furthermore, SPIO nanoparticles can be used to label cells via conjugation with antibodies or direct incubation in order to monitor specific cell viability, TERM construct function, and integration with the local tissue.126,128–130 Clinically, the most common MRI contrast agents are gadolinium-based, and are often directly injected systemically or into joints in order to increase signal contrast (shortens T1-relaxation time).131 Gadolinium-based contrast agents are less commonly incorporated into TERM constructs than SPIO nanoparticles. However, direct incorporation of gadolinium-based nanoparticles for bone TERM applications have been developed to enhance sensitivity to bone growth.132 For a comprehensive review on the use of contrast agents in MRI, please see the review by Wahsner and Gale et al.133
Robust analysis and interpretation of biomedical images
Image processing is an important technical component for getting high quality, quantitative data about a TERM construct. The standard image format for medical images is DICOM, which stands for Digital Imaging and Communications in Medicine. In addition to the image data itself, DICOM images often contain metadata in the form of headers, which are a list of information about the file typically containing information on the patient, study design, equipment used, and information relating to how the image was acquired.
A common step in image analysis is generating region of interest maps on the image itself, which can be done using a number of software packages. Several 2D and 3D image viewing and analysis platforms are available including ImageJ, Analyze, RadiAnt, Horos, Slicer, and PostDICOM. For a comparison of some common analysis packages, please see the review by Martin et al.134 Recent advances in AI and machine learning have led to the use of neural networks to automatically identify and mask regions of interest,135,136 however these software tools are still in the development phase and are not widely commercially available. Many of the research-oriented techniques outlined in Table 1 require substantial post-processing including filtering and fitting image signal data to mathematical models relating image signal to tissue parameters. This is often programmed using custom scripts using Matlab, Python, or R.
It is common to image a sample multiple times using different imaging techniques (multi-parametric imaging) or using different imaging modalities (multi-modal imaging) in order to assess multiple outcome measures of interest. This poses an challenging problem from a data analysis perspective as different imaging techniques, especially with multi-parametric imaging, have different limitations of resolution, which must be consolidated during image analysis. However, the best approach is to take into account differences in voxel size during image acquisition, and match or linearly scale voxel sizes between acquisitions. Also, an additional challenge during imaging processing is trying to match anatomic locations between scans (called image registration), which occurs when a patient moves in between scans, or during multi-modal imaging. Fortunately to overcome these issues, various image scaling and registration toolboxes have been developed to facilitate this aspect of imaging analysis.
There is a complicated relationship between the data measurements from which the images are derived and actual tissue parameters. This relationship can potentially be explored using in silico simulation, which can be used to optimize imaging parameters in order to maximize sensitivity or to help explain the relationship between imaging data and actual tissue structure.111 Furthermore, in vitro validation studies can be utilized in order to confirm these measurements reflect underlying tissue parameters of interest. For example, 3D printing has been used to fabricate MRI phantoms with ideal and histology informed muscle microstructure, in order to relate diffusion tensor imaging measurements to physical, tissue level microstructure (Fig. 6).137 Validation and optimization of imaging analysis and interpretation is vital to ensure physiologically relevant interpretation of quantitative measurements from medical imaging. For the most part, the research-focused imaging techniques reviewed are used sparingly in the clinical setting, as more rigorous validation is required to appropriately interpret these metrics. Therefore, as TERM constructs are being developed, it is necessary to innovate and validate novel imaging approaches that will facilitate accurate, noninvasive assessment of key outcome measures. If these tools are not developed in parallel, they risk not being approved by the FDA as appropriate, further complicating the translation of TERM constructs from benchtop to bedside. For a guide on requirements for using clinical imaging solutions in clinical trials, please see the Clinical Trial Imaging Endpoint Process Standards: Guidance for Industry.138
Fig. 6.
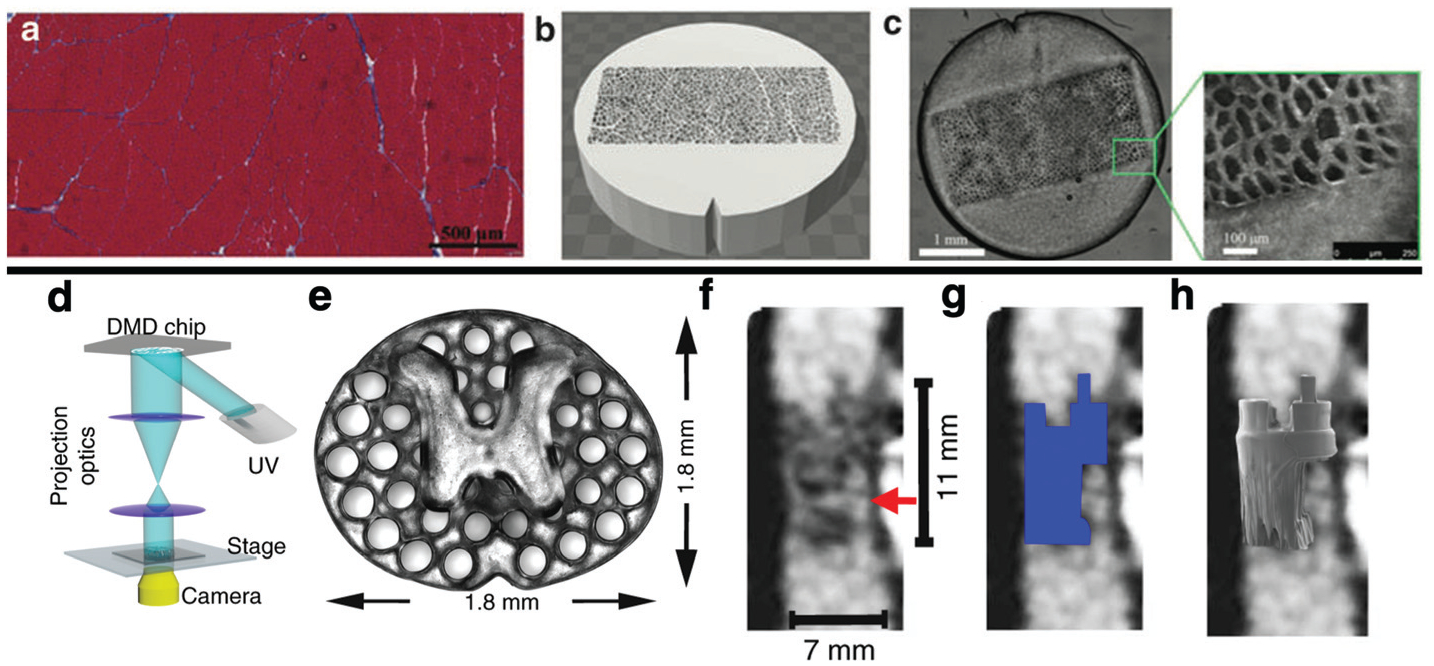
3D printing can be used to validate imaging techniques (Top). Histology of normal muscle (a) was used to inform the design of a phantom (b) with known geometric properties, which could be 3D printed (c) and scanned using MRI. This approach was used to relate measurements made using diffusion tensor MRI to known microstructural properties of the phantom.137 Biomedical imaging can also be used to inform the design of TERM constructs. A light based 3D printer (d) was used to print a scaffold (e) with x–y geometry informed by the axial distribution of white matter and grey matter in the spinal chord. Structural MRI of a complete spinal cord injury (f) can be used to inform the 3D geometry of the TERM scaffold (g), which can be precisely printed (h) to the precise dimensions of a patient’s lesion.139
Conclusions
Quantitative, sensitive imaging techniques are required for validating the state, integration, and functionality of TERM constructs in vivo. When designing TERM constructs, it is important to keep in mind the strengths and limitations of the imaging techniques available for specific outcome measures that pertain to that tissue. This may inform key decisions in construct design from the general shape of a construct to if a contrast agent needs to be directly incorporated to enhance sensitivity for a specific imaging modality.
In addition to being used to assess the aforementioned outcome measures of interest, imaging can be a valuable tool for designing patient specific TERM constructs. For example, Koffler and Zhu et al. recently utilized T1-weighted MRI of a patient’s spinal cord injury to develop a 3D model of the injury, from which a scaffold with the precise 3D geometry of the injury was 3D printed and implanted (Fig. 6).139 Studies such as this demonstrate how imaging techniques can be used in conjunction with the development novel of 3D biofabrication approaches, towards the ultimate goal of patient specific TERM constructs. Imaging can be used not only to inform the macroscale geometric properties (i.e. size) of a scaffold, but can potentially be used to inform other key design features of a TERM construct including microstructure, biological function, or mechanical properties. Thus, it is important to also understand how various medical imaging modalities can be used to inform construct design.
In order to exploit the strengths of multiple imaging modalities, multi-modal and multi-parametric imaging may provide enhanced sensitivity to specific outcome measures. These techniques, while requiring complicated registration and imaging analysis, can be used to take advantage of the strengths of different imaging modalities in order to provide the best quantitative information about a TERM construct. In order to ensure successful translation from benchtop to bedside, continued dialogue between the tissue engineering and the medical imaging communities is required in order to continue to innovate and rigorously validate approaches for highly accurate, sensitive, and quantitative assessment of TERM constructs.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Principles Of Tissue Engeniering, ed. Lanza R, Langer R, Vacanti J and Atala A, Elsevier, 5th edn, 2020. [Google Scholar]
- 2.Woodfield T, Lim K, Morouço P, Levato R, Malda J and Melchels F, in Comprehensive Biomaterials II, 2017, pp. 236–266. [Google Scholar]
- 3.Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ and Atala A, Nat. Biotechnol, 2016, 34, 312–319. [DOI] [PubMed] [Google Scholar]
- 4.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng GS, Sheikh F, Chien S and Chen S, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K and Chen S, Biomaterials, 2017, 124, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tappa K and Jammalamadaka U, J. Funct. Biomater, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, Ma X, Zhu W, Wang P, Miller KL, Stupin J, Koroleva-Maharajh A, Hairabedian A and Chen S, Biomaterials, 2019, 194, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Berry D, Moran A, He F, Tam T, Chen L and Chen S, Adv. Healthcare Mater, 2019, e1900977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butterfly iQ - Ultrasound, ultra-simplified., https://www.butterflynetwork.com/, (accessed 10 March 2020).
- 10.Ng A and Swanevelder J, Contin. Educ. Anaesthesia, Crit. Care Pain, 2011, 11, 186–192. [Google Scholar]
- 11.Fukunishi T, Best CA, Ong CS, Groehl T, Reinhardt J, Yi T, Miyachi H, Zhang H, Shinoka T, Breuer CK, Johnson J and Hibino N, Tissue Eng., Part A, 2018, 24, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukunishi T, Best CA, Sugiura T, Shoji T, Yi T, Udelsman B, Ohst D, Ong CS, Zhang H, Shinoka T, Breuer CK, Johnson J and Hibino N, PLoS One, 2016, 11(7), e0158555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao G, Kim H, Kim BS, Kong JS, Lee JY, Park BW, Chae S, Kim J, Ban K, Jang J, Park HJ and Cho DW, Appl. Phys. Rev, 2019, 6(4), 041402. [Google Scholar]
- 14.Solorio L, Babin BM, Patel RB, Mach J, Azar N and Exner AA, J. Controlled Release, 2010, 143, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice MA, Waters KR and Anseth KS, Acta Biomater, 2009, 5, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oe K, Miwa M, Nagamune K, Sakai Y, Lee SY, Niikura T, Iwakura T, Hasegawa T, Shibanuma N, Hata Y, Kuroda R and Kurosaka M, Tissue Eng., Part C, 2010, 16, 347–353. [DOI] [PubMed] [Google Scholar]
- 17.Mercado KP, Helguera M, Hocking DC and Dalecki D, Tissue Eng., Part C, 2015, 21, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uppal T and Mogra R, Australas. J. Ultrasound Med, 2010, 13, 32–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K and Wagner WR, Ann. Biomed. Eng, 2016, 176, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukunishi T, Ong CS, Yesantharao P, Best CA, Yi T, Zhang H, Mattson G, Boktor J, Nelson K, Shinoka T, Breuer CK, Johnson J and Hibino N, J. Tissue Eng. Regen. Med, 2019, 203–214. [DOI] [PubMed] [Google Scholar]
- 21.Wystrychowski W, McAllister TN, Zagalski K, Dusserre N, Cierpka L and L’Heureux N, J. Vasc. Surg, 2014, 60, 1353–1357. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, He Z, Li L, Liu G, Li Q, Yang D, Zhang Y and Li N, J. Cardiothorac. Surg, 2017, 12, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong CS, Fukunishi T, Liu RH, Nelson K, Zhang H, Wieczorek E, Palmieri M, Ueyama Y, Ferris E, Geist GE, Youngblood B, Johnson J and Hibino N, Tissue Eng., Part C, 2017, 23, 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey C, Cancer Imaging, 2015, 15, O19. [Google Scholar]
- 25.Unnikrishnan S and Klibanov AL, Am. J. Roentgenol, 2012, 199, 292–299. [DOI] [PubMed] [Google Scholar]
- 26.Xu DF, Qu GX, Yan SG and Cai XZ, Biomed Res. Int, 2018, 2018, 4606791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taljanovic MS, Gimber LH, Becker GW, Latt LD, Klauser AS, Melville DM, Gao L and Witte RS, Radiographics, 2017, 37, 855–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozturk A, Grajo JR, Dhyani M, Anthony BW and Samir AE, Abdom. Radiol, 2018, 43, 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K, Jeong CG and Hollister SJ, Acta Biomater, 2008, 4, 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong X, Annamalai RT, Kemerer TS, Deng CX and Stegemann JP, Biomaterials, 2018, 178, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng CX, Hong X and Stegemann JP, Tissue Eng., Part B, 2016, 22, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong X, Stegemann JP and Deng CX, Biomaterials, 2016, 88, 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Takanari K, Hong Y, Lee KW, Amoroso NJ, Wang Y, Wagner WR and Kim K, Biomaterials, 2013, 34, 2701–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barr RG, Ferraioli G, Palmeri ML, Goodman ZD, Garcia-Tsao G, Rubin J, Garra B, Myers RP, Wilson SR, Rubens D and Levine D, Radiology, 2015, 276, 845–861. [DOI] [PubMed] [Google Scholar]
- 35.Palmeri ML and Nightingale KR, Imaging Med, 2011, 3, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigrist RMS, Liau J, El Kaffas A, Chammas MC and Willmann JK, Theranostics, 2017, 7, 1303–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bushberg JT, Seibert JA, Leidholdt EM and Boone JM, The essential physics of medical imaging, Lippincott Williams & Wilkins, Philadelphia, 3rd edn, 2013. [Google Scholar]
- 38.Sladkova M, Cheng J, Palmer M, Chen S, Lin C, Xia W, Yu YE, Zhou B, Engqvist H and De Peppo GM, Tissue Eng., Part A, 2019, 25, 288–301. [DOI] [PubMed] [Google Scholar]
- 39.Mendes LF, Katagiri H, Tam WL, Chai YC, Geris L, Roberts SJ and Luyten FP, Stem Cell Res. Ther, 2018, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinibaldi R, Conti A, Sinjari B, Spadone S, Pecci R, Palombo M, Komlev VS, Ortore MG, Tromba G, Capuani S, Guidotti R, De Luca F, Caputi S, Traini T and Della Penna S, J. Tissue Eng. Regener. Med, 2018, 12, 750–761. [DOI] [PubMed] [Google Scholar]
- 41.Pearson JJ, Gerken N, Bae C, Lee KB, Satsangi A, McBride S, Appleford MR, Dean DD, Hollinger JO, Ong JL and Guda T, J. Biomed. Mater. Res. Part B Appl. Biomater, 2019, 1157–1166. [DOI] [PubMed] [Google Scholar]
- 42.Kopperdahl DL, Morgan EF and Keaveny TM, J. Orthop. Res, 2002, 20, 801–805. [DOI] [PubMed] [Google Scholar]
- 43.Unnikrishnan GU, Barest GD, Berry DB, Hussein AI and Morgan EF, J. Biomech. Eng, 2013, 135(10), 101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamezawa C, Numano T, Kawabata Y, Kanetaka H, Furuya M, Yokota K, Kato H, Yoneyama A, Hyodo K and Yashiro W, Appl. Phys. Express, 2020, 13(4), DOI: 10.35848/1882-0786/ab7e06. [DOI] [Google Scholar]
- 45.Cengiz IF, Oliveira JM and Reis RL, Biomater. Res, 2018, 22, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner ER, Parry J, Dadsetan M, Bravo D, Riester SM, Van Wijnen AJ, Yaszemski MJ and Kakar S, Connect. Tissue Res, 2018, 59, 542–549. [DOI] [PubMed] [Google Scholar]
- 47.Woloszyk A, Wolint P, Becker AS, Boss A, Fath W, Tian Y, Hoerstrup SP, Buschmann J and Emmert MY, Sci. Rep, 2019, 9, DOI: 10.1038/s41598-019-55411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Ketara S and Ford NL, Biomed. Phys. Eng. Express, 2020, 6, 035025. [DOI] [PubMed] [Google Scholar]
- 49.Karhula SS, Finnilä MA, Lammi MJ, Ylärinne JH, Kauppinen S, Rieppo L, Pritzker KPH, Nieminen HJ and Saarakkala S, PLoS One, 2019, 12(1), e0171075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin LH, Choi BH, Kim YJ, Oh HJ, Kim BJ, Yin XY and Min BH, Tissue Eng. Regen. Med, 2018, 15, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karhula SS, Finnilä MA, Freedman JD, Kauppinen S, Valkealahti M, Lehenkari P, Pritzker KPH, Nieminen HJ, Snyder BD, Grinstaff MW and Saarakkala S, Front. Phys, 2017, 5, 38. [Google Scholar]
- 52.Saukko AEA, Turunen MJ, Honkanen MKM, Lovric G, Tiitu V, Honkanen JTJ, Grinstaff MW, Jurvelin JS and Töyräs J, Sci. Rep, 2019, 9, 7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson BB, Stewart RC, Kawcak CE, Freedman JD, Patwa AN, Snyder BD, Goodrich LR and Grinstaff MW, Cartilage, 2018, DOI: 10.1177/1947603518812562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer AW, Guldberg RE and Levenston ME, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 19255–19260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coutu JM, Fatimi A, Berrahmoune S, Soulez G and Lerouge S, J. Biomed. Mater. Res. Part B Appl. Biomater, 2013, 101 B, 153–161. [DOI] [PubMed] [Google Scholar]
- 56.Lei K, Chen Y, Wang J, Peng X, Yu L and Ding J, Acta Biomater, 2017, 55, 396–409. [DOI] [PubMed] [Google Scholar]
- 57.Sandker MJ, Petit A, Redout EM, Siebelt M, Müller B, Bruin P, Meyboom R, Vermonden T, Hennink WE and Weinans H, Biomaterials, 2013, 34, 8002–8011. [DOI] [PubMed] [Google Scholar]
- 58.Ma Q, Lei K, Ding J, Yu L and Ding J, Polym. Chem, 2017, 8, 6665–6674. [Google Scholar]
- 59.Hong S, Carlson J, Lee H and Weissleder R, Adv. Healthcare Mater, 2016, 5, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei K, Ma Q, Yu L and Ding J, J. Mater. Chem. B, 2016, 4, 7793–7812. [DOI] [PubMed] [Google Scholar]
- 61.Hsu JC, Nieves LM, Betzer O, Sadan T, Noël PB, Popovtzer R and Cormode DP, WIREs Nanomed. Nanobiotechnol, 2020, DOI: 10.1002/wnan.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Bournonville S, Vangrunderbeeck S and Kerckhofs G, Contrast Media & Molecular Imaging, 2019, DOI: 10.1155/2019/8617406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shepherd DV, Shepherd JH, Best SM and Cameron RE, J. Mater. Sci. Mater. Med, 2018, 29, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meir R and Popovtzer R, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol, 2018, 10, e1480. [DOI] [PubMed] [Google Scholar]
- 65.Betzer O, Shwartz A, Motiei M, Kazimirsky G, Gispan I, Damti E, Brodie C, Yadid G and Popovtzer R, ACS Nano, 2014, 8, 9274–9285. [DOI] [PubMed] [Google Scholar]
- 66.Chhour P, Kim J, Benardo B, Tovar A, Mian S, Litt HI, Ferrari VA and Cormode DP, Bioconjugate Chem, 2017, 28, 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Betzer O, Meir R, Dreifuss T, Shamalov K, Motiei M, Shwartz A, Baranes K, Cohen CJ, Shraga-Heled N, Ofir R, Yadid G and Popovtzer R, Sci. Rep, 2015, 5, 15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meir R, Shamalov K, Betzer O, Motiei M, Horovitz-Fried M, Yehuda R, Popovtzer A, Popovtzer R and Cohen CJ, ACS Nano, 2015, 9, 6363–6372. [DOI] [PubMed] [Google Scholar]
- 69.Meir R, Betzer O, Motiei M, Kronfeld N, Brodie C and Popovtzer R, Nanomed. Nanotechnol. Biol. Med, 2017, 13, 421–429. [DOI] [PubMed] [Google Scholar]
- 70.Kim T, Lee N, Arifin DR, Shats I, Janowski M, Walczak P, Hyeon T and Bulte JWM, Adv. Funct. Mater, 2017, 27, 1604213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chhour P, Naha PC, O’Neill SM, Litt HI, Reilly MP, Ferrari VA and Cormode DP, Biomaterials, 2016, 87, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J, Chhour P, Hsu J, Litt HI, Ferrari VA, Popovtzer R and Cormode DP, Bioconjugate Chem, 2017, 28, 1581–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genant HK and Boyd D, Invest. Radiol, 1977, 12, 545–551. [DOI] [PubMed] [Google Scholar]
- 74.Chiro GD, Brooks RA, Kessler RM, Johnston GS, Jones AE, Herdt JR and Sheridan WT, Radiology, 1979, 131, 521–523. [DOI] [PubMed] [Google Scholar]
- 75.Millner MR, McDavid WD, Waggener RG, Dennis MJ, Payne WH and Sank VJ, Med. Phys, 1979, 6, 70–71. [DOI] [PubMed] [Google Scholar]
- 76.Flohr TG, McCollough CH, Bruder H, Petersilka M, Gruber K, Süss C, Grasruck M, Stierstorfer K, Krauss B, Raupach R, Primak AN, Küttner A, Achenbach S, Becker C, Kopp A and Ohnesorge BM, Eur. Radiol, 2006, 16, 256–268. [DOI] [PubMed] [Google Scholar]
- 77.Johnson TRC, AJR, Am. J. Roentgenol, 2012, 199, S3–S8. [DOI] [PubMed] [Google Scholar]
- 78.Si-Mohamed S, Bar-Ness D, Sigovan M, Tatard-Leitman V, Cormode DP, Naha PC, Coulon P, Rascle L, Roessl E, Rokni M, Altman A, Yagil Y, Boussel L and Douek P, Eur. Radiol. Exp, 2018, 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rajendran K, Löbker C, Schon BS, Bateman CJ, Younis RA, de Ruiter NJA, Chernoglazov AI, Ramyar M, Hooper GJ, Butler APH, Woodfield TBF and Anderson NG, Eur. Radiol, 2017, 27, 384–392. [DOI] [PubMed] [Google Scholar]
- 80.Cuccione E, Chhour P, Si-Mohamed S, Dumot C, Kim J, Hubert V, Crola C, Silva D, Vandamme M, Chereul E, Balegamire J, Chevalier Y, Berthezène Y, Boussel L, Douek P, Cormode DP and Wiart M, Nanotheranostics, 2020, 2020, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Badea CT, Clark DP, Holbrook M, Srivastava M, Mowery Y and Ghaghada KB, Phys. Med. Biol, 2019, 64, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ying J, Han Z, Zeng Y, Du Y, Pei S, Su L, Ruan D and Chen C, Am. J. Transl. Res, 2019, 11, 2028–2041. [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Z, Yan C, Yan S, Liu Q, Hou M, Xu Y and Guo R, Theranostics, 2018, 8, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Higgins CB, Herfkens R, Lipton MJ, Sievers R, Sheldon P, Kaufman L and Crooks LE, Am. J. Cardiol, 1983, 52, 184–188. [DOI] [PubMed] [Google Scholar]
- 85.Ferreira VM, Piechnik SK, Robson MD, Neubauer S and Karamitsos TD, in Journal of Thoracic Imaging, Wolters Kluwer Health, 2014, vol. 29, pp. 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y-XJ, Zhang Q, Li X, Chen W, Ahuja A and Yuan J, Quant. Imaging Med. Surg, 2015, 5, 858–85885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosenzweig DH, Fairag R, Mathieu AP, Li L, Eglin D, este M, Steffen T, Weber MH, Ouellet JA and Haglund L, Eur. Cells Mater, 2018, 36, 200–217. [DOI] [PubMed] [Google Scholar]
- 88.Mishra SK, Rana P, Khushu S and Gangenahalli G, Stem Cells Transl. Med, 2017, 6, 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stabler CL, Long RC, Sambanis A and Constantinidis I, Tissue Eng, 2005, 11, 404–414. [DOI] [PubMed] [Google Scholar]
- 90.Kotecha M, Klatt D and Magin RL, Tissue Eng., Part B, 2013, 19, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu B, Warnock G, Zaiss M, Lin C, Chen M, Zhou Z, Mu L, Nanz D, Tuura R and Delso G, EJNMMI Phys, 2016, 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vinogradov E, Sherry AD and Lenkinski RE, J. Magn. Reson, 2013, 229, 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu W, Chu C, Kuddannaya S, Yuan Y, Walczak P, Singh A, Song X and Bulte JWM, Adv. Funct. Mater, 2019, 29, 1903753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen M, Chen C, Shen Z, Zhang X, Chen Y, Lin F, Ma X, Zhuang C, Mao Y, Gan H, Chen P, Zong X and Wu R, Oncotarget, 2017, 8, 45759–45767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindeman LR, Randtke EA, High RA, Jones KM, Howison CM and Pagel MD, Magn. Reson. Med, 2018, 79, 2766–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han X, Huang J, To AKW, Lai JHC, Xiao P, Wu EX, Xu J and Chan KWY, Theranostics, 2020, 10, 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Detre JA, Leigh JS, Williams DS and Koretsky AP, Magn. Reson. Med, 1992, 23, 37–45. [DOI] [PubMed] [Google Scholar]
- 98.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E and Laval-Jeantet M, Radiology, 1986, 161, 401–407. [DOI] [PubMed] [Google Scholar]
- 99.Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J and Laval-Jeantet M, Radiology, 1988, 168, 497–505. [DOI] [PubMed] [Google Scholar]
- 100.Ribot EJ, Tournier C, Aid-Launais R, Koonjoo N, Oliveira H, Trotier AJ, Rey S, Wecker D, Letourneur D, Amedee Vilamitjana J and Miraux S, Sci. Rep, 2017, 7, 6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jahng GH, Li KL, Ostergaard L and Calamante F, Korean J. Radiol, 2014, 15, 554–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Essig M, Shiroishi MS, Nguyen TB, Saake M, Provenzale JM, Enterline D, Anzalone N, Dorfler A, Rovira À, Wintermark M and Law M, Am. J. Roentgenol, 2013, 200, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Le Bihan D, Neuroimage, 2019, 187, 56–67. [DOI] [PubMed] [Google Scholar]
- 104.Robson MD, Gatehouse PD, Bydder M and Bydder GM, J. Comput. Assist. Tomogr, 2003, 27, 825–846. [DOI] [PubMed] [Google Scholar]
- 105.Du J, Carl M, Diaz E, Takahashi A, Han E, Szeverenyi NM, Chung CB and Bydder GM, Magn. Reson. Med, 2010, 64, 834–842. [DOI] [PubMed] [Google Scholar]
- 106.Chang EY, Du J and Chung CB, J. Magn. Reson. Imaging, 2015, 41, 870–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Araujo ECA, Azzabou N, Vignaud A, Guillot G and Carlier PG, Magn. Reson. Med, 2017, 78, 997–1008. [DOI] [PubMed] [Google Scholar]
- 108.Williams A, Qian Y, Bear D and Chu CR, Osteoarthr. Cartil, 2010, 18, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan SJ, Wong J, Cheng X, Ma YJ, Chang EY, Du J and Shah SB, NMR Biomed, 2018, 31, e3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oudeman J, Nederveen AJ, Strijkers GJ, Maas M, Luijten PR and Froeling M, J. Magn. Reson. Imaging, 2016, 43, 773–788. [DOI] [PubMed] [Google Scholar]
- 111.Berry DB, Regner B, Galinsky V, Ward SR and Frank LR, Magn. Reson. Med, 2018, 80, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heemskerk AM, Drost MR, van Bochove GS, van Oosterhout MFM, Nicolay K and Strijkers GJ, Magn. Reson. Med, 2006, 56, 272–281. [DOI] [PubMed] [Google Scholar]
- 113.Sigmund EE, Novikov DS, Sui D, Ukpebor O, Baete S, Babb JS, Liu K, Feiweier T, Kwon J, McGorty K, Bencardino J and Fieremans E, NMR Biomed, 2014, 27, 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Winters KV, Reynaud O, Novikov DS, Fieremans E and Kim SG, Magn. Reson. Med, 2018, 80, 2094–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berry DB, Rodriguez–Soto AE, Englund EK, Shahidi B, Parra C, Frank LR, Kelly KR and Ward SR, JOR Spine, 2020, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu XY, Liang J, Wang Y, Zhong L, Zhao CY, Wei MG, Wang JJ, Sun XZ, Wang KQ, Duan JH, Chen C, Tu Y, Zhang S, Ming D and Li XH, J. Mater. Sci. Mater. Med, 2019, 30, 123. [DOI] [PubMed] [Google Scholar]
- 117.Tang A, Cloutier G, Szeverenyi NM and Sirlin CB, Am. J. Roentgenol, 2015, 205, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim W, Ferguson VL, Borden M and Neu CP, Ann. Biomed. Eng, 2016, 44, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Axel L and Dougherty L, Radiology, 1989, 171, 841–845. [DOI] [PubMed] [Google Scholar]
- 120.Mosher TJ and Smith MB, Magn. Reson. Med, 1990, 15, 334–339. [DOI] [PubMed] [Google Scholar]
- 121.Aletras AH, Ding S, Balaban RS and Wen H, J. Magn. Reson, 1999, 137, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Osman NF, Sampath S, Atalar E and Prince JL, Magn. Reson. Med, 2001, 46, 324–334. [DOI] [PubMed] [Google Scholar]
- 123.Chitiboi T and Axel L, J. Magn. Reson. Imaging, 2017, 46, 1263–1280. [DOI] [PubMed] [Google Scholar]
- 124.Bulte JWM and Kraitchman DL, NMR Biomed, 2004, 17, 484–499. [DOI] [PubMed] [Google Scholar]
- 125.Mahmoudi M, Zhao M, Matsuura Y, Laurent S, Yang PC, Bernstein D, Ruiz-Lozano P and Serpooshan V, BioImpacts, 2016, 6, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iyer SR, Xu S, Stains JP, Bennett CH and Lovering RM, Tissue Eng., Part B, 2017, 23, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, Bryant LH and Bulte JWM, Radiology, 2003, 228, 480–487. [DOI] [PubMed] [Google Scholar]
- 128.Chan JMS, Cheung MSH, Gibbs RGJ and Bhakoo KK, Clin. Transl. Med, 2017, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shevtsov MA, Nikolaev BP, Yakovleva LY, Marchenko YY, Dobrodumov AV, Mikhrina AL, Martynova MG, Bystrova OA, Yakovenko IV and Ischenko AM, Int. J. Nanomedicine, 2014, 9, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mesentier-Louro LA, Zaverucha-Do-Valle C, Da Silva AJ, Nascimento-Dos-Santos G, Gubert F, De Figueirêdo ABP, Torres AL, Paredes BD, Teixeira C, Tovar-Moll F, Mendez-Otero R and Santiago MF, PLoS One, 2014, 9, e110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clough TJ, Jiang L, Wong KL and Long NJ, Nat. Commun, 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mastrogiacomo S, Kownacka AE, Dou W, Burke BP, de Rosales RTM, Heerschap A, Jansen JA, Archibald SJ and Walboomers XF, Adv. Healthcare Mater, 2018, 7, 1800202. [DOI] [PubMed] [Google Scholar]
- 133.Wahsner J, Gale EM, Rodríguez-Rodríguez A and Caravan P, Chem. Rev, 2019, 119, 957–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martin CM, Roach VA, Nguyen N, Rice CL and Wilson TD, Anat. Sci. Educ, 2013, 6, 393–403. [DOI] [PubMed] [Google Scholar]
- 135.Akkus Z, Galimzianova A, Hoogi A, Rubin DL and Erickson BJ, J. Digit. Imaging, 2017, 30, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blackledge MD, Winfield JM, Miah A, Strauss D, Thway K, Morgan VA, Collins DJ, Koh DM, Leach MO and Messiou C, Front. Oncol, 2019, 9, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Berry DB, You S, Warner J, Frank LR, Chen S and Ward SR, Tissue Eng., Part A, 2017, 23, 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) and Center for Biologics Evaluation and Research (CBER), Clinical Trial Imaging Endpoint Process Standards Guidance for Industry Clinical/Medical Clinical Trial Imaging Endpoint Process Standards Guidance for Industry, 2018.
- 139.Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S and Tuszynski MH, Nat. Med, 2019, 25, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wong T, Lo LW, Fung PYE, Lai HYM, She HLH, Ng WKC, Kwok KMK and Lee CM, Insights Imaging, 2016, 7, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]


