Abstract
Unprocessed pro-Neuregulin 2 (pro-NRG2) accumulates on neuronal cell bodies at junctions between the endoplasmic reticulum and plasma membrane (ER-PM junctions). NMDA receptors (NMDARs) trigger NRG2 ectodomain shedding from these sites followed by activation of ErbB4 receptor tyrosine kinases, and ErbB4 signaling cell-autonomously downregulates intrinsic excitability of GABAergic interneurons by reducing voltage-gated sodium channel currents. NMDARs also promote dispersal of Kv2.1 clusters from ER-PM junctions and cause a hyperpolarizing shift in its voltage-dependent channel activation, suggesting that NRG2/ErbB4 and Kv2.1 work together to regulate intrinsic interneuron excitability in an activity-dependent manner. Here we explored the cellular processes underlying NMDAR-dependent NRG2 shedding in cultured rat hippocampal neurons. We report that NMDARs control shedding by two separate but converging mechanisms. First, NMDA treatment disrupts binding of pro-NRG2 to ER-PM junctions by post-translationally modifying conserved Ser/Thr residues in its intracellular domain. Second, using a mutant NRG2 protein that cannot be modified at these residues and that fails to accumulate at ER-PM junctions, we demonstrate that NMDARs also directly promote NRG2 shedding by ADAM-type matrix metalloproteinases. Using pharmacological and shRNA-mediated knock-down, and metalloproteinase overexpression, we unexpectedly find that ADAM10, but not ADAM17/TACE, is the major NRG2 sheddase acting downstream of NMDAR activation. Together, these findings reveal how NMDARs exert tight control over the NRG2/ErbB4 signaling pathway, and suggest that NRG2 and Kv2.1 are co-regulated components of a shared pathway that responds to elevated extracellular glutamate levels.
Keywords: Neuregulin, Kv2.1, ADAM10, ER-PM junction, sheddase, activity-dependent
INTRODUCTION
Neuregulins (NRG) are EGF-like ligands that signal via ErbB3/4 receptor tyrosine kinases. In the CNS, ErbB4 is the major neuronal NRG receptor while ErbB3 is expressed predominantly in glia [1]. The NRG/ErbB4 signaling pathway regulates a diverse array of processes in the developing and adult nervous system [2–5] and has been genetically associated with increased risk for schizophrenia [6,4,5,7]. Mouse genetic targeting studies of NRG1-NRG3 and the ErbB4 receptor have consistently shown that the NRG/ErbB4 signaling pathway regulates neuronal networks, critical period and synaptic plasticity, neurotransmitter pathways and behaviors relevant to traits affected in schizophrenia [8–23]. Most NRGs are translated as transmembrane proteins that undergo proteolytic processing, either to release their extracellular domains and to signal in paracrine or autocrine mode, or to remain membrane-attached and to signal in juxtacrine fashion [24–26]. NRG2 is a major ErbB receptor ligand in the postnatal brain [27,28] and is co-expressed with its neuronal receptor ErbB4 in cortical and hippocampal GABAergic interneurons where it signals in autocrine fashion [27,29]. In GABAergic interneurons, N-methyl-D-aspartate receptors (NMDARs) engage with NRG2/ErbB4 signaling in a negative feedback loop, with NMDAR activation promoting ectodomain shedding of pro-NRG2 and, in turn, ErbB4 receptors cell-autonomously downregulating surface expression and synaptic currents of GluN2B-containing NMDARs [29]. Similar effects of ErbB4 signaling on NMDAR currents in ErbB4+ GABAergic interneurons were recently reported using a NRG1 overexpressing mouse model [30]. Importantly, ErbB4 receptor activation also cell-autonomously reduces intrinsic interneuron excitability by downregulating voltage-gated sodium channels [31].
Contact sites between the endoplasmic reticulum and the plasma membrane (henceforth referred to as ER-PM junctions) are microdomains that regulate important and diverse cellular functions. For example, they are involved in lipid transfer via extended synaptotagmins and calcium entry via ORAI/STIM1 or junctophilins [32–34]. Neurons contain specialized ER-PM junctions characterized by large, flattened stacks of ER known as subsurface cisterns (SSCs) located on cell bodies and proximal dendrites [35]. Pro-NRG2 and pro-forms of single-pass transmembrane NRG1 isoforms accumulate at these SSC-type ER-PM junctions [36,29,37]. In cortical GABAergic interneurons, these pro-NRG2-containing ER-PM junctions are distinct from synaptic sites [29], whereas in the brain stem and spinal chord pro-NRG1 types I and II have been shown to accumulate at ER-PM junctions associated with C-boutons - large cholinergic synapses between V0C interneurons and α-motor neuron cell bodies [38,37]. Conversely, the dual-pass transmembrane pro-CRD-NRG1 and pro-NRG3 mostly target to axons and presynaptic terminals where they engage in forward and back-signaling with neuronal and glial targets [39–41,36,19]. At SSC-type neuronal ER-PM junctions, NRG2 co-localizes with micron-size clusters of the voltage-gated potassium channel Kv2.1, a predominant mediator of delayed rectifying K+ currents in neurons [42–44]. Like NRG2, Kv2.1 activity is dynamically regulated by NMDARs, which promotes Kv2.1 dispersal and results in a pronounced hyperpolarizing shift in voltage-dependent channel opening [45]. As both Kv2.1 and the NRG/ErbB4 signaling pathway are regulated by NMDARs and are involved in the control of intrinsic excitability, they conceivably represent separate components of a cellular homeostatic mechanism that downregulates intrinsic excitability during periods of high-frequency excitatory activity and that protects GABAergic interneurons during brief ischemic insults.
Here we explored the cellular processes underlying activity-dependent pro-NRG2 processing in cultured hippocampal neurons and report that NMDAR activation triggers rapid NRG2 shedding by two converging mechanisms, namely by downregulating pro-NRG2 clusters at ER-PM junctions and by directly promoting ADAM10-mediated ectodomain shedding. These findings enhance our understanding of the mechanistic basis underlying the functional link between NMDARs and the NRG2/ErbB4 signaling pathway in central neurons.
MATERIALS AND METHODS
Animals.
Sprague Dawley rats of either sex were used for tissue culture and protein biochemistry. Animals were treated in accordance with NIH Animal Welfare guidelines. All procedures were approved by the NICHD Animal Care and User Committee.
Antibodies.
NRG2: rabbit polyclonal antibody 7215 against the extracellular domain, used at 1–2 μg/ml; rabbit monoclonal antibody mAB11 against the ICD, used at 1 ug/ml; mouse monoclonal antibody 8D11 against the ECD [29]; Kv2.1: mouse monoclonal (clone K89/34; NeuroMab), used at 1 μg/ml; V5: mouse monoclonal (clone SV5-Pk1; Bio-Rad), used at 1–2 μg/ml; GFP: mouse monoclonal antibody (clone N86/6; NeuroMab), used at 1 ug/ml; ADAM10: rabbit monoclonal antibody (clone EPR5622; Abcam #124695), used at 1 μg/ml; ADAM17: rabbit polyclonal (Millipore/Sigma #19027), used at 1 μg/ml; MAP-2: guinea pig polyclonal antibody (Synaptic Systems #188004), used at 1:2,000; Tubulin: hFAB Rhodamine IgG (Bio-Rad #12004166), used at 1:5,000. Secondary antibodies conjugated to horseradish peroxidase for use in Western blotting were from Jackson Immunoresearch. Fluophore-conjugated secondary antibodies for immunofluorescence cytochemistry, including mouse IgG1- and IgG2a-selective antibodies used for co-labeling of NRG2 (with anti-V5) and Kv2.1 were from Thermo Fisher or Jackson Immunoresearch.
Drugs.
NMDA, GM6001, GI254023X, PMA, Cyclosporin A and Okadaic Acid were from Tocris. AP5, BACE-IV, Heparinase I-III enzymes and Complete™ protease inhibitor cocktail were from Millipore Sigma. Recombinant human NRG1β1 (EGF-like domain) was from R&D systems (HB-396).
Constructs.
All expression constructs were generated using Gateway cloning (Thermo Fisher) from open reading frames cloned into entry vectors pENTR223.1 (NRG2) or pENTR/D (NRG1, ADAM10 and ADAM17) that were transferred by recombination to Gateway destination vector pAAV(hSynI), a variant of pAAV_MCS (Agilent) driving cDNA expression from the human Synapsin I promoter. Human full-length NRG2 (IMAGE clone 100066341), modified to include a V5 epitope tag upstream of the EGF-like domain, served as the starting point to generate all NRG2 variants described in this study. Cleavage-resistant NRG2 was generated using a previously described strategy [36] by replacing a 840 bp SacI-KpnI restriction fragment with a synthetic DNA fragment (“gBlock“; Integrated DNA Technologies) that harbors the sequence encoding the extracellular juxtamembrane region of the ErbB4 isoform JM-b. Constructs NRG2_ΔCD and NRG2_8A were generated by replacing a 584 bp KpnI-AatII restriction fragment containing the C- and D-box sequences with a gBlock containing the ΔCD deletion or Ser/Thr -> Ala mutations (see also Fig. 5b). Carboxyl-terminally truncated crNRG2 variants P485* and E627* were generated by site-directed mutagenesis using QuikChange (Agilent). The c-tail variant of mouse Type II NRG1 was derived from the previously described corresponding a-tail variant in pENTR/D [36] by replacing a HindIII-EcoRV fragment with a gBlock that encodes the c-tail sequence. The open reading frame of rat ADAM10 was cloned by RT-PCR from E19 whole brain total RNA, and the rat ADAM17 cDNA was cloned from RNA isolated from adult lung tissue. For both ADAM10 and ADAM17, a V5 epitope tag was added at the very carboxyl-terminus. Potential target sequences for shRNA-mediated knockdown of rat ADAM10 and rat ADAM17 were identified using an online design tool available from Thermo Fisher (web address: https://rnaidesigner.thermofisher.com/rnaiexpress/). For each target, five high-scoring shRNA sequences were selected and cloned as double-stranded oligonucleotides between the EcoRI-BamHI sites of AAV vector pZacf (modified to co-express EGFP from the human Synapsin promoter). shRNAs were first tested by Western blotting of whole-cell lysates from 293 cells co-transfected with rat ADAM10 or ADAM17. This initial screen yielded highly effective shRNAs for ADAM10 (A10_349; target sequence: 5’-GGGTCTGTCGTTGATGGAAGA-3’) and for ADAM17 (A17_1358; target sequence: 5’- GCAGTAAACAGTCCATCTACA-3’); numbers indicate the position of the first nucleotide relative to the target open reading frame. These vectors were then packaged into AAV for transduction of cultured neurons (see below). The nontargeting control vector contained a sequence derived from the photinus pyralis luciferase gene. All constructs were confirmed by sequencing.
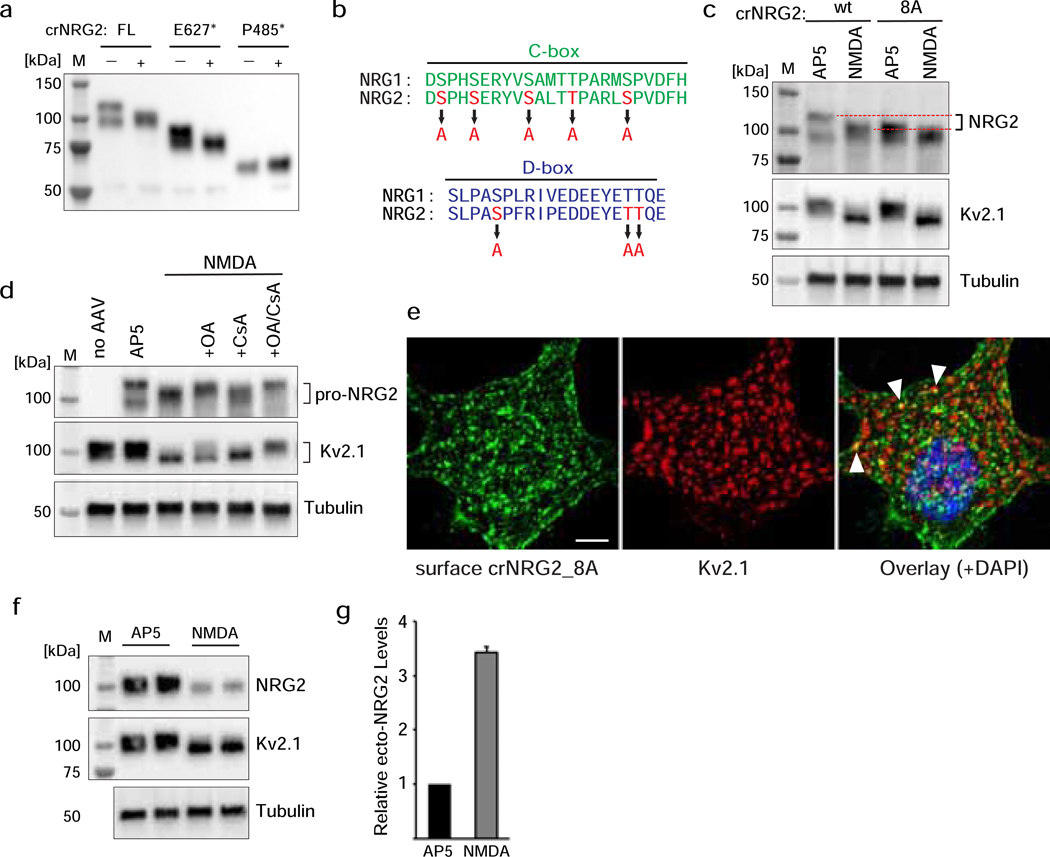
Fig. 5. Phosphorylation of conserved Ser/Thr residues in the NRG2 C/D-boxes regulate association with ER-PM junctions.
(a) Western blot of whole cell lysates from transduced hippocampal neurons illustrating increased electrophoretic mobility of full-length (FL) crNRG2 and crNRG2_E627*, but not crNRG2_P485*, after NMDAR stimulation with 20 μM glutamate for 20 min. (b) Conversion of Ser/Thr residues in the C- and D-boxes to Ala to generate constructs NRG2_8A and crNRG2_8A. (c) Western blot of crNRG2 vs. crNRG2_8A after 10-min treatment with AP5 or NMDAR. Note that the electrophoretic mobility of crNRG2_8A is increased compared to crNRG2 and similar to crNRG2 after NMDA treatment (red dotted lines). NMDA further increases the electrophoretic mobility of crNRG2_8A, albeit modestly. (d) Representative Western blot showing the effects of pre-treatment with Ser/Thr phosphatase inhibitors okadaic acid (500 nM; OA) or cyclosporin A (20 μM; CsA) on NMDA-mediated changes in crNRG2 (top) and Kv2.1 (middle) electrophoretic mobility. (e) Representative Z-projected confocal image of a cultured neuron expressing crNRG2_8A, surface-labeled with anti-V5 and then with anti-Kv2.1 following permeabilization. Arrowheads point to examples of some crNRG2_8A puncta that co-localize with Kv2.1, likely as a result of association with endogenous NRG2 puncta (see also Fig. S3). Scale bar = 5 μm. (f) Representative Western blot of whole cell lysates from transduced cultured neurons showing processing of pro-NRG2_8A in response to treatment with 50 μM NMDA (10 min). The panel includes Kv2.1 as positive control for the NMDA treatment and tubulin as loading control. (g) ELISA measurements of ecto-NRG2 in culture supernatants from NMDA-treated neurons expressing NRG2_8A. Data are normalized to AP5 controls and represent the mean ± SEM of 4 data points from 3 independent experiments.
Preparation of adeno-associated viruses (AAVs).
AAV-293 cells (Agilent), grown in DMEM medium with 10% FBS, were used for AAV production (serotype 1). Cells between passage 3–5 were seeded in 15 cm dishes and triple-transfected at 50–60% confluence with pHelper (Agilent), pAAV-RC1 (Cell Biolabs) and transfer vectors (see above). Medium was changed 2 h before transfection. For each dish, 15 μg DNA at equimolar ratios (6.6 μg pHelper, 4.2 ug pRC1 and 4.2 ug transfer vector) were first mixed with 60 μl polyethyleneimine (1 mg/ml) in 500 μl of DMEM medium (Thermo Fisher), incubated for 10 min and then added to the cells. Cells were fed 48 h and harvested 72 h after transfection by scraping into gradient buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris-Cl pH7.6). Cells were lysed by multiple freeze-thaw cycles, aided by repeated passages through a 23-gauge needle. Lysates were then incubated with benzonase (Millipore Sigma) for 1 h at 37°C and cleared by centrifugation (15 min at 4,000xg, 4°C). Supernatants were layered on top of an iodixanol step gradient (15, 25, 40 and 58% (v/v) in gradient buffer); the remaining volume was filled with gradient buffer. Ultracentrifugation was done at 48,000 rpm using a Type 70Ti fixed angle rotor (Beckman) for 2 h, 18°C. Viral particles were collected from the 40% iodixanol layer and stored at 4°C for immediate use or at −80°C for long-term storage. AAV preparations were titrated in cultured hippocampal neurons and used at dilutions that yielded widespread expression at moderate levels suitable for detection by Western blotting or low-to-moderate expression for immunocytochemistry and confocal microscopy.
Hippocampal cultures.
Dissociated hippocampal neurons were prepared from E19 Sprague-Dawley rat pups and propagated in defined Neurobasal/B27 medium (Thermo Fisher). Neurons were plated in 24-well plates, either directly on poly-D-lysine coated plastic for Western blotting, or on poly-D-lysine coated coverslips for immunofluorescence cytochemistry. Cultures were maintained in defined medium, and half of the medium was changed once every week. Cultures were transduced at DIV (days in vitro) 3 with AAVs for shRNAs and at DIV10 with AAVs for NRG or ADAM expression constructs. Shedding assays were performed in mature cultures (>DIV21) at least 2 days after medium change using the inner 8 wells of 24-well clusters to minimize well-to-well variability.
NRG2 shedding assay.
The NRG2 shedding assay was designed to simultaneously assess levels of pro-NRG2 protein by Western blotting of whole-cell lysates, and of ecto-NRG2 released into the cell culture medium by ELISA. Where necessary, neurons were pre-incubated for 10 min with AP5 or other inhibitors, as indicated. Because ecto-NRG2 is continuously released and accumulates in the conditioned culture supernatants over time, culture media was changed at the onset of the assay to ensure low baseline ecto-NRG2 levels in ELISA measurements. In preliminary experiments, we found that replacement of the conditioned medium with freshly prepared medium triggers unintended, albeit NMDAR-dependent, ectodomain shedding (Fig. S1). Washing neurons instead into pre-warmed conditioned supernatants from untransduced hippocampal neuron cultures largely eliminated this problem. Cultures were returned to the incubator for 10 min, unless noted otherwise. Culture supernatants were then collected for ELISA measurements of ecto-NRG2 levels, and neurons were quickly chilled by adding cold PBS, washed three times and directly lysed in 100 μl Western blotting sample buffer.
Western blotting.
Protein samples from cultured cells were size-fractionated on 4–15% Mini-Protean TGX precast gels (Bio-Rad) and electroblotted onto Nitrocellulose. Membranes were blocked with 3% BSA in Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBS/T), and incubated with primary antibodies in blocking solution for several hours at RT or overnight at 4°C. After several washes with TBS/T, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson Immunoresearch) for 1 h at RT. Signals were detected by chemiluminescence using a ChemiDoc MP imager (Bio-Rad) and quantified using Image Lab software (Bio-Rad). A rhodamine-conjugated anti-Tubulin antibody was used as a loading control.
ELISA measurements of ecto-NRG2 in conditioned cell culture supernatants.
An ELISA was developed to measure relative levels of ecto-NRG2 protein using anti-V5 for antigen capture and antibody 7215 against the extracellular domain of NRG2 for detection. Wells were coated for 1–2 h at RT with 1 μg anti-V5 antibody (clone SV5-Pk1; Bio-Rad), briefly washed in wash buffer (phosphate-buffered saline containing 0.1% Tween 20 and 1 mg/ml BSA) and then blocked for 1 hr with 10 mg/ml BSA in wash buffer. Culture supernatants were diluted 1:10 – 1:20 in wash buffer, added to wells (100 μl) and incubated overnight at 4°C. The next day, wells were washed four times with 300 μl ELISA wash buffer and then incubated for 1–2 h at RT with 1 μg/ml anti NRG2 antibody 7215 diluted in ELISA wash buffer. Wells were again washed and incubated for 1 h at RT with 1 μg/ml mouse monoclonal anti-rabbit IgG (Jackson Immunoresearch) and developed with the chromogenic substrate 3,3’,5,5’-Tetramethylbenzidine (TMB; Thermo Fisher). Absorption values were read at 405 nm using a Synergy H1 plate reader (Bio-Tek), with automatic background subtraction at 570 nm. Each assay included culture supernatant from untransduced cultures as blank controls. All samples were run as technical duplicates.
Confocal microscopy.
Neurons grown on 12-mm coverslips were fixed for 15 min with 4% paraformaldehyde (PFA) in PBS containing 4% sucrose. After extensive washing in PBS, coverslips were first blocked in PBS containing 10% normal goat serum (NGS) and then surface-labeled for 1 h at RT using anti-V5 (to detect NRG signals) in PBS containing 2% NGS. After washing with PBS, samples were permeabilized with 0.1% TX-100 in the presence of 10% NGS and then incubated with other primary and secondary antibodies as described above. Following 4–5 washes with PBS, neurons were incubated for 1 h at RT with fluophore-conjugated secondary antibodies in PBS containing 2% NGS and 0.1% TX-100 (Thermo Fisher or Jackson Immunoresearch; mouse IgG isotype-specific for anti-V5 [clone SV5-Pk1; IgG2a] and anti-Kv2.1 [clone K89/34; IgG1]). Following another 4–5 washes with PBS, coverslips were dipped in water and mounted in medium containing Mowiol and DABCO as antifade. Confocal Z-stack images of neuronal cell bodies and proximal dendrites were acquired at 2.5x digital zoom on a LSM780 confocal microscope (Zeiss) mounted with a 63x oil objective. Images were processed and analyzed using ImageJ/Fiji or ZEN software (Zeiss).
Morphometric analysis of crNRG2 puncta after NMDA treatment.
The effects of NMDAR stimulation on NRG2 clusters in the absence of proteolytic processing was assessed by measuring puncta size and number of crNRG2 puncta using ImageJ/Fiji. Following treatment with AP5 or NMDA, cultures were fixed and labeled for NRG2 using anti-V5 and for Kv2.1 as described above. Neurons were selected for imaging of NRG2 puncta on the basis of the subcellular distribution of their Kv2.1 immunoreactivity. In the AP5 condition, neurons with large and well-defined Kv2.1 clusters were chosen; in the NMDA condition, neurons with completely dispersed Kv2.1 immunoreactivity were chosen. The channel was then switched to NRG2, and Z-stacks were acquired under non-saturating conditions using the same laser intensity and gain across all neurons. Stacks were projected in Z and converted to binary images, and all identified objects larger than 0.05 μm2 were included in the analysis. Of note, earlier morphometric studies on endogenous NRG2 showed that NMDAR activation, in addition to reducing puncta size and number per cell, also reduces the number of ErbB4+ cells with NRG2 puncta [29]. However, as our AAV vectors do not contain a fluorescent reporter protein due to size limitations, we had no independent way of verifying successful transduction and therefore excluded neurons without NRG2 puncta from the analysis.
Colocalization between NRG variants and Kv2.1.
The colocalization module in the ZEN software was used to measure co-localization between NRG variants and Kv2.1 on a pixel-by-pixel basis. Images were acquired with minimal saturation in both NRG and Kv2.1 channels and fixed gain and laser intensities for the Kv2.1 channel across all samples. Cell bodies and proximal dendrites in Z-projected images were selected as regions of interest, and pixel intensity values were thresholded at 10 for NRGs and 80 for Kv2.1. Weighted colocalization coefficients were plotted to account for spatial as well as intensity correlations.
Statistics.
All statistical analyses were performed in Prism7 (GraphPad). 1-way ANOVA with Tukey’s multiple comparisons test was used for ELISA ectodomain shedding experiments, colocalization of NRG2 proteins with Kv2.1, and densitometric measurements of NRG2 shedding under ADAM10/17 overexpression conditions. An unpaired t-test was used to compare the effects of NMDA treatment on NRG2 puncta in the absence of ectodomain shedding (crNRG2 or wtNRG2 with GM6001 treatment). Error bars represent standard error of the mean (SEM).
RESULTS
NMDAR activation triggers rapid NRG2 processing in hippocampal neurons.
NRG2 ectodomain shedding can be assessed in cultured neurons by visualizing the proteolytic processing of pro-NRG2 in Western blots of whole-cell lysates, and measuring the released ecto-NRG2 in conditioned cell culture supernatants using ELISA. Levels of endogenous NRG2 protein in cultured neurons are below the limits of detection of these approaches, likely due to its restricted cellular expression in GABAergic interneurons that represent about ~15% of all neurons in typical hippocampal culture preparations, and to the sparse subcellular distribution of NRG2 [46,29]. As reported before, we employed AAV-mediated transduction of cultured neurons to overcome these limitations [29,36]. The high efficiency of viral gene delivery, together with low-to-moderate expression levels that results in the faithful targeting of pro-NRG2 to ER-PM junctions [36], enabled us to study the cellular processes underlying NMDAR-mediated NRG2 ectodomain shedding.
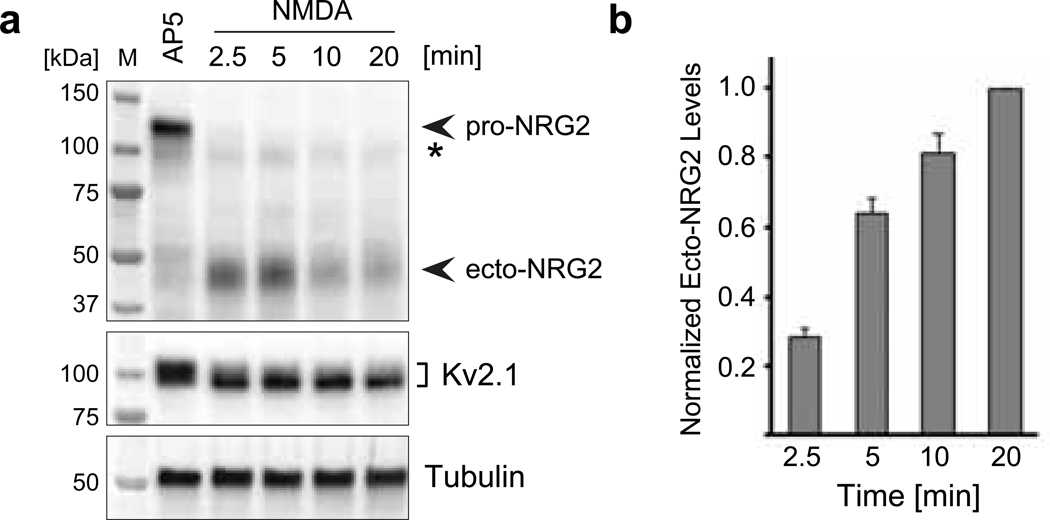
As shown in Fig. 1, NRG2 shedding in cultured hippocampal neurons in response to NMDA (50 μM) is remarkably fast and parallels the NMDAR-mediated increase in Kv2.1 electrophoretic mobility [45]. The bulk of pro-NRG2 was processed in as little as 2.5 min after the onset of treatment with 50 μM NMDA, and the ~120 kDa band was essentially undetectable after 10 mins. The resulting ecto-NRG2 fragment could be detected in the conditioned culture supernatants by ELISA and as a ~45 kDa band in the cellular fraction by Western blotting. Interestingly, there was a noticeable delay between the appearance of the processed ecto-NRG2 band in the Western blot and the emergence of ecto-NRG2 in the supernatant. Furthermore, the ecto-NRG2 band was strongest at early time points (2.5 and 5 min) and diminished afterwards, while ecto-NRG2 levels in the supernatants continuously increased throughout the duration of this assay (20 min). This temporal offset could stem from the transient binding of the ecto-NRG2 Ig-like domain to heparan sulfate proteoglycans in the extracellular matrix (ECM), or from binding of the NRG2 EGF-like motif to ErbB4 at 2.5 min and 10 min. We therefore tested whether accumulation of ecto-NRG2 in the cellular fraction is sensitive to pre-treatment with heparinase (to inhibit interactions with heparan sulfate proteoglycans) or with an excess of recombinant NRG1 EGF-like domain (to compete for ErbB receptor binding). We found that heparin digestion, but not competition for receptor binding, decreased the amount of ecto-NRG2 associated with the cellular fraction at 2.5 min at 10 min following NMDA treatment relative to untreated control cultures (Fig. S2). These findings indicate that the delayed accumulation of ecto-NRG2 in the supernatants upon pro-NRG2 processing is due to transient interactions with ECM, consistent with earlier studies on NRG1 showing that the Ig-like domain mediates interactions with the ECM, and that additional proteolytic processes mediate the release of its EGF-like domain from the ECM [47,48].
Fig. 1. Rapid NRG2 ectodomain shedding after NMDAR stimulation in cultured hippocampal neurons.
(a) Representative Western blot of whole-cell lysates prepared from AAV-transduced hippocampal neurons expressing wt NRG2 and treated with 100 μM AP5 or with 50 μM NMDA for the indicated times. The blot was probed with anti-NRG2 (Ab 7215) raised against its extracellular domain (top). It illustrates the rapid decrease of pro-NRG2 signals at ~120 kDa apparent molecular mass in response to NMDAR activation, and a concomitant increase in ecto-NRG2 levels at ~45 kDa apparent molecular mass. The weak band marked with an asterisk (*) is only detected in transduced neurons and likely reflects immature NRG2 protein. The blot was re-probed with anti-Kv2.1 to show the characteristic downward electrophoretic mobility shift of Kv2.1 protein in response to NMDAR stimulation (middle). Tubulin signals served as reference (bottom). (b) Corresponding ELISA measurements of ecto-NRG2 accumulation in conditioned cell culture supernatants demonstrating the parallel increase in levels of soluble ecto-NRG2. Data are normalized to NMDA at 20 min (by which time pro-NRG2 protein signals were consistently undetectable in Western blots) and represent the mean ± SEM from 3 independent experiments.
NMDAR activation promotes the downregulation of somatic pro-NRG2 puncta in the absence of ectodomain shedding
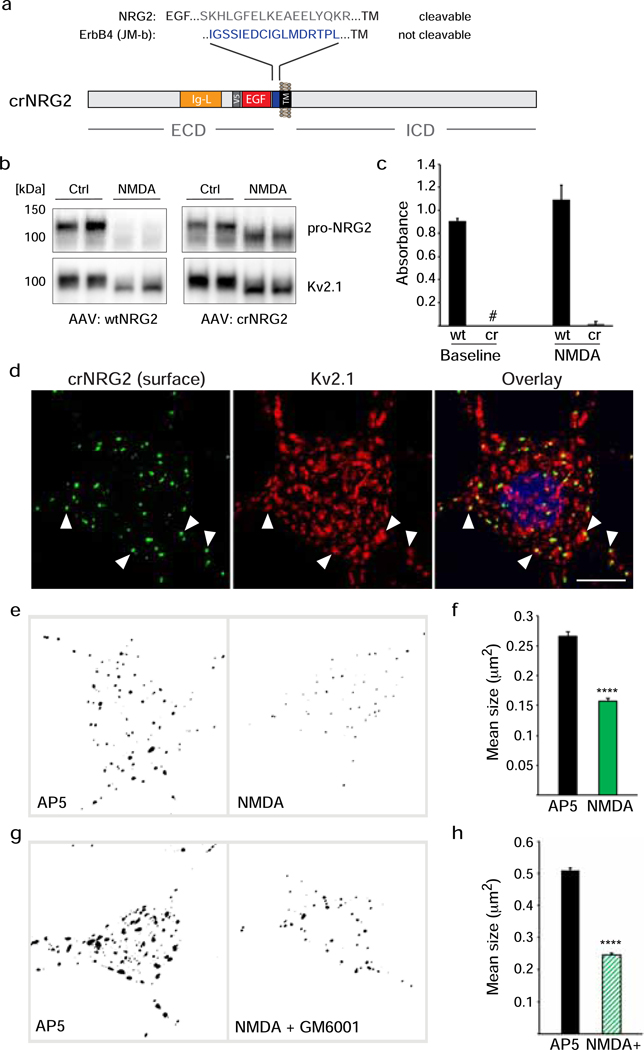
We considered two not mutually exclusive mechanisms by which NMDAR activity might regulate NRG2 shedding: 1) NMDARs could dissociate pro-NRG2 from neuronal SSC-type ER-PM junctions (henceforth simply referred to as ER-PM junctions) and thereby render the protein more accessible to sheddases, and 2) NMDARs could directly promote shedding by ADAM-type matrix metalloproteinases. To address the first possibility, we generated a cleavage-resistant NRG2 variant (crNRG2) by replacing its extracellular juxtamembrane sequence with a non-cleavable 17-amino acid linker (Fig. 2a), as described previously [36]. Western blot analysis of transduced hippocampal neurons confirmed that protein levels of crNRG2, unlike wtNRG2, were not diminished after a 10-min treatment with 50 μM NMDA (Fig. 2b). However, we observed an increase in the mobility of the crNRG2 band from ~120 kDa to ~100 kDa apparent molecular mass that paralleled a similar shift in the migration of the Kv2.1 protein; this finding will be explored in more detail further below. Furthermore, ecto-NRG2 was essentially undetectable in the conditioned cell culture supernatants from crNRG2-transduced neurons either at baseline (i.e., before treatment) or following NMDA treatment (Fig. 2c). We also confirmed by immunocytochemistry that rendering pro-NRG2 cleavage-resistant did not affect its accumulation at ER-PM junctions, as evidenced by its extensive colocalization with endogenous Kv2.1 clusters (Fig. 2d; colocalization co-efficient 0.563 ± 0.031; see also Fig. 3e).
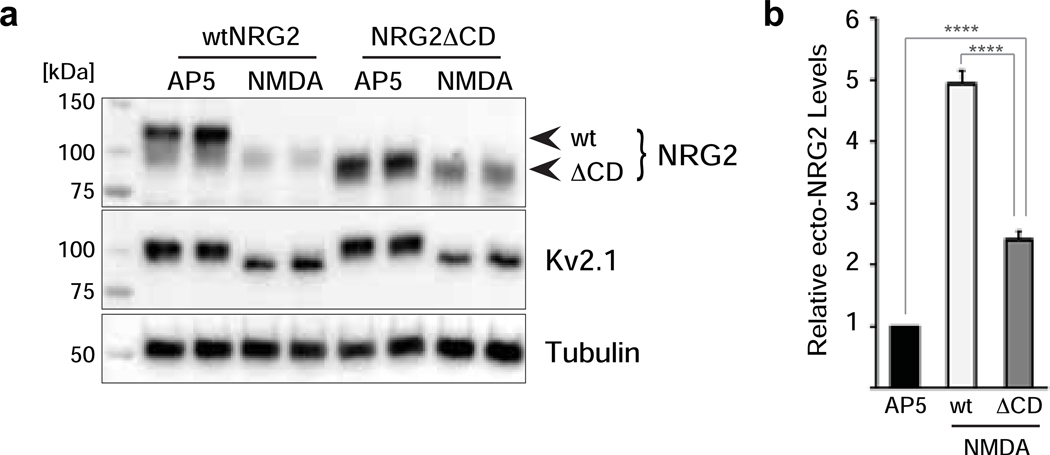
Fig. 2. Cleavage-resistant NRG2 reveals NMDAR-dependent downregulation of pro-NRG2 at ER-PM junctions in the absence of ectodomain shedding.
(a) Diagram of cleavage-resistant NRG2 (crNRG2), showing the replaced juxtamembrane extracellular sequence of wtNRG2 and the inserted non-cleavable juxtamembrane sequence of ErbB4 JM-b (see also [36]). Ig-like (Ig-L) and EGF-like domains are also indicated, as well as the location of a V5 epitope tag present in all NRG constructs described in this study that was used for ELISA and immunofluorescence cytochemistry. (b) Western blots of lysates prepared from hippocampal neurons expressing wtNRG2 (left) or crNRG2 (right), showing the effects of 10 min of 50 μM NMDA on pro-NRG2 protein levels. Note that NMDAR stimulation has little, if any, effect on crNRG2 protein levels (top) but causes a downward shift in its apparent electrophoretic mobility that parallels the shift in Kv2.1 mobility (below). (c) ELISA of ecto-NRG2 in the corresponding culture supernatants, demonstrating ectodomain shedding of wtNRG2 but not crNRG2. The (#) denotes that levels were below the limits of detection. Data represent the means of two biological replicates. (d) Representative confocal Z-projection of a hippocampal neuron expressing crNRG2 (surface-labeled with anti-V5), showing colocalization with endogenous Kv2.1 clusters. DAPI was included in the overlay image on the right. Arrowheads point to examples of closely associated signals for crNRG2 and Kv2.1. Scale bar = 10 μm (e) Representative binary images of crNRG2 puncta in hippocampal neurons following 10-min treatments with 100 μM AP5 (left) or 50 μM NMDA (right). (f) Quantitative analysis reveals reduction of mean crNRG2 puncta size after NMDAR stimulation (n=752 puncta from 11 neurons (AP5) and 397 puncta from 12 neurons (NMDA)). (g,h) Similar analysis of neurons expressing wtNRG2 and treated with AP5 or NMDA plus GM6001 (10 μM) (n=846 puncta from 12 neurons (AP5) and 566 puncta from 12 neurons (NMDA+GM6001)). ****, p<0.0001 (unpaired t-test).
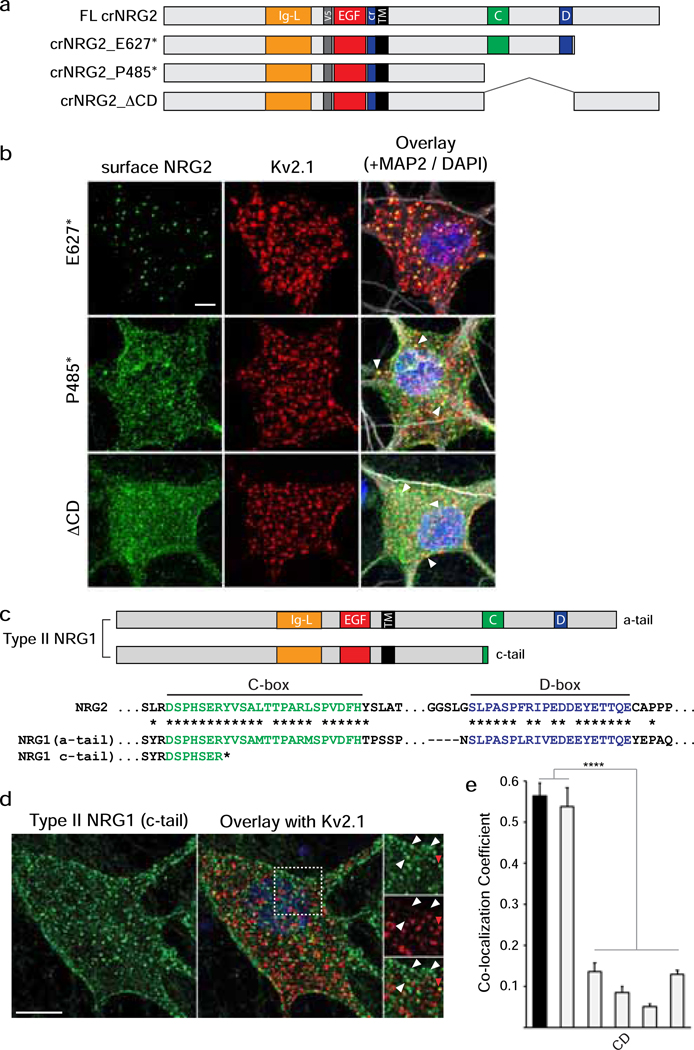
Fig. 3. NRG2 accumulates at ER-PM junctions via conserved sequence elements in its ICD.
(a) Schematic overview of NRG constructs used in this figure; motifs upstream of the TM are depicted as in Fig. 2a. Locations of conserved NRG2-ICD C/D-boxes are illustrated in green and blue, respectively. All deletions were introduced into crNRG2. (b) Representative Z-projected confocal images of neurons transduced with crNRG2_E627* (top), crNRG2_P485* (middle) or crNRG2_ΔCD (bottom). Cells were first surface-labeled with anti-V5 and subsequently labeled with anti-Kv2.1 and MAP-2 antibodies following permeabilization. Arrowheads in the crNRG2_E627* and crNRG2_P485* overlay images indicate puncta that overlap with Kv2.1 (see also Fig. S3). (c) Top, Schematic representation of a- and c-tail isoforms of type II NRG1. Note that c-tail NRG1 terminates upstream of the C-box. Bottom, Sequence comparison of the human NRG2 C- and D-boxes with the homologous sequences in the human NRG1 a-tail ICD. Conserved amino acids are marked by asterisks (*). The truncated c-tail is also shown. (d) Representative Z-projected confocal image of a transduced hippocampal neurons expressing type II NRG1 with the c-tail ICD and treated for 4 days with GM6001 (10 μM) and BACE-IV (1 μM) to block shedding. The image shows surface-labeled NRG1 using anti-V5 (left) and Kv2.1 labeled after permeabilization (right). The area marked by the bounding box in the overlay is magnified on the right and illustrates the lack of colocalization of most (white arrowheads) but not all (red arrowhead) NRG1 puncta with Kv2.1. Scale bar = 10 μM. (e) Quantitative analysis of colocalization of NRG2 variants and c-tail NRG1 with Kv2.1. Full-length crNRG2 (black) is included as reference. Data are plotted as weighted colocalization coefficients and represent the mean ± SEM (n=12 neurons per group). Note that the graph also includes data for the variant crNRG2_8A that is discussed in Fig. 5. ****, p<0.0001 (F (5,66) = 79.23; 1-way ANOVA with Tukey’s multiple comparison).
Next, we tested whether NMDAR stimulation causes the downregulation of crNRG2 puncta - independent of protein processing - by measuring their number and mean size in neurons treated for 10 min with 50 μM NMDA as compared to neurons treated with 100 μM AP5 to inhibit NMDARs. For technical reasons (see Methods), we had to limit our analysis to neurons in which crNRG2 puncta were still detectable after treatment, as opposed to neurons without puncta. Indeed, the number of crNRG2 puncta was lower in NMDA-treated neurons than in AP5-treated neurons (NMDA: 397 puncta [17–49/neuron] vs. AP5: 752 puncta; [25–116/neuron]; n=11 or 12 neurons), and their mean size was significantly smaller (NMDA: 0.157 μm2 ± 0.005 μm2 vs. AP5: 0.266 ± 0.008 μm2; Fig. 2e,f). Similar results were obtained in cultures transduced with wtNRG2 and treated with NMDA in the presence of the broad-spectrum matrix metalloproteinase inhibitor GM6001 to block shedding (10 μM; Fig. 2g,h). Similar to crNRG2, NMDAR activation reduced wtNRG2 puncta number (NMDA+GM6001: 566 puncta [range: 9–87/neuron] vs. AP5: 846 puncta [range: 11–173/neuron]) and mean size (NMDA+GM6001: 0.246 μm2 ± 0.009 μm2 vs. AP5: 0.509 ± 0.023 μm2; n=12 neurons). Since overall crNRG2 protein levels were not noticeably different between control and NMDA-treated cultures (see Fig. 2b), these findings strongly suggest that NMDAR stimulation reduces pro-NRG2 protein accumulation at ER-PM junctions independent of proteolytic processing.
NMDARs also directly promote NRG2 ectodomain shedding
To address whether NMDAR signaling stimulates pro-NRG2 shedding directly, we sought to generate a NRG2 variant that is deficient in targeting to ER-PM junctions. We began by introducing stop codons following proline 485 and glutamate 627 (Fig. 3a) which flank a region located in the intracellular domain (ICD) that harbors the “C” and “D” domains conserved between NRG2 and most NRG1 isoforms [28,26]. We reasoned that mis-targeted variants might be more susceptible to unregulated ectodomain shedding and therefore introduced the point mutations into crNRG2 to analyze their subcellular distribution by confocal microscopy. As shown in Fig. 3b (top), removal of the carboxyl-terminal 132 amino acids in crNRG2_E627* did not disrupt its targeting to ER-PM junctions, as evidenced by the presence of highly distinct puncta on cell bodies and dendrites that colocalized extensively with endogenous Kv2.1; its colocalization coefficient with Kv2.1 of 0.537±0.046 was very similar to full-length crNRG2 (0.563±0.031; p=0.979; Fig. 3e). By contrast, crNRG2_P485* was much more widely distributed throughout the somatodendritic domain (Fig. 3b; middle) and co-localized significantly less with Kv2.1 (0.136±0.021; p<0.0001; Fig. 3e). A third construct that lacked the region between P485 and E627 (crNRG2_ΔCD), but retained the C-terminal 132 amino acids (Fig. 3a and Fig. 3b; bottom), showed a similar distribution to crNRG2_P485* (colocalization coefficient: 0.148±0.013), suggesting that sequences between P485 and E627 that contain the C and D boxes are necessary for NRG2 accumulation at ER-PM junctions. Interestingly, although both crNRG2_P485* and crNRG2_ΔCD were clearly mis-targeted, we noticed that some punctate immunoreactivity overlapped with Kv2.1. Taking advantage of an antibody we generated previously against an epitope in the NRG2 ICD that is deleted in both NRG2 mutants [29], we found that these puncta colocalized with similar puncta of endogenous pro-NRG2, suggesting that they were indirectly targeted to ER-PM junctions by associating with endogenous pro-NRG2 clusters (Fig. S3). A similar result was obtained with a naturally occurring NRG1 (type II) variant harboring an alternative ICD sequence (“ c-tail”) that lacks the C- and D-boxes (Fig. 3c–d; [26]), which distributed broadly throughout the somatodendritic domain and failed to colocalize with Kv2.1 (colocalization coefficient: 0.051±0.007; Fig. 3e). Taken together, our results suggest that the region harboring the C- and D-boxes mediates interactions with ER-PM junctions in both NRG2 and single-pass NRG1 isoforms.
Next, we used a cleavable variant of NRG2_ΔCD to test whether NMDAR activation promotes NRG2 shedding in the absence of binding to ER-PM junctions. As shown in Fig. 4a, NMDA treatment (50 μM, 10 min) reduced pro-NRG2_ΔCD levels in whole-cell lysates as compared to controls in which NMDAR activity was blocked with 100 μM AP5, although the effect was not as complete as with wtNRG2. Since ectodomain shedding by ADAM-type metalloproteinases occurs primarily at the plasma membrane, we reasoned that the observed residual pro-NRG2_ΔCD signals after NMDA treatment might represent an intracellular pool of pro-NRG2_ΔCD that is unavailable for processing. We therefore analyzed surface vs intracellular expression of pro-NRG2_ΔCD by immunocytochemistry using anti-V5 under non-permeabilizing conditions followed by anti-NRG2 antibody 8D11 under permeabilizing conditions. As shown in the representative animated Z-stack available as Online Resource 1, we indeed found NRG2_ΔCD immunoreactivity on the plasma membrane as well as intracellularly. This is in stark contrast to endogenous wild-type NRG2 that gives rise to very little, if any, detectable immunoreactivity outside of its punctate accumulation at ER-PM junctions (Online Resource 2; [29]), supporting the idea that this intracellular pool represents the fraction of pro-NRG2_ΔCD that is resistant to NMDA treatment. It is currently unknown, however, to which extent intracellular accumulation of NRG2_ΔCD under steady-state conditions is a result of reduced surface delivery or increased internalization. Consistent with the Western blotting analysis of pro-NRG2_ΔCD processing, ELISA measurements revealed that shedding of ecto-NRG2_ΔCD into the culture supernatants in response to NMDA treatment increased by 2.4±0.16 fold relative to AP5-treated controls. By comparison, shedding of wtNRG2 in response to NMDAR activation was 4.93±0.23 fold over AP5-treated controls. These results show that NMDAR stimulation promotes NRG2 processing independently of its association with ER-PM junctions. Furthermore, the reduced size effect of NMDA treatment on NRG2_ΔCD ectodomain shedding suggests that NMDAR-mediated control over pro-NRG2 processing is due to promoting both pro-NRG2 dissociation from ER-PM junctions and proteolytic processing itself, consistent with the notion of two separate and converging mechanisms acting downstream of NMDAR activation.
Fig. 4. NMDAR activation promotes NRG2 shedding in the absence of ER-PM junction interactions.
(a) Stimulation with 50 μM NMDA for 10 min promotes processing of both wtNRG2 and, to a lesser extent, NRG2_ΔCD. Treatments were run in duplicates. Kv2.1 was included to show its electrophoretic mobility shift in response to NMDA treatment and tubulin signals served as an internal control. (b) ELISA measurements of ecto-NRG2 in culture supernatants from NMDA-treated neurons (50 μM) expressing wtNRG2 or NRG2_ΔCD, relative to their respective AP5-treated controls (100 μM). Values represent the mean ± SEM of 6 (wtNRG2) and 5 (NRG2_ΔCD) independent experiments. ****, p<0.0001 (F(2,16) = 242; 1-way ANOVA with Tukey’s multiple comparison).
Association with ER-PM junctions requires post-translational modification of conserved Ser/Thr residues located in the NRG2 C/D-box
We were intrigued by the apparent shift in electrophoretic mobility of crNRG2 that accompanied its dissociation from ER-PM junctions after NMDA treatment (see Fig. 2). This finding raised the possibility that similar mechanisms regulate the accumulation of pro-NRG2 and Kv2.1 at ER-PM junctions. To further test this hypothesis, we analyzed the mobility of crNRG2_E627* and crNRG2_P485* proteins in SDS-PAGE using cell lysates from AAV-transduced cultures subjected to NMDAR stimulation. As shown in Fig. 5a, we observed a downward shift for both full-length crNRG2 and crNRG2_E627* in SDS-PAGE, whereas the mobility of crNRG2_P485* was not affected. Together, these observations argued against the possibility that the change in apparent molecular mass was due to proteolysis near the carboxyl-terminus and instead suggested post-translational modification of residues within the NRG2 ICD region that includes the C- and D-boxes.
The shift in Kv2.1 electrophoretic mobility following NMDAR stimulation is caused by dephosphorylation of multiple serine residues located in its intracellular carboxyl-terminal tail [45,49]. Of note, the C- and D-boxes of NRG1 and NRG2 have five and four conserved Ser/Thr residues, respectively (Fig. 5b). A phosphorylation site prediction analysis using Netphos 3.1 revealed scores of 0.7 or higher for eight of them (max = 1). To test the potential role of phosphorylation of these residues in targeting of NRG2 to ER-PM junctions, we generated cleavable and cleavage-resistant mutants in which all of these Ser/Thr residues were converted to alanines (NRG2_8A; Fig. 5b). Western blot analysis revealed that, despite the almost identical molecular mass of their polypeptide chains, the electrophoretic mobility of crNRG2_8A was noticeably increased, with the protein running at ~105–110 kDa apparent molecular mass as compared to ~120 kDa for crNRG2. Interestingly, the electrophoretic mobility of crNRG2_8A was similar to the observed apparent molecular mass of crNRG2 following NMDAR stimulation (Fig. 5c). Furthermore, as shown in Fig. 5d, the shift was blocked by simultaneous inhibition of Ser/Thr phosphatases PP1/PP2A with okadaic acid (500 nM) and PP2B/calcineurin with cyclosporin A (20 μM).
We then analyzed the subcellular distribution of crNRG2_8A by confocal microscopy and found that the protein was widely distributed throughout the entire somatodendritic compartment and, for the most part, did not co-localize with Kv2.1 clusters (Fig. 5e), with a colocalization coefficient of 0.129±0.01 (see also Fig. 3e). Hence, the effect of either mutating these eight Ser/Thr residues or altogether removing NRG2 sequences between P485 and E627 on association of pro-NRG2 with ER-PM junctions was similar. Importantly, like NRG2_ΔCD (see Fig. 4), we found that proteolytic processing and ectodomain shedding of cleavable pro-NRG2_8A was augmented by NMDAR stimulation (Fig. 5f,g), consistent with the idea that NMDAR activity not only promotes dissociation of pro-NRG2 from ER-PM junctions but also directly stimulates ectodomain shedding.
NMDAR-dependent NRG2 shedding in neurons is mediated by ADAM10, and not by ADAM17/TACE
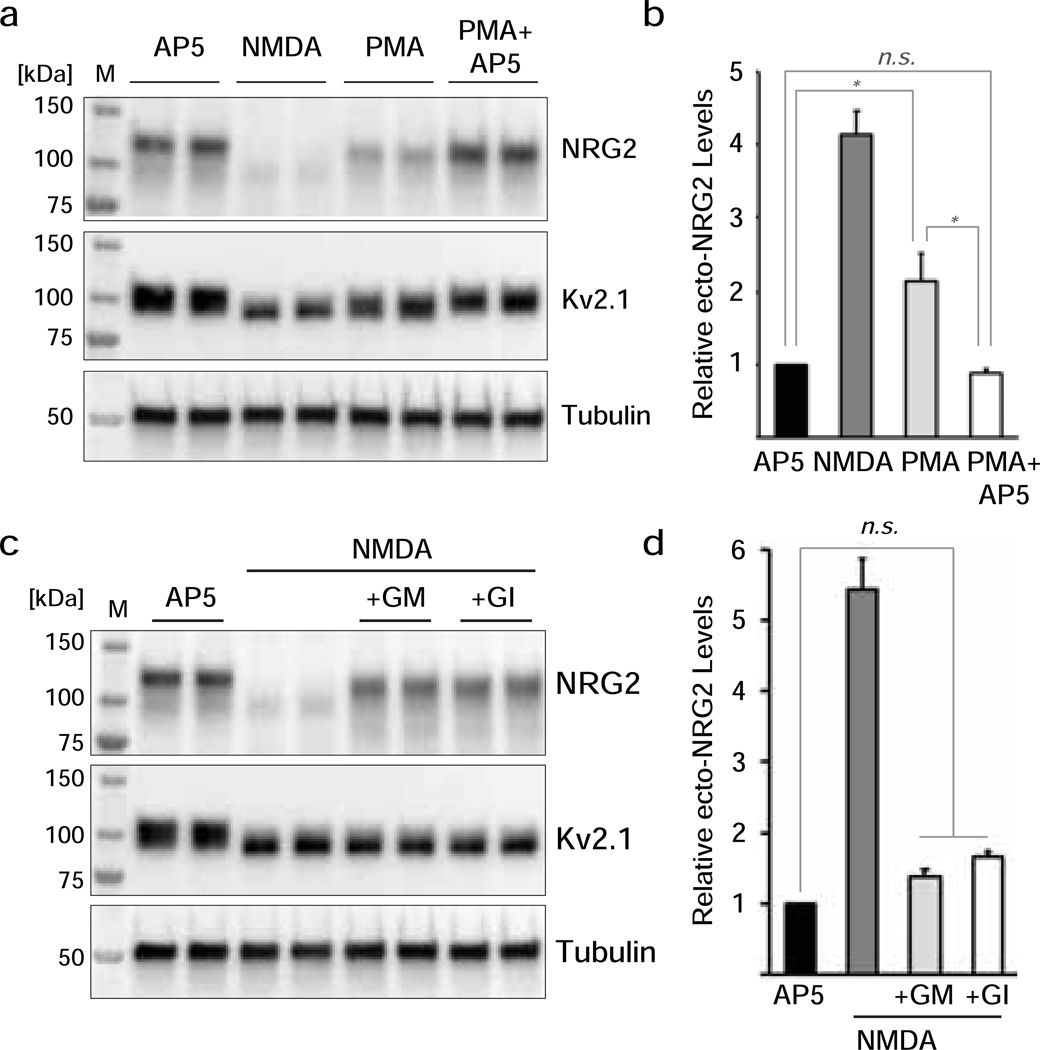
Next, we sought to identify the sheddase that mediates pro-NRG2 processing in neurons downstream of NMDAR activation. ADAM17/TACE (henceforth referred to as ADAM17) is considered an important processing enzyme for NRGs based on in-vitro and in-vivo studies on NRG1 [50–53]. PMA (phorbol 12-myristate-13-acetate) stimulation of protein kinase C has been used to promote shedding of type I NRG1 (ARIA) by ADAM17 from transfected CHO cells [24] and of type II NRG1 from immature (DIV7) cortical neurons [53]. We therefore asked whether PMA can stimulate pro-NRG2 processing downstream of NMDARs in our mature (>DIV21) hippocampal neuron culture system. As shown in Fig. 6a,b, a 10-min incubation with 0.2 μM PMA reduced pro-NRG2 levels in cell lysates and increased ecto-NRG2 levels in culture supernatants, although PMA was less potent than NMDA (PMA: 2.5±0.4 fold over AP5 control vs. NMDA: 4.2±0.4 fold over AP5 control). PMA-mediated shedding, however, was completely abolished when neurons were preincubated with AP5 to block NMDARs (PMA+AP5: 0.94±0.04). This result suggests that PMA augments NRG2 shedding indirectly by promoting NMDAR function, consistent with the well-documented effect of protein kinase C (PKC) activation on NMDAR trafficking and gating [54].
Fig. 6. NRG2 shedding by PMA requires NMDAR activity and the ADAM10 selective blocker GI254023X blocks NMDAR-mediated NRG2 shedding.
(a) Representative Western blot of whole cell lysates from transduced hippocampal neurons showing the effects of PMA (0.2 μM, 10 min), without or with 100 μM AP5 pre-treatment, on pro-NRG2 protein levels and electrophoretic mobility. AP5 and NMDA treatments are included as controls. (b) Corresponding ELISA measurements of ecto-NRG2 in culture supernatants treated as described in (a). Data are normalized to AP5 control and represent the mean ± SEM of 6 data points from 3 independent experiments. *, p<0.05; n.s., not significant (F(3,20) = 36.54; 1-way ANOVA with Tukey’s multiple comparison). (c) Representative Western blot of whole cell lysates from transduced hippocampal neurons showing inhibition of NMDAR-dependent pro-NRG2 processing in cultures that were pre-incubated with GM6001 (10 μM) or GI254023X (3 μM). (d) Corresponding ELISA measurements of ecto-NRG2 in culture supernatants treated as described in (d). Data are normalized to AP5 controls and represent the mean ± SEM of 6 data points from 3 independent experiments. n.s., not significant (F(3,20) = 85.27; 1-way ANOVA with Tukey’s multiple comparison).
ADAM10 is a major matrix metalloproteinase in the CNS but has received less attention as a sheddase for NRGs. Because ADAM10 has previously been implicated in NMDAR-mediated processing of other neuronal substrates [55,56], we investigated whether it also serves as an activity-dependent neuronal sheddase for pro-NRG2. We began by testing the effects of GI254023X, an inhibitor with 100-fold selectivity for ADAM10 (IC50: 5.3 μM) over ADAM17 (IC50: 541 μM) [57], on NMDAR-dependent NRG2 shedding. As shown in Fig. 6C, GI254023X (3 μM) blocked pro-NRG2 processing as potently as the broad-spectrum matrix metalloproteinase inhibitor GM6001 (10 μM). Interestingly, both compounds revealed a noticeable increase in electrophoretic mobility of the pro-NRG2 band after NMDAR stimulation, similar to results obtained with crNRG2 (see Fig. 2). This observation provides additional evidence for the existence of two separate mechanisms downstream of NMDAR activation that impinge on pro-NRG2. Likewise, GI254023X dramatically reduced NMDAR-dependent shedding of ecto-NRG2 into the culture medium (GI:1.62±0.08 vs NMDA: 4.81±0.64 fold over AP5 control), and comparable to GM6001 (1.4±0.12; Fig. 6d). Taken together, these pharmacological data suggest that ADAM10, but not ADAM17, acts as the major NRG2 sheddase downstream of NMDAR activation.
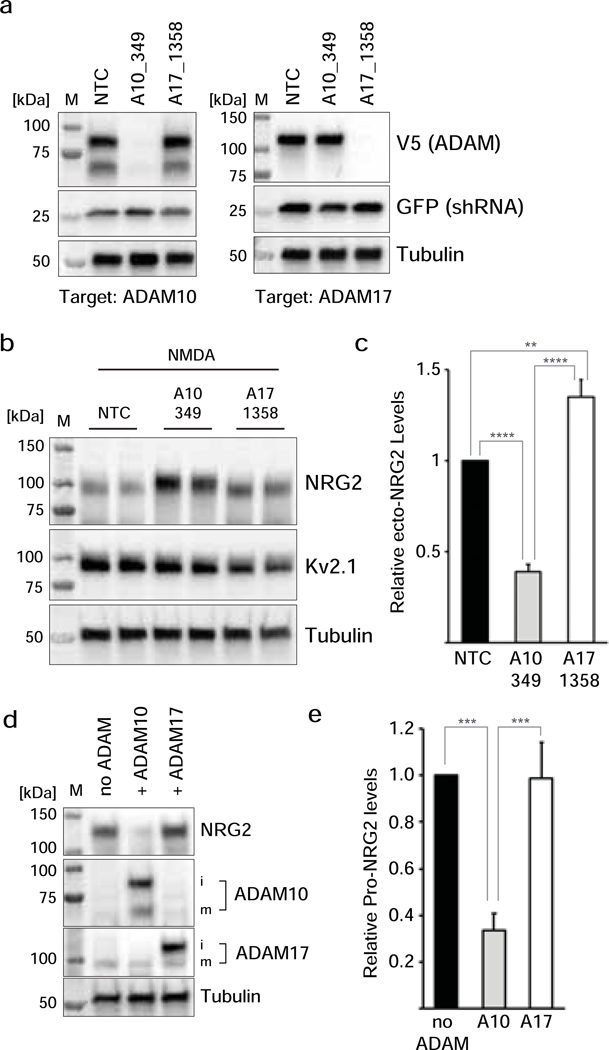
To further test the role of ADAM10 by an orthogonal genetic approach, we used shRNA-mediated knockdown of ADAM10 and ADAM17. As shown in Fig. 7a, we were able to generate shRNAs for both sheddases, termed A10_349 and A17_1358, that potently and selectively knocked down their respective targets. Cultured neurons were transduced with AAVs expressing wtNRG2 along with AAVs driving either a nontargeting shRNA (NTC), shRNA A10_349 or shRNA A17_1358, and then tested for pro-NRG2 processing and ecto-NRG2 levels in conditioned supernatants following a 10 min treatment with NMDA (50 μM). The results, shown in Fig. 7b,c, clearly demonstrate that knockdown of ADAM10 reduced NMDAR-dependent NRG2 processing and shedding of ecto-NRG2, whereas knockdown of ADAM17 moderately increased ecto-NRG2 levels (A10_349: 0.39±0.04 v. s A17_1358: 1.35±0.09; p<0.01). Because acute inhibition of ADAM10 by GI254023X almost abolished NMDAR-mediated NRG2 shedding, we propose that the residual release of ecto-NRG2 observed with A10_349 shRNA might be the result of incomplete ADAM10 knockdown and/or compensation by other ADAMs.
Fig. 7. ADAM10, but not ADAM17, mediates NRG2 shedding in response to NMDAR activation.
(a) Selective and potent shRNA-mediated knockdown of ADAM10 (shRNA A10_349) and of ADAM17 (shRNA A17_1358) in cultured hippocampal neurons. Neurons were transduced at DIV3 with AAVs driving a nontargeting control (NTC), shRNA A10_349 or shRNA A17_1358, and at DIV10 with AAVs expressing rat ADAM10 (left) or ADAM17 (right). (b,c) shRNA-mediated knockdown of ADAM10, but not of ADAM17, reduces NMDAR-mediated pro-NRG2 processing and ecto-NRG2 shedding. Values are normalized to the nontargeting control (NTC) and represent the mean ± SEM of 8 data points from 4 independent experiments. **, p<0.01; ****, p<0.0001 (F(2,21) = 67.14; 1-way ANOVA with Tukey’s multiple comparison). (d) AAV-driven cDNA expression of ADAM10, but not of ADAM17, in cultured hippocampal neurons decreases pro-NRG2 protein levels under baseline conditions (i.e., in the absence of exogenous NMDAR stimulation). Immature (i) and mature (m) forms of ADAM10 and ADAM17 are indicated. (e) Densitometric analysis of pro-NRG2 band intensities in control cultures and in cultures overexpressing ADAM10 or ADAM17. Data are normalized to controls and represent the mean ± SEM of 7 independent experiments. ***, p<0.001 (F(2,15) = 15.91; 1-way ANOVA with Tukey’s multiple comparison).
Because ADAM17 had been implicated in NRG1 processing in cultured cells and in the peripheral nervous system [52,51,53,58,59], we wanted to know if overexpression of ADAM17 could drive NRG2 processing in hippocampal neurons. We transduced neurons with wtNRG2 along with AAVs for ADAM10 or ADAM17, and analyzed pro-NRG2 protein levels at baseline (i.e., without NMDAR stimulation) by Western blotting. Consistent with our earlier results, we found that ADAM17 overexpression was without effect whereas ADAM10 reduced pro-NRG2 levels (ADAM10: 0.34±0.07 vs. ADAM17: 0.99±0.15; Fig. 7d,e). Taken together, our pharmacological, shRNA-mediated loss-of-function and ADAM over-expression experiments strongly suggest that ADAM10, rather than ADAM17, is the major neuronal sheddase processing pro-NRG2 in response to NMDAR activation.
DISCUSSION
The major finding of this study is that NMDAR activity triggers NRG2 shedding by two converging mechanisms. Firstly, NMDAR signaling downregulates pro-NRG2 clusters at ER-PM junctions by a process that involves dephosphorylation of conserved Ser/Thr residues in its ICD. Secondly, it directly promotes NRG2 shedding via ADAM10. Together, these results provide new insights into the mechanistic basis underlying the tight relationship between NMDAR activity and NRG2/ErbB4 signaling in GABAergic interneurons [29]. Furthermore, our findings adds NRG2 to the list of neuronal ADAM10 substrates whose shedding is augmented by NMDAR activity [55,56]; based on the extensive similarities in the subcellular distribution and NMDAR-dependent ectodomain shedding between NRG2 and single-pass NRG1 isoforms [36], we propose that type I and type II NRG1 isoforms are also processed by ADAM10 in neurons in response to NMDAR activation.
Activity-dependent modulation of Kv2.1 subcellular localization and channel activity is thought to homeostatically downregulate intrinsic excitability during periods of repetitive high-frequency firing and to protect neurons during brief ischemic insults, while strong oxidative stress conditions can trigger apoptotic Kv2.1-mediated K+ currents [60,45,61,62]. As NRG/ErbB4 signaling in GABAergic interneurons also regulates intrinsic excitability by downregulating voltage-gated sodium channels [31], we suggest that Kv2.1 and single-pass transmembrane NRGs are co-regulated components of a cellular mechanism that protects GABAergic interneurons from the metabolic stress associated with increased excitatory transmission and the excitotoxic effects of elevated extracellular glutamate levels. Consistent with the notion that NRGs dissociate from ER-PM junctions in response to cellular stress signals, it has recently been shown that NRG1 clusters at motor neuron C-bouton synapses are disrupted in response to acute stress or axotomy [38,63].
Rapid NRG2 shedding downstream of NMDAR activation.
NMDAR-mediated pro-NRG2 shedding in cultured neurons is remarkably fast, with the bulk of pro-NRG2 processed within 2.5 minutes. By comparison, shedding of type I NRG1/ARIA from CHO cells in response to PMA stimulation (presumably mediated by ADAM17) was reported to require hours, rather than minutes, to reach completion [24]. Our findings demonstrate that this potent NMDAR effect is the result of coordinated processes that liberate pro-NRG2 from ER-PM junctions and that promote its shedding following cleavage by ADAM10. However, while our experiments suggest that neuronal shedding of NRG2 and single-pass NRG1 isoforms in absence of NMDAR activity is low, they do not exclude the possibility that such slow constitutive shedding might occur in neurons independently of NMDAR activity.
NMDAR-mediated downregulation of pro-NRG2 and Kv2.1 at neuronal ER-PM junctions involves similar post-translational mechanisms.
We used a noncleavable variant of NRG2 to show that stimulation of NMDARs downregulates pro-NRG2 puncta in the absence of proteolytic processing and ectodomain shedding. This process is accompanied by an increase in its electrophoretic mobility from ~120 kDa to ~105 kDa, which is remarkably similar to the NMDAR-dependent shift in Kv2.1 electrophoretic mobility that results from dephosphorylation of multiple serine residues located in its carboxyl-terminal tail [64,65,49]. The shift in NRG2 electrophoretic mobility is also observed in detergent-solubilized rat cerebellar P2 membranes (not shown) and is blocked by Ser/Thr phosphatase inhibitors Okadaic acid and cyclosporin A. Importantly, this shift is not detected in NRG2 mutants that lack either the conserved C- and D-boxes or Ser/Thr residues located within these boxes, and neither mutant co-localizes with Kv2.1 at neuronal ER-PM junctions. These findings strongly suggest that, similar to Kv2.1, the accumulation of pro-NRG2 at ER-PM junctions depends on phosphorylation of at least some of the mutated Ser/Thr residues.
As this work was in preparation, two groups reported that Kv2.1 and Kv2.2 phosphorylation may regulate interactions with the ER vesicle-associated protein VAP by turning cryptic into functional FFAT sites that bind VAP proteins [66,67]. Like Kv2.1/Kv2.2, NRG2 lacks canonical FFAT sites in either the C- or D-boxes. Therefore, it will be interesting to investigate in future studies whether Ser/Thr phosphorylation can promote interactions between pro-NRG2 and VAP. However, it is worth noting that despite their similar subcellular distribution and regulation by NMDAR activity, pro-NRG2 and Kv2.1 are frequently found at non-overlapping sites within ER-PM junctions, with pro-NRG2 located at the center of doughnut-shaped Kv2.1 clusters (this study; [29]). Furthermore, endogenous pro-NRG2 only accumulates at a subset of Kv2.1-containing junctions (see, for example, Fig. 2). Therefore, it seems likely that additional factors regulate the accumulation of pro-NRG2 at ER-PM junctions. Interestingly, a recent ultrastructural study found that neuronal ER-PM junctions themselves are reversibly regulated by NMDAR activity [68]. Whether NMDAR-mediated downregulation of pro-NRG2 puncta at ER-PM junctions is caused by dephosphorylation, or if dephosphorylation indirectly results from the separation of SSCs from the plasma membrane, is an interesting question and should be addressed in future studies.
ADAM10 catalyzes NRG2 shedding in response to NMDAR activation.
Using a variety of pharmacological and genetic approaches, we have identified ADAM10, but not ADAM17, as a major NRG2 sheddase that functions downstream of NMDAR activation. This finding is consistent with the well-established role of ADAM10 in the processing of numerous neuronal substrates [69], and with the role of NMDAR activity in stimulating shedding of ADAM10 substrates nectin-1 and neuroligin-1 [55,56,69]. Interestingly, until this work, ADAM10 had received little attention as a NRG sheddase [70], unlike ADAM17 and BACE1 that were reported to play antagonistic roles in peripheral nerve myelination by processing type III NRG1 [71,51]. A recent study used PMA to implicate ADAM17 in the NMDAR-dependent shedding of type II NRG1 in neurons [53]. These findings need to be carefully evaluated considering the evidence presented here that PMA indirectly promotes NRG2 shedding by stimulating NMDAR activity (Fig. 6). Furthermore, the authors used TAPI-0 to inhibit shedding, but did not use either gene knockout or knockdown to selectively target ADAM10 and ADAM17 [53]. However, a prior study reported that TAPI-0 blocked shedding of the ErbB4 ligand betacellulin in cultured cells derived from ADAM17 mutant mice [72], suggesting that TAPI-0 is not selective for ADAM17. In our hands, PMA had a modest effect on NRG2 shedding. This, in principle, would be consistent with an involvement of PKC and would suggest that C1 kinase domain activation by phorbol esters is insufficient to recapitulate the full effects of endogenous second messengers. However, our finding that the PMA effect is blocked by AP5 strongly suggests that PMA exerts its effects via NMDARs, in agreement with the reported effects of PKC activation on NMDAR function [54].
How NMDARs promote ADAM10-mediated ectodomain shedding is not well understood. It is likely that the process involves Ca2+ influx through the NMDAR channel [55], consistent with other studies in cell lines showing that the calcium ionophore ionomycin augments ADAM10 shedding of numerous substrates [73,74]. However, neither the signaling pathway downstream of Ca2+ influx, nor the manner in which it regulates ADAM10 activity or its subcellular localization, has been identified. As ADAM10 is regulated at different levels, including its release from the ER where the bulk of the protein resides (Marcello et al., 2010), interactions with tetraspanins [75] and trafficking to plasma membranes by SAP-97 [76], it will be interesting to eventually delineate the cellular and molecular processes linking NMDAR and ADAM10 activity.
Relevance of co-regulation of NRG2 and Kv2.1 for neuroprotection, neural circuits and behavior
Until recently, neuronal NRG/ErbB signaling was thought to primarily occur across synapses or between axons and myelinating glia [5]. With the realization that many pro-NRGs (including pro-NRG2 and most single-pass transmembrane pro-NRG1 isoforms) accumulate at ER-PM junctions and are released in response to elevated extracellular glutamate levels [36,29,53], it appears reasonable to hypothesize that this mode of signaling is involved in the regulation of intrinsic excitability and mediation of the neuroprotective effects of NRG/ErbB signaling in cerebral ischemia (see, for example, [77–80]). Importantly, while Kv2.1 is broadly expressed throughout the CNS and regulates intrinsic excitability of many neuron types, including pyramidal neurons that lack ErbB4 [81], the combined effects of NMDAR signaling on sodium currents (via ErbB4) and potassium currents (via Kv2.1) might provide additional neuroprotection to GABAergic interneurons that are more resistant to ischemia than pyramidal neurons [82,83]. It should be noted, however, that it was previously reported that NRG1/ErbB4 signaling increased, rather than decreased, intrinsic excitability of parvalbumin (PV)-positive fast-spiking GABAergic interneurons in acute brain slices [84], although the same group failed to reproduce these findings in a subsequent study [85].
The physiological relevance of this signaling mode for normal neural circuit function is evident in NRG2 and ErbB4 mutant mice that show pronounced hyperactivity and other behavioral abnormalities [86,21]. As behavioral hyperactivity has also been reported in Kv2.1 deficient mice [87], it will be interesting to further explore how interactions between NRG2/ErbB4 signaling and Kv2.1 contribute to the homeostatic regulation of GABAergic neurons and potentially of neural circuit activity. Lastly, it will be important to dissect the relative contributions of axonal (type III NRG1 and NRG3) and somatic NRG isoforms (NRG2, types I and II NRG1) to the neuronal process reported to be regulated by NRG/ErbB4 signaling in the CNS, such as neuronal network synchrony [9,11–13,19,20] and visual cortical plasticity [16,17].
Supplementary Material
Acknowledgments and Support
We are grateful to Drs. Vincent Schram and Carolyn Smith from the Porter Neuroscience Center imaging core for expert assistance with confocal microscopy. This work was supported by the intramural research program of the Eunice Kennedy Shriver Institute of Child Health and Human Development (NICHD; ZIA- HD000711).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Gerecke KM, Wyss JM, Karavanova I, Buonanno A, Carroll SL (2001) ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J Comp Neurol 433 (1):86–100 [DOI] [PubMed] [Google Scholar]
- 2.Buonanno A, Fischbach GD (2001) Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol 11 (3):287–296. doi:S0959–4388(00)00210–5 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Birchmeier C, Nave KA (2008) Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia 56 (14):1491–1497. doi: 10.1002/glia.20753 [DOI] [PubMed] [Google Scholar]
- 4.Buonanno A (2010) The neuregulin signaling pathway and schizophrenia: from genes to synapses and neuralcircuits. Brain Res Bull 83 (3–4):122–131. doi:S0361–9230(10)00169–3 [pii] 10.1016/j.brainresbull.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mei L, Nave KA (2014) Neuregulin-ERBB Signaling in the Nervous System and Neuropsychiatric Diseases. Neuron 83 (1):27–49. doi: 10.1016/j.neuron.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mostaid MS, Lloyd D, Liberg B, Sundram S, Pereira A, Pantelis C, Karl T, Weickert CS, Everall IP, Bousman CA (2016) Neuregulin-1 and schizophrenia in the genome-wide association study era. Neurosci Biobehav Rev 68:387–409. doi: 10.1016/j.neubiorev.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 7.Chung DW, Volk DW, Arion D, Zhang Y, Sampson AR, Lewis DA (2016) Dysregulated ErbB4 Splicing in Schizophrenia: Selective Effects on Parvalbumin Expression. Am J Psychiatry 173 (1):60–68. doi: 10.1176/appi.ajp.2015.15020150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo RS, Li XM, Tao Y, Carpenter-Hyland E, Huang YZ, Weber J, Neiswender H, Dong XP, Wu J, Gassmann M, Lai C, Xiong WC, Gao TM, Mei L (2007) Neuregulin-1 enhances depolarization-induced GABA release. Neuron 54 (4):599–610. doi: 10.1016/j.neuron.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 9.Chen YJ, Johnson MA, Lieberman MD, Goodchild RE, Schobel S, Lewandowski N, Rosoklija G, Liu RC, Gingrich JA, Small S, Moore H, Dwork AJ, Talmage DA, Role LW (2008) Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci 28 (27):6872–6883. doi:28/27/6872 [pii] 10.1523/JNEUROSCI.1815-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon OB, Paredes D, Gonzalez CM, Neddens J, Hernandez L, Vullhorst D, Buonanno A (2008) Neuregulin-1 regulates LTP at CA1 hippocampal synapses through activation of dopamine D4 receptors. Proc Natl Acad Sci U S A 105 (40):15587–15592. doi:0805722105 [pii] 10.1073/pnas.0805722105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisahn A, Neddens J, Yan L, Buonanno A (2009) Neuregulin-1 Modulates Hippocampal Gamma Oscillations: Implications for Schizophrenia. Cereb Cortex 19 (3):612–618. doi: 10.1093/cercor/bhn107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson RH, Johnston A, Herman PA, Winzer-Serhan UH, Karavanova I, Vullhorst D, Fisahn A, Buonanno A (2012) Neuregulin and dopamine modulation of hippocampal gamma oscillations is dependent on dopamine D4 receptors. Proc Natl Acad Sci U S A 109 (32):13118–13123. doi:1201011109 [pii] 10.1073/pnas.1201011109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Pino I, Garcia-Frigola C, Dehorter N, Brotons-Mas JR, Alvarez-Salvado E, Martinez de Lagran M, Ciceri G,Gabaldon MV, Moratal D, Dierssen M, Canals S, Marin O, Rico B (2013) Erbb4 deletion from fast-spiking interneurons causes schizophrenia-like phenotypes. Neuron 79 (6):1152–1168. doi: 10.1016/j.neuron.2013.07.010 [DOI] [PubMed] [Google Scholar]
- 14.Yin DM, Chen YJ, Lu YS, Bean JC, Sathyamurthy A, Shen C, Liu X, Lin TW, Smith CA, Xiong WC, Mei L (2013) Reversal of behavioral deficits and synaptic dysfunction in mice overexpressing neuregulin 1. Neuron 78 (4):644–657. doi: 10.1016/j.neuron.2013.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loos M, Mueller T, Gouwenberg Y, Wijnands R, van der Loo RJ, Neuro BMPC, Birchmeier C, Smit AB, Spijker S (2014) Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry 76 (8):648–655. doi: 10.1016/j.biopsych.2014.02.011 [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Tran T, Murase S, Borrell A, Kirkwood A, Quinlan EM (2016) Neuregulin-Dependent Regulation of Fast-Spiking Interneuron Excitability Controls the Timing of the Critical Period. J Neurosci 36 (40):10285–10295. doi: 10.1523/JNEUROSCI.4242-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Ikrar T, Davis MF, Gong N, Zheng X, Luo ZD, Lai C, Mei L, Holmes TC, Gandhi SP, Xu X (2016) Neuregulin-1/ErbB4 Signaling Regulates Visual Cortical Plasticity. Neuron 92 (1):160–173. doi: 10.1016/j.neuron.2016.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong C, Akmentin W, Du C, Role LW, Talmage DA (2017) Axonal Type III Nrg1 Controls Glutamate Synapse Formation and GluA2 Trafficking in Hippocampal-Accumbens Connections. eNeuro 4 (1). doi: 10.1523/ENEURO.0232-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller T, Braud S, Juttner R, Voigt BC, Paulick K, Sheean ME, Klisch C, Gueneykaya D, Rathjen FG, Geiger JR, Poulet JF, Birchmeier C (2018) Neuregulin 3 promotes excitatory synapse formation on hippocampal interneurons. EMBO J 37 (17). doi: 10.15252/embj.201798858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Z, Robinson HL, Yin DM, Liu Y, Liu F, Wang H, Lin TW, Xing G, Gan L, Xiong WC, Mei L (2018) Dynamic ErbB4 Activity in Hippocampal-Prefrontal Synchrony and Top-Down Attention in Rodents. Neuron 98 (2):380–393. doi: 10.1016/j.neuron.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L, Shamir A, Skirzewski M, Leiva-Salcedo E, Kwon OB, Karavanova I, Paredes D, Malkesman O, Bailey KR, Vullhorst D, Crawley JN, Buonanno A (2018) Neuregulin-2 ablation results in dopamine dysregulation and severe behavioral phenotypes relevant to psychiatric disorders. Mol Psychiatry 23 (5):1233–1243. doi: 10.1038/mp.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang JM, Shen CJ, Chen XJ, Kong Y, Liu YS, Li XW, Chen Z, Gao TM, Li XM (2018) erbb4 Deficits in Chandelier Cells of the Medial Prefrontal Cortex Confer Cognitive Dysfunctions: Implications for Schizophrenia. Cereb Cortex. doi: 10.1093/cercor/bhy316 [DOI] [PubMed] [Google Scholar]
- 23.Deakin IH, Godlewska BR, Walker MA, Huang GJ, Schwab MH, Nave KA, Law AJ, Harrison PJ (2018) Altered hippocampal gene expression and structure in transgenic mice overexpressing neuregulin 1 (Nrg1) type I. Transl Psychiatry 8 (1):229. doi: 10.1038/s41398-018-0288-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb JA, Susanto ET, Fischbach GD (1998) The neuregulin precursor proARIA is processed to ARIA after expression on the cell surface by a protein kinase C-enhanced mechanism. Mol Cell Neurosci 11 (1–2):77–91. doi: 10.1006/mcne.1998.0676 [DOI] [PubMed] [Google Scholar]
- 25.Wang JY, Miller SJ, Falls DL (2001) The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem 276 (4):2841–2851. doi: 10.1074/jbc.M005700200 [DOI] [PubMed] [Google Scholar]
- 26.Falls DL (2003) Neuregulins: functions, forms, and signaling strategies. Experimental cell research 284 (1):14–30 [DOI] [PubMed] [Google Scholar]
- 27.Longart M, Liu Y, Karavanova I, Buonanno A (2004) Neuregulin-2 is developmentally regulated and targeted to dendrites of central neurons. J Comp Neurol 472 (2):156–172. doi: 10.1002/cne.20016 [DOI] [PubMed] [Google Scholar]
- 28.Carraway KL 3rd, Weber JL, Unger MJ, Ledesma J, Yu N, Gassmann M, Lai C (1997) Neuregulin-2, a new ligand of ErbB3/ErbB4-receptor tyrosine kinases. Nature 387 (6632):512–516. doi: 10.1038/387512a0 [DOI] [PubMed] [Google Scholar]
- 29.Vullhorst D, Mitchell RM, Keating C, Roychowdhury S, Karavanova I, Tao-Cheng JH, Buonanno A (2015) A negative feedback loop controls NMDA receptor function in cortical interneurons via neuregulin 2/ErbB4 signalling. Nat Commun 6:7222. doi: 10.1038/ncomms8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotzadimitriou D, Nissen W, Paizs M, Newton K, Harrison PJ, Paulsen O, Lamsa K (2018) Neuregulin 1 Type I Overexpression Is Associated with Reduced NMDA Receptor-Mediated Synaptic Signaling in Hippocampal Interneurons Expressing PV or CCK. eNeuro 5 (2). doi: 10.1523/ENEURO.0418-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen MJ, Leiva-Salcedo E, Buonanno A (2012) Neuregulin Directly Decreases Voltage-Gated Sodium Current in Hippocampal ErbB4-Expressing Interneurons. J Neurosci 32 (40):13889–13895. doi:32/40/13889 [pii] 10.1523/JNEUROSCI.1420-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saheki Y, De Camilli P (2017) Endoplasmic Reticulum-Plasma Membrane Contact Sites. Annu Rev Biochem 86:659–684. doi: 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- 33.Takeshima H, Hoshijima M, Song LS (2015) Ca microdomains organized by junctophilins. Cell Calcium 58:349–356. doi: 10.1016/j.ceca.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrasco S, Meyer T (2011) STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem 80:973–1000. doi: 10.1146/annurev-biochem-061609-165311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbluth J (1962) Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol 13:405–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vullhorst D, Ahmad T, Karavanova I, Keating C, Buonanno A (2017) Structural Similarities between Neuregulin 1–3 Isoforms Determine Their Subcellular Distribution and Signaling Mode in Central Neurons. J Neurosci 37 (21):5232–5249. doi: 10.1523/JNEUROSCI.2630-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallart-Palau X, Tarabal O, Casanovas A, Sabado J, Correa FJ, Hereu M, Piedrafita L, Caldero J, Esquerda JE (2014) Neuregulin-1 is concentrated in the postsynaptic subsurface cistern of C-bouton inputs to alpha-motoneurons and altered during motoneuron diseases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 28 (8):3618–3632. doi: 10.1096/fj.13-248583 [DOI] [PubMed] [Google Scholar]
- 38.Salvany S, Casanovas A, Tarabal O, Piedrafita L, Hernandez S, Santafe M, Soto-Bernardini MC, Caldero J, Schwab MH, Esquerda JE (2019) Localization and dynamic changes of neuregulin-1 at C-type synaptic boutons in association with motor neuron injury and repair. FASEB journal : official publication of the Federation of American Societies for Experimental Biology:fj201802329R. doi: 10.1096/fj.201802329R [DOI] [PubMed] [Google Scholar]
- 39.Wolpowitz D, Mason TB, Dietrich P, Mendelsohn M, Talmage DA, Role LW (2000) Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron 25 (1):79–91. doi:S0896–6273(00)80873–9 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Bao J, Wolpowitz D, Role LW, Talmage DA (2003) Back signaling by the Nrg-1 intracellular domain. J Cell Biol 161 (6):1133–1141. doi: 10.1083/jcb.200212085jcb.200212085 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock ML, Canetta SE, Role LW, Talmage DA (2008) Presynaptic type III neuregulin1-ErbB signaling targets {alpha}7 nicotinic acetylcholine receptors to axons. J Cell Biol 181 (3):511–521. doi:jcb.200710037 [pii] 10.1083/jcb.200710037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trimmer JS (1991) Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc Natl Acad Sci U S A 88 (23):10764–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakoshi H, Trimmer JS (1999) Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci 19 (5):1728–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malin SA, Nerbonne JM (2002) Delayed rectifier K+ currents, IK, are encoded by Kv2 alpha-subunits and regulate tonic firing in mammalian sympathetic neurons. J Neurosci 22 (23):10094–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS (2004) Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci 7 (7):711–718. doi: 10.1038/nn1260nn1260 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Benson DL, Watkins FH, Steward O, Banker G (1994) Characterization of GABAergic neurons in hippocampal cell cultures. Journal of neurocytology 23 (5):279–295 [DOI] [PubMed] [Google Scholar]
- 47.Loeb JA, Fischbach GD (1995) ARIA can be released from extracellular matrix through cleavage of a heparin-binding domain. J Cell Biol 130 (1):127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura H, Kawata M, Hamaguchi S, Ishikawa Y, Shiosaka S (2012) Processing of neuregulin-1 by neuropsin regulates GABAergic neuron to control neural plasticity of the mouse hippocampus. J Neurosci 32 (37):12657–12672. doi: 10.1523/JNEUROSCI.2542-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park KS, Mohapatra DP, Misonou H, Trimmer JS (2006) Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science 313 (5789):976–979. doi: 10.1126/science.1124254 [DOI] [PubMed] [Google Scholar]
- 50.Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A (2000) Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-alpha-converting enzyme. Mol Cell Neurosci 16 (5):631–648. doi: 10.1006/mcne.2000.0896 [DOI] [PubMed] [Google Scholar]
- 51.La Marca R, Cerri F, Horiuchi K, Bachi A, Feltri ML, Wrabetz L, Blobel CP, Quattrini A, Salzer JL, Taveggia C (2011) TACE (ADAM17) inhibits Schwann cell myelination. Nat Neurosci 14 (7):857–865. doi: 10.1038/nn.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleck D, van Bebber F, Colombo A, Galante C, Schwenk BM, Rabe L, Hampel H, Novak B, Kremmer E, Tahirovic S, Edbauer D, Lichtenthaler SF, Schmid B, Willem M, Haass C (2013) Dual cleavage of neuregulin 1 type III by BACE1 and ADAM17 liberates its EGF-like domain and allows paracrine signaling. J Neurosci 33 (18):7856–7869. doi: 10.1523/JNEUROSCI.3372-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwakura Y, Wang R, Inamura N, Araki K, Higashiyama S, Takei N, Nawa H (2017) Glutamate-dependent ectodomain shedding of neuregulin-1 type II precursors in rat forebrain neurons. PLoS One 12 (3):e0174780.doi: 10.1371/journal.pone.0174780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan JY, Skeberdis VA, Jover T, Grooms SY, Lin Y, Araneda RC, Zheng X, Bennett MV, Zukin RS (2001) Protein kinase C modulates NMDA receptor trafficking and gating. Nat Neurosci 4 (4):382–390. doi: 10.1038/86028 [DOI] [PubMed] [Google Scholar]
- 55.Kim J, Lilliehook C, Dudak A, Prox J, Saftig P, Federoff HJ, Lim ST (2010) Activity-dependent alpha-cleavage of nectin-1 is mediated by a disintegrin and metalloprotease 10 (ADAM10). J Biol Chem 285 (30):22919–22926. doi: 10.1074/jbc.M110.126649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki K, Hayashi Y, Nakahara S, Kumazaki H, Prox J, Horiuchi K, Zeng M, Tanimura S, Nishiyama Y, Osawa S, Sehara-Fujisawa A, Saftig P, Yokoshima S, Fukuyama T, Matsuki N, Koyama R, Tomita T, Iwatsubo T (2012) Activity-dependent proteolytic cleavage of neuroligin-1. Neuron 76 (2):410–422. doi: 10.1016/j.neuron.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 57.Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD (2005) Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen 8 (2):161–171 [DOI] [PubMed] [Google Scholar]
- 58.Parra LM, Hartmann M, Schubach S, Li Y, Herrlich P, Herrlich A (2015) Distinct Intracellular Domain Substrate Modifications Selectively Regulate Ectodomain Cleavage of NRG1 or CD44. Mol Cell Biol 35 (19):3381–3395. doi: 10.1128/MCB.00500-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP (2005) Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Developmental biology 283 (2):459–471. doi: 10.1016/j.ydbio.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 60.Du J, Haak LL, Phillips-Tansey E, Russell JT, McBain CJ (2000) Frequency-dependent regulation of rat hippocampal somato-dendritic excitability by the K+ channel subunit Kv2.1. J Physiol 522 Pt 1:19–31. doi:PHY_0063 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misonou H, Mohapatra DP, Menegola M, Trimmer JS (2005) Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci 25 (48):11184–11193. doi: 10.1523/JNEUROSCI.3370-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shah NH, Aizenman E (2014) Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl Stroke Res 5 (1):38–58. doi: 10.1007/s12975-013-0297-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casanovas A, Salvany S, Lahoz V, Tarabal O, Piedrafita L, Sabater R, Hernandez S, Caldero J, Esquerda JE (2017) Neuregulin 1-ErbB module in C-bouton synapses on somatic motor neurons: molecular compartmentation and response to peripheral nerve injury. Sci Rep 7:40155. doi: 10.1038/srep40155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murakoshi H, Shi G, Scannevin RH, Trimmer JS (1997) Phosphorylation of the Kv2.1 K+ channel alters voltage-dependent activation. Mol Pharmacol 52 (5):821–828 [DOI] [PubMed] [Google Scholar]
- 65.Misonou H, Trimmer JS (2004) Determinants of voltage-gated potassium channel surface expression and localization in Mammalian neurons. Crit Rev Biochem Mol Biol 39 (3):125–145. doi:39/3/125 [pii] 10.1080/10409230490475417 [DOI] [PubMed] [Google Scholar]
- 66.Johnson B, Leek AN, Sole L, Maverick EE, Levine TP, Tamkun MM (2018) Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proc Natl Acad Sci U S A 115 (31):E7331–E7340. doi: 10.1073/pnas.1805757115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirmiz M, Vierra NC, Palacio S, Trimmer JS (2018) Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum-plasma membrane junctions in mammalian brain neurons. J Neurosci 38 (35):7562–7584. doi: 10.1523/JNEUROSCI.0893-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao-Cheng JH (2018) Activity-dependent decrease in contact areas between subsurface cisterns and plasma membrane of hippocampal neurons. Mol Brain 11 (1):23. doi: 10.1186/s13041-018-0366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saftig P, Lichtenthaler SF (2015) The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog Neurobiol 135:1–20. doi: 10.1016/j.pneurobio.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 70.Willem M (2016) Proteolytic processing of Neuregulin-1. Brain Res Bull 126 (Pt 2):178–182. doi: 10.1016/j.brainresbull.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 71.Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C (2006) Control of peripheral nerve myelination by the beta-secretase BACE1. Science 314 (5799):664–666. doi: 10.1126/science.1132341 [DOI] [PubMed] [Google Scholar]
- 72.Sanderson MP, Abbott CA, Tada H, Seno M, Dempsey PJ, Dunbar AJ (2006) Hydrogen peroxide and endothelin-1 are novel activators of betacellulin ectodomain shedding. J Cell Biochem 99 (2):609–623. doi: 10.1002/jcb.20968 [DOI] [PubMed] [Google Scholar]
- 73.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP (2007) Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell 18 (1):176–188. doi: 10.1091/mbc.E06-01-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le Gall SM, Bobe P, Reiss K, Horiuchi K, Niu XD, Lundell D, Gibb DR, Conrad D, Saftig P, Blobel CP (2009) ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, L-selectin, and tumor necrosis factor alpha. Mol Biol Cell 20 (6):1785–1794. doi: 10.1091/mbc.E08-11-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matthews AL, Noy PJ, Reyat JS, Tomlinson MG (2017) Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: The emerging role of tetraspanins and rhomboids. Platelets 28 (4):333–341. doi: 10.1080/09537104.2016.1184751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, Borroni B, Cattabeni F, Sala C, Padovani A, Di Luca M (2007) Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J Neurosci 27 (7):1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shyu WC, Lin SZ, Chiang MF, Yang HI, Thajeb P, Li H (2004) Neuregulin-1 reduces ischemia-induced brain-damage in rats. Neurobiol Aging 25 (7):935–944. doi: 10.1016/j.neurobiolaging.2003.10.012 [DOI] [PubMed] [Google Scholar]
- 78.Guo WP, Wang J, Li RX, Peng YW (2006) Neuroprotective effects of neuregulin-1 in rat models of focal-cerebral ischemia. Brain Res 1087 (1):180–185. doi: 10.1016/j.brainres.2006.03.007 [DOI] [PubMed] [Google Scholar]
- 79.Guan YF, Wu CY, Fang YY, Zeng YN, Luo ZY, Li SJ, Li XW, Zhu XH, Mei L, Gao TM (2015) Neuregulin 1 protects against ischemic brain injury via ErbB4 receptors by increasing GABAergic transmission. Neuroscience 307:151–159. doi: 10.1016/j.neuroscience.2015.08.047 [DOI] [PubMed] [Google Scholar]
- 80.Zhang R, Liu C, Ji Y, Teng L, Guo Y (2018) Neuregulin-1beta Plays a Neuroprotective Role by Inhibiting theCdk5 Signaling Pathway after Cerebral Ischemia-Reperfusion Injury in Rats. J Mol Neurosci 66 (2):261–272. doi: 10.1007/s12031-018-1166-3 [DOI] [PubMed] [Google Scholar]
- 81.Vullhorst D, Neddens J, Karavanova I, Tricoire L, Petralia RS, McBain CJ, Buonanno A (2009) Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci 29 (39):12255–12264. doi:29/39/12255 [pii] 10.1523/JNEUROSCI.2454-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlander M, Hoyer S, Frotscher M (1988) Glutamate decarboxylase-immunoreactive neurons in the aging rat hippocampus are more resistant to ischemia than CA1 pyramidal cells. Neurosci Lett 91 (3):241–246 [DOI] [PubMed] [Google Scholar]
- 83.Frahm C, Haupt C, Witte OW (2004) GABA neurons survive focal ischemic injury. Neuroscience 127 (2):341–346. doi: 10.1016/j.neuroscience.2004.05.027 [DOI] [PubMed] [Google Scholar]
- 84.Li KX, Lu YM, Xu ZH, Zhang J, Zhu JM, Zhang JM, Cao SX, Chen XJ, Chen Z, Luo JH, Duan S, Li XM (2011) Neuregulin 1 regulates excitability of fast-spiking neurons through Kv1.1 and acts in epilepsy. Nat Neurosci 15 (2):267–273. doi:nn.3006 [pii] 10.1038/nn.3006 [DOI] [PubMed] [Google Scholar]
- 85.Yang JM, Zhang J, Chen XJ, Geng HY, Ye M, Spitzer NC, Luo JH, Duan SM, Li XM (2013) Development of GABA circuitry of fast-spiking basket interneurons in the medial prefrontal cortex of erbb4-mutant mice. J Neurosci 33 (50):19724–19733. doi: 10.1523/JNEUROSCI.1584-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shamir A, Kwon OB, Karavanova I, Vullhorst D, Leiva-Salcedo E, Janssen MJ, Buonanno A (2012) The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. J Neurosci 32 (9):2988–2997. doi:32/9/2988 [pii] 10.1523/JNEUROSCI.1899-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Speca DJ, Ogata G, Mandikian D, Bishop HI, Wiler SW, Eum K, Wenzel HJ, Doisy ET, Matt L, Campi KL, Golub MS, Nerbonne JM, Hell JW, Trainor BC, Sack JT, Schwartzkroin PA, Trimmer JS (2014) Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav 13 (4):394–408. doi: 10.1111/gbb.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.