Abstract
Melanoma is one of the most aggressive, potentially fatal forms of skin cancer, and has been shown to be associated with solar ultraviolet radiation-dependent initiation and progression. Despite remarkable recent advances with targeted- and immune- therapeutics, lasting and recurrence-free survival remain significant concerns. Therefore, additional novel mechanism-based approaches are needed for effective melanoma management. The sirtuin SIRT6 appears to have a pro-proliferative function in melanocytic cells. In this study, we determined the effects of genetic manipulation of SIRT6 in human melanoma cells, in vitro as well as in vivo. Our data demonstrated that CRISPR/Cas9-mediated knockout (KO) of SIRT6 in A375 melanoma cells resulted in a significant i) decrease in growth, viability, clonogenic survival, and ii) induction of G1-phase cell cycle arrest. Further, employing a RT2 Profiler PCR array containing 84 key transformation and tumorigenesis genes, we found that SIRT6 KO resulted in modulation of genes involved in angiogenesis, apoptosis, cellular senescence, epithelial-to-mesenchymal transition, hypoxia signaling, and telomere maintenance. Finally, we found significantly decreased tumorigenicity of SIRT6 KO A375 cells in athymic nude mice. Our data provide strong evidence that SIRT6 promotes melanoma cell survival, both in vitro and in vivo, and could be exploited as a target for melanoma management.
Graphical Abstract
SIRT6 has been shown to act as a tumor promoter or suppressor depending on the cancer type. This study supports the tumor promoter role of SIRT6 in melanoma, one of the deadliest forms of UV-related skin cancers. We found CRISPR/Cas9-mediated SIRT6 knockout (KO) imparted marked anti-proliferative effects in human melanoma cells in vitro and anti-tumorigenic response in vivo. Additionally, we found SIRT6 KO significantly affected several cancer-associated pathways, including apoptosis, cellular senescence, and epithelial-to-mesenchymal transition, suggesting SIRT6 has an important role in melanoma progression. Thus, SIRT6 inhibitory approaches could potentially be used in melanoma management and should be explored further.
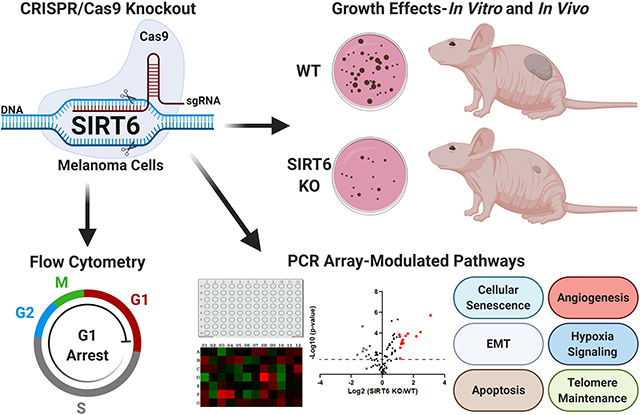
INTRODUCTION
Melanoma is one of the deadliest types of skin cancer, largely due to its ability to become aggressively metastatic when not diagnosed and treated at an early stage (1). It is estimated that during the course of 2020, approximately 100,350 new cases of melanoma will be diagnosed and 6,850 deaths will occur due to this malignancy (2). Solar ultraviolet (UV) radiation, a predominant environmental carcinogen, is a major risk factor for developing melanoma. Melanoma develops from melanin-producing melanocytes in the basal layer of the epidermis (3). UV irradiation causes cutaneous DNA damage leading to the formation of cyclobutane–pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts, which are the primary source of mutations in UV-induced melanoma (4). Current treatment options include surgery, cryotherapy, radiation therapy, or immunotherapies, as well as various chemotherapies, including drugs that target specific molecular pathways (i.e. BRAF or MEK inhibitors) (5–7). However, melanoma has been associated with the tendency to develop resistance to the available treatments, which can lead to a relapse of the cancer (7, 8). Additionally, although recently developed immunotherapies have been extremely promising, they are extremely expensive and may be associated with significant adverse effects. Therefore, it is crucial to identify new molecular targets and targeted therapies, which could be used alone or in combination with existing therapeutics for melanoma management.
The sirtuins are a family of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases (HDACs), with seven known members in mammals (SIRTs 1–7). Although SIRTs have structural similarities, their localization varies within the cells, influencing their unique roles (9–11). Overall, the sirtuins have been implicated in the regulation of several important molecular processes, including transcription, DNA repair, metabolism, and aging, (10, 12). Interestingly, studies have shown that the SIRTs can play both oncogenic and tumor suppressor roles, depending on the cell and tissue types (13–15). SIRT6, the focus of this study, is one of the less-studied sirtuins. It has been shown to interact with acetylated histone H3 lysines, such as acH3K9, acH3K18, and acH3K56 (16), and is generally found in the nucleus, where it plays roles in DNA repair, chromatin and transcription regulation, telomere maintenance, lipid metabolism and glycolysis (9, 17–19). A very limited number of studies suggest a potential oncogenic role of SIRT6 in skin (20, 21). In one such study by Ming et al, the authors showed that SIRT6 functions as an oncogene in non-melanoma skin cancer and exposure to UVB radiation leads to increased expression of SIRT6 in skin keratinocytes (21). The authors also found that in keratinocytes, SIRT6 promoted the expression of COX-2, an enzyme involved in inflammation, proliferation, and survival, via AMPK inhibition. In our lab, we have previously shown that SIRT6 was overexpressed in human melanoma cell lines compared to normal melanocytes as well as in melanoma tissues, and its inhibition via shRNA-mediated RNA interference resulted in a significant antiproliferative response in melanoma cells (22). Another study by Wang and colleagues supported our findings to show that SIRT6 contributed to melanoma growth in an autophagy-dependent manner (23). Together, these studies suggest a pro-proliferative role for SIRT6 in melanoma. However, additional investigations regarding the definitive role and mechanisms of SIRT6 in melanoma are needed to determine the therapeutic significance of SIRT6 in melanoma. This study was undertaken to determine the effects of SIRT6 KO in human melanoma cells in vitro and in vivo, and its associated downstream mechanisms.
MATERIALS AND METHODS
SIRT6 knockout cell line generation.
A375 (RRID:CVCL 0132) SIRT6 knockout cell pool (and WT cells) were purchased from Synthego and maintained in DMEM supplemented with 10% FBS using standard cell culture conditions (37°C, 5% CO2, humidified chamber). Cells were maintained at low passages and authenticated, as well as tested for mycoplasma (results were negative). The pool was generated using guide sequence CCUGAAGUCGGGGAUGCCAG. To grow single clones, the cell pool was plated in 96-well plates at an average of 0.5 cells/well and allowed to grow until a single, discrete clone could be seen in the well. Wells with single colonies were passaged and grown as standard cultures. Once enough cells were available, cells were collected and subjected to immunoblot as described below to determine SIRT6 protein expression. The clones with the best protein reduction were used in growth assays to determine the best clones for further experiments.
Trypan blue exclusion assay.
Cells were plated in triplicate in 6-well plates (TPP) and allowed to grow for 24–72 hours. The cells were then trypsinized and both live and dead cells were collected. A small aliquot was mixed 1:1 with trypan blue and counted using the Countess II FL Automated Cell Counter (ThermoFisher). Statistical significance was determined with t-test using Prism software (GraphPad Software).
Colony formation.
Cells were seeded at low density (300 cells per well) into 6-well plates in triplicate and allowed to grow for 8–12 days. Cells were then stained in 0.1% crystal violet in PBS:methanol (1:1), and destained using PBS rinses until background was clear before imaging.
Protein isolation and immunoblotting.
Proteins were isolated from collected cell pellets and subjected to SDS-PAGE and immunoblotting as described previously (22). Antibodies used for immunoblotting are outlined in Supporting Information (Table S1).
Cell cycle analysis.
Cells were seeded into 6-well plates (7×104 cells per well) and allowed to grow for 24 hours. Cells were then detached from the plates using trypsin, collected (both live and dead cells), rinsed, and stained with propidium iodide before analyzing on the BD Accuri C6 flow cytometer. Cell cycle profile was analyzed using ModFit software (Verity Software House). Data was averaged from 3 separate runs performed in triplicate. Statistical analysis was done using a 2-way ANOVA with Dunnet’s multiple comparison test using Prism software.
RNA isolation and RT-qPCR.
RNA was isolated using the Qiagen RNeasy Plus isolation kit per manufacturer’s protocol and quantified using the BioTek Synergy H1 multimode plate reader with Take3 plate. cDNA was made using MMLV-RT and Oligo dT primers (Promega) per manufacturer’s protocol and used in the qPCR array and for follow-up validations. Statistical significance was determined via multiple t-tests using the Holm-Sidak method to correct for multiple comparisons using Prism software.
PCR array.
The effects of SIRT6 KO were assessed using the Qiagen Human Cancer PathwayFinder RT2 Profiler PCR Array (#PAHS-033ZA) per the manufacturer’s instructions. Ct values for the genes were uploaded to the Qiagen GeneGlobe Data Analysis Center and analyzed using GAPDH and ACTB reference genes. The data analysis web portal calculated fold-change using the ΔΔCT method (Table 1). Also, statistically significant selected genes from the PCR array results (>2-fold in one group and ≥1.5-fold in other) were validated using RT-qPCR analysis. Primers pairs detailed in Supporting Information (Table S2) were retrieved from Primer Bank (24).
Mouse xenograft studies.
Animal studies were approved by the University of Wisconsin Animal Care and Use Committee and all measures were taken to reduce pain and/or discomfort to experimental animals. For cell implantation, 5×105 A375 WT or SIRT6 KO cells were subcutaneously injected into the right flank of each mouse (Envigo Nu/Nu Strain #088; n=12; 6 males, 6 females per group) and monitored until tumors were palpable. Tumors were then measured twice weekly using a Biopticon TumorImager and tumor volume was calculated by TumorManager software. At the end of the study, mice were euthanized and tumors were collected for further analysis. Statistical significance was determined via t-test using Prism software.
Immunohistochemistry.
After excision, tumors were fixed in 10% formalin for 48 hours and then embedded in paraffin and sectioned. Sections were deparaffinized and rehydrated using a standard xylene/ethanol series before epitope retrieval using IHC-Tek IHC Buffer and Epitope retrieval system. After blocking endogenous peroxidase activity, slides were blocked in normal serum and incubated overnight in primary antibody as outlined in Supporting Information (Table S1). The following day, slides were incubated in secondary antibody, ABC-HRP reagent (Vector Labs), and then Vector Red chromogen before counterstaining with hematoxylin. Slides were then dehydrated and coverslipped using Permount (Sigma) before imaging at 40x for Ki67 analysis. Cells with bright pink nuclei were counted as positive for Ki67, while blue/light purple cells were counted as negative. Percent of Ki67-positive cells were determined in each image and mean ±SEM for each group is shown. Statistical significance was determined via t-test using Prism software.
RESULTS AND DISCUSSION
CRISPR/Cas9-mediated SIRT6 knockout (KO) inhibits cellular growth, viability, clonogenic survival, and induces G1 arrest in A375 human melanoma cells
We employed an experimental strategy of CRISPR/Cas9-mediated knockout of SIRT6 in A375 human melanoma cells to determine the functional significance of SIRT6 in melanoma. We generated single clones from a CRISPR/Cas9-mediated SIRT6 KO A375 cell pool, selecting the best KO clones for further experiments (shown in Supporting Information; Fig. S1). As shown in Fig. 1A, we achieved significant SIRT6 protein KO in our selected clones. Next, we assessed the effects of SIRT6 KO on proliferative potential of cells using trypan blue exclusion and long-term clonogenic survival assays. Our data demonstrated that SIRT6 KO significantly decreased cell growth and viability in both of the selected clones (clones #6 and #9) (Fig. 1B). As seen in Fig. 1C, SIRT6 KO resulted in a marked decrease in the number of colonies, suggesting that SIRT6 KO results in a significant decrease in clonogenic survival of melanoma cells. Additionally, we wanted to determine if the observed anti-proliferative response following SIRT6 KO was associated with dysregulation in the cell cycle, since SIRT6 was shown to regulate melanoma growth via the IGF-AKT signaling pathway (23), which is known to cause G1 arrest via p21, p27, and Cdk2 (25). As measured by flow cytometric analyses (Figs. 1D–E), we found that SIRT6 KO resulted in an enhanced accumulation of cells in G1 phase, which was accompanied by reductions of cell populations in S and G2/M phases. These data support our previous findings (22), further validating the pro-proliferative function of SIRT6 in melanoma.
Figure 1.
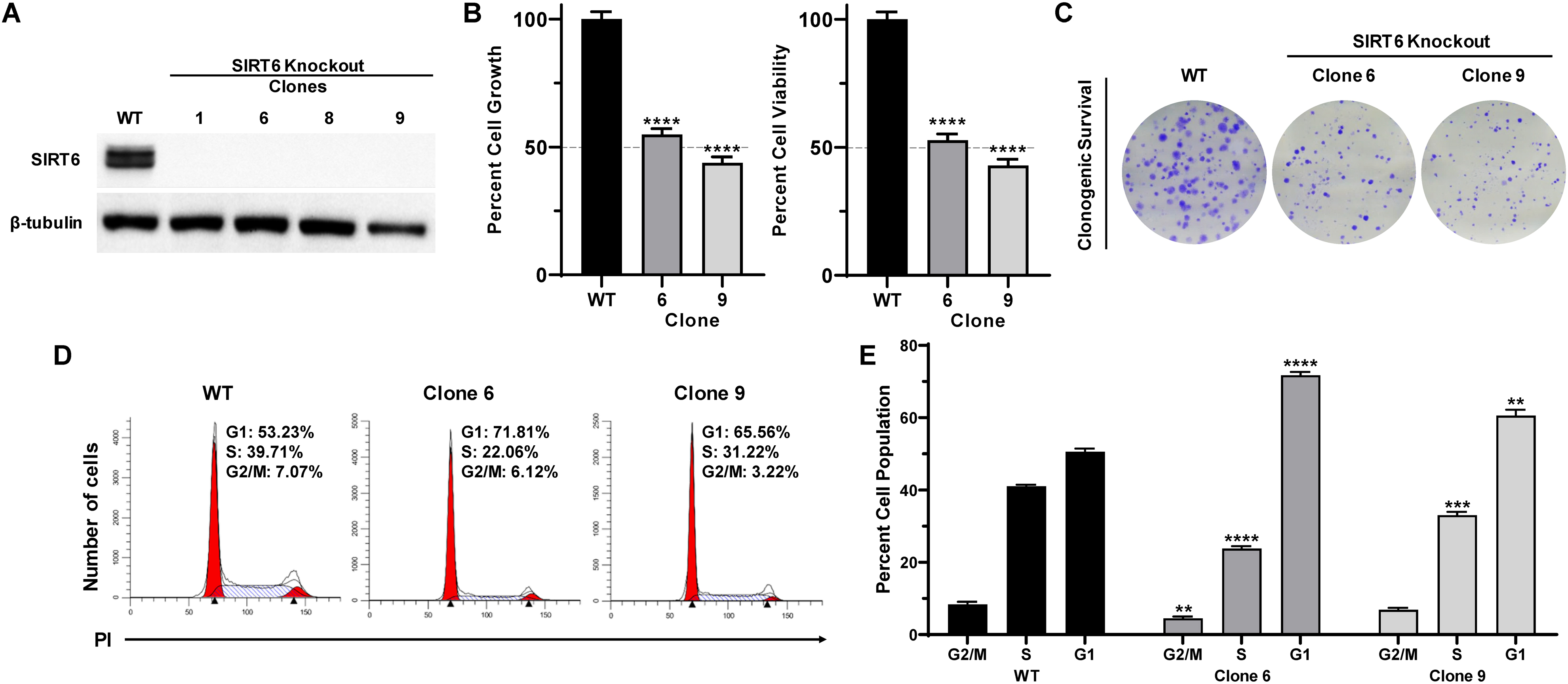
CRISPR/Cas9-mediated SIRT6 knockout inhibits growth and clonogenic survival, induces G1-phase arrest in human melanoma cells. A375 SIRT6 WT and KO clones were seeded and analyzed after 72 h, with different assays. (A) Cell lysates (with equal amount of protein) were subjected to immunoblot analysis to confirm SIRT6 KO. β-tubulin was used as a loading control. (B) Cell growth and viability were determined by trypan blue exclusion assay. Results are expressed as percentage of viable or total SIRT6 KO cells compared to WT. (C) To determine the clonogenic survival, an equal number of viable cells were plated into 6-well plates at low density. After ∼10 days, cells were fixed and stained with crystal violet. The images shown are representative of three experiments with similar results. (D) Cell cycle analysis was performed using propidium iodide (PI) staining, and data were analyzed with ModFit software, representative histograms are shown. (E) Mean percent of cells in each phase of the cell cycle are shown. The data are expressed as mean ±SEM of three experiments, done in triplicate. Statistical significance is determined by two-way ANOVA analyzed via GraphPad Prism Software and is denoted as **P≤0.01, ****P≤0.0001.
SIRT6 modulation via CRISPR/Cas9-mediated KO significantly altered multiple cancer pathways
To obtain an insight into the molecular mechanisms of the antiproliferative response of SIRT6 KO, we performed a PCR array analysis using the Qiagen Human Cancer PathwayFinder RT2 Profiler PCR Array, which allowed us to profile the expression of 84 key genes related to transformation and tumorigenesis using the two selected SIRT6 KO clones. Volcano plots outlining the distribution of the tested genes within the Cancer PathwayFinder qPCR Profiler Array are shown in Fig. 2A. As shown by the heat maps (Fig. 2B), SIRT6 KO caused a modulation in the expression of a number of important genes related to angiogenesis, apoptosis, cell cycle, cellular senescence, DNA damage repair, epithelial-to-mesenchymal transition (EMT), hypoxia signaling, metabolism, and telomere maintenance.
Figure 2.
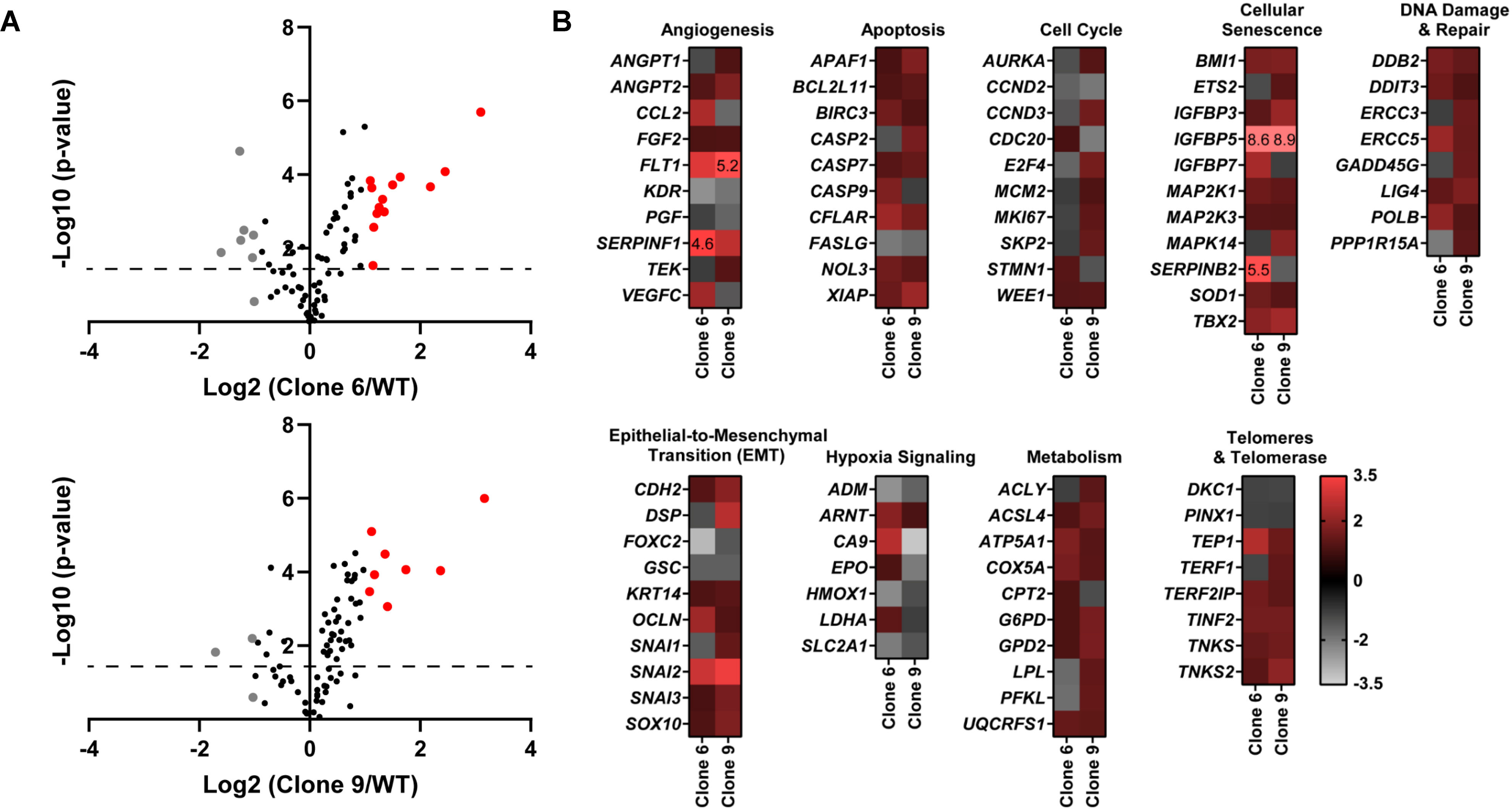
CRISPR/Cas9-mediated SIRT6 knockout causes alterations in multiple cancer-associated pathways. Qiagen Human Cancer PathwayFinder RT2 Profiler PCR Array was performed using A375 WT and SIRT6 KO cells (clones #6 and 9). (A) Volcano plots outlining the distribution of 84 tested genes within the Qiagen Human Cancer PathwayFinder RT2 Profiler PCR Array are displayed. Data was graphed log2 (ratio) of SIRT6 KO clone 6 (left) or SIRT6 KO clone 9 (right) vs WT against the -Log10 (p-value). Upregulated genes that reached the 2-fold cut-off are shown in red whereas the downregulated are depicted in grey, and black if no/negligible fold change was observed. The dashed line indicates the 0.05 p-value cut off. (B) Heat maps of each SIRT6 KO clone vs WT were generated to display gene fold changes classified by the cancer pathway identified in the array. Upregulated genes are shown in red whereas the downregulated are depicted in grey. To view clear differences in gene expression, the scale covers fold regulation of −3.5 (grey) to 3.5 (red). Gene expression values falling outside of this scale are indicated by the indicated fold change value in the relevant box.
The cut-off criteria chosen for further validation was for those genes significantly modulated >2-fold in one clone and ≥1.5-fold in the other clone. The key genes that met these criteria were ADM, CA9, CCL2, CFLAR, FLT1, IGFBP5, KDR, SERPINF1, SNAI2, TBX2 and TEP1. A graphical representation of this data is shown in Fig. 3A, with more details in Supporting Information (Table S3). Although most of the genes appear to have similar increases/decreases in expression in both clones tested in the PCR array, CA9 and CCL2 appeared to be differently regulated (i.e. increased in one clone but decreased in the other). However, further validation with RT-qPCR (Fig. 3B) found that both CA9 and CCL2, along with ADM, were downregulated in both SIRT6 KO clones. This also confirmed the significant upregulation of IGFBP5, FLT1 and SNAI2. Further, CFLAR, KDR, SERPINF1, TBX2 and TEP1 did not show significant modulation in either of the SIRT6 KO clone upon validation.
Figure 3.
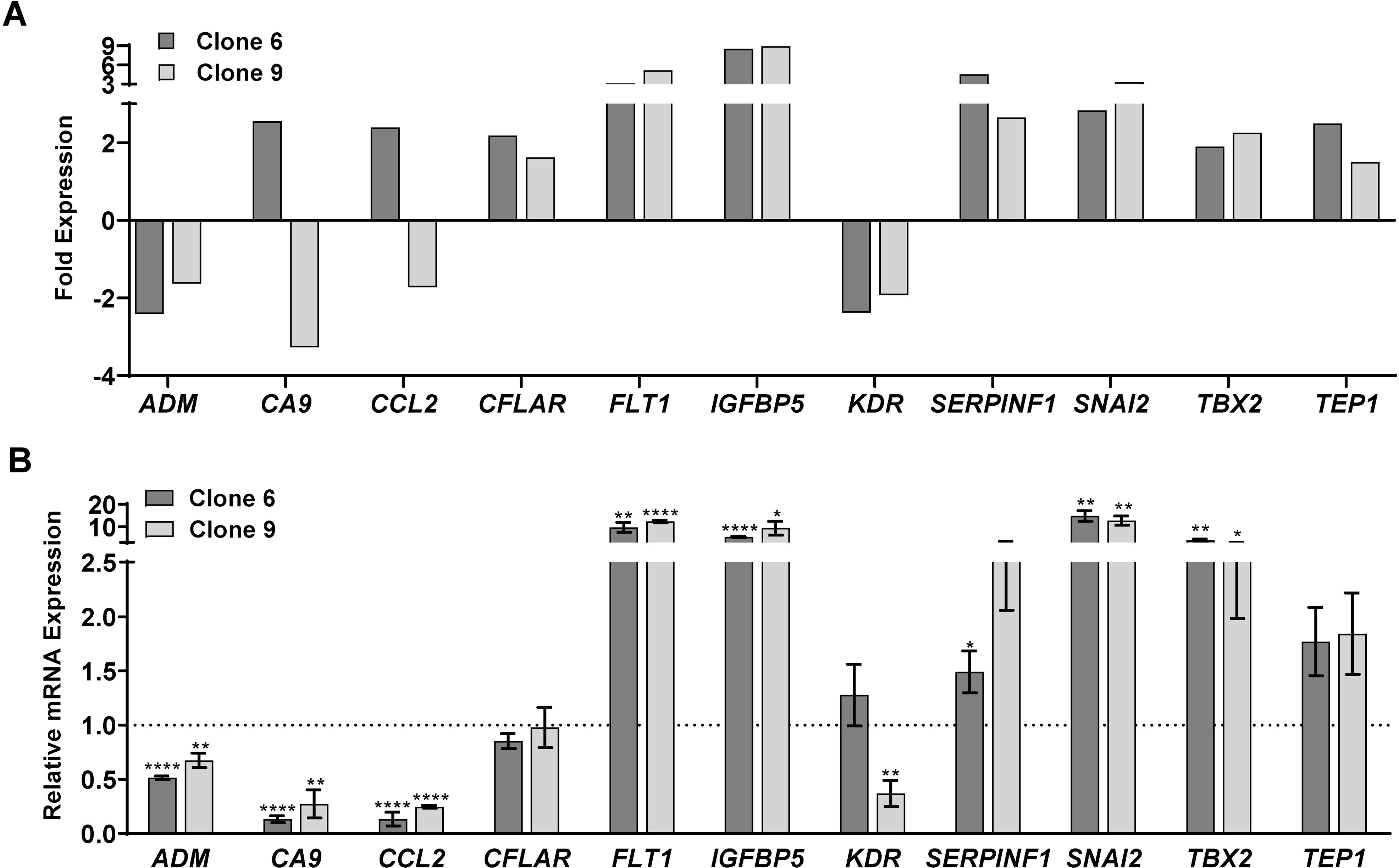
SIRT6 knockout by CRISPR/Cas9 results in alteration of key cancer-related genes. Key genes were found to be significantly modulated (>2-fold change in one clone and >1.5-fold change in the other, with statistical significance for both) after SIRT6 knockout. (A) Graphical representation of the 11 genes that were found differentially expressed upon SIRT6 KO. The data shown represent three biological replicates. (B) RT-qPCR analysis was performed to validate the key altered genes at mRNA levels in WT and SIRT6 KO cells as detailed in the Supporting Information. Data are represented as mean ± SEM of minimum three technical and two biological replicates, and were analyzed via GraphPad Prism software using multiple t-tests and the Holm-Sidak method for multiple comparisons, with significance indicated by * P≤0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Interestingly, in our PCR array analysis, we found that IGFBP5 (Insulin-Like Growth Factor Binding Protein 5) showed the maximum increase among the genes tested, with a greater than 8-fold increase in expression in both clones after SIRT6 KO. This is an important observation, since IGFBP5 is often dysregulated in human cancers (26), and a study by Wand et al. found that IGFBP5 functions as a tumor suppressor in human melanoma cells (27). Additionally, we found that SIRT6 KO caused a decrease in CA9 (Carbonic Anhydrase), a crucial regulator of pH that is overexpressed in several cancers. Intriguingly, a study by Martinez-Zaguilan et al. found that high pH environments significantly increased the invasiveness of human melanoma cells (28), suggesting the potential role for SIRT6 in EMT. We also found a downregulation of CCL2 (C-C Motif Chemokine Ligand 2), a known mediator of tumorigenesis in a variety of cancer types. CCL2 has been reported to enhance the survival and invasiveness of melanoma cells and favor melanoma growth in vivo (29), and use of CCL2-targeting antibodies and/or BRAF inhibitors have been shown to decrease tumor growth in mouse models of metastatic melanoma (30). Further, the observed decrease in ADM (adrenomedullin) in our experiments also supports the pro-proliferative function of SIRT6. ADM is expressed in tumor-associated macrophages in melanoma and plays a crucial role in both promoting angiogenesis and melanoma growth (31).
On the other hand, two of the genes found to be modulated upon SIRT6 KO did not support the pro-proliferative role of SIRT6. SNAI2 (Snail Family Transcriptional Repressor 2; also known as SLUG) was increased in SIRT6 KO clones. This is in accordance with the study by Kunming et al. demonstrating that SIRT6 overexpression reduced the transcription levels of SNAI2 in TGF-β1-treated A549 lung carcinoma cells (32). However, this does not support the antiproliferative response of SIRT6 KO seen in our study. Similarly, an increased level of FLT1 (fms related receptor tyrosine kinase 1; also known as VEGFR1) in SIRT6 KO clones was not in-line with our hypothesis that SIRT6 has a tumor-promoter function in melanoma, because FLT1 has been shown to stimulate tumor growth and metastasis (33). Therefore, the downstream effects of SIRT6 on SNAI2 and FLT1 need to be further investigated in additional experiments. Overall, our results based on PCR array data suggest that SIRT6 possesses an important role in melanoma progression, potentially acting as a tumor promoter.
CRISPR/Cas9-mediated SIRT6 KO results in marked decrease in melanoma tumor growth in vivo
To validate our in vitro data to in vivo situations, we conducted a tumor xenograft study with SIRT6 KO melanoma cells. We implanted A375 WT and A375 SIRT6 KO cells subcutaneously into athymic nude mice and followed their development over time. As shown in Fig. 4A, the A375 SIRT6 KO tumors were demonstrably smaller compared to A375 WT tumors at the end of the experiment, and demonstrated significantly decreased tumorigenicity, in terms of tumor volume (Fig. 4B) and tumor weight (Fig. 4C).
Figure 4.
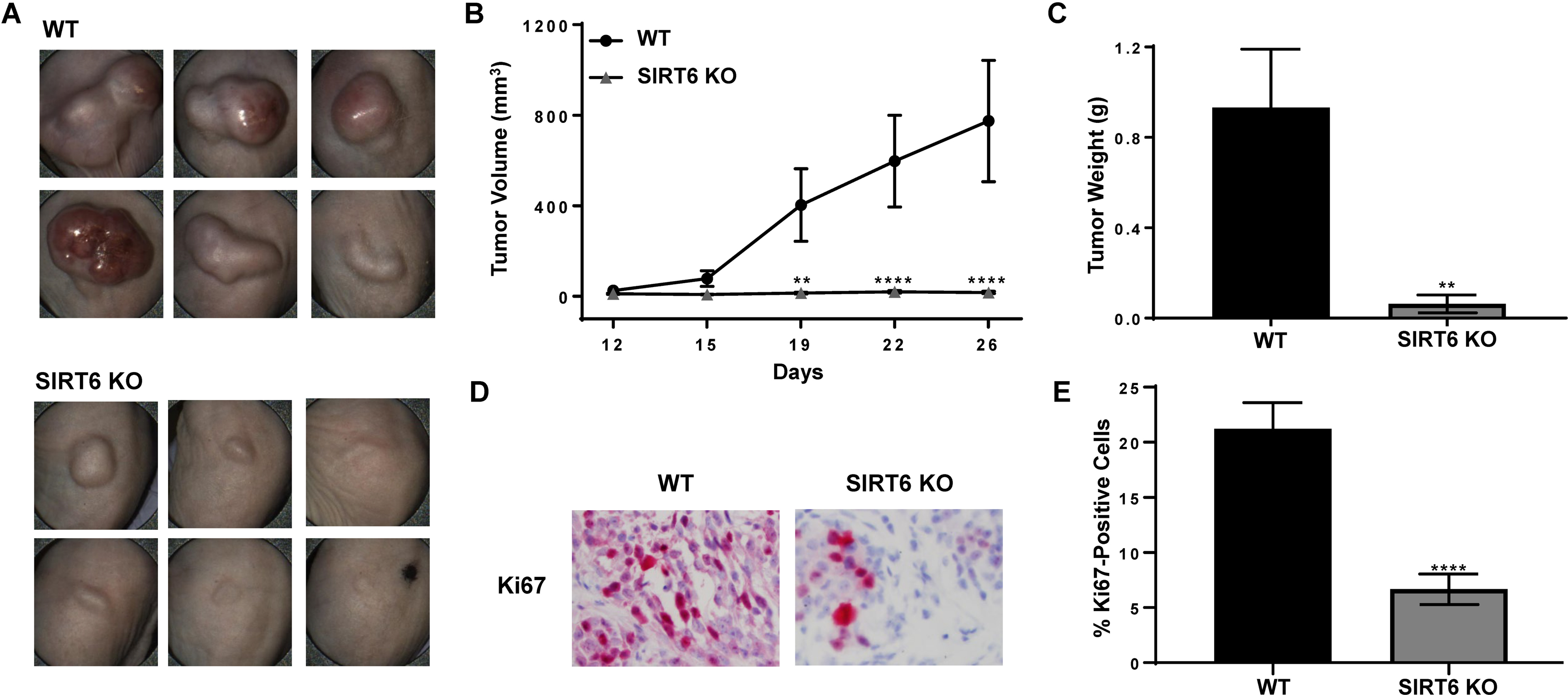
CRISPR/Cas9-mediated SIRT6 knockout results in marked decrease in melanoma cells-implanted tumors in athymic nude mice. A375 SIRT6 KO and WT cells were subcutaneously implanted and allowed to grow in athymic nude mice and tumor growth was assessed as described in Materials and Methods. (A) Pictures of tumors before resection were captured using the Biopticon TumorImager. (B) Tumor volume as calculated by the Biopticon TumorManager software and (C) final tumor weight after resection are shown. After excision, tumors were subjected to immunohistochemistry for Ki67. (D) Representative images and (E) percent positive cells are shown. Data is shown as mean ±SEM. Statistical analysis was performed using two-way ANOVA (B) or t-test (C and E) using GraphPad Prism Software, with significance denoted as **P<0.01, ****P<0.0001.
We found similar results with an additional clone, as well as in female mice (data not shown), suggesting that SIRT6 depletion consistently and reliably inhibits xenograft tumor progression in athymic nude mice. At the termination of the experiment, the tumors were excised and stained with Ki67 to determine the effects of SIRT6 KO on proliferation. As shown in Figs. 4D and 4E, the SIRT6 KO tumors showed significant reductions in the percent of Ki67-positive cells, indicating decreased proliferative indices of SIRT6 KO tumors. These results corroborate our in vitro data suggesting that SIRT6 promotes melanoma growth.
Thus, our study further underscores the tumor promoter function of SIRT6 in melanocytic cells, in vitro and in vivo. In order to further dissect the role of SIRT6 in melanoma, it is important to determine the association and interaction of SIRT6 with other melanoma driver pathways, including the PI3K/AKT and MAPK/ERK pathways. Additionally, as mentioned earlier, SIRT6 has been linked to the IGF-AKT signaling pathway (23), though further studies need to be conducted in order to understand the interplay between SIRT6 and these key pathways. To date, there is no study showing an association between SIRT6 and the MAPK/ERK signaling pathway in melanoma, although recent findings have revealed that the RAS/RAF/MEK/ERK signaling pathway is involved in the modulation of autophagy (34). This is relevant to our findings because we have shown in our previous studies that in melanoma cells, SIRT6 knockdown modulates key autophagy markers including BECN1, SQSTM1, ATG3, ATG7, ATG10 and GAA, as well as the reduced conversion of LC3 protein from its free form LC3-I (22). These results suggest that SIRT6 may positively regulate autophagy in melanoma cells. Thus, the exploration of an association between SIRT6 and autophagy through the RAS/RAF/MEK/ERK signaling pathway could provide new mechanistic information into interactions of SIRT6 with melanoma driver pathways. Indeed, detailed studies unraveling other pathways and mechanisms affected by SIRT6 in melanocytic cells could provide new information towards the development of novel treatments against melanoma.
One limitation of this study is that we have used a single CRISPR/Cas9-mediated SIRT6 knockout melanoma cell line. Although further work is needed to validate this work in other melanoma lines with varied genetic backgrounds, when combined with previously published data from our lab and others, there is strong evidence to support the oncogenic role of SIRT6 in melanoma. In combination with recent studies showing a potential role for SIRT6 in UV-induced DNA damage repair, our study provides a scientific basis for future investigations aimed at defining the connection between UV response and SIRT6 signaling during melanomagenesis and melanoma progression.
Supplementary Material
Figure S1. CRISPR/Cas9-mediated SIRT6 knockout results in reduced A375 cell growth in vitro. A375 SIRT6 Knockout (KO) cell pool was plated for isolation of individual colonies, and clones were tested for SIRT6 protein KO and growth effects. (A) Cell lysates from each clone (with equal amounts of protein) were subjected to immunoblot analysis to confirm SIRT6 KO. β-tubulin was used as a loading control. (B) Cell growth and viability of the 4 clones with the lowest SIRT6 expression was determined by trypan blue exclusion assay. Results are expressed as relative growth from 24h and percentage of viable SIRT6 KO cells compared to WT. (C) To determine the clonogenic survival, an equal number of viable cells were plated in triplicate into 6-well plates at low density. After ~10 days, cells were fixed with methanol and stained with crystal violet. The images shown are representative of three experiments with similar results. Statistical analysis was done by two-way ANOVA analyzed via GraphPad Prism Software. * P≤0.05.
Table S1. Antibodies used for immunoblot (IB) and immunohistochemistry (IHC) analyses.
Table S2. Primers used in this study.
Table S3. Significantly modulated genes.
ACKNOWLEDGEMENTS:
The authors would like to thank Ms. Anna Musarra for her technical help with the immunohistochemistry experiments in this manuscript. This work was partially supported by funding from the NIH (R01AR059130 and R01CA176748 to NA), and the Department of Veterans Affairs (VA Merit Review Awards I01CX001441 and 1I01BX004221; and a Research Career Scientist Award IK6BX003780 to NA). We also acknowledge the core facilities supported by the Skin Diseases Research Center (SDRC) Core Grant P30AR066524 from NIH/NIAMS.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article:
REFERENCES
- 1.Braeuer RR, Watson IR, Wu CJ, Mobley AK, Kamiya T, Shoshan E and Bar-Eli M (2014) Why is melanoma so metastatic? Pigment Cell Melanoma Res. 27, 19–36. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD and Jemal A (2020) Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30. [DOI] [PubMed] [Google Scholar]
- 3.Chhabra G, Garvey DR, Singh CK, Mintie CA and Ahmad N (2019) Effects and Mechanism of Nicotinamide Against UVA- and/or UVB-mediated DNA Damages in Normal Melanocytes. Photochem. Photobiol. 95, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhabra G, Ndiaye MA, Garcia-Peterson LM and Ahmad N (2017) Melanoma Chemoprevention: Current Status and Future Prospects. Photochem. Photobiol. 93, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maverakis E, Cornelius LA, Bowen GM, Phan T, Patel FB, Fitzmaurice S, He Y, Burrall B, Duong C, Kloxin AM, Sultani H, Wilken R, Martinez SR and Patel F (2015) Metastatic melanoma - a review of current and future treatment options. Acta Derm. Venereol. 95, 516–524. [DOI] [PubMed] [Google Scholar]
- 6.Hepner A, Salgues A, Anjos CAD, Sahade M, Camargo VP, Garicochea B, Shoushtari AN, Postow MA, Fernandes GS and Munhoz RR (2017) Treatment of advanced melanoma - A changing landscape. Rev. Assoc. Med. Bras. (1992) 63, 814–823. [DOI] [PubMed] [Google Scholar]
- 7.Naves LB, Dhand C, Venugopal JR, Rajamani L, Ramakrishna S and Almeida L (2017) Nanotechnology for the treatment of melanoma skin cancer. Prog. Biomater. 6, 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malissen N and Grob JJ (2018) Metastatic Melanoma: Recent Therapeutic Progress and Future Perspectives. Drugs 78, 1197–1209. [DOI] [PubMed] [Google Scholar]
- 9.Michishita E, Park JY, Burneskis JM, Barrett JC and Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, Mack NJ and Ahmad N (2018) The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 28, 643–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su S, Ndiaye M, Singh CK and Ahmad N (2020) Mitochondrial Sirtuins in Skin and Skin Cancers. Photochem. Photobiol, 96, 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George J, Nihal M, Singh CK and Ahmad N (2019) 4’-Bromo-resveratrol, a dual Sirtuin-1 and Sirtuin-3 inhibitor, inhibits melanoma cell growth through mitochondrial metabolic reprogramming. Mol. Carcinog. 58, 1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch-Presegue L and Vaquero A (2011) The dual role of sirtuins in cancer. Genes Cancer 2, 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George J, Nihal M, Singh CK, Zhong W, Liu X and Ahmad N (2016) Pro-Proliferative Function of Mitochondrial Sirtuin Deacetylase SIRT3 in Human Melanoma. J. Invest. Dermatol. 136, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilking MJ, Singh CK, Nihal M, Ndiaye MA and Ahmad N (2014) Sirtuin deacetylases: a new target for melanoma management. Cell Cycle 13, 2821–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang WW, Zeng Y, Wu B, Deiters A and Liu WR (2016) A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS Chem. Biol. 11, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kugel S and Mostoslavsky R (2014) Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM and Denu JM (2011) Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye X, Li M, Hou T, Gao T, Zhu WG and Yang Y (2017) Sirtuins in glucose and lipid metabolism. Oncotarget 8, 1845–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, Albinger-Hegyi A, Hoetzenecker W, Kolfschoten I, Wagner EF, Werner S and Dotto GP (2013) A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 32, 2248–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ming M, Han W, Zhao B, Sundaresan NR, Deng CX, Gupta MP and He YY (2014) SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 74, 5925–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Peterson LM, Ndiaye MA, Singh CK, Chhabra G, Huang W and Ahmad N (2017) SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer 8, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Guo W, Ma J, Dai W, Liu L, Guo S, Chen J, Wang H, Yang Y, Yi X, Wang G, Gao T, Zhu G and Li C (2018) Aberrant SIRT6 expression contributes to melanoma growth: Role of the autophagy paradox and IGF-AKT signaling. Autophagy 14, 518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Spandidos A, Wang H and Seed B (2012) PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40, D1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coqueret O (2003) New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 13, 65–70. [DOI] [PubMed] [Google Scholar]
- 26.Gullu G, Karabulut S and Akkiprik M (2012) Functional roles and clinical values of insulin-like growth factor-binding protein-5 in different types of cancers. Chin. J. Cancer 31, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q, Deng Y, Yang Y, Li Y, Ruan X, Xie F, Zhao H and Fang X (2015) Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget 6, 20636–20649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Zaguilan R, Seftor EA, Seftor RE, Chu YW, Gillies RJ and Hendrix MJ (1996) Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis 14, 176–186. [DOI] [PubMed] [Google Scholar]
- 29.Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, Rocchi S, Peyron JF, Lacour JP, Ballotti R and Bertolotto C (2011) Senescent cells develop a PARP-1 and nuclear factor-{kappa}B-associated secretome (PNAS). Genes Dev. 25, 1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, Haynes NM, Kinross K, Yagita H, Koya RC, Graeber TG, Ribas A, McArthur GA and Smyth MJ (2013) Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J. Clin. Invest. 123, 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Chen P, Huang Y, Bong R, Ding Y, Song N, Wang X, Song X and Luo Y (2011) Tumor-associated macrophages promote angiogenesis and melanoma growth via adrenomedullin in a paracrine and autocrine manner. Clin. Cancer Res. 17, 7230–7239. [DOI] [PubMed] [Google Scholar]
- 32.Tian K, Chen P, Liu Z, Si S, Zhang Q, Mou Y, Han L, Wang Q and Zhou X (2017) Sirtuin 6 inhibits epithelial to mesenchymal transition during idiopathic pulmonary fibrosis via inactivating TGF-beta1/Smad3 signaling. Oncotarget 8, 61011–61024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibuya M (2011) Involvement of Flt-1 (VEGF receptor-1) in cancer and preeclampsia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 87, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulda S and Kogel D (2015) Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene 34, 5105–5113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CRISPR/Cas9-mediated SIRT6 knockout results in reduced A375 cell growth in vitro. A375 SIRT6 Knockout (KO) cell pool was plated for isolation of individual colonies, and clones were tested for SIRT6 protein KO and growth effects. (A) Cell lysates from each clone (with equal amounts of protein) were subjected to immunoblot analysis to confirm SIRT6 KO. β-tubulin was used as a loading control. (B) Cell growth and viability of the 4 clones with the lowest SIRT6 expression was determined by trypan blue exclusion assay. Results are expressed as relative growth from 24h and percentage of viable SIRT6 KO cells compared to WT. (C) To determine the clonogenic survival, an equal number of viable cells were plated in triplicate into 6-well plates at low density. After ~10 days, cells were fixed with methanol and stained with crystal violet. The images shown are representative of three experiments with similar results. Statistical analysis was done by two-way ANOVA analyzed via GraphPad Prism Software. * P≤0.05.
Table S1. Antibodies used for immunoblot (IB) and immunohistochemistry (IHC) analyses.
Table S2. Primers used in this study.
Table S3. Significantly modulated genes.


