Abstract
Background
Epidemiological studies report increased comorbidity between depression and autoimmune diseases. The role of shared genetic influences in the observed comorbidity is unclear. We investigated the evidence for pleiotropy between these traits in the UK Biobank (UKB).
Methods
We defined autoimmune and depression cases using hospital episode statistics, self-reported conditions and medications, and mental health questionnaires. Pairwise comparisons of depression prevalence between autoimmune cases and controls, and vice versa, were performed. Cross-trait polygenic risk score (PRS) analyses tested for pleiotropy, i.e., whether PRSs for depression could predict autoimmune disease status, and vice versa.
Results
We identified 28,479 cases of autoimmune diseases (pooling across 14 traits) and 324,074 autoimmune controls, and 65,075 cases of depression and 232,552 depression controls. The prevalence of depression was significantly higher in autoimmune cases than in controls, and similarly, the prevalence of autoimmune disease was higher in depression cases than in controls. PRSs for myasthenia gravis and psoriasis were significantly higher in depression cases than in controls (p < 5.2 × 10−5, R2 ≤ 0.04%). PRSs for depression were significantly higher in inflammatory bowel disease, psoriasis, psoriatic arthritis, rheumatoid arthritis, and type 1 diabetes cases than in controls (p < 5.8 × 10−5, R2 range = 0.06%–0.27%), and lower in celiac disease cases than in controls (p < 5.4 × 10−7, R2 range = 0.11%–0.15%).
Conclusions
Consistent with the literature, depression was more common in individuals with autoimmune diseases than in controls, and vice versa. PRSs showed some evidence for involvement of shared genetic factors, but the modest R2 values suggest that shared genetic architecture accounts for a small proportion of the increased risk across traits.
Keywords: Autoimmune diseases; Depression; Genetics; Pleiotropy; Autoimmune diseases, Depression, Genetics, Pleiotropy, UK Biobank
There is evidence that individuals with a history of autoimmune disease are at greater risk for developing depression (1, 2, 3, 4) and that a history of depression increases risk for developing autoimmune diseases (5,6). The mechanisms driving the bidirectional relationship are poorly understood, but one contributory factor may be that these diseases share biological pathways. We and others have shown that there is no strong evidence for the involvement of human leukocyte antigen alleles in risk for depression, suggesting that the major histocompatibility complex does not harbor shared risk for depression and autoimmune diseases (6, 7, 8). However, genetic risk for autoimmune diseases occurs across the genome (9), and pleiotropic effects outside the major histocompatibility complex may be involved in shared risk for depression and autoimmune diseases.
Few studies have investigated evidence for genome-wide pleiotropy between depression and autoimmune diseases. Euesden et al. (10) found no evidence for association between polygenic risk scores (PRSs) for depression and risk for rheumatoid arthritis (RA), or vice versa. The Psychiatric Genomics Consortium indicated no evidence for significant genetic correlations (rG) between depression and nine autoimmune diseases (after multiple testing correction across 221 traits in total); the strongest correlation observed was between depression and inflammatory bowel disease (IBD) (rG = .07, uncorrected p = .01) (11). Recently, Liu et al. (6) found no association between PRSs for mental health disorders and risk for autoimmune diseases, and only a weak association between PRSs for autoimmune diseases and risk for mental health disorders.
We extend previous work, leveraging the UK Biobank (UKB) to test for pleiotropy between depression and autoimmune diseases with PRS methodology. Given the challenge of reliably defining complex traits using large-scale data, we took two approaches to defining autoimmune diseases and depression. We classified liberally defined cases, based on a single item endorsing diagnosis with an autoimmune disease, and strictly defined cases, based on multiple items. We used this approach to identify individuals affected by any of 14 autoimmune or autoinflammatory traits—collectively referred to as autoimmune diseases throughout. We took a similar approach to classifying depression by requiring a greater number of endorsements in strictly defined cases than in liberally defined cases. Liberally defined cases increase the sample size, while strictly defined cases will reduce the rate of misclassification. We performed cross-trait PRS analyses, testing for association between PRSs for autoimmune diseases and depression, and vice versa. Motivated by the observation of sex-dependent genetic correlations between schizophrenia and autoimmune diseases (12), and by higher prevalence of both depression and autoimmune diseases in females, we stratified PRS analyses by sex. Our study is one of the largest to explore pleiotropy between depression and autoimmune diseases and elucidates the contribution of shared genetic influences to the observed comorbidity.
Methods and Materials
Participants
The UKB is a prospective health study of 500,000 individuals in the United Kingdom. Participants were identified through National Health Service patient registers if they were 40 to 69 years of age during the recruitment phase (2006–2010) and living in proximity to an assessment center. Participants attended a baseline assessment and contributed health information via touchscreen questionnaires and verbal interviews (13). Subsets of participants completed repeat assessments: instance 1 comprised n = 20,335 between 2012 and 2013; instance 2 comprised n = 42,961 (interview) and n = 48,340 (touchscreen) in 2014; and instance 3 comprised n = 2843 (interview) and n = 3081 (touchscreen) in 2019. Participant data are linked to Hospital Episode Statistics containing episodes of inpatient care. Episodes are coded at admission using the ICD-10 (14). Inpatients are assigned one primary code (reason for admission) and a variable number of secondary codes. Additional data are available for psychiatric phenotyping, including an online Mental Health Questionnaire completed by 157,366 participants in 2017 (15). The UKB received ethical approval from the North West–Haydock Research Ethics Committee (reference 16/NW/0274). Participants provided electronic signed consent at recruitment (13).
Autoimmune Phenotyping
Guided by studies that investigated the epidemiological relationship between autoimmune diseases and depression (1,5) we identified cases for 14 autoimmune diseases: pernicious anemia (PA), autoimmune thyroid disease, type 1 diabetes (T1D), multiple sclerosis (MS), myasthenia gravis (MG), celiac disease, IBD (includes Crohn’s disease and ulcerative colitis), psoriasis, ankylosing spondylitis, polymyalgia rheumatica/giant cell arteritis, psoriatic arthritis (PsA), RA, Sjögren syndrome, and systemic lupus erythematosus (SLE).
Two sources of information were used to define autoimmune cases and controls: 1) Hospital Episode Statistics, in which primary and secondary ICD-10 diagnoses recorded between April 1997 to October 2016 were identified from the UKB Data Portal Record Repository; and 2) verbal interview, in which participants’ responses at baseline or instance 1 or 2 were used to determine self-endorsed medical conditions (past and current) and self-endorsed prescription medications (current). ICD-10 codes, self-endorsed conditions, and medications used to define each autoimmune disease are listed in Supplement 2.
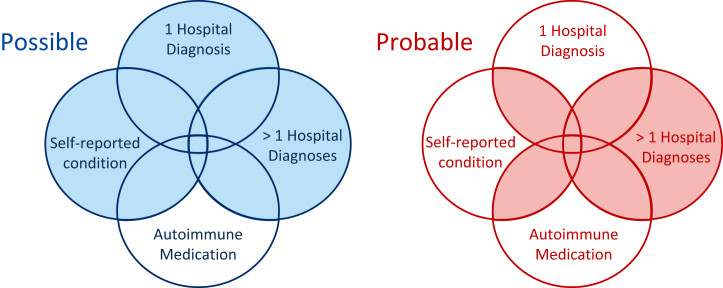
We took two approaches to defining autoimmune cases (Figure 1). To increase sample size, we created possible cases, comprising participants with an ICD-10 diagnosis or a self-endorsed condition. To increase validity, we used multiple criteria to define probable cases. Participants were coded as probable cases if at least two of ICD-10 diagnosis, self-endorsed condition, or medication were observed. More than one ICD-10 diagnosis for the corresponding autoimmune disease was also sufficient. A set of autoimmune controls was defined from participants with no ICD-10 diagnoses, self-endorsed conditions, or medications for all 14 autoimmune diseases. A single set of controls was used for all autoimmune diseases, given the known comorbidity between them.
Figure 1.
Autoimmune phenotyping approach. Cases are included in possible or probable if they fall within a shaded area. Autoimmune medication was used as a confirmatory but not as a primary source of information because several medications are not disease specific.
Depression Phenotyping
We created two depression case groups: strictly defined cases, termed “stringent depression,” and liberally defined cases, termed “any depression.” We have previously shown that single nucleotide polymorphism (SNP)–based heritability increases with multiple endorsements of depression (16). We classified stringent depression as participants endorsing at least three of the following depression measures: ICD-10 diagnoses (F32–F33.9), self-reported depression, self-reported antidepressant usage, single or recurrent depression [defined by Smith et al. (17) from responses to a questionnaire completed at baseline by 172,751 participants], or answering “yes” to the questionnaire: “Have you ever seen a GP/psychiatrist for nerves, anxiety, tension or depression?”
We classified “any depression” as participants who endorsed two or more depression measures, or if they met criteria for lifetime depression in the Composite International Diagnostic Interview assessed in the Mental Health Questionnaire (15). We classify cases defined from the Composite International Diagnostic Interview alone as “any depression” rather than as “stringent depression” because we previously observed lower SNP-based heritability in this group (h2SNP = 11%, SE = 0.008) compared with cases defined by three or more non–Composite International Diagnostic Interview measures of depression (h2SNP = 19%, SE = 0.018) (16).
Depression cases were screened for schizophrenia and bipolar according to any indication: ICD-10 diagnoses (F20–F29, F30–F31.9, F34–F39), self-endorsed conditions (schizophrenia, mania, bipolar disorder or manic depression) or self-endorsed antipsychotic usage reported at baseline or instance 1 or 2, bipolar type I (mania) or bipolar type II (hypomania) according to criteria adopted by Smith et al. (17), or indications of psychosis endorsed in the Mental Health Questionnaire. A single set of depression controls was defined from participants who did not meet the criteria for depression, schizophrenia, or bipolar disorder.
Derivation of depression, schizophrenia, and bipolar indications can be found in the Supplement from our previous publication (16).
Genetic Quality Control
The UKB performed preliminary quality control (QC) on genotype data assayed for all participants (13). Using genetic principal components (PCs) provided by the UKB, we performed 4-means clustering on the first two PCs to identify and retain individuals of European ancestry. QC was then performed using PLINK v1.9 (www.cog-genomics.org/plink/1.9) (18) to remove variants with missingness >0.02 (before individual QC), individuals with missingness >0.02, individuals whose self-reported sex was discordant from their genetic sex, variants with missingness >0.02 (after individual QC), variants departing from Hardy-Weinberg equilibrium (p < 10−8), and variants with minor allele frequency <0.01. Relatedness kinship estimates provided by the UKB were used to identify pairs of related individuals (KING r2 > .044) (19) and the GreedyRelated (20) algorithm used to remove 1 individual from each pair, preferentially retaining individuals that survived QC. FlashPCA2 (21) was used to generate PCs for the subset of individuals of European ancestry surviving QC. PRS analyses were performed using genotype data.
Statistical Analyses
We summarized sociodemographic data taken at baseline assessment: age, sex, socioeconomic status, body mass index, and current smoking status. We tested for significant differences in sociodemographic variables between cases and controls using Welch 2-sample t tests in R version 3.6 (R Foundation for Statistical Computing, Vienna, Austria). We tested for significant differences in 1) the prevalence of depression in autoimmune cases compared with autoimmune controls and 2) the prevalence of autoimmune diseases in depression cases compared with depression controls. These tests were performed for both probable/possible autoimmune cases and stringent/any depression, using 2-sample tests for equality of proportions in R version 3.6.
Summary Statistics for Autoimmune Diseases and Depression
We searched PubMed and the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics) for the latest genome-wide association study (GWAS) with publicly available summary statistics, using the name of the relevant trait (and GWAS or genome-wide association study on PubMed). We identified summary statistics for 8 of the 14 autoimmune diseases: celiac (22), IBD (23), MS (24), MG (25), psoriasis (26), PsA (27), RA (28), and SLE (29) (Table 1). For MG, psoriasis, and PsA, we contacted the authors of the primary GWASs to obtain access. Summary statistics from GWASs using the Immunochip (Illumina, San Diego, CA) were excluded, as it does not provide genome-wide coverage. For major depressive disorder (MDD), we used summary statistics from Wray et al. (11), excluding the UKB.
Table 1.
Genome-wide Association Study Summary Statistics Used to Generate Polygenic Risk Scores
| Trait | Year | Ancestry | Assembly | Cases | Controls |
|---|---|---|---|---|---|
| Celiac Disease | 2010 | European | GRCh37/hg19 | 4533 | 10,750 |
| Inflammatory Bowel Disease | 2015 | European | GRCh37/hg19 | 12,882 | 21,770 |
| Multiple Sclerosis | 2011 | European | GRCh37/hg19 | 9772 | 17,376 |
| Myasthenia Gravis | 2015 | European | GRCh37/hg19 | 1032 | 1998 |
| Psoriasis | 2017 | European | GRCh37/hg19 | 19,032 | 286,769 |
| Psoriatic Arthritis | 2018 | European | GRCh37/hg19 | 1430 | 1417 |
| Rheumatoid Arthritis | 2014 | European | GRCh37/hg19 | 14,361 | 43,923 |
| System Lupus Erythematosus | 2015 | European | GRCh37/hg19 | 7219 | 15,991 |
| Major Depressive Disorder | 2018 | European | GRCh37/hg19 | 116,404 | 314,990 |
PRS Analyses
PRS analyses were conducted using the PRSice-2 software (30). QC was performed on summary statistics to remove variants within the major histocompatibility complex (28.8–33.7 Mb), and default clumping settings were applied in PRSice-2 to remove variants in linkage disequilibrium (r2 > .1) with the lead variant within a 250-kb region.
To validate our phenotyping approach, we tested for association between PRSs for 8 autoimmune diseases and case-control status for the corresponding diseases (possible and probable cases), and between PRSs for MDD and case-control status for depression (any and stringent).
To investigate pleiotropy between autoimmune diseases and depression, we performed cross-trait analyses, testing for association between 1) PRSs for 8 autoimmune diseases and case-control status for depression (any and stringent cases) and 2) PRSs for MDD and case-control status for 14 autoimmune diseases (possible and probable cases). To test for sex-specific effects, we performed cross-trait analyses in males and females separately.
For each test, PRSs constructed at eight p-value thresholds (pT) (.001, .05, .1, .2, .3, .4, .5, and 1.0) were regressed on case-control status using logistic regression, adjusting for the following covariates: six PCs, genotyping batch, and assessment center (n = 128 variables). We report p values at the optimal pT for each test. To control for multiple testing across pT (×8) and across tests of association (autoimmune PRSs [×8] predicting any/stringent depression [×2] in men and women [×2], n = 32; and MDD PRSs predicting possible/probable [×2] autoimmune diseases [×14] in men and women [×2], n = 56), a Bonferroni correction was applied to give a p-value threshold for significance of 7.1 × 10−5 [.05 / 704 tests, 704 = 8 × (32 + 56)]. Where sex-specific associations were observed, sensitivity analyses were conducted to account for different sample sizes between sexes. We tested for interactions between sex and PRSs (at the optimal pT from sex-specific tests) in the full sample (phenotype ∼ sex + PRSs + [sex × PRSs] + covariates). We report R2 estimates transformed to the liability scale using the following population prevalences for outcome traits: PA = 0.1% (31), autoimmune thyroid disease = 2% (32), T1D = 0.3% (33), MS = 0.1% (24), MG = 0.02% (34), celiac = 1% (22), IBD = 0.5% (23), psoriasis = 2% (26), ankylosing spondylitis = 0.55% (35), polymyalgia rheumatica/giant cell arteritis = 0.85% (36), PsA = 0.5% (27), RA = 1% (37), Sjögren syndrome = 0.7% (38), SLE = 0.1% (39), and MDD = 15% (11).
AVENGEME (40) was used to estimate power to detect cross-trait PRS associations, assuming varying degrees of genetic correlation (rG) between corresponding traits (rG = .01–.5). Power was estimated for cross-trait analyses in which summary statistics for both traits were available (i.e., 8 autoimmune disorders and MDD) so that SNP-based heritability (required for power calculations in AVENGEME) could be estimated using linkage disequilibrium score regression (LDSC) (v1.0.1) (41) (Figure S1 in Supplement 1). Power was estimated using PRSs at the optimal pT identified in cross-trait association tests, and liberally defined sample sizes. Parameters used to estimate power are in Tables S1 and S2 in Supplement 1.
LDSC v1.0.1 (41) was used to estimate rG between the UKB depression phenotypes (any and stringent) and autoimmune diseases with publicly available summary statistics. To robustly apply LDSC, we limited the autoimmune diseases to those with sample sizes above 5000 in the primary GWAS [celiac (22), IBD (23), MS (24), psoriasis (26), RA (28), and SLE (29)]. To control for multiple testing across traits, a Bonferroni correction was applied to give a p-value threshold for significance of 4.1 × 10−3 in rG analyses (.05/12 tests).
Results
A total of 28,479 individuals were identified as possible cases across 14 autoimmune diseases, and a subset of 16,824 (59.1%) met the stringent criteria for probable cases (refer to Supplement 2 for representation of the overlap between criteria used to define cases). A total of 65,075 individuals met the criteria for any depression, 14,625 of whom met the criteria for stringent depression. Sociodemographic characteristics for autoimmune and depression cases and controls are summarized in Table 2. Overall, autoimmune and depression case groups contained a higher proportion of females, had lower socioeconomic status, had higher smoking prevalence, and had higher body mass index than their respective control groups (n = 324,074 autoimmune controls, n = 232,552 depression controls; all p values <5 × 10−21 in pairwise comparisons).
Table 2.
Sociodemographic Information for Autoimmune and Depression Cases and Controls
| Pop. Prev. |
Count |
UKB Prev. |
Age, Years, Mean (SD) |
Female |
TDI, Mean (SD) |
Current Smoker |
BMI, kg/m2, Mean (SD) |
Count |
UKB Prev. |
Age, Years, Mean (SD) |
Female |
TDI, Mean (SD) |
Current Smoker |
BMI, kg/m2, Mean (SD) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Possible |
Probable |
||||||||||||||
| Autoimmune Diseases | |||||||||||||||
| Circulatory System | |||||||||||||||
| Pernicious anemia | 0.10% | 1555 | 0.48% | 58.9 (7.61) | 71% | −0.85 (3.19) | 13% | 28.0 (5.55) | 423 | 0.13% | 60.1 (7.19) | 72% | −0.99 (3.23) | 12% | 28.5 (5.34) |
| Endocrine System | |||||||||||||||
| Autoimmune thyroid disease | 2.00% | 859 | 0.26% | 56.8 (7.75) | 85% | −1.11 (3.05) | 15% | 27.4 (5.11) | 607 | 0.19% | 57.0 (7.64) | 86% | −1.06 (3.03) | 16% | 27.8 (5.21) |
| Type 1 diabetes | 0.30% | 2751 | 0.84% | 58.3 (7.80) | 42% | −0.46 (3.39) | 13% | 30.1 (6.08) | 2292 | 0.70% | 58.0 (7.85) | 43% | −0.50 (3.39) | 12% | 29.9 (6.07) |
| Nervous System | |||||||||||||||
| Multiple sclerosis | 0.10% | 1683 | 0.52% | 55.4 (7.52) | 73% | −1.45 (3.01) | 16% | 26.9 (5.08) | 1154 | 0.35% | 55.5 (7.45) | 73% | −1.33 (3.06) | 17% | 26.9 (5.27) |
| Myasthenia gravis | 0.02% | 234 | 0.07% | 59.2 (7.39) | 56% | −0.94 (3.27) | 11% | 29.0 (5.44) | 147 | 0.05% | 60.1 (7.09) | 48% | −1.06 (3.32) | 12% | 29.3 (5.18) |
| Digestive System | |||||||||||||||
| Celiac | 1.00% | 2364 | 0.72% | 57.8 (7.79) | 67% | −1.49 (2.99) | 7% | 25.8 (4.57) | 1260 | 0.39% | 58.4 (7.66) | 68% | −1.42 (3.03) | 7% | 25.8 (4.60) |
| Inflammatory bowel disease | 0.50% | 5105 | 1.55% | 57.4 (7.92) | 51% | −1.27 (3.10) | 10% | 27.3 (4.70) | 3538 | 1.08% | 57.3 (7.98) | 50% | −1.28 (3.07) | 9% | 27.2 (4.63) |
| Skin | |||||||||||||||
| Psoriasis | 2.00% | 5459 | 1.66% | 56.8 (8.01) | 46% | −1.05 (3.22) | 15% | 28.4 (5.17) | 2759 | 0.84% | 57.1 (8.04) | 42% | −0.90 (3.28) | 16% | 28.7 (5.26) |
| Musculoskeletal System and Connective Tissue | |||||||||||||||
| Ankylosing spondylitis | 0.55% | 1344 | 0.41% | 57.9 (7.47) | 38% | −1.02 (3.21) | 13% | 27.8 (4.80) | 413 | 0.13% | 57.4 (7.57) | 26% | −1.06 (3.22) | 12% | 27.9 (4.85) |
| Polymyalgia rheumatica/GCA | 0.85% | 1627 | 0.50% | 63.1 (5.16) | 67% | −1.70 (2.79) | 9% | 28.3 (5.08) | 898 | 0.28% | 63.7 (4.58) | 68% | −1.67 (2.75) | 9% | 28.4 (5.24) |
| Psoriatic arthritis | 0.50% | 1107 | 0.34% | 56.6 (7.48) | 50% | −1.16 (3.18) | 11% | 29.0 (5.43) | 779 | 0.24% | 56.6 (7.52) | 51% | −1.06 (3.23) | 11% | 29.3 (5.59) |
| Rheumatoid arthritis | 1.00% | 6360 | 1.92% | 59.5 (7.06) | 67% | −0.91 (3.29) | 13% | 28.5 (5.53) | 3451 | 1.05% | 59.6 (7.01) | 70% | −1.08 (3.18) | 12% | 28.2 (5.50) |
| Sjögren syndrome | 0.70% | 647 | 0.20% | 59.3 (7.10) | 90% | −1.20 (3.03) | 6% | 27.1 (5.72) | 389 | 0.12% | 59.2 (7.26) | 89% | −1.19 (3.10) | 7% | 27.0 (5.55) |
| Systemic lupus erythematosus | 0.10% | 624 | 0.19% | 56.7 (8.18) | 84% | −1.02 (3.21) | 14% | 27.3 (5.65) | 362 | 0.11% | 56.6 (8.00) | 86% | −0.99 (3.15) | 14% | 27.3 (5.82) |
| Any Autoimmune Disease | NA | 28,479 | 8.08% | 58.1 (7.73) | 58% | −1.11 (3.17) | 12% | 27.9 (5.28) | 16,824 | 4.94% | 58.1 (7.73) | 57% | −1.10 (3.17) | 12% | 28.0 (5.34) |
| Autoimmune Controls |
NA |
324,074 |
NA |
56.4 (8.06) |
52% |
−1.50 (2.98) |
10% |
27.2 (4.64) |
324,074 |
NA |
56.4 (8.06) |
52% |
−1.50 (2.98) |
10% |
27.2 (4.64) |
| Depression |
Any |
Stringent |
|||||||||||||
| Depression Cases | 15% | 65,075 | 21.86% | 55.4 (7.86) | 67% | −1.09 (3.14) | 13% | 27.8 (5.32) | 14,625 | 5.92% | 56.1 (7.88) | 67% | −0.58 (3.35) | 19% | 28.9 (5.77) |
| Depression Controls | NA | 232,552 | NA | 57.1 (8.10) | 47% | −1.65 (2.89) | 9% | 27.2 (4.53) | 232,552 | NA | 57.1 (8.10) | 47% | −1.65 (2.89) | 9% | 27.2 (4.53) |
Negative scores on the TDI indicate less deprivation.
BMI, body mass index; GCA, giant cell arteritis; NA, not applicable; Pop. Prev., population prevalence estimate; TDI, Townsend deprivation index; UKB Prev., prevalence of cases in the UK Biobank as a proportion of autoimmune/depression controls.
The prevalence of any depression was significantly higher in autoimmune cases than in autoimmune controls (p = 6 × 10−177 for possible cases of any autoimmune disease vs. controls, p = 2 × 10−124 for probable cases of any autoimmune disease vs. controls). The prevalence of stringent depression was significantly higher in autoimmune cases than in autoimmune controls (p = 3 × 10−207 for possible cases of any autoimmune disease vs. controls, p = 6 × 10−163 for probable cases of any autoimmune disease vs. controls) (Table 3).
Table 3.
Prevalence of Depression Within Autoimmune Cases Compared With Autoimmune Controls, Stratified by Possible/Probable for Autoimmune Diseases and Any/Stringent for Depression Cases
| Autoimmune Trait | Depression Prevalence in Autoimmune Traits |
|||
|---|---|---|---|---|
| Any Depression Prevalence |
Stringent Depression Prevalence |
|||
| Possible Autoimmune | Probable Autoimmune | Possible Autoimmune | Probable Autoimmune | |
| Autoimmune Controls | 20.6% | 5.1% | ||
| Any Autoimmune Disease | 28.9% (6 × 10−177) | 29.5% (2 × 10−124) | 10.8% (3 × 10−207) | 11.5% (6 × 10−163) |
| Circulatory System | ||||
| Pernicious anemia | 35.8% (4 × 10−36) | 33.7% (4 × 10−8) | 15.2% (1 × 10−39) | 16.6% (4 × 10−15) |
| Endocrine System | ||||
| Autoimmune thyroid disease | 35.1% (6 × 10−19) | 34.7% (6 × 10−13) | 13.7% (1 × 10−16) | 13.4% (3 × 10−11) |
| Type 1 diabetes | 31.1% (3 × 10−31) | 31.3% (1 × 10−27) | 14.1% (5 × 10−59) | 13.5% (5 × 10−43) |
| Nervous System | ||||
| Multiple sclerosis | 39.3% (6 × 10−56) | 42.5% (3 × 10−51) | 16.9% (5 × 10−54) | 20.0% (8 × 10−57) |
| Myasthenia gravis | 33.3% (5 × 10−5) | 29.2% (3 × 10−2) | 14.1% (5 × 10−6) | 14.8% (4 × 10−5) |
| Digestive System | ||||
| Celiac | 27.3% (3 × 10−12) | 29.8% (2 × 10−12) | 8.4% (4 × 10−8) | 9.5% (7 × 10−8) |
| Inflammatory bowel disease | 24.9% (6 × 10−11) | 24.9% (3 × 10−8) | 8.9% (8 × 10−22) | 9.1% (2 × 10−17) |
| Skin | ||||
| Psoriasis | 26.8% (2 × 10−22) | 28.3% (1 × 10−17) | 8.7% (4 × 10−20) | 9.9% (4 × 10−18) |
| Musculoskeletal System and Connective Tissue | ||||
| Ankylosing spondylitis | 26.9% (5 × 10−7) | 28.4% (5 × 10−4) | 9.6% (7 × 10−9) | 10.5% (9 × 10−5) |
| Polymyalgia rheumatica/GCA | 29.3% (2 × 10−13) | 27.8% (5 × 10−6) | 10.8% (3 × 10−15) | 10.6% (1 × 10−8) |
| Psoriatic arthritis | 31.8% (3 × 10−15) | 34.3% (5 × 10−16) | 10.2% (1 × 10−8) | 13.0% (2 × 10−13) |
| Rheumatoid arthritis | 30.6% (5 × 10−62) | 29.4% (6 × 10−28) | 12.7% (3 × 10−92) | 12.1% (1 × 10−45) |
| Sjögren syndrome | 40.7% (4 × 10−25) | 41.9% (1 × 10−17) | 18.9% (2 × 10−28) | 18.9% (7 × 10−18) |
| Systemic lupus erythematosus | 40.2% (2 × 10−25) | 44.5% (2 × 10−22) | 18.8% (6 × 10−30) | 22.2% (8 × 10−27) |
The p values from pairwise comparisons of depression prevalence in autoimmune cases compared with autoimmune controls are shown in parentheses.
GCA, giant cell arteritis.
The prevalence of possible cases of any autoimmune disease was significantly higher in depression cases than in depression controls (p = 6 × 10−177 for any depression vs. controls, p = 3 × 10−207 for stringent depression vs. controls). The prevalence of probable cases of any autoimmune disease was significantly higher in depression cases than in depression controls (p = 2 × 10−124 for any depression vs. controls, p = 6 × 10−163 for stringent depression vs. controls) (Table 4).
Table 4.
Prevalence of Autoimmune Diseases Within Depression Cases Compared With Depression Controls, Stratified by Possible/Probable for Autoimmune Diseases and Any/Stringent for Depression Cases
| Autoimmune Trait | Autoimmune Prevalence in Depression |
|||||
|---|---|---|---|---|---|---|
| Possible Autoimmune Prevalence |
Probable Autoimmune Prevalence |
|||||
| Any Depression | Stringent Depression | Depression Controls | Any Depression | Stringent Depression | Depression Controls | |
| Any Autoimmune Disease | 10.48% (6 × 10−177) | 14.27% (3 × 10−207) | 6.94% | 6.59% (2 × 10−124) | 9.48% (6 × 10−163) | 4.19% |
| Circulatory System | ||||||
| Pernicious anemia | 0.77% (4 × 10−36) | 1.17% (1 × 10−39) | 0.36% | 0.19% (4 × 10−8) | 0.35% (4 × 10−15) | 0.10% |
| Endocrine System | ||||||
| Autoimmune thyroid disease | 0.41% (6 × 10−19) | 0.58% (1 × 10−16) | 0.20% | 0.28% (6 × 10−13) | 0.39% (3 × 10−11) | 0.14% |
| Type 1 diabetes | 1.20% (3 × 10−31) | 2.07% (5 × 10−59) | 0.69% | 1.02% (1 × 10−27) | 1.65% (5 × 10−43) | 0.58% |
| Nervous System | ||||||
| Multiple sclerosis | 0.87% (6 × 10−56) | 1.31% (5 × 10−54) | 0.35% | 0.63% (3 × 10−51) | 1.02% (8 × 10−57) | 0.22% |
| Myasthenia gravis | 0.11% (5 × 10−5) | 0.17% (5 × 10−6) | 0.05% | 0.06% (3 × 10−2) | 0.12% (4 × 10−5) | 0.04% |
| Digestive System | ||||||
| Celiac | 0.92% (3 × 10−12) | 1.08% (4 × 10−8) | 0.64% | 0.54% (2 × 10−12) | 0.64% (7 × 10−8) | 0.33% |
| Inflammatory bowel disease | 1.82% (6 × 10−11) | 2.56% (8 × 10−22) | 1.43% | 1.28% (3 × 10−8) | 1.85% (2 × 10−17) | 1.00% |
| Skin | ||||||
| Psoriasis | 2.05% (2 × 10−22) | 2.55% (4 × 10−20) | 1.46% | 1.10% (1 × 10−17) | 1.47% (4 × 10−18) | 0.73% |
| Musculoskeletal System and Connective Tissue | ||||||
| Ankylosing spondylitis | 0.52% (5 × 10−7) | 0.72% (7 × 10−9) | 0.37% | 0.17% (5 × 10−4) | 0.25% (9 × 10−5) | 0.11% |
| Polymyalgia rheumatica/GCA | 0.66% (2 × 10−13) | 0.93% (3 × 10−15) | 0.42% | 0.35% (5 × 10−6) | 0.51% (1 × 10−8) | 0.23% |
| Psoriatic arthritis | 0.50% (3 × 10−15) | 0.58% (1 × 10−8) | 0.28% | 0.38% (5 × 10−16) | 0.51% (2 × 10−13) | 0.19% |
| Rheumatoid arthritis | 2.66% (5 × 10−62) | 4.14% (3 × 10−92) | 1.59% | 1.42% (6 × 10−28) | 2.23% (1 × 10−45) | 0.89% |
| Sjögren syndrome | 0.34% (4 × 10−25) | 0.55% (2 × 10−28) | 0.13% | 0.21% (1 × 10−17) | 0.33% (7 × 10−18) | 0.08% |
| Systemic lupus erythematosus | 0.36% (2 × 10−25) | 0.59% (6 × 10−30) | 0.14% | 0.23% (2 × 10−22) | 0.39% (8 × 10−27) | 0.08% |
The p values from pairwise comparisons of autoimmune prevalence in depression cases compared with depression controls are shown in parentheses.
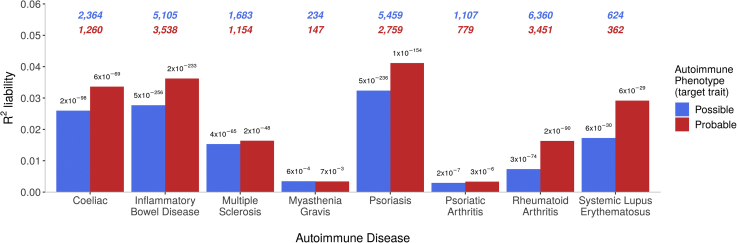
Testing for same-trait PRS associations, PRSs for MDD were significantly associated with any depression case status (p < 5 × 10−324, R2 = 1.48%) and stringent depression case status (p = 2 × 10−228, R2 = 2.23%). PRSs for autoimmune diseases were significantly associated with both possible and probable case-control status for the corresponding diseases (Figure 2). The variance in liability, R2, explained by PRSs was higher in strictly defined compared with liberally defined phenotypes. Most results were highly significant (p < 6 × 10−29), except for MG (p < 7 × 10−3), which had the smallest sample size of 234 cases, and PsA (p < 3 × 10−6), in which the discovery GWAS had only 1430 cases.
Figure 2.
Variances in autoimmune liability explained by polygenic risk scores for the corresponding autoimmune diseases. The number of cases are shown at the top of the plot (possible = blue, probable = red). The p values are shown atop each bar.
Power analyses showed that in the prediction of any depression from autoimmune PRSs, there was 80% power to detect associations assuming modest levels of underlying genetic correlation (rG); rG < .05 for celiac, MS, psoriasis, and SLE; rG < .1 for IBD, PsA, and RA; and rG < .17 for MG (Figure S2 in Supplement 1). In the prediction of possible autoimmune diseases from depression PRSs, there was 80% power to detect associations assuming rG < .05 for celiac and IBD, rG < .1 for psoriasis and RA, and rG < .15 for PsA and SLE. There were two exceptions, MS and MG, in which the underlying rG would need to approach ∼.3 to achieve 80% power (Figure S3 in Supplement 1).
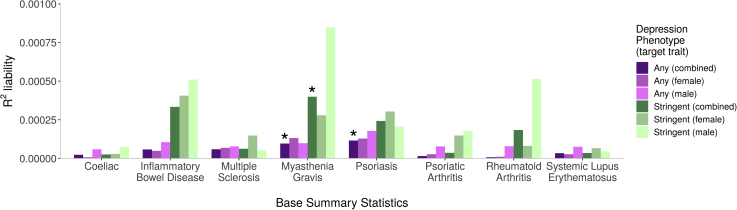
In the prediction of depression from autoimmune PRSs (Figure 3), PRSs for MG were significantly associated with case status for any depression (p = 5.2 × 10−5, R2 = 0.01%) and stringent depression (p = 1.6 × 10−5, R2 = 0.04%). PRSs for psoriasis were significantly associated with case status for any depression (p = 8.7 × 10−6, R2 = 0.01%). No other autoimmune PRSs predicted depression case-control status, and no sex-specific analyses met the Bonferroni-corrected threshold. The R2 values for variance explained in depression by autoimmune PRSs were all very low, at <0.1%, and substantially lower than the R2 for autoimmune diseases (Figure 2).
Figure 3.
Variances in depression liability explained by polygenic risk score for autoimmune diseases (x-axis). Asterisks denote associations with p values <7.1 × 10−5, meeting Bonferroni correction. Number of cases for depression phenotypes: any (combined) = 65,075; any (female) = 43,413; any (male) = 21,662; stringent (combined) = 14,625; stringent (female) = 9738; stringent (male) = 4887.
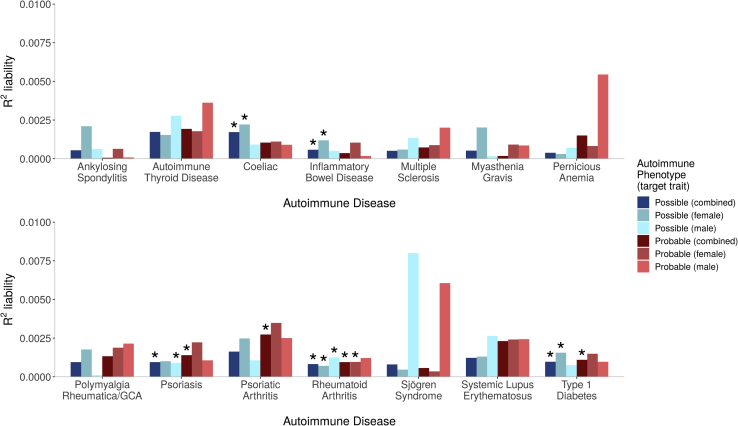
In the prediction of autoimmune diseases from depression PRSs, genetic liability for MDD was significantly associated with six autoimmune diseases: celiac, IBD, psoriasis, PsA, RA, and T1D (all p values <5.8 × 10−5, R2 range between 0.06% and 0.27%) (Figure 4). For three, the association with MDD was observed in probable and possible cases (psoriasis, RA, and T1D). For celiac and IBD, the association was only in possible cases. For PsA, the association was only in probable cases. For all significant associations, higher PRSs increased risk for the outcome phenotype, except for celiac, in which higher MDD PRSs were associated with reduced risk (p = 6 × 10−8, R2 = 0.17%, beta = −0.11, SE = 0.02, in the combined sample).
Figure 4.
Variances in autoimmune liability (x-axes) explained by polygenic risk scores for major depressive disorder. Asterisks denote associations with p values <7.1 × 10−5, meeting Bonferroni correction. Number of cases for the autoimmune diseases are given in Table 2. GCA, giant cell arteritis.
In the prediction of autoimmune diseases from depression PRSs, sex-specific associations were observed, primarily in female autoimmune cases (celiac, IBD, T1D, and RA, all p < 4.5 × 10−5). Association in males was observed in psoriasis (possible cases, p = 5.8 × 10−5) and in RA (possible cases, p = 1.6 × 10−5). The most consistent results were observed in RA, in which the sample size was largest, with five of the six analyses reaching Bonferroni threshold (all p < 4.5 × 10−5, R2 range between 0.07% and 0.1%). However, there was no evidence for a significant interaction between sex and PRSs in the combined samples of men and women (all p > .02), indicating that sex-specific associations were generally influenced by sample size.
Full results of each test are shown in Tables S3 to S6 and Figures S4 to S7 in Supplement 1.
Significant genetic correlations (rG) were observed between IBD and any depression (rG = .11; 95% confidence interval, .03–.18; p = 3.8 × 10−3) and stringent depression (rG = .16; 95% confidence interval, .07–.24; p = 3.0 × 10−4) and between psoriasis and stringent depression (rG = .16; 95% confidence interval, .06–.26; p = 1.1 × 10−3). No other traits met the Bonferroni-corrected threshold for significance in rG analyses (Table S7 in Supplement 1).
Discussion
Motivated by epidemiological findings of a bidirectional relationship between depression and autoimmune diseases, we tested for evidence of pleiotropy between these traits, adopting both liberal and strict phenotyping to define cases in the UKB. We showed modest association of PRSs from autoimmune diseases with MDD, and slightly stronger associations of MDD PRSs with autoimmune diseases. These observations suggest that only a minor component of observed comorbidity is due to shared genetics between depression and autoimmune diseases.
We made three key observations: 1) phenotypic variance explained by PRSs for corresponding traits was higher in strictly defined than liberally defined cases, indicating that more rigorous phenotyping improved the validity of autoimmune and depression cases; 2) the phenotypic overlap between depression and autoimmune diseases was consistent with the literature, in which depression was more common in individuals with autoimmune diseases, and vice versa; and 3) cross-trait PRS analyses identified significant associations between depression and some autoimmune diseases, but with effect sizes indicating the existence of a shared biological component of modest effect on the observed comorbidity.
Our phenotyping approach used both strictly defined and liberally defined cases, integrating the multiple sources of UKB data. PRSs for 8 autoimmune diseases predicted case-control status, increasing confidence in the robustness of case definition. The phenotypic variance explained was higher in strictly defined cases, potentially reflecting greater specificity; identifying individuals with multiple endorsements for a disease reduces the probability of misclassifying controls as cases. Conversely, the criteria for liberally defined cases increases sample size but may induce misclassification.
For each of the autoimmune diseases considered, cases had higher frequencies of depression than controls, recapitulating the effect observed in epidemiological studies. Similarly, the prevalence of each autoimmune disease was significantly higher in depression cases than in controls. Prevalence estimates reported here are cross-sectional, and we lack information on the temporal relationship between traits.
Cross-trait PRS analyses identified significant associations, although observed effect sizes were small, ranging between R2 = 0.01% and 0.27%. Compared with the substantially higher phenotypic variance explained by PRSs in corresponding traits, the small effect sizes observed in cross-trait PRS analyses provide a useful contrast, indicating only a small contribution of shared genetic influences in the observed comorbidities. However, this was not universally true—MDD PRSs captured nearly the same amount of variance in probable PsA (0.27%) as the PRSs for PsA (0.29%). For all significant associations, higher PRSs increased risk for the outcome phenotype. Interestingly, there was one exception, in which higher MDD PRSs were associated with reduced risk for celiac disease. This is intriguing, given the positive phenotypic correlation between depression and celiac disease, and may warrant further investigation.
For three traits, we observed significant associations in liberally defined but not strictly defined cases (psoriasis PRSs were associated with any depression, and MDD PRSs were associated with possible celiac and IBD). In contrast, MDD PRSs were associated with probable, but not possible, PsA, suggesting misclassification in possible cases. Misclassification bias may vary across diseases; some autoimmune diseases may be more prone to misclassification with other autoimmune diseases, while other diagnoses may misclassify with noninflammatory conditions. For example, osteoarthritis (noninflammatory) may misclassify as PsA in the absence of multiple-item endorsement to increase diagnosis validity.
Cross-trait PRS analyses identified some sex-dependent associations. MDD PRSs were associated with psoriasis in males and MDD PRSs were associated with celiac, IBD, and T1D in females. However, sensitivity analyses revealed no evidence for significant interactions between PRSs and sex in the combined sample, indicating that sex-dependent associations were generally driven by different sample sizes in sex-stratified analyses. RA was the most common autoimmune disease and showed the most consistency in cross-trait associations; MDD PRSs were significantly associated with RA in all case groups, except for probable males. This is in contrast with Euesden et al.(10), who found no evidence for association between depression PRSs and risk for RA, but in a smaller sample of 226 cases. Liu et al. (6) also found no evidence for association between composite mental health disorder PRSs and risk for autoimmune diseases, but also in a smaller sample of 1383 individuals with any of seven autoimmune diseases. However, composite PRSs for autoimmune diseases did show weak association with case-control status in a sample of 43,902 individuals with any of six mental health disorders. This highlights the importance of sample size, and our study benefits from the scale of the UKB, in which power calculations indicated our investigation was able to detect modest pleiotropic effects.
In contrast to the small, but significant, cross-trait PRS associations observed between depression and several autoimmune diseases, we only observed significant genetic correlations between depression and two autoimmune diseases: IBD and psoriasis. The PRS methodology, which exploits the use of individual-level data, may have increased power to detect weak genetic effects compared with LDSC, which uses only summary statistics.
The weak genetic contribution suggests that another mechanism may be driving or contributing to the bidirectional relationship between autoimmune diseases and depression (42). Inflammatory factors underlying some cases of depression could provide a common biological pathogenesis with autoimmune diseases. Lynall et al. (43) observed increased immune cell counts in depression cases compared with controls, and identified a subgroup of cases with elevated inflammatory markers who presented with more severe depression than uninflamed cases. Environmental risk factors such as body mass index and childhood maltreatment increase risk of both depression and autoimmune diseases and would contribute to the bidirectional effect (44,45). Similarly, some treatments for depression (antidepressants) and autoimmune diseases (steroids) are obesogenic and may increase comorbidity. Diagnosis with autoimmune disease increases risk of depression due to psychological factors in adjusting to a chronic disorder and changes in behavior such as reduced exercise. Health-related behaviors that are elevated in depression (smoking, poor diet, reduced physical activity) increase risk for autoimmune diseases. These mechanisms may not be independent of joint genetic contributors. For example, shared inflammatory mechanisms would lead to horizontal pleiotropy, in which genetic variants directly affect both disorders, and vertical pleiotropy can arise through environmental risk factors in which genetic variation influences one trait through mediation on another trait. The mechanisms underpinning the observed cross-trait PRS associations may warrant further investigation, potentially using Mendelian randomization to investigate whether MDD risk alleles have a causal effect on autoimmune diseases, and vice versa. It is also interesting to speculate that associations could be driven by phenotypic hitchhiking, in which a GWAS for one trait (e.g., MDD) ascertains patients with comorbid diseases (e.g., autoimmune), potentially inducing cross-trait correlations. Disentangling pleiotropy from phenotypic hitchhiking may warrant further investigation.
Limitations
A healthy volunteer bias has been observed in the UKB (46) and is a noted limitation of the study. However, it has been proposed that this bias may attenuate, but not invalidate, exposure-outcome relationships (47). A further limitation of the ability to extrapolate our results is the lack of representation in individuals of diverse ancestries. The literature has demonstrated attenuation in PRS analyses in which training and target samples come from different ancestral populations (48), highlighting the need to perform GWASs in diverse ancestries. This limitation may have broader implications than would otherwise be the case for some conditions, such as SLE, that disproportionately affect individuals of African and Asian ancestry.
Although every effort has been made to address the potential for misclassification through the criteria for multiple-item endorsements in strictly defined cases, the approach remains imperfect. For example, limited sample size led us to combine thyroiditis and Graves’ disease, which have opposing thyroid function, under the broader classification of autoimmune thyroid disease.
Power calculations showed that for some rare autoimmune diseases, larger samples would be required to reject the presence of a weak genetic correlation with depression. We also observed low SNP-based heritability using published summary statistics for MS, which reduced power to detect pleiotropic effects. The Bonferroni correction applied to cross-trait PRS analyses was conservative because the eight PRS p value thresholds included in each test of association are correlated, although it is difficult to determine the appropriate correction, and we chose to be strict rather than liberal.
Conclusions
We identified cases and controls for depression and 14 autoimmune diseases in the UKB, using both strict and liberal phenotyping. PRS analyses indicated that strict phenotyping improved the validity of cases, demonstrating that multiple UKB variables can be leveraged to increase specificity. Consistent with the literature, we found that depression was enriched in autoimmune cases, and vice versa. Despite having power to detect subtle pleiotropic effects, we found little evidence that shared genetic factors have a meaningful influence on the observed co-occurrence of depression and autoimmune diseases in the UK Biobank. The limited shared genetic component will make only a modest contribution to the bidirectional disease risks, and shared environmental factors, including health-related characteristics and stressful life events, may be important. Future studies leveraging phenotypic, genetic, diagnostic, treatment and environmental risk factors may be necessary to unpick the mechanisms contributing to shared risks for autoimmune diseases and depression. In particular, future research should consider the psychological impacts of autoimmune disease while remaining cognizant of the need to consider and treat the two diseases in parallel.
Acknowledgments and Disclosures
This work was supported by the UK Medical Research Council (Ph.D. studentship to KPG; Grant No. MR/N015746/1). This work represents independent research part-funded by the National Institute for Health Research Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health and Social Care.
We thank participants and scientists involved in making the UK Biobank resource available (http://www.ukbiobank.ac.uk/). The UK Biobank received ethical approval from the North West – Haydock Research Ethics Committee (reference 16/NW/0274). This study was conducted under application number 18177. We thank the research participants and employees of 23andMe for making this work possible. The MDD GWAS summary statistics results from 23andMe were available through a Data Transfer Agreement between 23andMe, Inc., and King’s College, London. Only summary statistics were shared with no individual level data. 23andMe participants provided informed consent and participated in the research online. The 23andMe protocol was approved by an external Association for the Accreditation of Human Research Protection Programs accredited Institutional Review Board, Ethical and Independent Review Services. Participants were included in the analysis on the basis of consent status as checked at the time data analyses were initiated. Statistical analyses were carried out on the King's Health Partners High Performance Compute Cluster funded with capital equipment grants from the GSTT Charity (Grant No. TR130505) and Maudsley Charity (Grant No. 980). We thank Nick Dand, Satveer Mahil, and Catherine Smith of King’s College London for their contribution in identifying medications used in the treatment of psoriasis in the UK Biobank. Data are available from the UK Biobank subject to standard procedures (www.ukbiobank.ac.uk). The full genome-wide association study summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#publication for more information and to apply to access the data. The code used during this study is available at GitHub (https://github.com/kglanville/pleiotropy_autoimmune_depression_ukb).
KPG, CML, JG, and PFO contributed to conceptualization and study design. KPG performed analysis and drafted the manuscript. CML, JG, PFO, and JRIC contributed to analytical consultation and interpretation. JRIC and KPG performed UKB data curation and management. JRIC and KPG performed genetic data preparation. CML, JG, and PFO were project supervisors. All authors critically edited the article.
CML is a member of the SAB for Myriad Neuroscience. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.03.002.
Supplementary Material
References
- 1.Benros M.E., Waltoft B.L., Nordentoft M., Ostergaard S.D., Eaton W.W., Krogh J., et al. Autoimmune diseases and severe infections as risk factors for mood disorders: A nationwide study. JAMA Psychiatry. 2013;70:812–820. doi: 10.1001/jamapsychiatry.2013.1111. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R.J., Freedberg K.A., Clouse R.E., Lustman P.J. The prevalence of comorbid depression in adults with diabetes. Diabetes Care. 2001;24:1069–1078. doi: 10.2337/diacare.24.6.1069. [DOI] [PubMed] [Google Scholar]
- 3.Patten S.B., Beck C.A., Williams J.V.A., Barbui C., Metz L.M. Major depression in multiple sclerosis: A population-based perspective. Neurology. 2003;61:1524–1527. doi: 10.1212/01.wnl.0000095964.34294.b4. [DOI] [PubMed] [Google Scholar]
- 4.Kurina L., Goldacre M., Yeates D., Gill L.W. Depression and anxiety in people with inflammatory bowel disease. J Epidemiol Commun Health. 2001;55:716–720. doi: 10.1136/jech.55.10.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson N.W., Gustafsson L.N., Okkels N., Taha F., Cole S.W., Munk-Jorgensen P., et al. Depression and the risk of autoimmune disease: A nationally representative, prospective longitudinal study. Psychol Med. 2015;45:3559–3569. doi: 10.1017/S0033291715001488. [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Nudel R., Thompson W.K., Appadurai V., Schork A.J., Buil A., et al. Genetic factors underlying the bidirectional relationship between autoimmune and mental disorders – Findings from a Danish population-based study. Brain Behav Immun. 2021;91:10–23. doi: 10.1016/j.bbi.2020.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Glanville K.P., Coleman J.R.I., Hanscombe K.B., Euesden J., Choi S.W., Purves K.L., et al. Classical human leukocyte antigen alleles and C4 haplotypes are not significantly associated with depression. Biol Psychiatry. 2020;87:419–430. doi: 10.1016/j.biopsych.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nudel R., Benros M.E., Krebs M.D., Allesøe R.L., Lemvigh C.K., Bybjerg-Grauholm J., et al. Immunity and mental illness: Findings from a Danish population-based immunogenetic study of seven psychiatric and neurodevelopmental disorders. Eur J Hum Genet. 2019;27:1445–1455. doi: 10.1038/s41431-019-0402-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X., Daly M. What have we learned from six years of GWAS in autoimmune diseases, and what is next? Cur Opin Immunol. 2012;24:571–575. doi: 10.1016/j.coi.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Euesden J., Danese A., Lewis C.M., Maughan B. A bidirectional relationship between depression and the autoimmune disorders - New perspectives from the National Child Development Study. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pouget J.G., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Han B., Wu Y., Mignot E., Ollila H.M., et al. Cross-disorder analysis of schizophrenia and 19 immune-mediated diseases identifies shared genetic risk. Hum Mol Genet. 2019;28:3498–3513. doi: 10.1093/hmg/ddz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization . 10th ed. World Health Organization; Geneva, Switzerland: 1992. International Classification of Diseases. [Google Scholar]
- 15.Davis K.A.S., Coleman J.R.I., Adams M., Allen N., Breen G., Cullen B., et al. Mental health in UK Biobank – Development, implementation and results from an online questionnaire completed by 157 366 participants: A reanalysis. BJPsych Open. 2020;6:E18. doi: 10.1192/bjo.2019.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glanville K.P., Coleman J.R.I., Howard D.M., Pain O., Hanscombe K.B., Jermy B., et al. Multiple measures of depression to enhance validity of major depressive disorder in the UK Biobank. BJPsych Open. 2021;7:E44. doi: 10.1192/bjo.2020.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D.J., Nicholl B.I., Cullen B., Martin D., Ul-Haq Z., Evans J., et al. Prevalence and characteristics of probable major depression and bipolar disorder within UK Biobank: Cross-sectional study of 172,751 participants. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi S.W. choishingwan/GreedyRelated: Function update. 2020. https://github.com/choishingwan/GreedyRelated/tree/1.2.0 Available at: Accessed March 5, 2019.
- 21.Abraham G., Qiu Y., Inouye M. FlashPCA2: Principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 22.Dubois P.C.A., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The International Multiple Sclerosis Genetics Consortium & The Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renton A.E., Pliner H.A., Provenzano C., Evoli A., Ricciardi R., Nalls M.A., et al. A genome-wide association study of myasthenia gravis. JAMA Neurol. 2015;72:396–404. doi: 10.1001/jamaneurol.2014.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsoi L.C., Stuart P.E., Tian C., Gudjonsson J.E., Das S., Zawistowski M., et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Comnmun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aterido A., Cañete J.D., Tornero J., Ferrándiz C., Pinto J.A., Gratacós J., et al. Genetic variation at the glycosaminoglycan metabolism pathway contributes to the risk of psoriatic arthritis but not psoriasis. Ann Rheum Dis. 2019;78:355–364. doi: 10.1136/annrheumdis-2018-214158. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y., Di Wu, Trynka G., Raj T., Terao C., Ikari K., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi S.W., O’Reilly P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andres E., Serraj K. Optimal management of pernicious anemia. J Blood Med. 2012;3:97–103. doi: 10.2147/JBM.S25620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmonds M.J., Gough S.C.L. Unravelling the genetic complexity of autoimmune thyroid disease: HLA, CTLA-4 and beyond. Clin Exp Immunol. 2004;136:1–10. doi: 10.1111/j.1365-2249.2004.02424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradfield J.P., Qu H.-Q., Wang K., Zhang H., Sleiman P.M., Kim C.E., et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spillane J., Higham E., Kullmann D.M. Myasthenia gravis. BMJ. 2012;345:e8497. doi: 10.1136/bmj.e8497. [DOI] [PubMed] [Google Scholar]
- 35.International Genetics of Ankylosing Spondylitis Consortium (IGAS) Cortes A., Hadler J., Pointon J.P., Robinson P.C., Karaderi T., et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Partington R.J., Muller S., Helliwell T., Mallen C.D., Sultan A.A. Incidence, prevalence and treatment burden of polymyalgia rheumatica in the UK over two decades: A population-based study. Ann Rheum Dis. 2018;77:1750–1756. doi: 10.1136/annrheumdis-2018-213883. [DOI] [PubMed] [Google Scholar]
- 37.Humphreys J.H., Verstappen S.M.M., Hyrich K.L., Chipping J.R., Marshall T., Symmons D.P.M. The incidence of rheumatoid arthritis in the UK: Comparisons using the 2010 ACR/EULAR classification criteria and the 1987 ACR classification criteria. Results from the Norfolk Arthritis Register. Ann Rheum Dis. 2013;72:1315–1320. doi: 10.1136/annrheumdis-2012-201960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lessard C.J., Li H., Adrianto I., Ice J.A., Rasmussen A., Grundahl K.M., et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren's syndrome. Nat Genet. 2013;45:1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees F., Doherty M., Grainge M., Davenport G., Lanyon P., Zhang W. The incidence and prevalence of systemic lupus erythematosus in the UK, 1999–2012. Ann Rheum Dis. 2016;75:136–141. doi: 10.1136/annrheumdis-2014-206334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palla L., Dudbridge F. A fast method that uses polygenic scores to estimate the variance explained by genome-wide marker panels and the proportion of variants affecting a trait. Am J Hum Genet. 2015;97:250–259. doi: 10.1016/j.ajhg.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gold S.M., Köhler-Forsberg O., Moss-Morris R., Mehnert A., Miranda J.J., Bullinger M., et al. Comorbid depression in medical diseases. Nat Rev Dis Primers. 2020;6:69. doi: 10.1038/s41572-020-0200-2. [DOI] [PubMed] [Google Scholar]
- 43.Lynall M.E., Turner L., Bhatti J., Cavanagh J., de Boer P., Mondelli V., et al. Peripheral blood cell-stratified subgroups of inflamed depression. Biol Psychiatry. 2020;88:185–196. doi: 10.1016/j.biopsych.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Dube S.R., Fairweather D., Pearson W.S., Felitti V.J., Anda R.F., Croft J.B. Cumulative childhood stress and autoimmune diseases in adults. Psychosom Med. 2009;71:243–250. doi: 10.1097/PSY.0b013e3181907888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes K., Bellis M.A., Hardcastle K.A., Sethi D., Butchart A., Mikton C., et al. The effect of multiple adverse childhood experiences on health: A systematic review and meta-analysis. Lancet Public Health. 2017;2:e356–e366. doi: 10.1016/S2468-2667(17)30118-4. [DOI] [PubMed] [Google Scholar]
- 46.Fry, Littlejohns T.J., Sudlow C., Doherty N., Adamska L., Sprosen T., et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batty G.D., Gale C., Kivimaki M., Deary I., Bell S. Generalisability of results from UK Biobank: Comparison with a pooling of 18 cohort studies. medRxiv. 2019 doi: 10.1101/19004705. [DOI] [Google Scholar]
- 48.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Comnmun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.