Abstract
Previous studies have indicated important roles for NIMA-related kinase 1 (NEK1) in modulating DNA damage checkpoints and DNA repair capacity. To broadly assess the contributions of NEK1 to genotoxic stress and mitochondrial functions, we characterised several relevant phenotypes of NEK1 CRISPR knockout (KO) and wild-type (WT) HAP1 cells. Our studies revealed that NEK1 KO cells resulted in increased apoptosis and hypersensitivity to the alkylator methyl methanesulfonate, the radiomimetic bleomycin and UVC light, yet increased resistance to the crosslinker cisplatin. Mitochondrial functionalities were also altered in NEK1 KO cells, with phenotypes of reduced mitophagy, increased total mitochondria, elevated levels of reactive oxygen species, impaired complex I activity and higher amounts of mitochondrial DNA damage. RNA-seq transcriptome analysis coupled with quantitative real-time PCR studies comparing NEK1 KO cells with NEK1 overexpressing cells revealed that the expression of genes involved in DNA repair pathways, such as base excision repair, nucleotide excision repair and double-strand break repair, are altered in a way that might influence genotoxin resistance. Together, our studies underline and further support that NEK1 serves as a hub signalling kinase in response to DNA damage, modulating DNA repair capacity, mitochondrial activity and cell fate determination.
Introduction
The NIMA-related kinases (NEKs) represent the third largest family of kinases and are broadly involved in cell cycle regulation. They engage a wide variety of substrates and operate in different biological processes (1). Cell cycle progression is monitored by a series of checkpoints in response to DNA damage, and some NEKs are checkpoint targets, inhibited by DNA damage (2,3), and have integral roles in DNA damage response (DDR) signalling (4). Among the NEKs, NEK1 has been the most studied and has been connected to several human diseases, such as polycystic kidney (5), amyotrophic lateral sclerosis (6–8), glioma (9), Wilms tumour (10), thyroid cancer (11) and prostate cancer (12,13). NEK1 has clear roles in regulating the progression of the cell cycle (14), and due to its prominence in the meiotic phase, the kinase operates in gametogenesis (15), spermatogenesis (16), and helps in the assembly of the meiotic spindle (15). Additionally, it has been associated with many DDR pathways, interacting directly with various repair proteins (14).
The first evidence of NEK1’s involvement in DNA damage and repair arose from a yeast two-hybrid screen, which identifies protein interactions unbiasedly (17). Using the regulatory and catalytic domain of NEK1 as a bait, we identified proteins involved in cell cycle checkpoints and double-strand break (DSB) repair. The most prominent interactors were: MRE11, a nuclease component of the MRN complex (MRE11/NBS1/RAD50) that binds damaged DNA and signals DSBs (18); ATRX (alpha thalassaemia/mental retardation syndrome X-linked), a protein i.e. functionally like RAD54 as a chromatin remodeller in homologous recombination (HR) (15); and 53BP1, a mediator of the DNA damage checkpoint (19). At the molecular level, it has been shown that NEK1 stabilises the complex between ATR (ATM and Rad3-related) and ATRIP (ATR interacting protein), two central components of the DDR to replication stress, through phosphorylation (20).
NEK1 has also been shown to be important in the process of cell apoptosis. The onset of apoptosis is initiated by the release of cytochrome C from mitochondria (21). Permeabilisation of mitochondria is seminal for the regulation of apoptosis induced by cytotoxic stress and is modulated by the mitochondrial permeability transition pore, composed of the outer mitochondrial membrane protein VDAC1 (voltage-dependent anion-selective channel protein 1) (16,22). One of the key regulators for the VDAC1 channel is NEK1. The communication between NEK1 and VDAC1 is central to cell survival and appears to be an important element of the DDR as well as the apoptotic pathway (23–25).
Distinct DNA repair pathways have evolved to cope with specific forms of genetic injury. Some lesions may be removed by simple reversal, such as directed by the repair protein MGMT (O6-methylguanine-DNA methyltransferase). Other lesions that distort the DNA helix are handled by nucleotide excision repair (NER) (26), whereas base excision repair (BER) (27,28) copes with most simple base modifications and mismatch repair (MMR) deals with mismatched bases that are erroneously introduced during replication. DNA DSBs are usually resolved by HR (27,28) or non-homologous end-joining (29,30). Other types of genomic injury, such as DNA interstrand crosslinks, require the involvement of multiple pathways for resolution, such as NER, HR and translesion synthesis (29,31).
Defects in mitochondrial proteins, mutations in non-mitochondrial genes or impairment of nuclear-to-mitochondrial communication can underlie mitochondrial dysfunction. Moreover, several predominantly nuclear DNA repair proteins are also present in mitochondria (32). All mitochondrial DNA (mtDNA) repair enzymes are encoded by nuclear genes and are translocated to the mitochondria, making signalling between the nucleus and mitochondria imperative (33,34). The objective of the present study was to understand the mechanisms by which NEK1 regulates mitochondrial activity, and the response to genotoxic stress.
Materials and Methods
Cell culture and transfection
Wild-type (WT) HAP1 and NEK1 knockout (KO) human HAP1 cell lines were obtained from Horizon Discovery Group plc. HAP1 is a near-haploid human cell line derived from KBM-7 (35). The KO cells were edited by CRISPR/Cas9 and contain a 2 bp deletion in a coding exon of NEK1 (Supplementary Figure 1, available at Mutagenesis Online). Cells were maintained in a humid incubator with 5% CO2 at 37°C and cultivated in Iscove’s Modified Dulbecco’s Medium (Gibco Thermo Fisher Scientific, Waltham, MA, USA) enriched with 10% certified foetal bovine serum (Gibco) and supplemented with penicillin/streptomycin (100 units/ml, Gibco). To create the transient NEK1 overexpression (OVER) cell line, HAP1 cells were transfected with 10 ng of FLAG-NEK1 (Isoform 1) using Lipofectamine® 3000 Reagent (Thermo Fisher Scientific) in OPTI-MEM reduced-serum media, according to the manufacturer’s protocol. All experiments (e.g. RNA isolation) were performed after 48 h of transfection.
Western blotting
Whole cell extracts were prepared using RIPA buffer (Thermo Fisher Scientific). The supernatant was transferred to a new tube, and protein concentrations were determined using either a Bradford assay (Bio-Rad) or the BCA protein assay kit (Thermo Fisher Scientific) to generate the standard curve. After quantitation, protein samples (as indicated) were separated on a polyacrylamide–SDS gel and transferred to a nitrocellulose membrane (Amersham, GE Healthcare, Chicago, IL). Immunoblot analysis was performed with the primary antibodies: mouse anti‐NEK1 (1:500; Santa Cruz Biotechnology), rabbit anti‐β-actin (1:1000; Abcam, Cambridge, UK), mouse anti‐vinculin (1:700; Abcam, Cambridge, UK), mouse anti-cleaved caspase-9 (1:1000; Cell Signaling), mouse anti-caspase-9 (1:1000; Cell Signaling), rabbit anti-cleaved caspase-3 (1:1000; Cell Signaling), rabbit anti-caspase-3 (1:1000; Cell Signaling), rabbit anti-Bax (1:1000; Cell Signaling), rabbit anti-Bcl-xL (1:1000; Cell Signaling), rabbit anti-Bcl-2 (1:1000; Cell Signaling), rabbit anti-APE1 (1:3000; GeneTex), rabbit anti-DNA POLδ (1:1000; GeneTex), rabbit anti-PCNA (1:2500; GeneTex), rabbit anti-PARP (1:2500; GeneTex), rabbit anti-XPA (1:2000; GeneTex), rabbit anti-XPB (1:1000; GeneTex), rabbit anti-XPA (1:2000; GeneTex), rabbit anti-XPD (1:1000; GeneTex), rabbit anti-RPA70 (1:1000; GeneTex) and rabbit anti-ERCC1 (1:1000; GeneTex), diluted in blocking solution (2% bovine serum albumin and 0.02% sodium azide in tris‐buffered saline). Protein bands were detected by peroxidase‐conjugated antibodies, and blots were developed utilising the enhanced chemiluminescence kit, ECL western blotting system (Amersham).
DNA-damaging agent exposure and cell viability
HAP1 cells (WT and KO) were plated at 2500 cells per well in a 96-well plate, grown overnight at 37°C, and then treated the next day with methyl methanesulfonate (MMS), ultraviolet radiation-short-wave C (UVC), bleomycin or cisplatin; the chemical agents were obtained from Sigma (St Louis, MO, USA). We used these agents since each one is related to a specific DNA repair pathway: MMS methylates DNA generating BER substrates, UVC produces DNA photoproducts repaired by the NER subways, bleomycin creates complex lesions and DSBs primarily resolved by double strand break (DSBR) pathways, and cisplatin is a bifunctional alkylating agent that forms mostly intrastrand crosslinks in DNA that are repaired by NER (36–39). The following concentration ranges were used: MMS (25–200 µM), UVC (5–25 J/m2), bleomycin (6–62 µM) and cisplatin (3–33 µM). After incubation, cell viability was evaluated using the CCK-8 viability assay (Dojindo Molecular Technologies). In brief, 10 µl of the CCK-8 solution was added to the culture, and the plate was placed back in the 37°C incubator for 3 h. The optical density was then measured at 450 nm using a microplate reader, and the results plotted relative to the untreated control.
RNA-seq
Total RNA was extracted from WT, KO and OVER cells in triplicate using AllPrep RNA/Protein Kit (Qiagen); for overexpression cells, RNA was collected immediately after transfection. The construction and analysis of the RNA-seq studies were performed by Novogene Corporation Inc. (Sacramento, CA). Downstream analysis was performed using a combination of programs including STAR, HTseq, Cufflink and wrapped scripts. Alignments were parsed using the Tophat program, and differential expressions were determined through DESeq2/edgeR. KEGG (Kyoto Encyclopedia of Genes and Genomes) is a database resource for understanding high-level functions and utilities of the biological system (http://www.genome.jp/kegg/). We used a cluster profiler R package to test the statistical enrichment of differentially expressed genes in KEGG pathways.
Quantitative real-time PCR
Total RNA was extracted from cells using AllPrep RNA/Protein Kit (Qiagen). 2 µg RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative real-time PCR (qRT-PCR) was performed with a 7500 real-time PCR system (Applied Biosystems). Each real-time RT-PCR reaction (in 20 µl) contained Taqman Fast Advanced MasterMix (Applied Biosystems), cDNA and the primes (TaqMan® Array Plate—custom) for controls and DNA repair genes (PCNA, APEX1, APEX2, FEN1, PARP1, POLB, POLD, OGG1, LIG3, LIG1, XPA, XPC, RPA, DDB1, DDB2, ERCC1, ERCC2, MSH2, MSH6, PMS2, MRE11, XRCC5, BRCA1, ATM and ATR). For qRT-PCR, the cells were grown in normal conditions. The cycling conditions consisted of a single initial step of 20 s at 95°C, followed by 40 cycles of 3 s at 95°C and 30 s at 60°C. PCR amplifications were performed in triplicates for each sample. Gene expression levels were quantified in relation to the expression of four different controls (18s rRNA, GAPDH, HPRT and GUSB), using an optimised comparative Ct (ΔΔCt) method, obtaining fold-change values that reflect the expression of the target gene.
Mitochondrial parameters
Cells were plated to reach 80% confluency after 24 h at 37°C, the solutions were heated to 37°C, cells were harvested, centrifuged, washed in phosphate-buffered saline and resuspended in appropriate solutions depending on the measurement. To determine mitochondrial content, 100 nM MitoTracker Green (Life Technologies) was used for 15 min at 37°C. To determine membrane potential, cells were incubated with 40 nM tetramethylrhodamine methyl ester (TMRM, Life Technologies) for 15 min at 37°C. To determine levels of mitochondrial reactive oxygen species (ROS), cells were resuspended in a solution containing 5 µM MitoSOX Red (Life Technologies) and incubated for 30 min at 37°C, and for cellular ROS, cells were resuspended in a solution containing 3 µM dihydroethidium (hydroethidine) (DHE, Life Technologies) incubated for 30 min at 37°C. Mitophagy was detected by Mitophagy Detection Kit (Dojindo Laboratories, Kumamoto, Japan). Samples will be prepared according to the manufacturer’s instructions. Fluorescence of all samples was determined by flow cytometry (BD Accuri C6) and the cells were plated in 12-well plates; the assays were performed in biological and experimental triplicates. For the analysis of mitochondrial OXPHOS Complex I (NADH dehydrogenase) enzyme activity, we used the Complex I Enzyme Activity Assay Kit (Abcam, ab109721), prepared according to the manufacturer’s instructions. Complex I activity was determined following the oxidation of NADH to NAD+ and the simultaneous reduction of a dye which leads to increased absorbance at OD = 450 nm.
Relative mtDNA copy number
Total cellular DNA was isolated using the DNAeasy Mini Kit (Qiagen, Hilden, Germany), and DNA concentrations were determined spectrophotometrically with Nanodrop. The relative copy number was performed through NovaQUANT™ Human Mitochondrial to Nuclear DNA Ratio Kit (catalog # 72620 EMD Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Equal amounts of total DNA were assayed by quantitative PCR (qPCR) with the SYBR Green mix (Applied Biosystems, Thermo Fisher Scientific). Samples were analysed at least in triplicates in StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Thermo Fisher Scientific). As described in the protocol of the manufacturer, the kit provides pre-aliquoted plates that contain a set of four optimised PCR primer pairs targeting two nuclear and two mitochondrial genes: nuclear genes (BECN1 [GenBank: AC016889.28 (89304–80432)] and NEB [GenBank: NG_009382.2 (16079–16195)]) and mitochondrial genes (ND1 [GenBank KP875569 (3314–3467)] and ND6 [GenBank: KP875569 (14341–14436)]).
A single assay was run using four wells, whereby each well individually testing one of the four gene targets. Resultant Ct values were obtained from the Real-Time instrument and were used to represent the level of each gene, averaging the copy number calculated from the ND1/BECN1 pair and the ND6/NEB pair. To calculate copy number, first calculate N = 2−ΔCt where ΔCt1 = CtND1 − CtBECN1 and ΔCt2 = CtND6 − CtNEB, then to the average of 2−ΔCt1 and 2−ΔCt2. In this way, ratios are calculated and then averaged to represent the mtDNA copy number per cell.
Long-extension PCR
The total DNA of cells was extracted using the DNeasy Blood and Tissue kit (Qiagen) following the specification of the manufacturer. The amplification of short nuclear and of the long mitochondrial fragment was performed using AccuPrime Taq DNA Polymerase High Fidelity kit (Invitrogen). The nuclear gene (177 bp) amplified was HPRT1 (Hypoxanthine Phosphoribosyltransferase 1) using the oligos Forward: 5′-TGACATGTGCCGCCTGCGAG-3′ and Reverse: 5′-GTGGTCGCTTTCCGTGCCGA-3′. The mitochondrial fragment (16 258 bp) was amplified using the oligos Forward: 5′-TGAGGCCAAATATCATTCTGAGGGGC-3′ and Reverse: 5′-TTTCATCATGCGGAGATGTTGGATGG-3′. Cells were incubated in normal conditions or exposed to MMS (75 µM) for 2 h. The intensity (area of the PCR product bands) was measured using ImageJ software and the quantification of the long fragment was divided by the short fragment.
Statistical analysis
The statistical analysis represents the mean of three independent replicates of each experiment. Data were compared using one-way analysis of variance or Tukey’s multiple comparison test or with Student t-test unpaired two-tailed analysis in GraphPad Prism version 8 software (GraphPad Software, San Diego, CA). For all graphs, data are shown as the mean ± SEM; statistically significant differences are represented by * and are considered to be significant if P < 0.05. Fisher’s exact tests were used to test for statistical overrepresentation of KEGG pathways among lists of differentially expressed genes.
Results
NEK1 KO results in increase apoptosis and selective genotoxic hypersensitivity
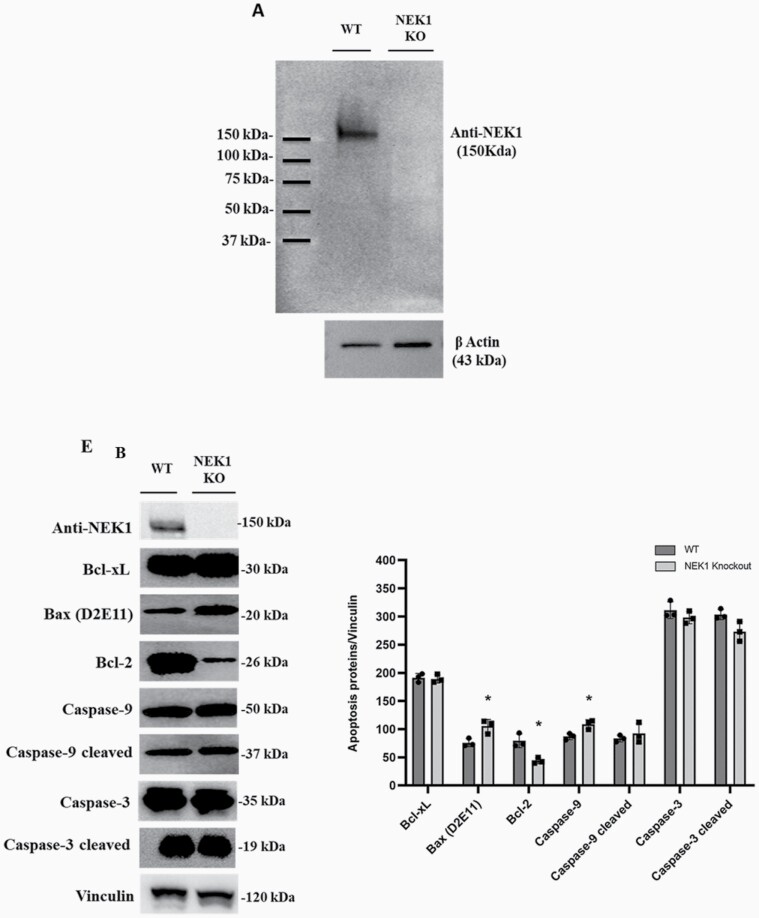
HAP1 control (WT) and NEK1 KO human cell lines were obtained from the Horizon Discovery Group plc, with the KO cells being edited by CRISPR/Cas to contain a 2 bp deletion in a coding exon of the NEK1 gene (Supplementary Figure 1, available at Mutagenesis Online and Figure 1A). To confirm the inactivation of NEK1 gene expression, whole cell extracts were prepared from both cell lines and western blotting was performed. First, no NEK1 protein was observed in the KO cell extracts, consistent with complete gene inactivation (Figure 1A). Second, we also carried out immunodetection of anti-apoptotic and pro-apoptotic proteins (Figure 1B). Anti-apoptotic members, such as Bcl-2, act to prevent apoptosis by sequestering caspases or preventing the release of mitochondrial apoptogenic factors. In HAP1 NEK1 KO cells, we observed a significant decrease in Bcl-2 protein expression (P < 0.0001), implying a reduced capacity to prevent cell death. Conversely, pro-apoptotic members like Bax and Caspase 9 (P < 0.0001) showed a slight and significant increase in KO cells compared with WT, supporting an involvement of NEK1 in suppressing the apoptotic process. We mention that we did not observe significant differences in Caspase 3 expression in KO cells relative to controls. Although Caspase 3 is often a crucial factor in apoptosis, studies have proposed that caspases such as 8 or 9, which have long prodomains, can also activate apoptosis through the activation of Caspase 7 and not Caspase 3. Furthermore, the existence of other proteases similar to Caspase 3 cannot be ruled out, therefore, activation of Caspase 3 is not in itself the only evidence for apoptotic events and cell death (40–43).
Figure 1.
Effect of the HAP1 NEK1 KO cells in apoptosis proteins. (A) Western blotting revealed with anti-NEK1 and β-actin antibodies. (B) Apoptotic proteins expression in WT and NEK1 KO cells. Protein level expression (arbitrary unit) and normalised by vinculin. The significance level was represented by *P < 0.0001; all experiments and data are representative of three experiments and biological replicates.
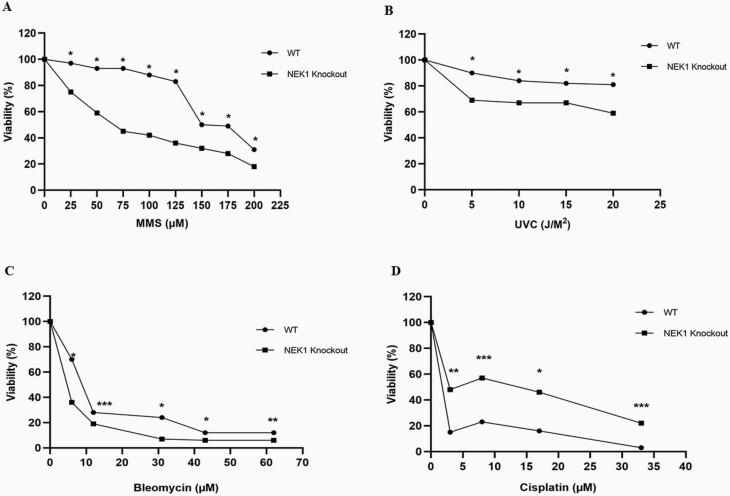
To determine the effect of NEK1 deficiency on genotoxin agent sensitivity, we performed CCK-8 viability assays using cells treated with different concentrations/doses of the alkylator MMS, UVC light (ultraviolet C), the radiomimetic bleomycin or the crosslinker cisplatin. Our viability analysis revealed that NEK1 KO cells exhibit increased sensitivity to MMS, UVC and bleomycin relative to WT cells (Figure 2A, B and C, respectively), suggesting a modulatory role in diverse DDRs. Treatment of HAP1 WT cells with cisplatin resulted in reduced viability in comparison with the KO cells (Figure 2D), indicating slightly elevated cisplatin resistance of NEK1-deficient cells.
Figure 2.
Cellular sensitivity of WT and NEK1 KO cell line. (A) MMS, (B) UVC, (C) bleomycin and (D) cisplatin exposed. Cell viability (relative to the untreated sample) was determined using the CCK-8 viability assay at the indicated doses of genotoxin. Each value represents the average and standard deviation of three data points. The paired t-test was performed to compare WT vs KO cells, and statistically significant differences are indicated: *P < 0.001; **P < 0.01; ***P < 0.05.
NEK1 deficiency alters mitochondrial functions
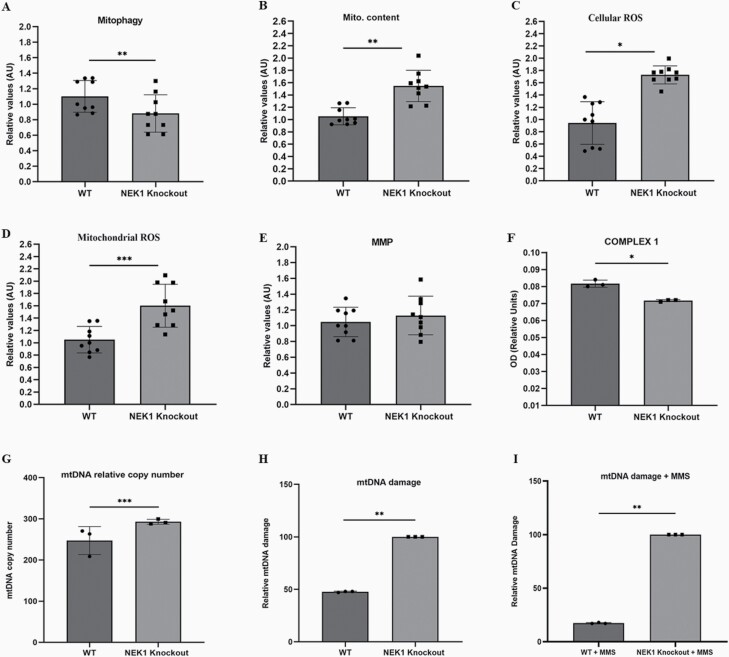
Mitochondrial dysfunction is frequently reflected by changes in the mitochondria number, structure and function (44). We explored the relationship between NEK1 expression and parameters of mitochondrial functionality. Specifically, we performed assays to measure: (i) mitophagy using a Mitophagy Detection Kit, (ii) cellular ROS by DHE, (iii) mitochondrial ROS by MitoSOX, (iv) mitochondrial content using MitoTracker and (v) membrane potential (MMP) using the drug TMRM (Figure 3). As shown in Figure 3, NEK1 KO cells exhibit a decrease in mitophagy (panel A) and an increase in the number of total mitochondria (panel B). NEK1 deficiency is also associated with increased cellular and mitochondrial ROS (panels C and D). The increase in mitochondria could reflect the reduced clearance rate from mitophagy, culminating in a high number of defective mitochondria and increased ROS stress. To determine the efficiency of the mitochondrial electron transport chain, we analysed the activity of complex I (NADH dehydrogenase) in WT and NEK1 KO cells. We observed that NEK1-deficient HAP1 cells exhibit reduced complex I activity (panel F), a phenotype not seen in WT cells, where complex I activity remained high. A specific defect in complex I, which is generally considered to be a main source of ROS, may explain the increased production of mitochondrial ROS in the KO cells.
Figure 3.
NEK1 KO cells exhibit mitochondrial dysfunction. (A) Flow cytometry was used to quantify relative mitophagy (Mitophagy Detection Kit). (B) Mitochondrial content (MitoTracker Green). (C) Cellular ROS (DHE). (D) Mitochondrial ROS (MitoSOX). (E) Mitochondrial membrane potential (TMRM) and (F) mitochondrial complex I activity were measured according to the manufacturer’s instructions (catalog nos. ab109721 Abcam, USA). Each bar represents mean ± SE, n = 3 biologically independent experiments (one-way analysis of variance). (G) The number of mitochondrial DNA copy number by real-time qPCR. (H) The mtDNA damage of NEK1 KO and WT cells was analysed by long-extension PCR and the ratio of the amplification is shown in the graph in basal conditions. (I) The mtDNA damage of cells treated with 75 µM MMS treatment for 2 h was evaluated by long-extension PCR. Results show the difference in mtDNA damage of cells with MMS treatment from the mtDNA damage under basal conditions. The statistical analysis is a comparison to the control of WT cells. Both graphs show analyses from n = 3 independent experiments. *P < 0.001; **P < 0.01; ***P < 0.05.
MtDNA copy number and damage are increased in NEK1 KO cells
Given the mitochondrial abnormalities seen in the NEK1 KO HAP1 cells (see above), the effect of NEK1 deficiency on the relative mtDNA copy number and mtDNA damage was investigated (Figure 3). To determine mtDNA copy number, we used NovaQUANT™ Human Mitochondrial to Nuclear DNA Ratio Kit, which entails real-time PCR with ND1, BECN1, ND6 and NEB primers as the respective markers. Here, we observed that NEK1 KO resulted in increased levels of mtDNA (panel G), which could be explained by impaired mitophagy or NEK1 possibly controlling DNA replication in addition to controlling mitochondrial numbers (i.e. number of mitochondria per cell), thereby regulating mtDNA content. For mtDNA damage, we using long-extension PCR, an assay that compares control amplification (short-range) to test amplification (long-range) within the mitochondrial genome. NEK1 absence decreased the ratio of test: control mtDNA amplification in comparison to WT, suggesting increased DNA damage that interferes with the long-range PCR (panel H). Lastly, we measured mtDNA damage in WT and NEK1 KO HAP1 cells following treatment with 75 µM MMS for 2 h (panel I), observing that NEK1-deficient cells exposed to MMS possess greater mtDNA damage compared with WT cells, perhaps reflective of reduced DNA repair capacity.
Differentially expressed genes and pathways in cells with altered NEK1 expression
Since the collective results indicate that NEK1 is a multifunctional protein involved in apoptosis, and the response to DNA damage, we carried out RNA-seq analysis of NEK1 KO and WT HAP1 cells, focussing specifically on DNA repair and mitochondrial genes. To expand our investigation into the consequences of altered NEK1 expression, with the hypothesis that increased expression might have an opposite effect as observed for KO, we included a NEK1 overexpressing HAP1 cell line (OVER) (Supplementary Figure 2, available at Mutagenesis Online). A 48 h transfection was performed for NEK1 overexpression, so that the vast majority of mRNAs are at their stationary levels, as recent scientific evidence shows that the half-life of most regulated mRNAs in the cell is on the time scale of minutes (45). Moreover, NEK1 overexpression at the protein level has been reported in patients with thyroid cancer and correlated with aggressive characteristics, and is associated with tumour grade and low patient survival for glioma (9,11). Increased expression of NEK1 also promotes persistent phosphorylation of VDAC1 in renal carcinoma cells, preventing apoptosis under genotoxic insults (46), highlighting the importance of understanding the consequences of elevated NEK1 presence.
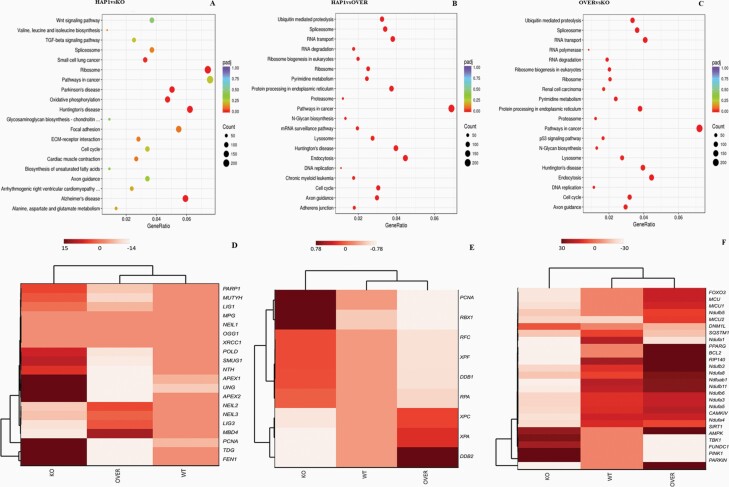
Employing KEGG enrichment analysis (47), where the KEGG pathway is used to understand high-level functions of genes and biological systems, we identified the top 20 pathways that were significantly enriched among the padj-adjusted P-value (P < 0.05, Figure 4A–C). We can see that WT HAP1 vs NEK1 KO (panel A) has a large number of statistically significant (red circles) differentially expressed genes mainly for neurological disease pathways (Alzheimer, Parkinson and Huntington), the ribosome and oxidative phosphorylation, consistent with the mitochondrial defects observed in the NEK1-deficient cells. For WT HAP1 vs NEK1 OVER (panel B) and NEK1 OVER vs NEK1 KO (panel C), many pathways are significantly dysregulated, most notably cancer-related pathways, cell cycle, DNA replication and RNA degradation and transport pathways. An enrichment analysis was carried out, in addition to KEEG, including GO Enrichment (BP: biological process), DO Enrichment and Reactome Database Enrichment, revealing other functions and/or biological pathways that are significantly associated with differential expression of NEK1 (Supplementary Table 1, available at Mutagenesis Online).
Figure 4.
The characteristics of KEGG pathways enrichment scatter plot and hierarchical clustering heat maps in NEK1 gene expression. (A) HAP1 WT vs NEK1 KO cells in different pathways related to biological systems. (B) HAP1 WT vs NEK1 OVER cells in different pathways related to biological systems. (C) NEK1 OVER vs NEK1 KO cells in different pathways related to biological systems. The number of genes expressed in the pathway is indicated by the circle area, and the circle colour represents the ranges of the corrected P-value. Hierarchical clustering heat maps of RNA-seq data from WT, NEK1 KO and NEK1 overexpression (OVER) cells. Counts were normalised with DESEQ2 and then transformed via log2 prior to clustering and visualisation. (D) Gene cluster involved BER. (E) Gene cluster involved NER. (F) Gene cluster involved mitochondrial function.
DDR and mitochondrial pathway components in NEK1 KO and OVER cells
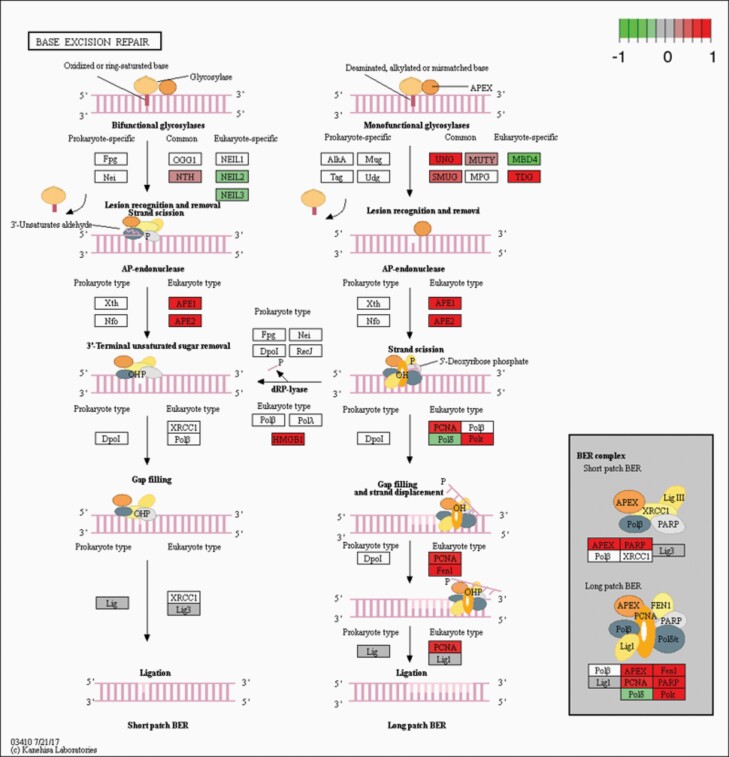
Given the patterns of genotoxin sensitivities and mitochondrial abnormalities described herein, we focussed on the expression of genes within relevant pathways. Since initial analysis found that WT cells tended to express very low levels of many of the genes of interest (0 to negative, weakers colour; Figure 4D–F), we focussed our comparison on the RNA-seq results of NEK1 KO vs OVER cells. As a major system in MMS resistance, we found several changes in components of the BER pathway. Specifically, NEK1 KO vs NEK1 OVER cells showed that UNG, SMUG1, TDG, MUTYH, APEX1, APEX2, POLκ, PCNA, FEN1 and PARP1 genes were upregulated in the absence of the kinase (P = 1.92E−09, padj = 0.0004), and that NEIL2, NEIL3, MBD4 and POLδ were downregulated, while LIG1 and LIG3 showed unchanged expression, but with values of 0 or close to 0 (Figures 4D and 5, and Supplementary Table 2, available at Mutagenesis Online); the opposite of this was seen when comparing OVER to KO (Supplementary Figure 3A, available at Mutagenesis Online).
Figure 5.
KEGG analysis in the BER pathway. NEK1 absence induced changes to the BER pathway, by upregulation. The figures show the complete pathway as annotated in KEGG, but shading (green, grey and red) is applied only to those RNAs detected in our dataset that also pass DESEQ2 statistical filters and map to a unique ENTREZ ID. Colour corresponds to the magnitude and direction of log2 fold change (control), with green (negative fold change) indicating genes that are downregulation and with red (positive fold change) indicating genes that are upregulation.
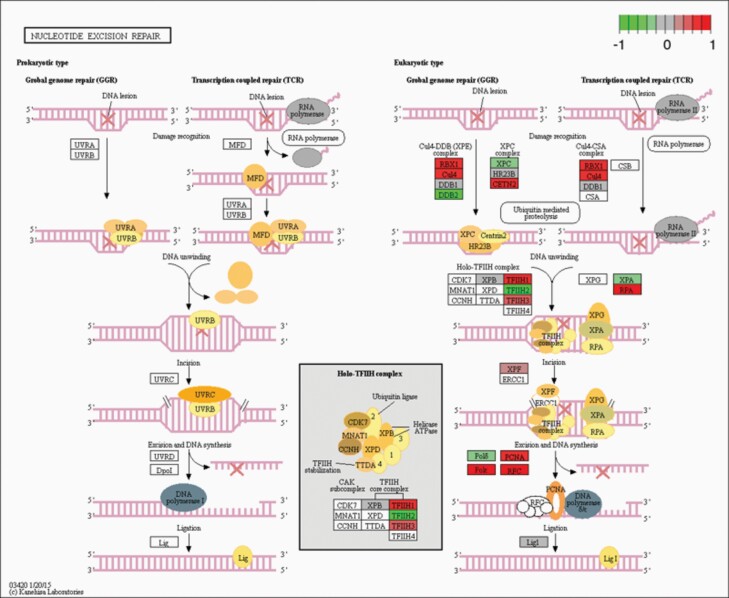
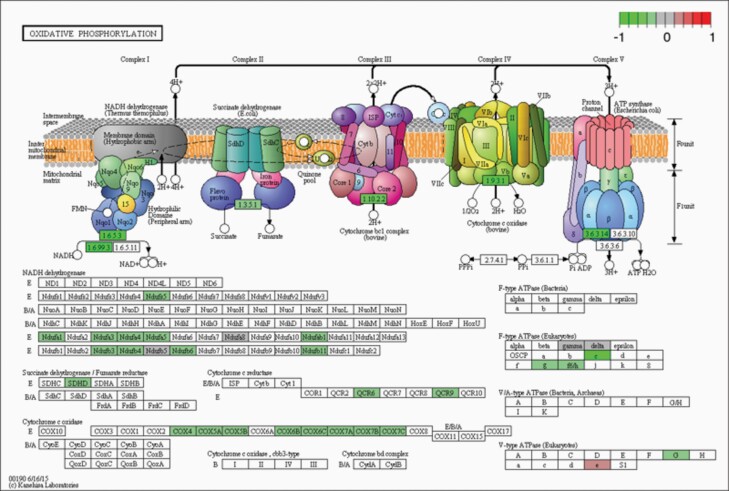
In light of the UVC sensitivity, we also looked at the NER pathway. Some genes belonging to the NER pathway were indeed significantly altered in NEK1 KO cells (P = 9.03E−07, padj = 2.84E−09). In particular, NEK1 KO vs NEK1 OVER showed that the CD4, RBX1, PCNA, RPA, RFC, TFIIH1, TFIIH3 and XPF genes (Figures 4E and 6, and Supplementary Table 2, available at Mutagenesis Online) were upregulated, whereas XPA, XPC, DDB2 and TFHII2 showed downregulation. For NEK1 OVER vs NEK1 KO cells, the converse results are reported in Supplementary Figure 3B, available at Mutagenesis Online. Lastly, we observed downregulation in the NEK1 KO cells (relative to the WT cells) of many genes involved in mitochondrial functions (Figure 4F). Some of the mitochondrial genes (Ndufs5, Ndufa1, 3–5, Ndufab1 and Ndufb3, 4, 6 and 11) are specifically related to the complex I oxidative phosphorylation pathway (Figure 7 and Supplementary Table 3, available at Mutagenesis Online). Reduced expression in components of the oxidative phosphorylation pathway in NEK1 KO cells may explain the elevated concentrations of ROS.
Figure 6.
KEGG analysis in the NER pathway. NEK1 absence induced changes to the NER pathway, by upregulation. The figures show the complete pathway as annotated in KEGG, but shading (green, grey and red) is applied only to those RNAs detected in our dataset that also pass DESEQ2 statistical filters and map to a unique ENTREZ ID. Colour corresponds to the magnitude and direction of log2 fold change (control), with green (negative fold change) indicating genes that are downregulation and with red (positive fold change) indicating genes that are upregulation.
Figure 7.
KEGG analysis in oxidative phosphorylation. NEK1 absence induced changes to the complex I, by downregulation. The figures show the complete pathway as annotated in KEGG, but shading (green, grey and red) is applied only to those RNAs detected in our dataset that also pass DESEQ2 statistical filters and map to a unique ENTREZ ID. Colour corresponds to the magnitude and direction of log2 fold change (control), with green (negative fold change) indicating genes that are downregulation and with red (positive fold change) indicating genes that are upregulation.
Validation of the RNA-seq data
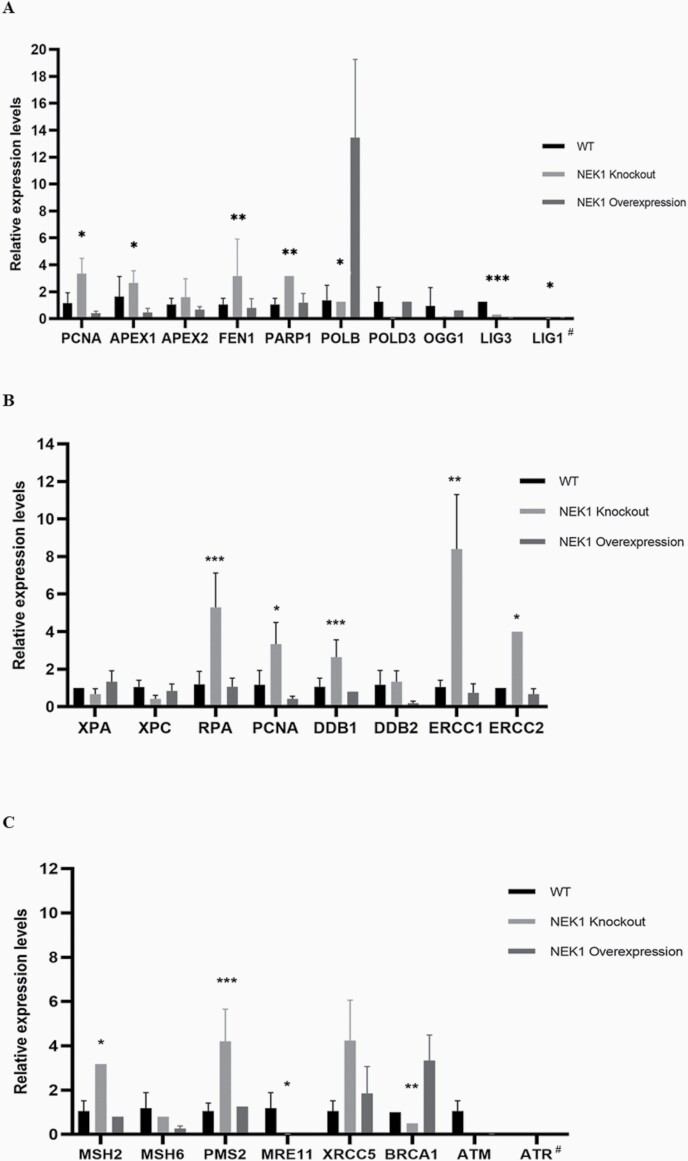
To validate the findings of the RNA-seq experiments, we performed qRT-PCR for select genes of the different DNA repair pathways (Figure 8). We were able to confirm that certain genes in the BER pathway have a higher expression in NEK1 KO cells relative to OVER cells, such as APEX1, PCNA, PARP1 and FEN1 (variation of 3 fold change, respectively, panel A), while also confirming the low expression of POLδ (0.013, fold change, panel A). For LIG1 and 3, which were unchanged in the RNA-seq evaluation, they showed a fold-change value of 0.011 and 0.055 (KO vs OVER), respectively, consistent with unaltered expression; interestingly, perhaps reflective of a need for precise NEK1 levels, LIG3 expression was decreased in both KO and OVER cells in comparison to WT cells. We also separately evaluated POLβ and OGG1, and found these two genes to be lowly expressed (1, and 0.10, fold change, panel A) in the KO cells relative to the OVER.
Figure 8.
Validation of RNA-seq data by quantitative RT-PCR. (A) Fold change of mRNA genes of BER pathway targeted by WT, KO and OVER. (B) Fold change of mRNA genes of NER pathway targeted by WT, KO and OVER in normal condition for (C) fold change of mRNA genes of DNA repair pathway targeted by WT, KO and OVER. The relative amount of each mRNA tested was normalised with 18s rRNA, GAPDH, HPRT and GUSB. The significance level was represented by *P < 0.05; **P < 0.01; ***P < 0.001. #Values below 0.0.
As for genes involved in the NER pathway (panel B), we confirmed the high expression of RPA and PCNA (variation of 5, and 3 fold change, respectively), and the low expression of XPC and XPA (variation of 0.41 and 0.66 fold change, respectively) in the KO cells (in many cases relative to both WT and OVER cells). We also evaluated the ERCC1 gene, which together with XPF forms a critical nuclease complex in NER and interstrand crosslink repair (39,40). Our data indicate a high expression of ERCC1 (variation 8, fold change) in NEK1 KO cells relative to OVER cells, an outcome that mirrors the positive regulation of XPF observed in the RNA-seq experiments. In the case of DDB1 and ERCC2, qRT-PCR indicated increased expression in the KO cells (variation of 3 and 4, fold change, respectively), where the RNA-seq studies found DDB1 to be unchanged and ERCC2 (a.k.a., XPD) to be invalid.
To evaluate the expression of key genes in other DDR pathways, we performed qRT-PCR against MMR (MSH2, MSH6 and PMS2), DSBR (MRE11, XRCC5 and BRCA1) and major DDR signalling kinases (ATM and ATR) (panel C). In KO cells (relative to OVER cells), MSH2 and PMS2 are upregulated (3 and 4, fold change), suggesting that the main components of the MMR recognition complex remain highly active. Conversely, the KO cells, as well as in some cases the OVER cells, show low expression of MRE11, ATM and ATR (0.010, 0.001 and 0.006 fold change), key elements in DSB signaling and repair, suggesting as seen for LIG3 a need for an optimal level of NEK1 for the regulation of these genes.
In addition to validating the RNA-seq experiments with complementing qPCR studies, we quantified the level of some proteins involved in BER (DNA POLβ, PARP1, LIG1, FEN1, APEX1 and PCNA) and NER (XPB, XPD, RPA, ERCC1 and XPA) by western blot (Supplementary Figure 4A and B, available at Mutagenesis Online). In this analysis, only a positive relationship between gene expression and protein expression was observed for APEX1, RPA and ERCC1. While it is often assumed that there will be a correlation between mRNA and protein levels, numerous investigations have found that there are many post-transcriptional mechanisms that can determine protein stability and their half-lives.
Discussion
The individual members of the NEK family have in many cases been shown to play important roles in cell cycle progression and the DDR. For example, NEK1 (48), NEK4 (49), NEK5 (50), NEK7 (51), NEK8 (52), NEK10 (53) and NEK11 (54) have been reported to operate in responses to oxidative DNA damage, DSBs, DNA crosslinks and replicative stress.
When NEK1 is mutated or when its activity is insufficient, cells are more likely to die or fail to proliferate when they normally should (55). Our results indicate that NEK1 KO HAP1 cells exhibit an altered expression of proteins involved in anti and pro-apoptotic processes (Figure 1B). Specifically, the expression of Bcl-2, who’s high expression has been associated with impaired apoptosis and disorderly proliferation, as seen in tumours (56), is lower in KO cells. Conversely, Bax protein, the first identified pro-apoptotic member of the Bcl-2 protein family (57), who’s reduction is critical to the progression of cancer (58) and who’s increased expression is central to chemotherapeutic drug efficacy (59), is upregulated in NEK1-deficient HAP1 cells. The inversed expression responses of Bcl-2 (down) and Bax (up), in combination with the expected replicative stress, likely contribute mechanistically to the apoptotic proneness of the NEK1 KO cells. Prior studies have also demonstrated that NEK1 participates in modulating cell death via the phosphorylation of the outer mitochondrial membrane protein, VDAC1 (24,25).
In the present study, we showed that NEK1 KO cells were hypersensitive to genotoxic agents such as MMS, UVC light and bleomycin (Figure 2A, B and C). MMS is a monofunctional alkylating agent that methylates DNA, mostly generating substrates for the BER pathway. UVC light produces primarily DNA photoproducts, such as cyclobutane pyrimidine dimers and 6-4 photolesions, damage forms that are typically repaired by the sub-pathways of NER, i.e. global-genome (GG-NER) and transcription-coupled (TC-NER) (36). Bleomycin acts as a radiomimetic, creating multiply damaged sites involving abasic lesions and single-strand breaks, often leading to the formation of DSBs (38). Consistently, prior studies using genetic deletion or siRNA/RNAi techniques have found that NEK1 deficiency in mouse or human HK2 and Hek293t cells results in hypersensitivity to several genotoxins, including MMS, hydrogen peroxide and ionising radiation (23,48,55). Notably, a portion of NEK1 protein was found to relocalise from the cytoplasm to sites of DNA damage in the nucleus, implying a direct role for the kinase in mitigating the response to the various genotoxic insults (23,55,60). NEK1 has also been observed to be overexpressed in renal cell carcinoma cells, and this increased expression results in hyperresistance to genotoxic agents such as MMS, UV irradiation, etoposide and 5-fluorouracil (46). Collectively, the data strongly support a role for NEK1 in mitigating proper cell cycle checkpoint control and efficient DNA damage repair.
One inconsistency with our results and prior work is the observation herein that NEK1 KO in HAP1 cells results in cis-Pt resistance (Figure 2D). A previous study had shown that NEK1 silencing in HEK293T and U87 cells results in increased sensitivity to cis-Pt and reduced resolution of interstrand crosslinks [8]. One possible explanation is the near-haploid nature of HAP1 cells, a feature that would not allow HR events to resolve complex intermediates formed when a replication fork collides with a cis-Pt adduct. Of course, disparate DNA repair capacity profiles among different cell types might also explain any variability in genotoxin agent sensitivity. One relevant finding of our RNA-seq experiments was the reduced expression of XPA and XPC in the NEK1 KO cells. XPC is critical to initial damage recognition during the GG-NER response (61), and collaborates with XPA to facilitate this repair mechanism (62–64). It could be that reduced GG-NER, a major system for coping with cis-Pt adducts, results in an increased reliance on TC-NER for crosslink repair in the haploid NEK1 KO cells, perhaps resulting in more efficient damage resolution and elevated resistance to cis-Pt (65).
Notably, many members of the NEK family have been shown to be involved in the DDR. For example, NEK4 (49) depleted cells show decreased activation of histone γ-H2AX and impaired recruitment of DNA-PKcs after damage induction; NEK5 (50) depletion results in increased DNA damage; NEK7 (51) deficient cells exhibit elevated aberrations at telomeres, longer-lasting foci of γH2AX and 53BP1, and increased cell death after oxidative damage to telomeric DNA; NEK8 (66) deficiency causes DSB accumulation and replicative stress, possibly through the regulation of CDK activity; NEK10 (53) acts as a positive regulator of ERK1/2 after UV irradiation, potentially working together to regulate cell cycle and apoptosis; and NEK11 (54) has been connected to DNA replication and DDRs through its rapid activation.
In normal, uninjured cells, NEK1 is located mainly in the cytoplasm, but it has been reported that a portion of the protein is located in mitochondria (24). In addition, NEK1 mutant cells have been found to exhibit an increase in mitochondrial membrane permeability, and ectopic expression of mutant NEK1 results in the loss of VDAC1 phosphorylation and consequent mitochondrial-mediated cell death (24,25). A recent study also showed that activation of NEK1 via phosphorylation by TLK1 contributes to the phosphorylation and stability of VDAC1 and thus to permeability and mitochondrial integrity (67). These data indicate that the TLK1/NEK1 axis contributes to the interruption of the cell cycle and activation of the DDR to mediate survival after damage, where the intrinsic apoptotic cascade occurs with high DNA damage that cannot be repaired efficiently. Our study found that NEK1 KO HAP1 cells exhibit a decrease in mitophagy (Figure 3A), likely leading to an increase in the number of mitochondria relative to WT cells (Figure 3B). Additionally, NEK1 deficiency was associated with an increase in cellular and mitochondrial ROS (Figure 3C and D), presumably due to the impaired mitophagy and increased number of defective mitochondria. Moreover, NEK1 KO cells harboured defective complex I activity (Figure 3F), a major site of ROS production and a likely source of the elevated ROS levels. As for mtDNA, we identified an increase in the number of copies of mtDNA (Figure 3G) and a reduction in mtDNA integrity, showing greater damage in NEK1 KO cells (Figure 3H), supporting either a direct or indirect role of the kinase in regulating mitochondrial phenotypes and function.
Other members of the NEK family have also been found in mitochondria. For example, our group showed that stable NEK5 expression has protective effects against Hek293-T cells death after thapsigargin treatment, and NEK5 silenced cells as well as cells expressing a ‘kinase-dead’ version, displayed an increase in ROS formation after thapsigargin treatment (68,69). More recently, our group showed that silenced NEK10 results in fragmented mitochondria, changes in ROS levels, impaired mitochondrial respiration and a decreased ratio of mtDNA amplification, possibly due to DNA damage (70). The mtDNA copy number was also analysed, and interestingly, NEK10 depletion increased mtDNA levels, suggesting that the kinase is involved in controlling mtDNA content (70). The potential contribution of other NEK family members, particularly in light of the emerging nuclear-mitochondrial crosstalk, warrants further investigation.
As for the RNA-seq results, since our cellular studies focussed on the role of NEK1 in survival, stress resistance and mitochondrial functionality, we concentrated on the effects of altered NEK1 expression on related pathways. In the case of BER, we observed upregulation in APEX1, APEX2, POLκ, PCNA, FEN1 and PARP1, yet downregulation of NEIL2, NEIL3 and POLδ in the KO cells relative to the OVER cells (Figure 5). This mixed response of the BER genes does not clearly explain the MMS hypersensitivity, but might indicate that imbalances in BER contribute to altered responses to genotoxic stress (71). Since UV photoproducts are major substrates of NER, the confirmed lower expression of XPA and XPC likely explains (Figure 6), at least in part, the increased sensitivity of the NEK1 KO cells to UV irradiation. In our evaluation of other key pathway factors (Figure 8), we observed a downregulation of MRE11 and ATM, central factors in the response to DSBs, and perhaps a mechanistic reason behind the bleomycin sensitivity, given its radiomimetic genotoxic behaviour. We also observed a negative regulation for ATR, an important protein that acts in response to a variety of DNA lesions that interfere with DNA replication (59), in addition, we detected an upregulation in the KO cells of MSH2 and PMS2, key MMR recognition factors. Lastly, we observed a significant decrease in the levels of genes that make up mitochondrial complex I, i.e. Ndufs5, Ndufa1, 3–5, Ndufab1 and Ndufb3, 4, 6 and 11, perhaps explaining the altered oxidative phosphorylation pathway (KEGG analysis, Figure 7) and the elevated concentrations of ROS. While the RNA-seq work provides mostly foundational work for the future, it does provide potential mechanistic insights into the cellular phenotypes of NEK1 deficiency reported herein. Finally, the collective studies support the mounting evidence that NEK1, and many of the family members, play critical roles in damage repair, mitochondrial homeostasis and ultimately cellular health, at both the proteome and now transcriptome level.
Supplementary data
Supplementary data are available at Mutagenesis Online.
Supplementary Fig. 1. Certificate of analysis—NEK1 knockout cell line.
Supplementary Fig. 2. Western blot of WT, KO and OVER NEK1 cells using anti-NEK1. The antibody anti-vinculin was used as a loading control. The graph’s quantifications are shown from n = 3 independent experiments.
Supplementary Fig. 3. KEGG analysis showed changes in the repair pathway and oxidative phosphorylation—complex I in NEK1 OVER and WT cells. (A) NEK1 OVER induced changes to the base excision repair pathway, by downregulation. (B) NEK1 OVER induced changes to the nucleotide excision repair pathway, by downregulation. (C) NEK1 WT vs NEK1 KO induced changes to complex I, by upregulation. The figures show the complete pathway as annotatedin KEGG, but shading (green, grey and red) is applied onlyto those RNAs detected in our dataset that also pass DESEQ2 statistical filters and map to a unique ENTREZ ID. Colour correspondsto the magnitude and direction of log2 fold change (control), with green (negative fold change) indicating genes that aredownregulation and with red (positive fold change) indicating genes that are upregulation.
Supplementary Fig. 4. DNA repair proteins expression in NEK1 WT, KO and NEK1 OVER cells. (A) BER proteins level expression (arbitrary unit), DNA POLδ, PARP, DNA Lig1, FEN1, APE1 and PCNA. (B) NER proteins level expression (arbitrary unit), XPB, XPD, RPA70, ERCC1 and XPA. Expression level (arbitrary unit) normalised with anti-tubulin. Data in graphics are means of triplicate experiments ± SD. The significance level for KO vs OVER was represented **P < 0.01 and ***P < 0.001.
Supplementary Table 1. Enrichment analysis (GO Enrichment, DO Enrichment and Reactome Database Enrichment) for NEK1 expression.
Supplementary Table 2. Expression of the genes analysed for the BER and NER pathways for the NEK1 KO and OVER cells.
Supplementary Table 3. Expression of the genes analysed for the oxidative phosphorylation for the NEK1 KO and WT cells.
Acknowledgements
The authors gratefully acknowledge the NIH/NIA, National Institutes of Health/National Institute of Aging, the financial support from the Foundation for Research of the State of São Paulo (Fapesp), Laboratory of Cancer Molecular Genetics (GEMOCA), Faculty of Medical Sciences and Unicamp and our group from the Laboratory of Signaling Mechanisms (LMS), Instituto de Biologia, Unicamp.
Funding
This study was supported by Fundação de Amparo à Pesquisa do Estado São Paulo (FAPESP), process numbers 2016/02040-8, 2017/21067-7 to M.B.M., and Grant Temático 2017/03489-1 to J.K. and Grant Temático 2014/15982-6 to C.F.M.M.; Coordenação de Aperfeiçoamento do Pessoal de Ensino Superior (CAPES, Funding code 01, Brasíia, DF, Brazil) and Conselho Nacional de Pesquisa e Desenvolvimento (CNPq; Grant 308868/2018‐8 to C.F.M.M., Grant 302534/2017-2 to J.K.; Brasília, DF, Brazil). The work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Conflict of interest statement: The author declares that there is no conflict of interests.
Author Contributions
All authors reviewed the literature, contributed to specific parts of the manuscript and read and improved the final version. M.B.M. performed and analysed all the experiments with the assistance of A.M.P., V.A.B., D.M.W. and J.K. M.B.M., A.M.P., V.A.B., D.M.W. and J.K. elaborated the final version of the paper, and D.M.W. and J.K. coordinated the study.
References
- 1. Meirelles, G. V., Perez, A. M., de Souza, E. E., Basei, F. L., Papa, P. F., Melo Hanchuk, T. D., Cardoso, V. B. and Kobarg, J. (2014) “Stop Ne(c)king around”: how interactomics contributes to functionally characterize Nek family kinases. World J. Biol. Chem., 5, 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fletcher, L., Cerniglia, G. J., Nigg, E. A., Yend, T. J. and Muschel, R. J. (2004) Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat. Res., 162, 128–135. [DOI] [PubMed] [Google Scholar]
- 3. Lee, M. Y., Kim, H. J., Kim, M. A., et al. (2008) Nek6 is involved in G2/M phase cell cycle arrest through DNA damage-induced phosphorylation. Cell Cycle., 7, 2705–2709. doi: 10.4161/cc.7.17.6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fry, A. M., O’Regan, L., Sabir, S. R. and Bayliss, R. (2012) Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci., 125, 4423–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Upadhya, P., Birkenmeier, E. H., Birkenmeier, C. S. and Barker, J. E. (2000) Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. U. S. A., 97, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cirulli, E. T., Lasseigne, B. N., Petrovski, S., et al. ; FALS Sequencing Consortium. (2015) Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science, 347, 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kenna, K. P., van Doormaal, P. T., Dekker, A. M., et al. ; SLAGEN Consortium. (2016) NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet., 48, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner, D., Müller, K., Wieland, T., et al. (2016) NEK1 mutations in familial amyotrophic lateral sclerosis. Brain., 139, e28. doi: 10.1093/brain/aww033 [DOI] [PubMed] [Google Scholar]
- 9. Zhu, J., Cai, Y., Liu, P. and Zhao, W. (2016) Frequent Nek1 overexpression in human gliomas. Biochem. Biophys. Res. Commun., 476, 522–527. doi: 10.1016/j.bbrc.2016.05.156 [DOI] [PubMed] [Google Scholar]
- 10. Cabral de Almeida Cardoso, L., Rodriguez-Laguna, L., Del Carmen Crespo, M., et al. ; GT-CSGP Working Group. (2015) Array CGH analysis of paired blood and tumor samples from patients with sporadic Wilms tumor. PLoS One, 10, e0136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melo-Hanchuk, T. D., Martins, M. B., Cunha, L. L., Soares, F. A., Ward, L. S., Vassallo, J. and Kobarg, J. (2020) Expression of the NEK family in normal and cancer tissue: an immunohistochemical study. BMC Cancer, 20, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh, V., Jaiswal, P. K., Ghosh, I., Koul, H. K., Yu, X. and De Benedetti, A. (2019) The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett., 453, 131–141. [DOI] [PubMed] [Google Scholar]
- 13. Singh, V., Jaiswal, P. K., Ghosh, I., Koul, H. K., Yu, X. and De Benedetti, A. (2019) Targeting the TLK1/NEK1 DDR axis with Thioridazine suppresses outgrowth of androgen independent prostate tumors. Int. J. Cancer, 145, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melo-Hanchuk, T. D., Slepicka, P. F., Meirelles, G. V., et al. (2017) NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin. Sci. Rep., 7, 5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dronkert, M. L., Beverloo, H. B., Johnson, R. D., Hoeijmakers, J. H., Jasin, M. and Kanaar, R. (2000) Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Veenman, L., Shandalov, Y. and Gavish, M. (2008) VDAC activation by the 18 kDa translocator protein (TSPO), implications for apoptosis. J. Bioenerg. Biomembr., 40, 199–205. [DOI] [PubMed] [Google Scholar]
- 17. Surpili, M. J., Delben, T. M. and Kobarg, J. (2003) Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry, 42, 15369–15376. [DOI] [PubMed] [Google Scholar]
- 18. Huang, J. (2002) Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res., 30, 667–674. doi: 10.1093/nar/30.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang, B., Matsuoka, S., Carpenter, P. B. and Elledge, S. J. (2002) 53BP1, a mediator of the DNA damage checkpoint. Science, 298, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 20. Liu, S., Ho, C. K., Ouyang, J. and Zou, L. (2013) Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc. Natl. Acad. Sci. U. S. A., 110, 2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldstein, J. C., Waterhouse, N. J., Juin, P., Evan, G. I. and Green, D. R. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol., 2, 156–162. [DOI] [PubMed] [Google Scholar]
- 22. Tsujimoto, Y. and Shimizu, S. (2002) The voltage-dependent anion channel: an essential player in apoptosis. Biochimie., 84, 187–193. doi: 10.1016/S0300-9084(02)01370-6 [DOI] [PubMed] [Google Scholar]
- 23. Chen, Y., Chen, P. L., Chen, C. F., Jiang, X. and Riley, D. J. (2008) Never-in-mitosis related kinase 1 functions in DNA damage response and checkpoint control. Cell Cycle, 7, 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen, Y., Craigen, W. J. and Riley, D. J. (2009) Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle, 8, 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen, Y., Gaczynska, M., Osmulski, P., Polci, R. and Riley, D. J. (2010) Phosphorylation by Nek1 regulates opening and closing of voltage dependent anion channel 1. Biochem. Biophys. Res. Commun., 394, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chaim, I. A., Gardner, A., Wu, J., Iyama, T., Wilson, D. M. and Samson, L. D. (2017) A novel role for transcription-coupled nucleotide excision repair for the in vivo repair of 3,N4-ethenocytosine. Nucleic Acids Res., 45, 3242–3252. doi: 10.1093/nar/gkx015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Safdar, A., Khrapko, K., Flynn, J. M., et al. (2016) Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice. Skelet. Muscle, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Berquist, B. R., Singh, D. K., Fan, J., et al. (2010) Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res., 38, 5023–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moraes, M. C. S., Neto, J. B. C. and Menck, C. F. M. (2012) DNA repair mechanisms protect our genome from carcinogenesis. Front. Biosci., 17, 1362–1388. doi: 10.2741/3992 [DOI] [PubMed] [Google Scholar]
- 30. Altieri, F., Grillo, C., Maceroni, M. and Chichiarelli, S. (2008) DNA damage and repair: from molecular mechanisms to health implications. Antioxid. Redox Signal., 10, 891–937. [DOI] [PubMed] [Google Scholar]
- 31. McCabe, K. M., Olson, S. B. and Moses, R. E. (2009) DNA interstrand crosslink repair in mammalian cells. J. Cell. Physiol., 220, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fang, E. F., Scheibye-Knudsen, M., Chua, K. F., Mattson, M. P., Croteau, D. L. and Bohr, V. A. (2016) Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol., 17, 308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boesch, P., Weber-Lotfi, F., Ibrahim, N., Tarasenko, V., Cosset, A., Paulus, F., Lightowlers, R. N. and Dietrich, A. (2011) DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta, 1813, 186–200. [DOI] [PubMed] [Google Scholar]
- 34. Alexeyev, M., Shokolenko, I., Wilson, G. and LeDoux, S. (2013) The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb. Perspect. Biol., 5, a012641. doi: 10.1101/cshperspect.a012641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carette, J. E., Raaben, M., Wong, A. C., et al. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature, 477, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lundin, C., North, M., Erixon, K., Walters, K., Jenssen, D., Goldman, A. S. and Helleday, T. (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res., 33, 3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slupphaug, G., Kavli, B. and Krokan, H. E. (2003) The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res., 531, 231–251. [DOI] [PubMed] [Google Scholar]
- 38. Hecht, S. M. (2000) Bleomycin: new perspectives on the mechanism of action. J. Nat. Prod., 63, 158–168. [DOI] [PubMed] [Google Scholar]
- 39. Basu, A. and Krishnamurthy, S. (2010) Cellular responses to cisplatin-induced DNA damage. J. Nucleic Acids., 2010, 201367. doi: 10.4061/2010/201367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cohen, G. M. (1997) Caspases: the executioners of apoptosis. Biochem. J., 326 (Pt 1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fraser, A. and Evan, G. (1996) A license to kill. Cell., 85, 781–784. doi: 10.1016/S0092-8674(00)81005-3 [DOI] [PubMed] [Google Scholar]
- 42. Srinivasula, S. M., Fernandes-Alnemri, T., Zangrilli, J., et al. (1996) The Ced-3/interleukin 1β converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2α are substrates for the apoptotic mediator CPP32. J. Biol. Chem., 271, 27099–27106. doi: 10.1074/jbc.271.43.27099 [DOI] [PubMed] [Google Scholar]
- 43. Porter, A. G. and Jänicke, R. U. (1999) Emerging roles of caspase-3 in apoptosis. Cell Death Differ., 6, 99–104. [DOI] [PubMed] [Google Scholar]
- 44. Capetanaki, Y. (2002) Desmin cytoskeleton: a potential regulator of muscle mitochondrial behavior and function. Trends Cardiovasc. Med., 12, 339–348. doi: 10.1016/S1050-1738(02)00184-6 [DOI] [PubMed] [Google Scholar]
- 45. Chan, L. Y., Mugler, C. F., Heinrich, S., Vallotton, P. and Weis, K. (2017) Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability, November 7, 2017. biorXiv, 7, e32536. doi: 10.1101/214775, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 46. Chen, Y., Chen, C. F., Polci, R., Wei, R., Riley, D. J. and Chen, P. L. (2014) Increased Nek1 expression in renal cell carcinoma cells is associated with decreased sensitivity to DNA-damaging treatment. Oncotarget., 5, 4283–4294. doi: 10.18632/oncotarget.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pelegrini, A. L., Moura, D. J., Brenner, B. L., Ledur, P. F., Maques, G. P., Henriques, J. A., Saffi, J. and Lenz, G. (2010) Nek1 silencing slows down DNA repair and blocks DNA damage-induced cell cycle arrest. Mutagenesis, 25, 447–454. [DOI] [PubMed] [Google Scholar]
- 49. Nguyen, C. L., Possemato, R., Bauerlein, E. L., Xie, A., Scully, R. and Hahn, W. C. (2012) Nek4 regulates entry into replicative senescence and the response to DNA damage in human fibroblasts. Mol. Cell. Biol., 32, 3963–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Melo-Hanchuk, T. D., Slepicka, P. F., Pelegrini, A. L., Menck, C. F. M. and Kobarg, J. (2019) NEK5 interacts with topoisomerase IIβ and is involved in the DNA damage response induced by etoposide. J. Cell. Biochem., 120, 16853–16866. [DOI] [PubMed] [Google Scholar]
- 51. Tan, R., Nakajima, S., Wang, Q., et al. (2017) Nek7 protects telomeres from oxidative DNA damage by phosphorylation and stabilization of TRF1. Mol. Cell., 65, 818–831.e5. doi: 10.1016/j.molcel.2017.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi, H. J., Lin, J. R., Vannier, J. B., et al. (2013) NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell, 51, 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moniz, L. S. and Stambolic, V. (2011) Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol. Cell. Biol., 31, 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Noguchi, K., Fukazawa, H., Murakami, Y. and Uehara, Y. (2002) Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses. J. Biol. Chem., 277, 39655–39665. [DOI] [PubMed] [Google Scholar]
- 55. Polci, R., Peng, A., Chen, P. L., Riley, D. J. and Chen, Y. (2004) NIMA-related protein kinase 1 is involved early in the ionizing radiation-induced DNA damage response. Cancer Res., 64, 8800–8803. [DOI] [PubMed] [Google Scholar]
- 56. Gross, A., McDonnell, J. M. and Korsmeyer, S. J. (1999) BCL-2 family members and the mitochondria in apoptosis. Genes Dev., 13, 1899–1911. [DOI] [PubMed] [Google Scholar]
- 57. Oltval, Z. N., Milliman, C. L. and Korsmeyer, S. J.(1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell., 74, 609–619. doi: 10.1016/0092-8674(93)90509-O [DOI] [PubMed] [Google Scholar]
- 58. Roth, W. and Reed, J. C. (2002) Apoptosis and cancer: when Bax is TRAILing away. Nat. Med., 8, 216–218. doi: 10.1038/nm0302-216 [DOI] [PubMed] [Google Scholar]
- 59. Westphal, D., Kluck, R. M. and Dewson, G. (2014) Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ., 21, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen, Y., Chen, C. F., Riley, D. J. and Chen, P. L. (2011) Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle., 10, 655–663. doi: 10.4161/cc.10.4.14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pascucci, B., D’Errico, M., Parlanti, E., Giovannini, S. and Dogliotti, E. (2011) Role of nucleotide excision repair proteins in oxidative DNA damage repair: an updating. Biochemistry (Mosc.), 76, 4–15. [DOI] [PubMed] [Google Scholar]
- 62. Pâques, F. and Haber, J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kobaisi, F., Fayyad, N., Rezvani, H. R., Fayyad-Kazan, M., Sulpice, E., Badran, B., Fayyad-Kazan, H., Gidrol, X. and Rachidi, W. (2019) Signaling pathways, chemical and biological modulators of nucleotide excision repair: the faithful shield against UV genotoxicity. Oxid. Med. Cell. Longev., 2019, 4654206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gregersen, L. H. and Svejstrup, J. Q. (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci., 43, 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Furuta, T., Ueda, T., Aune, G., Sarasin, A., Kraemer, K. H. and Pommier, Y. (2002) Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res., 62, 4899–4902. [PubMed] [Google Scholar]
- 66. Choi, H. J., Lin, J. R., Vannier, J. B., et al. (2013) NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell, 51, 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singh, V., Khalil, M. I. and De Benedetti, A. (2020) The TLK1/Nek1 axis contributes to mitochondrial integrity and apoptosis prevention via phosphorylation of VDAC1. Cell Cycle., 19, 363–375. doi: 10.1080/15384101.2019.1711317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hanchuk, T. D. M. and Kobarg, J. (2017) Nek5 association with mitochondria proteins and functions: is it a Nek family characteristic? J. Cell Signal., 2, 1. doi: 10.4172/2576-1471.1000133 [DOI] [Google Scholar]
- 69. Melo Hanchuk, T. D., Papa, P. F., La Guardia, P. G., Vercesi, A. E. and Kobarg, J. (2015) Nek5 interacts with mitochondrial proteins and interferes negatively in mitochondrial mediated cell death and respiration. Cell. Signal., 27, 1168–1177. [DOI] [PubMed] [Google Scholar]
- 70. de Oliveira, A. P., Basei, F. L., Slepicka, P. F., et al. (2020) NEK10 interactome and depletion reveal new roles in mitochondria. Proteome Sci, 18,1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dianov, G. L. and Hübscher, U. (2013) Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res., 41, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.