Abstract
Aims: Catecholamine metabolism via monoamine oxidase (MAO) contributes to cardiac injury in models of ischemia and diabetes, but the pathogenic mechanisms involved are unclear. MAO deaminates norepinephrine (NE) and dopamine to produce H2O2 and highly reactive “catecholaldehydes,” which may be toxic to mitochondria due to the localization of MAO to the outer mitochondrial membrane. We performed a comprehensive analysis of catecholamine metabolism and its impact on mitochondrial energetics in atrial myocardium obtained from patients with and without type 2 diabetes.
Results: Content and maximal activity of MAO-A and MAO-B were higher in the myocardium of patients with diabetes and they were associated with body mass index. Metabolomic analysis of atrial tissue from these patients showed decreased catecholamine levels in the myocardium, supporting an increased flux through MAOs. Catecholaldehyde-modified protein adducts were more abundant in myocardial tissue extracts from patients with diabetes and were confirmed to be MAO dependent. NE treatment suppressed mitochondrial ATP production in permeabilized myofibers from patients with diabetes in an MAO-dependent manner. Aldehyde dehydrogenase (ALDH) activity was substantially decreased in atrial myocardium from these patients, and metabolomics confirmed lower levels of ALDH-catalyzed catecholamine metabolites. Proteomic analysis of catechol-modified proteins in isolated cardiac mitochondria from these patients identified >300 mitochondrial proteins to be potential targets of these unique carbonyls.
Innovation and Conclusion: These findings illustrate a unique form of carbonyl toxicity driven by MAO-mediated metabolism of catecholamines, and they reveal pathogenic factors underlying cardiometabolic disease. Importantly, they suggest that pharmacotherapies targeting aldehyde stress and catecholamine metabolism in heart may be beneficial in patients with diabetes and cardiac disease. Antioxid. Redox Signal. 35, 235–251.
Keywords: catecholamines, monoamine oxidase, human heart, mitochondria, diabetes, aldehydes
Innovation
The body of work in this article comprises metabolomic, proteomic, and functional analysis of catecholamine metabolism by monoamine oxidases (MAOs) in the human heart. Our findings suggest that MAO-mediated catabolism of norepinephrine and dopamine imposes a unique form of carbonyl stress on cardiac mitochondria in the form of catecholaldehydes. We propose that these catecholaldehydes are always being produced and metabolized in the heart, but with obesity and diabetes, the combined effect of increased sympathetic tone, increased cardiac MAO activity, and decreased aldehyde scavenging forces a “detoxification bottleneck” in the myocardium, which causes derangements in mitochondrial oxidative phosphorylation and contributes to electro-mechanical dysfunction.
Introduction
Following the initial reports of the Framingham Study decades ago, persistent efforts have been made to uncover pathophysiological mechanisms underlying cardiac complications associated with diabetes (38). Myocardial tissue remodeling and electro-mechanical dysfunction underlie most of these complications, and the role of cardiomyocyte bioenergetics and redox imbalance in these processes has recently come to the forefront. Mitochondria are at the nexus of cardiomyocyte energetics and redox balance, which is why disruptions in energy production and mitochondrial sources of reactive oxygen species (ROS) have been explored as pathogenic factors contributing to cardiometabolic disease (8, 49). Established sources of mitochondrial ROS include NADPH oxidase and sites within the electron transport system (ETS), whereas lipid peroxidation of the mitochondrial membrane is a source of reactive aldehydes (e.g., 4-hydroxy-nonenal [HNE] and malondialdehyde [MDA]) (7, 24, 30). More recently, monoamine oxidase (MAO) has emerged as a significant source of both H2O2 and aldehydes within the myocardium (3, 36).
Two isoforms, MAO-A and MAO-B, located on the outer membrane of the mitochondrion, metabolize norepinephrine (NE), dopamine (DA) to produce ammonia, H2O2, and the highly reactive and toxic catecholaldehydes 3,4-dihydroxyphenylglycolaldehyde (DOPEGAL) and 3,4-dihydroxyphenylacetaldehyde (DOPAL), respectively (21, 63). Although short-lived, the toxicity of catecholaldehydes stems from their ability to form covalent aldehyde protein adducts (47, 58, 59). Catecholaldehydes are primarily detoxified via oxidation (aldehyde dehydrogenase [ALDH]) or reduction (aldose reductase [AR], AKR1B1) (20, 21, 22).
A pathogenic role for MAO has recently been described in animal models of heart failure (HF), ischemia, and, more recently, diabetic cardiomyopathy (6, 17, 37, 57). Although these studies have laid the groundwork for establishing MAO as a pathogenic factor, the translational significance of these studies and the pathophysiologic mechanisms by which MAO may be acting remain uncertain. Therefore, we performed a comprehensive metabolomic, proteomic, and functional analysis of catecholamine metabolism in atrial myocardium of age-matched patients with or without type 2 diabetes, with particular emphasis placed on MAO activity and its impact on mitochondrial metabolism and energetics. Given that MAO-derived catecholamine metabolites (i.e., catecholaldehydes) are highly reactive, we tested the hypothesis that in patients with diabetes, increased MAO activity in the heart contributes to mitochondrial dysfunction by disrupting oxidative phosphorylation (OxPHOS).
Results
MAO expression and activity is upregulated in atrial myocardium of patients with diabetes
For this study, samples of right atrial myocardium were analyzed from 96 patients, 51 with diabetes and 45 with normal glycemic control (i.e., nondiabetes). Demographic and clinical variables are shown in Supplementary Table S1. No differences existed between groups in sex, mean age, or mean body mass index (BMI). Although there was a trend for more African Americans in the diabetes group, it was not statistically significant. No significant differences in preoperative medications were observed between groups, with exception of the angiotensin-converting enzyme inhibitors/angiotensin II receptor blockers and the antihyperglycemics (Supplementary Table S1). None of the patients included within the study were taking MAO inhibitors.
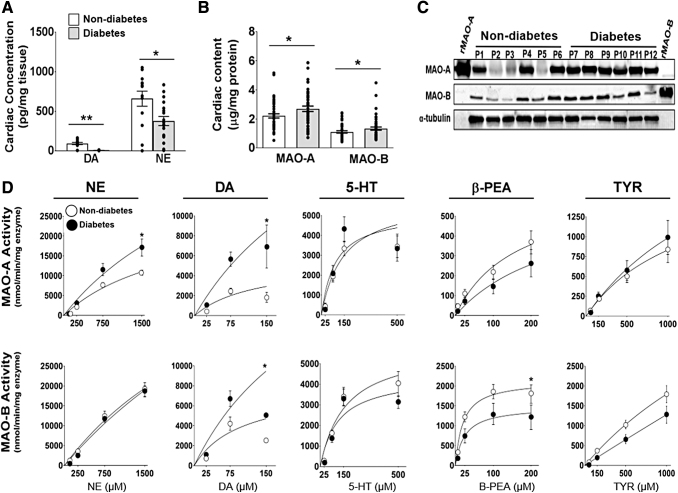
Since MAO metabolizes intracellular catecholamines, we first measured catecholamine levels within the myocardium. NE was found to be the predominant catecholamine within the samples, and patients with diabetes showed significantly lower concentrations of DA (p < 0.001) and NE (p = 0.012) in their myocardium, as compared with patients without diabetes (Fig. 1A). Recent work in rodent models showed that MAO expression increases in the heart and brain with insulin resistance/diabetes (41, 57), which could account for the lower NE and DA concentrations in the patients with diabetes. Therefore, we measured MAO enzyme content and activity in myocardial extracts. Results from the quantitative enzyme-linked immunosorbent assay (ELISA) revealed that the predominant MAO isoform within atrial myocardium was MAO-A. In univariable analysis, the content of both MAO isoforms was significantly increased within the myocardium from patients with diabetes (p < 0.05, Fig. 1B, C and Supplementary Fig. S1). After adjusting for age, sex, and race, multivariable analysis revealed that only MAO-A was significantly higher in patients with diabetes (p = 0.024) (Supplementary Table S2).
FIG. 1.
Cardiac MAO protein expression and enzymatic activity in human atrial myocardium. LC-MS analysis of DA and NE concentrations in human atrial myocardium (A) from patients with type 2 diabetes and nondiabetes (n = 15/metabolic group). MAO-A and MAO-B concentrations measured by using MAO isoform specific antibodies via (B) ELISA (n = 45–55/metabolic group) and (C) Western blot, α-tubulin was used as the loading control (n = 6/metabolic group). Kinetic traces of MAO activity (D) using 5 biogenic substrates: NE, DA, serotonin, βPEA, and TYR. MAO activity was measured by monitoring the increase in fluorescence of resorufin at Ex/Em: 571/585. MAO-A activity is shown on the top panel and MAO-B activity is on the bottom panel (n = 20/metabolic group). Data are represented as ± SEM. (A, B, D) was analyzed by using unpaired t-test. *p < 0.05, **p < 0.005 compared with nondiabetes. 5-HT, serotonin; βPEA, β-phenylethylamine; DA, dopamine; ELISA, enzyme-linked immunosorbent assay; LC-MS, liquid chromatography–mass spectrometry; MAO, monoamine oxidase; NE, norepinephrine; SEM, standard error of the mean; TYR, tyramine.
To further characterize MAO, kinetic assays were performed by using clorgyline (MAO-A inhibitor) and selegiline (MAO-B inhibitor) to distinguish isoform-specific activity. Assay specificity for each isozyme is shown with these inhibitors in Supplementary Figure S1. MAO kinetics differed by diabetes status, and the effect varied depending on the monoamine substrate being tested. The observed Vmax for both MAO-A and MAO-B isoforms were higher in patients with type 2 diabetes when DA was used as the substrate. MAO-A, but not MAO-B, displayed a higher observed Vmax when NE was used (Fig. 1D), which is consistent with the known selectivity of MAO-A for NE. Multivariable analysis confirmed that MAO-A activity supported NE (Table 1) and DA (Supplementary Table S3) was significantly increased within myocardium from patients with diabetes. With β-phenylethylamine (βPEA), the observed Vmax of MAO-A and MAO-B appeared to be lower in patients with diabetes (Fig. 1D). No significant effect was observed between metabolic groups when 5-HT or tyramine (TYR) was used as substrates. Taken together, these results indicate that not only MAO expression is increased but also MAO activity is altered in a substrate-specific manner in the heart during diabetes.
Table 1.
Monoamine Oxidase Isoenzyme Activity (Norepinephrine as Substrate) and Risk for Diabetes
| Diabetes | Nondiabetes | p | |
|---|---|---|---|
| MAO-A total [pmol/(min·mg) enzyme] | |||
| Mean ± standard deviation | 5943 ± 3092 | 2673 ± 1527 | 0.010a |
| ptrend < 0.0018b | |||
| MAO-B total [pmol/(min·mg) enzyme] | |||
| Mean ± standard deviation | 6759 ± 3000 | 4044 ± 3228 | 0.010a |
| ptrend < 0.010b | |||
p-Value computed by using Friedman's nonparametric test for central tendency, adjusting for age, sex, and race.
p-Value computed by using the likelihood ratio trend test, adjusting for age, sex, and race.
MAO, monoamine oxidase.
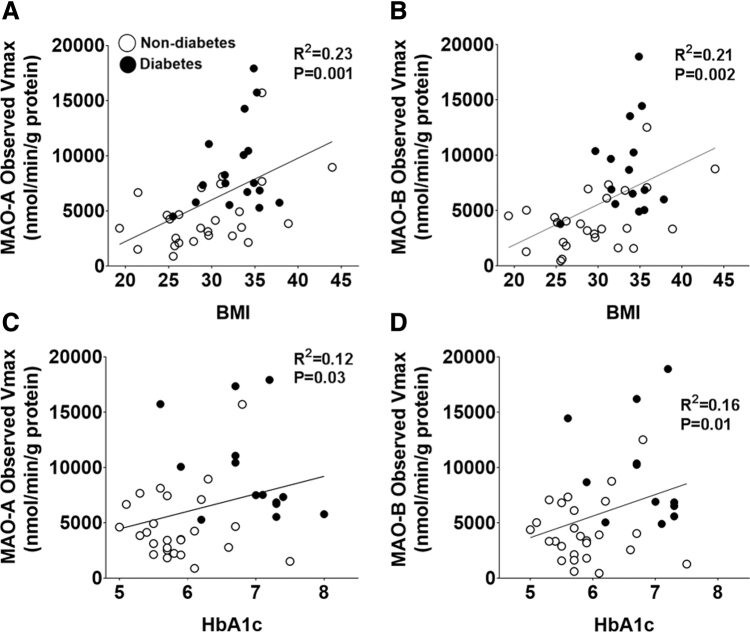
Irrespective of metabolic status, more than 70% of the patients within the cohort were determined to be either overweight or obese (BMI 25–33.3). Therefore, we sought to determine whether there was an association between MAO and metabolic parameters, and we determined that BMI has a stronger association with DA-supported maximal activity of both isoforms (MAO-A: R2 = 0.23, p = 0.001 and MAO-B: R2 = 0.21, p = 0.002) (Fig. 2A, B) compared with glycated hemoglobin (HbA1c) levels (Fig. 2C, D). These results suggest that obesity may be a more important factor underlying the increase in MAO compared with HbA1c.
FIG. 2.
Maximal MAO activity and association with metabolic parameters. Shown are pairwise correlations between BMI and observed Vmax of (A) MAO-A and (B) MAO-B. In (C, D) are pairwise correlations between HbA1c and observed Vmax of MAO-A and MAO-B, respectively. For this analysis, MAO activity was measured by using DA. n = 26 for nondiabetes and n = 16 for diabetes. Pairwise correlations were performed by using linear regression analysis. BMI, body mass index; HbA1c, glycated hemoglobin.
MAO activity disrupts mitochondrial OxPHOS in myocardium in patients with diabetes
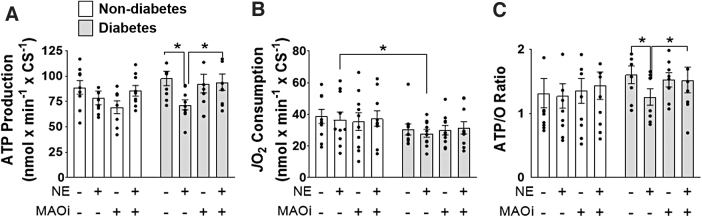
Metabolism of TYR and DA by MAO has been reported to cause suppression of mitochondrial respiration in isolated rat brain mitochondria, but whether catecholamine metabolism by MAO disrupts mitochondrial bioenergetics within the heart is not known (5, 16). To address this question, we measured mitochondrial OxPHOS (ATP production/O2 consumption) by using permeabilized myofibers (Pmfbs) prepared from fresh samples of atrial myocardium. In Pmfbs isolated from patients without diabetes, incubation with NE did not significantly alter ATP production or O2 consumption with the fatty acid palmitoyl-l-carnitine (Fig. 3A, B). The ADP-stimulated respiration supported by palmitoyl-carnitine was decreased in Pmfbs from patients with diabetes (Fig. 3B), consistent with what was previously observed (3). Metabolism of NE by MAO significantly decreased ATP production (p < 0.05) in Pmfbs from patients with type 2 diabetes, whereas O2 consumption was not significantly affected (Fig. 3A, B), corresponding to a decreased ATP/O ratio (Fig. 3C). This effect was confirmed to be driven by MAO metabolism of NE, as concurrent treatment with MAO inhibitors attenuated the effect.
FIG. 3.
MAO-derived byproducts disrupt oxPHOS. Simultaneous (A) ATP production, (B) O2 consumption, and (C) ATP/O ratio were measured in permeabilized fibers prepared from atrial myocardium treated with: no MAO substrate or inhibitors, 75 μM NE, MAO inhibitors, or MAO inhibitors +75 μM NE. n = 10/metabolic group with 4 fibers from each patient. (A–C) were analyzed by using one-way ANOVA followed by unpaired t-tests. *p < 0.05 with unpaired t-tests. ND, nondiabetes; OxPHOS, oxidative phosphorylation.
Metabolism of catecholamines by MAO generates catechol-protein adducts in extracts of atrial myocardium
DOPAL and DOPEGAL are quickly metabolized by aldehyde dehydrogenase 2 (ALDH2) and AR into acetic acid (dihydroxyphenylacetic acid [DOPAC]) and alcohol (dihydroxyphenylglycol [DHPG]), respectively, which are their primary routes of metabolism (Fig. 4A). However, when these detoxification pathways are overwhelmed, catecholaldehydes have been shown to form catechol-protein adducts (2, 18, 21, 58). To our knowledge, there has been no previous study examining the formation of catecholaldehydes or catechol-modified protein adducts in myocardial tissue. Therefore, we first measured unbound DOPAL and DOPAC in plasma and myocardial extracts by using quantitative tandem mass spectrometry (MS/MS) approach. In plasma, free DOPAL level was increased in patients with type 2 diabetes, whereas DOPAC levels were found to be similar between the metabolic groups (Supplementary Fig. S2). Within myocardium, DOPAL and DOPAC concentrations were below the detectable limit of the instruments used.
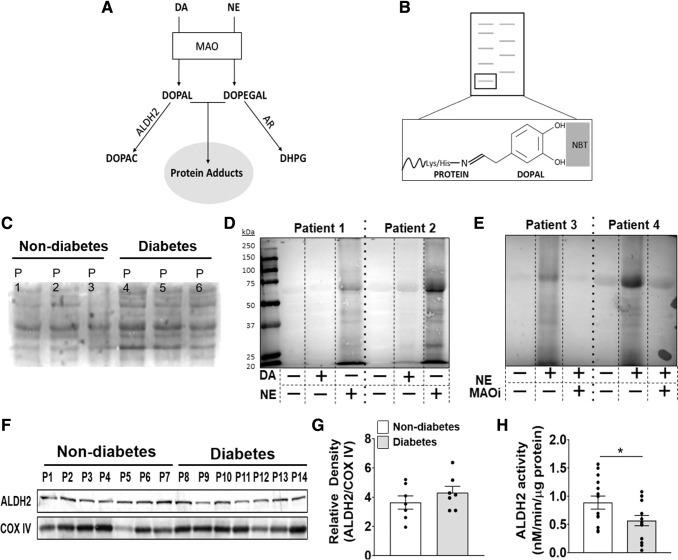
FIG. 4.
Detection and metabolism of catecholaldehydes in human atrial myocardium. Pathway of catecholamine metabolism into catecholaldehydes (by MAO) and then into acetic acid (DOPAC) or alcohol (DHPG) by ALDH2 or AR, respectively (A). If metabolism by ALDH2 or AR is compromised, catecholaldehydes can form carbonyl adducts with proteins. Shown in (B) is a representative image of catechol-protein adduct detection by NBT staining. In the presence of catechol-modified proteins, NBT stains proteins blue on nitrocellulose membrane. Representative NBT stains of cardiac lysate untreated (C), after overnight incubation with DA (25 μM) or NE (75 μM) (D), and overnight incubation with MAO inhibitors ± NE (75 μM) (E). Western blot analysis (F) and densitometric analysis (G) of ALDH2 in myocardium. COX IV was used as a loading control to calculate relative density. n = 7/metabolic group. ALDH2 activity (H) measured by monitoring the increase in absorbance of NADH at 450 nm in myocardial samples. n = 14/metabolic group. (H) Was analyzed by using unpaired t-test. *p < 0.05. All data are shown as mean ± SEM. ALDH2, aldehyde dehydrogenase 2; AR, aldose reductase; COX IV, complex IV; DHPG, dihydroxyphenylglycol; DOPAC, dihydroxyphenylacetic acid; MAOi, monoamine oxidase inhibitors; NBT, nitroblue tetrazolium.
This was not surprising, however, because owing to their high reactivity, free catecholaldehyes are unlikely to be present in myocardium. Therefore, we refined a technique using nitroblue tetrazolium (NBT), to determine whether myocardial tissue contains catechol-modified protein adducts (51). NBT is a dye that selectively reacts with the catechol moiety on proteins to form a stable blue conjugate on nitrocellulose membrane (Fig. 4B). Myocardial extracts all contain catechol-modified protein adducts at baseline, with substantially greater levels within the myocardium from patients with diabetes (Fig. 4C). Extracts exposed to NE, and to a lesser extent, DA, revealed an increase in catechol-modified protein adducts (Fig. 4D). The catechol modifications were determined to be dependent on MAO activity, which strongly suggests that MAO-derived catecholaldehydes are responsible for this modification (Fig. 4E and Supplementary Fig. S4C).
Within the mitochondria, aldehydes formed from lipid peroxidation (HNE, MDA) are metabolized by ALDH2, the same enzyme that primarily metabolizes DOPAL and, to a lesser extent, DOPEGAL (18, 46, 48). Studies have shown that HNE and DOPAL, at high concentrations, can allosterically inhibit ALDH2 activity, thereby creating a “bottleneck” for aldehyde detoxification (25, 33). Further, a recent study from our lab revealed that myocardium from patients with type 2 diabetes contain greater amounts of HNE compared with patients without diabetes, suggesting basal aldehyde stress within the cardiac mitochondrial microenvironment of these patients (39). In animal models of diabetes, the attenuation of ALDH2 activity leads to an increase in aldehyde load and contributes to left ventricular dysfunction (27, 64). Since ALDH2 is involved in catecholaldehyde metabolism, ALDH2 content and activity was measured in atrial myocardium. Interestingly, although there were no differences in ALDH2 enzyme content between the metabolic groups as determined by immunoblot (Fig. 4F–G and Supplementary Fig. S4D–E), ALDH2 activity was significantly reduced (p < 0.039) in patients with type 2 diabetes (Fig. 4H). Pairwise analysis of these data revealed that increases in ALDH2 activity were weakly associated with increases in BMI (Supplementary Fig. S3A) whereas a decrease in activity was associated with HbA1c (Supplementary Fig. S3B), suggesting that hyperglycemia negatively impacts the activity of this important detoxification enzyme. To determine whether AR was similarly impacted by diabetes, we measured AR activity in a subset of the patients and to our surprise, we observed an increase in the mean activity of AR in patients with diabetes, although it failed to reach statistical significance with this sample size (Supplementary Fig. S3C).
Proteomic analysis reveals catechol modifications of numerous mitochondrial enzymes in human cardiac mitochondria
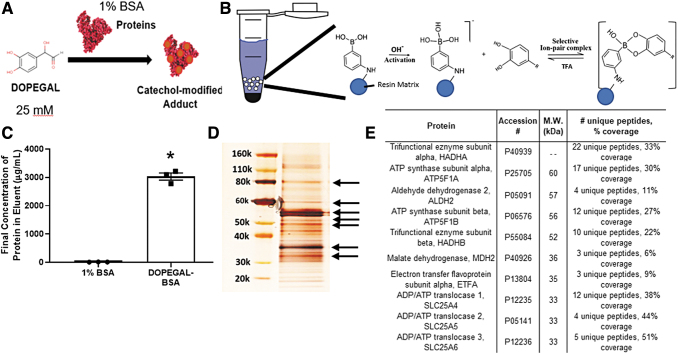
To gain further insight on the potential effects of catecholaldehyde formation in human cardiac mitochondria, we modified a method that was previously developed and validated for enrichment of DA quino-protein adducts from brain tissue (44). This method employs the tight binding capacity and selectivity of aminophenylboronic acid (APBA) for catechols, allowing for capture and enrichment of catechol-modified proteins in biological material. To validate this method, we generated samples of DOPEGAL from NE by using recombinant MAO-A, and we prepared mixtures of DOPEGAL adducts of bovine serum albumin (BSA) (Fig. 5A). This DOPEGAL-BSA reference sample was subjected to APBA resin binding, wash, and elution as shown in Figure 5B. After elution with trifluoroacetic acid (TFA), only DOPEGAL-modified BSA, but not unmodified BSA, was eluted from the resin (Fig. 5C), demonstrating the catechol-modification requirement for proteins to bind to this resin.
FIG. 5.
Catechol-modified protein extraction and detection in human cardiac mitochondria. Formation of catechol-modified BSA mixture is shown in (A), via reacting DOPEGAL with a 1% BSA solution in phosphate-buffered saline. (A) Schematic of the reaction between APBA resin and catechol-modified proteins is shown in (B). Solvent-accessible catechol moieties covalently bind to the phenylboronic moiety on the resin at basic pH; this reaction is reversed at acidic pH with use of TFA, dissociating the proteins from the resin. Binding is dependent on catechol modification, as only DOPEGAL-modified, but not -unmodified BSA, is captured by the resin (C). Catechol-modified mitochondrial proteins were then extracted by incubating pooled samples of isolated human atrial mitochondria with the APBA resin. After silver stain, protein bands with the highest optical density were excised from the gel (D, denoted by arrows). In (E) is an abbreviated table of the most abundant mitochondrial proteins identified in those bands at the indicated M.W., using LC/MS. (C) Was analyzed by using unpaired t-tests. *p < 0.00001 versus BSA alone. APBA, aminophenylboronic acid; BSA, bovine serum albumin; TFA, trifluoroacetic acid.
We then sought to determine which mitochondrial proteins may be modified by catechols in the human atrial samples, recognizing that even in the basal condition there are likely numerous mitochondrial enzymes/proteins that may be modified. Mitochondria isolated from atrial myocardium were then pooled from multiple patients (regardless of diabetes status), and catechol-modified proteins were enriched from these samples by using the APBA method. After sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), silver stain of these extracts revealed many bands ranging from 20 to 120 kDa. Six of the largest bands were excised, and proteomics analysis revealed >300 mitochondrial proteins, most of which are involved in tricarboxylic acid (TCA) cycle, fatty acid oxidation, and OxPHOS. A short list of the proteins representing the most abundant unique peptides captured at the molecular weights indicated by the arrows in Figure 5D are shown in Figure 5E. The full annotated proteomics file can be found online in the Proteomics IDEntifications Database (https://www.ebi.ac.uk/pride), identifier PXD021759, 10.6019/PXD021759.
Metabolomic profiling shows pathways of catecholamine synthesis, and metabolism is altered in hearts with diabetes
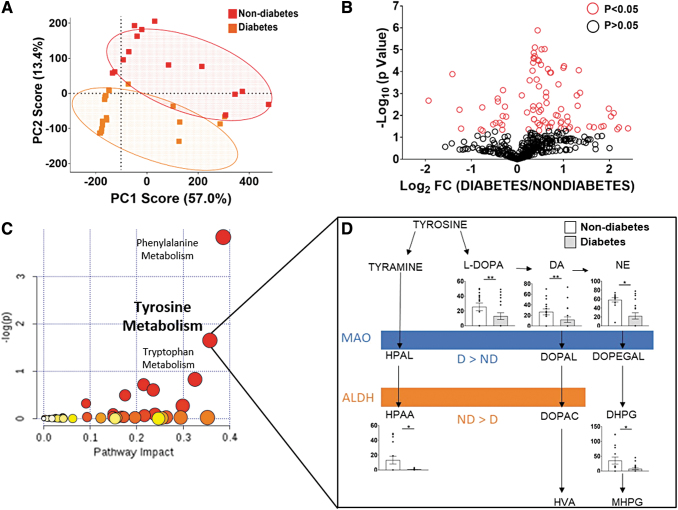
To examine whether the changes in MAO and ALDH2 corresponded to alterations in total catecholamine flux within the myocardium, global metabolomics analysis was performed on atrial tissue from a subset of patients in each group. Liquid chromatography–mass spectrometry (LC/MS) peak profiling revealed 592 unique metabolite features across all samples. Principal component analysis (PCA) results showed defined clustering by diabetes status (Fig. 6A). After analysis of the 592 metabolites, 110 features were found to be statistically different between metabolic groups (Fig. 6B). Based on m/z and retention times, the metabolites were queried, identified by using the human metabolomic database (HMDB), and compiled into MetaboAnalyst™ for pathway analysis. Results showed that the three most significant pathways that were significantly different between diabetic status were phenylalanine, tyrosine and tryptophan metabolism pathways (Fig. 6C). The tyrosine metabolism pathway is broad and responsible for the synthesis and metabolism of TYR, DA, and NE and it includes associated MAO and ALDH byproducts. Based on further MS/MS analysis, we identified and confirmed three metabolites within the tyrosine/catecholamine pathway that were significantly different in the myocardial tissue from the patients with diabetes (Supplementary Table S4).
FIG. 6.
Metabolomics pathway analysis of human atrial myocardium. LC-MS analysis of metabolic profiles was performed by using cardiac lysate. (A) Is a representative PCA scores plot showing clusters based on metabolic status. (B) After alignment and data extraction, 592 metabolite features were detected, of which 110 were found to be statistically significant between metabolic groups (p ≤ 0.05). (C) Metaboanalyst pathway analysis revealed that five of the metabolites were involved within the tyrosine metabolism pathway. (D) The five metabolites shown in the abbreviated tyrosine pathway were confirmed by comparison of the observed mass fragmentation patterns and retention times to those published in the Human Metabolome Database. Peak areas of the identified metabolites are compared and shown in D. All data are shown as mean ± SEM and analyzed by using unpaired t-tests. *p < 0.05, **p < 0.005 compared with nondiabetes. D, diabetes; DOPAL, 3,4-dihydroxyphenylacetaldehyde; DOPEGAL, 3,4-dihydroxyphenylglycolaldehyde; HPAA, hydroxphenylacetic acid; HPAL, hydroxyphenylacetaldehyde; HVA, homovanillic acid; MHPG, 3-methoxy-4-hydroxyphenylglycol; PCA, principal component analysis.
Tyrosine can be converted to either TYR or l-DOPA, the product of tyrosine hydroxylase. TYR is then converted to its associated catecholaldehyde, hydroxyphenylacetaldehyde (HPAL), by MAO. Samples from patients with diabetes had much lower levels of hydroxyphenylacetic acid (HPAA) (p < 0.05), the metabolite of HPAL produced by ALDH, which is consistent with reduced ALDH activity in these patients. l-DOPA, the precursor of DA and NE synthesis (p < 0.05), and DA (p < 0.005) and NE (p < 0.05) were also decreased with diabetes. Although no differences were observed within DA metabolism, DHPG, the alcohol metabolite of DOPEGAL, was substantially decreased in myocardial tissue from patients with diabetes (p < 0.05) (Fig. 6D).
Discussion
This translational study provides direct evidence that within the myocardium of patients with diabetes, MAO activity increases, whereas ALDH activity decreases, and this has potentially profound impacts on redox balance and mitochondrial OxPHOS. Global metabolomics confirmed our biochemical analyses, suggesting that increased MAO and decreased ALDH activities in myocardium from patients with diabetes are shunting catcholaldehydes (DOPAL and DOPEGAL) into alternative metabolic pathways, including formation of toxic catechol-modified protein adducts in mitochondria.
In a previous study of human myocardium, MAO expression was measured in atrial samples from a small cohort of patients with established HF, or HF and diabetes combined, and no significant differences in MAO-A or MAO-B expression were found between groups (19). However, that group used real-time PCR and immunohistochemistry in fixed myocardial tissue, whereas we performed immunoblot and ELISA analysis after validating the specificity of the MAO-A and MAO-B antibodies by using immunoblot with recombinant forms of each enzyme (Fig. 1C). We then developed a quantitative ELISA method with these antibodies to determine isoform-specific concentrations in this tissue and by using MAO isoenzyme-specific inhibitors performed a comprehensive kinetic analysis of MAO-A and MAO-B activity in a large cohort of patients. Given the high degree of variability (>10-fold) in MAO enzyme content (Fig. 1B), it is unsurprising that studies in small patient cohorts fail to show significant differences. A more interesting and potentially important finding in our MAO analysis is that both MAO-A and MAO-B are catalytically altered in patients with diabetes. By normalizing the enzymatic activity to the content (via ELISA), we were able to determine that there are altered kinetics at the level of the enzyme itself. To our knowledge, this is the first time that the kinetics of these critical enzymes have been assessed in the human myocardium. These findings also suggest that MAO catalytic activity is regulated at the post-translational level (e.g., phosphorylation, acetylation, carbonylation, etc.), and that some post-translational modification of MAO belies the increased catalytic activity in the myocardium with diabetes.
Interestingly, when comparing MAO isoform activity with BMI, a significant correlation was only observed within the nondiabetes patient group. This finding suggests that in normoglycemic patients, a positive relationship exists between obesity and MAO activity; however, in hyperglycemic patients regardless of obesity, MAO-dependent metabolism of catecholamines remains elevated. Although obesity and overnutrition have been associated with increased sympathetic activity (1, 29), the release of and sensitivity to insulin may act as a “brake” on sympathetic drive. This is supported by a study in a brain-specific insulin-receptor knockout mouse model, which had increased brain-specific MAO activity, catecholamine metabolism, and interestingly, depression-like behavior (41).
We found that metabolism of NE by MAO decreased mitochondrial ATP production in myocardium from patients with type 2 diabetes. This decrease in ATP production without a simultaneous change in oxygen consumption implies a decreased efficiency of the phosphorylation system (e.g., ATP-synthase, adenine nucleotide translocase, ANT), as opposed to a disruption in electron flux within the ETS. Since the effect of NE was mitigated by MAO inhibitors, it is most likely due to the reactivity of MAO-derived byproducts. Numerous oxidative post-translational modifications of the ATP synthase have been reported to affect ATP synthase efficiency (32, 62, 65). Using the APBA resin to pull down catechol-modified proteins, our proteomic analysis confirmed that multiple isoforms of ANT and ATP-synthase are, indeed, potential targets of catecholaldehydes (Fig. 5E). Not only these enzymes but also many other enzymes critical to fatty acid oxidation, TCA cycle, mitochondrial fission/fusion, and redox status are all potentially catechol modified. Although these proteomic findings are preliminary and do not definitively confirm that these catechol modifications are inhibitory, or that they necessarily result in altered protein function, they are, nevertheless, an important first step in this line of investigation. Future studies are warranted to elucidate mechanisms by which MAO-derived byproducts disrupt ATP production (e.g., specific residues within enzymes and their corresponding effects), and the extent to which these byproducts contribute to cardiomyocyte electromechanical dysfunction (Fig. 7).
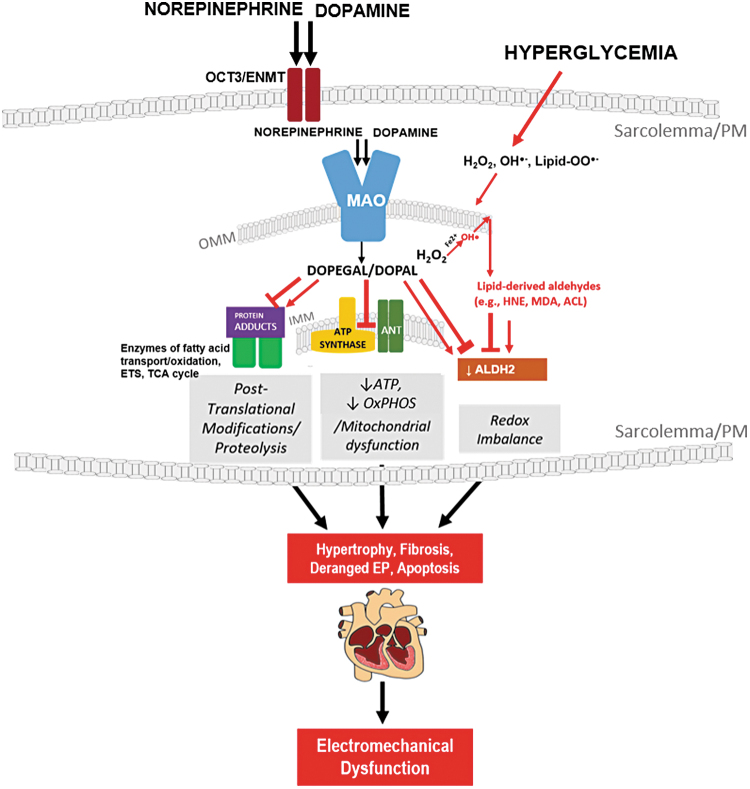
FIG. 7.
Pathophysiological effects of MAO-derived byproducts within myocardium from patients with type 2 diabetes. MAO expression and activity are increased in the myocardium of patients with type 2 diabetes, suggesting increased flux of catecholamines through MAO. In the myocardium, MAO-derived byproducts form catechol-modified protein adducts (post-translational modifications) on enzymes responsible for fatty acid oxidation, tricarboxylic acid cycle, and OxPHOS (energy disruption). In patients with diabetes, the compromised ALDH2 activity would exacerbate these toxicities by creating a “detoxification bottleneck.” Chronic carbonyl stress due to hyperglycemia and insulin resistance may contribute to hypertrophy, fibrosis, and EP derangements, ultimately leading to electromechanical dysfunction in the myocardium of patients with diabetes. ANT, adenine nucleotide translocase; EP, electrophysiology; IMM, inner-mitochondrial membrane; OCT3/ENMT, organic cation transporter-3/extraneuronal monoamine transporter; OMM, outer mitochondrial membrane.
Although this study represents the first time that catecholaldehydes have been studied in the human heart, these unique carbonyls have long been suspected to be pathogenic in models of neurodegenerative disease, specifically in the pathogenesis of Parkinson's disease. DOPAL has been shown to induce protein aggregation and cell death, and to inhibit tyrosine hydroxylase, the enzyme responsible for DA and NE biosynthesis (10, 11, 47, 59). Further, these earlier studies established that DOPAL and DOPEGAL induce mitochondrial-mediated cell death via opening of the permeability transition pore (PTP) in dopaminergic cell models (11, 42, 46). Taking into consideration that subunits of the ATP-synthase, which is a major component of the PTP, were found to be present in our catechol-modified mitochondrial proteomic analysis, it is plausible that the decrease in ATP production observed in Pmfbs exposed to NE was due to transient opening of the PTP induced by catecholaldehydes. However, preliminary studies in our laboratory using isolated cardiac mitochondria from wild-type and cardiomyocyte-specific MAO-A knockout mice have shown that NE does not stimulate premature PTP opening whatsoever. Indeed, MAO-mediated catabolism of NE may even modestly prevent PTP opening (data not shown). Rigorous studies along these lines of investigation are ongoing in our lab.
Our study also revealed ALDH2 activity to be compromised within myocardium from patients with type 2 diabetes. This mitochondrial-localized enzyme has a critical role in phase 2 detoxification and carbonyl metabolism. Numerous studies of cardiac disease, including experimental models of diabetes, have illustrated the importance of ALDH2 in preserving myocardial structure and function (27, 60). Further, therapeutic strategies that increase ALDH2 activity (Alda-1) or sequester aldehydes have shown promise in diabetes-associated cardiac complications (60, 64). ALDH2 is among the many enzymes responsible for metabolism of DOPAL and DOPEGAL. Although the free levels of these catecholaldehydes were undetectable in the myocardial samples in our hands (i.e., below the detectable limits of our MS instruments), we found that catechol-modified protein adducts are more abundant in myocardial tissue from patients with diabetes (Fig. 4C). These findings strongly suggest that an aldehyde “detoxification bottleneck” may be present in the heart with diabetes, and that catecholaldehydes are diverted more toward catechol modification of proteins, thereby contributing to cardiotoxicity via carbonyl stress and redox imbalance in the myocardium of these patients as we have previously reported (4).
Limitations
Although the diabetes and nondiabetes groups were well matched with respect to demographics and clinical variables, there are additional variables that have not been captured in our dataset, which may possibly be contributing to our findings. Specifically, although the patients were diagnosed as having diabetes, we were unable to capture an index of insulin resistance in this cohort (i.e., HOMA-IR), a condition known to be associated with altered sympathetic activity (34, 56). In addition, we cannot completely exclude the potential influence of cardiovascular disease comorbidities in our dataset, although cardiac disease prevalence was evenly matched between groups (Supplementary Table S1).
The potential for antihyperglycemic drugs to have contributed to the increased cardiac MAO in patients with diabetes cannot be completely excluded. The majority of patients with diabetes within our cohort (73%) were treated with antihyperglycemics, including insulin, which has been shown to reduce circulating catecholamine levels in untreated, long-term diabetes (14, 15). Further, a significant number of patients with diabetes (26%) were treated with metformin, which has been proposed to inhibit the activities of α1/2-adrenoceptors, thereby possibly preventing NE release/concentration within the heart (23, 31, 45).
Owing to the high degree of sympathetic innervation in the atrium, it is likely that some MAO content/activity could be coming from ganglionic nerve terminals. However, we take great care to cleanly dissect myocardium from right atrial appendage samples. Further, the fact that in Pmfbs, which are permeabilized and largely void of nonmyocardial tissue, there is a significant effect of NE on mitochondrial ATP production in patients with diabetes, leads us to conclude that there is, indeed, a localized effect of MAO in myofibers that impacts mitochondrial OxPHOS. We also cannot exclude the possibility that the decreased catecholamine levels in the myocardium of patients with diabetes may be due to a dysregulation in NE release and re-uptake, which may be due to potential influence of cardiac autonomic neuropathy. Within the heart, the majority of catecholamines released from the presynaptic cleft are returned to the nerve terminal via catecholamine-specific transporters, including norepinephrine transporter (NET) and dopamine transporter. A smaller portion of catecholamines are transported into non-neuronal cells, including the myocardium, through isoforms of the organic cation transporter-3 and extraneuronal monoamine transporters, while the remaining catecholamines enter the general circulation (i.e., spillover). In experimental models of type 2 diabetes, several groups have shown an attenuation in NET function, increased NE spillover, and decreased NE release (9, 40, 43). We were unable to measure catecholamine transport and re-uptake parameters within our patient cohort.
Proteomic analysis of catechol-modified proteins was performed on mitochondria pooled from patients with and without diabetes for several reasons. First, mitochondrial yield from aged human right atrial appendages is low, despite our best technical skills. Sometimes, the samples have a lot of extracellular matrix and fibrous material, and this lowers the ability to protect/preserve intact mitochondrial fractions during extraction steps. Next, although the APBA resin captures the catecholaldehyde adducts with fairly strong affinity, there is material lost in the binding that further decreases sample yield. Although this method prevented us from identifying potential differences in the identity and quantity of catechol-modified proteins between groups, we, nevertheless, provide evidence for the first time that such modifications exist and remain a valid and active area of future investigation. Additional studies in human cardiac tissue are warranted and would be beneficial in discerning molecular characteristics (e.g., specific amino acids, structure) of the modifications caused by catecholaldehydes.
Finally, we were not able to directly quantify absolute concentration of catecholaldehydes (DOPAL, DOPEGAL) in myocardial extracts and only captured adducts formed on proteins via NBT staining and APBA resin enrichment. There are several reasons for this. First, catecholaldehydes in tissue are highly reactive and they spontaneously and rapidly react with solvent-accessible Lys and Cys residues on proteins or are further metabolized via phase 2 enzymes (26, 53). In addition, these adducts are proving to be very challenging to measure with MS, due to their instability and reactivity. For example, DOPEGAL is not easily synthesized and it rapidly self-polymerizes and undergoes rearrangement; thus, it is difficult to quantify. Despite these challenges, we are confident that with continued refinement in our LC-MS/MS and organic chemistry methods we expect to have a sensitive and reproducible detection method in place very soon.
Conclusions/perspectives
This study provides evidence that catecholamine flux is significantly enhanced, whereas carbonyl detoxification is diminished in human myocardium with diabetes, and that MAO-derived catecholaldehydes contribute to carbonyl stress and mitochondrial dysfunction in this tissue. Our findings suggest that these catecholaldehydes are always being produced and metabolized in the heart, but in obesity/diabetes, the combined effect of increased sympathetic tone, increased cardiac MAO activity, and decreased aldehyde-scavenging enzyme activity forces a “detoxification bottleneck” in the myocardium, which causes derangements in mitochondrial OxPHOS and contributes to electro-mechanical dysfunction (Fig. 7). This same pathogenic process undoubtedly occurs in the etiology of HF, and it rapidly occurs during reperfusion injury as reported quite recently in a very interesting study (54). Although much more work is necessary to completely understand the reactivity and cardiotoxicity of catecholaldehydes, these initial findings provide insight into a pathogenic mechanism by which MAO contributes to cardiac disease in diabetes (i.e., atrial fibrillation, cardiomyopathy). It is anticipated that these results will inform future studies in experimental models concerning the role of MAO in cardiac diseases. Finally, our study suggests that targeting catecholamine metabolism and aldehyde detoxification in heart may have therapeutic benefit in patients with type 2 diabetes.
Materials and Methods
Chemicals and reagents used in this study were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise noted.
Patient enrollment, blood and atrial tissue collection
All experiments involving human subjects received approval by the Institutional Review Board of the Brody School of Medicine at East Carolina University and the University of Iowa. Patients undergoing nonemergent elective coronary artery bypass graft (CABG) or CABG/valve surgery were enrolled between January 2011 and July 2018. Inclusion criteria included patients between the age of 50 and 70 without a previous history of cardiac surgery or arrhythmia. Informed consent was obtained from each patient to perform all biochemical analysis on the cardiac tissue provided. A fasted blood draw was performed before the initiation of anesthetics. Before placement on cardiopulmonary bypass, a portion of myocardium was biopsied in each patient after establishing a purse-string suture around the right atrial appendage. Myocardial tissue was dissected, rinsed in ice-cold phosphate-buffered saline (PBS), and used immediately for mitochondrial experiments. A separate portion of myocardium was snap frozen and maintained at −80°C for biochemical and metabolomics analysis.
Measurement of DA and NE using LC-MS
After homogenization, samples were centrifuged at 14,000 g for 30 min. The supernatant was placed under nitrogen flow, maintained at 50°C, and evaporated to dryness. The residue was resuspended in 1:1 methanol: H2O. Standards of catecholamines (10 to 1500 ng/mL) were prepared in 1:1 H2O: methanol and maintained at −20°C before use. Quantitative analysis of catecholamines was performed by using an Exion LC/AB SCIEX 3200 triple quadropole mass spectrometer (AB SCIEX, Framingham, MA). LC was performed by using a Hypersil Gold aQ (3 × 50 mm, 1.9 μm) column (Thermo Fisher Scientific, Waltham, MA). The following gradient solvents were utilized: mobile phase A: 95:5 water:ACN (acetonitrile) with 0.1% formic acid and B: 100% methanol with a flow rate of 0.3 mL/min. A 15 min total run time was used with a linear gradient from 97% B over 8 min, was held for 1 min, decreased to 0% B for 1 min, and equilibrated for 5 min. A 10 μL injection volume was used from each sample.
Quantitative analysis of MAO-A and MAO-B enzyme content in myocardium
Standards and samples were added to immunolon-coated 96-well assay plates and incubated overnight at 4°C. The plates were washed with PBS +0.05% Tween-20 and blocked with 5% BSA at 37°C. Samples were incubated with primary antibodies for MAO-A or MAO-B (Abcam, Cambridge, MA) followed by incubation with horseradish peroxidase-conjugated secondary antibody. Amplex Red (Thermo Fisher Scientific) was added, and the fluorescence was measured at Ex/Em 567/590 nm.
For MAO-A and MAO-B Western blots, 20 μg of sample protein lysate was loaded onto a 10% acrylamide gel and subject to SDS-PAGE. The proteins were transferred to a PVDF membrane and blocked with 5% BSA in TBS-Tween. The membranes were incubated with primary antibodies specific for each MAO isoform. The membranes were washed, probed with secondary antibodies, and imaged by using the Odyssey imaging system (Li-Cor Bioscience, Lincoln, NE).
MAO enzymatic activity in myocardial tissue
MAO activity was determined by measuring H2O2 production in the presence of Amplex UltraRed (Thermo Fisher Scientific). Each assay was measured in 1.5 mL by using Amplex Red (10 μM), horseradish peroxidase (1 U/mL), superoxide dismutase (25 U/mL), and 15 μL of myocardial lysate. Enzymatic activities were normalized to the concentration of isozyme obtained from the ELISA and reported as nmol/(min·mg) enzyme.
Preparation of permeabilized muscle fibers
After excision of myocardium from the atrial sample, ∼300 mg myocardium was placed in ice-cold Buffer X (7.23 mM K2EGTA, 2.77 mM CaK2EGTA, 20 mM imidazole, 0.5 mM DTT, 20 mM taurine, 5.7 mM ATP, 14.3 mM phosphocreatine, 6.56 mM MgCl2·6H2O, 50 mM KMES, pH 7.1) followed by a 30-min incubation with 3 μg/mL of collagenase type I (Worthington, Lakewood, NJ) at 4°C. Using fine-tip tweezers, muscle fiber bundles were separated along the longitudinal axis and permeabilized in 10 μg/mL saponin for 30 min. The fibers were then incubated in the respiratory buffer containing blebbistatin until used for respiratory experiments (<1 h). To prevent extra-mitochondrial production of ATP, P1,P5-di(adenosine-5′) pentaphosphate (AP5a), an adenylate kinase inhibitor, was added. Experiments were performed by using Buffer Z-lite (10 mM K-MES, 30 mM KCl, 10 mM KH2PO4, 5 mM MgCl2·6H2O, 0.5 mg/mL BSA, 20 mM creatine), 1 mM EGTA, 200 μM NADP+, 4 mM glucose, 1.7 U/mL of glucose-6-phosphate, and 3.4 U/mL of hexokinase. The recordings were normalized for 10 min followed by the addition of 50 μM palmitoyl-l-carnitine and 2 mM malate. After 3 min, 500 μM ADP was added to drive maximal respiration. After each experiment, myofibers were collected and stored at −20°C in a freezing buffer (Buffer X, glycerol, protease inhibitor cocktail).
Normalization of mitochondrial content in permeabilized muscle fibers
To estimate mitochondrial content within Pmfbs, citrate synthase activity was measured based on published methods (55). Fibers were removed from freezing buffer, homogenized 1:20 (w/v) in Tris-buffer (100 mM Tris-base, 2 mM EDTA pH 7.5), and centrifuged for 10 min at 10,000 g at 4°C. Homogenates were diluted 1:10 with Tris-buffer and added onto a clear polysterene 96-well plate. A reaction cocktail including 5,5′-dithiobis(2-nitrobenzoic acid), acetyl-CoA, and oxaloacetate was added to each well and absorbance was measured at 412 nm every minute for 10 min. Citrate synthase activity was calculated by using the extinction coefficient of 13.6 L/(mol·cm), and the results were used to normalize respiratory experiments.
Mitochondrial analysis in permeabilized human cardiac myofibers
Previous methods published from our lab were used to prepare permeabilized muscle fibers from human atrial myocardium (Pmfb) (3). Simultaneous and real-time ATP/O2 measurements were measured by using a customized system of a coupled spectrofluorometer (HORIBA Scientific, Edison, NJ) to an Oroboros O2K Oxygraph (Oroboros, Innsbruck, Austria) via lightguides. The glucose-6-phosphate dehydrogenase/hexokinase coupled enzyme system links the conversion of glucose, via mitochondrial ATP production, to the reduction of NADP+ to NADPH, which is autofluorescent at Ex/Em 345/460. The rate of ATP production was calculated based on a standard curve of fluorescence corresponding to known ATP concentrations. Oxygen flux was quantified by using Oroboros DatLab software. Experimental conditions are as outlined in the Figure legends.
Detection and proteomic analysis of catecholaldehyde-modified proteins
NBT is a redox-cycling dye that binds to catechol-protein adducts within a nitrocellulose membrane and stains the proteins blue, allowing the visualization of catechol-modified proteins (51, 52). Myocardial lysate alone, or after having been incubated with catecholamines ± MAO inhibitors, was subjected to SDS-PAGE. The gel, filter paper, and nitrocellulose membrane were incubated in Bjerrum Schaefer-Nielsen transfer buffer (48 mM Tris-Base, 39 mM glycine, 20% methanol) and transferred at 10 V for 70 min. The membrane was incubated in NBT solution (0.24 mM NBT in 2 M potassium glycine buffer) in the dark for 40 min followed by imaging by using a ChemiDoc Imaging System (Bio-Rad, Hercules, CA). Extraction of catechol-modified protein adducts from isolated cardiac mitochondrial samples was performed by using APBA-agarose method, using a partially modified method that was validated and previously published (44, 47). Proteomic analysis was performed on the mitochondrial proteins extracted from the APBA resin by staff at the University of Iowa-Carver College of Medicine's Proteomics Core Facility (PCF), using LC-MS/MS and tandem MS on the Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an Easy-nLC-1200™ System (Thermo Fisher Scientific). Spectral searches were performed by using both Mascot version 2.6.2 (Matrix Science, Inc., Boston, MA) and Byonic search engines ver. 2.8.2 (Protein Metrics, Cupertino, CA), with database queries, as outlined in the Supplementary Materials.
ALDH2 activity and protein detection in human atrial myocardium
ALDH2 is the mitochondrial isoform of ALDH, and it is responsible for metabolizing catecholaldehydes into weak acids. ALDH2 activity was measured based on previous methods and determined by monitoring the ALDH2 conversion of NAD+ to NADH reflected by an increase in absorbance at 450 nm. Assays were performed by using 50 mM sodium pyrophosphate buffer (pH 8.0), 200 μg of cardiac protein lysate, 2.5 mM NAD+, and 2 μM rotenone. The reaction was initiated with the addition of 10 mM acetaldehyde, and absorbance was recorded every 30 s for 15 min. The detection of ALDH2 within myocardial lysates was performed by using Western blot techniques previously noted by using a primary antibody targeting ALDH2 (Abcam).
AR activity in human atrial myocardium
AR is another major pathway for the metabolism of DOPEGAL. With NADPH as a cofactor, AR converts DOPEGAL to DHPG (Figs. 4A and 6). AR enzymatic activity in myocardial lysates was measured spectrophotometrically by monitoring the oxidation of NADPH to NADP at 340 nm (12). A reaction volume of 200 μL was used, which consisted of 0.1 M potassium phosphate buffer (pH 6.0), 200 μg cardiac protein lysate, and 0.15 mM NADPH. The reaction was initiated with the addition of 10 mM glyceraldehyde, and absorbance was monitored every 30 s for 5 min.
Mitochondrial isolation from human atrial myocardium
Mitochondria from atrial samples were isolated with methods adapted from previous studies (28). Briefly, fresh atrial myocardium was dissected from endocardial side of the atrial appendage sample, minced on ice for 4 min, and subjected to a brief trypsin digestion. Trypsin inhibitor was added after 2 min, and the mixture was placed in a 50 mL conical tube and allowed to settle. The supernatant was removed, and the minced tissue was resuspended in mitochondrial isolation medium (MIM) (in mM: 300 sucrose, 10 Na-HEPES, 0.2 EDTA) and homogenized by using a prechilled Dounce homogenizer. The mixture was homogenized slowly with 10–12 strokes. The homogenate underwent differential centrifugation steps, and the pellets of mitochondria were resuspended in MIM + BSA.
Synthesis of DOPEGAL and DOPEGAL-albumin adducts
DOPEGAL was prepared by a previously described method (50). In a 15 mL Falcon tube, 9.5 mL of 10 mM potassium phosphate buffer (pH 7.5), 1.0 mL of 100 mM sodium bisulfite, 0.5 mL of a 250 mM solution of NE (Sigma-Aldrich), and 100 μL (500 μg) of recombinant MAO (Corning Life Sciences, Tewksbury, MA) were combined. The reaction vessel cap was perforated to expose the mixture to oxygen and gently shaken at 30°C for 10 h. After the reaction was stopped by ultracentrifugation at 100,000 g for 30 min, the supernatant was collected, and the MAO pellet was resuspended in potassium phosphate buffer and frozen for later reuse. To make DOPEGAL-BSA adducts, a 50 μL aliquot of the supernatant was incubated with 1 mL of a 3% BSA solution in 50 mM sodium pyrophosphate buffer (pH 8.8) overnight at 4°C.
Extraction of catechol-modified proteins from isolated cardiac mitochondria via m-APBA resin
Using a commercially available APBA-agarose resin (Sigma-Aldrich), catechol-modified protein adducts were isolated by using a previously described method with slight modifications (44, 47). APBA suspension (500 μL) was washed three times in 50 mM potassium phosphate buffer (pH 7.4) and centrifuged at 6600 g for 2 min. Protein from human cardiac mitochondrial preps (700 μg) in 50 mM sodium pyrophosphate buffer was added to the APBA resin and incubated overnight at 4°C while rocking. The APBA agarose was then washed three times with 1 mL of 1:1 ACN:50 mM sodium pyrophosphate buffer three times, followed by a wash with 1 mL of 5 mM sodium pyrophosphate buffer, and then a wash with ddH2O. Every wash was proceeded by centrifugation at 9000 g for 5 min and discarding the supernatant. Catechol-modified proteins were then eluted from the APBA resin by incubating with 100 μL 1:1 ACN: 1% TFA solution for an hour at room temperature while rocking. After centrifuging the suspension (9000 g for 5 min), the supernatant was collected and immediately neutralized with 30 μL of 1 M potassium phosphate buffer (pH 7.4).
Proteomic analysis of APBA extracts prepared from isolated human atrial mitochondria
Mitochondrial protein extracts eluted from the APBA column were then washed 5 times with cold PBS. Overall, 1 × SDS sample-loading buffer was added to the beads, and the mixture was boiled for 5 min. The gel was stained with Silver Stain kit (Thermo Fisher Scientific), and the bands corresponding to those indicated with arrows in Figure 5 were manually excised, minced into 1 mm3 pieces, and further treated with ACN, to effectively “dry” the gel segments and then reduced in 50 μL of 10 mM DTT at 56°C for 60 min. After this, gel-trapped proteins were alkylated with 55 mM chloroacetamide (CAM) for 30 min at room temperature. The gel pieces were washed and incubated with trypsin at 37°C for 16 h. The combined peptide extracts were lyophilized and resuspended in 15 μL of LC buffer A and analyzed in conjunction with LC-MS/MS on the orbitrap.
Liquid chromatography tandem mass spectrometry
Mass spectrometry data were collected by using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an Easy-nLC-1200 System (Thermo Fisher Scientific). Typically, 3–4 μL of reconstituted digest was loaded on a 2.5 cm C18 trap (New Objective, Inc., Woburn, MA) coupled to an analytical column through a microcross assembly (IDEX Health & Science, Oak Harbor, WA). Peptides were desalted on the trap by using 16 μL mobile phase A in 4 min. The waste valve was then blocked, and a gradient began flowing at 4 μL/min through a self-packed analytical column; 10 cm in length × 75 μm id.
Peptides were separated in-line with the mass spectrometer by using a 70 min gradient composed of linear and static segments, wherein buffer A is 0.1% formic acid and B is 95% ACN, 0.1% formic acid. The gradient begins first holds at 4% for 3 min and then makes the following transitions (%B, min): (2, 0), (35, 46), (60, 56), (98, 62), and (98, 70).
MS/MS on the LUMOS Orbitrap
Data acquisitions begin with a survey scan (m/z 380–1800) acquired on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) at a resolution of 120,000 in the off axis Orbitrap segment (MS1) with Automatic Gain Control (AGC) set to 3E06 and a maximum injection time of 50 ms. The MS1 scans were acquired every 3 s during the 70 min gradient described earlier. The most abundant precursors were selected among two to six charge state ions at a 1E05 AGC and 70 ms maximum injection time. Ions were isolated with a 1.6 Th window by using the multi-segment quadrupole and subject to dynamic exclusion for 30 s if they were targeted twice in the prior 30 s. Selected ions were then sequentially subjected to both collision-induced dissociation (CID) and high energy collision-induced dissociation (HCD) activation conditions in the IT and the ion routing multipole (IRM), respectively. The AGC target for CID was 4.0E04, 35% collision energy, with an activation Q of 0.25 and a 75 ms maximum fill time. Targeted precursors were also fragmented by HCD at 30% collision energy in the IRM. HCD fragment ions were analyzed by using the Orbitrap (AGC 1.2E05, maximum injection time 110 ms, and resolution set to 30,000 at 400 Th). Both MS2 channels were recorded as centroid, and the MS1 survey scans were recorded in profile mode.
Proteomic searches
Initial spectral searches were performed with both Mascot version 2.6.2 (Matrix Science, Inc.) and Byonic search engines ver. 2.8.2 (Protein Metrics). Search databases were composed of the Uniprot KB for species 9606 (Human) downloaded October 24, 2017 containing 92645 sequences and Uniprot KB for taxonomy 562 (Escherichia coli) downloaded on November 8, 2017 containing 10079 sequences. For Byonic searches, these two databases were directly concatenated. In either search, an equal number of decoy entries were created and searched simultaneously by reversing the original entries in the target databases. Precursor mass tolerance was set to 10 ppm, and fragments acquired in the Linear Trap and the Orbitrap were searched at 4 Da and 10 ppm, respectively. A fixed 57 Da modification was assumed for cysteine residues (from CAM treatment), whereas variable oxidation modifications were allowed at methionine. The false discovery rate (FDR) was maintained at 1% by tracking matches to the decoy database.
Both Mascot and Byonic search results were combined and validated by using Scaffold ver. 4.8.5 (Proteome Software, Portland, OR). Protein assignments required a minimum of two peptides established at 70% probability (Local FDR algorithm) and an overall <1% protein FDR (assigned by Protein Prophet). Approximately 600 protein families (including common contaminants) were assigned. Proteins were annotated with GO terms from goa_uniprot_all.gaf downloaded on May 3, 2017. The complete data file has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021759 and 10.6019/PXD021759.
Metabolomic profiling of myocardial samples
An Eksigent 425 microLC/SCIEX 5600+ time-of-flight mass spectrometer (SCIEX) was used to measure metabolites within myocardial samples. Samples were prepared the same as the NE and DA LC-MS preparation. A Halo C18 0.5 × 50 mm 2.7 μm column was used for separation of the metabolites with the following solvent compositions: mobile phase A: 95:5 water:ACN with 0.1% formic acid and B: ACN with 0.1% formic acid. A linear gradient was utilized with a flow rate of 10 μL/min where the gradient started with 10% B for 2 min, increased to 90% B for 15 min, held for 5 min, dropped to 10% B over 2 min, and equilibrated for 10 min for a total run time of 30 min. Injection volume for samples was 5 μL.
Data were acquired for MS and MS/MS analysis by using independent data acquisition for the top 20 ions in the positive ionization mode. The scan range for MS was 80–1250 Da. PCA and t-tests were conducted by using MarkerView 1.3.1. Statistically significant masses were tentatively identified by using the Human Metabolome Database (hmdb.ca) and confirmed with MS-MS or a standard. Pathway analysis was then conducted via Metaboanalyst (www.metaboanalyst.ca) on database identified peaks (13, 61).
Statistical analysis
In the human clinical data analysis, categorical variables are presented as frequency and percentage and continuous variables were presented as mean ± standard deviation. Fisher exact and χ2 procedures were used to compute statistical significance of group comparisons for categorical variables. The Deuchler-Wilcoxon test was used for continuous variables. For the multivariable analysis, quartiles of MAO-A and MAO-B activity were analyzed by using a robust Poisson regression model with relative risk of diabetes as the measure of association. Missing values for all clinical and biochemical variables (<5%) were imputed by using the iterative expectation-maximization algorithm (3, 39). p-Value in multivariable analysis was computed by using Friedman's nonparametric test for central tendency while adjusting for age, sex and race. Ptrend was computed by using likelihood ratio trend test, adjusting for age and sex. SAS Version 9.4 (SAS, Cary, NC) was used for all analyses of human biochemical and clinical variables. For MAO kinetic measurements, best-fit curves were implemented by using nonlinear regression analysis and rates of MAO activity were compared between metabolic groups at each titration timepoint.
The LC-MS, ELISA, enzyme activity, and immunoblot densitometry were analyzed by using unpaired Student's t-tests to compare values between metabolic groups. Multiple paired Student's t-test were used to compare differences in treatment groups of mitochondrial respiration experiments. Metabolomic data were analyzed by using PCA followed by Student's t-test to compare peak values between metabolic groups. Data were presented as mean ± standard error of the mean, and values of p < 0.05 were considered statistically significant. All analysis was performed by using GraphPad Prism 8 (GraphPad Software, La Jolla, California).
Supplementary Material
Acknowledgments
The authors would like to thank the clinical staff of the East Carolina Heart Institute for their kind assistance with patient recruitment, blood and tissue collection. They would also like to acknowledge use of resources at the Carver College of Medicine's Proteomics Core Facility (PCF) at the University of Iowa where mass spectrometry data were collected. Dr. R. Marshall Pope directs the PCF, which is supported by an endowment from the Carver Foundation and by a Thermo Lumos awarded by an HHMI grant to Dr. Kevin Campbell.
Abbreviations Used
- 5-HT
serotonin
- βPEA
β-phenylethylamine
- ACN
acetonitrile
- ALDH
aldehyde dehydrogenase
- ALDH2
aldehyde dehydrogenase 2
- ANT
adenine nucleotide translocase
- AP5a
P1,P5-di(adenosine-5′) pentaphosphate
- APBA
aminophenylboronic acid
- AR
aldose reductase
- BMI
body mass index
- BSA
bovine serum albumin
- CABG
coronary artery bypass graft
- CAM
chloroacetamide
- CID
collision-induced dissociation
- COX IV
complex IV
- DA
dopamine
- DHPG
dihydroxyphenylglycol
- DOPAC
dihydroxyphenylacetic acid
- DOPAL
3,4-dihydroxyphenylacetaldehyde
- DOPEGAL
3,4-dihydroxyphenylglycolaldehyde
- ELISA
enzyme-linked immunosorbent assay
- EP
electrophysiology
- ETS
electron transport system
- FDR
false discovery rate
- HbA1c
glycated hemoglobin
- HCD
high energy collision-induced dissociation
- HF
heart failure
- HMDB
human metabolomic database
- HNE
4-hydroxy-nonenal
- HOMA-IR
homeostatic model assessment for insulin resistance
- HPAA
hydroxyphenylacetic acid
- HPAL
hydroxyphenylacetaldehyde
- HVA
homovanillic acid
- IMM
inner-mitochondrial membrane
- IRM
ion routing multipole
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MAO
monoamine oxidase MAOi, monoamine oxidase inhibitors
- MDA
malondialdehyde
- MHPG
3-methoxy-4-hydroxyphenylglycol
- MIM
mitochondrial isolation medium
- NBT
nitroblue tetrazolium
- ND
nondiabetes
- NE
norepinephrine
- NET
norepinephrine transporter
- OCT3/ENMT
organic cation transporter-3/extraneuronal monoamine transporter
- OMM
outer mitochondrial membrane
- OxPHOS
oxidative phosphorylation
- PBS
phosphate-buffered saline
- PCA
principal component analysis
- Pmfbs
permeabilized myofibers
- PTP
permeability transition pore
- ROS
reactive oxygen species
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SEM
standard error of the mean
- TCA
tricarboxylic acid
- TFA
trifluoroacetic acid
- TYR
tyramine
Authors' Contributions
M.M.N. helped design the study, performed experiments, analyzed data, and prepared the article. L.A.K. and J.T.E were responsible for statistical and epidemiological analysis of the data. K.A.K., C.N.B., Q.S., J.A.D., S.A.A., H.A., and J.R. assisted with data collection and provided input on the article. T.B.M. played a critical role in the preparation and proteomic analysis of catechol-modified mitochondrial proteins. E.J.A. designed the study, assisted with article preparation, and is the guarantor of the data and content presented.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by National Institutes of Health grant R01HL122863 and R21AG057006 (Ethan J. Anderson).
Supplementary Material
References
- 1. Amador N, Guizar JM, Malacara JM, Perez-Luque E, and Paniagua R. Sympathetic activity and response to ACE inhibitor (enalapril) in normotensive obese and non-obese subjects. Arch Med Res 35: 54–58, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Anderson DG, Florang VR, Schamp JH, Buettner GR, and Doorn JA. Antioxidant-mediated modulation of protein reactivity for 3,4-dihydroxyphenylacetaldehyde, a toxic dopamine metabolite. Chem Res Toxicol 29: 1098–1107, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson EJ, Efird JT, Davies SW, O'Neal WT, Darden TM, Thayne KA, Katunga LA, Kindell LC, Ferguson TB, Anderson CA, Chitwood WR, Koutlas TC, Williams JM, Rodriguez E, and Kypson AP. Monoamine oxidase is a major determinant of redox balance in human atrial myocardium and is associated with postoperative atrial fibrillation. J Am Heart Assoc 3: e000713, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson EJ, Kypson AP, Rodriguez E, Anderson CA, Lehr EJ, and Neufer PD. Substrate-specific derangements in mitochondrial metabolism and redox balance in the atrium of the type 2 diabetic human heart. J Am Coll Cardiol 54: 1891–1898, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berman SB and Hastings TG. Dopamine oxidation alters mitochondrial respiration and induces permeability transition in brain mitochondria. J Neurochem 73: 1127–1137, 1999 [DOI] [PubMed] [Google Scholar]
- 6. Bianchi P, Kunduzova O, Masini E, Cambon C, Bani D, Raimondi L, Seguelas MH, Nistri S, Colucci W, Leducq N, and Parini A. Oxidative stress by monoamine oxidase mediates receptor-independent cardiomyocyte apoptosis by serotonin and postischemic myocardial injury. Circulation 112: 3297–3305, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Boveris A. Determination of the production of superoxide radicals and hydrogen peroxide in mitochondria. Methods Enzymol 105: 429–435, 1984 [DOI] [PubMed] [Google Scholar]
- 8. Bugger H, Abel ED. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia 57: 660–671, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgdorf C, Richardt D, Kurz T, Adler S, Notzold A, Kraatz EG, Sievers HH, and Richardt G. Norepinephrine release is reduced in cardiac tissue of Type 2 diabetic patients. Diabetologia 46: 520–523, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Burke WJ, Kristal BS, Yu BP, Li SW, and Lin TS. Norepinephrine transmitter metabolite generates free radicals and activates mitochondrial permeability transition: a mechanism for DOPEGAL-induced apoptosis. Brain Res 787: 328–332, 1998 [DOI] [PubMed] [Google Scholar]
- 11. Burke WJ, Vijaya BK, Pandey N, Panneton WM, Gan Q, Franko MW, O'Dell M, Li SW, Pan Y, Chung HD, and Galvin JE. Aggregation of a-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol 115: 193–203, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, and Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321: 1493–1495, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chong J and Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34: 4313–4314, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christensen NJ. Plasma norepinephrine and epinephrine in untreated diabetics, during fasting and after insulin administration. Diabetes 23: 1–8, 1974 [DOI] [PubMed] [Google Scholar]
- 15. Christensen NJ. Plasma catecholamines in long-term diabetes with and without neuropathy and in hypophysectomized subjects. J Clin Invest 51: 779–787, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen G and Kesler N. Monoamine oxidase and mitochondrial respiration. J Neurochem 73: 2310–2315, 1999 [DOI] [PubMed] [Google Scholar]
- 17. Deshwal S, Forkink M, Hu C, Buonincontri G, Antonucci S, Di Sante M, Murphy MP, Paolocci N, Mochly-Rosen D, Krieg T, Di Lisa F, and Kaludercic N. Monoamine oxidase-dependent endoplasmic reticulum-mitochondria dysfunction and mast cell degranulation lead to adverse cardiac remodeling in diabetes. Cell Death Differ 25: 1671–1685, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doorn JA, Florang VR, Schamp JH, and Vanle BC. Aldehyde dehydrogenase inhibition generates a reactive dopamine metabolite autotoxic to dopamine neurons. Parkinsonism Relat Disord 20: S73–S75, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duicu OM, Lighezan R, Sturza A, Balica R, Vaduva A, Feier H, Gaspar M, Ionac A, Noveanu L, Borza C, Muntean DM, and Mornos C. Assessment of mitochondrial dysfunction and monoamine oxidase contribution to oxidative stress in human diabetic hearts. Oxid Med Cell Longev 2016: 8470394, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edmondson DE. Hydrogen peroxide produced by mitochondrial monoamine oxidase catalysis: biological implications. Curr Pharm Des 20: 155–160, 2014 [DOI] [PubMed] [Google Scholar]
- 21. Eisenhofer G, Kopin IJ, and Goldstein DS. Catecholamine metabolism: a contemporary view with implications for physiology and medicine. Pharmacol Rev 56: 331–349, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Erwin VG and Deitrich RA. Brain aldehyde dehydrogenase: localization, purification, and properties. J Biol Chem 241: 3533–3539, 1966 [PubMed] [Google Scholar]
- 23. Esteghamati A, Eskandari D, Mirmiranpour H, Noshad S, Mousavizadeh M, Hedayati M, and Nakhjavani M. Effects of metformin on markers of oxidative stress and antioxidant reserve in patients with newly diagnosed type 2 diabetes: a randomized clinical trial. Clin Nutr 32: 179–185, 2013 [DOI] [PubMed] [Google Scholar]
- 24. Esterbauer H, Schaur RJ, and Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11: 81–128, 1991 [DOI] [PubMed] [Google Scholar]
- 25. Florang VR, Rees JN, Brogden NK, Anderson DG, Hurley TD, and Doorn JA. Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology 28: 76–82, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Goldstein DS, Kopin IJ, and Sharabi Y. Catecholamine autotoxicity. implications for pharmacology and therapeutics of parkinson disease and related disorders. Pharmacol Ther 144: 268–282, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomes KM, Bechara LR, Lima VM, Ribeiro MA, Campos JC, Dourado PM, Kowaltowski AJ, Mochly-Rosen D, and Ferriera JC. Aldehydic load and aldehyde dehydrogenase 2 profile during the progression of post-myocardial infarction cardiomyopathy: benefits of alda-1. Int J Cardiol 179: 129–138, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gostimskaya I and Galkin A.. Preparation of highly coupled rat heart mitochondria. J Vis Exp 2010. DOI: 10.3791/2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grassi G, Seravalle G, Cattaneo BM, Bolla GB, Lanfanchi A, Colombo M, Giannattasio C, Brunani A, Cavagnini F, and Mancia G. Sympathetic activation in obese normotensive subjects. Hypertension 25: 560–563, 1995 [DOI] [PubMed] [Google Scholar]
- 30. Griendling KK, Minieri CA, Ollerenshaw JD, and Alexander RW. Angiotensin II stimulates NADH and NADPH oxidase activity in cultured vascular smooth muscle cells. Circ Res 74: 1141–1148, 1994 [DOI] [PubMed] [Google Scholar]
- 31. Huerta de la Cruz S, Medina-Terol GJ, Beltran-Ornelas JH, Gomez CB, Morato-Valderrama Y, Sanchez-Lopez A, and Centurion D. Pharmacological evidence that metformin blocks the vasopressor responses mediated by stimulation of a1- and a2-adrenorceptors in pithed rats. Eur J Pharmacol 820: 130–137, 2018 [DOI] [PubMed] [Google Scholar]
- 32. Ichihara S, Suzuki Y, Chang J, Kuzuya K, Inoue C, Kitamura Y, and Oikawa S. Involvement of oxidative modification of proteins related to ATP synthesis in the left ventricles of hamsters with cardiomyopathy. Sci Rep 7: 9243, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jinsmaa Y, Florang VR, Rees JN, Anderson DG, Strack S, and Doorn JA. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol 22: 835–841, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jung CH, Kim BY, Kim CH, Kang SK, Jung SH, and Mok JO. Association of serum adipocytokine levels with cardiac autonomic neuropathy in type 2 diabetic patients. Cardiovasc Diabetol 11: 24, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, and Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J Biol Chem 281: 15110–15120, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Kaludercic N, Mialet-Perez J, Paolocci N, Parini A, and Di Lisa F. Monoamine oxidases as sources of oxidants in the heart. J Mol Cell Cardiol 73: 34–42, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaludercic N, Takimoto E, Nagayama T, Feng N, Lai EW, Bedja D, Chen K, Gabrielson KL, Blakely RD, Shih JC, Pacak K, Kass DA, Di Lisa F, and Paolocci N. Monoamine oxidase A-mediated enhanced catabolism of norepinephrine contributes to adverse remodeling and pump failure in hearts with pressure overload. Circ Res 106: 193–202, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kannel WB, Hjortland M, and Castelli WP. Role of diabetes in congestive heart failure: the Framingham study. Am J Cardiol 34: 29–34, 1974 [DOI] [PubMed] [Google Scholar]
- 39. Katunga LA, Gudimella P, Efird JT, Abernathy S, Mattox TA, Beatty C, Darden TM, Thayne KA, Alwair H, Kypson AP, Virag JA, and Anderson EJ. Obesity in a model of gpx4 haploinsufficiency uncovers a causal role for lipid-derived aldehydes in human metabolic disease and cardiomyopathy. Mol Metab 4: 493–506, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kiyono Y, Kajiyama S, Fujiwara H, Kanegawa N, and Saji H. Influence of the polyol pathway on norepinephrine transporter reduction in diabetic cardiac sympathetic nerves: implications for heterogenous accumulation of MIBG. Eur J Nucl Med Mol Imaging 32: 993–997, 2005 [DOI] [PubMed] [Google Scholar]
- 41. Kleinridders A, Cai W, Cappellucci L, Ghazarian A, Collins WR, Vienberg SG, Pothos EN, and Kahn CR. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc Natl Acad Sci U S A 112: 3463–3468, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kristal BS, Conway AD, Brown AM, Jain JC, Ulluci PA, Li SW, and Burke WJ. Selective dopaminergic vulnerability: 3,4-dihydroxyphenylacetaldehyde targets mitochondria. Free Radic Biol Med 30: 924–931, 2001 [DOI] [PubMed] [Google Scholar]
- 43. Kusmic C, Morbelli S, Marini C, Matteucci M, Cappellini C, Pomposelli E, Marzullo P, L'abbate A, and Sambuceti G. Whole-body evaluation of MIBG tissue extraction in a mouse model of long-lasting type II diabetes and its relationship with norepinephrine transport protein concentration. J Nucl Med 49: 1701–1706, 2008 [DOI] [PubMed] [Google Scholar]
- 44. LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, and Selkoe DJ. Dopamine covalently modifies and functionally inactivates Parkin. Nat Med 11: 1214–1221, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Lee JM and Peuler JD. Acute vasorelaxant effects of metformin and attenuation by stimulation of sympathetic agonist release. Life Sci 64: 57–63, 1999 [DOI] [PubMed] [Google Scholar]
- 46. Marchitti SA, Deitrich RA, and Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev 59: 125–150, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Mexas LM, Florang VR, and Doorn JA. Inhibition and covalent modification of tyrosine hydroxylase by 3,4-dihydroxyphenylacetaldehyde, a toxic dopamine metabolite. Neurotoxicology 32: 471–477, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitchell DY and Petersen DR. The oxidation of alpha-beta unsaturated aldehydic products of lipid peroxidation by rat liver aldehyde dehydrogenases. Toxicol Appl Pharmacol 87: 403–410, 1987 [DOI] [PubMed] [Google Scholar]
- 49. Murphy E, Ardehali H, Balaban RS, DiLisa F, Dorn GW, Kitsis RN, Otsu K, Ping P, Rizzuto R, Sack MN, Wallace D, and Youle RJ. Mitochondrial function, biology, and role in disease: a scientific statement from the American heart association. Circ Res 118: 1960–1991, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nilsson GE and Tottmar O. Biogenic aldehydes in brain: on their preparation and reactions with rat brain tissue. J Neurochem 48: 1566–1572, 1987 [DOI] [PubMed] [Google Scholar]
- 51. Paz MA, Fluckiger R, Boak A, Kagan HM, and Gallop PM. Specific detection of quinoproteins by redox-cycling staining. J Biol Chem 266: 689–692, 1991 [PubMed] [Google Scholar]
- 52. Rees JN, Florang VR, Anderson DG, and Doorn JA. Lipid peroxidation products inhibit dopamine catabolism yielding aberrant levels of a reactive intermediate. Chem Res Toxicol 20: 1536–1542, 2007 [DOI] [PubMed] [Google Scholar]
- 53. Rees JN, Florang VR, Eckert LL, and Doorn JA. Protein reactivity of 3,4-dihydroxyphenylacetaldehyde, a toxic dopamine metabolite, is dependent on both the aldehyde and catechol. Chem Res Toxicol 22: 1256–1263, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Santin Y, Fazal L, Sainte-Marie Y, Sicard P, Maggiorani D, Tortosa F, Yucel YY, Teyssedre L, Rouquette J, Marcellin M, Vindis C, Shih JC, Lairez O, Burlet-Schiltz O, Parini A, Lezoualc'h F, and Mialet-Perez J. Mitochondrial 4-HNE derived from MAO-A promotes mitoCa(2+) overload in chronic postischemic cardiac remodeling. Cell Death Differ 27: 1907–1923, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Srere PA. Citrate synthase. Methods Enzymol 13: 3–5, 1969 [Google Scholar]
- 56. Straznicky NE, Grima MT, Sari CI, Eikelis N, Lambert EA, Nestel PJ, Esler MD, Dixon JB, Chopra R, Tilbrook AJ, Schlaich MP, and Lambert GW. Neuroadrenergic dysfunction along the diabetes continuum: a comparative study in obese metabolic syndrome subjects. Diabetes 61: 2506–2516, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Umbarkar P, Singh S, Arkat S, Bodhankar SL, Lohidasan S, Sitasawad SL. Monoamine oxidase-A is an important source of oxidative stress and promotes cardiac dysfunction, apoptosis, and fibrosis in diabetic cardiomyopathy. Free Radic Biol Med 87: 263–273, 2015 [DOI] [PubMed] [Google Scholar]
- 58. Vanle BC, Florang VR, Murry DJ, Aguirre AL, and Doorn JA. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by the dopamine metabolite, 3,4-dihydroxyphenylacetaldehyde. Biochem Biophysl Res Commun 492: 275–281, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vermeer LM, Florang VR, and Doorn JA. Catechol and aldehyde moieties of 3,4-dihydroxyphenylacetaldehyde contribute to tyrosine hydroxylase inhibition and neurotoxicity. Brain Res 1474: 100–109, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang C, Fan F, Cao Q, Shen C, Zhu H, Wang P, Zhao X, Sun X, Dong Z, Ma X, Liu X, Han S, Wu C, Zou Y, Hu K, Ge J, and Sun A. Mitochondrial aldehyde dehydrogenase 2 deficiency aggravates energy metabolism disturbance and diastolic dysfunction in diabetic mice. J Mol Med 94: 1229–1240, 2016 [DOI] [PubMed] [Google Scholar]
- 61. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, and Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46: D608–D617, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yarian CS, Rebrin I, and Sohal RS. Aconitase and ATP synthase are targets of malondialdehyde modification and undergo an age-related decrease in activity in mouse heart mitochondria. Biochem Biophys Res Commun 330: 151–156, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Youdim MB, Edmonson DE, and Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nat Rev Neurosci 7: 295–309, 2006 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y, Babcock SA, Hu N, Maris JR, Wang H, and Ren J. Mitochondrial aldehyde dehydrogenase (ALDH2) protects against streptozotocin-induced diabetic cardiomyopathy: role of GSK3B and mitochondrial function. BMC Med 10: 40, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65. Zhao Y, Miriyala S, Miao L, Mitov M, Schnell D, Dhar SK, Cai J, Klein JB, Sultana R, Butterfield DA, Vore M, Batinic-Haberle I, Bondada S, St. and Clair DK. Redox proteomic identification of HNE-bound mitochondrial proteins in cardiac tissues reveals a systemic effect on energy metabolism after doxorubicin treatment. Free Radic Biol Med 72: 55–65, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.