Abstract
Less than a year following the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak, variants of concern have emerged in the form of variant Alpha (B.1.1.7, the British variant) and Beta (B.1.351, the South Africa variant). Due to their high infectivity and morbidity, it has become clear that it is crucial to quickly and effectively detect these and other variants. Here, we report improved primers-probe sets for reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) for SARS-CoV-2 detection including a rapid, cost-effective, and direct RT-qPCR method for detection of the two variants of concern (Alpha, B.1.1.7 and Beta, B.1.351). All the developed primers-probe sets were fully characterized, demonstrating sensitive and specific detection. These primer-probe sets were also successfully employed on wastewater samples aimed at detecting and even quantifying new variants in a geographical area, even prior to the reports by the medical testing. The novel primers-probe sets presented here will enable proper responses for pandemic containment, particularly considering the emergence of variants of concern.
Keywords: Reverse transcriptase polymerase chain reaction, Wastewater-based epidemiological monitoring, SARS-CoV-2, Molecular probes, Variants of concern
1. Introduction
The SARS-CoV-2 world pandemic erupted in early 2020 with rising numbers in morbidity and mortality. SARS-CoV-2 was recognized as an RNA virus, therefore detection methods emerging for immediate response were mainly based on of reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) (Lu et al., 2020). In RT-qPCR, RNA is extracted, undergoes reverse transcription for DNA strand generation, followed by PCR amplification and TaqMan probes fluorescence detection. To date, RT-qPCR is the most common methodology for SARS-CoV-2 diagnostics (Vogels et al., 2020).
Starting in September 2020, new variants of concern of SARS-CoV-2 virus began to be detected in patients (Mercatelli and Giorgi, 2020). Amongst them, the variants termed the Alpha Variant (British, B.1.1.7) and the Beta variant (South Africa, B.1.351) became dominant compared to the original SARS-CoV-2 virus (Wang et al., 2021). Due to their higher infection rate and high morbidity, identification of these variants in the population became essential. This led for a search for a diagnostic tool that could quickly and efficiently distinguish between the variants to help evaluate their distribution. Such “variant mapping” provides much needed information to enforce appropriate policy for pandemic containment.
Currently, three methodologies have been developed for SARS-CoV-2 variant diagnostics. The first methodology, that is still the one being mainly employed, is the next generation sequencing (NGS) approach (Andrés et al., 2020; Khan et al., 2020). In NGS the entire variant's genome is sequenced. Despite the importance of this technique for identification of new variants, it is time-consuming (three to five days minimum) and requires significant financial means and data analysis (Korber et al., 2020) (https://www.protocols.io/about).
Additional detection methods are based on RT-qPCR and include a “drop-out” signal, available in commercial kits (such as TaqPath COVID-19 diagnostic tests, Thermo Scientific, Helix® COVID-19 Test) or utilizing a method published in a recent study (Vogels et al., 2021). These methodologies use RT-qPCR with two different markers, a double signal manifest for the original SARS-CoV-2 virus, while only a single signal manifest for the targeted variant. Another detection methodology uses the characterization of ΔCt between one detection signal and another, amongst the different variants (Kovacova et al., 2021). These current RT-qPCR approaches for variant detection, though significantly faster and more cost-effective than the NGS methodology, focus on indirect detection and may result in false/inconclusive identification. Therefore, there is still a great need for detection methods with improved specificity, sensitivity and speed of detection of SARS-CoV-2 variants. Such advanced methodologies will be amenable for clinical diagnostics as well as for environmental-derived quantification, greatly improving wastewater and population level epidemiological investigation. Recent commercial developments such as PerkinElmer® or GT molecular™ offers detection kits for Alpha and Beta variants of concern, however the design specifics are not available and therefore cannot be reviewed.
Urban sewage treatment is carried out through wastewater treatment plants that are usually separated from industrial sewage. Therefore, wastewater sampling is often used as an epidemiological tool, as it can provide a snapshot of the microbial lineages and their diversity in the population (Martin et al., 2020). Although wastewater samples are not unbiased and present some challenges when monitored, many studies and countries have adopted this methodology as it provides a more reliable representation of population morbidity status compared to volunteered clinical tests (reviewed in Ahmed et al., 2020b; Hart and Halden, 2020; Hill et al., 2020; Polo et al., 2020).
In the present study, we developed primer-probe sets for an improved and sensitive detection of SARS-CoV-2. Moreover, we developed a RT-qPCR assay for the direct detection of the SARS-CoV-2 Alpha variant, B.1.1.7 and Beta variant, B.1.351. Our design was tested on S gene deletion and non-deletion DNA templates and on RNA originating from wastewater samples in order to assess the sensitivity and specificity of the described primer sets.
2. Methods
2.1. Primers and probes design
The original sequence of SARS-CoV-2 (NC_045512.2) was taken from NCBI database. Alpha, B.1.1.7 variant (EPI_ISL_742238) and Beta, B.1.351 variant (EPI_ISL_736935) sequences were taken from GISAID database (Shu and McCauley, 2017). The probe design focused on the S gene 21724-21828 bp location that includes the Alpha variant deletion 69–70 or S gene 22243-22331 bp location that includes the Beta variant deletion 241–243. All primers and probes were purchased through Integrated DNA Technologies (IDT). ZEN Quencher was added to the probes as a second, internal quencher in qPCR 5′-nuclease assay. To allow a possibility for duplex assay, S1 probe was assigned a 6-carboxy-fluorescein (FAM) fluorophore and SΔ69 probe was assigned to Yakima Yellow (YakYel) fluorophore. SΔ241 probe was assigned with FAM as well.
2.2. RT-qPCR
RT-qPCR was executed using One Step PrimeScript III RT-qPCR mix using standard manufacture protocol (RR600 TAKARA, Japan). Each reaction mixture contains primers (0.5 μM each), probe (0.2 μM each), ROX reference dye and 5 μL of DNA or RNA (dH2O was added to a final volume of 20 μL reaction volume). RT-qPCR amplification was executed using Step One Plus real-time PCR system (Applied Biosystems, Thermo Scientific). In addition to what is described above, in each run, all RT-qPCR experiments included quality controls. The first control was using water samples instead of DNA/RNA (Non template control (NTC)). The second control, used for RNA extractions, was MS2 phage detection (Dreier et al., 2005).
2.3. Calibration curves and limit of detection determination
Calibration curves were performed on a known-positive DNA gene block. For N gene detection, standard CDC positive control plasmid was purchased from Integrated DNA Technologies (IDT). For S gene detection, two different gene blocks were used. The first gene block, containing SARS-CoV-2 S gene sequence as reported for Wuhan-Hu-1 (NC_045512.2): TACCCTGACAAAGTTTTCAGATCCTCAGTTTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATACATGTCTCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTTCCACTGAGAAGTCTAACATAATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTTTTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTATTCTAGTGCGAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTAATTTCAAAAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTATTAATTTAGTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAACATCACTAGGTTTCAAACTTTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCTGGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGG. The second gene block containing S gene sequence matching the reported 69–70 deletion of the Alpha variant (B.1.1.7) as well as the reported 241–243 deletion of the Beta variant (B.1.351): TACCCTGACAAAGTTTTCAGATCCTCAGTTTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATCTCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTTCCACTGAGAAGTCTAACATAATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTTTTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTATTCTAGTGCGAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTAATTTCAAAAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTATTAATTTAGTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAACATCACTAGGTTTCAAACTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCTGGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGG. Calibration of S1 probe was performed using S gene sequence from NC_045512.2, while calibration of SΔ69 probe and SΔ241 was performed using S gene sequence with the relevant deletions. Serial dilutions for the relevant gene block were prepared based on copy number calculations. The resulting Ct values were plotted against the log copy number of the N or S gene template. Each concentration was examined by six repetitions and a standard deviation was calculated. Linear regression was performed between the log copy number and the Ct values from the RT-qPCR results.
2.4. Wastewater RNA extraction
For wastewater sampling, 24 h composite sewage samples from the wastewater treatment plant (WWTP) were immediately transferred to the lab under chilled conditions. The samples were kept at 4 °C until processed. Direct or concentrated RNA was extracted twice according to manufacture protocol as described in the NucleoSpin RNA extraction kit (Macherey Nagel, Germany). The MS2 phage (105 copies) was added to the lysis buffer in each RNA extraction as internal control. RNA was eluted with 50 μL of RNase free water and kept at −80 °C.
2.5. Complex matrix detection
RNA extracted from wastewater sample, pre-determined as negative for SARS-CoV-2 using standard CDC's detection sets, was supplemented with known concentrations of a desired gene block. The samples underwent the same RT-qPCR conditions as described for the calibration curves. Matrix pre-determined as negative, was constantly verified as negative in each assay without spiking. In each set, eight repetitions were performed for each viral concentration or control. Ct results were plotted to represent the new probes limit of detection in a complex environment.
2.6. Wastewater concentration
A volume of 2 to 5 L of composite wastewater samples collected from the WWTP were shaken and mixed for 2 min manually. The samples were left standing for 15 min to ensure large particle settlement. The samples were then pumped at a rate of 10 L/min through a dialysis filter with a pore size 3–30 nm (NUFiltration©, Israel). The filter was backwashed with 0.07–0.15 L DW and collected directly to new 0.25 L bottle. After each sample concentration procedure, 2.5 L of 0.01% hypochlorite solution was passed through the system followed by a 10 min wash with DW to ensure no hypochlorite traces and new dialysis filter was placed for new batch concentration.
3. Results and discussion
3.1. Improved primers and probes for SARS-CoV-2 N gene detection
When the SARS-CoV-2 emerged, the recommended detection method was through RT-qPCR analysis; a methodology that is still the preferred method. Several different published primers-probe sets were used, aiming at different genes of the SARS-CoV-2 virus (such as N gene, E gene, S gene) (Corman et al., 2020; Lu et al., 2020; Vogels et al., 2020). Initially, the Centers for Disease Control and Prevention (CDC)'s, recommendation was to carry out RT-qPCR analysis using 3 sets of primers and probes, N1, N2 and N3, all targeting the N gene. Apart from clinical testing, these CDC recommended primer sets were also examined on wastewater samples and demonstrated positive SARS-CoV-2 detection (Ahmed et al., 2020a; Bar-Or et al., 2020; Medema et al., 2020; La Rosa et al., 2020; Trottier et al., 2020; Westhaus et al., 2021; Wu et al., 2020a, 2020b). However, after careful scrutiny of these published sequences, we detected possible problems with annealing stability and thus suggested a modification of the published primer-probe to improve annealing stability.
Since the 3′ end of a primer sequence is the transcription initiation point for the polymerase, it is preferable the 3′ end of a primer will possess G/C for more stable annealing (3 hydrogen bonds, compare to only 2 between A/T pair). Thus, looking to improve the available sets of primers, when there was no G/C at the 3′ end, the primers were shifted to include G/C. In addition, to improve the N3 set, the probe sequence was shifted as well in order to maintain close proximity to the forward primer. Based on this approach, all 3 of the recommended primers-probe sets were improved (Fig. 1 a and b). Furthermore, we also generate a fourth primers-probe termed “N4 new”.
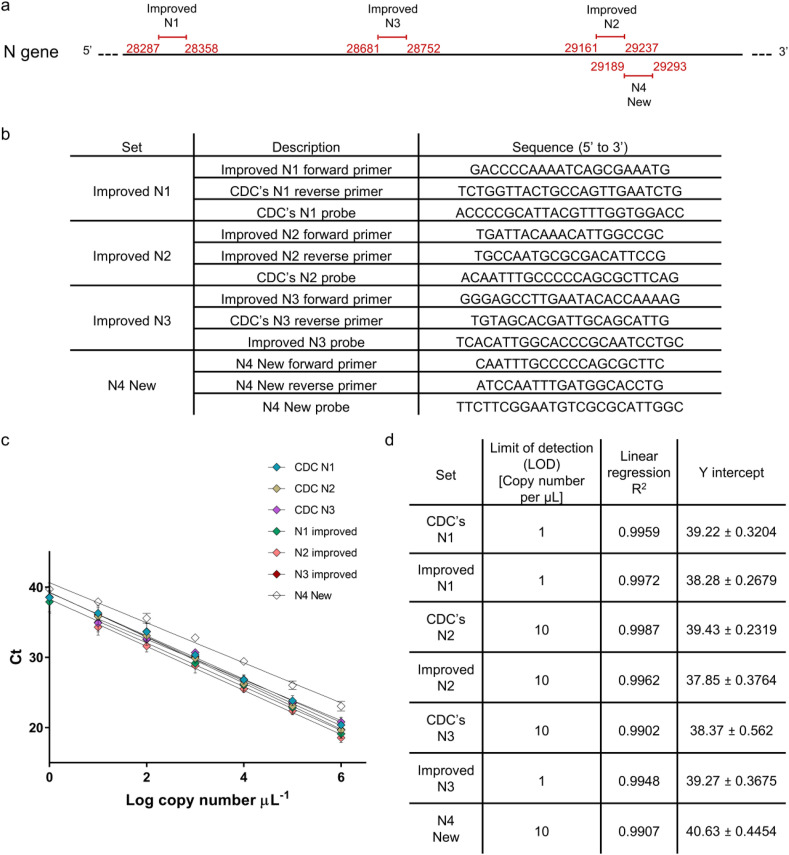
Fig. 1.
N gene detection designs and characterization. (a) Improved and new primers-probe sets design along SARS-CoV-2 N gene. Numbers in red indicate bp region amplified in RT-qPCR with the relevant primers-probe set. (b) List of Improved and new primers-probe sets sequences. (c) Calibration curves for all examined primers-probe sets for N gene detection. Resulted Ct value from RT-qPCR plotted against Log copy number tested. Error bars present standard deviation for six replicates. (d) List of Limit of detection, Linear regression R2, and Y intercept values extracted from the resulted calibration curves. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Once genetated, all primers-probe sets were thoroughly characterized and compared to the previously published CDC's sets (N1, N2, and N3). Using dsDNA templates, a calibration curve was generated for each set (Fig. 1c). A detection range of between 106 copies and 100 copies per μL was tested for each set. Employing linear regression on all calibration curves resulted in a good fit and allowed the determination of limit of detection (LOD) for each set (Fig. 1d). Comparing each set to its improved version, LOD for CDC's N1 vs. Improved N1 and CDC's N2 vs. Improved N2 were the same (1 and 10 copies per μL respectively). For CDC's N3 vs. Improved N3, a significant improvement was observed when LOD was reduced from 10 copies per μL to 1 copy per μL. The newly designed N4 set demonstrated LOD of 10 copies per μL.
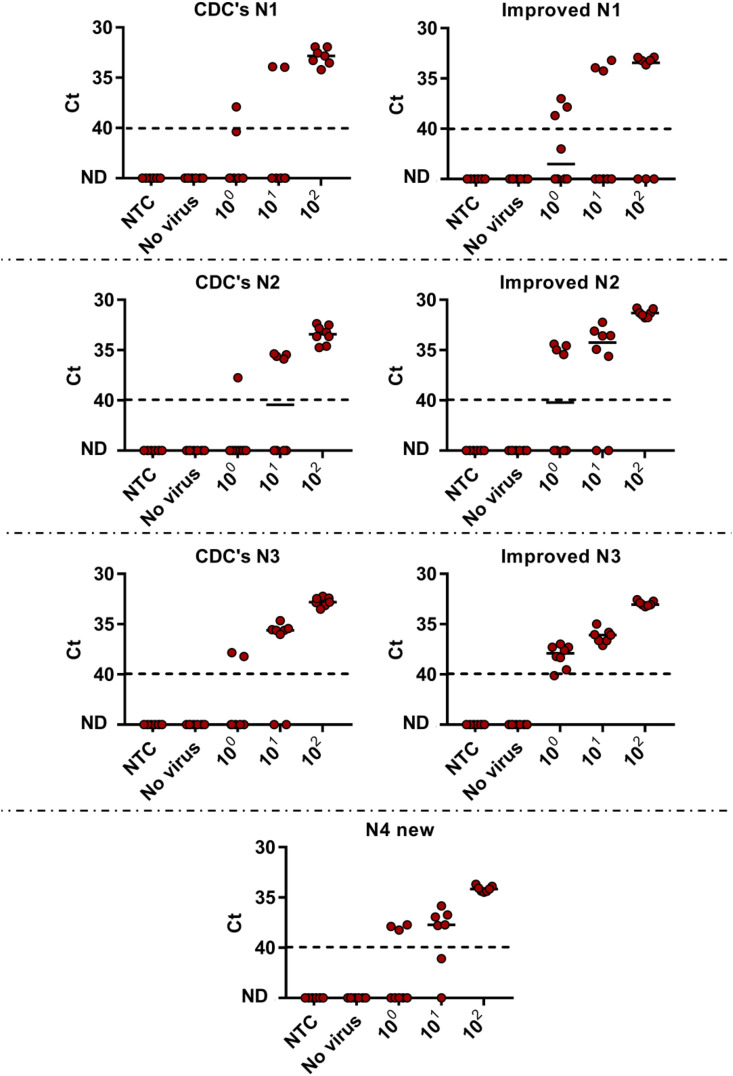
Following basic characterization, further confirmation was sought after using a more complex environment for the RT-qPCR reaction. Detection of SARS-CoV-2 in wastewater is important with regards to the development of a quick early warning system for virus detection during the global pandemic (Bar-Or et al., 2020). Thus to test this ability, wastewater matrix was collected from wastewater treatment plant in the city of Binyamina, Israel. All primers-probe sets were employed on the wastewater samples that had been pre-determined as negative for SARS-CoV-2 when examined using dsDNA template copies (Fig. 2 ). As can be seen in Fig. 2, results for the CDC's detection sets (N1, N2, N3) were somewhat similar to previously published detection results (Vogels et al., 2020).
Fig. 2.
Lower detection limit of N gene examined primers–probe sets in wastewater matrix. RNA extracted from negative detection wastewater sample (No virus) served as wastewater matrix and spiked with known concentrations of SARS-CoV-2. Matrix was spiked with different concentration of N gene template (100–102 copies per μL) or Non-Template Control (NTC, water). ND - not detected. Solid lines indicate the median and dashed lines indicate the detection limit as decided by clinical guidelines (Ct > 40).
LOD is determined as the lowest detected copy number per μL in 90% of the tested cases. Given the complexity of the wastewater matrix, a reduction in LOD could be observed for all sets, CDC and Improved sets, except for Improved N3 set. Compared to the calibration curves, while all the sets LOD rose to 100 copies per μL (compared to 10 or 1 copies concluded before), the Improved N3 was the only set that remained with a stable LOD value of 1 copy per μL. Comparison between the CDC's sets and improved sets in a controlled wastewater sample, revealed that each improved set demonstrated better detection abilities than the original CDC's set, as expected. A hierarchy of sensitivity was given to the examined N gene detection sets as follows (best to poorest): Improved N3 > Improved N2 > N4 new > Improved N1 > CDC's N3 > CDC's N2 > CDC's N1. Thus the best set observed was Improved N3 with the ability to detect 1 copy per μL at 100% of the cases, an improved result when compared to other primers-probe sets reported in literature (Vogels et al., 2020).
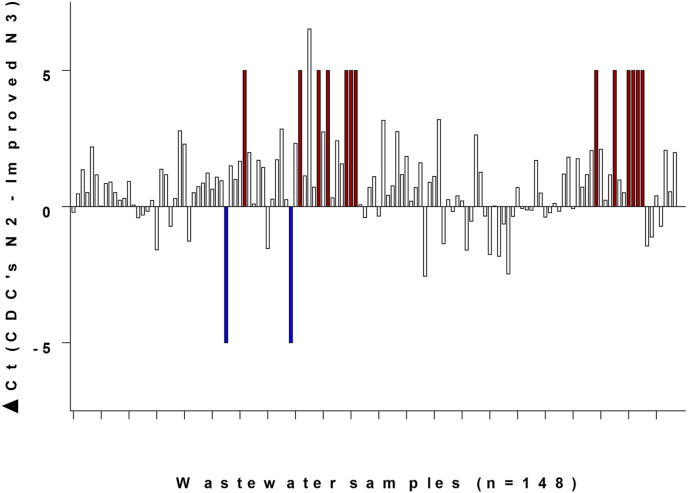
Based on all characterizations performed, the Improved N3 set demonstrated the best detection abilities out of all examined sets. It was then validated on various wastewater samples. Together with Improved N3 set, the CDC's N2 set was also added as a formerly approved control to each experiment (CDC's N3 performed better than CDC's N2, however N3 set was not recommended by the CDC after a few months of usage and only N1 and N2 remained as officially approved). Out of 148 different wastewater samples, 17 were not detected by either set. For the remaining 131 samples, a ΔCt was calculated between CDC's N2 Ct and Improved N3 Ct (Fig. 3 ). A positive ΔCt value means a better detection by the Improved N3 set, while a negative value indicates better detection by CDC's N2. Out of 131 wastewater samples with a calculated ΔCt, the majority showed positive ΔCt in favor of the Improved N3 set, indicating better detection by this set (Fig. 3). Moreover, in 13 samples Improved N3 detected SARS-CoV-2 while CDC's N2 did not detect any signal. Only 2 samples showed the reverse, where CDC's N2 detected a signal and Improved N3 did not. Matching the characterization results, Fig. 3 shows the ability of the improved N3 set to better detect SARS-CoV-2 in wastewater, as it systematically provided lower Ct values. Consequently, the Improved N3 primers-probe set is recommended and provides more sensitive virus detection and can be employed on various samples.
Fig. 3.
SARS-CoV-2 detection in wastewater using primers-probe sets Improved N3 and CDC's N2. For each sample out of 148 different samples in September–December 2020, ΔCt was calculated. ΔCt is comprised from reduction of the resulted Ct for Improved N3 from the resulted Ct for CDC's N2. For samples where CDC's N2 resulted in a signal while Improved N3 did not, meaning only CDC's N2 detected, an arbitrary value of −5 was assigned and colored in blue. For samples where Improved N3 resulted in a signal while CDC's N2 did not, meaning only Improved N3 detected, an arbitrary value of +5 was assigned and colored in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. RT-qPCR detection of variants of concern Alpha, B.1.1.7 and Beta, B.1.351
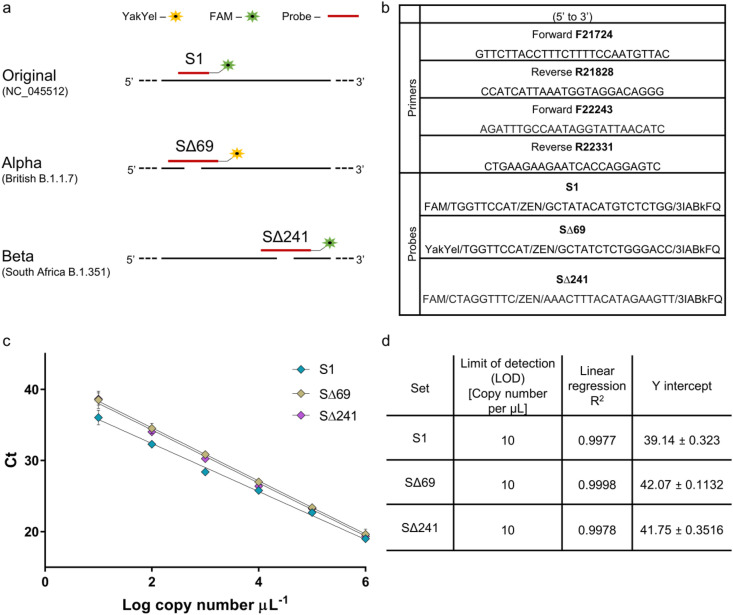
Existing detection methods for SARS-CoV-2 variants are either through NGS sequencing or indirect RT-qPCR assays. To improve rapid detection, we focused on the development of a direct RT-qPCR assay for detection of variants of concern, the Alpha variant, B.1.1.7 and Beta variant, B.1.351. These were deemed the most urgent variants in need for fast detection in Israel. Our design for RT-qPCR detection assays of these two variants (Fig. 4 ) is based on the differences in the S gene from the original SARS-CoV-2 sequence published (NC_045512.2). The Alpha variant S gene contains a deletion known as Δ69-70 and the Beta variant S gene contains a deletion known as Δ241-243. Accordingly, our designed detection-probe set focused on these regions and presented in Fig. 4.
Fig. 4.
S gene detection designs and characterization for variants of concern. (a) Designed detection sets for differentiation and identification of SARS-CoV-2 Alpha variant, B.1.1.7 and Beta variant, B.1.351. Design was based on sequence differences in the S gene between the variants. (b) List of primers and probes sequences for variants detection. (c) Calibration curves for primers-probe sets for variants of concern detection. Resulted Ct value from RT-qPCR plotted against Log copy number tested. Error bars present standard deviation for six replicates. (d) List of Limit of detection, Linear regression R2, and Y intercept values extracted from the resulted calibration curves. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For Alpha detection, the designed set is located at the S gene 21724-21828 bp of the original sequence. Within this range, the original SARS-CoV-2 and Alpha variant sequences are completely identical, apart from 6 nucleotides deletions (Fig. 4a). Our main attempt was to create two separate detections to the amplified area, one corresponding to the original sequence (when using S1 probe) and the other corresponding to the Alpha variant (when using the SΔ69 probe). Using designated primers (Fig. 4b) to amplify the specified region surrounding the 6 nucleotides differences, amplification will be generated regardless of the variant. The probes can thus be used in a single duplex assay via separate wavelengths, where a signal signifies a direct detection of either the original sequence, of Alpha variant, or a combination of the two if exist.
For Beta variant detection, the designated detection region was chosen further along the S gene when compared to the Alpha variant detection region. Focusing on S gene 22243-22331 bp of the original sequence, the original SARS-CoV-2 sequence is identical to the Beta variant sequence except for a 9 nucleotides deletion (Fig. 4a). Using a detection set comprised of two primers meant to amplify the target region, a single probe (SΔ241 probe) was designed for the detection of the Beta variant. The SΔ241 probe is meant to correspond only to the deletion of 9 nucleotides in the specified region characterizing the Beta variant, therefore it will signal detection only when the Beta variant is present and will not correspond to the original or the Alpha variant sequence.
To ensure functionality, the described sets of primers and probes underwent characterization. Initially, a calibration curve was generated for primers with the relevant probe separately, using dsDNA as a template. A detection range of between 106 copies and 100 copies per μL was tested for each set (Fig. 4c). Linear regression performed for the three probes demonstrated strong correlation and also validated the usage of the probe on the amplified fragment. An LOD could be determined for each primer-probe set and was identified as 101 copies per μL for all three sets (Fig. 4d).
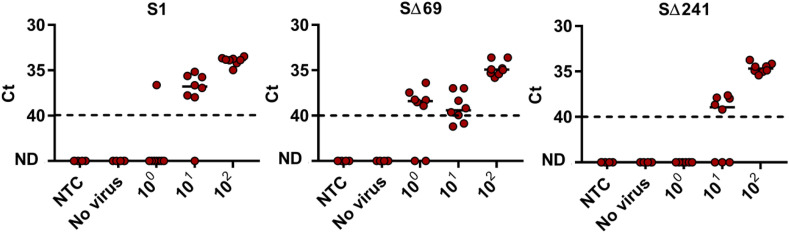
As with the initial N gene detection sets, basic characterization was followed with further confirmation to the described methodology using a more complex environment for the RT-qPCR reaction. Here as well, wastewater from Beer Sheva, Israel was used as the complex environment. All three probes were employed on wastewater samples pre-determined as negative for SARS-CoV-2 with various dsDNA template copies (Fig. 5 ). As can be seen in Fig. 5, despite the wastewater matrix, SΔ69 maintained a LOD of 101 copies per μL. Unlike SΔ69 set, S1 and SΔ241 sets’ LOD increased 10-fold to 102 copies per μL, meaning that the complexity of the wastewater matrix influenced detection performance. Despite the fact that LOD increased for two out of the three sets, all three detection sets have a LOD of 102 or lower, which corresponds to other utilized, functional RT-qPCR SARS-CoV-2 detection sets currently being used (Vogels et al., 2020).
Fig. 5.
Lower detection limit of SΔ69, SΔ241 and S1 primers–probe sets in wastewater matrix. RNA extracted from negative detection wastewater sample (No virus) served as wastewater matrix and spiked with known concentrations of SARS-CoV-2. Matrix was spiked with different concentration of S gene template (100–102 copies per μL) or Non-Template Control (NTC, water). For SΔ69 set and SΔ241 set, the S gene deletion template corresponded to Δ69–70 deletion site in the Alpha strain and Δ241–243 deletion site in the Beta strain. For S1 set, the S gene template corresponded to NC_045512.2 original sequence. ND - not detected. Solid lines indicate the median and dashed lines indicate the detection limit as decided by clinical guidelines (Ct > 40).
To examine the probes specificity and rule out possible false-positive cases, each set was tested with a negative control. For S1 set, the negative control was comprised of a dsDNA template with the S gene with Δ69–70 and Δ241–243 nucleotides deletions. While for SΔ69 set and SΔ241 set, the original S gene sequence was used as negative control. As expected, none of the probes manifested a signal in the presence of a negative control and non-specific detection was not observed (Table 1 ).
Table 1.
Detection sets results when employed on positive or negative template and wastewater samples.
| WWTP | Date of sampling | MS2 Ct | Improved N3 Ct | CDC's N2 Ct | S1 Ct | SΔ69 (Alpha) Ct | SΔ241 (Beta) Ct |
|---|---|---|---|---|---|---|---|
| S gene Original template | – | – | NDa | NDa | 28.61 | NDa | NDa |
| S gene deletion template | – | – | NDa | NDa | NDa | 30.29 | 29.94 |
| Rahat | 09.02.21 | 27.8 | 31.48 | 32.19 | NDa | 33.96 | NDa |
| ShafDan | 08.02.21 | 27.51 | 31.07 | 31.26 | NDa | 32.92 | NDa |
| Haifa | 14.02.21 | 27.82 | 32.74 | 32.95 | 36.24 | ND | NDa |
| Natanya | 15.02.21 | 27.55 | 29.38 | 28.33 | NDa | 32.44 | NDa |
| Tzfat | 15.02.21 | 27.35 | 28.96 | 27.46 | NDa | 31.46 | NDa |
| Al-Hamra | 29.03.21 | 29.43 | 33.64 | 32.26 | NDa | 32.11 | NDa |
| Ar'ara | 29.03.21 | 29.36 | 32.63 | 31.7 | NDa | 30.11 | NDa |
| Lehavim | 23.02.21 | 27.73 | 28.95 | 27.82 | NDa | 32.13 | NDa |
| Beer-Sheva | 25.02.21 | 27.82 | 29.3 | 28.91 | NDa | 32.55 | NDa |
*WWTP-Wastewater treatment plant.
ND-Not Detected.
Finally, the three sets, S1, SΔ69 and SΔ241 were tested on wastewater samples collected in February and March 2021 from different regions in Israel, (Table 1). The CDC's N2 detection set was used as standard detection reference that can correspond to each of the variants (Lu et al., 2020) as was the Improved N3 set. All samples were positive for N gene detection by the CDC's N2 and Improved N3 detection sets, meaning all samples contained SARS-CoV-2. Examining detection ability of variants using this method showed that none of the samples were positive for the Beta variant using the SΔ241 set. This indicated that the Beta variant was absent from all wastewater samples. On the other hand, all regions apart from one resulted in detection by the SΔ69 set while the S1 set had no signal. Such a result not only indicated that Alpha variant is present in most regions in Israel, but also that there was no detectible trace of the original SARS-CoV-2 NC_045512.2. Haifa was the only region where S1 was detected, while SΔ69 was not, indicating the absence of Alpha variant and presence of the original NC_045512.2. Our results correlated well with reports of the Alpha variant, B.1.1.7 outbreak in Israel.
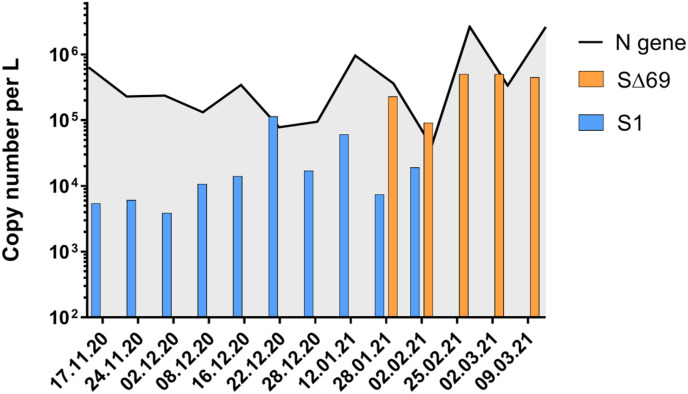
Following the positive detection of Alpha variant in wastewater in Israel, we wanted to learn when did the variant's outbreak occur. Since wastewater samples from the city of Beer Sheva had been collected throughout the SARS-CoV-2 outbreak, this allowed us to examine the time period from November 2020 until March 2021. All wastewater samples were subjected to N gene, S1 and SΔ69 detection (Fig. 6 ). As can be seen in Fig. 6, the original SARS-CoV-2 NC_045512.2 was present without any trace of the Alpha variant until mid-January 2021. By the end of January 2021, the Alpha variant had emerged in the wastewater and was already more abundant than the original NC_045512.2. By the end of February 2021, there was no trace of the original NC_045512.2, and the Alpha variant was the only variant detected. These results demonstrated the power of the novel detection sets ability for discovery of variants of concern. Moreover, considering that unlike clinical samples, wastewater samples can contain several variants at the same time, these probes enabled us to quantify and monitor several variants in a single wastewater sample, which is an imperative ability.
Fig. 6.
SARS-CoV-2 variants detection in Beer Sheva wastewater over time. Samples collected between November 2020 and March 2021 from WWTP were tested for N gene (black line), S1 (blue) and SΔ69 (orange) detection. Consist of levels for SARS-CoV-2 overall detection (N gene), whereas Alpha strain detection manifested by the end of January 2021 and became dominant over the original variant within a month. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
An interesting observation presented in Fig. 6, was that the N gene detection constantly produced lower Ct values compared to S1 detection. Upon the appearance of the Alpha variant the N gene and SΔ69 detection gaps were drastically reduced. This may imply different gene expression distributions or different durability of the RNA segments in the wastewater (Emanuel et al., 2021; Kim et al., 2020). As it is known that the Alpha variant has a more potent expression than the original NC_045512.2 (Brown et al., 2021; Parker et al., 2021), the fact that positive SΔ69 detection is closer to the N gene detection than the proximity of positive S1 detection compared to N gene detection is understandable. However, these observations need further study and validation. In the meantime, this phenomenon may also affect the “drop-out” assays resulting in false-positives, reinforcing the need in direct detection. Overall, the displayed results indicated that the developed assay can be employed and will provide essential, direct detection abilities for Alpha variant, B.1.1.7 and Beta variant, B.1.351 variants of concern.
4. Conclusions
The ongoing concern regarding the COVID-19 pandemic and the emergence of new variants with higher infection rate and morbidity, create great global concern. RT-qPCR is the most abundant method for the detection of SARS-CoV-2, yet improved detection is needed for lowering detection limits. Following thorough examination of existing primers-probe detection sets, we designed several improved sets. All improved sets were characterized and compared to previously published sets. The designed detection set named Improved N3, demonstrated a superior ability and better limit of detection. The improved set was also successfully employed on SARS-CoV-2 positive wastewater samples.
With regards to the highly dominant variants of concern, Alpha, B.1.1.7 and Beta, B.1.351, current detection diagnostic tools are expensive, time-consuming or indirect. We therefore developed specific RT-qPCR assay for the direct detection of these two variants. In addition, to direct identification, the developed assay is also capable for variant differentiation and quantification. For Alpha variant detection, a single set comprised of two new primers and two new probes focusing on the characterized deletion area known as Δ69–70, were designed and validated. In addition, an RT-qPCR direct detection assay was developed for the Beta variant using two new primers focusing on a characterized deletion area known as Δ241–243, and a third probe was designed and validated. Variant detection sets were fully characterized and employed on various wastewater samples for proof-of-concept. In addition to the improved ability for positive detection of the dominant Alpha variant, the developed assay showed an improved differentiation ability, especially when samples contained both the original SARS-CoV-2 NC_045512.2 and Alpha variant. Interestingly at the time of sampling Beta variant was characterized as a functional detection set, however it did not correspond to the presence of the Beta variant in Israel. The presented primers-probe sets may be used as described here, or even combined in the future in different combinatorial approaches for rapid, cost-effective and direct detection of the two variants.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We would like to acknowledge funding from Ben Gurion University, The Corona Challenge Covid-19 (https://in.bgu.ac.il/en/corona-challenge/Pages/default.aspx) and funding from the Israeli ministry of Health. In addition, we thank the Israeli Ministry of Health for funding and providing us with the sewage samples appearing in Table 1. We thank the Wastewater treatment plants authorities in the city of Beer-Sheva, Israel. We gratefully acknowledge GISAID database for access to SARS-CoV-2 variants sequences. We gratefully acknowledge Esti Kramarsky-Winter assistance for comments and scientific editing of the manuscript.
Abbreviations
- Severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2)
- Reverse transcriptase quantitative polymerase chain reaction
(RT-qPCR)
- Next generation sequencing
(NGS)
- Wastewater treatment plant
(WWTP)
- 6-carboxy-fluorescein
(FAM)
- Yakima Yellow
(YakYel)
- Non-template control
(NTC)
- Centers for Disease Control and Prevention
(CDC)
- Limit of detection
(LOD)
- Threshold cycle
(Ct)
- Not detected
(ND)
Author contributions
K.Y. and E.O. share equal contribution to this manuscript. K.Y. designed sequence, conceived, performed and analyzed experiments and authored this manuscript. E.O. designed sequence, conceived and analyzed experiments and authored this manuscript. M. S., S. L., N.P. and N.S.B. took part in experiments execution. A.K. conceived experiments, supervised, provided research facilities and edited the manuscript.
Funding sources
We would like to acknowledge funding from Ben Gurion University, The Corona Challenge Covid-19 (https://in.bgu.ac.il/en/corona-challenge/Pages/default.aspx) and funding from the Israeli ministry of Health.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Heal. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés C., Garcia-Cehic D., Gregori J., Piñana M., Rodriguez-Frias F., Guerrero-Murillo M., Esperalba J., Rando A., Goterris L., Codina M.G., Quer S., Martín M.C., Campins M., Ferrer R., Almirante B., Esteban J.I., Pumarola T., Antón A., Quer J. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg. Microb. Infect. 2020;9:1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Bitkover E., Paitan Y., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. 2020. medRxiv. 1–11. [DOI] [PMC free article] [PubMed]
- Brown J.C., Goldhill D.H., Zhou J., Peacock T.P., Frise R., Goonawardane N., Baillon L., Kugathasan R., Pinto A.L., McKay P.F., Hassard J., Moshe M., Singanayagam A., Burgoyne T., Investigators the A. Consortium P.V., Barclay W.S. Increased transmission of SARS-CoV-2 lineage B . 1 . 1 . 7 ( VOC 2020212/01 ) is not accounted for by a replicative advantage in primary airway cells or antibody escape. 2021. bioRxiv. 1–13. [DOI]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Luisa Schmidt M., Gjc Mulders D., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C., Victor C.M., Olfert L., Marco K., Richard M., Adam M., Daniel C.K., Tobias B., Sebastian B., Julia S., Marie Luisa S., Daphne Gjc M., Bart H.L., der Veer Bas V., den Brink Sharon V., Lisa W., Gabriel G., Jean-Louis R., Joanna E., Maria Z., Malik P., Herman G., Chantal R. Detection of 2019 -nCoV by RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier J., Störmer M., Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel W., Kirstin M., Vedran F., Asija D., Theresa G.L., Roberto A., Filippos K., David K., Katja H., Salah A., Christopher B., Karen H., Anja R., Ivano L., Andranik I., Tommaso M., Simone D.G., Jan P., Samantha P., Meyer Thomas F., Alexander M.M., Daniela N., Andreas H., Matthias S., Altuna A., Nikolaus R., Christian D., Markus L. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience. 2021:102151. doi: 10.1016/j.isci.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Zamyadi A., Deere D., Vanrolleghem P.A., Crosbie N.D. SARS-CoV-2 known and unknowns, implications for the water sector and wastewater-based epidemiology to support national responses worldwide: early review of global experiences with the COVID-19 pandemic. Water Qual. Res. J. 2020:1–11. http://iwaponline.com/wqrj/article-pdf/doi/10.2166/wqrj.2020.100/695667/wqrj2020100.pdf [Google Scholar]
- Khan M.I., Khan Z.A., Baig M.H., Ahmad I., Farouk A.E.A., Song Y.G., Dong J.J. Comparative genome analysis of novel coronavirus (SARS-CoV-2) from different geographical locations and the effect of mutations on major target proteins: an in silico insight. PLoS One. 2020;15:1–18. doi: 10.1371/journal.pone.0238344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043%0ASUMMARY. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacova V., Boršová K., Paul E.D., Radvanszka M., Hajdu R., Čabanová V., Sláviková M., Ličková M., Ľ Lukáčiková, Belák A., Roussier L., Kostičová M., Líšková A., Mad’arová L., Štefkovičová M., Reizigová L., Nováková E., Sabaka P., Koščálová A., Brejová B., Vinař T., Nosek J., Čekan P., Klempa B. A novel, room temperature-stable, multiplexed RT-qPCR assay to distinguish lineage B.1.1.7 from the remaining SARS-CoV-2 lineages. 2021. http://medrxiv.org/content/early/2021/02/12/2021.02.09.21251168.abstract medRxiv. 1–28. [DOI] [PMC free article] [PubMed]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Javier, Klapsa Dimitra, Wilton Thomas, Zambon Maria, Bentley Emma, Bujaki Erika, Fritzsche Martin, Mate Ryan, Majumdar Manasi. Tracking SARS-CoV-2 in sewage: Evidence of changes in virus variant predominance during COVID-19 pandemic javier. Viruses. 2020;12(10):1144. doi: 10.3390/v12101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1–13. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.D., Lindsey B.B., Shah D.R., Hsu S., Keeley A.J., Partridge D.G., Leary S., Cope A., State A., Jhonson K., Ali N., Raghei R., Heffer J., Smith N., Zhang P., Gallis M., Louka S.F., Whiteley M., Foulkes B.H., Christou S., Wolverson P., Pohare M., Hansford S.E., Green L.R., Evans C., Raza M., Wang D., Gaudieri S., Mallal S., Consortium TC-19 GU (COG-U, de Silva TI Altered subgenomic RNA expression in SARS-CoV-2 B.1.1.7 infections. 2021. bioRxiv. 1–21. [DOI]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 2020;186:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22:2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Heal. 2020;10:100157. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O El, Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B., Breban M., Alpert T., Petrone M.E., Watkins A.E., Hodcroft E.B., Mason C.E., Khullar G., Metti J., Dudley J.T., MacKay M.J., Nash M., Wang J., Liu C., Hui P., Murphy S., Neal C., Laszlo E., Landry M.L., Muyombwe A., Downing R., Razeq J., Neher R.A., Fauver J.R., Grubaugh N.D. PCR assay to enhance global surveillance for SARS-CoV-2 variants of concern. medRxiv. 2021;351:2021. doi: 10.1101/2021.01.28.21250486. 01.28.21250486. [DOI] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Nair M.S., Huang Y., Ho D.D. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021. [DOI] [PubMed]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. 2020. medRxiv. 1–11. [DOI] [PMC free article] [PubMed]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. In: Gilbert Jack A., editor. Vol. 5. 2020. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases; pp. 1–9.https://msystems.asm.org/content/5/4/e00614-20 mSystems. [DOI] [PMC free article] [PubMed] [Google Scholar]