Abstract
Bardet-Biedl syndrome (BBS) is a rare autosomal recessive disorder caused by mutations in genes encoding components of the primary cilium and is characterized by hyperphagic obesity. To investigate the molecular basis of obesity in human BBS, we developed a cellular model of BBS using induced pluripotent stem cell–derived (iPSC-derived) hypothalamic arcuate-like neurons. BBS mutations BBS1M390R and BBS10C91fsX95 did not affect neuronal differentiation efficiency but caused morphological defects, including impaired neurite outgrowth and longer primary cilia. Single-cell RNA sequencing of BBS1M390R hypothalamic neurons identified several downregulated pathways, including insulin and cAMP signaling and axon guidance. Additional studies demonstrated that BBS1M390R and BBS10C91fsX95 mutations impaired insulin signaling in both human fibroblasts and iPSC-derived neurons. Overexpression of intact BBS10 fully restored insulin signaling by restoring insulin receptor tyrosine phosphorylation in BBS10C91fsX95 neurons. Moreover, mutations in BBS1 and BBS10 impaired leptin-mediated p-STAT3 activation in iPSC-derived hypothalamic neurons. Correction of the BBS mutation by CRISPR rescued leptin signaling. POMC expression and neuropeptide production were decreased in BBS1M390R and BBS10C91fsX95 iPSC–derived hypothalamic neurons. In the aggregate, these data provide insights into the anatomic and functional mechanisms by which components of the BBSome in CNS primary cilia mediate effects on energy homeostasis.
Keywords: Stem cells
Keywords: Obesity
Introduction
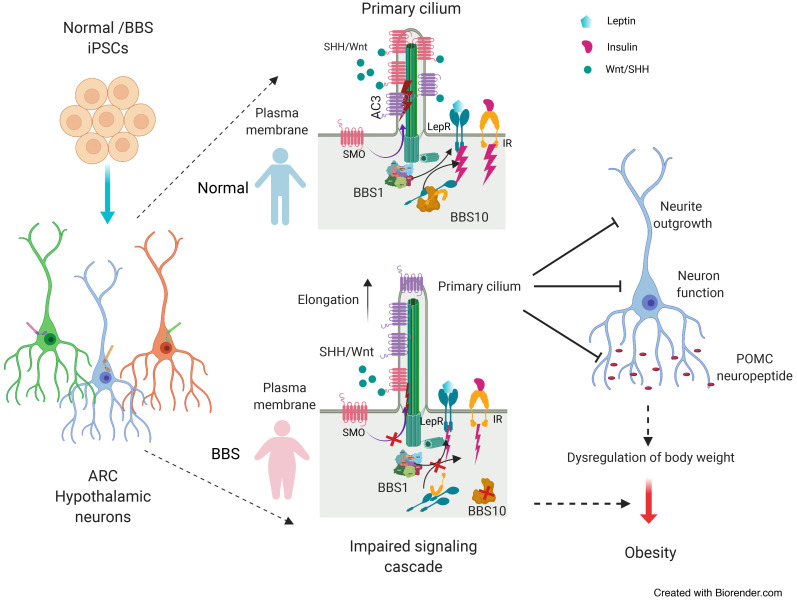
Bardet-Biedl syndrome (BBS) is an autosomal recessive disorder characterized by severe, early-onset obesity, as well as an array of other diagnostic features typical of ciliopathies including polydactyly, retinal degeneration, renal cysts, and intellectual impairment (1). To date, 21 genes have been implicated in specific instances of BBS, and 16 have been confirmed as components of the basal body of the primary cilium (2). Eight BBS-associated genes, BBS1, -2, -4, -5, -7, -8, -9, and -18, encode proteins that form a complex denominated the BBSome (3). The BBSome mediates transport of molecular cargo to the base of the primary cilium via the Golgi or recycling endosomes (4), as well as ciliary exit by directing intraflagellar transport–loaded cargo out of the primary cilium (5, 6). Ciliary cargo includes the receptors for sonic hedgehog (SHH), Wnt, PDGFα, and other signaling molecules that are essential for embryonic development, cell growth, cell migration, and cell survival (7, 8). Proteins associated with the primary cilium are also important for the regulation of energy homeostasis; in Alström syndrome, autosomal recessive mutations in the ALMS1 gene — encoding a ciliary basal body protein — also cause obesity (9). Loss-of-function mutations in adenylate cyclase 3 (ACIII, encoded by ADCY3) or melanocortin 4 receptor (MC4R), both of which localize to neuronal primary cilia, cause severe obesity in humans (10–14). In mice, systemic congenital deletion of Bbs gene Alms1, Adcy3, or Mc4r recapitulates the obesity phenotype (15–21). Hypomorphism of ciliary genes Ift88, Kif3a, and Rpgrip1l results in increased adiposity in mice (22–24). Deletions of BBS-associated genes also cause obesity in mice; Bbs1M390R/M390R, Bbs2–/–, Bbs4–/–, Bbs6–/–, and Bbs7–/– mice are hyperphagic and obese (17–20).
The mechanisms by which BBS proteins affect body weight regulation are not well understood. BBS2 or BBS4 deletion in mice is associated with failure of G protein–coupled receptors (GPCRs) that regulate energy balance such as somatostatin (SST) receptor 3 (SSTR3) and melanin-concentrating hormone receptor 1 (MCHR1) to localize to the primary cilium in the central nervous system (25). Similarly, BBS1 deletion in mice prevents localization of the neuropeptide Y 2 receptor (NPY2R) to the primary cilium in the arcuate hypothalamus (ARH), and BBS1 has been implicated in trafficking of the obesity-associated serotonin 2C receptor (5-HT2CR) in the primary cilium (26). BBS1, a BBSome structural protein that facilitates recruitment of membrane-bound proteins to the primary cilium (3), also interacts with isoform B of the leptin receptor (LEPRB), which binds RPGRIP1L at the transition zone of the primary cilium (23, 27–29). The BBS1 mutation M390R prevents the BBS1-LEPRB interaction, suggesting that BBS1 may participate in leptin signaling by influencing leptin receptor trafficking to the vicinity of the primary cilium (27). In mouse embryonic fibroblasts (MEFs) in which Bbs1 is knocked down, or in human primary fibroblasts homozygous for BBS1M390R, localization of the insulin receptor (IR) to the cell membrane is reduced (30). Consistent with these findings, preobese Bbs2–/–, Bbs4–/–, and Bbs6–/– mice are hyperinsulinemic (30).
BBS10 mutations account for approximately 20% of instances of clinical BBS. Among BBS proteins, BBS6, BBS10, and BBS12 are chaperonin-like molecules that mediate the assembly of the BBSome in physical conjunction with the CCT/TRiC family of chaperonins (31). BBS10 per se regulates the formation of the BBS-chaperonin complex, which is a critical step in BBSome assembly (32). Interestingly, BBS patients segregating for deleterious variants in BBS6, BBS10, and BBS12 display more severe phenotypes compared with individuals segregating for other BBS mutations (33). Specifically, patients with BBS10 mutations have significantly higher BMIs and greater insulin resistance than those with BBS1 mutations (34). The BBS10 C91fsX95 allele studied here accounts for 26%–48% of clinical BBS10 mutations (33).
BBS broadly impacts the development of the central nervous system; MRI scans of BBS patients show distinct tissue- and region-specific brain abnormalities, including reduced white matter in most brain regions and reduced gray matter in the caudate/putamen (dorsal striatum), and hypoplasia of the hypothalamus and thalamus (35). Cells within the hypothalamus play a critical role in the control of body weight and systemic glucose homeostasis (36). In the ARH, neurons expressing proopiomelanocortin (POMC) and those coexpressing neuropeptide Y (NPY) and Agouti-related protein (AgRP) sense peripheral hormones such as leptin, insulin, and ghrelin, and inhibit or increase food intake and energy expenditure (37). Conditional ablation of ciliary proteins encoded by Tg737, Kif3a, or Rpgrip1l in POMC neurons results in obesity (22, 38). In mice, congenital ablation of Bbs1 in POMC or AgRP neurons also increases adiposity (26). In Bbs1M390R/M390R mice, cilia of ependymal cells lining the third ventricle at the ARH are elongated with swollen distal tips, suggesting that BBS1 is also important for maintaining normal ciliary structure in the hypothalamus (20).
Based on the studies enumerated above, it is likely that cilia are dysfunctional in BBS patient hypothalamic cells. The ability to generate brain region–specific neurons from patient-specific induced pluripotent stem cells (iPSCs) now enables analysis of molecular and structural phenotypes in processes important to the control of food intake and energy expenditure (39). And, single-cell RNA sequencing (scRNA-seq) has been widely used in developmental biology, oncology, immunology, and metabolic disease research (40–44) to identify cell-type-specific regulatory relationships among genes (41–43). It is particularly useful for resolving cellular heterogeneity in human iPSC–derived functional cell types. Here, we used these technologies to examine specific aspects of the molecular neurobiology of obesity in human BBS and demonstrated that both BBS1 and BBS10 regulate ciliary morphology, neurite outgrowth, POMC neuropeptide production, and metabolic signaling pathways in a cellular context that is independent of obesity.
Results
Generation of BBS and control iPSCs.
Two BBS fibroblast lines (GM05948 and GM05950) were obtained from Coriell. Ten skin biopsies (for the establishment of fibroblast lines) were obtained from BBS and healthy control subjects attending the Naomi Berrie Diabetes Center of the Columbia University Irving Medical Center under an IRB protocol (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI146287DS1). The diagnosis of BBS was ascertained primarily by a physician to whom they had been referred for evaluation of associated retinopathy. Their clinical phenotypes were consistent with the diagnosis of BBS and included obesity, retinitis pigmentosa, polydactyly, anosmia, renal cysts, and intellectual impairment (Supplemental Figure 1, A and B). Clinical diagnoses were confirmed by Asper Ophthalmics genetic testing or whole-exome sequencing, which were further validated by dideoxy sequencing (Supplemental Figure 1C). The genotype of each subject is indicated in Supplemental Table 1. Fibroblasts derived from skin biopsies from BBS and control subjects were reprogrammed into iPSCs using retroviruses expressing OCT4, SOX2, KLF4, and c-Myc (45). The studies reported here used the following cell lines: BBS1A (iPSCs derived from patient 1085 with a homozygous BBS1 M390R mutation), BBS1B (iPSCs derived from patient 1097 with a homozygous BBS1 M390R mutation), BBS10A (iPSCs derived from the GM05948 primary fibroblast line with a homozygous mutation in BBS10 [C91fsX95]), BBS10B (iPSCs derived from GM05950 fibroblasts segregating for a compound heterozygous mutation in BBS10 [S303RfsX3]) (Supplemental Figure 1C), as well as control 1 and control 2. The generation of iPSC lines BBS1A, BBS1B, BBS10B, control 1, and control 2 was previously published (35), and BBS10A is described here. Immunocytochemistry (ICC) analysis revealed that BBS10A expressed pluripotency markers (Supplemental Figure 2A). Teratomas generated by BBS10A iPSCs included all 3 germ layers. BBS10A had a normal karyotype (Supplemental Figure 2, B and C). Both control and BBS iPSC lines expressed endogenous pluripotency markers and showed complete silencing of exogenous genes (viral transgenes) (Supplemental Figure 3). We have attempted CRISPR gene manipulation in all 4 BBS lines. However, only 1 isogenic control iPSC line for BBS1B was successfully generated. In this line, the M390R mutations in both alleles were corrected as confirmed by TOPO cloning (Supplemental Figure 4, A and B). The corrected BBS1B isogenic control line (referred to as c-BBS1B) had a normal karyotype and was pluripotent (Supplemental Figure 4, C–E).
BBS mutations increase ciliary length in iPSC-derived TUJ1+ neurons.
Elongated, swollen primary cilia on ependymal cells lining the third ventricle have been observed in Bbs1M390R/M390R, Bbs2–/–, Bbs4–/–, and Bbs6–/– mice, indicating that BBS proteins are essential for maintaining normal ciliary structure (20, 46). Here, ACIII, ARL13B, and γ-tubulin were used as markers in ICC analysis to determine ciliary length. In human fibroblast cultures, 40%–50% of cells were ciliated (Supplemental Figure 5A) and there was no difference among control, BBS1, and BBS10 mutant cells in the percentage of ciliated cells or ciliary length (Supplemental Figure 5, B–E).
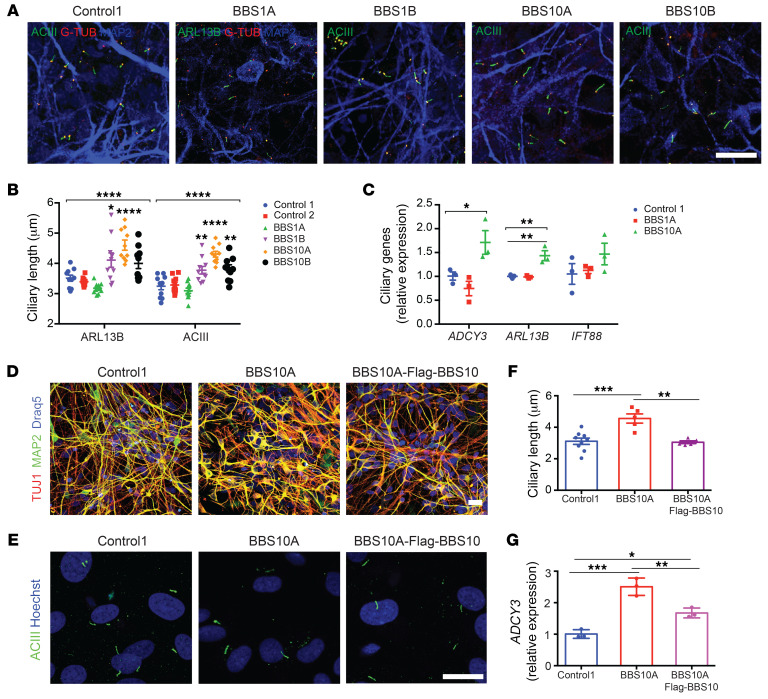
Generic TUJ1+ iPSC neurons were used in circumstances where high cell density (cilia morphology) or early developmental status (neuronal development and neurite outgrowth) was necessary. Both control and BBS iPSCs were converted into neurons using dual SMAD inhibition (Supplemental Figure 6A) (47, 48). On day 30, these cells became TUJ1+ neurons, and there was no difference in percentages of NCAM+ neurons and MAP2 expression among the control and BBS iPSC lines (Supplemental Figure 6, B–D). Whole-cell current clamps revealed that control, BBS1, and BBS10 iPSC–derived neurons generated comparable action potentials after current stimulation (Supplemental Figure 6E). Control, BBS1, and BBS10 mutant iPSC–derived neurons were all ciliated (Figure 1A), but in BBS1B, BBS10A, and BBS10B mutant neurons, cilia were approximately 14% to 28% longer than those in control iPSC–derived neurons (Figure 1B). The mRNA levels of ciliary genes ADCY3 and ARL13B were different among control and BBS iPSC–derived neurons (Figure 1C). Exposure to SHH and FGF8 resulted in further lengthening of the primary cilium in BBS10A mutant neurons by 50%, but not in BBS1 mutant or control neurons, suggesting divergent roles of BBS1 and BBS10 mutations in SHH/FGF8 signaling (Supplemental Figure 7).
Figure 1. BBS mutations increase ciliary length in iPSC-derived neurons.
(A) Immunocytochemistry (ICC) staining of primary cilia in iPSC-derived neurons. Neurons were stained for MAP2 as well as ciliary markers: adenylate cyclase III (ACIII, basal body and axoneme), ARL13B (axoneme), and γ-tubulin (G-TUB, basal body). Scale bar: 20 μm. (B) Quantification of ciliary length in control and BBS iPSC–derived neurons in A (n = 10 for each line). *P < 0.05, **P < 0.01, ****P < 0.0001 by 1-way ANOVA followed by Bonferroni’s multiple-comparison test (compared with control 1). (C) Expression of ciliary genes in control and BBS iPSC–derived neurons (n = 3). *P < 0.05, **P < 0.01 by 1-way ANOVA followed by Tukey’s multiple-comparison test. (D) ICC staining of control 1, BBS10A, and BBS10A-FLAG-BBS10 iPSC–derived neurons. TUJ1 and MAP2 are neuronal markers. Scale bar: 20 μm. (E) ICC staining of primary cilia in FACS-isolated NCAM+ neurons. Cells were stained with anti-ACIII and Hoechst. Scale bar: 10 μm. (F) Quantification of ciliary length in E (n = 9, 5, and 6, left to right). (G) Ectopic expression of FLAG-BBS10 partially reverses ADCY3 expression in BBS10A iPSC–derived neurons (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA followed by Tukey’s multiple-comparison test (F and G).
In order to confirm that increased ciliary length was due to the BBS hypomorphic mutations, FLAG-BBS10 was overexpressed in the BBS10A mutant line using lentivirus (Supplemental Figure 8). Overexpression of FLAG-BBS10 did not affect neuronal differentiation compared with control and BBS10A mutant lines (Figure 1D). Anti-ACIII staining of FACS-ioslated NCAM+ neurons showed that overexpression of intact BBS10 normalized cilia length in BBS10A mutant neurons (Figure 1, E and F). Gene expression analysis revealed that overexpression of FLAG-BBS10 restored BBS10 expression in the BBS10A mutant line and also partially restored ADCY3 expression (Figure 1G and Supplemental Figure 8D). These data indicate that the BBS10 mutation elongates cilia and increases ADCY3 in iPSC-derived neurons.
Molecular signatures of BBS1 mutant and its isogenic control hypothalamic neurons as obtained by scRNA-seq.
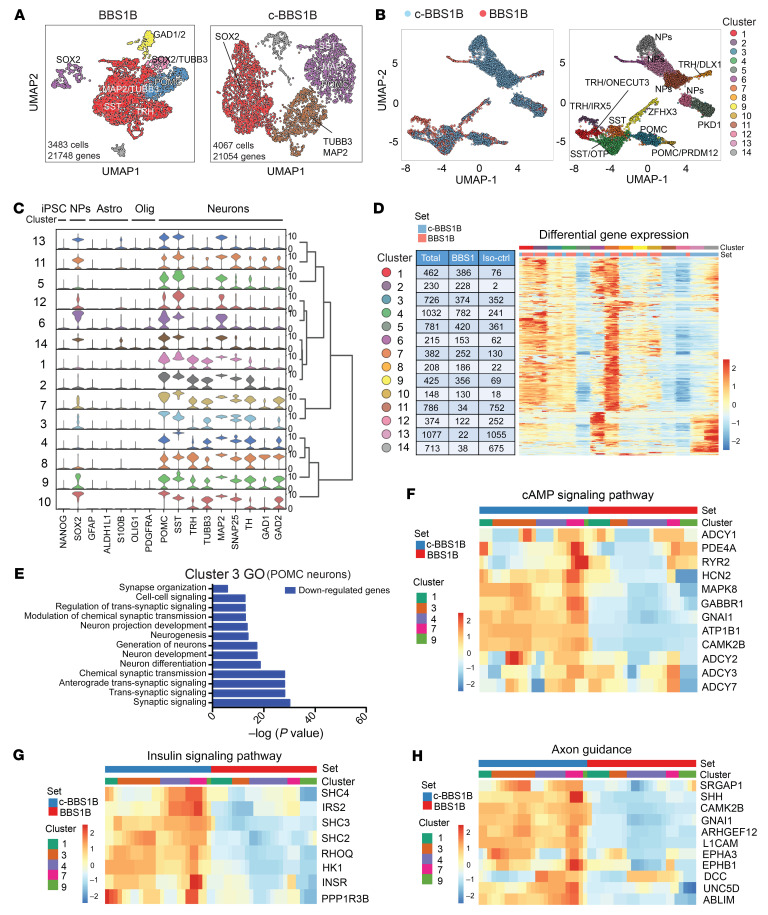
BBS1B and c-BBS1B ARH-like neurons were generated as previously described (35) and subjected to scRNA-seq using the 10× Genomics Chromium platform. For each line, we obtained more than 3,400 valid cells (≥200 expressed genes) after filtering, with over 21,000 genes detected in 3 or more cells (Figure 2A). Data from BBS1B and c-BBS1B iPSC–derived hypothalamic neurons were integrated and sorted into 14 clusters based on their signature genes (Figure 2B, Supplemental Figure 9A, and Supplemental Table 5). Clusters 1–4 and 7–10 share very similar gene expression profiles and were considered differentiated neurons based on the presence of neuronal markers (including TUBB3, SNAP25, TH, GAD1, and GAD2) (Figure 2C). POMC was highly expressed in clusters 3 and 10, and SST was expressed in clusters 4 and 7 (Figure 2C and Supplemental Figure 9B). Thyrotropin-releasing hormone (TRH) was expressed in clusters 1, 2, and 8. The remaining clusters (expressing MAP2 but not TUBB3) show characteristics of neuronal progenitors (Figure 2, B and C). In particular, clusters 6 and 11–14 were enriched for the neuron-progenitor marker SOX2 (Figure 2C). Stem cell, astrocyte, or oligodendrocyte markers such as NANOG, GFAP, ALDH1L1, S100B, OLIG1, and PDGFRA were either absent or expressed at very low levels (Figure 2C). Overall, the scRNA-seq analysis confirmed the hypothalamic neuron lineage and the anticipated heterogeneity of the human iPSC–derived ARH-like neurons.
Figure 2. scRNA-seq analysis of BBS iPSC–derived hypothalamic neurons.
(A) Uniform manifold approximation and projection (UMAP) of BBS1B and isogenic control (c-BBS1B) iPSC–derived hypothalamic neurons. (B) UMAP of BBS1B and c-BBS1B neurons after integration and identification of 14 clusters. Marker genes and cell types for each cluster are indicated. (C) Violin plots of cell-type-specific markers for all 14 clusters and hierarchical clustering. Signature genes for iPSCs, neuronal progenitors (NPs), astrocytes (Astro), oligodendrocytes (Olig), and neurons are included. (D) Heatmap of differentially expressed genes in all clusters. Number of cells from which line within each cluster is indicated in the table. (E) Gene Ontology analysis of downregulated genes (BBS1B/c-BBS1B) identified in cluster 3. (F–H) Heatmaps of genes of pathways highlighted in KEGG pathway analysis. Genes involved in cAMP signaling pathway (F), insulin signaling pathway (G), and axon guidance (H) were plotted against cell set and cluster ID.
Next, we performed differential expression analysis of each cluster between the BBS1B mutant and c-BBS1B lines to explore cell type–specific effects of BBS hypomorphism (Supplemental Table 6). As shown in the differential gene expression heatmap, all clusters contained differentially expressed (by BBS1 genotype) genes except cluster 2 that contained only a couple of cells from the c-BBS1B line (Figure 2D and Supplemental Figure 9A). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for genes that were either significantly up- or downregulated in BBS1B versus c-BBS1B neurons. Specifically, GO analysis of downregulated genes in cluster 3 revealed that synaptic organization, trans-synaptic and synaptic signaling, chemical synaptic transmission, neurogenesis, and neuronal development were negatively impacted in the BBS1B mutant POMC+ neurons (Figure 2E). Neuronal projection development and synaptic functions were also negatively impacted in BBS1B mutant cells of the other neuronal clusters, 1, 4, and 7–10 (Supplemental Figure 10). KEGG pathway analysis (Supplemental Figure 11) revealed that transcripts related to cAMP signaling (Figure 2F), insulin signaling (Figure 2G), axon guidance (Figure 2H), type 2 diabetes mellitus, and Wnt signaling (Supplemental Figure 12) were diminished in BBS1B mutant neurons in clusters 1, 3, 7, and 9.
BBS mutations impact neurite outgrowth and diminish Wnt and SHH signaling in iPSC-derived neurons.
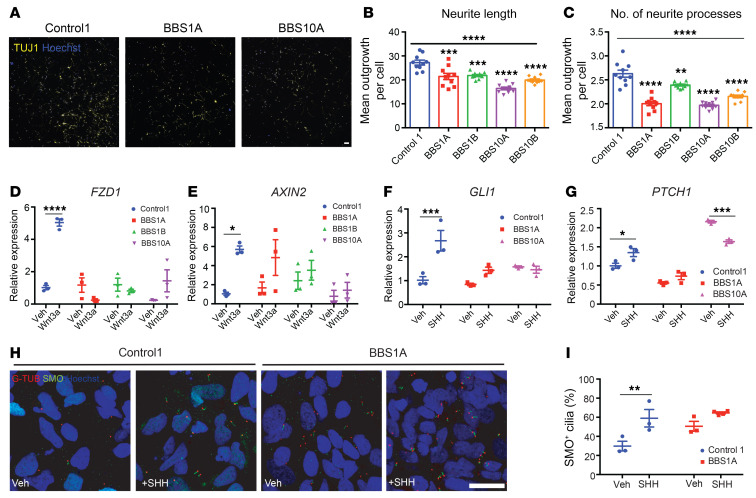
Neuroanatomic alterations in BBS patients include ventriculomegaly, thinning of the cerebral cortex, and reduction in size of the hippocampus and corpus striatum (35). Based on the gene expression results of the iPSC-derived hypothalamic neurons described above, we investigated the impact of BBS hypomorphism on neurogenesis in iPSC-derived TUJ1+ neurons. Because BBS mutations did not affect the efficiency of iPSC differentiation into TUJ1+ neurons, we investigated the numbers of processes and lengths of neurites 2 days after replating of day 30 control and BBS iPSC–derived neurons. BBS1 and BBS10 iPSC–derived neurons showed reductions in both number of processes and neurite length (Figure 3, A–C).
Figure 3. BBS mutations disturb neurite outgrowth and impair Wnt and SHH signaling in TUJ1+ iPSC–derived neurons.
(A–C) Neurite outgrowth assay of control and BBS iPSC–derived neurons on day 30 of differentiation. (A) TUJ1 staining of control and BBS mutant cultures. Scale bar: 50 μm. Mean neurite length (B) and average number of neurite processes (C) were calculated using the neurite outgrowth tool in MetaMorph software based on TUJ1 and Hoechst staining (n = 10 independent wells, 2,500 cells/well). **P < 0.01, ***P < 0.001, ****P < 0.0001 by 1-way ANOVA followed by Bonferroni’s multiple-comparison test (vs. control 1); the asterisks above the horizontal bars in B and C are for the 1-way ANOVA that permitted the pair-wise testing. (D and E) Wnt signaling is impaired in BBS iPSC–derived neurons. Control 1, BBS1A, BBS1B, and BBS10A iPSC–derived neurons (day 30) were treated with vehicle (Veh) or 100 ng/mL Wnt3a for 16 hours. Frizzled 1 (FZD1) and AXIN2 mRNA levels were determined by qPCR (n = 3). (F and G) SHH signaling is reduced in BBS iPSC–derived neurons. Control and BBS iPSC–derived neurons (day 30) were treated with vehicle or 100 ng/mL SHH for 16 hours. GLI1 and PTCH1 mRNA levels were analyzed by qPCR (n = 3). (H) Smoothened (SMO) staining in SHH-treated control and BBS iPSC–derived neurons. Control and BBS1A iPSC–derived neurons were treated with vehicle or 100 ng/mL SHH overnight. Neurons were fixed and stained with anti-SMO and anti–γ-tubulin (G-TUB, basal body) antibodies for cilia and Hoechst for nuclei. Scale bar: 20μm. (I) Quantification of SMO+ cilia in E. Hoechst was used as nuclear marker. SMO+ cilia percentage was calculated by (SMO+ cells/Hoechst+ cells) × 100 (n = 3 independent images). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by 2-way ANOVA followed by Bonferroni’s multiple-comparison test (D–G and I).
Wnt and SHH signaling are critical for neurogenesis and neuronal migration during brain development (38, 49–52). The trafficking and localization of SHH receptors Smoothened (SMO) and Patched 1 (PTCH1) is tightly regulated at the primary cilium, and Wnt signaling is activated near the base of the primary cilium (53). Wnt and SHH signaling were further investigated in TUJ1+ neurons. Wnt3a exposure strongly induced the expression of Wnt downstream targets FZD1 and AXIN2 in control neurons but not in BBS mutant lines (Figure 3, D and E). Similarly, exposure of control neurons to SHH increased the expression of its target genes GLI1 and PTCH1, a response that was blunted in BBS10A neurons but partially preserved in BBS1A neurons (Figure 3, F and G). In support of this, the scRNA-seq and bulk RNA-seq of iPSC-derived hypothalamic neurons also revealed that Wnt signaling was downregulated in BBS1B (Supplemental Figure 12B) and BBS10B lines (Supplemental Figure 13, A and B). Both SMO and PTCH1 were reduced in BBS1B and BBS10B hypothalamic neurons based on RNA-seq data (Supplemental Figure 13, C and D). The number of SMO-positive cilia following SHH induction was significantly reduced in BBS mutant–derived compared with control iPSC–derived neurons (Figure 3, H and I). Blunted neurite outgrowth and defective Wnt and SHH signaling further implicate BBS proteins in important aspects of neuronal development.
BBS mutations impair insulin signaling through interactions with the IR.
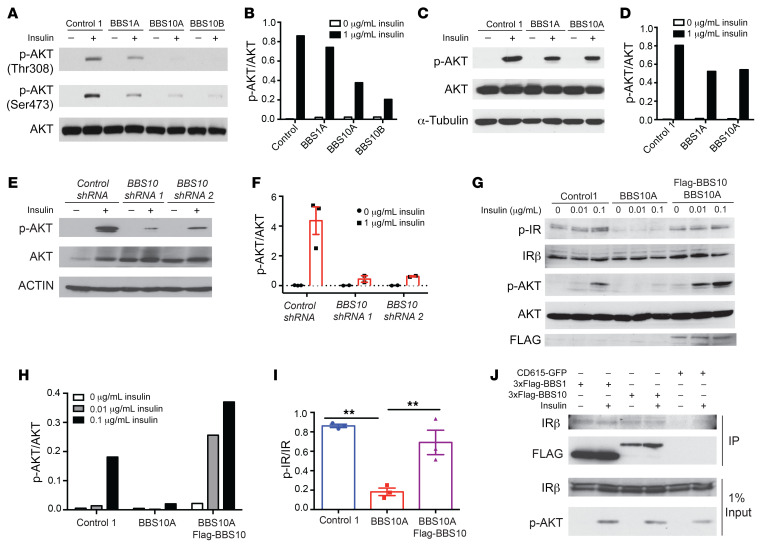
Insulin signaling mediates aspects of energy and glucose homeostasis centrally and peripherally (54, 55). In the hypothalamus, insulin acts in a manner similar to that of leptin to reduce food intake by activating POMC neurons and inhibiting NPY/AGRP neurons (54, 56). Deletion of Bbs2, -4, or -6 results in hyperinsulinemia in mice independently of obesity, and the M390R homozygous mutation in BBS1 fibroblasts results in perturbed IR trafficking to the cell membrane (30). It has been reported that IGF1 receptor function may be dependent on primary cilia in 3T3-L1 preadipocytes (57). To determine whether BBS proteins participate in insulin signaling in humans, we assessed insulin sensitivity by quantifying AKT phosphorylation (p-AKT) in response to insulin in BBS fibroblasts. By Western blotting, insulin-induced AKT phosphorylation at Thr308 was reduced by 50% and 80%–90% in BBS1 and BBS10 fibroblasts, respectively, compared with fibroblasts from control subjects (Figure 4, A and B).
Figure 4. BBS1 and BBS10 bind to the insulin receptor and influence insulin signaling.
(A) BBS mutations disrupt insulin signaling in human fibroblasts. Western blot (WB) analysis of insulin signaling as indicated by phosphorylation of AKT in control and BBS fibroblasts. Fibroblasts were serum starved overnight and treated with 0 or 1 μg/mL insulin for 30 minutes. AKT, p-AKT Thr308, and p-AKT Ser473 were probed. (B) Quantification of p-AKT (Thr308)/AKT from A (n = 1). (C) BBS mutations abrogate insulin signaling in iPSC-derived TUJ1+ neurons. Control, BBS1A, and BBS10A iPSC–derived neurons (day 30) were serum starved overnight and treated with 0 or 1 μg/mL insulin for 30 minutes. p-AKT Ser473, AKT, and α-tubulin (A-TUB) were probed. (D) Quantification of p-AKT/AKT from WB in C (n = 1). (E) Diminished insulin signaling in neurons derived from 2 BBS10 shRNA-knocked-down iPSC lines as indicated. iPSC-derived TUJ1+ neurons (day 30) were serum starved overnight and treated with 0 or 1 μg/mL insulin for 30 minutes. p-AKT Ser473, AKT, and actin were examined by WB. (F) Quantification of p-AKT/AKT from WB in E (n = 2–3). (G) BBS10 disrupts insulin signaling by disturbing phosphorylation of the insulin receptor (IR) in day 12 neuronal progenitors (NPs). WB analysis of insulin signaling molecules as indicated in control, BBS10A, and BBS10A-FLAG-BBS10 transgenic NPs after 30-minute treatment with 0.01 and 0.1 μg/mL insulin. FLAG was used to confirm the overexpression of WT BBS10. (H and I) Quantification of p-AKT/AKT (H) (n = 1) and p-IR/IR (I) (n = 3) in D. **P < 0.01 by 1-way ANOVA followed by Tukey’s multiple-comparison test. (J) Coimmunoprecipitation (IP) confirms the interaction between BBS proteins and the IR. 293FT cells were transfected with CD615-GFP or CD615-3×FLAG-BBS1-GFP or CD615-3×FLAG-BBS10-GFP for 48 hours before insulin (1 μg/mL) treatment (30 minutes). IRβ, p-AKT Ser473, and FLAG were probed in FLAG IP samples and total input.
In iPSC-derived TUJ1+ neurons, insulin-stimulated p-AKT levels were decreased by approximately 40% in neurons of both BBS1 and BBS10 mutant genotypes (Figure 4, C and D). To assess the role of BBS proteins in insulin signaling, we knocked down BBS10 expression in control iPSCs using lentiviral shRNA (Supplemental Figure 14). In neurons generated from iPSCs in which BBS10 was knocked down by 40%–60%, insulin-induced phosphorylation of AKT was reduced by 70%–90% (Figure 4, E and F). In the BBS10A mutant iPSC–derived neuronal progenitors, the insulin-induced p-AKT level was decreased 80%–90% and fully rescued by overexpression of intact BBS10 (Figure 4, G and H). Tyrosine phosphorylation of the IR was also reduced 75% in the BBS10 mutant neuronal progenitors, a molecular phenotype that was fully rescued by overexpressing intact BBS10 (Figure 4, G and I). To determine whether BBS proteins directly interact with the IR, we performed coimmunoprecipitation in 293FT cells overexpressing transgenic FLAG-BBS1 or FLAG-BBS10. BBS1 or BBS10 coimmunoprecipitated with the IR, irrespective of the presence of ambient insulin (Figure 4J). Thus, BBS1 and BBS10 may regulate insulin signaling by direct protein-protein interaction with the IR.
BBS mutations curtail leptin signaling in iPSC-derived hypothalamic neurons independently of obesity.
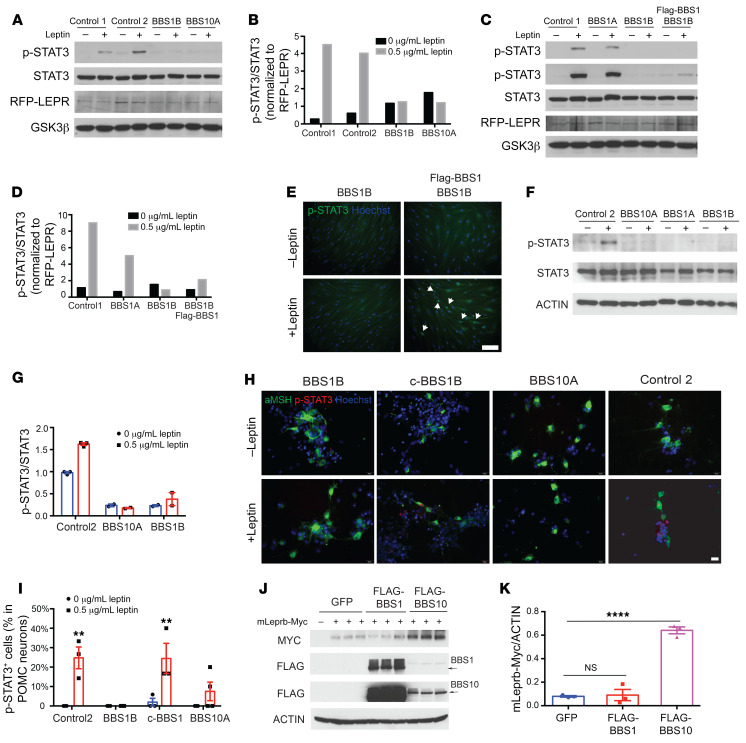
It is challenging to parse defects in leptin signaling that are independent of the obesity exhibited in various Bbs-knockout mice (17, 18, 20, 31). Human fibroblasts do not express LEPRB and therefore do not increase p-STAT3 levels upon leptin exposure in vitro (Supplemental Figure 15, A and B). We induced ectopic expression of RFP-tagged LEPRB (RFP-LEPRB) in both control and BBS human fibroblasts using a lentivirus (Supplemental Figure 15, C and D). By Western blot analysis, RFP-LEPRB was expressed in control and BBS fibroblast lines (Figure 5A). Leptin-induced p-STAT3 levels were reduced approximately 50% in leptin-stimulated RFP-LEPRB–transfected BBS1A fibroblasts, and completely blunted in transfected BBS1B and BBS10A fibroblasts compared with the control (Figure 5, A and B, and Supplemental Figure 15, E and F). p-STAT3 immunostaining in leptin-treated transgenic fibroblasts revealed fewer leptin-induced p-STAT3 positive cells in BBS10A compared with control fibroblasts (Supplemental Figure 15G), consistent with diminished leptin sensitivity of the BBS1B and BBS10A primary fibroblast lines. Overexpression of intact BBS1 in BBS1B fibroblasts partially restored leptin signaling, as reflected in partially restored p-STAT3 protein levels (Figure 5, C and D), and increased p-STAT3–positive cells (Figure 5E). Leptin signaling was also assessed in iPSC-derived hypothalamic neurons, which constitute an in vitro model without any influences of systemic adiposity. In day 34 differentiated hypothalamic neurons, we found that leptin increased p-STAT3 in the control cells but this increase was weaker or undetectable in BBS1 and BBS10 mutant lines (Figure 5, F and G). Furthermore, POMC-expressing isogenic control c-BBS1B and control neurons responded to leptin (p-STAT3 immunostaining), while both BBS1B and BBS10A mutant lines did not increase p-STAT3 in response to leptin exposure (Figure 5H). Correction of the BBS1 M390R mutation by CRISPR fully restored the leptin response in c-BBS1B POMC neurons compared with the BBS1B line (Figure 5, H and I). Collectively, these experiments suggest that BBS mutations contribute to diminished leptin sensing in the central nervous system independently of intercurrent increased adiposity.
Figure 5. BBS mutations impair leptin signaling in RFP-LEPR transgenic human fibroblasts and iPSC-derived hypothalamic neurons.
(A–D) Leptin signaling is impaired in RFP-LEPR transgenic BBS fibroblasts. Control 1, control 2, BBS1A, BBS1B, BBS10A, and BBS1B + FLAG-BBS1 RFP-LEPR transgenic fibroblasts were treated with 0 or 0.5 μg/mL leptin (30 minutes) after overnight serum starvation. Leptin signaling was assessed by Western blot (WB) of p-STAT3 levels. STAT3, RFP, and GSK3β were also probed (A and C). In C, the 2 p-STAT3 blot images are from short (top) and long time exposures. (B and D) Quantification of p-STAT3/STAT3 from A and C. p-STAT3/STAT3 was further normalized to RFP-LEPR/GSK3β (n = 1). (E) Leptin signaling can be rescued in BBS1B RFP-LEPR transgenic fibroblasts by overexpressing FLAG-BBS1. BBS1B and BBS1B + FLAG-BBS1 RFP-LEPR transgenic fibroblasts were treated as in C and stained with anti–p-STAT3 and Hoechst. Arrows indicate p-STAT3–positive cells. Scale bar: 200 μm. (F and G) Leptin signaling is disturbed in BBS iPSC–derived hypothalamic neurons. Day 34 iPSC-derived hypothalamic neurons were serum starved overnight and treated with 0 or 1 μg/mL leptin for 30 minutes. (F) p-STAT3, STAT3, and actin were examined by WB. (G) Quantification of WB in F (n = 2–3). (H and I) Leptin signaling, as measured by immunostaining of p-STAT3, is disrupted in BBS iPSC–derived hypothalamic neurons. (H) Day 34 neurons were treated with leptin and stained with anti-αMSH, anti–p-STAT3, and Hoechst. Scale bar: 20 μm. (I) Quantification of p-STAT3+ POMC neurons in H (n = 3). **P < 0.01 by 2-way ANOVA followed by Bonferroni’s multiple-comparison test. (J) Overexpression of BBS10 increases total LEPR proteins. 293FT cells were cotransfected with mouse LEPRB-Myc (mLeprb-Myc) and GFP or FLAG-BBS1 or FLAG-BBS10 for 24 hours. Myc, FLAG, and actin were probed. Arrows indicate FLAG-BBS1 and FLAG-BBS10 bands. (K) Quantification of mLeprb-Myc in J. ****P < 0.0001 by 1-way ANOVA followed by Tukey’s multiple-comparison test. NS, no significant difference.
In order to further assess the role of BBS proteins in the regulation of leptin signaling, 293FT cells were cotransfected with mouse LEPRB Myc-tagged in the C-terminus, and WT human FLAG-BBS1 and -BBS10, and assayed for the total amount of LEPRB protein by Western blotting. Cells cotransfected with FLAG-BBS10 contained 3- to 4-fold more LEPRB protein than GFP- or FLAG-BBS1–cotransfected cells, suggesting that BBS10 enhances LEPRB translation and/or reduces protein degradation (Figure 5, J and K). BBS10 proteins may participate in aspects of leptin signaling by effects on the stability of LEPRB. BBS1 mutations are reported to impair leptin signaling by disrupting the trafficking of LEPR (27). Our findings suggest that BBS10 regulates leptin signaling by promoting molecular stability or trafficking of LEPR in iPSC-derived neurons.
BBS1 M390R mutation increases body weight and reduces hypothalamic POMC production.
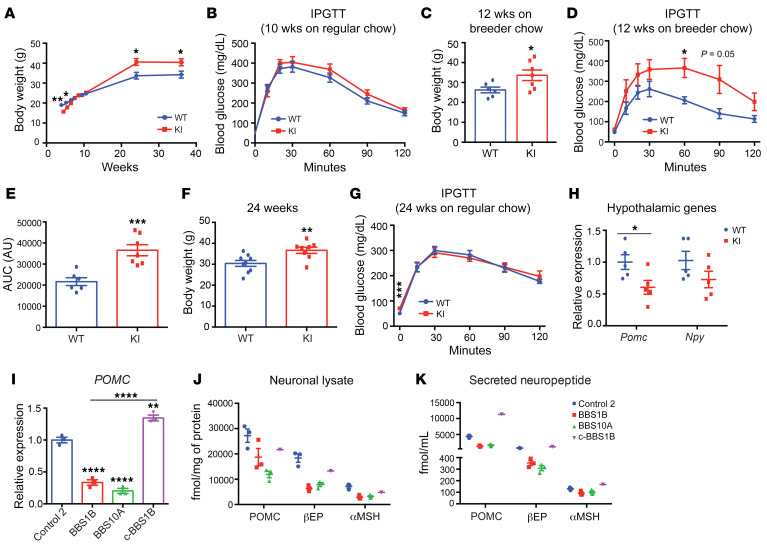
Previous reports claimed that hypothalamic Pomc mRNA levels were decreased in several BBS mouse models (19, 27). To further explore the potential effects of the BBS1 M390R mutation on POMC expression, we studied BBS1M390R-knockin (KI) mice (20). On regular (6% fat) chow, KI mice displayed lower body weight during the first 6 weeks of age compared with littermate control mice (Figure 6A). At 10 weeks of age, the KI mice had the same body weight as control mice and performed comparably during the glucose tolerance test (Figure 6B). However, at 12 weeks of age the KI mice fed breeder chow (10% fat) had increased body weight and diminished glucose tolerance compared with control mice (Figure 6, C–E). By 24 weeks of age, KI mice weighed 15% more than control littermates (Figure 6F). Fasting glucose in 24-week-old KI mice was higher than control mice (Figure 6G). However, the glucose tolerance of these KI mice was not affected (Figure 6G). Finally, in hypothalami of 25-week-old KI mice, the Pomc transcript level was reduced by approximately 40%, while Npy expression was unaffected (Figure 6H). The reduced hypothalamic Pomc expression in KI mice is consistent with studies of other BBS mice (19, 27).
Figure 6. BBS1M390R mutation reduces POMC expression in both mouse hypothalamus and human iPSC–derived hypothalamic neurons.
(A) Body weight curve of male WT and BBS1M390R-knockin (KI) mice (n = 9, 10). (B) Intraperitoneal glucose tolerance test (IPGTT) of 10-week-old male mice on regular chow diet (n = 9, 10). (C and D) Body weight (C) and IPGTT (D) of 12-week-old male mice fed breeder chow ad libitum (n = 6, 7). (E) The glucose area under the curve (AUC) in WT and KI mice as shown in D. ***P < 0.001. (F) Body weight of 24-week-old WT and KI mice on regular chow diet (n = 9, 8). (G) IPGTT of 24-week-old WT and KI mice on regular chow diet (n = 9, 8). (H) qPCR analysis of Pomc and Npy expression in hypothalamus of WT and KI mice (24-week-old males) after 16-hour fasting followed by 4-hour refeeding (n = 5, 5). *P < 0.05, **P < 0.01 by 2-tailed Student’s t test (A–H). (I) qPCR analysis of POMC expression in day 35 iPSC-derived hypothalamic neurons (n = 3). **P < 0.01, ****P < 0.0001 by 1-way ANOVA followed by Tukey’s multiple-comparison test. (J and K) Amount of neuropeptide produced in neuronal lysates (J) and in cultured medium (16-hour culture, K) from control (n = 3), BBS1B (n = 3), BBS10A (n = 3), and c-BBS1B (n = 1) iPSC–derived hypothalamic neurons. POMC, αMSH, and βEP concentrations were measured with ELISA and radioimmunoassay and normalized to total protein.
As previously reported, there were no gross differences in the percentage of NKX2.1+ and POMC+ hypothalamic neurons derived from control and BBS iPSCs (39). POMC mRNA levels were reduced in BBS1B and BBS10A mutant iPSC–derived hypothalamic neurons, while correcting the BBS mutation by CRISPR rescued POMC expression in c-BBS1B cells (Figure 6I). BBS1B and BBS10A mutant human iPSC–derived hypothalamic neurons had lower amounts (protein) of POMC, β-endorphin (βEP, derivative of POMC), and a trend toward decreased α-melanocyte–stimulating hormone (αMSH) in neuronal lysates and in the growth medium compared with the control and c-BBS1B isogenic lines (Figure 6, J and K), suggesting that BBS proteins regulate POMC production.
Discussion
We examined the molecular pathogenetic impact of the homozygous BBS1 M390R and BBS10 C91fsX95 mutations and the heterozygous BBS10 S303RfsX3 mutation. Patients segregating for BBS10 mutations are generally more severely clinically affected than those segregating for BBS1 mutations (34). BBS10 is not a structural component of the BBSome but participates in the initial steps of the BBSome assembly by facilitating the interactions of BBSome structural components with canonical CCT chaperonins (31). These distinctions notwithstanding, we found that both BBS genes regulate ciliary structure, neurite anatomy, POMC expression, and responses to leptin and insulin. These findings are concordant with the anatomic and functional consequences of BBS mutations in human subjects with BBS1 or BBS10 mutations (34, 35).
In agreement with reports of abnormal ciliary structures in various BBS mouse models (20, 44), ciliary length in human stem cell–derived BBS mutant TUJ1+ neurons — and especially in BBS10 mutant neurons — is significantly increased. Inhibition of ACIII activity promotes ciliary lengthening in cultured synoviocytes (57). The upregulation of ADCY3 expression in the BBS10 mutant neurons suggests that BBS10 may influence ciliary length via an ACIII-mediated mechanism. scRNA-seq identified differential expression of genes involved in the cAMP signaling pathway between BBS and its isogenic control neurons. Previous human and mouse genetic studies have implicated ACIII in body weight regulation (10–12). It is possible that BBS proteins regulate body weight at least partially through an ACIII-dependent cAMP signaling pathway.
The scRNA-seq analyses of BBS1B and c-BBS1B iPSC–derived neurons confirmed their hypothalamic lineage and revealed the heterogeneity of these differentiated cells, as reflected by the identification of 14 cell clusters after integration of both data sets. The ARH-like neuronal differentiation protocol employed here yields predominantly POMC neurons, but also smaller fractions of other neuronal subtypes such as SST and TRH, which have been previously detected in the mouse ARH using scRNA-seq (58). Such heterogeneity of human pluripotent stem cell–derived neurons has also been observed by other investigators (42, 59). The application of scRNA-seq, however, enabled us to identify signaling and molecular pathways that were affected in specific neuron types.
Using ICC, no gross differences in neuronal differentiation efficiency were observed between BBS mutant– and control iPSC–derived TUJ1+ neurons. However, neurite outgrowth (number of mean processes and neurite length) was reduced in both BBS1 and BBS10 iPSC–derived neurons, suggesting a defective neurite outgrowth or disturbed neuronal development in BBS lines. Wnt and SHH signaling pathways are important in the control of neuronal projection development (60, 61). Thus, the neurite outgrowth phenotype may be resulting from the deficiencies in Wnt and SHH signaling pathways in BBS mutant cells, as suggested by our studies. GO and KEGG pathway analysis of scRNA-seq data of BBS1B and c-BBS1B highlight signaling related to axon guidance, neuronal projection development, and synaptic organization and transmission. In Drosophila, increased insulin signaling by knocking down FoxO and PTEN during metamorphic neuronal remodeling promotes neuronal growth and neurite branching (62). Moreover, leptin plays a neurotrophic role to promote neurite outgrowth of ARH neurons in neonatal mice (63). Therefore, the diminished insulin and leptin signaling in BBS mutant neurons shown in our cellular assays may contribute to reduced projection density.
Insulin signaling was impaired both in BBS1 and BBS10 mutant human fibroblasts, and more severely in BBS10 cells. scRNA-seq data also indicated that expression of genes involved in the insulin signaling pathway were significantly reduced in BBS1B line compared with its isogenic control line. The differences in insulin sensitivity between the BBS1 and BBS10 mutant fibroblasts are consistent with the observed insulin resistance assessed by HOMA-IR in human subjects with BBS10 mutations. The severity of resistance in BBS10 patients is greater than that in BBS1 patients (34). BBS1 and BBS10 coimmunoprecipitate with the IR and the BBS10 mutation decreases IR autophosphorylation in response to insulin. Previous studies reported direct interactions between the IR and BBS1/BBS17 but not BBS10 (30). The physical interaction between BBS10 and IR could be indirectly mediated by BBS1 or BBS17.
Leptin signaling was reduced in BBS1 and BBS10 mutant human iPSC–derived hypothalamic neurons; genetic correction restored leptin signaling in these neurons. In POMC neuron–specific Bbs1-knockout mice, the number of leptin-induced p-STAT3–positive POMC neurons was greatly reduced at 8-weeks of age, prior to the onset of obesity (26), supporting the contention that impaired leptin signaling is a primary consequence of hypomorphism for specific BBS genes. Because BBS1 and BBS10 are important for BBSome-associated ciliary trafficking, and the BBSome and transition zone of the primary cilium have been implicated in LEPRB trafficking at the base of the cilium, it is plausible that BBS1 and BBS10 affect leptin sensitivity by effects on LEPRB trafficking to the primary cilium (27, 31). In addition to LEPRB trafficking, BBS10 may enhance LEPRB protein stability and/or reduce its degradation, as overexpression of intact BBS10 in 293FT cells increased the amount of coexpressed HA-LEPR protein. Since BBS10 is a chaperonin protein that is required for the formation of BBSomes (31, 32), whether BBSome assembly is actually disrupted in these human iPSC–derived neurons requires further investigation. Moreover, whether BBS10 acts alone in influencing LEPR stability or also requires the participation of other chaperonin-like BBS members such as BBS6 and BBS12 is unclear at this point.
Decreased numbers of POMC neurons and levels of Pomc mRNA expression have been reported in hypothalami of Bbs2-, Bbs4-, and Bbs6-knockout mice, suggesting that Bbs mutations may affect neurogenesis and/or survival of POMC neurons (19, 27). In Bbs1M390R-KI mice, hypothalamic Pomc mRNA was reduced compared with littermate controls. BBS1B and BBS10A mutant human iPSC–derived hypothalamic neurons were also characterized by reduced POMC mRNA expression as well as lower amounts of processed αMSH and βEP in protein lysates and growth medium. These findings suggest that BBS proteins participate in the regulation of POMC production and processing.
In summary, we assessed the cellular and molecular consequences of BBS mutations in patient-specific iPSC-derived hypothalamic neurons in an effort to identify mechanisms by which these molecules regulate ciliary processes related to weight homeostasis.
Methods
Research subjects and cell lines.
BBS diagnoses were ascertained by a physician in the Department of Ophthalmology, Columbia University Irving Medical Center, to whom they had been referred for evaluation of retinal phenotypes. The skin biopsies from these patients were obtained at the Naomi Berrie Diabetes Center. These lines are designated Berrie Center skin biopsy–derived fibroblast lines 1085 (BBS1A) and 1097 (BBS1B), as well as the other 8 fibroblast lines (Supplemental Table 1 and https://www.eglilab.com/cell-line-repository). They were screened for the 2 most prevalent mutations found in BBS subjects: BBS1M390R (c.1169T>G) and BBS10C91fsX95 (c.271dupT) with PCR and Sanger sequencing. Both BBS1A and BBS1B are homozygous for the BBS1 M390R mutation (Supplemental Table 1). The other 2 BBS fibroblast lines were obtained from the NIGMS Human Genetic Cell Repository of the Coriell Institute for Medical Research: GM05948 (BBS10A) and GM05950 (BBS10B) (Supplemental Table 1). Genomic DNA was isolated from cultured fibroblasts or blood with a QIAmp DNA Mini Kit (QIAGEN). The mutations in the 2 Coriell fibroblast lines, BBS10A and BBS10B, were identified by Asper Biotech and further confirmed by dideoxy sequencing (Genewiz). See Supplemental Table 2 for genotyping primers.
Animal studies.
The BBS1M390R-KI mice were obtained from Val Sheffield’s lab (University of Iowa, Iowa City, Iowa, USA) (20). Mice were maintained at 22°C to 24°C with a 12-hour light/12-hour dark cycle (7 am to 7 pm) in a pathogen-free barrier facility. Only male mice were included in this study. All mice were fed ad libitum regular chow diet (6% fat, Purina LabDiet 5053) or breeder chow diet (10% fat, Purina LabDiet 5058). Glucose tolerance tests were performed after 16 hours of overnight fasting with intraperitoneal injection of 20% glucose (2 g glucose/kg body mass). Body weights were measured weekly. Body composition was determined by TD-NMR using an EchoMRI-100H body composition analyzer (EchoMRI). Mice were sacrificed after a 16-hour fast. NSG mice were purchased from The Jackson Laboratory (stock no. 005557) and fed regular chow ad libitum.
Information on all antibodies used in this study can be found in Supplemental Table 4. Reagents, if not otherwise specified, were all from Thermo Fisher Scientific.
Generation and characterization of iPSCs from human fibroblasts.
Primary fibroblasts were converted into iPSCs using retroviruses as described previously (39). All iPSC lines were cultured and characterized as previously described (39). Quantitative PCR (qPCR) analysis was used to confirm retroviral gene silencing and expression of endogenous pluripotency genes of all iPSC lines. Pluripotency was further confirmed by teratoma assay (39). For karyotyping, control and BBS iPSC lines were cultured in T25 flasks according to Cell Line Genetics instructions and karyotyped by Cell Line Genetics.
Generation of c-BBS1B iPSCs using CRISPR/Cas9.
BBS1 guide RNA (gRNA) was designed using the software from Feng Zhang’s lab at MIT (http://crispr.mit.edu). The sequences of the gRNA and single-strand DNA (ssDNA) oligonucleotides were BBS1M390RgRNA, CACCTCGAGTGGTCCTGATGAGG and BBS1ssDNA oligo, TTCCCCAACTAAACTCTGACGTCTCCACATAGGATGCAGTGACCAGCCTTTGCTTTGGCCGGTACGGGCGGGAGGACAACACACT CATCATGACCACTCGAGGTGAGTGGAGTCAGACCTGGCAAGGGCTTTGAAGTCGGGAGTGAAGGGACAGGCCTGCTTCTGGGGAAAGAGGAGGAG. The gRNA was subcloned into the pGS-U6-gRNA plasmid by Genscript. The pCas9_GFP plasmid was purchased from Addgene (plasmid 44719). After the BBS iPSCs became 90% confluent on a 6-well MEF plate (2 wells), each well was transfected with Lipofectamine 3000 supplemented with 2.5 μg pCas9_GFP plus 2.5 μg pGS-U6-gRNA in 2 mL hESC medium. Forty-eight to 72 hours after transfection, GFP+ cells were isolated by FACS and plated onto 6-well MEF plates with human embryonic stem cell (hESC) medium plus 10 μM Rock inhibitor (Rocki) Y-27632 (Selleckchem). Flow cytometry data were analyzed using BD FACSDiva 8.0.1 software. After 7–14 days, individual hESC colonies were picked for clonal expansion and screened with Sanger sequencing (Genewiz) of the PCR products amplified around the targeted genomic loci and confirmed via TOPO cloning (TOPO TA Cloning Kit). The isogenic control line was further characterized for pluripotency and karyotype.
Neuronal differentiation of iPSCs.
Human iPSCs were cultured and maintained in hESC medium on MEFs (111,000 cells/well in 6-well plates) (39, 64). Upon reaching confluence, iPSCs were cultured for 4 days in embryoid body (EB) medium (hESC medium without bFGF) plus 2 SMAD inhibitors — 10 μM SB43154 and 2.5 μM LDN-193189 (Selleckchem) (39). For days 5–8, EB medium was replaced with N2 medium in steps — from 75% to 50% to 25% and to 0%, while maintaining SB43154 and LDN-193189 at constant concentrations (39, 47). Neuronal progenitors were detached from plates on day 12 after incubating in Trypsin LE for 4 minutes at 37°C. Neuronal progenitors were washed twice with N2 medium and expanded for 2–3 passages in N2 medium plus SB43154 and LDN-193189 on poly-L-ornithine– and laminin-coated (PO/LA-coated) plates (39). For further neuronal differentiation, cells were cultured on PO/LA-coated plates at 50,000 cells per well of a 24-well or 4-well plate, and at 500,000 cells per well of a 6-well plate. After culturing 2–3 weeks in N2 medium supplemented with 10 μM DAPT (Selleckchem), B27, and 20 ng/mL BDNF (R&D Systems), TUJ1+ differentiated neurons were obtained.
ARH-like neurons were differentiated using a previously described protocol based on dual SMAD inhibition plus SHH activation and inhibition of Notch signaling (39, 64). After day 30, differentiated neurons were maintained in BrainPhys Neuronal Medium (Stemcell Technologies) for another 1–2 weeks for functional maturation.
Plasmids and lentivirus production.
Lentiviral cloning and expression vectors pCDH-UbC-MCS-EF1-Hygro (CD615B-1) and pCDH-EF1-MCS-T2A-Puro (CD520A-1) were purchased from System Biosciences Inc. (SBI). Human cDNA clones for LEPR transcript variant 1 (catalog HG10322-M), BBS1 (catalog HG10498-M), and BBS10 (catalog HG15095-G) were purchased from SBI. Fluorescence-based expression vectors pEGFP-N3 (Clontech) and pCAG-DsRed (Addgene) were used in this study. Genes for GFP and RFP were subcloned into the CD520 and CD615 vectors with BamHI and NotI digestion. CD520-RFP-LEPR was generated as described elsewhere (23). 3×FLAG-BBS1 cDNA was PCR amplified from the BBS1 cDNA plasmid using the primers 5′-TCTAGAtctagaGCGAAGATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATCACGATGCCGCTGCGTCCTCATCGGA-3′ and 5′-GGATCCggatccCAGCAGCTCAGGTCACAGGCGG-3′ and was further subcloned into the CD615-GFP plasmid in the Xba I and Bam HI restriction sites to construct the CD615-3×FLAG-BBS1-GFP plasmid. Primers 5′-ATAtctagaAGATATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATCACGATTTAAGTTCTATGGCCGCTGCAGG-3′ and 5′-TTAgctagcCTTCTGATGTGATAGTTTATCTTCTG-3′ were used to amplify 3×FLAG-BBS10 from the BBS10 ORF plasmid using CloneAmp HiFi PCR premix (Takara), which was further subcloned into the CD520A-GFP plasmid via XbaI and Nhe I restriction sites. CD615-GFP and CD520-3×FLAG-BBS10-EGFP were digested with XbaI and BstBI to generate CD615-3×FLAG-BBS10-EGFP. Plasmid amplification was performed using the QIAGEN Plasmid Maxi Kit (catalog 12162). To prepare lentiviral stocks, HEK293 cells were cultured to 90% confluence in 10-cm-diameter tissue culture dishes and transfected with 10 μg lentiviral expression vector (CD520-RFP-LEPR, CD615-3×FLAG-BBS1, or CD615-3×FLAG-BBS10) and 20 μg lentiviral packing vector mix with 45 μL Lipofectamine 2000 in Opti-MEM Reduced Serum Medium (catalog 31985088). After 6 hours of incubation at 37°C and 5% CO2, the Opti-MEM was replaced with 12 mL of fibroblast medium: 500 mL DMEM containing 4.5 g/mL glucose (catalog 11960044), 50 mL FBS (catalog 16000044), and 0.5–1 μg/mL penicillin/streptomycin from a 100× stock solution. To remove cell debris from suspensions containing lentiviral particles, 48 hours after transfection, the supernatant containing the lentiviral particles was filtered through a 0.45-μm filter (Millipore).
Generation of human transgenic cell lines.
Primary human fibroblast lines were cultured and maintained in fibroblast medium. At confluence, 2 mL of the lentiviral supernatant harvested from HEK293 cells was added to each well in 6-well plates. After 2 days, the lentivirus supernatant was removed and fibroblast medium with 1 μg/mL puromycin or 50 μg/mL hygromycin (Life Technologies) was added until we obtained stable transgenic fibroblast lines that express RFP-LEPR or express both RFP-LEPR and CD615-3×FLAG-BBS1-GFP. To construct human transgenic iPSC lines, 2 mL of the lentiviral supernatant was added to each well of the 6-well–cultured iPSCs (>90% confluent). After 48 hours, the lentiviral medium was replaced with hESC medium plus puromycin (1 μg/mL) or hygromycin (50–200 μg/mL) until iPSC lines stably expressing CD615-3×FLAG-BBS10-GFP were obtained.
BBS10 knockdown.
BBS10 shRNA lentiviral particles were purchased from Sigma-Aldrich. The sequences of the 2 shRNAs selected were CCGGCCTCAGAAAGTTCACAATCAACTCGAGTTGATTGTGAACTTTCTGAGGTTTTTTG (TRCN0000167181) and CCGGGCATTTATACCACACTCTATACTCGAGTATAGAGTGTGGTATAAATGCTTTTTTG (TRCN0000167794) (SHCLNV-NM_024685). The nonmammalian-targeting shRNA (catalog SHC002V, Sigma-Aldrich) was used as a negative control. iPSC cultures were established on 24-well MEF feeder cells. At 90% confluence, 10 μL shRNA lentiviral particles were added to wells filled with 0.5 mL hESC medium. After incubation for 48 hours with the shRNA lentivirus, the medium was removed and fresh hESC medium containing 1 μg/mL puromycin was added for another 24–48 hours. New MEFs were added to each well to help the survival of the iPSCs. The selection step was repeated until pure, puromycin-resistant iPSCs were obtained.
Neurite outgrowth assay.
Day 30 iPSC-derived neurons were dissociated from plates after 5-minute Trypsin LE treatments and collected in N2/B27/Rocki medium. After centrifugation at 124g for 5 minutes, supernatant was removed, and the cell pellet was washed once with N2/B27 medium. Neurons were resuspended with 2 mL N2/B27/Rocki medium and passed through a 5-mL round-bottom polystyrene tube with cell strainer cap (Thermo Fisher Scientific) to obtain single-cell suspensions. Cells were counted with a hemocytometer and 2,500 neurons were added to each well on PO/LA-coated 96-well plates (transparent bottom black plate). Forty-eight hours later, neurons were fixed with 4% paraformaldehyde for 10 minutes and stained with anti-TUJ1 and anti-neurofilament antibodies and Hoechst. Whole-well images were obtained with a TROPHOS Plate RUNNER. Images were further analyzed by MetaMorph software (Molecular Devices) to quantify neurite length, neuron body size, number of processes, and count the number of TUJ1+ cells for each channel.
qPCR analysis.
RNA was purified with the RNeasy Mini (QIAGEN) or Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. cDNA was generated with the GoScript Reverse Transcription kit (Promega) with 1 μg RNA from each sample. Twenty microliters of cDNA was diluted with 180 μL H2O. A GoTaq qPCR kit (Promega) was used for qPCR analysis. See Supplemental Table 3 for qPCR primers.
Western blot and immunoprecipitation.
Human fibroblasts or iPSCs were starved in 6-well plates or 10-cm tissue culture dishes in serum-free DMEM overnight. They were then treated with insulin (1 μg/mL) or leptin (0.5 μg/mL) or vehicle (PBS) for 30 minutes at 37°C and 5% CO2. After treatment, medium was removed and cells were rinsed once with cold PBS. For iPSC-derived neuronal progenitors or neurons, we cultured cells in neurobasal medium (catalog 21103049) supplemented with B27 minus insulin overnight. Variable doses of insulin or 1 μg/mL leptin, or vehicle for each, were then added to the neurobasal medium and the cells were incubated at 37°C and 5% CO2 for 30 minutes. Neuronal progenitors were cultured on PO/LA-coated plates and transfected with the RFP-LEPR–carrying plasmid using the TransIT-Neural transfection reagent (Mirus Bio) for 48 hours before leptin treatment. After aspiration of the medium, cells were rinsed with cold PBS. Lysis buffer (250 μL; 20 mM Tris, pH 7.4, 150 mM NaCl, 2% Nonidet P-40, 1 mM EDTA, pH 8.0, 10% glycerol, 0.5% sodium deoxycholate, 0.2% semi-dehydroascorbate, supplemented with phosphatase and proteinase inhibitors, catalog 78440) was added to each well to prepare protein lysates. Fifteen or 30 μg of protein from fibroblasts, iPSCs, neuronal progenitors, or neurons were loaded in each lane of a 4%–12% NuPAGE gel (Thermo Fisher Scientific, catalog NP0335BOX). 293FT cells were cotransfected with Myc-Lepr(b) plasmid (65) together with CD615-GFP, CD615-3×FLAG-BBS1-GFP, or CD615-3×FLAG-BBS10-GFP with Lipofectamine 2000. Total protein (40 μg) from neuronal samples or 293FT cell lysates were used. For immunoprecipitations, 293FT cells were transfected on 10-cm tissue culture dishes with CD615-3×FLAG-BBS1-GFP or CD615-3×FLAG-BBS10-GFP or CD615-GFP with Lipofectamine 2000. After 24 hours, 293FT cells were switched into serum-free DMEM overnight before insulin treatment. Insulin (1 μg/mL) was added to 293FT cells for 30 minutes at 37°C and 5% CO2. Cells were rinsed with cold PBS before being lysed with IP buffer (50 mM Tris pH 7.9, 150 mM NaCl, 10% glycerol, supplemented with protease inhibitors). For coimmunoprecipitations, 1 mg protein lysate was incubated with 30 μL anti-FLAG antibody–conjugated agarose beads (Sigma-Aldrich) overnight at 4°C. Beads were washed 4 times with IP buffer and precipitates were eluted by boiling in nonreducing sample buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 10% glycerol, and 1% SDS). See complete unedited blots in the supplemental material.
ICC/immunohistochemistry.
Fibroblasts were cultured on 12-mm-diameter round glass coverslips (Neuvitro) and fasted for 24–48 hours in serum-free DMEM before fixation, while iPSC-derived neurons were also cultured on PO/LA-coated glass coverslips for further cilia imaging. Day 34 iPSC-derived TUJ1+ neurons were dissociated with Trypsin LE and harvested into N2 medium. Cell pellets were further resuspended in 150 μL PBS with 0.5% BSA and stained with PE-conjugated anti–human CD56 antibody (1:100, BioLegend) for 20 minutes in the dark. Cells without antibody staining were used as negative control. CD56+ cells were isolated by FACS with a BD Biosciences FACSAria II cell sorter and replated on PO/LA-coated glass coverslips at 2,500 cells/well on 4-well plates for further cilia imaging. Cultured cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature. For p-STAT3 staining, cold methanol treatment was used for an additional 3–5 minutes at 4°C after fixation. The following staining procedure is the same as described previously (39). Teratomas were embedded in paraffin and cut into 8-μm sections. These teratoma sections were stained with hematoxylin and eosin (H&E) (39). See Supplemental Table 4 for antibody information. Images were acquired with an Olympus IX71 epifluorescence microscope with Olympus DP30BW black-and-white digital camera for fluorescence, and DP72 digital color camera for H&E staining. Some images were acquired with a Zeiss LSM5 Pascal microscope. Ciliary length was quantified based on the distance between the basal body and the tip of the axoneme (38).
Neuropeptide assays.
Neuropeptide assays on iPSC-derived hypothalamic neurons were performed as previously described (39). Briefly, all neurons were cultured in 12-well plates and initiated with 300,000 cells on day 12. For neuronal lysate samples, medium was aspirated, and the differentiated neurons rinsed once with PBS, after which 250 μL of 0.1N HCl was added to each well. Cells were harvested with a cell scraper and the cell lysates were transferred into 1.7-mL Eppendorf tubes. These samples were sonicated on ice with maximum power for 5 minutes. After centrifugation at 10,000g for 10 minutes at 4°C, the supernatants were collected for POMC, αMSH, and βEP assays. Results are presented in fmol/mL. Protein concentrations in neuronal lysates were determined using a Pierce BCA protein assay kit. Neuropeptide concentrations were normalized to the amount of protein in each well for each cell line.
Single-cell isolation, scRNA-seq, and data analysis.
On day 32 of differentiation, neurons were dissociated using the cell dissociation enzyme Accutase. Briefly, for each cell line, 1 mL of Accutase was added to 1 well of 6-well plate and incubated at 37°C for 2 minutes. Next, the Accutase was carefully removed, replaced with 1 mL of BrainPhys Neuronal Medium supplemented with N2 Supplement-A (Stemcell Technologies) and B27 supplement minus vitamin A (N2/B27 BrainPhys Neuronal Medium), and immediately dissociated by gently pipetting (15–20 times with a P1000) using low-retention and wide-orifice tips (Rainin). The cell suspension was transferred into a 50-mL Falcon tube. The well was rinsed with an additional 6 mL of N2/B27 BrainPhys Neuronal Medium in order to collect remaining cells and this was subsequently added to the tube. The suspension was centrifugated for 5 minutes at 300g at room temperature. After centrifugation, the pellet was resuspended in 1 mL of N2/B27 BrainPhys Neuronal Medium by gentle pipetting (10 times with P1000) using low-retention and wide-orifice tips, and then transferred to a low-binding microcentrifuge tube (Thermo Fisher Scientific). The suspension was then spun for 5 minutes at 300g and 4°C. After centrifugation, the pellet was resuspended in 100 or 200 μL of N2/B27 BrainPhys Neuronal Medium initially by gentle pipetting with a P200 using low-retention and wide-orifice tips 20–25 times followed by low-retention and regular-orifice tips (Rainin) 10 times. Then, the suspension was diluted 1:2 with fresh medium and passed through a 40-μm cell strainer (Thermo Fisher Scientific) to obtain single-cell suspensions. Trypan blue was used to determine number of viable cells. A viability of 60%–80% was obtained for all the samples. All single-cell suspensions were kept on ice until use. Libraries were prepared using the Chromium Single Cell 3′ Reagent Kit v3 (10× Genomics) and sequenced on an Illumina NovaSeq 6000.
Cell Ranger (v3.1.0, 10× Genomics) was employed with default parameters to process the scRNA-seq data (66). The human reference genome (GRCh38-3.0.0) was used for the alignment of reads obtained from 10× Genomics. The Cell Ranger summaries indicated that both data sets passed the quality controls, with valid barcodes greater than 97%, mean reads per cell greater than 66,000, and median genes per cell greater than 3,700. The filtered gene-barcode matrix from the Cell Ranger outputs was used for subsequent analysis.
The raw counts of scRNA-seq data were filtered and normalized using the RISC package (v1.0.0) (67). Genes expressed in fewer than 3 cells were discarded, and cells expressing fewer than 200 genes were removed. At the end, approximately 4,000 valid cells were obtained for c-BBS1B, and approximately 3,500 valid cells for the BBS1B M390R mutant, with more than 20,000 genes detected for each sample. The count normalization was performed independently for each sample, with counts log transformed and weighted by sequencing depth. The batch effects between the 2 data sets were corrected using the RISC package in the R environment (v3.6.3) by projecting all the cells from both data sets into a common reference space. The top principal components (PCs = 18) were used to build the reference space for the integrated data, containing 7,550 valid cells and approximately 19,700 genes. We also used these PCs for cell clustering, based on the Louvain method of the igraph package (v1.2.5), with neighbors set to 20 (https://igraph.org/). In total, 14 clusters were obtained for the integrated data. Cell types were identified according to the expression patterns of cell-type marker genes that were significantly more highly expressed in one cluster versus the others. The scanpy package (v1.5.1) was utilized to generate the stacked violin plot for cell-type markers in the Python environment (v 3.8.5) (68).
Differentially expressed (DE) genes of the integrated data were determined using a negative binomial generalized linear model in the RISC package, at an adjusted P value of less than 0.05 and log2(fold change) greater than 0.25 or less than –0.25. Heatmaps of DE genes were generated using the R pheatmap package (v1.0.12); the violin plots of DE genes were prepared by the Python seaborn package (v0.10.1) (https://doi.org/10.5281/zenodo.883859). GO and KEGG pathway analyses were performed using the ToppGene Suite (https://toppgene.cchmc.org/enrichment.jsp), with significance set at a P value of less than 0.05. The scRNA-seq data have been deposited in the NCBI’s Gene Expression Omnibus (GEO GSE164891).
Electrophysiology study of iPSC-derived neurons.
Patch-clamp recordings of iPSC-derived neurons were conducted as previously described (39).
Statistics.
Unless otherwise indicated, all graphical data are presented as mean ± SEM. Statistical analyses were performed with Prism 6.0 software (GraphPad). Two-tailed Student’s t tests were used for unpaired comparisons between 2 groups, and 1-way ANOVA for comparisons among 3 or more groups followed by Tukey’s multiple-comparison test. Two-way ANOVA was performed to assess influences of 2 variables, followed, when appropriate, by Bonferroni’s multiple-comparison test. Pairwise 2-tailed t tests were used for assessment of differences between specific data points. P values of less than 0.05 were considered significant.
Study approval.
All human subjects provided written informed consent prior to their participation in this study. Human subject research was reviewed and approved by the Columbia Stem Cell Committee and the Columbia University IRB (protocol IRB-AAAK6905). All animal studies were approved by the Columbia University IACUC.
Author contributions
LW, DE, and RLL designed this study. LW performed the experiments, data analysis, and generated the initial draft of the manuscript. CAD helped with iPSC line characterization and manuscript composition. GS contributed to data analysis and manuscript composition. YL and DZ performed the scRNA-seq data analysis and helped draft the manuscript. SP and SW helped with the neuropeptide assays and analysis. DJW performed the electrophysiology studies. RG and SHT provided skin biopsies and blood cells from the BBS subjects; SHT ascertained the diagnosis of BBS in the samples. L Sui and LCB helped with cell culture experiments. CAL and L Shang helped with mouse work. YZ provided the mouse Myc-ObRb-HA plasmid and suggestions on leptin signaling studies. MCDR and CAD helped with neuronal differentiation and sample preparation for scRNA-seq. HJG contributed to the scRNA-seq data analysis.
Supplementary Material
Acknowledgments
We thank Haiqing Hua and Aiqun Li for discussions related to this work; Yao Li and Jing Yang for help with collecting the biopsy samples; Val Sheffield for BBSM390RKI mice; Kathryn Anderson for the anti-SMO antibody (Memorial Sloan Kettering, New York, New York, USA); Tamara Caspary for the anti-ARL13B antibody (Emory University, Atlanta, Georgia, USA); and Lianqun Yang of the Diabetes and Endocrinology Research Center Pathology Core at Columbia University for histological analysis. Some of the studies reported here were conducted in the Genomics and High Throughput Screening Shared Resource at Columbia University (NIH/NCI P30CA013696). This research was supported by NIH grant R01 DK52431-23, the New York Stem Cell Foundation, a NYSTEM IIRP awards (SDHDOH01-C32590GG-3450000 and C026184 to DE), the Rudin Foundation, the Russell Berrie Foundation Program in Cellular Therapies, the Diabetes (P30 DK63608-16) and Obesity Research (P30 DK26687-41) Center of Columbia University, R01DK093920, U54OD020351, R24EY028758, U01 EY030580, R24EY027285, 5P30EY019007, R01EY018213, R01EY024698, R01EY026682, R21AG050437, New York State (N13G-275), and Foundation Fighting Blindness New York Regional Research Center Grants (PPA-1218-0751-COLU) 7T32DK00755925 and 1K01DK123199.
Version 1. 02/25/2021
In-Press Preview
Version 2. 04/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(8):e146287.https://doi.org/10.1172/JCI146287.
See the related Commentary at The BBSome: a nexus controlling energy metabolism in the brain.
Contributor Information
Liheng Wang, Email: lw2381@cumc.columbia.edu.
Yang Liu, Email: yang.liu@einsteinmed.org.
George Stratigopoulos, Email: gs2172@columbia.edu.
Lina Sui, Email: sln1981163@gmail.com.
Yiying Zhang, Email: yz84@columbia.edu.
Charles A. Leduc, Email: cal8@columbia.edu.
Hannah J. Glover, Email: hg2553@cumc.columbia.edu.
Maria Caterina De Rosa, Email: md3291@cumc.columbia.edu.
Lisa C. Burnett, Email: lcburnett@levotx.com.
Damian J. Williams, Email: dw2471@columbia.edu.
Linshan Shang, Email: shang133@umn.edu.
Robin Goland, Email: rsg2@columbia.edu.
Stephen H. Tsang, Email: sht2@columbia.edu.
Dieter Egli, Email: de2220@cumc.columbia.edu.
Deyou Zheng, Email: deyou.zheng@einstein.yu.edu.
Claudia A. Doege, Email: cad2114@cumc.columbia.edu.
Rudolph L. Leibel, Email: RL232@cumc.columbia.edu.
References
- 1.Tsang SH, et al. Ciliopathy: Bardet-Biedl syndrome. Adv Exp Med Biol. 2018;1085:171–174. doi: 10.1007/978-3-319-95046-4_33. [DOI] [PubMed] [Google Scholar]
- 2.M’Hamdi O, et al. Update on the genetics of Bardet-Biedl syndrome. Mol Syndromol. 2014;5(2):51–56. doi: 10.1159/000357054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141(7):1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingfield JL, et al. Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 2018;62(6):753–763. doi: 10.1042/EBC20180030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, Lechtreck KF. The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci U S A. 2018;115(5):E934–E943. doi: 10.1073/pnas.1713226115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snow JJ, et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6(11):1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 7.Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19(3):220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satir P, Christensen ST. Structure and function of mammalian cilia. Histochem Cell Biol. 2008;129(6):687–693. doi: 10.1007/s00418-008-0416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collin GB, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alström syndrome. Nat Genet. 2002;31(1):74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- 10.Grarup N, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018;50(2):172–174. doi: 10.1038/s41588-017-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saeed S, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat Genet. 2018;50(2):175–179. doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 12.Siljee JE, et al. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50(2):180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaisse C, et al. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20(2):113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 14.Yeo GS, et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet. 1998;20(2):111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, et al. Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS One. 2009;4(9):e6979. doi: 10.1371/journal.pone.0006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collin GB, et al. Alms1-disrupted mice recapitulate human Alström syndrome. Hum Mol Genet. 2005;14(16):2323–2333. doi: 10.1093/hmg/ddi235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, et al. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci U S A. 2011;108(51):20678–20683. doi: 10.1073/pnas.1113220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, et al. BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J Cell Sci. 2013;126(pt 11):2372–2380. doi: 10.1242/jcs.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahmouni K, et al. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J Clin Invest. 2008;118(4):1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci U S A. 2007;104(49):19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh DJ, et al. Response of melanocortin-4 receptor-deficient mice to anorectic and orexigenic peptides. Nat Genet. 1999;21(1):119–122. doi: 10.1038/5070. [DOI] [PubMed] [Google Scholar]
- 22.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17(18):1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stratigopoulos G, et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 2014;19(5):767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;126(5):1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berbari NF, et al. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105(11):4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo DF, et al. The BBSome in POMC and AgRP neurons is necessary for body weight regulation and sorting of metabolic receptors. Diabetes. 2019;68(8):1591–1603. doi: 10.2337/db18-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo S, et al. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet. 2009;18(7):1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratigopoulos G, et al. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J Biol Chem. 2011;286(3):2155–2170. doi: 10.1074/jbc.M110.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo DF, et al. The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet. 2016;12(2):e1005890. doi: 10.1371/journal.pgen.1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starks RD, et al. Regulation of insulin receptor trafficking by Bardet Biedl syndrome proteins. PLoS Genet. 2015;11(6):e1005311. doi: 10.1371/journal.pgen.1005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo S, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010;107(4):1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, et al. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem. 2012;287(24):20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Satta M, et al. Bardet-Biedl syndrome as a chaperonopathy: dissecting the major role of chaperonin-like BBS proteins (BBS6-BBS10-BBS12) Front Mol Biosci. 2017;4:55. doi: 10.3389/fmolb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuillan PP, et al. Patients with Bardet-Biedl syndrome have hyperleptinemia suggestive of leptin resistance. J Clin Endocrinol Metab. 2011;96(3):E528–E535. doi: 10.1210/jc.2010-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keppler-Noreuil KM, et al. Brain tissue- and region-specific abnormalities on volumetric MRI scans in 21 patients with Bardet-Biedl syndrome (BBS) BMC Med Genet. 2011;12:101. doi: 10.1186/1471-2350-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz MW, et al. Central nervous system control of food intake. Nature. 2000;404(6778):661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 37.Barsh GS, Schwartz MW. Genetic approaches to studying energy balance: perception and integration. Nat Rev Genet. 2002;3(8):589–600. doi: 10.1038/nrg862. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, et al. Ciliary gene RPGRIP1L is required for hypothalamic arcuate neuron development. JCI Insight. 2019;4(3):123337. doi: 10.1172/jci.insight.123337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, et al. Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J Clin Invest. 2015;125(2):796–808. doi: 10.1172/JCI79220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artegiani B, et al. A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 2017;21(11):3271–3284. doi: 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- 41.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 42.Lang C, et al. Single-cell sequencing of iPSC-dopamine neurons reconstructs disease progression and identifies HDAC4 as a regulator of parkinson cell phenotypes. Cell Stem Cell. 2019;24(1):93–106. doi: 10.1016/j.stem.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xin Y, et al. RNA Sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24(4):608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 46.Swiderski RE, et al. Structural defects in cilia of the choroid plexus, subfornical organ and ventricular ependyma are associated with ventriculomegaly. Fluids Barriers CNS. 2012;9(1):22. doi: 10.1186/2045-8118-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crawford TQ, Roelink H. The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev Dyn. 2007;236(3):886–892. doi: 10.1002/dvdy.21083. [DOI] [PubMed] [Google Scholar]
- 49.Bocchi R, et al. Perturbed Wnt signaling leads to neuronal migration delay, altered interhemispheric connections and impaired social behavior. Nat Commun. 2017;8(1):1158. doi: 10.1038/s41467-017-01046-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valvezan AJ, Klein PS. GSK-3 and Wnt signaling in neurogenesis and bipolar disorder. Front Mol Neurosci. 2012;5:1. doi: 10.3389/fnmol.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belgacem YH, et al. The many hats of sonic hedgehog signaling in nervous system development and disease. J Dev Biol. 2016;4(4):E35. doi: 10.3390/jdb4040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breunig JJ, et al. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc Natl Acad Sci U S A. 2008;105(35):13127–13132. doi: 10.1073/pnas.0804558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh EC, Katsanis N. Context-dependent regulation of Wnt signaling through the primary cilium. J Am Soc Nephrol. 2013;24(1):10–18. doi: 10.1681/ASN.2012050526. [DOI] [PubMed] [Google Scholar]
- 54.Baskin DG, et al. Insulin and leptin: dual adiposity signals to the brain for the regulation of food intake and body weight. Brain Res. 1999;848(1–2):114–123. doi: 10.1016/s0006-8993(99)01974-5. [DOI] [PubMed] [Google Scholar]
- 55.Plum L, et al. Central insulin action in energy and glucose homeostasis. J Clin Invest. 2006;116(7):1761–1766. doi: 10.1172/JCI29063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Air EL, et al. Small molecule insulin mimetics reduce food intake and body weight and prevent development of obesity. Nat Med. 2002;8(2):179–183. doi: 10.1038/nm0202-179. [DOI] [PubMed] [Google Scholar]
- 57.Zhu D, et al. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. J Cell Sci. 2009;122(pt 15):2760–2768. doi: 10.1242/jcs.046276. [DOI] [PubMed] [Google Scholar]
- 58.Campbell JN, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci. 2017;20(3):484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merkle FT, et al. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015;142(4):633–643. doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berretta A, et al. Sonic hedgehog stimulates neurite outgrowth in a mechanical stretch model of reactive-astrogliosis. Sci Rep. 2016;6:21896. doi: 10.1038/srep21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prajerova I, et al. Distinct effects of sonic hedgehog and Wnt-7a on differentiation of neonatal neural stem/progenitor cells in vitro. Neuroscience. 2010;171(3):693–711. doi: 10.1016/j.neuroscience.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 62.Gu T, et al. Insulin signaling regulates neurite growth during metamorphic neuronal remodeling. Biol Open. 2014;3(1):81–93. doi: 10.1242/bio.20136437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouret SG, et al. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304(5667):108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, et al. Efficient generation of hypothalamic neurons from human pluripotent stem cells. Curr Protoc Hum Genet. 2016;90:21 5.1–21.5.14. doi: 10.1002/cphg.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gan L, et al. TNF-α up-regulates protein level and cell surface expression of the leptin receptor by stimulating its export via a PKC-dependent mechanism. Endocrinology. 2012;153(12):5821–5833. doi: 10.1210/en.2012-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. doi: 10.1101/483297. Liu Y, et al. RISC: robust integration of single-cell RNA-seq datasets with different extents of cell cluster overlap [preprint]. Posted on bioRxiv November 29, 2018. [DOI]
- 68.Wolf FA, et al. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.