Abstract
Bardet-Biedl syndrome (BBS) is a syndromic ciliopathy that has obesity as a cardinal feature. BBS is caused by mutations in BBS genes. BBS proteins control primary cilia function, and BBS mutations therefore lead to dysfunctional primary cilia. Obesity in patients with BBS is mainly caused by hyperphagia due to dysregulated neuronal function in the brain, in particular in the hypothalamus. However, the mechanism by which mutations in BBS genes result in dysfunction in hypothalamic neurons is not well understood. In this issue of the JCI, Wang et al. used BBS and non-BBS patient–derived induced pluripotent stem cells to generate neurons and hypothalamic neurons. Using this human model system, the authors demonstrated that mutations in BBS genes affected primary cilia function, neuronal morphology, and signaling pathways regulating the function of hypothalamic neurons, which control energy homeostasis. This study provides important insights into the mechanisms of BBS-induced obesity.
Primary cilia dysfunction in BBS
Primary cilia are short membrane protrusions, emanating from the surface of most vertebrate cells. They function as cellular antennae, receiving information from the environment and locally transducing the information into a cellular response. Various signaling pathways can exert their function via the primary cilium, including SHH, Wnt, and G protein–coupled cAMP signaling. Notably, the function of primary cilia depends on an array of proteins that maintain the structure and signaling dynamics in the cilium (1).
Mutations that encode defective ciliary proteins often lead to ciliopathies, which are multisystemic diseases affecting many organs. Bardet-Biedl syndrome (BBS) is a syndromic ciliopathy that is characterized by retinal degradation, obesity, cystic kidneys, and polydactyly (2). So far, mutations in 21 genes have been associated with BBS. Eight of these genes encode the core BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBIP10), which form a protein complex termed the BBSome (1). The BBSome does not directly contribute to ciliary formation, but rather controls the dynamic localization of different proteins in the cilium, in particular G protein–coupled receptors (GPCRs) like the melanin concentrating hormone receptor (MCHR1), the neuropeptide Y receptor (NPYR2), or the somatostatin receptor 3 (SSTR3) (3). These GPCRs signal either through Gαi/o to inhibit cAMP synthesis or Gαs to activate cAMP synthesis via adenylate cyclases (ACs). In fact, ACIII is used as marker for neuronal primary cilia (4), indicating that cAMP signaling plays a prominent role in controlling primary cilia function in neurons (Figure 1B).
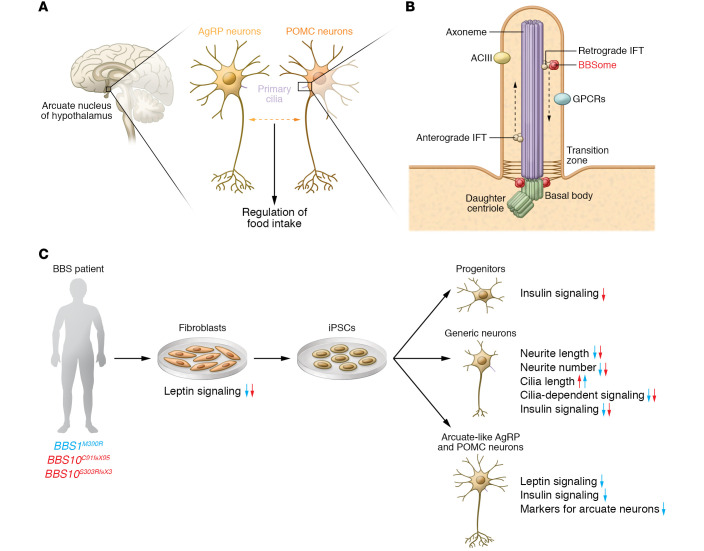
Figure 1. Bardet-Biedl syndrome proteins control primary cilia function in hypothalamic neurons.
(A) In the brain, the arcuate nucleus of the hypothalamus is central for controlling energy homeostasis in the body. Here, the interplay of proopiomelanocortin (POMC) and Agouti-related protein (AgRP) neurons, which require primary cilia for their normal function, regulates food intake. (B) Primary cilia originate from the basal body, a modified mother centriole, which is part of the centrosome. The basal body serves as a nucleation site for microtubules, which form the core structure of all cilia, the axoneme. Along the axoneme, the intraflagellar transport (IFT) machinery transports proteins anterogradely to the ciliary tip or retrogradely to the ciliary base via the molecular motors kinesin-2 and dynein-2, respectively. The transition zone at the ciliary base prevents free diffusion of proteins from the cell body into the cilium. The BBSome is a central component, which, together with the IFT, regulates the protein content in the cilium. (C) Wang et al. (14) differentiated generic neurons as well as POMC and AgRP neurons from BBS and non-BBS patient–derived iPSCs (via fibroblasts). Using this human in vitro system, the authors uncovered the effect of mutations in BBSome-encoding genes on neuronal differentiation and on the central signaling pathways regulating the function of POMC and AgRP neurons in energy homeostasis. Arrows indicate decrease or increase. Note that some aspects were only investigated in cells with BBS1 (blue arrows) or BBS10 (red arrows) mutations.
Hypothalamic neuronal dysfunction linking BBS to obesity
Energy metabolism is the process of generating energy from nutrients. A chronic imbalance between energy intake and energy expenditure leads to obesity. In mouse models, missense or loss-of-function mutations in several BBS genes recapitulate obesity as a hallmark of BBS (5–8). Specific deletion of Bbs1 in proopiomelanocortin (POMC) or Agouti-related protein (AgRP) neurons, which reside in the arcuate nucleus of the hypothalamus, the energy control center in the brain, leads to an increase in food intake and, thereby, to obesity (refs. 9, 10 and Figure 1A). These data suggest that obesity in BBS patients is caused by BBSome-dependent dysfunction of POMC or AgRP neurons. Still, the molecular and pathophysiological changes in hypothalamic neurons caused by mutations in BBS genes are not well understood, and have not yet been studied in a human model system.
Recent data suggest that altered GPCR/cAMP signaling contributes to disease development. Loss of Bbs1 dysregulates the localization of NPY2R and serotonin 5-HT2C receptor to cilia and the plasma membrane, respectively (10). Downstream of NPY2R and serotonin 5-HT2C, ACIII drives cAMP signaling. Because mutations or genetic loss of ADCY3, which encodes ACIII, causes monogenic severe obesity and increases the risk for type 2 diabetes, cAMP likely plays a key role in developing disease. (11, 12). Indeed, obesity and type 2 diabetes have been attributed to ACIII function in neurons, in particular to its ciliary localization and function (13).
Modeling BBS mutations in patient-derived neurons
To investigate the impact of BBS mutations in a human model system, Wang et al. (14) generated neurons from induced pluripotent stem cells (iPSCs) derived from fibroblasts of patients with mutations in BBS1 or BBS10 (Figure 1C). In generic iPSC-derived human neurons, BBS mutations led to morphological defects, such as reduced neurite length and number as well as increased length of primary cilia. In addition, ciliary signaling pathways, such as SHH and Wnt signaling, were impaired in patient-derived neurons (14). Because the developmental maturity stage of iPSC-derived neurons generally corresponds to neurons in the prenatal brain (15), these results suggest that BBS proteins play an important role in neuronal development processes that are likely to influence the anatomical organization and function of neurons in later life. Importantly, Wang et al. (14) succeeded in generating hypothalamic arcuate-like neurons from patient-derived iPSCs, thus establishing an in vitro model to study the effects of BBS mutations specifically in human POMC and AgRP neurons. Comparison of the gene expression profile of hypothalamic neurons, generated from a patient-derived iPSC line with a BBS1 mutation and from isogenic control cells, using single-cell RNA sequencing revealed a number of signaling pathways that were downregulated in specific subclusters, i.e., pathways controlling axon guidance as well as insulin, leptin, and cAMP signaling pathways. The authors then used fibroblasts, neuronal progenitors, and generic and hypothalamic neurons to demonstrate that both insulin and leptin signaling pathways were indeed impaired in cells carrying a BBS mutation (Figure 1C). These deficits could be restored by expressing wild-type BBS protein or by correcting the mutation using CRISPR/Cas. Taken together, these results point toward the BBSome playing a role in neuronal development and in the regulation of neuronal signaling pathways crucial for energy homeostasis (14).
Is obesity in BBS caused by ciliary dysfunction?
The importance of primary cilia in regulating energy homeostasis in the brain by controlling food intake has been demonstrated in a number of studies (reviewed in ref. 16), suggesting that ciliary dysfunction is the primary cause of obesity in BBS. However, although the BBSome is well recognized for its ciliary function, it also regulates protein trafficking to the plasma membrane. Among those proteins are the leptin receptor, the insulin receptor, and the serotonin 5-HT2C receptor (9, 10, 17). Thus, when analyzing the pathophysiology of BBS, ciliary versus nonciliary phenotypes have to be carefully dissected. Strikingly, leptin, insulin, and 5 HT2C receptors, whose localization is controlled by BBS proteins outside the cilium, are also key players in the regulation of energy homeostasis. Whether BBSome dysfunction also contributes to impaired receptor trafficking to the plasma membrane associated with common obesity has to be investigated in future studies. Another point that should be explored further is whether ciliary dysfunction affects energy homeostasis and function of hypothalamic neurons directly and/or during neuronal development. Induction and patterning of the hypothalamus are controlled by SHH and Wnt signaling, which are dependent on primary cilia (18). Evidence that points to developmental defects as a primary cause stems from mouse models, in which ciliary function was abolished in POMC neurons either in the developing or in the adult brain. Strikingly, mice stayed lean when ciliary function was abolished in adult POMC neurons, but became obese when ciliary function was inhibited during development. This was accompanied by altered neurogenesis and differentiation, which reduced the complexity of neuronal morphology and changed axonal projections of developing POMC neurons (19, 20). In BBS patients, MRI scans show anatomical abnormalities in several brain regions, including hypoplasia of the hypothalamus, further suggesting that mutations in BBS genes alter processes crucial for normal brain development (21). However, the effect of loss of function in BBSome-encoding genes on hypothalamic development has not yet been fully addressed.
Studying ciliopathies in human cell and organoid models
In the last decade, there has been an enormous advance in establishing 3D models of various human brain regions. These so-called organoids partially recapitulate early developmental processes. Recent studies have established hypothalamic organoids, containing a range of hypothalamic neurons, including POMC and/or AgRP neurons (22, 23). One of these studies even generated 3D structures of the hypothalamus in association with the anterior pituitary (22). While these 3D in vitro models do not show a clear organization into the various hypothalamic nuclei, they still open the possibility to study the effect of mutations in BBSome-encoding genes on the assembly and generation of the hypothalamic-pituitary axis, and on the differentiation mechanisms of arcuate nucleus neurons.
Another syndromic ciliopathy that resembles BBS and displays obesity is Alström syndrome (AS). AS is due to mutations in ALMS1 (24). The ALMS1 protein is part of the centrosome, which is important for primary cilia, as cilia originate from the basal body, a modified mother centriole that is part of the centrosome (Figure 1). ALMS1-mutant mice are hyperphagic, indicating that defects in hypothalamic neurons are involved in the development of obesity in AS. Recently, iPSCs carrying ALMS1 mutations have been generated (25). To reveal mechanistic insights about the common denominators underlying neuronal dysfunction in the hypothalamus in BBS and AS, iPSC-derived human neurons, combined with 2D and 3D cultures, will likely be the future.
Acknowledgments
The SB and DW labs were supported by grants from the Deutsche Forschungsgemeinschaft (DFG): SPP1926, SPP1726, TRR83/SFB, FOR2743, SFB1454 (all to DW), under Germany’s Excellence Strategy – EXC2151 – 390873048 (to DW), and SFB1089 (to SB).
Version 1. 04/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(8):e148903. https://doi.org/10.1172/JCI148903.
Contributor Information
Sandra Blaess, Email: sandra.blaess@uni-bonn.de.
Dagmar Wachten, Email: dwachten@uni-bonn.de.
References
- 1.Nachury MV, Mick DU. Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol. 2019;20(7):389–405. doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forsythe E, Beales PL. Bardet–Biedl syndrome. Eur J Hum Genet. 2013;21(1):8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malicki J, Avidor-Reiss T. From the cytoplasm into the cilium: bon voyage. Organogenesis. 2014;10(1):138–157. doi: 10.4161/org.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop GA, et al. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505(5):562–571. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, et al. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci U S A. 2011;108(51):20678–20683. doi: 10.1073/pnas.1113220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, et al. BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J Cell Sci. 2013;126(11):2372–2380. doi: 10.1242/jcs.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci U S A. 2007;104(49):19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahmouni K, et al. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J Clin Invest. 2008;118(4):1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo D-F, et al. The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet. 2016;12(2):e1005890. doi: 10.1371/journal.pgen.1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo D-F, et al. The BBSome in POMC and AgRP neurons is necessary for body weight regulation and sorting of metabolic receptors. Diabetes. 2019;68(8):1591–1603. doi: 10.2337/db18-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao H, et al. Disruption of type 3 adenylyl cyclase expression in the hypothalamus leads to obesity. Integr Obes Diabetes. 2016;2(2):225–228. doi: 10.15761/IOD.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grarup N, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018;50(2):172–174. doi: 10.1038/s41588-017-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siljee JE, et al. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50(2):180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. Bardet-Biedl syndrome proteins regulate intracellular signaling and neuronal function in patient-specific iPSC-derived neurons. J Clin Invest. 2021;131(8):e146287. doi: 10.1172/JCI146287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke EE, et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat Commun. 2020;11(1):462. doi: 10.1038/s41467-019-14266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engle SE, et al. Cilia signaling and obesity. Semin Cell Dev Biol. 2020;110:43–50. doi: 10.1016/j.semcdb.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starks RD, et al. Regulation of insulin receptor trafficking by Bardet Biedl syndrome proteins. PLoS Genet. 2015;11(6):e1005311. doi: 10.1371/journal.pgen.1005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedont JL, et al. Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip Rev Dev Biol. 2015;4(5):445–468. doi: 10.1002/wdev.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, et al. Ciliary gene RPGRIP1L is required for hypothalamic arcuate neuron development. JCI Insight. 2019;4(3):e123337. doi: 10.1172/jci.insight.123337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CH, et al. Primary cilia mediate early life programming of adiposity through lysosomal regulation in the developing mouse hypothalamus. Nat Commun. 2020;11(1):5772. doi: 10.1038/s41467-020-19638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keppler-Noreuil KM, et al. Brain tissue- and region-specific abnormalities on volumetric MRI scans in 21 patients with Bardet-Biedl syndrome (BBS) BMC Med Genet. 2011;12(1):101. doi: 10.1186/1471-2350-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasai T, et al. Hypothalamic contribution to pituitary functions is recapitulated in vitro using 3D-cultured human iPS cells. Cell Reports. 2020;30(1):18–24. doi: 10.1016/j.celrep.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collin GB, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alström syndrome. Nat Genet. 2002;31(1):74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- 25.Ji X, et al. Generation of an induced pluripotent stem cell line from an Alström Syndrome patient with ALMS1 mutation (c.3902C>A, c.6436C>T) and a gene correction isogenic iPSC line. Stem Cell Res. 2020;49:102089. doi: 10.1016/j.scr.2020.102089. [DOI] [PubMed] [Google Scholar]