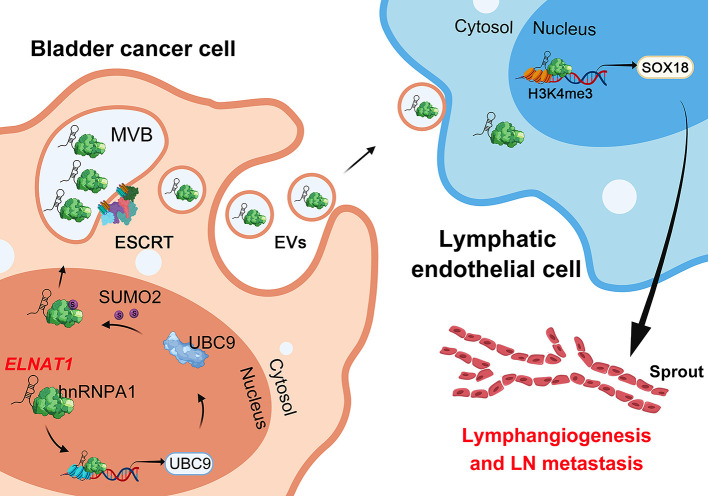
Abstract
Small ubiquitin-like modifier (SUMO) binding (termed SUMOylation) emerged as the inducer for the sorting of bioactive molecules into extracellular vesicles (EVs), triggering lymphangiogenesis and further driving tumor lymph node (LN) metastasis, but the precise mechanisms remain largely unclear. Here, we show that bladder cancer (BCa) cell–secreted EVs mediated intercellular communication with human lymphatic endothelial cells (HLECs) through transmission of the long noncoding RNA ELNAT1 and promoted lymphangiogenesis and LN metastasis in a SUMOylation-dependent manner in both cultured BCa cell lines and mouse models. Mechanistically, ELNAT1 induced UBC9 overexpression to catalyze the SUMOylation of hnRNPA1 at the lysine 113 residue, which mediated recognition of ELNAT1 by the endosomal sorting complex required for transport (ESCRT) and facilitated its packaging into EVs. EV-mediated ELNAT1 was specifically transmitted into HLECs and epigenetically activated SOX18 transcription to induce lymphangiogenesis. Importantly, blocking the SUMOylation of tumor cells by downregulating UBC9 expression markedly reduced lymphatic metastasis in EV-mediated, ELNAT1-treated BCa in vivo. Clinically, EV-mediated ELNAT1 was correlated with LN metastasis and a poor prognosis for patients with BCa. These findings highlight a molecular mechanism whereby the EV-mediated ELNAT1/UBC9/SOX18 regulatory axis promotes lymphangiogenesis and LN metastasis in BCa in a SUMOylation-dependent manner and implicate ELNAT1 as an attractive therapeutic target for LN metastatic BCa.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Extracellular matrix, Lymph
Introduction
Bladder cancer (BCa) is one of the most prevalent malignancies in the genitourinary system (1). As seen in the infiltration of muscle, BCa develops into 2 distinct subtypes: non–muscle-invasive BCa (NMIBC) (75%) and muscle-invasive BCa (MIBC) (25%), with MIBC posing a higher risk of metastasis (2). Lymph node (LN) metastasis is considered the main metastatic route and the leading reason for a poor prognosis in BCa, with a decrease in the 5-year survival rate of patients from 77.6% to 18.6% (3). A previous study revealed that lymphangiogenesis is the pivotal and rate-limiting step in tumor LN metastasis (4), as it favors tumor cells for invasion of the lymphatic system because of the incomplete basement membranes of neonatal lymphatic vessels (5). Although the crucial role of lymphangiogenesis has been well established in BCa LN metastasis, its regulatory mechanism still requires further elucidation.
Extracellular vesicles (EVs) are endogenous double-layered membrane vesicles that serve as molecular cargo carriers to regulate intercellular communication that can result in LN metastasis of multiple cancers (6, 7). EVs possess highly specific fusogenic properties and uptake machinery for recipient cells, enabling targeted regulation of the tumor and tumor microenvironment (TME), which is crucial for the tumor cell–TME crosstalk that leads to tumor metastasis (8, 9). Tumor cell–secreted EVs express distinct integrins that specifically target organ-specific cells and promote metastatic organotropism (10). Nonetheless, the underlying mechanism of EV-induced LN metastasis remains largely unknown and requires further investigation.
Small ubiquitin-like modifier (SUMO) binding (termed SUMOylation), a crucial posttranslational modification, regulates intracellular transportation and signaling transduction by mediating protein stability and subcellular localization (11, 12). Recently, SUMOylation has been reported to play an essential role in EV packaging by mediating the recognition of molecules via the endosomal sorting complex required for transport (ESCRT) and facilitating their loading into a multivesicular body (MVB) (13, 14). SUMOylated hnRNPA2B1 selectively packages bioactive molecules into EVs via the recognition of specific miRNAs (15). SUMOylation mediates α‑synuclein encapsulation into EVs with the assistance of autophagy-related 5 (ATG5) (16), suggesting that SUMOylation is an important regulator of the sorting of molecules into EVs. However, the regulators and mechanisms triggering SUMOylation to induce EV packaging are unknown.
Long noncoding RNAs (lncRNAs), defined as a series of RNAs longer than 200 nt, play an important role in tumor progression (17). In the present study, we identified the lncRNA SNHG16, termed EV-mediated LN-associated transcript 1 (ELNAT1), which was upregulated in BCa-secreted EVs and associated positively with LN metastasis. We observed that overexpression of ELNAT1-mediated ubiquitin carrier protein 9 (UBC9) promoted the SUMOylation of lysine 113 on hnRNPA1 (hnRNPA1K113), thus enhancing ELNAT1 packaging into EVs, which epigenetically activated SOX18 transcription to induce tumor lymphangiogenesis and LN metastasis. Moreover, EV-mediated ELNAT1 showed markedly higher diagnostic efficiency for BCa LN metastasis than did urine cytology or FISH. These findings demonstrate a molecular mechanism whereby the EV-mediated ELNAT1/UBC9/SOX18 regulatory axis promotes SUMOylation-dependent lymphangiogenesis and LN metastasis of BCa and indicate that ELNAT1 may be a feasible therapeutic target for BCa LN metastasis.
Results
Identification of SUMOylation-associated oncogenic lncRNAs in BCa LN metastasis.
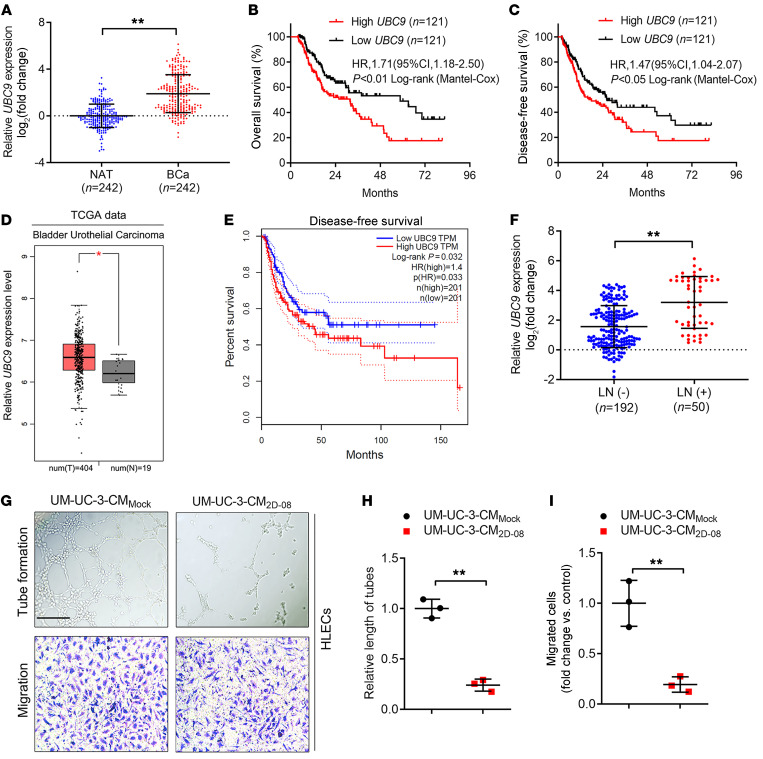
Given that SUMOylation is known to play a pivotal role in initiating or sustaining tumorigenesis (18), we demonstrated that several core small ubiquitin–related modifier (SUMO) pathway components, including UBC9, SUMO2, and SUMO3, were overexpressed and correlated with a poor prognosis in patients with BCa (Figure 1, A–C, and Supplemental Figure 1, A–D), consistent with the data from The Cancer Genome Atlas (TCGA) database (Figure 1, D and E, and Supplemental Figure 1, E–H). Moreover, we found a close correlation between UBC9 and SUMO3 overexpression and LN metastasis in a cohort of 242 patients with BCa (Figure 1F and Supplemental Figure 1I). Notably, IHC revealed that the expression of UBC9 and SUMO3 were positively associated with microlymphatic vessel density (MLD) in the intratumoral and peritumoral regions (Supplemental Figure 1, J and K). As shown in Figure 1, G–I, and Supplemental Figure 1, L–N, blocking SUMOylation by its specific inhibitor (2D-08) markedly impeded the promotional effect of BCa cells in inducing the tube formation and migration of human lymphatic endothelial cells (HLECs), indicating that SUMOylation might contribute to BCa lymphangiogenesis.
Figure 1. SUMOylation is involved in LN metastasis of BCa.
(A) qRT-PCR analysis of the expression of UBC9, one of the core components of SUMOylation, in BCa tissues and paired NATs from a cohort of 242 patients with BCa. The nonparametric Mann-Whitney U test was used to assess statistical significance. (B and C) Kaplan-Meier curves for OS and DFS of patients with BCa with low versus high UBC9 expression levels (the cutoff value is the median). (D) UBC9 expression levels in patients with BCa compared with expression levels in controls from TGCA database. (E) Kaplan-Meier survival analysis of patients with BCa according to UBC9 expression levels from TCGA database (cutoff value is the best cutoff). (F) qRT-PCR analysis of UBC9 expression in LN-positive and LN-negative BCa tissues (n = 242). The nonparametric Mann-Whitney U test was used to assess statistical significance. (G–I) Representative images (G) and quantification of tube formation (H) and Transwell migration (I) of HLECs incubated with culture media from the indicated UM-UC-3 cells treated with PBS or the SUMOylation inhibitor, 2D-08. Scale bar: 100 μm. A 2-tailed Student’s t test was used to assess statistical significance. Error bars showed the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
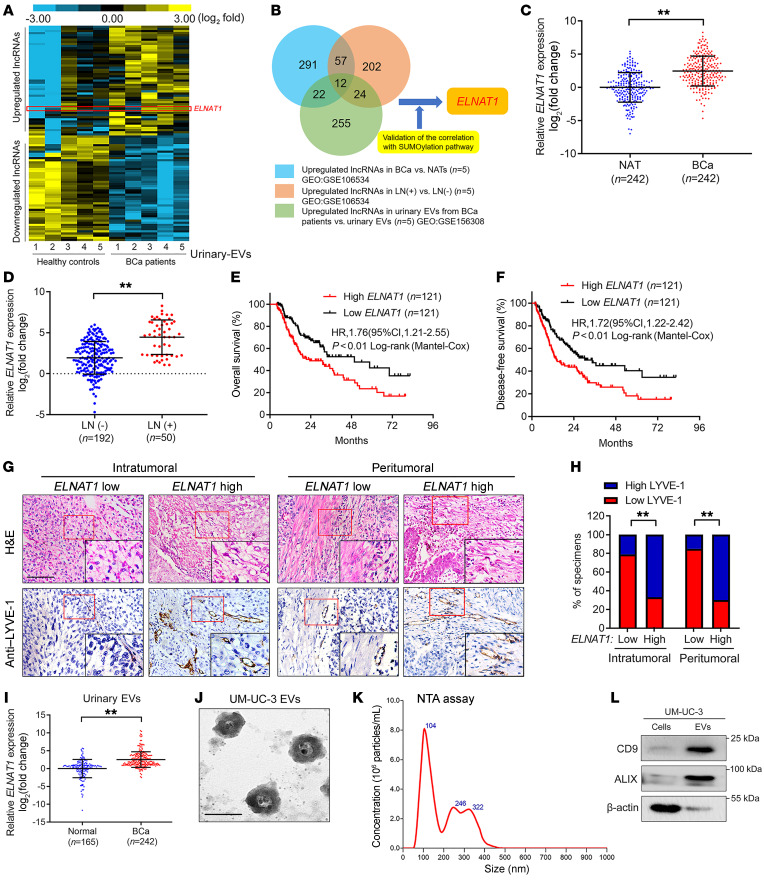
We previously demonstrated that EV-mediated lncRNA transportation is a crucial process that occurs through signal transduction between tumor cells and the TME (19). Thus, in the present study, we performed 3 rounds of sequencing to identify crucial SUMOylation-associated oncogenic, EV-mediated lncRNAs involved in BCa LN metastasis. First, we performed next-generation sequencing (NGS) to determine the global expression profiles of lncRNAs in urinary EVs from 5 patients with MIBC and 5 healthy volunteers (Gene Expression Omnibus [GEO] GSE156308). Supplemental Table 1 shows the characteristics of the participants. Statistical analysis revealed that 255 lncRNAs were upregulated by more than 2-fold in urinary EVs from patients with MIBC compared with urinary EVs from healthy volunteers (Figure 2A). Second, we intersected these lncRNAs with the results of NGS performed on 5 MIBC tissues and paired normal adjacent tissues (NATs) and in another 5 LN-positive BCa tissues paired with 5 LN-negative BCa tissues to further identify the EV-mediated lncRNAs required for LN metastasis of BCa (GEO GSE106534). From the 255 lncRNAs validated in the first round of experiments, we further selected 12 lncRNAs that were consistently upregulated in both urinary EVs of patients with MIBC and LN-positive BCa tissues (Figure 2B and Supplemental Table 2). Third, we further detected the relationship of these 12 lncRNAs with SUMOylation to show that ELNAT1 (SNHG16: ENSG00000163597, RefSeq accession number NR_038108.1) dramatically promoted the expression of SUMO pathway components (Supplemental Figure 1, O and P). We performed the 5′ and 3′ rapid amplification of cDNA ends (RACE) assay and identified the lncRNA ELNAT1 as a 2538 nt intergenic transcript encoded by a gene on human chromosome 17q25.1 (Supplemental Figure 2, A–E). Analysis of TCGA database revealed that ELNAT1 was consistently upregulated in tumor tissues compared with the normal control and was positively correlated with LN metastasis in human cancers (Supplemental Figure 3, A–G), suggesting that ELNAT1 is a vital oncogene involved in LN metastasis. Moreover, the higher expression levels of ELNAT1 were associated with a poor prognosis in various cancers (Supplemental Figure 3, H–L). Therefore, we selected ELNAT1 for further study.
Figure 2. EV-mediated ELNAT1 overexpression correlates with LN metastasis of BCa.
(A) Heatmap of the lncRNAs differentially expressed in urinary EVs from patients with BCa and healthy participants. (B) Schematic illustration of the screening of lncRNAs co-upregulated in urinary EVs from patients with BCa and LN-positive BCa tissues. (C and D) qRT-PCR analysis of ELNAT1 expression in BCa tissues and NATs (C) and in LN-positive and LN-negative BCa tissues (D) from a cohort of 242 patients with BCa. The nonparametric Mann-Whitney U test was used to assess statistical significance. (E and F) Kaplan-Meier curves for the OS and DFS of BCa patients with low versus high ELNAT1 expression (cutoff value is the median). (G and H) Representative IHC images and percentages for lymphatic vessel density in BCa tissues according to ELNAT1 expression levels. Scale bar: 50 μm. Original magnification, ×4 (insets in G). The χ2 test was used to assess statistical significance. (I) qRT-PCR analysis of ELNAT1 expression in urinary EVs from 242 patients with BCa and 165 healthy participants. The nonparametric Mann-Whitney U test was used to assess statistical significance. (J and K) TEM and NTA identified the characteristics of UM-UC-3-EVs. Scale bar: 100 nm. (L) Western blot analysis of EV markers in cell lysates or UM-UC-3-EVs. Error bars show the SD of 3 independent experiments. **P < 0.01.
ELNAT1 overexpression correlates with BCa LN metastasis.
To determine the clinical relevance of ELNAT1 in BCa, we performed quantitative real-time PCR (qRT-PCR) and ISH analyses to examine ELNAT1 expression in a larger clinical cohort of patients with BCa. As shown in Figure 2, C and D, and Supplemental Figure 4, A and B, ELNAT1 expression was dramatically higher in BCa tissues than in NATs and in patients with BCa with LN metastasis compared with patients without LN metastasis. We found that paired metastatic LNs had higher ELNAT1 expression than did BCa primary tumors (Supplemental Figure 4C), implying that ELNAT1 is a key component of LN metastatic cells. Moreover, ELNAT1 overexpression was associated with a poor prognosis for patients with BCa (Figure 2, E and F), which was consistent with the results of TCGA analysis (Supplemental Figure 4D). Notably, ISH assays indicated that ELNAT1 expression was markedly upregulated in LN-positive BCa tissues, slightly increased in LN‑negative BCa tissues, but rarely detected in NATs (Supplemental Figure 4, E and F). ELNAT1 expression was positively correlated with lymphatic vessel density in both intratumoral and peritumoral regions (Figure 2, G and H), suggesting that ELNAT1 is widely involved in the lymphangiogenesis of BCa. Collectively, our results demonstrate that ELNAT1 plays a vital role in LN metastasis of BCa.
ELNAT1 is overexpressed in BCa-secreted EVs.
Given that extracellular lncRNAs mainly have crucial biological functions in the mediation of cell-to-cell interactions and contribute to tumor LN metastasis (20), we further examined ELNAT1 expression in EVs isolated from urine samples from patients with BCa and healthy volunteers. The results showed that ELNAT1 was notably overexpressed in urinary EVs from patients with BCa (Figure 2I). Moreover, we isolated EVs from BCa cell culture medium. EVs with a double-layer membrane structure and a size distribution of 30–150 nm were characterized by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) (Figure 2, J and K). We detected high expression levels of the EV protein markers CD9 and ALIX (Figure 2L). As shown in Supplemental Figure 5A, we detected ELNAT1 overexpression in BCa cells and their corresponding EVs compared with expression in normal bladder epithelial cells (SV-HUC-1). Moreover, ELNAT1 expression was higher in BCa cell–secreted small EVs (~30–150 nm in size) compared with its intracellular expression, but was hardly detected in the soluble fraction or large EVs (~150–1000 nm in size) (Supplemental Figure 5, A–C), suggesting that ELNAT1 may exert its function preferentially under the encapsulation by small EVs instead of large EVs or the soluble fraction. Additionally, we found that ELNAT1 expression in UM-UC-3 and T24 cell–secreted EVs was markedly upregulated by transfection with ELNAT1-overexpressing plasmids and downregulated by knocking down ELNAT1 in UM-UC-3 and T24 cells (Supplemental Figure 5, D–G), indicating that alteration of cellular ELNAT1 expression affects EV-mediated ELNAT1 expression. Taken together, these findings demonstrated that ELNAT1 was enriched in BCa cell–secreted EVs.
EV-mediated ELNAT1 facilitates lymphangiogenesis in vitro.
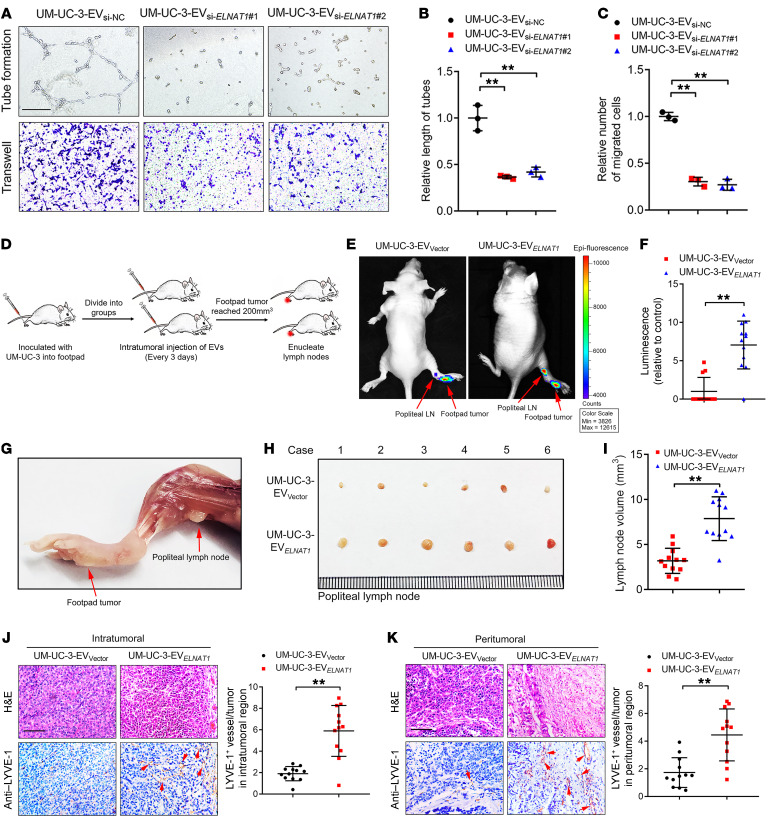
Tumor-associated lymphangiogenesis, which is an independent prognostic factor in BCa, is correlated with LN metastasis (21). To determine whether EV‑mediated ELNAT1 promotes lymphangiogenesis in vitro, we analyzed HLECs incubated with BCa cell–secreted EVs for tube formation and migration. As shown in Supplemental Figure 5, H–J, the tube formation and migratory ability of HLECs was markedly enhanced when treated with EVs secreted by UM-UC-3 and T24 cells, and slightly increased after incubation with EVs secreted by RT112 and UM-UC-1 cells, whereas no change was observed after treatment with RT4 cell–secreted EVs. Moreover, ELNAT1 knockdown abolished the ability of UM-UC-3 and T24 cell–secreted EVs (UM-UC-3-EVsi-ELNAT1 or T24-EVsi-ELNAT1) to induce tube formation and migration of HLECs (Figure 3, A–C, and Supplemental Figure 6, A–C). Conversely, EVs secreted by ELNAT1-overexpressing UM-UC-3 and T24 cells (UM-UC-3-EVELNAT1 or T24-EVELNAT1) notably enhanced the tube formation and migratory ability of HLECs compared with the control (Supplemental Figure 6, D–I). Taken together, these results demonstrate that EV-mediated ELNAT1 induced lymphangiogenesis in vitro.
Figure 3. EV-mediated ELNAT1 facilitates lymphangiogenesis and lymphatic metastasis of BCa in vitro and in vivo.
(A–C) Representative images (A) and quantification of tube formation (B) and Transwell migration (C) of HLECs treated with UM-UC-3-EVsi-NC, UM-UC-3-EVsi-ELNAT1#1, or UM-UC-3-EVsi-ELNAT1#2. Scale bar: 100 μm. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (D) Schematic representation for establishing the nude mouse model of popliteal LN metastasis. (E and F) Representative bioluminescence images and quantification of metastatic popliteal LNs from nude mice in the UM-UC-3-EVVector and UM-UC-3-EVELNAT1 groups (n = 12). The red arrows indicate the footpad tumor and metastatic popliteal LN. A 2-tailed Student’s t test was used to assess statistical significance. (G–I) Representative images of popliteal LNs and quantification of the LN volume for the UM-UC-3-EVVector and UM-UC-3-EVELNAT1 groups (n = 12). A 2-tailed Student’s t test was used to assess statistical significance. (J and K) Representative IHC images and quantification of lymphatic vessels in the intratumoral and peritumoral regions of footpad tumors (n = 12). Scale bars: 50 μm. A 2-tailed Student’s t test was used to determine statistical significance. Error bars show the SD of 3 independent experiments. **P < 0.01.
EV-mediated ELNAT1 promotes LN metastasis in vivo.
To further investigate the effects of EV-mediated ELNAT1 on LN metastasis in vivo, we constructed a popliteal LN metastasis model as described previously (22, 23). Mice were divided randomly into 2 groups (n = 12) and received intratumoral injections of EVs secreted by vector- or ELNAT1-transfected UM‑UC‑3 cells (UM-UC-3-EVVector or UM‑UC‑3‑EVELNAT1) every 3 days. The tumors and popliteal LNs were harvested when the primary tumor size reached 200 mm3 (Figure 3D). Strikingly, we observed that UM-UC-3-EVELNAT1 facilitated the metastasis of UM-UC-3 cells to popliteal LNs compared with the UM-UC-3-EVVector cell group, as determined by the in vivo imaging system (IVIS) (Figure 3, E and F). Moreover, we observed a larger popliteal LN volume and an increased LN metastasis rate in the UM-UC-3-EVELNAT1 group than in the UM-UC-3-EVVector group (Figure 3, G–I, and Supplemental Table 3). As lymphangiogenesis represents a pivotal step of LN metastasis (4), we further assessed the effect of EV-mediated ELNAT1 on lymphangiogenesis in vivo. Importantly, the UM-UC-3-EVELNAT1 group dramatically increased lymphatic vessel endothelial hyaluronan receptor 1–positive (LYVE-1–positive) lymphatic vessels in both the intratumoral and peritumoral regions of footpad tumors (Figure 3, J and K), indicating that EV-mediated ELNAT1 induced BCa lymphangiogenesis. Collectively, these results indicate that EV-mediated ELNAT1 facilitates the lymphangiogenesis and LN metastasis of BCa in vivo.
ELNAT1 directly interacts with hnRNPA1.
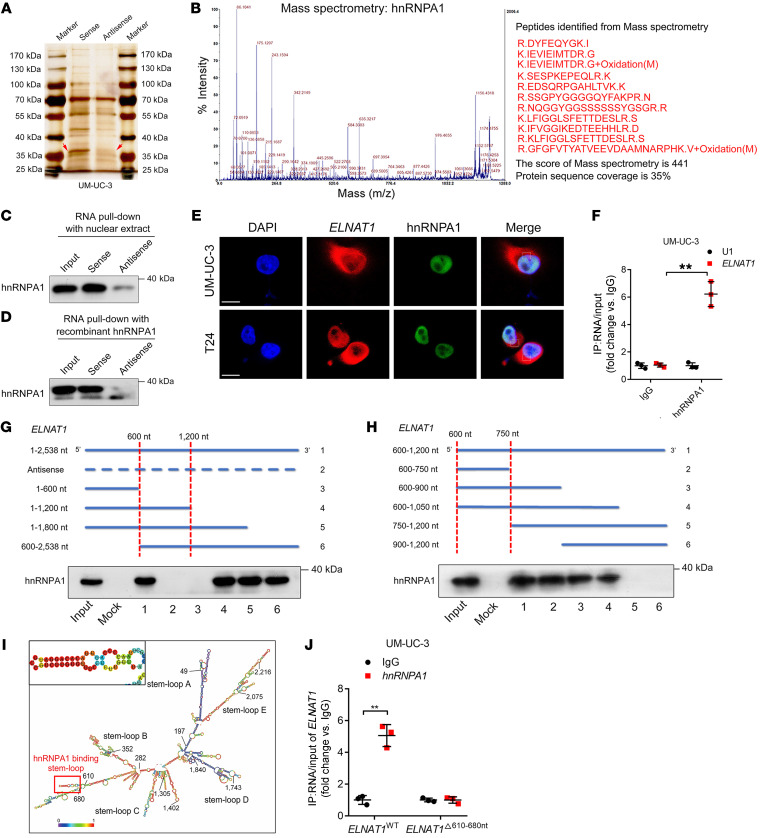
Since the molecular functions of lncRNAs are correlated with their subcellular localization (24), we conducted FISH and subcellular fractionation assays to detect the subcellular location of ELNAT1 and found that ELNAT1 was located in both the cytoplasm and nucleus of UM-UC-3 and T24 cells (Supplemental Figure 7, A and B). Moreover, in vitro RNA pull-down assays with biotinylated ELNAT1 and an antisense control revealed an evident band, with a molecular weight ranging from 35 to 40 kDa (Figure 4A). Mass spectrometry (MS) and Western blot analyses revealed that HnRNPA1 was the most abundant ELNAT1-interacting protein (Figure 4, B–D). Consistently, fluorescence staining confirmed the colocalization of ELNAT1 and hnRNPA1 in both UM-UC-3 and T24 cells (Figure 4E). RNA immunoprecipitation (RIP) assays showed ELNAT1 enrichment by endogenous hnRNPA1 (Figure 4F), further validating the interaction between ELNAT1 and hnRNPA1. In addition, sequential deletion experiments showed that the 600–750 nt region of ELNAT1 was required for hnRNPA1 interaction (Figure 4, G and H). Sequence analysis by POSTAR2 (25) predicted that a stem-loop structure in the 610–680 nt region of ELNAT1 was potentially recognized by hnRNPA1 (Figure 4I and Supplemental Figure 7C). Deletion of the 610–680 nt region in ELNAT1 impaired its enrichment by hnRNPA1 (Figure 4J), suggesting that these specific sequences are crucial to ELNAT1‑hnRNPA1 interaction.
Figure 4. ELNAT1 directly interacts with hnRNPA1.
(A) RNA pull-down assay of ELNAT1 in UM-UC-3 cells. (B) MS analysis of the proteins from the RNA pull-down assay. (C and D) RNA pull-down with nuclear extract (C) and Western blotting with purified recombinant hnRNPA1 (D) were performed to evaluate the interaction between ELNAT1 and hnRNPA1. (E) Immunofluorescence was performed to assess the colocalization of ELNAT1 and hnRNPA1 in UM-UC-3 and T24 cells. Scale bars: 5 μm. (F) RIP assay using anti-hnRNPA1 to assess the enrichment of ELNAT1 by hnRNPA1. IgG was used as a negative control; U1 was used as a nonspecific control. A 2-tailed Student’s t test was performed to determine statistical significance. (G and H) RNA pull-down assays using serial deletions of ELNAT1 were performed to evaluate the regions required for the binding of ELNAT1 and hnRNPA1. (I) Prediction for the stem-loop structures of hnRNPA1-binding sites in ELNAT1. (J) RIP assay after deletion of 610–680 nt of ELNAT1 in UM-UC-3 cells. A 2-tailed Student’s t test was used to assess statistical significance. Error bars show the SD of 3 independent experiments. **P < 0.01.
ELNAT1 upregulates the SUMOylation-related E2 conjugating enzyme UBC9.
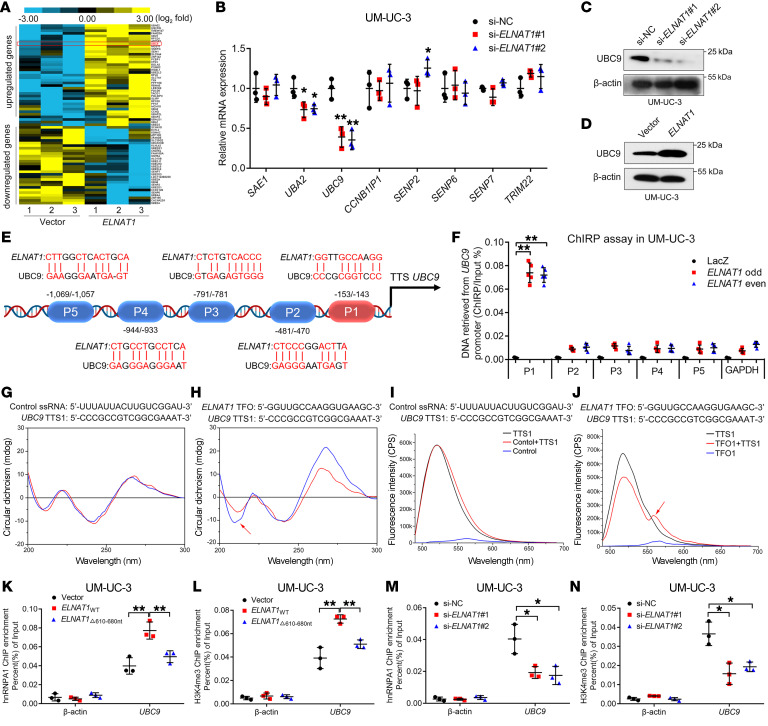
To explore the molecular mechanisms underlying ELNAT1-induced lymphatic metastasis in BCa, we profiled ELNAT1-overexpressing BCa cells and control cells using NGS (Figure 5A, GEO GSE156461). Since SUMOylation modification has been shown to regulate the recognition of specific RNAs and participate in the process of RNA sorting into EVs (15), we sought to identify the SUMOylation-associated target genes of ELNAT1. Among 925 genes that are regulated by ELNAT1 (P < 0.05, fold change >1.5), we found that UBC9 was the most markedly altered SUMOylation-related gene by qRT-PCR and Western blot analyses (Figure 5, B–D, Supplemental Figure 7D, and Supplemental Table 4). To further investigate the molecular mechanisms underlying ELNAT1-induced transcriptional activation of UBC9 expression in BCa, we performed luciferase assays using serial luciferase constructs containing truncated UBC9 promoter sequences (–2000 to +200 bp). Our results revealed that ELNAT1 overexpression increased the transcription activities of the –200 to +1 bp sequence in the UBC9 promoter (Supplemental Figure 7, E and F). Then, chromatin isolation by RNA purification (ChIRP) assays showed that ELNAT1 physiologically interacted with P1 (–153 to –143 bp) sequences in the UBC9 promoter region (Figure 5, E and F, and Supplemental Figure 7G). Moreover, LongTarget, a tool for predicting lncRNA-DNA–binding motifs (26), identified 5 potential triplex-forming oligonucleotides (TFOs) within ELNAT1 and the paired triplex target sites (TTS) in the UBC9 promoter. Each binding motif was subjected to circular dichroism (CD) spectroscopy and fluorescence resonance energy transfer (FRET) analysis (Supplemental Table 5). CD spectroscopy verified a notable positive peak at 270 to 280 nm and a negative peak at 210 nm in the ELNAT1/UBC9 TTS1 group, with a similar finding in the FENDRR/PITX2 positive control group (Figure 5, G and H, and Supplemental Figure 7H). FRET analysis revealed that the fluorescence intensity changed dramatically from 520 nm to 570–580 nm in the ELNAT1/UBC9 TTS1 group compared with fluorescence intensity the control single-stranded RNA/UBC9 (ssRNA/UBC9) TTS1 group, which was in accordance with the results in the FENDRR/PITX2 positive control group (Figure 5, I and J, and Supplemental Figure 7I). Furthermore, we sought to determine whether hnRNPA1 contributed to ELNAT1-induced transcriptional activation of UBC9 by regulating histone methylation at the UBC9 promoter. ChIP analysis showed that overexpression of ELNAT1 increased the enrichment of hnRNPA1 and H3K4me3 at the UBC9 promoter and that enrichment was inhibited by deletion of the ELNAT1-binding site of hnRNPA1 (Figure 5, K and L, and Supplemental Figure 7, J and K). Meanwhile, ELNAT1 silencing dramatically reduced hnRNPA1 occupancy and H3K4me3 methylation at the UBC9 promoter in UM-UC-3 and T24 cells (Figure 5, M and N, and Supplemental Figure 7, L and M), indicating that ELNAT1 regulated the transcription of UBC9 by forming a triplex structure with its promoter sequence and inducing hnRNPA1-associated H3K4me3 modification.
Figure 5. ELNAT1 forms a DNA-RNA triplex with the UBC9 promoter to enhance H3K4me3 modification by recruiting hnRNPA1.
(A) Heatmap of differentially expressed genes after overexpression of ELNAT1 in the indicated BCa cells. (B) qRT-PCR analysis of the SUMOylation-related genes after ELNAT1 knockdown in UM-UC-3 cells. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (C and D) Western blot analysis of UBC9 expression in UM-UC-3 cells after silencing (C) or overexpressing (D) ELNAT1. siNC, negative control. (E) Schematic presentation of the predicted ELNAT1-binding sites in the UBC9 promoter. (F) ChIRP analysis of ELNAT1-associated chromatin in UM-UC-3 cells. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. P1, –143 to –153 bp in the UBC9 promoter; P2, –470 to –481 bp in the UBC9 promoter; P3, –781 to –791 bp in the UBC9 promoter; P4, –933 to –944 bp in the UBC9 promoter; P5, –1057 to –1069 bp in the UBC9 promoter. (G and H) CD spectrum of TFOs in ELNAT1 with TTSs in the UBC9 promoter. The control ssRNA with a TTS in the UBC9 promoter was used as a negative control. (I and J) FRET analysis of TFOs in ELNAT1 with TTSs in the UBC9 promoter. The control ssRNA with a TTS in the UBC9 promoter was used as a negative control. (K–N) ChIP-qPCR analysis of hnRNPA1 occupancy and H3K4me3 status in the UBC9 promoter in the indicated UM-UC-3 cells. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. Error bars showed the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
UBC9-induced SUMOylation of hnRNPA1 packages ELNAT1 into EVs.
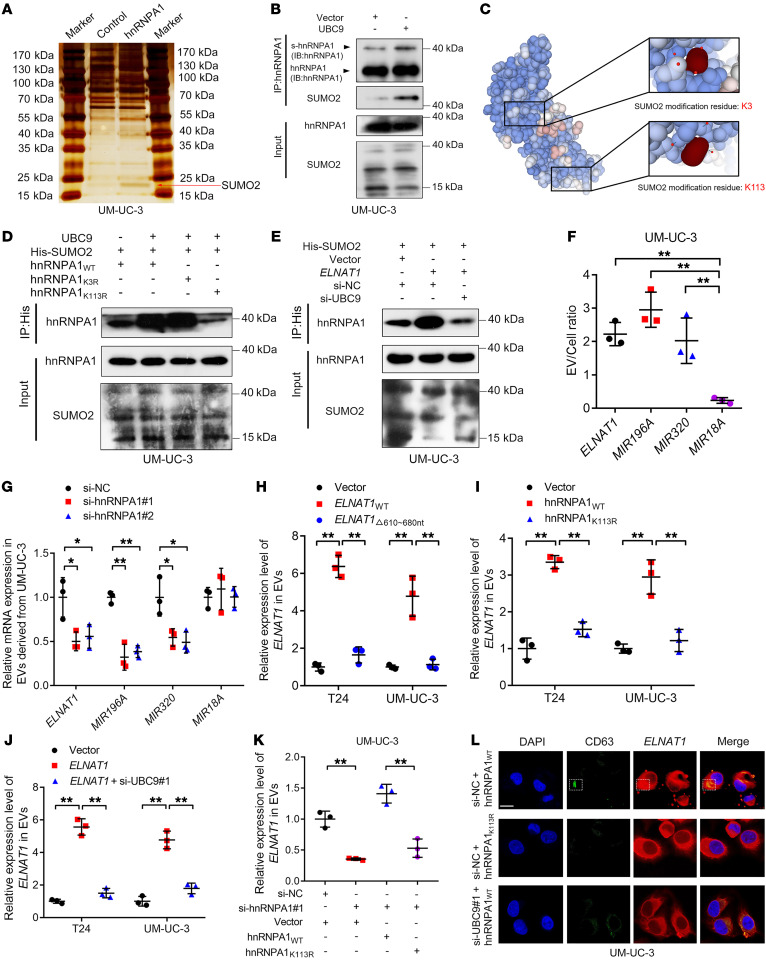
It has been well established that UBC9 can catalyze the SUMOylation of target proteins to regulate their interaction with biomolecules and cellular transportation (15, 27). The observation that ELNAT1 directly interacts with hnRNPA1 to upregulate UBC9 expression prompted us to hypothesize that UBC9 overexpression might stimulate the SUMOylation of hnRNPA1 to promote the packaging of ELNAT1 into EVs. To confirm this hypothesis, we performed a co‑IP assay and observed that an obvious 15–25 kDa band was specifically enriched by hnRNPA1 (Figure 6A), which was identified as SUMO2 by MS (Supplemental Figure 8, A and B). Moreover, IP assays revealed that UBC9 overexpression enhanced the SUMO2 conjunction of hnRNPA1, suggesting that SUMOylation of hnRNPA1 was induced by UBC9 (Figure 6B). To evaluate the specific modification sites of SUMOylation in hnRNPA1, we used GPS-SUMO (28), a tool for SUMOylation site analysis, to obtain 2 potential SUMO2 conjunction residues of hnRNPA1 — lysine 3 (K3) and lysine 113 (K113) — which were replaced with arginine (R) (hnRNPA1K3R, hnRNPA1K113R; Figure 6C and Supplemental Figure 8, C–F) and subjected to co-IP assays to show that the hnRNPA1K113R substitution, but not hnRNPA1K3R, abolished the SUMOylation of hnRNPA1 (Figure 6D). Moreover, we demonstrated that ELNAT1 overexpression upregulated the SUMOylation of hnRNPA1K113, which was abolished by knocking down UBC9 (Figure 6E), suggesting that ELNAT1-induced UBC9 overexpression promotes the SUMOylation of hnRNPA1 at the K113 residue.
Figure 6. ELNAT1 is packaged into EVs by UBC9-induced SUMOylation of hnRNPA1.
(A) Co-IP assay using anti-hnRNPA1 in UM-UC-3 cells. Red arrow indicates SUMO2. (B) Co-IP assay using anti-hnRNPA1 was performed to assess the conjunction of SUMO2 on hnRNPA1 in UM-UC-3 cells after UBC9 overexpression. (C) Schematic representation showing the predicted SUMO2 conjunct residues on hnRNPA1. (D) A co-IP assay using anti-His was performed to evaluate the conjunction of His-labeled SUMO2 on hnRNPA1 in UM-UC-3 cells after hnRNPA1K3R or hnRNPA1K113R mutation. (E) An IP assay was performed to evaluate His-labeled SUMO2 conjunction on hnRNPA1 in ELNAT1-overexpressing UM-UC-3 cells after UBC9 knockdown. (F) EV/cell ratio of RNAs in UM-UC-3 cells. (G) qRT-PCR was performed to analyze RNA expression in EVs secreted by UM-UC-3 cells after hnRNPA1 knockdown. (H) Assessment of ELNAT1 expression in BCa cell–secreted EVs after deletion of 610–680 nt of ELNAT1. (I) Analysis of ELNAT1 expression in BCa cell–secreted EVs after hnRNPA1K113R mutation. (J) Evaluation of ELNAT1 expression in EVs secreted by ELNAT1-overexpressing BCa cells after UBC9 knockdown. (K) ELNAT1 expression in EVs secreted by hnRNPA1-knockdown UM-UC-3 cells after hnRNPA1WT or hnRNPA1k113R overexpression was assessed by qRT-PCR. (L) Representative immunofluorescence images showing the accumulation of ELNAT1 into CD63-indicated MVBs in UM-UC-3 cells after hnRNPA1K113R mutation or UBC9 knockdown. Scale bar: 5 μm. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance in F–K. Error bars showed the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
Next, we explored whether ELNAT1 was packaged into BCa cell–secreted EVs by SUMOylated hnRNPA1. First, we found that ELNAT1 exhibited an EV-to-cell ratio comparable to that of miR-196a and miR-320 (Figure 6F and Supplemental Figure 9A), which were previously reported to be loaded into EVs by hnRNPA1 (29, 30). We found that HnRNPA1 silencing markedly inhibited ELNAT1 enrichment in EVs secreted by BCa cells (Figure 6G and Supplemental Figure 9B), suggesting that sorting of ELNAT1 into EVs depends on hnRNPA1. Furthermore, truncated ELNAT1, which has deletion of the 610–680 nt sequences that contain the hnRNPA1-binding sites, was predominantly retained in BCa cells rather than secreted into EVs (Figure 6H and Supplemental Figure 9C), confirming that ELNAT1 is loaded into EVs through interaction with hnRNPA1.
Since we demonstrated that hnRNPA1 was SUMOylated in BCa cells, we further evaluated whether SUMOylation contributed to hnRNPA1-mediated EV encapsulation of ELNAT1. The SUMOylation-defective mutant of hnRNPA1 or UBC9 inhibition in BCa cells notably abolished the ELNAT1 enrichment in ELNAT1-transduced BCa cell–secreted EVs (Figure 6, I and J, and Supplemental Figure 9D). HnRNPA1K113R transfection failed to restore the downregulation of EV-mediated ELNAT1 after silencing of hnRNPA1 compared with silencing of hnRNPA1WT (Figure 6K and Supplemental Figure 9E). Importantly, confocal microscopy revealed that ELNAT1 accumulation into CD63-indicated MVBs, the precursors of EVs, was markedly downregulated after UBC9 silencing or hnRNPA1K113R mutation (Figure 6L), indicating that the sorting of ELNAT1 into EVs was regulated by the SUMOylation of hnRNPA1. Additionally, we further sought to determine whether the lysine 113 mutation of hnRNPA1 affects its interaction with ELNAT1, thus impairing ELNAT1 packaging into EVs. The results showed that ELNAT1 remained directly bound to hnRNPA1 after mutating lysine 113 on hnRNPA1 (Supplemental Figure 9, F–H), indicating that the mutation of lysine 113 on hnRNPA1 had no affect on its interaction with ELNAT1. Together, these results demonstrate that ELNAT1 was packaged into BCa cell–secreted EVs by UBC9-induced SUMOylation of hnRNPA1.
EV-mediated ELNAT1 is delivered to HLECs to induce lymphangiogenesis.
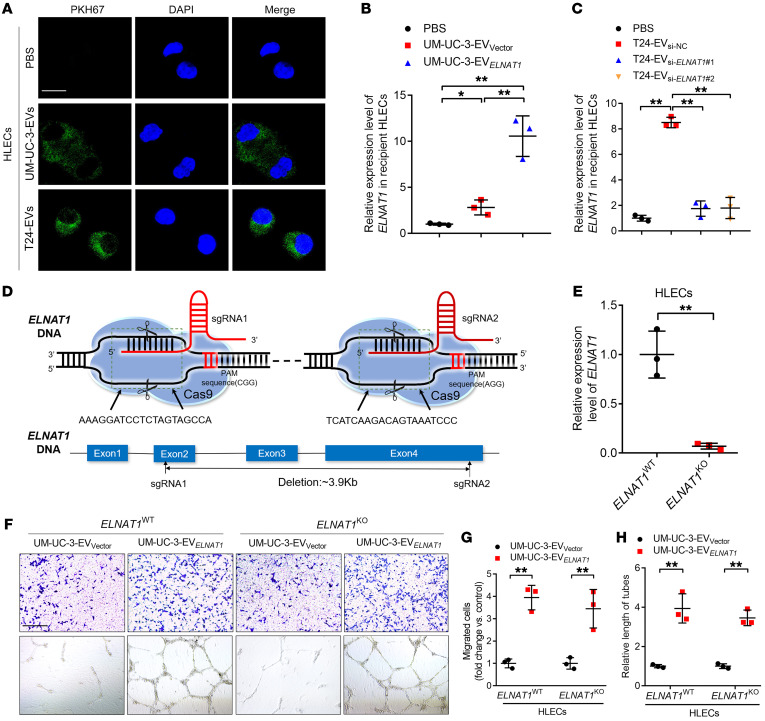
Since our results indicated that EV-mediated ELNAT1 promoted lymphangiogenesis, we evaluated the internalization of EV-mediated ELNAT1 by HLECs. Confocal microscopy revealed punctate fluorescence intensity in HLECs incubated with PKH67-labeled EVs (Figure 7A), indicating internalization of BCa-secreted EVs by HLECs. Moreover, incubation with UM-UC-3-EVELNAT1 and T24-EVELNAT1 markedly upregulated ELNAT1 expression in HLECs, whereas downregulated ELNAT1 expression in EVs secreted by UM-UC-3 and T24 cells impaired the ability of the EVs to induce ELNAT1 overexpression in HLECs (Figure 7, B and C, and Supplemental Figure 9, I and J).
Figure 7. EV-mediated ELNAT1 is internalized by HLECs to induce lymphangiogenesis.
(A) Representative immunofluorescence images of PBS- or PKH67-labeled (green), EV-treated HLECs. Scale bar: 5 μm. (B and C) qRT-PCR analysis of ELNAT1 expression in HLECs treated with PBS, UM-UC-3-EVVector, UM-UC-3-EVELNAT1, or T24-EVsi-NC, T24-EVsi-ELNAT1#1, T24-EVsi-ELNAT1#2. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (D) Schematic illustration of ELNAT1 deletion in HLECs using the CRISPR/Cas9 approach. (E) qRT-PCR analysis validated the successful KO of ELNAT1 in HLECs. A 2-tailed Student’s t test was used to assess statistical significance. (F–H) Representative images (F) and quantification of Transwell migration (G) and tube formation (H) of ELNAT1WT and ELNAT1KO HLECs treated with UM-UC-3-EVVector or UM-UC-3-EVELNAT1. Scale bar: 100 μm. A 2-tailed Student’s t test was performed to assess statistical significance. Error bars showed the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
To exclude the possibility that the lymphangiogenesis was induced by activation of endogenous ELNAT1 in HLECs, ELNAT1-KO HLECs (HLECsELNAT1-KO) were constructed (Figure 7, D and E). We studied the effects of EV-mediated ELNAT1 on both HLECsELNAT1-WT and HLECsELNAT1-KO. Consistent with HLECsELNAT1-WT, we observed that the tube formation and migratory ability of HLECsELNAT1-KO was enhanced by EV-mediated ELNAT1 overexpression, while knocking down ELNAT1 inhibited the ability of BCa cell–secreted EVs to induce tube formation and migration of HLECsELNAT1-KO (Figure 7, F–H, and Supplemental Figure 10, A–I), suggesting that BCa cell–secreted EVs promoted lymphangiogenesis by transporting EV-mediated ELNAT1 rather than transcriptionally activating endogenous ELNAT1. Taken together, these results demonstrate that EV-mediated ELNAT1 was internalized by HLECs to induce BCa lymphangiogenesis.
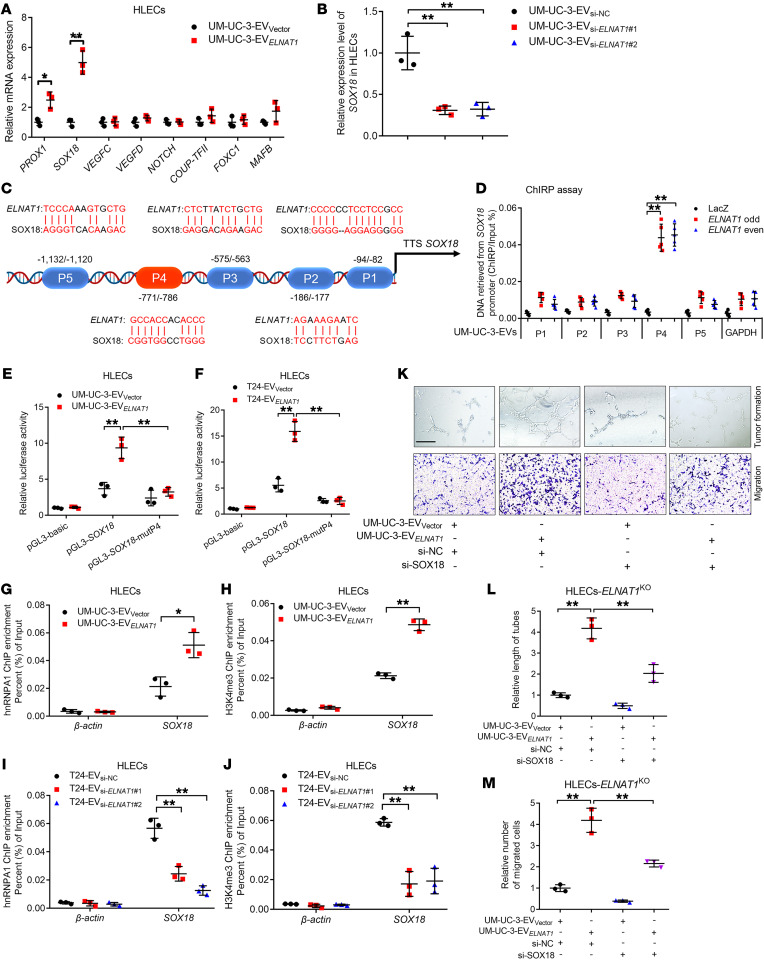
EV-mediated ELNAT1 upregulates SOX18 expression in HLECs.
Next, we analyzed the expression of lymphangiogenesis-related genes in HLECs treated with EV-mediated ELNAT1. Our results showed that SRY-box transcription factor 18 (SOX18) was the most obvious gene that positively associated with EV-mediated ELNAT1 expression (Figure 8, A and B, and Supplemental Figure 11, A and B). EV-mediated ELNAT1 downregulation markedly decreased SOX18 expression, whereas overexpressing EV-mediated ELNAT1 promoted SOX18 expression in HLECs compared with the control (Supplemental Figure 11, C–F). It has been proposed that SOX18 represents a crucial regulator for the budding of new lymphatic vessels through the induction of genes that contribute to the phenotype of lymphatic vessels, including vascular endothelial growth factor C (VEGF-C) and prospero-related homeobox transcription factor 1 (PROX1) (31, 32). To further explore the mechanisms of EV-mediated ELNAT1–upregulated SOX18 expression in HLECs, a series of truncated SOX18 promoters ranging from –2000 to +200 bp relative to the transcriptional start site were cloned into the luciferase reporter genes. As shown in Supplemental Figure 11, G and H, luciferase assays revealed that EV-mediated ELNAT1 enhanced transcriptional activity when the –800 to –400 bp region of the SOX18 promoter was introduced into HLECs. Moreover, ChIRP assays demonstrated that EV-mediated ELNAT1 directly interacted with –771 to –786 bp of the SOX18 promoter (refers to SOX18-P4) in HLECs (Figure 8, C and D, and Supplemental Figure 11I). Mutation of the SOX18-P4 region reduced the luciferase activity induced by EV-mediated ELNAT1 (Figure 8, E and F), suggesting that SOX18-P4 was crucial for the EV-mediated ELNAT1–induced SOX18 upregulation in HLECs. Furthermore, we found that the enrichment of hnRNPA1 and H3K4me3 at the SOX18 promoter was markedly associated with EV-mediated ELNAT1 expression (Figure 8, G–J, and Supplemental Figure 12, A–D), indicating that EV-mediated ELNAT1 increased hnRNPA1-induced H3K4me3 levels at the SOX18 promoter. Moreover, we assessed whether SOX18 was indispensable for EV-mediated ELNAT1-induced lymphangiogenesis. EV-mediated ELNAT1 overexpression enhanced the tube formation and migratory ability of HLECs, while downregulating SOX18-impaired, EV-mediated ELNAT1–induced lymphangiogenesis (Figure 8, K–M, and Supplemental Figure 12, E-G), indicating that SOX18 was required for EV-mediated ELNAT1 to drive BCa lymphangiogenesis in vitro. Together, these results revealed that EV-mediated ELNAT1 promoted BCa lymphangiogenesis via transcriptional upregulation of SOX18 expression in HLECs.
Figure 8. EV-mediated ELNAT1 upregulates SOX18 expression in HLECs.
(A) qRT-PCR analysis of lymphangiogenesis-related gene expression in UM-UC-3-EVVector– or UM-UC-3-EVELNAT1–treated HLECs. A 2-tailed Student’s t test was used to assess statistical significance. (B) qRT-PCR analysis of SOX18 expression in HLECs treated with UM-UC-3-EVsi-NC, UM-UC-3-EVsi-ELNAT1#1, or UM-UC-3-EVsi-ELNAT1#2. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (C) Schematic representation of the predicted EV-mediated ELNAT1-binding sites in the SOX18 promoter in HLECs. (D) ChIRP analysis of EV-mediated ELNAT1-associated chromatin in UM-UC-3-EVs–treated HLECs. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (E and F) Luciferase assays were performed to evaluate the WT or ELNAT1-binding site–mutated SOX18 promoter in HLECs treated with UM-UC-3-EVVector, UM-UC-3-EVELNAT1, T24-EVVector, or T24-EVELNAT1. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (G–J) ChIP-qPCR analysis of hnRNPA1 occupancy and H3K4me3 status in the SOX18 promoter in HLECs treated with UM-UC-3-EVVector, UM-UC-3-EVELNAT1, T24-EVsi-NC, T24-EVsi-ELNAT1#1, or T24-EVsi-ELNAT1#2. A 2-tailed Student’s t test or 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (K–M) Representative images (K) and quantification of tube formation (L) and Transwell migration (M) of UM-UC-3-EVVector– or UM-UC-3-EVELNAT1–treated ELNAT1KO HLECs transfected with si-NC or si-SOX18. Scale bar: 100 μm. A 1-way ANOVA followed by Dunnett’s test was used to determine statistical significance. Error bars show the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
Blocking SUMOylation suppresses EV-mediated ELNAT1–induced LN metastasis.
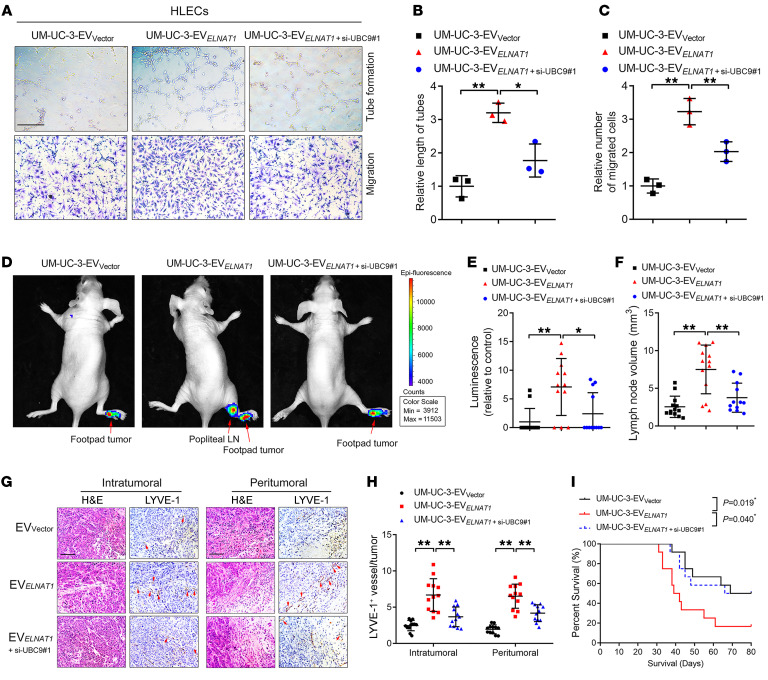
EV-mediated ELNAT1 functions in a SUMOylation-dependent manner; therefore, we sought to determine whether blocking UBC9-induced SUMOylation could inhibit EV-mediated ELNAT1–induced lymphangiogenesis and LN metastasis in BCa. We demonstrated that ELNAT1 overexpression notably promoted BCa cell–secreted EVs to induce lymphangiogenesis in vitro, whereas silencing UBC9 reversed this effect (Figure 9, A–C, and Supplemental Figure 12, H–J), indicating that UBC9-induced SUMOylation contributed to EV-mediated ELNAT1–induced lymphangiogenesis. Importantly, IVIS demonstrated that UM-UC-3-EVELNAT1 enhanced popliteal LN metastasis in vivo, whereas silencing UBC9 inhibited this effect (Figure 9, D and E). The UM-UC-3-EVELNAT1+si-UBC9#1 group had a lower volume of popliteal LNs than did the UM-UC-3-EVELNAT1 group (Figure 9F). Moreover, UM-UC-3-EVELNAT1 increased the number of lymphatic vessels in footpad tumors of mice compared with the UM‑UC‑3‑EVVector group, while downregulating UBC9 expression resulted in an incremental decrease in the number of EV-mediated ELNAT1–induced lymphatic vessels (Figure 9, G and H), suggesting that blocking UBC9 impairs the effect of EV-mediated ELNAT1 on lymphangiogenesis in vivo. Moreover, we observed a reduction in the rate of LN metastasis in the UM-UC-3-EVELNAT1+si-UBC9#1 group compared with metastasis rates in the UM-UC-3-EVELNAT1 group (Supplemental Table 6), which was accompanied by longer survival times (Figure 9I). Taken together, these results indicate that inhibition of UBC9-induced SUMOylation suppresses EV-mediated ELNAT1–induced lymphangiogenesis and LN metastasis in BCa.
Figure 9. Blocking SUMOylation suppresses EV-mediated ELNAT1–induced LN metastasis.
(A–C) Representative images (A) and quantification of tube formation (B) and Transwell migration (C) of HLECs treated with EVs secreted by control or ELNAT1-overexpressing UM-UC-3 cells transfected with si-NC or si-UBC9#1. Scale bar: 100 μm. A 1-way ANOVA followed by Dunnett’s test was used to assess the statistical significance. (D and E) Representative bioluminescence images (D) and quantification (E) of popliteal metastatic LNs from nude mice treated with EVs secreted by control or ELNAT1-overexpressing UM-UC-3 cells transfected with si-NC or si-UBC9#1 (n = 12). The red arrows indicate footpad tumors and metastatic LNs. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (F) Quantification of popliteal LN volume (n = 12). A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (G and H) Representative IHC images (G) and quantification (H) of lymphatic vessels in footpad tumors (n = 12). Scale bars: 50 μm. A 1-way ANOVA followed by Dunnett’s test was used to assess statistical significance. (I) Kaplan-Meier curves show the survival of nude mice treated with EVs secreted by control or ELNAT1-overexpressing UM-UC-3 cells transfected with si-NC or si-UBC9#1. Error bars show the SD of 3 independent experiments. *P < 0.05 and **P < 0.01.
Clinical relevance of EV-mediated ELNAT1 in patients with BCa.
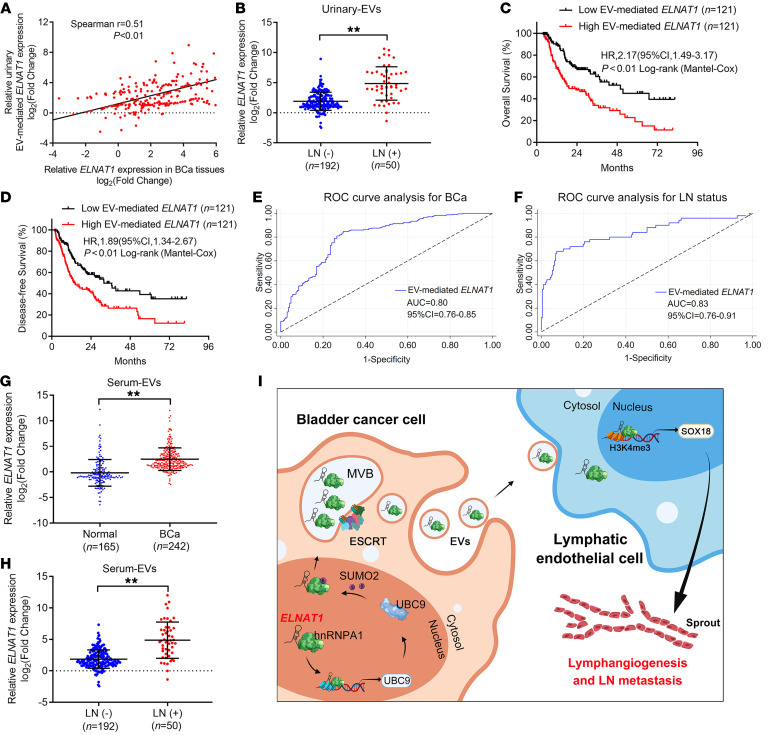
EV-mediated lncRNAs are considered promising early diagnostic biomarkers and potential therapeutic targets in BCa (33, 34). Thus, it is important to determine the clinical relevance of EV-mediated ELNAT1 in LN metastasis of BCa. First, we found a positive correlation of ELNAT1 expression between urinary-EVs from patients with BCa and paired BCa tissues, implicating EV-mediated ELNAT1 as an essential participant in the regulation of ELNAT1 in BCa (Figure 10A). Since we showed that ELNAT1 was overexpressed in LN-positive BCa, we next explored whether EV-mediated ELNAT1 is clinically relevant to BCa LN metastasis. Strikingly, we found that urinary EV–mediated ELNAT1 overexpression positively correlated with LN metastasis of BCa (Figure 10B, Supplemental Figure 12K, and Supplemental Table 7). Moreover, Kaplan-Meier analysis showed that BCa patients with higher EV-mediated ELNAT1 expression had shorter overall survival (OS) and disease-free survival (DFS) rates (Figure 10, C and D). Univariate and multivariate analysis revealed that EV-mediated ELNAT1 was an independent factor for the poor prognosis of patients with BCa, indicating its potential role as a therapeutic target for BCa (Supplemental Tables 8 and 9). Additionally, we performed receiver operating characteristic (ROC) analysis to assess the diagnostic performance of EV-mediated ELNAT1 and compared the results with urine cytology and FISH, the standard noninvasive diagnostic interventions for BCa (35, 36). ROC analysis showed that urinary EV–mediated ELNAT1 could effectively distinguish patients with BCa from healthy controls (AUC: 0.80; 95% CI: 0.76–0.80; Figure 10E and Supplemental Table 10). Remarkably, we found that urinary EV–mediated ELNAT1 was highly accurate in the diagnosis of BCa LN metastasis (AUC: 0.83; 95% CI: 0.76–0.91) compared with that of urine cytology or FISH (Figure 10F and Supplemental Table 10). Furthermore, we also showed that 63% of patients with BCa evaluated as LN negative by CT were correctly predicted as being LN positive by the detection of urinary EV–mediated ELNAT1 (Supplemental Table 11), suggesting that urinary EV–mediated ELNAT1 may be a better alternative to diagnose BCa LN metastasis. We also consistently observed higher levels of EV-mediated ELNAT1 expression in serum from patients with BCa than in serum from heathy controls (Figure 10G). EV-mediated ELNAT1 expression was upregulated in serum from BCa patients with LN metastasis compared with serum from those without LN metastasis (Figure 10H). Taken together, our findings reveal that EV-mediated ELNAT1 plays an essential role in LN-metastatic BCa (Figure 10I).
Figure 10. EV-mediated ELNAT1 is associated with LN metastasis of BCa.
(A) Correlation analysis of ELNAT1 expression in tumor tissues and urinary EVs from a cohort of 242 patients with BCa. (B) qRT-PCR analysis of ELNAT1 expression in urinary EVs obtained from a cohort of 242 patients with BCa with or without LN metastasis. The nonparametric Mann-Whitney U test was used to assess statistical significance. (C and D) Kaplan-Meier survival analysis of patients with BCa according to EV-mediated ELNAT1 expression levels (cutoff value is the median). (E and F) ROC curves for the efficiency of urinary EV–mediated ELNAT1 in diagnosing BCa and LN metastasis. (G and H) qRT-PCR analysis of ELNAT1 expression in serum EVs obtained from 242 patients with BCa and 165 healthy volunteers (G) and patients with LN-positive or LN-negative BCa (H). The nonparametric Mann-Whitney U test was used to assess statistical significance. (I) Proposed model of how BCa-secreted EV-mediated ELNAT1 induces the hnRNPA1/UBC9/SOX18 axis to promote lymphangiogenesis and LN metastasis of BCa. Error bars show the SD of 3 independent experiments. **P < 0.01.
Discussion
SUMOylation regulates the sorting of RNAs into EVs to serve as an essential mediator in intercellular signal transduction, thus contributing to the crosstalk between tumor cells and the TME (37, 38). Nevertheless, the regulatory role of SUMOylation in packaging specific lncRNAs into EVs and its association with tumor LN metastasis remain largely unknown. In this study, we showed that the lncRNA ELNAT1 mediated the SUMOylation of hnRNPA1, thereby regulating the sorting of ELNAT1 into EVs and correlated with BCa LN metastasis. ELNAT1 upregulated UBC9 expression by binding to the –153 to –143 bp region of the UBC9 promoter and recruiting hnRNPA1 to induce H3K4me3 modification, which in turn caused the SUMOylation of hnRNPA1 at the K113 residue to package ELNAT1 into EVs. Subsequently, EV-mediated ELNAT1 directly formed a DNA-RNA triplex with the SOX18 promoter and increased hnRNPA1-induced H3K4me3 modification, thereby transcriptionally activating SOX18 expression and facilitating BCa LN metastasis. Our findings highlight the regulatory mechanism by which EV-mediated ELNAT1 induced BCa LN metastasis in a SUMOylation-dependent manner and identify EV-mediated ELNAT1 as a potential therapeutic target for BCa.
Lymphangiogenesis induced by SOX18 transcriptional activation has been considered the most essential step in tumor LN metastasis (5). SOX18 is the earliest molecular hallmark of endothelial cells during embryonic development and plays an essential role in the formation of new lymphatic vessels (31, 32). Although the decisive role of SOX18 in lymphangiogenesis is well characterized, the mechanisms of EV-mediated SOX18 expression in HLECs are unclear. Here, we demonstrated a regulatory mechanism by which EV-mediated ELNAT1 activated SOX18 transcription through direct binding to the SOX18 promoter and recruited hnRNPA1 to induce H3K4me3 modification. Downregulation of SOX18 reversed EV-mediated ELNAT1–induced lymphangiogenesis and LN metastasis of BCa. These findings reveal the crucial mechanism by which EV-mediated ELNAT1 promotes LN metastasis and identify EV-mediated ELNAT1 as a feasible therapeutic target in BCa.
SUMOylation regulates the function of proteins by influencing their subcellular localization, protein interaction, and transcriptional activity (39, 40). UBC9 is the unique E2 ligase of SUMOylation that catalyzes the conjunction of SUMOs to lysine residues of the substrates (27, 41). However, the mechanism governing UBC9-mediated SUMOylation is largely unknown. In this study, we demonstrated that EV-mediated ELNAT1 formed a direct DNA-RNA triplex with the UBC9 promoter to promote hnRNPA1-induced H3K4me3 modification and further induced SUMOylation of hnRNPA1 at the K113 residue. Moreover, EV-mediated ELNAT1-induced SUMOylation dramatically promoted the lymphangiogenesis and LN metastasis of BCa both in vitro and in vivo, suggesting that EV-mediated ELNAT1 functions as a crucial regulator of SUMOylation-induced LN metastasis of BCa. Recent studies have shown that targeting the SUMOylation pathway is an effective intervention for the treatment of various cancers (42). Anacardic acid was found to suppress the SUMOylation pathway by targeting the SUMO-activating enzyme E1 and showed great efficiency in the treatment of B cell lymphoma (43). Additionally, melatonin enhanced the sensitivity of brain cells to chemotherapy through disturbance of the SUMOylation-mediated nuclear translocation of Nestin (44). Therefore, our study demonstrating the essential role of EV-mediated ELNAT1 in regulating the SUMOylation pathway points to EV-mediated ELNAT1 as a potential therapeutic target for BCa LN metastasis.
Urine cytology is the current, albeit insufficiently sensitive, standard intervention for the noninvasive diagnosis of BCa, which exhibits dissatisfied sensitivity in the diagnosis of BCa (35). More recently, FISH, which is a more sensitive diagnostic tool, partly compensates for the deficiency of urine cytology testing but has been limited by its low specificity (45). And recently detected molecules in EVs obtained from body fluids, including urine, have been widely recognized as promising biomarkers with high efficacy in tumor diagnosis (34, 46). In the present study, we found that urinary EV–mediated ELNAT1 offered slightly improved specificity in the diagnosis of BCa compared with FISH but showed no obvious difference in sensitivity, given the higher sensitivity of FISH in the diagnosis of high-grade BCa. Interestingly, our use of EV-mediated ELNAT1 to distinguish LN‑positive from LN-negative BCa resulted in satisfactory sensitivity and specificity, which markedly improved accuracy in diagnosing LN-metastatic BCa compared with FISH and urine cytology. Currently, CT and MRI are the most commonly recommended approaches for preoperative nodal staging in patients with BCa based on the size and shape of the LNs, but these technologies have limited accuracy in diagnosing microscopic metastasis (47). Here, we found that 63% of patients with LN-negative BCa by preoperative imaging were correctly predicted as having LN-positive BCa by the detection of urinary EV–mediated ELNAT1. Our findings highlight the clinical relevance of urinary EV–mediated ELNAT1 detection in assessing LN status and support the idea that urinary EV–mediated ELNAT1 may represent a better alternative for the diagnosis of BCa LN metastasis.
In summary, we demonstrated that EV-mediated ELNAT1 promoted lymphangiogenesis and LN metastasis of BCa in a SUMOylation-dependent manner. A full elucidation of the precise mechanism of EV-mediated ELNAT1 in activating the hnRNPA1/UBC9/SOX18 axis to induce BCa LN metastasis will not only increase our knowledge of EV-mediated LN metastasis but also enable the development of an effective therapeutic strategy to treat BCa.
Methods
Clinical samples.
All formalin-fixed, paraffin-embedded tissue samples were obtained from patients with pathologically confirmed BCa (confirmed by 2 independent pathologists) who underwent surgery at Sun Yat-sen Memorial Hospital of Sun Yat-sen University. The urine samples were obtained from the same BCa patients and another 165 healthy participants. Patient information, including characteristics, clinical stage, and pathological classification is summarized in Supplemental Tables 1 and 7.
Cell lines and cell culture.
The human BCa cell lines (UM-UC-1, RT112, RT4, UM-UC-3, T24, and 5637) and human normal bladder epithelial cells (SV-HUC-1) were purchased from the American Type Culture Collection (ATCC). HLECs were obtained from ScienCell Research Laboratories (catalog 2500). All cell lines were cultured in 5% CO2 at 37°C in a humidified incubator. DMEM (Gibco, Thermo Fisher Scientific) containing 10% FBS was used to culture UM-UC-3 and T24 cells, whereas RPMI 1640 (Gibco, Thermo Fisher Scientific) was used to culture UM-UC-1, RT112, and 5637 cells. McCoy’s 5A medium (Gibco, Thermo Fisher Scientific) and F-12K medium (Hyclone), both containing 10% FBS, were used to culture RT4 and SV-HUC-1 cells. HLECs were cultured in endothelial cell medium (ECM) with 5% FBS (ScienCell).
ISH and IHC analysis.
To measure the expression of ELNAT1 in formalin-fixed, paraffin-embedded tissues, ISH analysis was performed with the double-digoxin–labeled (5′ and 3′) ELNAT1-targeted probes and the scrambled probe. Briefly, the slides were dewaxed with dimethylbenzene and rehydrated with gradient alcohol. Then, proteinase K was added to thoroughly digest the sections, after which the slides were hybridized with the ELNAT1 probe at 37°C overnight. Subsequently, the slides were incubated with the anti-digoxin antibody at 37°C for 2 hours, followed by staining using the 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (BCIP/NBT) color substrate solution and counterstaining with nuclear fast red. A Nikon Eclipse Ti microscope (Nikon) was used to capture the images. The probes for ISH analysis are listed in Supplemental Table 12.
IHC was conducted to further analyze the formalin-fixed and paraffin-embedded tissues obtained from patients with BCa and nude mice. All tissue sections were first processed at 60°C for 2 hours and dewaxed with dimethylbenzene, followed by hydration with different concentrations of alcohol. Then, the antigen was repaired with EDTA and the catalase blocked with 3% hydrogen peroxide. Subsequently, the sections were blocked with goat serum for 15 minutes, followed by incubation with the primary antibodies at 4°C overnight. Finally, the sections were incubated with secondary antibodies, and DAB and hematoxylin were used to mark the antigen and counterstain the nuclei, respectively.
Statistical significance for ISH and IHC analyses was assessed using the H-score, which was calculated as follows: H-score = ∑ (P × I), with P denoting the percentage of stained cells and I the intensity of the staining. Staining intensity was graded as follows: 0 (absent), 1 (weak), 2 (moderate), and 3 (strong).
Isolation of EVs.
For the isolation of BCa cell–secreted EVs in cultured media, supernatants of BCa cells that were cultured in their respective media containing 10% EV-free FBS for 48 hours were collected. Then, the samples were centrifuged successively at 2000g for 10 minutes, 10,000g for 30 minutes, and 120,000g for 70 minutes at 4°C to obtain deposited EVs. The extracted EVs in the bottom of the tube were then resuspended in PBS and saved in a –80°C refrigerator for further analysis. For the isolation of EVs from urine and serum samples, urine and blood were collected from all participants and extracted according to the same differential centrifugation method described above.
All isolated EVs were further quantified according to protein content using the BCA Protein Assay Kit (Thermo Fisher Scientific, catalog 23227) following the manufacturer’s instructions (48).
EV internalization analysis.
To verify that BCa cell–secreted EVs were internalized by HLECs, internalization experiments were conducted by labeling the isolated EVs with PKH67 green fluorescent dye. Next, 10 μg/mL PKH67-labeled EVs were incubated with HLECs for 12 hours at 37°C in a humidified incubator at 5% CO2. Subsequently, the HLECs were fixed with formaldehyde and stained with DAPI. The Zeiss confocal microscope system was used to observe HLEC EV internalization and capture images.
Mouse popliteal lymphatic metastasis model.
To explore the role of EV-mediated ELNAT1 in the LN metastasis of BCa, a mouse popliteal lymphatic metastasis model was constructed. Four- to 6-week-old BALB/c nude mice were obtained and kept at the animal center of Sun Yat-sen University for the duration of the experiment. Briefly, 1 × 106 UM-UC-3 cells labeled with luciferase were harvested and resuspended in PBS for injection into the footpads of nude mice. Subsequently, 10 μg isolated EVs from the indicated BCa cells supplemented with 50 μL PBS or its equivalent were injected intratumorally into the mice every 3 days, following the widely used protocol (49). Popliteal LN metastasis was monitored weekly by IVIS until the primary tumor size reached 200 mm3. Then, the footpad tumors and popliteal LNs of the nude mice were dissected for further analysis by IHC. The Nikon Eclipse Ti microscope was used to visualize the sections.
RNA pull-down analysis.
The RNA pull-down assay was performed to detect ELNAT1 binding proteins in BCa cells. First, biotinylated full-length ELNAT1 and antisense sequences were acquired using the Transcript Aid T7 High Yield Transcription Kit (Thermo Fisher Scientific, catalog K0441) according to the manufacturer’s instructions. Then, the Pierce Magnetic RNA-Protein Pull-down Kit (Thermo Fisher Scientific, catalog 20164) was used to perform the pull-down assays according to the manufacturer protocols, in which the biotinylated ELNAT1 and antisense were incubated with BCa cells lysate to pull down the binding proteins. Finally, the binding proteins were eluted for analysis by silver staining or Western blotting, and the differential band was further analyzed with a MALDI-TOF instrument (Bruker Daltonics).
ChIRP analysis.
Following the Magna ChIRP RNA Interactome Kit protocol (MilliporeSigma, catalog 17-10494), ChIP assays followed by qRT-PCR analysis were performed to detect interaction between ELNAT1 and the target gene promoters. A total of 2 × 107 BCa cells or HLECs treated with 10 μg/mL of the indicated EVs per group were fixed with 1% glutaraldehyde and lysed in cell lysis buffer. The cell lysate was then sonicated into 100 to 200 bp fragments in an ultrasonic processor at 4°C for 1 hour, followed by a further incubation with the biotinylated ELNAT1 probes (Supplemental Table 12) at 4°C overnight. Subsequently, the pretreated beads were added to extract the DNA for qRT-PCR analysis.
Bioinformatics analysis.
TCGA data were obtained from Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html). The SUMO2 conjunction site of hnRNPA1 was predicted by GPS-SUMO (http://sumosp.biocuckoo.org). The structure model of ELNAT1 was obtained from RNAalifold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAalifold.cgi). The enrichment of the binding sequences of hnRNPA1 was predicted by POSTAR2 (http://lulab.life.tsinghua.edu.cn/postar2/).
Antibodies.
The following antibodies were used in this study: anti-UBC9 (Abcam, ab75854); anti-SUMO3 (Abcam, ab203570); anti–LYVE-1 (Abcam, ab218535); anti–β-actin (MilliporeSigma, A5441); anti-CD9 (Cell Signaling Technology, 13403); anti-ALIX (Cell Signaling Technology, 92880); anti-hnRNPA1 (Abcam, ab5832); anti-SUMO2 (Abcam, ab233222); anti-His (Abcam, ab5000); anti-H3K4me3 (Abcam, ab1012); anti–rabbit IgG-HRP (Cell Signaling Technology, 7074); anti-SOX18 (Abcam, ab109194); anti–mouse IgG-HRP (Cell Signaling Technology, 7076); anti–rabbit IgG-HRP (Proteintech, SA00001-2); anti–mouse IgG-HRP (Proteintech, SA00001-1).
Additional details on the methods for electron microscopic analysis, lentivirus infection, cell transfection, RACE, RNA extraction, qRT-PCR analysis, Western blotting, FISH, immunofluorescence (IF), nuclear fractionation, tube formation assays, Transwell assays, RIP assays, serial deletion analysis, dual-luciferase reporter experiments, FRET spectroscopy, CD spectroscopy, ChIP analysis, IP assays, co-IP assays, and CRISPR/Cas9-mediated gene deletion are provided in the Supplemental Methods.
Statistics.
All experiments in the present study were performed independently 3 separate times. Quantitative data are presented as the mean ± SD. A χ2 test was performed to compare nonparametric variables. A 2-tailed Student’s t test or 1-way ANOVA was used for the comparison of parametric variables. The Kaplan-Meier method was used to assess the survival of patients and animals. All data analysis was performed using SPSS, version 13.0 (IBM), and a P value of less than 0.05 was considered statistically significant. Statistical significance for ISH and IHC analyses was assessed using the H-score, which was calculated as follows: H score = ∑ (P × I), with P denoting the percentage of stained cells and I the intensity of the staining. Staining intensity was graded as follows: 0 (absent), 1 (weak), 2 (moderate), and 3 (strong).
Study approval.
All tissues and urine samples used in this study were obtained from patients and healthy volunteers with their informed consent or that of their appropriate surrogates, and the use of these samples was approved by the ethics committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (approval number: 2013[61]). All animal experiments were performed with the approval of the IACUC of Sun Yat-Sen University (approval number: 2013[61]). The mouse popliteal lymphatic metastasis model was constructed with the approval of the IACUC of Sun Yat-Sen University.
Author contributions
CC and TL designed the study. CC, HZ, and YK performed the in vitro and in vivo experiments. Y. Luo, BG, and YZ performed the data analyses. HH and WH performed the clinical data analyses. Y. Li, YK, and JH performed the IF and IHC experiments. MA and Y. Li conducted the Western blot analyses. CC, HZ, and Y. Luo wrote the manuscript. All authors read and approved the final manuscript. The order of the co–first authors was determined on the basis of their relative contributions.
Supplementary Material
Acknowledgments
The authors thank J.X. Zhang of the Department of Medical Statistics and Epidemiology at Sun Yat-sen University for statistical advice and research discussions. This study was funded by the National Key Research and Development Program of China (grants 2018YFA0902803 and 2017YFC1308600); the National Natural Science Foundation of China (grants 81825016, 81802530, 81830082, 81672395, 81871945, 81772719, 81772728, 82072639, and 91740119); the Natural Science Foundation of Guangdong Province (grants 2020A1515010815, 2018B010109006, and 2017A020215072); the Outstanding Youth Foundation of Guangdong Province (grant 2021B1515020091); the Science and Technology Planning Project of Guangzhou, China (grants 202002030388, 201803010049, 2017B020227007, and 201704020097); the Yixian Youth project of Sun Yat-sen Memorial Hospital (grant YXQH201812); and the Young Teacher Training Funding program of Sun Yat-sen University (grant 19ykpy121).
Version 1. 03/04/2021
In-Press Preview
Version 2. 04/15/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(8):e146431.https://doi.org/10.1172/JCI146431.
Contributor Information
Changhao Chen, Email: harevyee@qq.com.
Hanhao Zheng, Email: 1846733924@qq.com.
Yuming Luo, Email: 1205171747@qq.com.
Yao Kong, Email: 850464917@qq.com.
Mingjie An, Email: 644743230@qq.com.
Yuting Li, Email: li-yut@qq.com.
Wang He, Email: hewang525344@126.com.
Bowen Gao, Email: 345621094@qq.com.
Yue Zhao, Email: 344246728@qq.com.
Hao Huang, Email: huanghao8911@qq.com.
Jian Huang, Email: huangj8@mail.sysu.edu.cn.
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cumberbatch MGK, et al. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74(6):784–795. doi: 10.1016/j.eururo.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Leveridge MJ, et al. Radical cystectomy and adjuvant chemotherapy for bladder cancer in the elderly: a population-based study. Urology. 2015;85(4):791–798. doi: 10.1016/j.urology.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 4.Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124(3):922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escobedo N, Oliver G. Lymphangiogenesis: origin, specification, and cell fate determination. Annu Rev Cell Dev Biol. 2016;32:677–691. doi: 10.1146/annurev-cellbio-111315-124944. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinbichler TB, et al. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–181. doi: 10.1016/j.semcancer.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu M, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues G, et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat Cell Biol. 2019;21(11):1403–1412. doi: 10.1038/s41556-019-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu H, et al. SENP6-mediated M18BP1 deSUMOylation regulates CENP-A centromeric localization. Cell Res. 2019;29(3):254–257. doi: 10.1038/s41422-018-0139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou L, et al. SUMOylation stabilizes hSSB1 and enhances the recruitment of NBS1 to DNA damage sites. Signal Transduct Target Ther. 2020;5(1):80. doi: 10.1038/s41392-020-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larios J, et al. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol. 2020;219(3):e201904113. doi: 10.1083/jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercier V, et al. Endosomal membrane tension regulates ESCRT-III-dependent intra-lumenal vesicle formation. Nat Cell Biol. 2020;22(8):947–959. doi: 10.1038/s41556-020-0546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunadt M, et al. Extracellular vesicle sorting of α-Synuclein is regulated by sumoylation. Acta Neuropathol. 2015;129(5):695–713. doi: 10.1007/s00401-015-1408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flippot R, et al. Long non-coding RNAs in genitourinary malignancies: a whole new world. Nat Rev Urol. 2019;16(8):484–504. doi: 10.1038/s41585-019-0195-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8(6):631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y, et al. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18(1):37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He W, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128(2):861–875. doi: 10.1172/JCI96218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CH, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9(1):3826. doi: 10.1038/s41467-018-06152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong Y, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19(1):82. doi: 10.1186/s12943-020-01205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchese FP, et al. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, et al. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 2019;47(D1):D203–D211. doi: 10.1093/nar/gky830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He S, et al. LongTarget: a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics. 2015;31(2):178–186. doi: 10.1093/bioinformatics/btu643. [DOI] [PubMed] [Google Scholar]
- 27.Jakobs A, et al. Ubc9 fusion-directed SUMOylation identifies constitutive and inducible SUMOylation. Nucleic Acids Res. 2007;35(17):e109. doi: 10.1093/nar/gkm617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42(W1):W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20(1):12. doi: 10.1186/s13059-018-1604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X, et al. Chronic myelogenous leukemia cells remodel the bone marrow niche via exosome-mediated transfer of miR-320. Theranostics. 2019;9(19):5642–5656. doi: 10.7150/thno.34813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moustaqil M, et al. Homodimerization regulates an endothelial specific signature of the SOX18 transcription factor. Nucleic Acids Res. 2018;46(21):11381–11395. doi: 10.1093/nar/gky897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francois M, et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456(7222):643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- 33.Zhan Y, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17(1):142. doi: 10.1186/s12943-018-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBleu VS, Kalluri R. Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer. 2020;6(9):767–774. doi: 10.1016/j.trecan.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Talukdar S, et al. Noninvasive approaches for detecting and monitoring bladder cancer. Expert Rev Anticancer Ther. 2015;15(3):283–294. doi: 10.1586/14737140.2015.989838. [DOI] [PubMed] [Google Scholar]
- 36.Marin-Aguilera M, et al. Utility of fluorescence in situ hybridization as a non-invasive technique in the diagnosis of upper urinary tract urothelial carcinoma. Eur Urol. 2007;51(2):409–415. doi: 10.1016/j.eururo.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 37.Dassler-Plenker J, et al. Communication in tiny packages: exosomes as means of tumor-stroma communication. Biochim Biophys Acta Rev Cancer. 2020;1873(2):188340. doi: 10.1016/j.bbcan.2020.188340. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 39.Yu F, et al. SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 sumoylation. Cell Res. 2018;28(6):670–685. doi: 10.1038/s41422-018-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell. 2018;71(3):409–418. doi: 10.1016/j.molcel.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21(3):349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Eifler K, Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40(12):779–793. doi: 10.1016/j.tibs.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoellein A, et al. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood. 2014;124(13):2081–2090. doi: 10.1182/blood-2014-06-584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee H, et al. Melatonin disturbs SUMOylation-mediated crosstalk between c-Myc and nestin via MT1 activation and promotes the sensitivity of paclitaxel in brain cancer stem cells. J Pineal Res. 2018;65(2):e12496. doi: 10.1111/jpi.12496. [DOI] [PubMed] [Google Scholar]
- 45.Placer J, et al. Clinical utility of a multiprobe FISH assay in voided urine specimens for the detection of bladder cancer and its recurrences, compared with urinary cytology. Eur Urol. 2002;42(6):547–552. doi: 10.1016/S0302-2838(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhou B, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5(1):144. doi: 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birkhauser FD, et al. Combined ultrasmall superparamagnetic particles of iron oxide-enhanced and diffusion-weighted magnetic resonance imaging facilitates detection of metastases in normal-sized pelvic lymph nodes of patients with bladder and prostate cancer. Eur Urol. 2013;64(6):953–960. doi: 10.1016/j.eururo.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 48.Yue Y, et al. Interleukin-10 deficiency alters endothelial progenitor cell-derived exosome reparative effect on myocardial repair via integrin-linked kinase enrichment. Circ Res. 2020;126(3):315–329. doi: 10.1161/CIRCRESAHA.119.315829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atayde VD, et al. Exosome secretion by the parasitic protozoan leishmania within the sand fly midgut. Cell Rep. 2015;13(5):957–967. doi: 10.1016/j.celrep.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.