Abstract
Aberrant lipid metabolism promotes the development of skeletal muscle insulin resistance, but the exact identity of lipid-mediated mechanisms relevant to human obesity remains unclear. A comprehensive lipidomic analysis of primary myocytes from individuals who were insulin-sensitive and lean (LN) or insulin-resistant with obesity (OB) revealed several species of lysophospholipids (lyso-PLs) that were differentially abundant. These changes coincided with greater expression of lysophosphatidylcholine acyltransferase 3 (LPCAT3), an enzyme involved in phospholipid transacylation (Lands cycle). Strikingly, mice with skeletal muscle–specific knockout of LPCAT3 (LPCAT3-MKO) exhibited greater muscle lysophosphatidylcholine/phosphatidylcholine, concomitant with improved skeletal muscle insulin sensitivity. Conversely, skeletal muscle–specific overexpression of LPCAT3 (LPCAT3-MKI) promoted glucose intolerance. The absence of LPCAT3 reduced phospholipid packing of cellular membranes and increased plasma membrane lipid clustering, suggesting that LPCAT3 affects insulin receptor phosphorylation by modulating plasma membrane lipid organization. In conclusion, obesity accelerates the skeletal muscle Lands cycle, whose consequence might induce the disruption of plasma membrane organization that suppresses muscle insulin action.
Keywords: Metabolism, Muscle Biology
Keywords: Insulin signaling, Skeletal muscle
Introduction
Type 2 diabetes is the 7th leading cause of death in the United States (1) and is a major risk factor for cardiovascular disease, the leading cause of death (2). Skeletal muscle is the site of the largest glucose disposal in humans (3, 4). Insulin resistance in skeletal muscle is a necessary precursor to type 2 diabetes (5) and can be triggered by aberrant lipid metabolism (6–8). Several classes of lipids have been implicated in initiating cellular signals that suppress insulin action, but there has not been a clear consensus that these molecules are upregulated in skeletal muscle insulin resistance that occurs in the human population (9–14).
Accurate measurement of the muscle lipidome is confounded by the intramyofibrillar adipocytes, which are particularly abundant in muscle biopsy samples from individuals with obesity. Human skeletal muscle cells (HSkMCs) are primary myoblasts that can be isolated, propagated, and differentiated from muscle biopsies. This in vitro system provides a unique model to study the skeletal muscle lipidome and signaling pathways free of contaminating cell types and circulating factors that affect muscle metabolism. Importantly, these HSkMCs are known to retain their insulin sensitivity phenotype ex vivo, providing a platform to study mechanisms directly relevant to human physiology (15, 16).
In this study, we harvested HSkMCs from subjects who were insulin-sensitive and lean (LN) or insulin-resistant with obesity (OB) (subject characteristics are shown in Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI135963DS1). We then propagated and differentiated these samples for analyses of the muscle lipidome, gene expression profile, insulin signaling, and membrane properties (Figure 1A). This approach led us to examine the lysophospholipid (lyso-PL) remodeling pathway (Lands cycle) as a potential diet-responsive mechanism that regulates skeletal muscle insulin action. Here, we provide evidence that implicates this pathway in the pathogenesis of diet-induced skeletal muscle insulin resistance. Genetic or pharmacologic suppression of this pathway was sufficient to enhance skeletal muscle insulin action in vitro and in vivo.
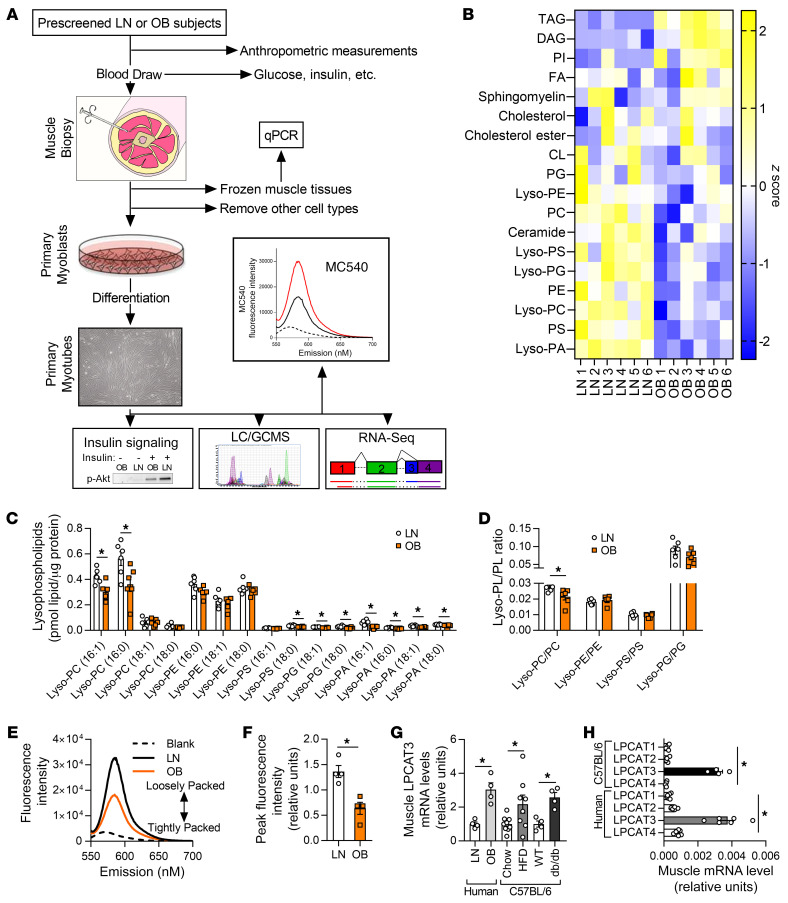
Figure 1. Lipidomic analyses of skeletal muscle samples from human subjects who were lean or with obesity.
(A) A schematic of the workflow for the clinical study. LC/GCMS, liquid chromatography/gas chromatography-mass spectrometry. (B–D) Lipidomic analysis of HSkMCs from subjects who were insulin-sensitive and lean (LN) or insulin-resistant with obesity (OB). (B) Heatmap of lipid content by class. CL, cardiolipin; DAG, diacylglycerol; FA, fatty acid; Lyso-PA, lysophosphatidic acid; Lyso-PC, lysophosphatidylcholine; Lyso-PE, lysophosphatidylethanolamine; Lyso-PG, lysophosphatidylglycerol; Lyso-PS, lysophosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; TAG, triacylglycerol. (C) Species of lysophospholipids. (D) Lysophospholipid/phospholipid ratio (n = 6). (E and F) Quantification of MC540 fluorescence in LN and OB HSkMCs (n = 4). (G) LPCAT3 mRNA in muscle biopsies from LN or OB subjects (left), in skeletal muscle of WT mice fed standard chow or high-fat diet (HFD) (middle), and in skeletal muscle of WT or db/db mice (right) (LN and OB: n = 4; chow: n = 9; HFD: n = 8; WT and db/db: n = 4). (H) Expression of all isoforms of LPCAT in muscle samples from mouse (n = 4) or human (n = 6) skeletal muscle. Two-tailed t tests (C, D, F, and G) or 1-way ANOVA followed by post hoc multiple comparisons (H) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
Results
Human primary muscle cells from OB subjects were more insulin resistant in comparison with LN controls (15). A global lipidomic analysis of these LN and OB myotubes revealed many classes of lipids that were differentially abundant (Figure 1B and Supplemental Figure 1, A–L). Among these, several species of lyso-PLs, intermediates of the Lands cycle (17), were lower in OB HSkMCs compared with LN (Figure 1C), a finding not previously described for an insulin-resistant state. While species of many classes of lyso-PLs were reduced with obesity, the ratio of lyso-PL to its parent phospholipid was significantly lower for only lysophosphatidylcholine (lyso-PC)/phosphatidylcholine (PC) (Figure 1D). Previous studies suggest that altering the lyso-PC content of cell membranes is sufficient to alter the physical properties of membranes (18, 19). Consistent with this notion, phospholipid packing of LN myotubes and that of OB myotubes were remarkably different, with OB cells exhibiting more tightly packed membrane head groups compared with LN (Figure 1, E and F). This occurred in the absence of changes in the phospholipid acyl chain saturation index (Supplemental Figure 1M).
What is the molecular mechanism by which obesity promotes a lower abundance of lyso-PL in skeletal muscle? LN and OB HSkMCs used for the lipidomic analyses were cultured ex vivo for several weeks in identical media conditions. Thus, differences in the lipidome of these samples are likely the result of genetic and/or epigenetic influences, instead of hormonal or neuronal inputs that alter cells in vivo. We reasoned that such differential programming might be expected to manifest in gene expression profiles. A whole transcriptome sequencing of LN and OB myotubes revealed that lyso-PC acyltransferase 3 (LPCAT3), an enzyme of the Lands cycle, was more highly expressed with obesity (GEO series accession number GSE162265). These findings were recapitulated in muscle biopsy samples (not myotubes) from LN and OB individuals as well as muscle tissue from WT mice fed a high-fat diet (HFD) or db/db mice (Figure 1G). The Lands cycle represents a series of phospholipid-remodeling reactions by which acyl chains become transacylated (17). Of the 13 lyso-PL acyltransferase enzymes (20), LPCAT3 has the highest affinity for 16:0 and 18:0 lyso-PC, consistent with the specificity of reduced lyso-PC/PC ratio (21, 22). Silencing of LPCAT3 in fibroblasts has been shown to increase Akt phosphorylation (23), while incubation of the same cells with 16:0/20:4 PC decreased Akt phosphorylation as a result of plasma membrane–specific effects (24). Mice with a liver-specific deletion of LPCAT3 exhibit enhanced ordering of membranes (25). In both human and mouse skeletal muscle, LPCAT3 is very highly expressed compared with other isoforms of LPCAT (Figure 1H), and skeletal muscle LPCAT3 expression is directly correlated to circulating insulin in 106 mouse strains (data not shown; biweight midcorrelation = 0.296, P = 0.0016) (26).
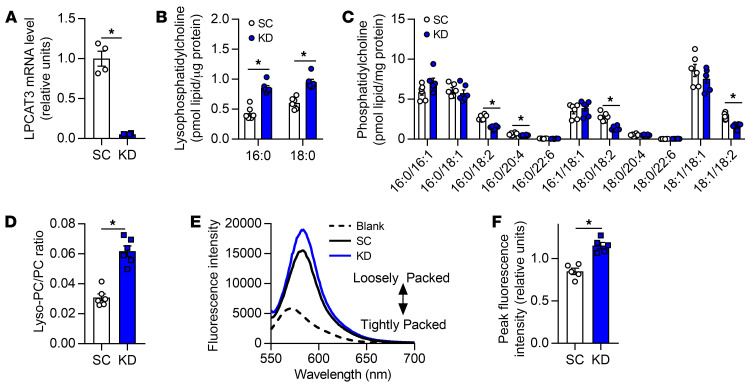
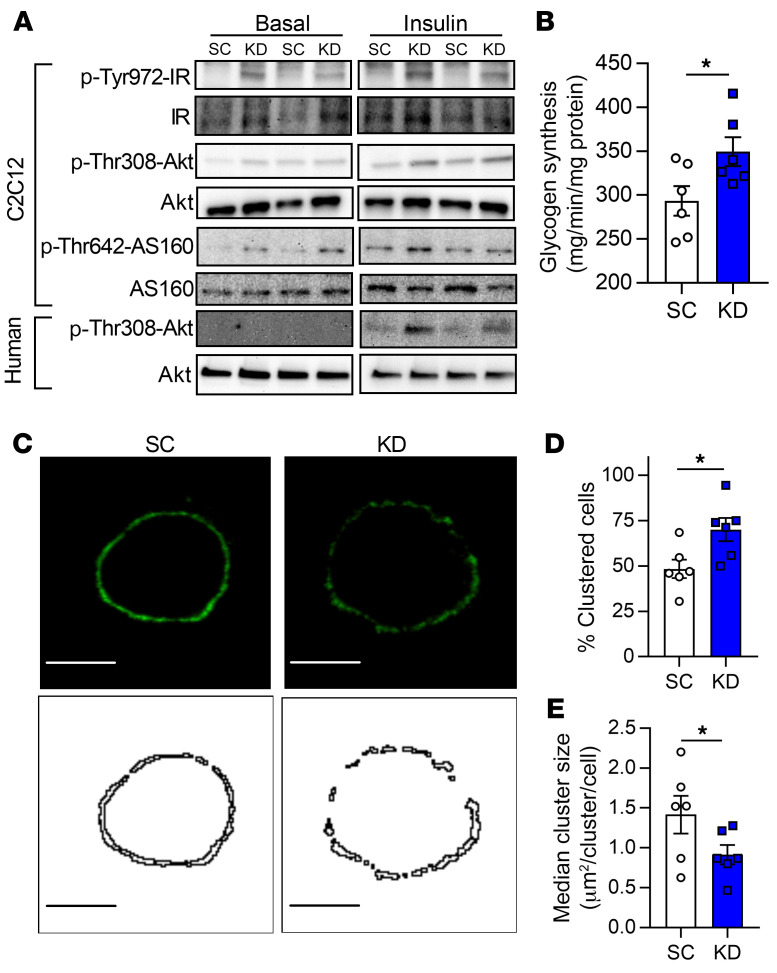
To study the role of LPCAT3 in skeletal muscle insulin action, we performed a lentivirus-mediated shRNA knockdown of LPCAT3 (scrambled, SC; LPCAT3 knockdown, KD) in C2C12 myotubes (Figure 2A). LPCAT3 knockdown did not affect protein content for MyoD, various MHC isoforms, and respiratory complexes (Supplemental Figure 2, A–C), suggesting that the deletion of LPCAT3 has no effect on myotube lineage or mitochondrial density. Targeted lipidomic analyses revealed that LPCAT3 knockdown increased lyso-PC and decreased PC (Figure 2, B and C), substrates and products of the LPCAT3-mediated reaction, respectively (27, 28). Together, these differences were sufficient to elevate the lyso-PC/PC ratio with LPCAT3 deletion (Figure 2D). Similar effects were seen with lipid species composed of an ethanolamine head group (Supplemental Figure 2, D–F), while the phospholipid saturation index increased with LPCAT3 knockdown (Supplemental Figure 2G). Analogous to differences observed in LN and OB HSkMCs, LPCAT3 deletion reduced phospholipid head group packing (Figure 2, E and F). We then incubated SC and KD cells in a submaximal concentration of insulin to assess insulin signaling events. Strikingly, inhibition of LPCAT3 robustly enhanced insulin signaling with or without insulin (Figure 3A and Supplemental Figure 2H). Notably, the increase occurred at the level of the insulin receptor (IR), a node that is localized in the phospholipid-rich plasma membrane. Consequently, LPCAT3 deletion enhanced insulin-stimulated glycogen synthesis (Figure 3B), suggesting that this intervention increases skeletal muscle insulin sensitivity in vitro (because of low GLUT4:GLUT1 stoichiometry, insulin-stimulated glucose uptake is not an ideal surrogate for insulin sensitivity in C2C12 myotubes). LPCAT3 knockdown also enhanced insulin signaling in HSkMCs from subjects with obesity (Figure 3A).
Figure 2. LPCAT3 knockdown alters skeletal muscle membrane phospholipid composition in vitro.
(A) LPCAT3 mRNA levels in myoblasts infected with lentiviruses expressing shRNA for scrambled (SC) or LPCAT3 sequences (KD) and differentiated into myotubes (n = 4). (B–D) Lipids were extracted from C2C12 myotubes for analysis between SC and KD cells. Quantification of lyso-PC (B), PC (C), and total lyso-PC/PC (n = 6) (D). (E and F) Quantification of MC540 fluorescence in SC and KD myotubes (n = 5). Two-tailed t tests were performed. All data are represented as mean ± SEM. *P ≤ 0.05.
Figure 3. LPCAT3 inhibition enhances skeletal muscle insulin sensitivity in vitro.
Myoblasts were infected with lentivirus encoding scrambled (SC) or LPCAT3-knockdown (KD) sequences and differentiated into myotubes. (A) Phosphorylation and total protein of IR, Akt, and AS160 were measured via Western blot with insulin (0.6 nM) or without insulin in C2C12 myotubes (top) and human primary skeletal muscle cells (bottom) (image is representative of 3 experiments). (B) Glycogen synthesis was quantified in C2C12 cells incubated with insulin (12 nM) (n = 6). (C–E) GM-1–enriched microdomains were labeled in SC and KD myotubes that had been rounded up. (C) Plasma membrane GM-1 localization was visualized (top: fluorescence images; bottom: binary images). Scale bars: 10 μM. (D) Cells were scored as clustered or nonclustered between SC and KD myotubes. (E) Particle size was measured for each cell in 6 separate experiments, and the median for each experiment was used as a representative of that experiment (n = 35–50 per experiment, 6 separate experiments). Two-tailed t tests were performed. All data are represented as mean ± SEM. *P ≤ 0.05.
The organization and clustering of plasma membrane microdomains are linked to the induction of tyrosine kinase signaling events, such as IR signaling (29–31). Because LPCAT3 deletion promoted enhanced insulin signaling at the level of IR phosphorylation, we visualized the organization of plasma membrane microdomains with labeling and patching of plasma membrane GM-1, a known marker of microdomains. Indeed, a greater proportion of C2C12 cells with LPCAT3 knockdown exhibited clustering of GM-1–enriched microdomains (Figure 3C, top, and Figure 3D). Furthermore, LPCAT3 deletion decreased the size of these clusters (Figure 3C, bottom, and Figure 3E), with no differences in total fluorescence from each cell (Supplemental Figure 2I). These data indicate that LPCAT3 inhibition induces a reorganization of plasma membrane microdomains, potentially explaining increased IR phosphorylation.
CI-976 is a pan–lysophospholipid acyltransferase inhibitor (32, 33) that has the ability to disrupt the Lands cycle, similarly to LPCAT3 deletion. To examine the possibility that the insulin-sensitizing effect of LPCAT3 knockdown is attributable to an unknown function of LPCAT3 outside of the Lands cycle, we studied C2C12 myotubes with or without CI-976. Consistent with our findings with LPCAT3 knockdown (Figure 2, E and F), preincubation of WT C2C12 myotubes with CI-976 robustly decreased phospholipid headgroup packing (Figure 4, A and B) and increased insulin-stimulated Akt phosphorylation (Figure 4, C and D). We also administered CI-976 to WT (C57BL/6J) mice that had been fed HFD. Strikingly, mice injected with CI-976 exhibited increased glucose tolerance compared with vehicle-treated controls without body weights being altered (Figure 4, E–G). This evidence supports our findings that inhibition of the Lands cycle may increase skeletal muscle insulin sensitivity.
Figure 4. Administration of CI-976 in vitro and in vivo.
(A–D) C2C12 myoblasts were differentiated into myotubes with either CI-976 or vehicle. (A and B) Quantification of MC540 fluorescence (n = 6). (C and D) Western blot and quantification of Thr308 phosphorylation and total Akt in the presence (12 nM) and absence of insulin (n = 14; P = 0.024, main effect of insulin). (E–G) WT C57BL6/J mice were fed with HFD for 6 weeks, with subcutaneous injection for the last 7 days with vehicle (1:9 ethanol/polyethylene glycol) or 30 mg/kg CI-976. Body mass (E), intraperitoneal glucose tolerance excursion curves (F), and AUC (G) from glucose tolerance tests on vehicle-treated or CI-976–injected mice (n = 8 per group). Two-tailed t tests (B, E, and G) or 2-way ANOVA with Šidák’s multiple-comparison test (D and F) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
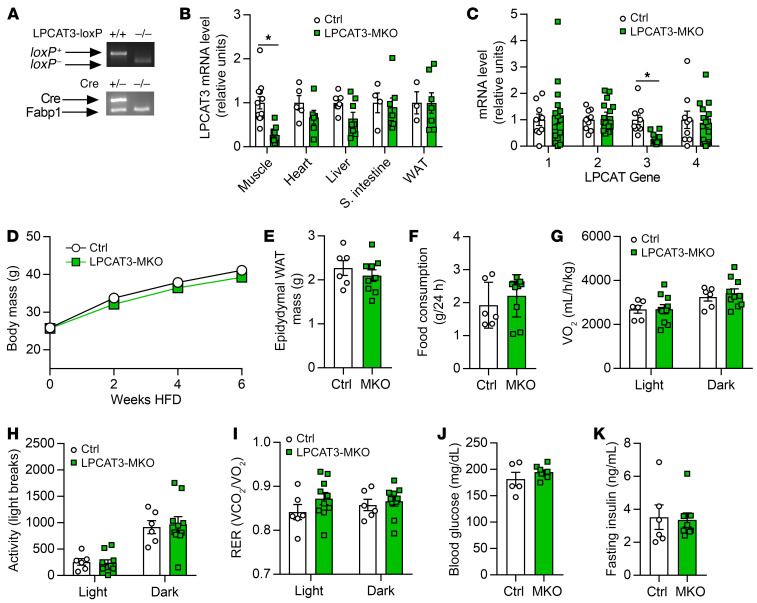
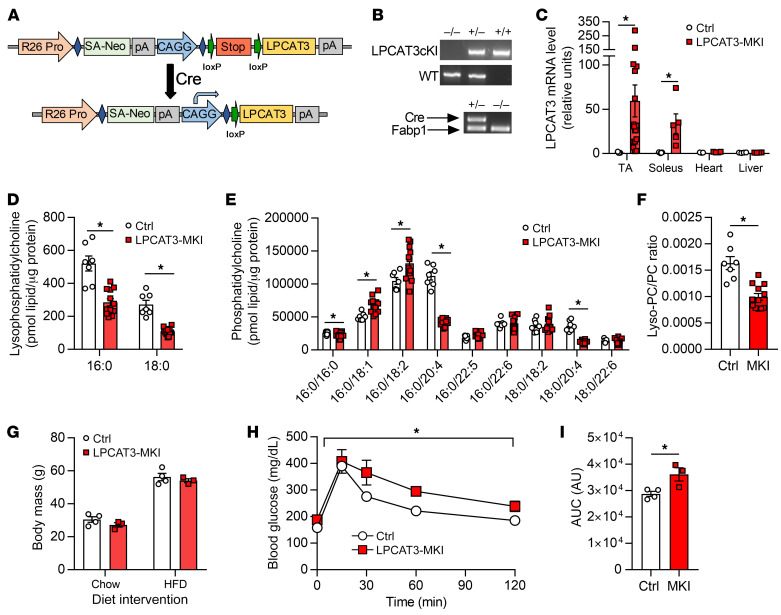
As effects of CI-976 administration in vivo cannot be attributed to effects on skeletal muscle alone, we performed skeletal muscle–specific deletion of LPCAT3 in vivo. Mice with tamoxifen-inducible skeletal muscle–specific knockout of LPCAT3 (LPCAT3-MKO) were generated by crossing of the HSA-MerCreMer mice (34) with mice with conditional knockout of LPCAT3 (exon 3 of the Lpcat3 gene flanked with loxP sites) (25) (Figure 5A). This strategy successfully yielded mice with suppressed LPCAT3 expression in skeletal muscle without affecting other tissues (Figure 5B), and without compensatory upregulation of other members of the LPCAT family (Figure 5C and Supplemental Figure 3A). Control and LPCAT3-MKO mice gained weight equally when fed HFD (Figure 5D) with no difference in adipose tissue weight at the end of diet intervention (Figure 5E). Food consumption, whole-body oxygen consumption, spontaneous activity, and respiratory exchange ratio were similarly not different between groups (Figure 5, F–I). Fasting glucose and insulin, and glucose tolerance (Figure 5, J and K, and Figure 6A), were unchanged, but circulating insulin during the glucose tolerance test was substantially lower in LPCAT3-MKO mice compared with the control group (Figure 6B).
Figure 5. Whole-body phenotyping of LPCAT3-MKO mice.
(A) Mice with tamoxifen-inducible skeletal muscle–specific Cre recombinase (HSA-MerCreMer+/–) were crossed with mice with loxP sites flanking exon 3 of the Lpcat3 gene (LPCAT3cKO+/+) to generate skeletal muscle–specific inducible knockout of LPCAT3 (LPCAT3cKO+/+; HSA-MerCreMer+/–) (LPCAT3-MKO). Littermates (LPCAT3cKO+/+; HSA-MerCreMer–/–) (Ctrl) were used as control mice for all experiments. (B) LPCAT3 mRNA in tibialis anterior (Muscle), heart, liver, small intestine, and inguinal white adipose tissue (WAT) (muscle: Ctrl n = 12, MKO n = 15; heart: Ctrl n = 5, MKO n = 6; liver: Ctrl n = 7, MKO n = 8; small intestine: Ctrl n = 4, MKO n = 7; WAT: Ctrl n = 3, MKO n = 7). (C) mRNA of all LPCAT isoforms in tibialis anterior muscles of Ctrl and LPCAT3-MKO mice (Ctrl n = 9, MKO n = 14). (D) Body mass during HFD feeding in Ctrl and LPCAT3-MKO mice (Ctrl n = 8, MKO n = 11). (E) Epididymal WAT mass (Ctrl n = 6, MKO n = 9). (F–I) Ctrl and LPCAT3-MKO mice were placed in metabolic chambers for measurement of food consumption (F), VO2 (G), activity (H), and respiratory exchange ratio (RER) (I) (Ctrl n = 6, MKO n = 10). (J) Fasting glucose (Ctrl n = 5, MKO n = 9). (K) Fasting insulin (Ctrl n = 6, MKO n = 9). All data except those in A are from HFD-fed mice. Two-tailed t tests (B, C, E, F, J, and K) or 2-way ANOVA with Šidák’s multiple-comparison test (D and G–I) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
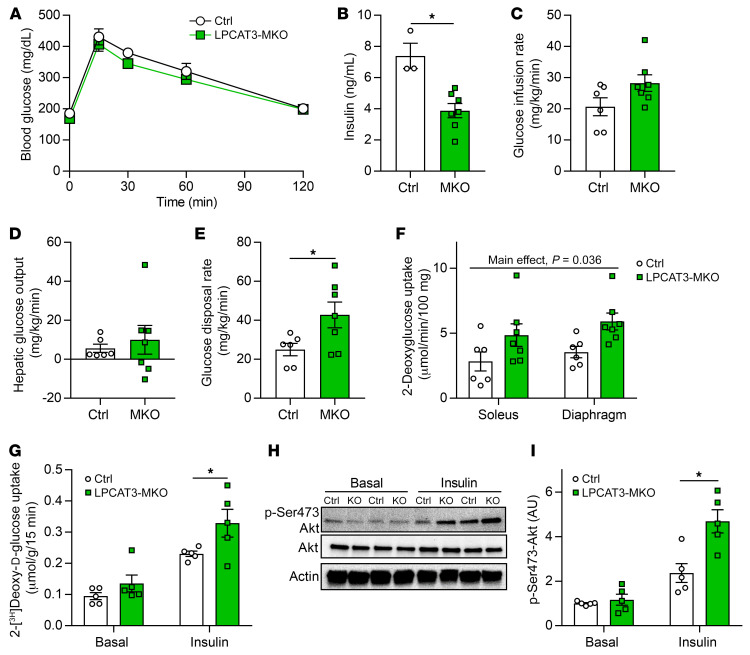
Figure 6. LPCAT3-MKO mice are protected from diet-induced skeletal muscle insulin resistance.
(A) Intraperitoneal glucose tolerance test (Ctrl n = 6, MKO n = 8). (B) Serum insulin at the 30-minute time point of the glucose tolerance test (Ctrl n = 3, MKO n = 8). (C–F) Hyperinsulinemic-euglycemic clamps were performed in conscious unrestrained mice (Ctrl n = 6, MKO n = 7). (C) Glucose infusion rate required to maintain constant blood glucose of 150 mg/dL during clamp phase. (D) Hepatic glucose output during the clamp phase. (E) Rate of whole-body glucose disposal during the clamp phase. (F) 14C-2-deoxyglucose uptake quantification in soleus and diaphragm muscles during the clamped state. (G–I) Soleus muscles were dissected and incubated with or without 200 μU/mL of insulin. (G) Ex vivo 2-deoxyglucose uptake (n = 5). (H and I) Ser473 phosphorylation and total Akt (n = 5). All data are from HFD-fed mice. Two-way ANOVA with Šidák’s multiple-comparison test (A, F, G, and I) or 2-tailed t tests (B–E) were performed. All data are represented as mean ± SEM. *P ≤ 0.05.
To evaluate whether improved glycemic efficiency was attributable to greater skeletal muscle insulin sensitivity, we subjected HFD-fed control and LPCAT3-MKO mice to hyperinsulinemic-euglycemic clamping experiments. Similar to results from the glucose tolerance test, control and LPCAT3-MKO mice had similar glucose infusion rates (Figure 6C), suggesting comparable systemic insulin sensitivity. Likewise, hepatic glucose output was not different between the groups (Figure 6D). In contrast, glucose disposal rate was approximately 2-fold higher in LPCAT3-MKO mice compared with control, indicating increased peripheral insulin sensitivity (Figure 6E). Furthermore, infusion of 14C-2-deoxyglucose during the insulin-stimulated clamp phase elevated uptake in soleus and diaphragm muscles from LPCAT3-MKO mice compared with control (Figure 6F). We further validated elevated muscle insulin sensitivity by performing the 3H-2-deoxyglucose uptake assay ex vivo. Indeed, insulin-stimulated skeletal muscle glucose uptake was robustly enhanced in LPCAT3-MKO mice compared with control (Figure 6G). The increase in glucose uptake coincided with augmented insulin-stimulated Akt phosphorylation (Figure 6, H and I), similar to C2C12 and human primary myotubes (Figure 3). These results suggest that inhibition of muscle LPCAT3 increases skeletal muscle insulin sensitivity in vivo.
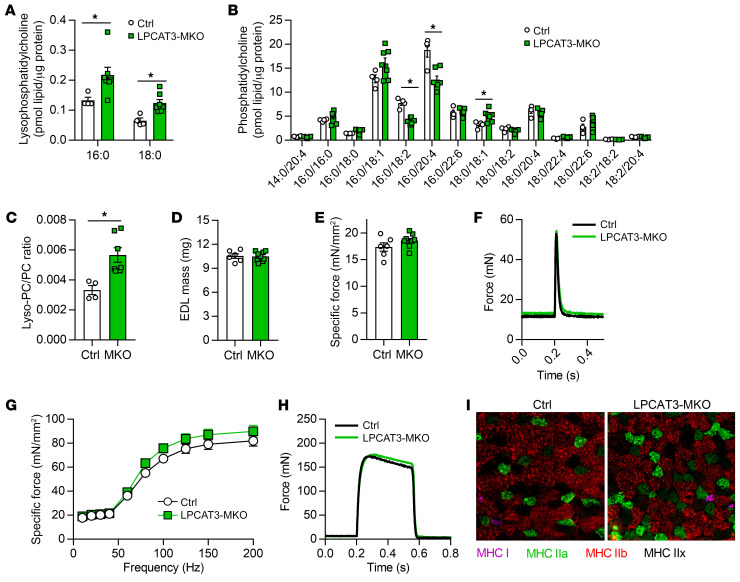
Similar to results from LPCAT3 knockdown in vitro, muscles from LPCAT3-MKO mice had elevated lyso-PC (16:0 and 18:0) (Figure 7A) and lower levels of PC species known to be the main products of the LPCAT3 reaction (16:0/18:2 and 16:0/22:4) (Figure 7B) (22, 35, 36). As a result, the lyso-PC/PC ratio was approximately 2-fold greater in LPCAT3-MKO mice compared with control (Figure 7C). In contrast, the ratio of lysophosphatidylethanolamine (lyso-PE) to phosphatidylethanolamine (PE) or phospholipid saturation index was unaltered between control and LPCAT3-MKO muscles (Supplemental Figure 3, B–E), similar to the lipidome in LN and OB HSkMCs (Figure 1 and Supplemental Figure 1). Muscles from control and LPCAT3-MKO mice did not differ in mass, length, force-generating capacity, fiber-type distribution, or content of proteins in the electron transport chain (Figure 7, D–I, and Supplemental Figure 3, F–H).
Figure 7. Skeletal muscle lipid composition and contractility in LPCAT3-MKO mice.
(A–C) Lipids were extracted from gastrocnemius muscles of Ctrl and LPCAT3-MKO mice for mass spectrometric analysis. Quantification of lyso-PC (A), PC (B), and total lyso-PC/PC (C) (Ctrl n = 4, MKO n = 7). (D–I) Extensor digitorum longus (EDL) muscles of Ctrl and LPCAT3-MKO mice were dissected for measurement of mass (D), force produced with a pulse stimulation (E and F), force produced with tetanic stimulation ranging from 10 to 200 Hz (G and H) (H, force tracing at 200 Hz stimulation) (Ctrl n = 6, MKO n = 9), and skeletal muscle fiber type (MHC I, pink; MHC IIa, green; MHC IIb, red; MHC IIx, negative) (I). Two-tailed t tests (A–E) or 2-way ANOVA with Šidák’s multiple-comparison test (G) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
We also explored whether acceleration of the Lands cycle would be sufficient to impair glucose handling. For these experiments, we generated mice with skeletal muscle–specific LPCAT3 overexpression (LPCAT3-MKI). A mouse LPCAT3 cDNA sequence was inserted into the Rosa26 locus preceded by a stop codon, which was flanked with loxP sites to generate mice with conditional LPCAT3 knockin (LPCAT3cKI+/–) (Figure 8A). LPCAT3cKI+/+ mice were crossed with the HSA-MerCreMer+/– mice to generate the control (LPCAT3cKI+/– HSA-MerCreMer–/–) or LPCAT3-MKI (LPCAT3cKI+/– HSA-MerCreMer+/–) mice (Figure 8B). This strategy successfully resulted in LPCAT3 overexpression specific to skeletal muscle (Figure 8C), which was sufficient to accelerate the Lands cycle to reduce lyso-PC (Figure 8D), increase some species of PC (Figure 8E), and reduce the lyso-PC/PC ratio (Figure 8F). LPCAT3-MKI mice had similar body weights to littermate controls when fed standard chow or HFD (Figure 8G). Importantly, LPCAT3-MKI mice were found to be more glucose intolerant compared with controls (Figure 8, H and I). Thus, acceleration of skeletal muscle Lands cycle is sufficient to impair systemic glucose handling.
Figure 8. Generation and characterization of mice with skeletal muscle–specific overexpression of LPCAT3 (LPCAT3-MKI).
(A) Schematic of mouse conditional LPCAT3 knockin (LPCAT3cKI+/+) with insertion of mouse LPCAT3 cDNA sequence in the Rosa26 locus preceded by a stop codon flanked with loxP sites. (B) Genotyping of LPCAT3cKI+/+ mice crossed with HSA-MCM+/– mice to generate LPCAT3cKI+/–; HSA-MCM–/– (Ctrl) or LPCAT3cKI+/–; HSA-MCM+/– (LPCAT3-MKI) mice. (C) Quantitative RT-PCR of LPCAT3 in tibialis anterior (TA) (Ctrl n = 9, MKI n = 17), soleus (Ctrl n = 5, MKI n = 5), heart (Ctrl n = 3, MKI n = 4), and liver (Ctrl n = 4, MKI n = 4). (D–I) Lipids were extracted from TA muscles and quantified for lyso-PC (D), PC (E), and lyso-PC/PC ratio (F) (Ctrl n = 7, MKI n = 12). (G) Body mass of Ctrl and LPCAT3-MKI mice fed a standard chow diet or after 12 weeks of HFD feeding. (H and I) Intraperitoneal glucose tolerance test (H) and AUC (I) from glucose tolerance test between Ctrl and LPCAT3-MKI mice after HFD feeding (Ctrl n = 4, MKI n = 3). Two-tailed t tests (C–G and I) or 2-way ANOVA (H) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
How does the inhibition of the Lands cycle promote greater insulin action in skeletal muscle? LPCAT3 deficiency enhanced insulin signaling at the level of IR, which was concomitant with altered plasma membrane lipid organization (Figure 3), suggesting that changes in plasma membrane properties may mediate the insulin-sensitizing effects. Membrane organization is vital to insulin action, as IR is localized to highly ordered microdomains on the plasma membrane (37, 38). The interaction between caveolae and IR enhances insulin signaling in other cell types (39, 40). Mice that lack caveolin-3 (cav3), a skeletal muscle–specific scaffolding protein critical in the formation of caveolae on the plasma membrane, exhibit skeletal muscle insulin resistance due to plasma membrane–specific effects on the IR (41, 42). Overexpression of dominant-negative cav3 leads to decreased glucose uptake and glycogen synthesis in C2C12 cells, which is attributed to decreased Akt phosphorylation (43–45). Conversely, an increase in WT cav3 expression is sufficient to enhance Akt phosphorylation and glucose uptake (46). Indeed, LPCAT3 knockdown substantially increased cav3 content in C2C12 myotubes (Figure 9A). To examine the possibility that the absence of LPCAT3 increases the abundance of lipids in caveolae, we isolated membrane fractions from C2C12 myotubes with or without LPCAT3 deletion and subjected them to further purification by density gradient ultracentrifugation.
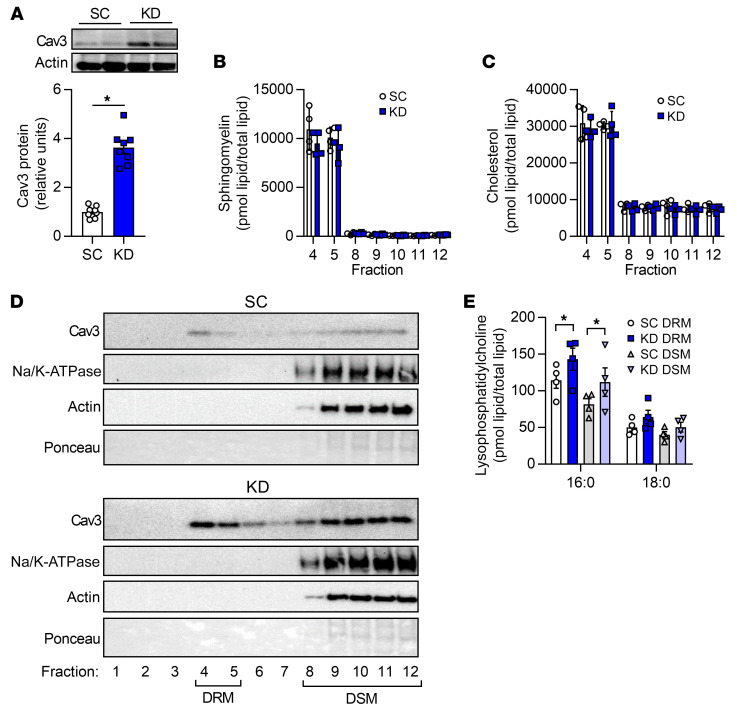
Figure 9. LPCAT3 deletion alters plasma membrane organization.
C2C12 cells were infected with lentiviruses expressing shRNA for scrambled (SC) or LPCAT3 (KD) sequences and differentiated into myotubes. (A) Caveolin-3 (cav3) protein content. (B and C) Detergent-resistant membranes (DRMs; fractions 4 and 5) and detergent-soluble membranes (DSMs; fractions 8–12) were isolated, and lipids were extracted for quantification of sphingomyelin (B) and cholesterol (C) (n = 4). (D) Cav3, Na/K-ATPase, actin, and total protein content were assessed via Western blot in all fractions from the sucrose gradient (image is representative of 4 experiments). (E) Lyso-PC levels in DSM and DRM isolations (n = 4). Two-tailed t tests (A–C) or 2-way ANOVA with Šidák’s multiple-comparison test (E) was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
Cholesterol and sphingomyelin are 2 classes of lipids that are more highly abundant in the detergent-resistant membrane (DRM; i.e., ordered membrane) fraction compared with the detergent-soluble membrane (DSM) fraction (47). Experiments in WT C2C12 myotubes indicated that fractions 4 and 5 have substantial amounts of total lipid (Supplemental Figure 4A). These fractions were enriched in sphingomyelin and cholesterol, which are known to be conducive to more highly ordered membranes (Supplemental Figure 4, B and C), with a relatively low abundance of lipids involved in the LPCAT3-mediated reaction (Supplemental Figure 4, D–G). LPCAT3 knockdown did not appear to alter the overall content of lipid in the DRM fraction, nor did it affect enrichment of sphingomyelin and cholesterol (Figure 9, B and C). Even though the DRM fraction is known to contain very little protein (48), we detected substantial cav3 in both SC and KD myotubes (Figure 9D). While LPCAT3 deletion did not affect the proportion of cav3 in the DRM fraction, it is noteworthy that elevated cav3 content with LPCAT3 knockdown (Figure 9A) is reflected in the DRM fraction. This is particularly interesting considering there was no enrichment of sphingomyelin or cholesterol in the DRM fraction, which may have been expected given that these lipids induce sequestration of cav3 into caveolae. Consistent with this notion, LPCAT3 deletion was sufficient to elevate lyso-PC in the DRM as well as DSM fractions of the membrane (Figure 9E), with minimal effect on PC species (Supplemental Figure 5A). Similar results were exhibited with lyso-PE and PE (Supplemental Figure 5, B and C). The saturation index of phospholipids was slightly increased in both DRM and DSM fractions, which may also contribute to the increase in the plasma membrane lipid clustering (Supplemental Figure 5D). Thus, LPCAT3 deletion promotes the accumulation of lyso-PC in the DRM fraction, which may contribute to membrane organization.
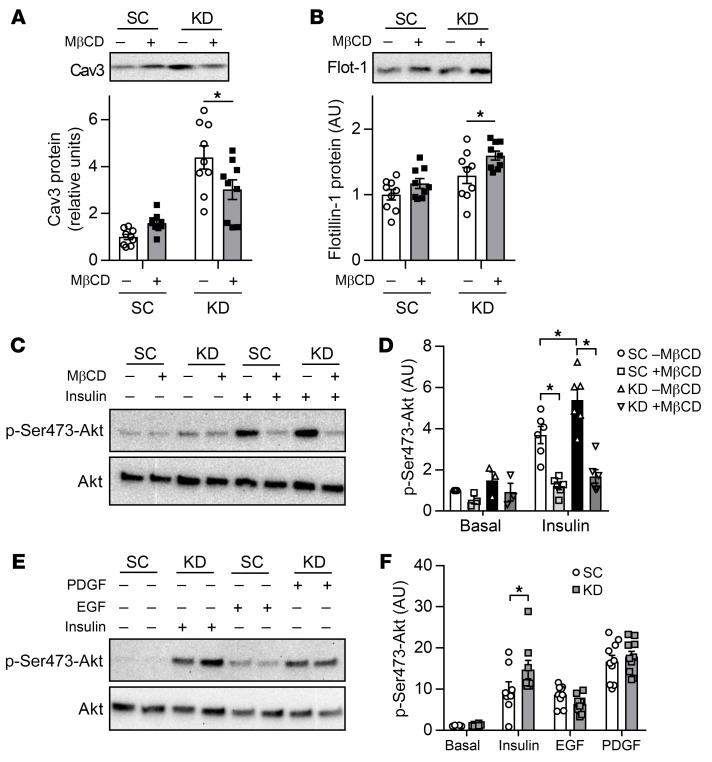
To test our hypothesis that an increase in membrane organization mediates the insulin-sensitizing effect of LPCAT3 deletion, we incubated C2C12 myotubes with methyl-β-cyclodextrin (MβCD), a cholesterol-depleting compound that disrupts plasma membrane microdomains (49). Indeed, incubation of cells with MβCD decreased cav3 protein content (Figure 10A) without decreasing the abundance of flotillin-1 (Figure 10B), a protein associated with noncaveolar microdomains. MβCD treatment normalized insulin-stimulated Akt phosphorylation with LPCAT3 deletion to control levels (Figure 10, C and D). Finally, we also examined whether alteration of plasma membrane microdomains induced by LPCAT3 deletion would affect other cell surface receptor–mediated signaling events. For these experiments, we incubated C2C12 myotubes with epidermal growth factor (EGF) or platelet-derived growth factor (PDGF) in addition to insulin with or without LPCAT3 knockdown. Both EGF and PDGF receptors are localized at the plasma membrane and activate Akt independently of IR (50, 51). Strikingly, LPCAT3 deletion did not augment Akt phosphorylation induced by EGF or PDGF (Figure 10, E and F), suggesting that the effects of LPCAT3 deletion are selective. These findings are consistent with the notion that LPCAT3 deletion enhances insulin signaling by its effect on plasma membrane organization that is relatively specific to IR.
Figure 10. Disruption of caveolae is sufficient to normalize insulin signaling with LPCAT3 knockdown.
C2C12 myoblasts were infected with lentivirus expressing shRNA for scrambled (SC) or LPCAT3-knockdown (KD) sequences and were differentiated into myotubes. (A–D) Myotubes were incubated with or without 10 mM of methyl-β-cyclodextrin (MβCD) for 1 hour. (A and B) MβCD successfully depletes cav3 (P < 0.001, main effect of LPCAT3 knockdown) but not flotillin-1 (P = 0.003, main effect of LPCAT3 knockdown; P = 0.01, main effect of MβCD) (n = 9). (C and D) Cells were incubated in the presence (0.6 nM) or absence of insulin and were blotted for total or Ser473 phosphorylation of Akt (n = 3 basal, n = 6 insulin). (E and F) C2C12 myotubes were incubated in the absence or presence of either PDGF (2.5 ng/mL), EGF (100 ng/mL), or insulin (12 nM) and were blotted for total or Ser473 phosphorylation of Akt (basal and insulin, n = 8; PDGF and EGF, n = 10). Two-way ANOVA with Šidák’s multiple-comparison test was performed. All data are represented as mean ± SEM. *P ≤ 0.05.
Discussion
Obesity promotes aberrant lipid metabolism in various tissues, including skeletal muscle, where it dampens its ability to respond to circulating insulin and increase glucose uptake. Studies in model organisms have led to the identification of lipotoxic lipids that might promote insulin resistance in various tissues (52, 53), but some studies were unable to validate these mechanisms in human muscles (54, 55). To gain a global understanding of changes that occur in muscle lipid metabolism with human obesity, we conducted lipidomic analyses on muscle samples from LN and OB subjects. Obesity was associated with decreases in various species of lysophospholipids, an observation that, to our knowledge, had never been reported previously. Many of these lipids are generated by the enzymes of the Lands cycle, which removes fatty acyl chains at the sn-2 position of phospholipids to generate lysophospholipids. We propose a mechanism by which obesity accelerates the skeletal muscle Lands cycle to promote insulin resistance.
The acceleration of muscle phospholipid transacylation was apparently driven by increased LPCAT3 expression, likely attributable to diet-induced activation of LXRs and PPARs (22, 56, 57). Indeed, we found that skeletal muscle–specific overexpression of LPCAT3 was sufficient to augment glucose intolerance induced by HFD feeding. In contrast, inhibition of LPCAT3 enhances insulin signaling at the level of IR to improve skeletal muscle insulin sensitivity. We believe that the insulin-sensitizing effect of Lands cycle inhibition is mediated by its effect on the plasma membrane lipid organization (Figure 11). Consistent with this notion, LPCAT3 deletion and/or CI-976 treatment was sufficient to alter membrane phospholipid packing and GM1 microdomain clustering, cav3 content, and lipid composition of detergent-resistant and -soluble membranes. Furthermore, disruption of cholesterol-rich microdomains was sufficient to eliminate the insulin-sensitizing effect of LPCAT3 inhibition. Interventions that interfere with the plasma membrane organization would be predicted to have effects on other cellular events, but the deletion of LPCAT3 did not alter EGF or PDGF sensitivity, suggesting some specificity for IR. LPCAT3 deletion did not appear to have an overly adverse effect on skeletal muscle, including mass, fiber type, or force-generating capacity. It would be of substantial interest to pursue implications of altered Lands cycle and/or plasma membrane organization in the context of other cellular events. In particular, a recent study suggests that plasma membrane sn-1,2-diacylglycerol accumulation promotes PKCε activation, increased IR-Thr1160 phosphorylation, and decreased IR-Tyr1162 phosphorylation in liver (58). Thus, it would be of great interest to examine how changes in the Lands cycle alter IR phosphorylation by interacting with this mechanism in muscle.
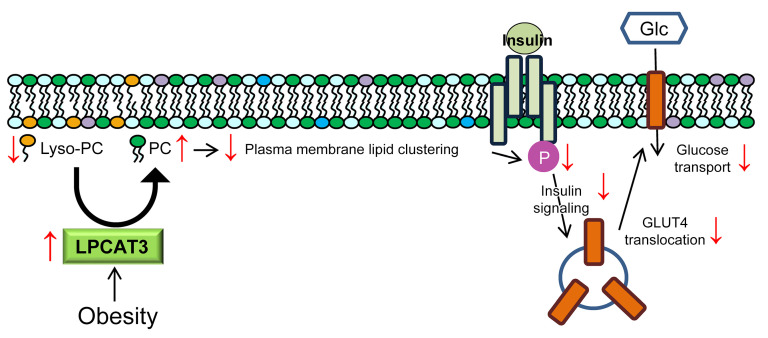
Figure 11. A proposed mechanism of action by which LPCAT3 promotes diet-induced skeletal muscle insulin resistance.
Obesity leads to acceleration of the Lands cycle through upregulation of LPCAT3. Increased LPCAT3 activity decreases lyso-PC and increases PC content, which leads to decreased microdomain clustering on the plasma membrane. Disorganization of plasma membrane lipid organization leads to attenuated insulin receptor signaling and decreased insulin-stimulated glucose uptake.
Observations in this study open up a potential opportunity to pharmacologically target this pathway to enhance skeletal muscle insulin sensitivity. It is noteworthy that the current study partly drew its conclusions from lipidomic analyses and loss-of-function studies performed in human samples, suggesting that this mechanism may be directly involved in the pathogenesis of skeletal muscle insulin resistance in human obesity. We are also interested in examining whether obesity induces similar changes in plasma membrane organization of other tissues to promote pathology.
Methods
Human subjects.
All participants were prescreened and/or self-reported to be free of any known metabolic diseases or heart conditions, were not tobacco users, were not taking any medications known to alter metabolism, and were sedentary. Six lean subjects without diabetes (LN: BMI < 25 kg/m2) and 6 subjects with severe obesity (OB: BMI > 40 kg/m2) were studied (all White females). The subjects were instructed not to exercise for approximately 48 hours before the muscle biopsy. A fasting blood sample (glucose and insulin) and muscle biopsy from the vastus lateralis were collected. A portion of the biopsy sample was frozen immediately, and another portion was used to isolate primary muscle cells.
Cell culture.
Primary human skeletal muscle cells (HSkMCs) were isolated from fresh muscle biopsies as previously described (15, 59). HSkMCs were cultured in growth medium containing low-glucose DMEM, 10% FBS, 0.5 mg/mL BSA, 0.5 mg/mL fetuin, 10 ng/mL human EGF, 1 μM dexamethasone, and 0.1% penicillin-streptomycin. HSkMCs were differentiated in low-glucose DMEM, 2% horse serum, 0.5 mg/mL BSA, 0.5 mg/mL fetuin, and 0.1% penicillin-streptomycin.
C2C12 myoblasts were grown in high-glucose DMEM (4.5 g/L glucose, with l-glutamine; Gibco 11965-092) supplemented with 10% FBS (heat-inactivated, certified, US origin; Gibco 10082-147), and 0.1% penicillin-streptomycin (10,000 U/mL; Gibco 15140122). C2C12 cells were differentiated into myotubes with low-glucose DMEM (1 g/L glucose, with l-glutamine and 110 mg/L sodium pyruvate; Gibco 11885-084) supplemented with 2% horse serum (defined; VWR 16777), and 0.1% penicillin-streptomycin. For experiments with CI-976, C2C12 myoblasts were differentiated with either 10 μM of CI-976 or equal-volume DMSO (vehicle). For experiments with MβCD, cells were incubated with 10 mM (1320 g/mol) MβCD for 1 hour directly dissolved into medium. For experiments with PDGF or EGF, C2C12 myotubes were incubated with 2.5 ng/mL PDGF, 100 ng/mL EGF, or 12 nM insulin for 10 minutes. Before all experiments, cells were serum-starved for 3 hours in low-glucose DMEM containing 1% BSA and 0.1% penicillin-streptomycin.
Quantitative reverse transcription PCR.
Samples were homogenized in TRIzol reagent (Life Technologies) to extract total RNA. One microgram RNA was reverse-transcribed using an IScript cDNA synthesis kit (Bio-Rad). Reverse transcription PCR (RT-PCR) was performed with the Viia 7 Real-Time PCR System (Life Technologies) using SYBR Green reagent (Life Technologies). All data were normalized to ribosomal L32 gene expression, and primer sequences are provided (Supplemental Table 2).
Mass spectrometry.
Global lipidomic analyses of LN and OB HSkMCs were performed at the Mass Spectrometry Resource at the Washington University School of Medicine (15). Extracted lipids with internal standards were analyzed with a Thermo Vantage triple-quadrupole mass spectrometer or a Thermo Trace GC Ultra mass spectrometer. Targeted lipidomic analyses for C2C12 myotubes and mouse skeletal muscles were conducted in the Metabolomics Core at the University of Utah (60–62). Extracted lipids with internal standards were analyzed with an Agilent triple-quadrupole mass spectrometer. The quantity of each lipid species was normalized to total lipid content for DRM/DSM experiments or to the total protein content for all others. The phospholipid saturation index was quantified by multiplication of the relative abundance of each phospholipid species by the total number of double bonds in the acyl chains of that species.
Merocyanine 540.
Merocyanine 540 (MC540) measurements were taken as previously described (63). In short, skeletal muscle cells (C2C12 cells and HSkMCs) were fully differentiated, and 2 million were used for measurements. Cells were washed with HBSS (Gibco 14025092) before resuspension in a cuvette with HBSS. MC540 in DMSO was added at a final concentration of 0.2 μM, and after a 10-minute dark incubation, an emission scan was performed ranging from 550 to 750 nm with fluorescence excitation set at 540 nm on a PTI QuantaMaster 6000 Fluorimeter.
Lentivirus-mediated knockdown of LPCAT3.
LPCAT3 expression was decreased using pLKO.1 lentiviral-RNAi system. Plasmids encoding shRNA for mouse LPCAT3 (shLPCAT3: TRCN0000121437) were obtained from MilliporeSigma. Packaging vector psPAX2 (ID 12260), envelope vector pMD2.G (ID 12259), and scrambled shRNA plasmid (SC: ID 1864) were obtained from Addgene. HEK293T cells in 10 cm dishes were transfected using 50 μL 0.1% polyethylenimine, 200 μL 0.15 M sodium chloride, and 500 μL Opti-MEM (with HEPES, 2.4 g/L sodium bicarbonate, and l-glutamine; Gibco 31985) with 2.66 μg of psPAX2, 0.75 μg of pMD2.G, and 3 μg of either scrambled or LPCAT3 shRNA plasmid. After 48 hours, growth medium was collected, filtered using 0.22 μm vacuum filters, and used to treat undifferentiated HSkMCs or C2C12 cells for 48 hours. To ensure that only cells infected with shRNA vectors were viable, cells were selected with puromycin throughout differentiation.
Western blot.
Whole muscle or cells were homogenized and Western blots were performed as previously described (59). Protein homogenates were analyzed for abundance of phosphorylated Tyr972–insulin receptor (Invitrogen, 44-800G), insulin receptor-β (Cell Signaling, 3020S), phosphorylated Thr308-Akt (Cell Signaling, 9275S), phosphorylated Ser472-Akt (Cell Signaling, 9271L), Akt (Cell Signaling, 9272S), phosphorylated Thr642-AS160 (Cell Signaling, 8881), AS160 (MilliporeSigma, 07-741), MyoD (Developmental Studies Hybridoma Bank [DSHB], D7F2), mitochondrial complexes I–V (Abcam, ab110413), MHC type I (DSHB, A4.840), MHC type IIa (DSHB, SC-71), MHC type IIx (DSHB, 6H1), MHC type IIb (DSHB, BF-F3), MHC neo (DSHB, N1.551), MHC emb (DSHB, BF-G6), caveolin-3 (BD Biosciences, 610-420), Na/K-ATPase (Cell Signaling, 3010S), flotillin-1 (Cell Signaling, 3253), and actin (MilliporeSigma, A2066).
Glycogen synthesis.
The glycogen synthesis rate was quantified as previously described (64, 65). Briefly, cells were incubated in medium containing d-[U-14C]glucose with insulin (12 nM) or without insulin for 2 hours at 37°C. Cells were then washed with ice-cold PBS and homogenized for 1 hour with 0.05% SDS. Part of the lysate was used for a protein assay, and the other was combined with 2 mg carrier glycogen and incubated at 100°C for 1 hour. Ice-cold ethanol (100%) was added to the boiled samples before overnight rocking at 4°C. Samples were then centrifuged at 11,000g for 15 minutes at 4°C to pellet glycogen. Pellets were resuspended in deionized H2O, and glycogen synthesis was calculated with liquid scintillation.
Generation of mice with skeletal muscle–specific LPCAT3 knockout or overexpression.
Conditional-LPCAT3-knockout (LPCAT3cKO+/+) mice were previously generated by flanking of exon 3 of the Lpcat3 gene with loxP sites (25). LPCAT3cKO+/+ mice were then crossed with tamoxifen-inducible, skeletal muscle–specific Cre recombinase (HSA-MerCreMer+/-) (34) mice to generate LPCAT3cKO+/+; HSA-MerCreMer-/- (control) and LPCAT3cKO+/+; HSA-MerCreMer+/- (skeletal muscle–specific LPCAT3 knockout; LPCAT3-MKO) mice. Conditional-LPCAT3-knockin (LPCAT3cKI+/+) mice were generated by insertion of a mouse LPCAT3 cDNA that was preceded by a lox-STOP-lox sequence into the Rosa26 locus of the genome. LPCAT3cKI+/+ mice were then crossed with HSA-MerCreMer+/– mice to generate LPCAT3cKI+/–; HSA-MerCreMer–/– (control) and LPCAT3cKI+/-; HSA-MerCreMer+/- (skeletal muscle–specific LPCAT3 knockin; LPCAT3-MKI) mice. Tamoxifen-injected (7.5 μg/g body mass, 5 consecutive days) control and LPCAT3-MKO/LPCAT3-MKI littermates were used for all experiments. Mice were maintained on a 12-hour light/12-hour dark cycle in a temperature-controlled room. Before all terminal experiments and tissue harvesting, mice were given an intraperitoneal injection of 80 mg/kg ketamine and 10 mg/kg xylazine.
Glucose tolerance test.
Intraperitoneal glucose tolerance tests were performed by injection of 1 mg glucose per gram body mass. Mice were fasted for 4 hours before glucose injection. Blood glucose was measured before glucose injection and 15, 30, 60, and 120 minutes after injection via tail bleed with a handheld glucometer (Bayer Contour 7151H). In a separate set of experiments, mice were injected with 1 mg glucose per gram body mass, and blood was taken from the facial vein at the 30-minute time point for insulin quantification.
Serum insulin and glucose quantification.
Blood was collected from the facial vein either before anesthesia or at the 30-minute time point of the glucose tolerance test. Blood was then placed at room temperature for 20 minutes to allow for clotting before centrifugation at 2000g for 10 minutes at 4°C. The supernatant (serum) was placed in a separate tube and stored at –80°C until analysis.
Serum glucose was quantified using a colorimetric assay. A glucose standard curve was generated (MilliporeSigma, G6918), and serum samples were mixed with a PGO enzyme (MilliporeSigma, P7119) and colorimetric substrate (MilliporeSigma, F5803) and measured at OD450 on a plate reader. Serum insulin was quantified with an insulin mouse serum kit (CisBio, 62IN3PEF) using fluorescence resonance energy transfer on a plate reader (Varioskan LUX, Thermo Fisher Scientific).
Hyperinsulinemic-euglycemic clamp.
Hyperinsulinemic-euglycemic clamping was performed as previously described (66–68). Briefly, unrestrained mice were able to move freely while being continuously infused with insulin (2 mU/kg/min) and a variable infusion of 50% dextrose to allow for steady-state blood glucose of approximately 150 mg/dL. Constant infusion of 3H-glucose throughout the experiment and for 90 minutes before the clamp allowed for the quantification of glucose kinetics. At the end of a 2-hour clamp, 14C-2-deoxyglucose (13 μCi/mouse) was administered during steady-state conditions.
Ex vivo skeletal muscle [3H]2-deoxy-d-glucose uptake.
Ex vivo glucose uptake was measured in the soleus muscle as previously described (69, 70). In brief, soleus muscles were dissected and placed in a recovery buffer (KHB with 0.1% BSA, 8 mM glucose, and 2 mM mannitol) at 37°C for 10 minutes. After incubation in recovery buffer, muscles were moved to preincubation buffer (KHB with 0.1% BSA, 2 mM sodium pyruvate, and 6 mM mannitol) with or without 200 μU/mL insulin for 15 minutes. After preincubation, muscles were placed in incubation buffer (KHB with 0.1% BSA, 9 mM [14C]mannitol, 1 mM [3H]2-deoxyglucose) with or without 200 μU/mL insulin for 15 minutes. Contralateral muscles were used for basal or insulin-stimulated measurements. After incubation, muscles were blotted dry on ice-cold filter paper, snap-frozen, and stored at –80°C until analyzed with liquid scintillation counting.
Muscle force generation.
Force-generating properties of extensor digitorum longus (EDL) muscles were measured as previously described (71). Briefly, EDL muscles were sutured at each tendon, and muscles were suspended at optimal length (Lo), which was determined by pulse stimulation. After Lo was identified, muscles were stimulated (0.35 seconds, pulse width 0.2 milliseconds) at frequencies ranging from 10 to 200 Hz. Muscle length and mass were measured to quantify cross-sectional area (72–74) for force normalization.
Muscle immunohistochemistry.
Frozen hind-limb muscle samples (tibialis anterior or EDL) embedded in OCT compound were sectioned at 10 μm using a cryostat (Microtome Plus). After 1 hour of blocking with M.O.M. mouse IgG Blocking Reagent (Vector Laboratories, MKB-2213), myofiber sections were incubated for 1 hour with concentrated BA.D5, SC.71, and BF.F3 (all 1:100; DSHB) and laminin (1:200; MilliporeSigma, L9393) in 2.5% normal horse serum. To visualize laminin (for fiber border), myosin heavy chain I (MHC I), MHC IIa, and MHC IIb, slides were incubated for 1 hour with the following secondary antibodies, respectively: AMCA (1:250; Vector Laboratories, CI-1000), Alexa Fluor 647 (1:250; Invitrogen, A21242), Alexa Fluor 488 (1:500; Invitrogen, A21121), and Alexa Fluor 555 (1:500; Invitrogen, A21426). Negatively stained fibers were considered MHC IIx. After staining, slides were coverslipped with mounting medium (Vector Laboratories, H-1000). Stained slides were imaged with a fully automated wide-field light microscope (Nikon Corp.) with a 10× objective lens. Images were captured using a high-sensitivity Andor Clara CCD camera.
Administration of CI-976 in vivo.
CI-976 was administered as previously described (75). Briefly, C57BL/6J mice were fed a high-fat diet (Envigo, TD.88137) for 6 weeks before 7 consecutive days of subcutaneous injection of either vehicle (1:9 vol/vol of ethanol/polyethylene glycol; Rigaku Reagents, 1008414) or CI-976 at a final concentration of 30 mg/kg.
GM-1 labeling and imaging.
GM-1 clusters were labeled using a Vybrant Alexa Fluor 488 Lipid Raft Labeling Kit (Thermo Fisher Scientific, V34404) as previously described (76). Briefly, 2 million myotubes were incubated in 1 mL in ice-cold starvation medium with 0.8 μg/mL fluorescent cholera toxin subunit B conjugate (CT-B) for 10 minutes. CT-B conjugates were then cross-linked with an anti–CT-B antibody (1:200) in ice-cold starvation medium for 15 minutes. Cells were fixed for 1 hour at 4°C in ice-cold 4% paraformaldehyde in PBS in the dark. Between each step, cells were washed twice in ice-cold PBS. Cells were imaged on an Olympus FV1000 confocal microscope (2.5×, HV: 600; offset: 30). Images were processed using NIH ImageJ. All images were background-subtracted with a rolling ball radius of 50 pixels. Images were blindly scored as exhibiting clustering of microdomains or nonclustering. Images were then subjected to color thresholding using the Otsu method (77, 78) (designed for thresholding images for cluster analyses) and made binary. A particle analysis of all particles that were larger than 0.1 μm2 was performed to determine the average cluster size for each cell that was imaged (79). For each experiment, 35–50 cells per group were analyzed and the median was taken as a representative of that experiment.
Detergent-resistant membrane isolation.
Detergent-resistant membranes (DRMs) and detergent-soluble membranes (DSMs) were isolated as previously described (79). Briefly, 2 × 15 cm plates of cells were scraped in ice-cold PBS and then pelleted. Cells were resuspended in 1 mL of cold homogenization buffer (Mes-buffered saline [MBS], 1% Triton X wt/vol, and protease and phosphatase inhibitor) and passed through a 23-gauge needle 6 times before incubation at 4°C for 30 minutes. MBS was added to the homogenate until a volume of 2.5 mL was reached, then mixed with 2.5 mL of 90% sucrose in MBS, and 4 mL of this mixture was added to an ultracentrifuge tube (Beckman Coulter, 344061). A sucrose gradient was generated by addition of 4 mL of 35% sucrose followed by 4 mL of 5% sucrose. Samples were then centrifuged at 100,000g at 4°C for 20 hours in a swinging bucket rotor (Beckman L8-M Ultracentrifuge, SW28 Rotor).
Statistics.
Statistical analysis was performed using Prism 7 software (GraphPad). Two-tailed Student’s t tests were performed with data composed of 2 groups, and 2-way ANOVA followed by Šidák’s multiple-comparison test was performed for more than 2 groups. All data are mean ± SEM, and statistical significance was set at P less than 0.05.
Study approval.
The experimental protocol was approved by the Internal Review Board for Human Research at East Carolina University. Informed consent was obtained prior to inclusion in the study.
Animal experiments were approved by the University of Utah Institutional Animal Care and Use Committee.
Author contributions
PJF and KF contributed to study concept design and wrote the manuscript. JAH performed human muscle biopsies. JMJ and MJD contributed to study design and data analysis. XR, PT, TDG, JMM, and PJF developed and maintained the mouse models. PJF and WLH performed hyperinsulinemic-euglycemic clamp procedure and analysis. JAM, JEC, HS, and JT performed mass spectrometry analyses. PJF, KF, and SRS performed analyses of the physical properties of phospholipid membranes. ARPV and PJF performed analysis of muscle force production. PS performed muscle histology measurements. PJF performed all biochemical assays and metabolic phenotyping measurements. KCK and AJL performed correlation analyses with 106 mouse strains. JAH, PT, JT, JEC, MJD, SRS, and WLH edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK107397, DK109888, AG063077 (to KF); DK108333 and DK112826 (to WLH); HL030568 and HL136618 (to PT); AT008375 and ES031378 (to SRS); HL028481 and HL030568 (to AJL); AG050781 (to MJD); HL125695 (to JMM); and DK056112 (to JAH); American Heart Association grants 18PRE33960491 (to ARPV) and 19PRE34380991 (to JMJ); and the Larry H. and Gail Miller Family Foundation (to PJF). The University of Utah Metabolomics, Mass Spectrometry, and Proteomics Core is supported by NIH grants S10-OD016232, S10-OD021505, and U54-DK110858. The Washington University Biomedical MS Resource is supported by US Public Health Services grants P41-GM103422, P30-DK020579, and P30-DK056341.
Version 1. 02/16/2021
In-Press Preview
Version 2. 04/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(8):e135963.https://doi.org/10.1172/JCI135963.
See the related Commentary at Remodeling glycerophospholipids affects obesity-related insulin signaling in skeletal muscle.
Contributor Information
Patrick J. Ferrara, Email: patrick.ferrara@health.utah.edu.
Xin Rong, Email: rongxin1215@gmail.com.
J. Alan Maschek, Email: alan.maschek@pharm.utah.edu.
Anthony R.P. Verkerke, Email: anthony.verkerke@health.utah.edu.
Piyarat Siripoksup, Email: p.siripoksup@utah.edu.
Haowei Song, Email: songhw@gmail.com.
Thomas D. Green, Email: GREENT@ecu.edu.
Karthickeyan C. Krishnan, Email: c.karthickeyan@gmail.com.
Jordan M. Johnson, Email: jordan.johnson@health.utah.edu.
John Turk, Email: jturk@wustl.edu.
Joseph A. Houmard, Email: HOUMARDJ@ecu.edu.
Aldons J. Lusis, Email: jlusis@mednet.ucla.edu.
Micah J. Drummond, Email: micah.drummond@hsc.utah.edu.
Joseph M. McClung, Email: mcclungj@ecu.edu.
James E. Cox, Email: jcox@genetics.utah.edu.
Peter Tontonoz, Email: ptontonoz@mednet.ucla.edu.
William L. Holland, Email: will.holland@hsc.utah.edu.
Katsuhiko Funai, Email: kfunai@health.utah.edu.
References
- 1.Heron M. Deaths: leading causes for 2017. Natl Vital Stat Rep. 2019;68(6):1–77. [PubMed] [Google Scholar]
- 2.Huo X, et al. Risk of non-fatal cardiovascular diseases in early-onset versus late-onset type 2 diabetes in China: a cross-sectional study. Lancet Diabetes Endocrinol. 2016;4(2):115–124. doi: 10.1016/S2213-8587(15)00508-2. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA, et al. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30(12):1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 4.Thiebaud D, et al. The effect of graded doses of insulin on total glucose uptake, glucose oxidation, and glucose storage in man. Diabetes. 1982;31(11):957–963. doi: 10.2337/diacare.31.11.957. [DOI] [PubMed] [Google Scholar]
- 5.DeFronzo RA, Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009;32(suppl 2):S157–S163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube JJ, et al. Effects of acute lipid overload on skeletal muscle insulin resistance, metabolic flexibility, and mitochondrial performance. Am J Physiol Endocrinol Metab. 2014;307(12):E1117–E1124. doi: 10.1152/ajpendo.00257.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manco M, et al. Insulin resistance directly correlates with increased saturated fatty acids in skeletal muscle triglycerides. Metabolism. 2000;49(2):220–224. doi: 10.1016/S0026-0495(00)91377-5. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, et al. Saturated, but not n-6 polyunsaturated, fatty acids induce insulin resistance: role of intramuscular accumulation of lipid metabolites. J Appl Physiol (1985) 2006;100(5):1467–1474. doi: 10.1152/japplphysiol.01438.2005. [DOI] [PubMed] [Google Scholar]
- 9.Chavez JA, Summers SA. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys. 2003;419(2):101–109. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Bergman BC, et al. Localisation and composition of skeletal muscle diacylglycerol predicts insulin resistance in humans. Diabetologia. 2012;55(4):1140–1150. doi: 10.1007/s00125-011-2419-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perreault L, et al. Intracellular localization of diacylglycerols and sphingolipids influences insulin sensitivity and mitochondrial function in human skeletal muscle. JCI Insight. 2018;3(3):96805. doi: 10.1172/jci.insight.96805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams JM, 2nd, et al. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes. 2004;53(1):25–31. doi: 10.2337/diabetes.53.1.25. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz-Peiffer C, et al. Ceramide generation is sufficient to account for the inhibition of the insulin-stimulated PKB pathway in C2C12 skeletal muscle cells pretreated with palmitate. J Biol Chem. 1999;274(34):24202–24210. doi: 10.1074/jbc.274.34.24202. [DOI] [PubMed] [Google Scholar]
- 14.Itani SI, et al. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. 2002;51(7):2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- 15.Paran CW, et al. Reduced efficiency of sarcolipin-dependent respiration in myocytes from humans with severe obesity. Obesity (Silver Spring) 2015;23(7):1440–1449. doi: 10.1002/oby.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulver MW, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2(4):251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lands WE. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231(2):883–888. [PubMed] [Google Scholar]
- 18.Fink KL, Gross RW. Modulation of canine myocardial sarcolemmal membrane fluidity by amphiphilic compounds. Circ Res. 1984;55(5):585–594. doi: 10.1161/01.RES.55.5.585. [DOI] [PubMed] [Google Scholar]
- 19.Bing RJ, et al. Membrane function and vascular reactivity. Biosci Rep. 1993;13(2):61–67. doi: 10.1007/BF01145958. [DOI] [PubMed] [Google Scholar]
- 20.Hishikawa D, et al. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J Lipid Res. 2014;55(5):799–807. doi: 10.1194/jlr.R046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazachkov M, et al. Substrate preferences of a lysophosphatidylcholine acyltransferase highlight its role in phospholipid remodeling. Lipids. 2008;43(10):895–902. doi: 10.1007/s11745-008-3233-y. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, et al. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J Biol Chem. 2008;283(13):8258–8265. doi: 10.1074/jbc.M710422200. [DOI] [PubMed] [Google Scholar]
- 23.Pein H, et al. Vitamin A regulates Akt signaling through the phospholipid fatty acid composition. FASEB J. 2017;31(10):4566–4577. doi: 10.1096/fj.201700078R. [DOI] [PubMed] [Google Scholar]
- 24.Koeberle A, et al. Arachidonoyl-phosphatidylcholine oscillates during the cell cycle and counteracts proliferation by suppressing Akt membrane binding. Proc Natl Acad Sci U S A. 2013;110(7):2546–2551. doi: 10.1073/pnas.1216182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong X, et al. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. Elife. 2015;4:e06557. doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks BW, et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21(2):334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eto M, et al. Lysophosphatidylcholine acyltransferase 3 is the key enzyme for incorporating arachidonic acid into glycerophospholipids during adipocyte differentiation. Int J Mol Sci. 2012;13(12):16267–16280. doi: 10.3390/ijms131216267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin SA, et al. Measurement of lysophospholipid acyltransferase activities using substrate competition. J Lipid Res. 2014;55(4):782–791. doi: 10.1194/jlr.D044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, et al. Expression and characterization of recombinant caveolin. Purification by polyhistidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J Biol Chem. 1996;271(1):568–573. doi: 10.1074/jbc.271.1.568. [DOI] [PubMed] [Google Scholar]
- 30.Murata M, et al. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci U S A. 1995;92(22):10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janes PW, et al. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J Cell Biol. 1999;147(2):447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers K, Brown WJ. Characterization of a novel CI-976-sensitive lysophospholipid acyltransferase that is associated with the Golgi complex. Biochem Biophys Res Commun. 2004;313(3):681–686. doi: 10.1016/j.bbrc.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Chambers K, et al. A unique lysophospholipid acyltransferase (LPAT) antagonist, CI-976, affects secretory and endocytic membrane trafficking pathways. J Cell Sci. 2005;118(Pt 14):3061–3071. doi: 10.1242/jcs.02435. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy JJ, et al. Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet Muscle. 2012;2(1):8. doi: 10.1186/2044-5040-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hishikawa D, et al. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc Natl Acad Sci U S A. 2008;105(8):2830–2835. doi: 10.1073/pnas.0712245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gijon MA, et al. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283(44):30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vainio S, et al. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 2002;3(1):95–100. doi: 10.1093/embo-reports/kvf010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foti M, et al. The neck of caveolae is a distinct plasma membrane subdomain that concentrates insulin receptors in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 2007;104(4):1242–1247. doi: 10.1073/pnas.0610523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabayama K, et al. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proc Natl Acad Sci U S A. 2007;104(34):13678–13683. doi: 10.1073/pnas.0703650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn-Obercyger M, et al. A high-cholesterol diet increases the association between caveolae and insulin receptors in rat liver. J Lipid Res. 2009;50(1):98–107. doi: 10.1194/jlr.M800441-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Capozza F, et al. Caveolin-3 knockout mice show increased adiposity and whole body insulin resistance, with ligand-induced insulin receptor instability in skeletal muscle. Am J Physiol Cell Physiol. 2005;288(6):C1317–C1331. doi: 10.1152/ajpcell.00489.2004. [DOI] [PubMed] [Google Scholar]
- 42.Oshikawa J, et al. Insulin resistance in skeletal muscles of caveolin-3-null mice. Proc Natl Acad Sci U S A. 2004;101(34):12670–12675. doi: 10.1073/pnas.0402053101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng YF, et al. The Caveolin-3 P104L mutation of LGMD-1C leads to disordered glucose metabolism in muscle cells. Biochem Biophys Res Commun. 2017;486(2):218–223. doi: 10.1016/j.bbrc.2017.02.072. [DOI] [PubMed] [Google Scholar]
- 44.Shang L, et al. The caveolin-3 P104L mutation in LGMD-1C patients inhibits non-insulin-stimulated glucose metabolism and growth but promotes myocyte proliferation. Cell Biol Int. 2019;43(6):669–677. doi: 10.1002/cbin.11144. [DOI] [PubMed] [Google Scholar]
- 45.Huang Y, et al. Effect of type 2 diabetes mellitus caveolin-3 K15N mutation on glycometabolism. Exp Ther Med. 2019;18(4):2531–2539. doi: 10.3892/etm.2019.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shang L, et al. Caveolin-3 promotes glycometabolism, growth and proliferation in muscle cells. PLoS One. 2017;12(12):0189004. doi: 10.1371/journal.pone.0189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 48.Galbiati F, et al. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem. 2001;276(24):21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- 49.Uhles S, et al. Isoform-specific insulin receptor signaling involves different plasma membrane domains. J Cell Biol. 2003;163(6):1327–1337. doi: 10.1083/jcb.200306093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa A, et al. PDGF enhances store-operated Ca2+ entry by upregulating STIM1/Orai1 via activation of Akt/mTOR in human pulmonary arterial smooth muscle cells. Am J Physiol Cell Physiol. 2012;302(2):C405–C411. doi: 10.1152/ajpcell.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi K, et al. Zinc promotes proliferation and activation of myogenic cells via the PI3K/Akt and ERK signaling cascade. Exp Cell Res. 2015;333(2):228–237. doi: 10.1016/j.yexcr.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106(2):171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chavez JA, Summers SA. A ceramide-centric view of insulin resistance. Cell Metab. 2012;15(5):585–594. doi: 10.1016/j.cmet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Kien CL, et al. A lipidomics analysis of the relationship between dietary fatty acid composition and insulin sensitivity in young adults. Diabetes. 2013;62(4):1054–1063. doi: 10.2337/db12-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timmers S, et al. Augmenting muscle diacylglycerol and triacylglycerol content by blocking fatty acid oxidation does not impede insulin sensitivity. Proc Natl Acad Sci U S A. 2012;109(29):11711–11716. doi: 10.1073/pnas.1206868109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demeure O, et al. Regulation of LPCAT3 by LXR. Gene. 2011;470(1–2):7–11. doi: 10.1016/j.gene.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Yamazaki T, et al. Peroxisome proliferators attenuate free arachidonic acid pool in the kidney through inducing lysophospholipid acyltransferases. J Pharmacol Sci. 2009;111(2):201–210. doi: 10.1254/jphs.09162FP. [DOI] [PubMed] [Google Scholar]
- 58.Lyu K, et al. A membrane-bound diacylglycerol species induces PKCϵ-mediated hepatic insulin resistance. Cell Metab. 2020;32(4):654–664. doi: 10.1016/j.cmet.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heden TD, et al. Greater oxidative capacity in primary myotubes from endurance-trained women. Med Sci Sports Exerc. 2017;49(11):2151–2157. doi: 10.1249/MSS.0000000000001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heden TD, et al. Mitochondrial PE potentiates respiratory enzymes to amplify skeletal muscle aerobic capacity. Sci Adv. 2019;5(9):eaax8352. doi: 10.1126/sciadv.aax8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verkerke ARP, et al. Phospholipid methylation regulates muscle metabolic rate through Ca2+ transport efficiency. Nat Metab. 2019;1(9):876–885. doi: 10.1038/s42255-019-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson JM, et al. Alternative splicing of UCP1 by non-cell-autonomous action of PEMT. Mol Metab. 2020;31:55–66. doi: 10.1016/j.molmet.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeczycki TN, et al. Increasing levels of cardiolipin differentially influence packing of phospholipids found in the mitochondrial inner membrane. Biochem Biophys Res Commun. 2014;450(1):366–371. doi: 10.1016/j.bbrc.2014.05.133. [DOI] [PubMed] [Google Scholar]
- 64.Park S, et al. Electrical pulse stimulation induces differential responses in insulin action in myotubes from severely obese individuals. J Physiol. 2019;597(2):449–466. doi: 10.1113/JP276990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Khalili L, et al. Insulin action in cultured human skeletal muscle cells during differentiation: assessment of cell surface GLUT4 and GLUT1 content. Cell Mol Life Sci. 2003;60(5):991–998. doi: 10.1007/s00018-003-3001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holland WL, et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med. 2011;17(1):55–63. doi: 10.1038/nm.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma AX, et al. Glucagon receptor antagonism improves glucose metabolism and cardiac function by promoting AMP-mediated protein kinase in diabetic mice. Cell Rep. 2018;22(7):1760–1773. doi: 10.1016/j.celrep.2018.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Funai K, et al. Skeletal muscle phospholipid metabolism regulates insulin sensitivity and contractile function. Diabetes. 2016;65(2):358–370. doi: 10.2337/db15-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Funai K, et al. Muscle lipogenesis balances insulin sensitivity and strength through calcium signaling. J Clin Invest. 2013;123(3):1229–1240. doi: 10.1172/JCI65726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrara PJ, et al. Hypothermia decreases O2 cost for ex vivo contraction in mouse skeletal muscle. Med Sci Sports Exerc. 2018;50(10):2015–2023. doi: 10.1249/MSS.0000000000001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mendez J, Keys A. Density and composition of mammalian muscle. Metabolism. 1960;9:184–188. [Google Scholar]
- 73.Moorwood C, et al. Isometric and eccentric force generation assessment of skeletal muscles isolated from murine models of muscular dystrophies. J Vis Exp. 2013;(71):50036. doi: 10.3791/50036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brooks SV, Faulkner JA. Contractile properties of skeletal muscles from young, adult and aged mice. J Physiol. 1988;404:71–82. doi: 10.1113/jphysiol.1988.sp017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krause BR, et al. In vivo evidence that the lipid-regulating activity of the ACAT inhibitor CI-976 in rats is due to inhibition of both intestinal and liver ACAT. J Lipid Res. 1993;34(2):279–294. doi: 10.1016/S0022-2275(20)40755-2. [DOI] [PubMed] [Google Scholar]
- 76.Rockett BD, et al. Fish oil increases raft size and membrane order of B cells accompanied by differential effects on function. J Lipid Res. 2012;53(4):674–685. doi: 10.1194/jlr.M021782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 78.Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging. 2004;13(1):146–168. doi: 10.1117/1.1631315. [DOI] [Google Scholar]
- 79.Shaikh SR, et al. Docosahexaenoic acid modifies the clustering and size of lipid rafts and the lateral organization and surface expression of MHC class I of EL4 cells. J Nutr. 2009;139(9):1632–1639. doi: 10.3945/jn.109.108720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.