Abstract
BACKGROUND
Rejection is the primary barrier to broader implementation of vascularized composite allografts (VCAs), including face and limb transplants. The immunologic pathways activated in face transplant rejection have not been fully characterized.
METHODS
Using skin biopsies prospectively collected over 9 years from 7 face transplant patients, we studied rejection by gene expression profiling, histology, immunostaining, and T cell receptor sequencing.
RESULTS
Grade 1 rejection did not differ significantly from nonrejection, suggesting that it does not represent a pathologic state. In grade 2, there was a balanced upregulation of both proinflammatory T cell activation pathways and antiinflammatory checkpoint and immunomodulatory pathways, with a net result of no tissue injury. In grade 3, IFN-γ–driven inflammation, antigen-presenting cell activation, and infiltration of the skin by proliferative T cells bearing markers of antigen-specific activation and cytotoxicity tipped the balance toward tissue injury. Rejection of VCAs and solid organ transplants had both distinct and common features. VCA rejection was uniquely associated with upregulation of immunoregulatory genes, including SOCS1; induction of lipid antigen–presenting CD1 proteins; and infiltration by T cells predicted to recognize CD1b and CD1c.
CONCLUSION
Our findings suggest that the distinct features of VCA rejection reflect the unique immunobiology of skin and that enhancing cutaneous immunoregulatory networks may be a useful strategy in combatting rejection.
Trial registration
ClinicalTrials.gov NCT01281267.
FUNDING
Assistant Secretary of Defense and Health Affairs, through Reconstructive Transplant Research (W81XWH-17-1-0278, W81XWH-16-1-0647, W81XWH-16-1-0689, W81XWH-18-1-0784, W81XWH-1-810798); American Society of Transplantation’s Transplantation and Immunology Research Network Fellowship Research Grant; Plastic Surgery Foundation Fellowship from the American Society of Plastic Surgeons; Novo Nordisk Foundation (NNF15OC0014092); Lundbeck Foundation; Aage Bangs Foundation; A.P. Moller Foundation for the Advancement of Medical Science; NIH UL1 RR025758.
Keywords: Immunology, Transplantation
Keywords: Adaptive immunity, Cellular immune response
Introduction
At least 40 human face transplants have been reported worldwide since the first procedure was performed in 2005. In the intervening years, face transplantation has proven to be an effective and life-transforming reconstructive technique (1, 2). It is estimated that approximately 7 million people each year in the United States could benefit from vascularized composite allografts (VCAs), such as face and limb transplants, to repair defects from trauma, tumor resections, or limb loss (3). Despite promising functional and aesthetic restoration, vascularized composite allotransplantation has yet to reach wide implementation in large part because of the challenges associated with diagnosing and treating rejection.
VCA procedures transfer large areas of donor skin that are highly populated by donor-derived professional antigen-presenting cells (APCs), including Langerhans cells, dermal dendritic cells, and macrophages (4), as well as resident memory T cells with both pro- and antiinflammatory properties (5, 6). The transfer of skin and its resident immune cell populations may underlie the high frequency of rejection episodes in face transplant recipients. At least 1 episode of skin acute cellular rejection (ACR) has been reported during the first year after transplant in all face transplant recipients (7), compared with 10%–15% in kidney or heart recipients (8) on a similar immunosuppressive regimen. Moreover, episodes of ACR after the first year post-transplant are common in face transplants but rare in solid organ transplants (9). Although most ACR episodes in vascularized composite allografts are reversible with prompt treatment, reports suggest that the number of ACR episodes may be predictive of future chronic allograft rejection (10) and graft loss (11). A deeper understanding of the immunopathology of face transplant ACR is needed to establish treatment and preventative therapies specifically targeted for these patients. In this study, we addressed this knowledge gap by gene expression profiling, histology, immunostaining, and T cell receptor (TCR) sequencing. We studied all grades of ACR that were prospectively diagnosed in one of the most well established vascularized composite allotransplantation centers worldwide to ensure that our studies included the spectrum of ACR encountered in clinical practice. We also compared rejection with non–transplant-related inflammatory skin diseases to identify the genes uniquely associated with face transplant rejection.
Results
Patient characteristics and biopsy details.
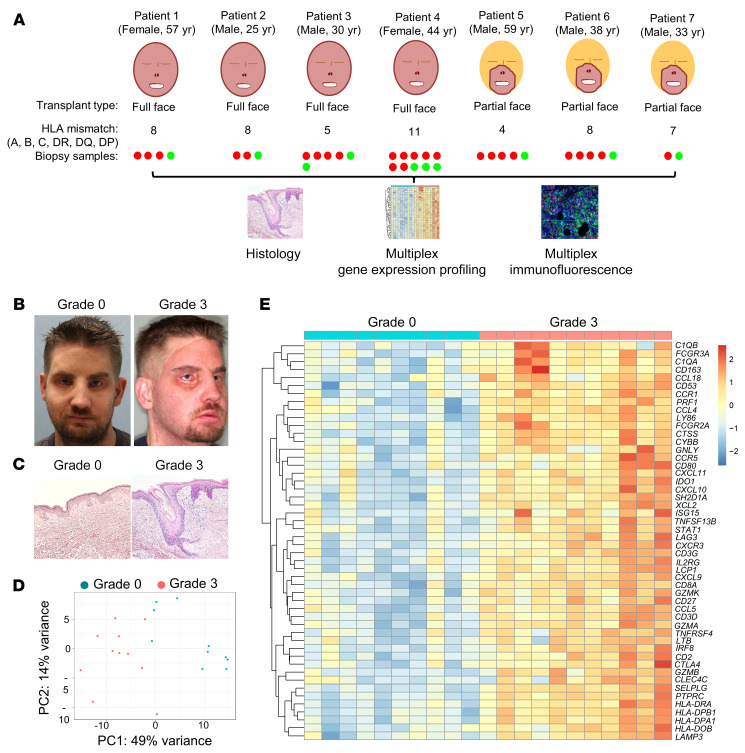
Between April 2009 and February 2014, 7 patients received face transplants at the Brigham and Women’s Hospital. All patients received transplants across multiple HLA mismatches between donor and recipient (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI135166DS1). There were 5 male and 2 female recipients; all were White. We identified 38 face transplant skin biopsies from these patients that fulfilled our inclusion criteria (see Methods). These biopsies were examined using multiplex gene expression profiling, immunostaining, histologic examination, and high-throughput TCR sequencing. Histologic analysis was performed using the 2007 Banff classification (12) (grade 0 = no rejection; grade 1 = mild rejection; grade 2 = moderate rejection; grade 3 = severe rejection). All patients had more than 1 biopsy included with at least 1 biopsy collected during ACR and 1 biopsy collected during nonrejection (Figure 1, A–C, and Supplemental Table 2). The mean follow-up time was 76.2 months (range 48–114 months) after transplant. All episodes of ACR were successfully treated (treatments for each ACR episode are shown in Supplemental Table 2). There were no graft losses or patient deaths. For gene expression analyses, we studied the same archived formalin-fixed paraffin-embedded (FFPE) skin biopsy used for histologic assessment of rejection; we used NanoString-based gene expression profiling because of its superior ability to quantitatively measure RNA expression in FFPE samples (13).
Figure 1. Human face transplant rejection has a distinct gene expression signature.
(A) Design of the study. Skin biopsies from 7 face transplant patients collected during episodes of acute cellular rejection (red) and nonrejection (green) were analyzed using histologic examination, multiplex gene expression profiling, and immunostaining. (B) Clinical photographs of a recipient of a full face transplant during nonrejection (grade 0) and severe acute cellular rejection (grade 3), demonstrating edema and erythema of the transplanted face. (C) Representative examples of H&E staining of a face transplant skin biopsy graded as nonrejection (grade 0, minimal inflammatory infiltrates), and a second biopsy graded as severe acute cellular rejection (grade 3, dermal inflammatory infiltrates with apoptotic keratinocytes). (D) Unsupervised principal component analysis clustered grade 3 rejection biopsies (n = 11) separately from grade 0 samples (n = 10). (E) Heatmap of the top 50 genes differentially expressed in grade 3 compared with grade 0 biopsies (log2 fold change >1; adjusted P value <0.05). Differentially expressed genes (DEGs) were obtained using normalized gene expression counts as input and the Wald significance test. Each column represents a facial allograft biopsy. Gene values are row scaled. The full list of differentially expressed genes and associated statistics are shown in Supplemental Table 3.
Severe ACR in face transplants has a distinct gene expression signature.
We compared the gene expression profiles of grade 0 biopsies with those obtained during grade 3 rejection to identify the molecular changes associated with severe ACR. Unsupervised principal component analysis (PCA) clustered grade 3 biopsies according to similarities in their patterns of expression in 769 genes, irrespective of the immunosuppressive regimen the patient was receiving at the time of biopsy. There was a clear separation of grade 3 biopsies from grade 0 along the first principal component (Figure 1D). We next used unsupervised PCA with labeling of individual patients to rule out the possibility that the inclusion of multiple samples from each patient might account for the observed molecular clustering. The analysis revealed that the biopsies clustered on the basis of rejection status, and that there was no clustering based on an individual patient (Supplemental Figure 1).
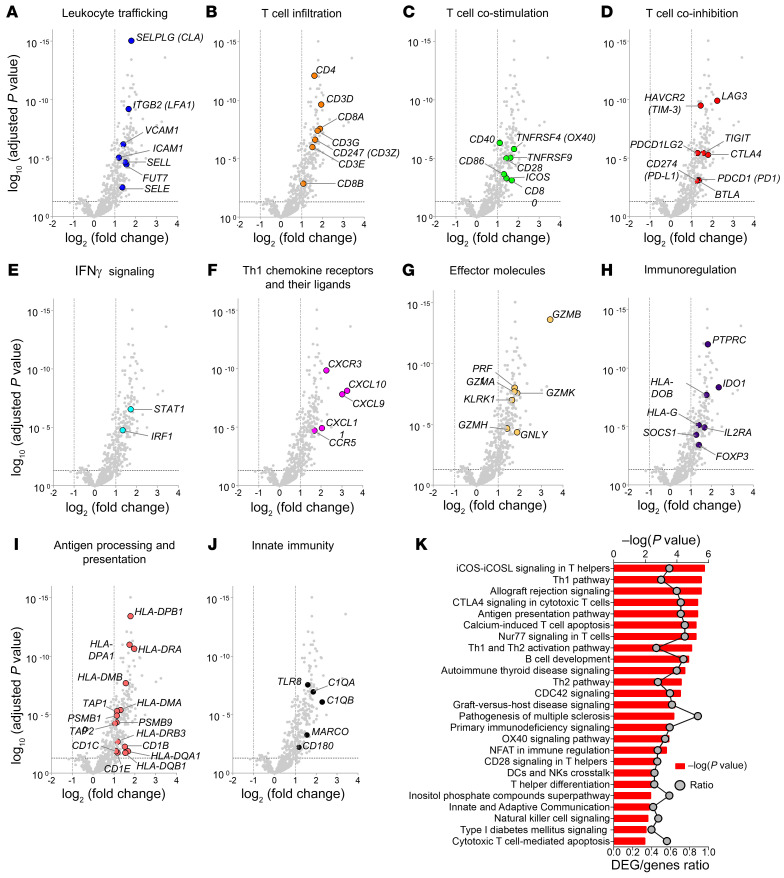
A total of 202 genes were differentially expressed in grade 3 biopsies compared with grade 0 (log2 fold change >1; adjusted P value <0.05) (Supplemental Table 3). The top 50 differentially expressed genes (DEGs) are shown in Figure 1E. The single most upregulated gene was GZMB (log2 fold change = 3.41 compared with grade 0). Many of the top upregulated genes encoded proteins associated with leukocyte trafficking (Figure 2A), T cell infiltration (Figure 2B), T cell costimulation (Figure 2C), IFN-γ signaling (Figure 2E), Th1 polarization (Figure 2F), and effector molecules (Figure 2G). Genes associated with antigen processing and presentation were prominent among upregulated genes (Figure 2I), as were genes associated with innate immunity (Figure 2J). Interestingly, grade 3 ACR biopsies had increased expression of T cell coinhibitory receptor genes, including CTLA4, LAG3, TIGIT, BTLA, PDCD1, CD274, PDCD1LG2, and HAVCR2 (Figure 2D), and genes associated with immunoregulation, including IDO1, IL2RA, FOXP3, SOCS1, HLA-DOB, HLA-G, and PTPRC (Figure 2H), suggesting that regulatory processes are induced within face transplants during severe rejection.
Figure 2. Effector T cell molecules, T cell cosignaling, and IFN-γ signaling molecules are upregulated in acute cellular rejection of face transplants.
(A–J) Volcano plots showing genes differentially expressed in grade 3 rejection biopsies (n = 11) versus grade 0 nonrejection biopsies (n = 10). DEGs were obtained using normalized gene expression counts as input and the Wald significance test. Each dot represents an individual gene. Horizontal dashed lines represent an adjusted P value cutoff of –log10 (0.05); vertical dashed lines represent log2 fold change of –1 and +1. Synonymous gene symbols, according to NCBI Gene, are provided in parentheses. All volcano plots illustrate identical data, but each highlights selected genes associated with leukocyte trafficking (A); T cell infiltration (B); T cell costimulation (C); T cell coinhibition (D); IFN-γ signaling (E); Th1 chemokine receptors and their ligands (F); effector molecules (G); immunoregulation (H); antigen processing and presentation (I); or innate immunity (J). (K) The top 25 canonical pathways overrepresented in 202 DEGs in grade 3 rejection biopsies in relation to grade 0 are shown. The significance of the association between gene expression and canonical pathways was estimated by the P value (depicted in bar graphs; primary y axis), and the ratio value reflects its strength (depicted as line graphs; secondary y axis). P values were determined using Fisher’s exact test with multiple testing adjustments according to the Benjamini-Hochberg false discovery rate method.
We next performed functional pathway analysis of the 202 DEGs in grade 3 biopsies compared with grade 0 using Ingenuity Pathway Analysis (IPA). Consistent with the changes in gene expression, the top 5 canonical pathways were iCOS/iCOSL signaling in Th cells, the Th1 pathway, allograft rejection signaling, CTLA-4 signaling in cytotoxic T cells, and the antigen presentation pathway (Figure 2K and Supplemental Table 4). We further explored the biological processes that occurred within the allografts during grade 3 ACR using Gene Ontology (GO) analysis. The top 5 biological processes most strongly associated with the DEGs in grade 3 ACR were lymphocyte costimulation, T cell costimulation, immune system process, leukocyte activation, and antigen processing and presentation of exogenous antigen (Supplemental Table 5). Next, the IPA upstream regulator analysis was used to identify potential upstream regulators of gene expression changes and ascertain their activated or inhibited status. The top cytokine upstream regulators inferred to be activated during grade 3 ACR were IFN-γ, TNF, and IL-2 (Supplemental Table 6).
The biopsies included in this study were collected at various times after transplant (range 22–2520 days following transplantation). To address the potential confounding effects of the time of biopsy from transplantation, we performed an additional analysis comparing severe ACR and nonrejection samples collected at early (<730 days) and late (>730 days) time points after transplant. Unsupervised PCA revealed that the biopsies clustered together based on the rejection status, irrespective of the time of biopsy collection (Supplemental Figure 2). This finding suggests that the severe ACR signature reflects the biology of rejection and is independent of time from transplantation.
Mild ACR and nonrejection samples are not significantly different.
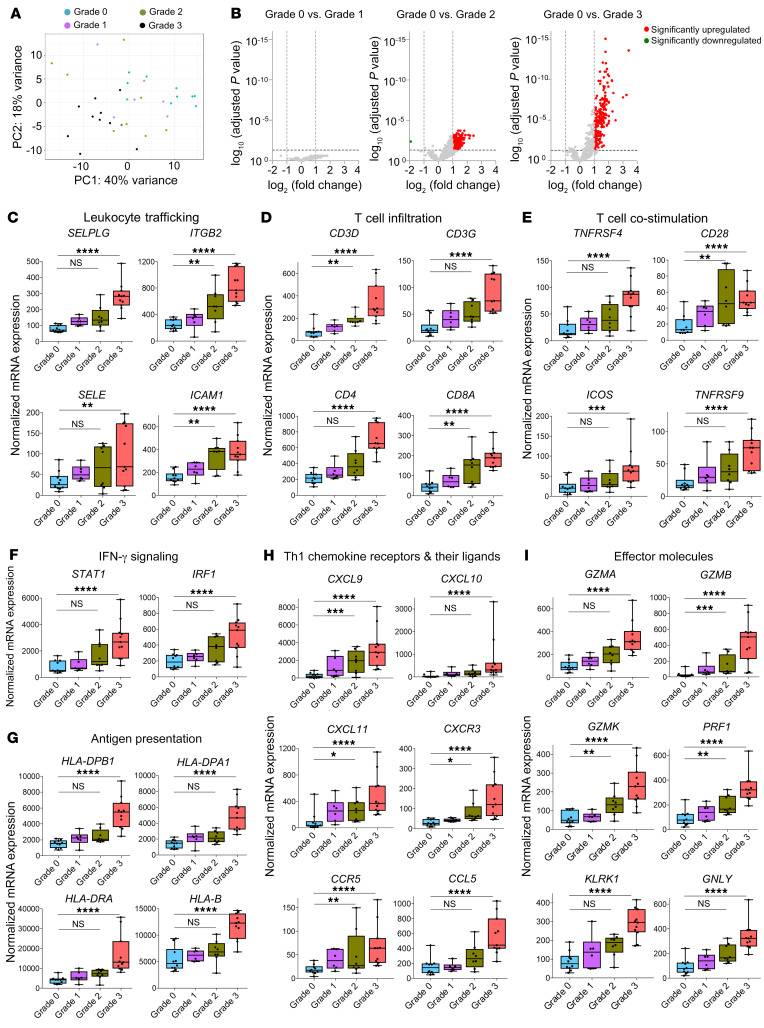
Mild ACR (grade 1) is the most commonly reported rejection grade in VCAs such as face transplants (14). According to the 2007 Banff schema, the difference between no rejection (grade 0) and mild rejection (grade 1) is “rare perivascular inflammation” versus “mild perivascular inflammation,” respectively. The terms “rare” and “mild” are not defined by an objective set of parameters. Healthy adult human skin contains 1 million memory T cells per square centimeter, and many of these T cells are located in a perivascular distribution under noninflamed conditions (5). The perivascular T cell presence in VCA biopsies could, therefore, represent either normal skin resident T cell presence or true pathogenic immune activation. To discriminate between these possibilities, we carried out an unsupervised PCA of normalized gene expression counts of biopsies collected during nonrejection and mild (grade 1), moderate (grade 2), and severe (grade 3) ACR. There was no clear separation between mild ACR and nonrejection biopsies (Figure 3A). Subsequent differential expression analysis between nonrejection and grade 1 biopsies revealed no DEGs (Figure 3B, left), suggesting there was no significant difference between nonrejection and grade 1 ACR at the gene expression level.
Figure 3. Comparison of acute rejection stages demonstrates that distinct gene expression patterns develop in grade 2 and grade 3 rejections.
(A) Unsupervised principal component analysis clustered all grade 3 samples (n = 11) separately from grade 0 biopsies (n = 10), but grade 1 (n = 6) and grade 2 biopsies (n = 8) were molecularly heterogeneous. (B) Volcano plots showing DEGs in grade 1, 2, and 3 rejections in relation to grade 0 samples. DEGs were obtained using normalized gene expression counts as input and the Wald significance test. (C–I) Box plots of normalized expression values of genes associated with leukocyte trafficking (C); T cell infiltration (D); T cell costimulation (E); IFN-γ signaling (F); antigen presentation (G); Th1 chemokine receptors and their ligands (H); and effector molecules (I). Horizontal lines represent median values, with whiskers extending to the farthest data points. Adjusted *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. The full lists of DEGs and associated statistics are shown in Supplemental Tables 3 and 7.
Progression to severe ACR involves a stepwise, coordinated induction of APC- and T cell–associated genes.
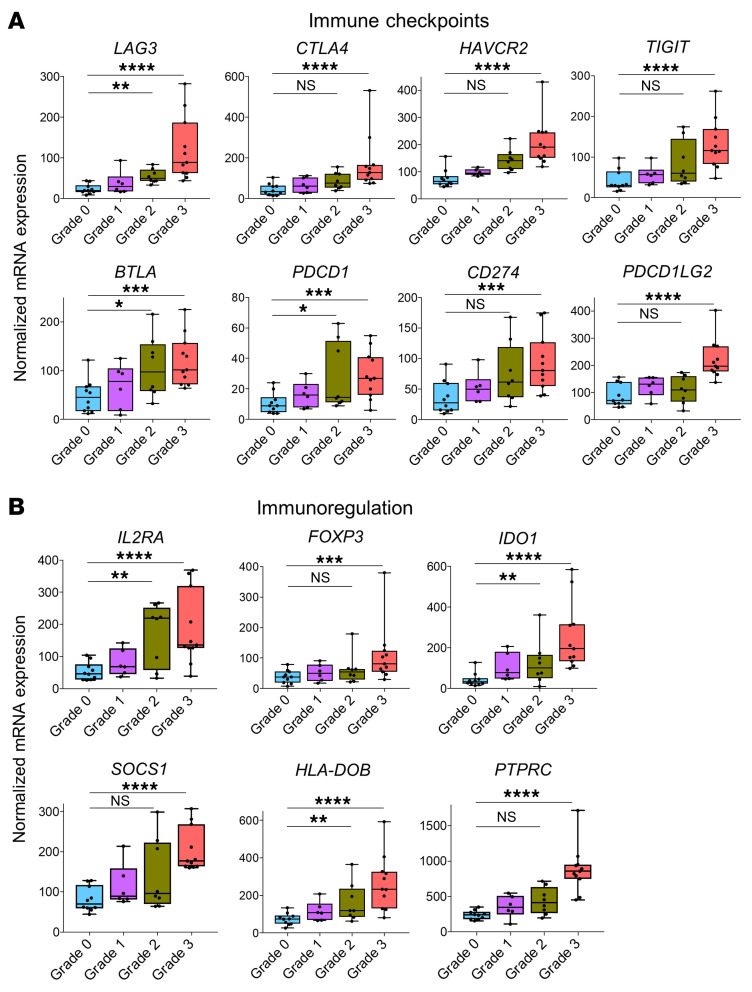
Although there were no DEGs in grade 1 ACR compared with grade 0, the number of DEGs identified increased with the transition to grade 2 and grade 3 (153 DEGs in grade 2 vs. grade 0; 202 DEGs in grade 3 vs. grade 0; Figure 3B, middle and right, and Supplemental Table 7). There was a stepwise increase in the expression of genes related to leukocyte trafficking, T cell infiltration, T cell costimulation, IFN-γ signaling, antigen presentation, Th1 polarization, and effector molecules (Figure 3, C–I). In accordance with these changes in gene expression, pathway analyses identified progressive enrichment of T cell–associated pathways with increasing severity of rejection (Figure 3 and Supplemental Figure 3). Remarkably, there was progressive upregulation of genes associated with proinflammatory immune activation (Figure 3, C–I) as well as those associated with antiinflammatory immune checkpoints and immunoregulation (Figure 4) with increasing ACR severity. Although IFN-γ signaling is known to induce expression of PDCD1and IDO1, we found it intriguing that other immunoregulatory pathways were also upregulated.
Figure 4. Immune checkpoint molecules and immunoregulatory genes are upregulated in face transplant rejection.
Box plots of normalized expression values of genes associated with immune checkpoints (A) and genes associated with regulation of the immune response (B). Normalized gene expression values are shown for biopsies collected during grade 0 (n = 10), grade 1 (n = 6), grade 2 (n = 8), and grade 3 rejection (n = 11). Horizontal lines represent median values, with whiskers extending to the farthest data points. Adjusted *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. The full lists of DEGs and associated statistics are shown in Supplemental Tables 3 and 7.
To confirm our findings, we performed a second, independent analysis of our results using pattern analysis (see Methods). We identified 273 genes with monotonically increasing expression with rejection severity from grade 0 to grade 3 (Supplemental Figure 4 and Supplemental Table 8). In agreement with our initial analyses, GO analysis showed that these 273 genes were enriched for the following biological processes: T cell costimulation, lymphocyte costimulation, immune system process, positive regulation of leukocyte cell-cell adhesion, and leukocyte cell-cell adhesion (Supplemental Table 9).
Severe ACR is characterized by the expression of effector molecules and further induction of immunoregulatory pathways.
Keratinocyte cell death (apoptosis, dyskeratosis, and/or keratinolysis) is the primary histologic feature that distinguishes severe from moderate ACR according to the Banff classification. We hypothesized that grade 3 biopsies displayed an increased expression of cytotoxic effector genes, in agreement with the cell death observed histologically. To examine this, we identified grade 3–specific genes. A set of 107 genes was uniquely differentially expressed in grade 3 ACR (but not in grade 2) when compared with grade 0 (Figure 5A and Supplemental Table 10). Among these were 7 genes associated with cytotoxicity (GZMA, GZMH, GZMM, GNLY, KLRB1, KLRD1, KLRK1; Figure 5B and Supplemental Table 10). Sixteen genes associated with antigen processing and presentation were also uniquely upregulated in grade 3 ACR (HLA-B, HLA-C, HLA-DMB, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, TAP1, TAP2, PSMB8, PSMB9, PSMB10, CD1B, CD1C, CD1E). Notably, a number of genes associated with T cell coinhibition and immunoregulation (CTLA4, FOXP3, HAVCR2, CD247, TIGIT, PDCD1LG2, SOCS1, PTPRC) were exclusively overexpressed in grade 3 rejection (Figure 5B and Supplemental Table 10).
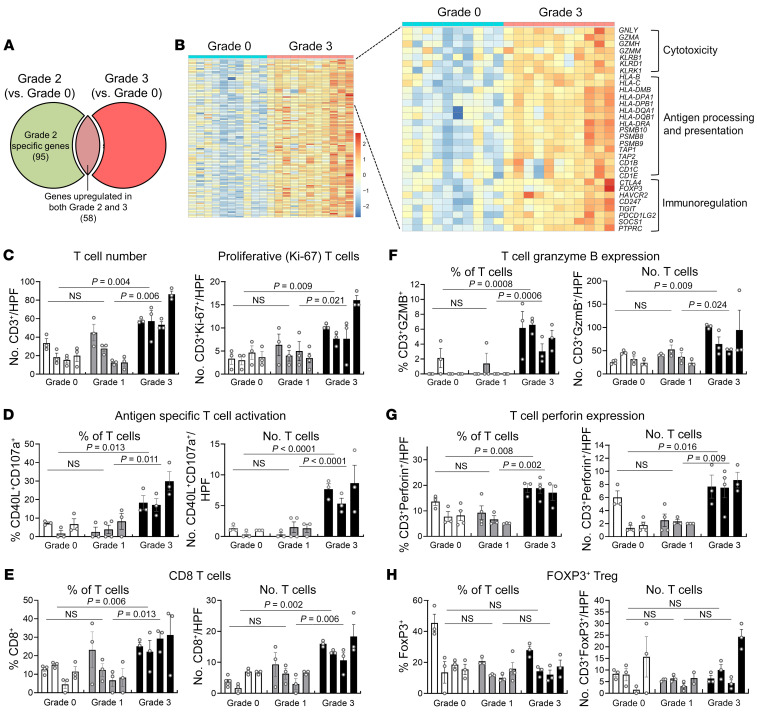
Figure 5. Severe rejection is characterized by upregulation of cytotoxicity, antigen presentation, and immunoregulation genes and infiltration of skin by proliferating T cells expressing markers of antigen-specific activation and cytotoxic effector molecules.
(A) Venn diagram showing shared and unique DEGs in grade 2 (n = 8) and grade 3 (n =11) biopsies (when compared with grade 0 samples). DEGs were obtained using normalized gene expression counts as input and the Wald significance test. One hundred seven genes were exclusively differentially expressed in grade 3 rejection. (B) Heatmap of 107 genes that are uniquely differentially expressed in grade 3 rejection, which included genes associated with cytotoxicity, antigen processing and presentation, and immunoregulation. Each column represents a biopsy. Gene expressions values are row scaled. (C–H) Multiplex immunostaining of grade 0, 1, and 3 rejection demonstrates the infiltration of proliferative, activated, and cytotoxic T cells in grade 3 skin biopsies but no significant differences between grades 0 and 1. Bars represent individual donors, and error bars represent the mean and SEM of at least 3 separate measurements per donor. (C) The numbers of total (CD3+) and proliferative (CD3+Ki-67+) T cells per ×200 high-power field (HPF) are shown. (D–H) The relative percentages (left) and absolute numbers per HPF (right) of T cells expressing markers of antigen-specific activation (CD40L+CD107a+) (D), CD8 (E), granzyme B (F), perforin (G), and the Treg marker FOXP3 (H) are shown. Significance was determined by nested 1-way ANOVA and Tukey’s post hoc test for comparison between grades.
Stage 3 rejection biopsies are infiltrated by increased numbers of proliferative T cells expressing cytotoxic effector molecules and markers of antigen-specific T cell activation.
We carried out multiplex immunostaining to further characterize the rejection process and confirm our findings at the protein level (Figure 5, C–H). The number of total and proliferative (Ki-67+) T cells was increased in grade 3 rejection, but there was no significant difference between grades 0 and 1 (Figure 5C). T cells undergoing antigen-specific activation were identified by immunostaining for CD40 ligand (CD40L) and CD107a. CD40L+CD107a+ T cells were significantly increased in grade 3 but not grade 1 rejection (Figure 5D). CD8+, granzyme-expressing, and perforin-expressing T cells were also significantly increased in grade 3 rejection, but there was no significant difference in these cells in grade 0 versus grade 1 rejection (Figure 5, E–G). Finally, despite the observed increase in expression of the FOXP3 gene, there were no significant changes in the number of FOXP3+ regulatory T cells in grade 3 (Figure 5H).
T cells are the major source of cytotoxic injury in grade 3 rejection.
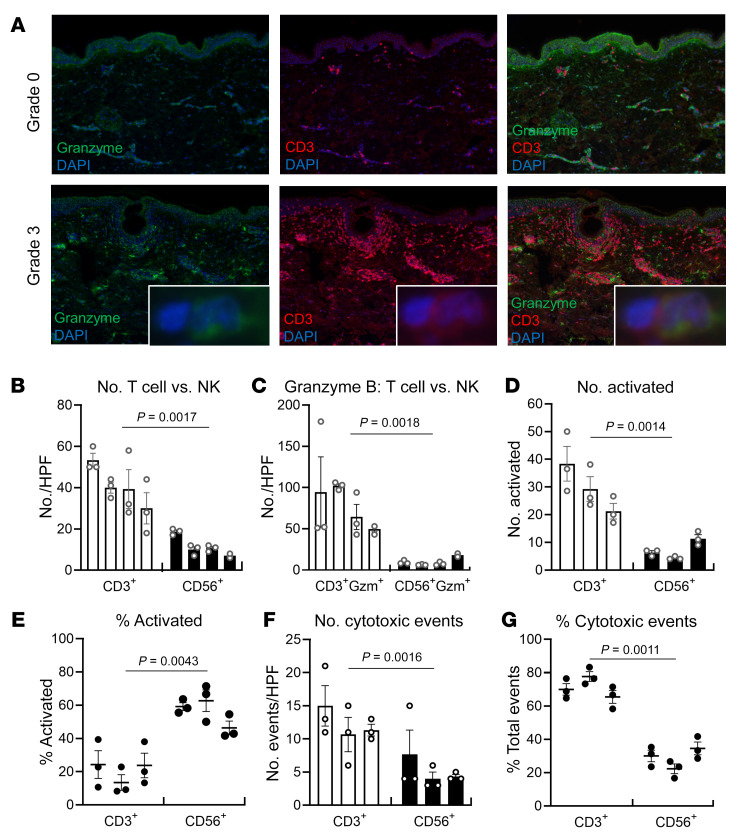
Multiplex immunostaining demonstrated increased expression of granzyme B in grade 3 rejection, but not all granzyme-producing cells were CD3+ T cells (Figure 6A). Natural killer (NK) cells are rich sources of cytotoxic molecules and can participate in organ transplant rejection (15). We carried out additional studies to evaluate the relative contributions of T cells and NK cells to cytotoxic injury. There were significantly more T cells than NK cells in skin biopsies of grade 3 rejection, and the majority of granzyme B was produced by T cells (Figure 6, B and C). Activated (CD40L+) T cells outnumbered activated (CD107a+) NK cells 4-fold (Figure 6D), although NK cells were relatively more activated than T cells (mean 56% of NK cells were activated compared with mean 21% of T cells; Figure 6E). To evaluate the relative contributions of T cells and NK cells to cytotoxic injury, we immunostained for caspase-8, a marker of cells undergoing cytotoxic death (16, 17), and determined whether T cells or NK cells were juxtaposed with dying cells. T cells mediated significantly more cytotoxic events than NK cells and were responsible for a mean 71% of cytotoxic events (Figure 6, F and G).
Figure 6. T cells are the major source of cytotoxic injury in grade 3 rejection.
(A) T cells are a major but not exclusive source of granzyme B. Multiplex immunostaining demonstrated increased expression of granzyme B in grade 3 rejection, but not all granzyme-producing cells were CD3+ T cells (original magnification, ×100). (B–G) Quantitative multiplex immunostaining was carried out to evaluate the relative contributions of T cells versus NK cells to cytotoxic injury in grade 3 rejection. Bars represent individual donors, and error bars represent the mean and SEM of at least 3 separate measurements per donor. (B) There were significantly more T cells than NK cells in skin biopsies of grade 3 rejection. The numbers of CD3+ T cells and CD56+ NK cells per ×200 HPF are shown. (C) The majority of granzyme B was produced by T cells. The numbers of granzyme-positive CD56+ NK cells and CD3+ T cells per ×200 HPF are shown. (D) There were significantly more activated T cells than activated NK cells. The numbers of CD3+CD40L+ (activated T) cells and CD56+CD107a+ (activated NK) cells per ×200 HPF are shown. (E) NK cells had higher frequencies of activation. The percentages of total CD3+ T cells expressing CD40L (left) and CD56+ NK cells expressing CD107a (right) are shown. (F and G) T cells mediated significantly more cytotoxic events than NK cells. Cells undergoing cytotoxic cell death were identified by immunostaining for caspase-8, and the number (F) and relative frequency (G) of events in which T cells (left) or NK cells (right) were juxtaposed with dying cells were enumerated. Significance was determined by nested t tests.
Face transplant rejection has both common and unique signatures compared with solid organ transplant rejection.
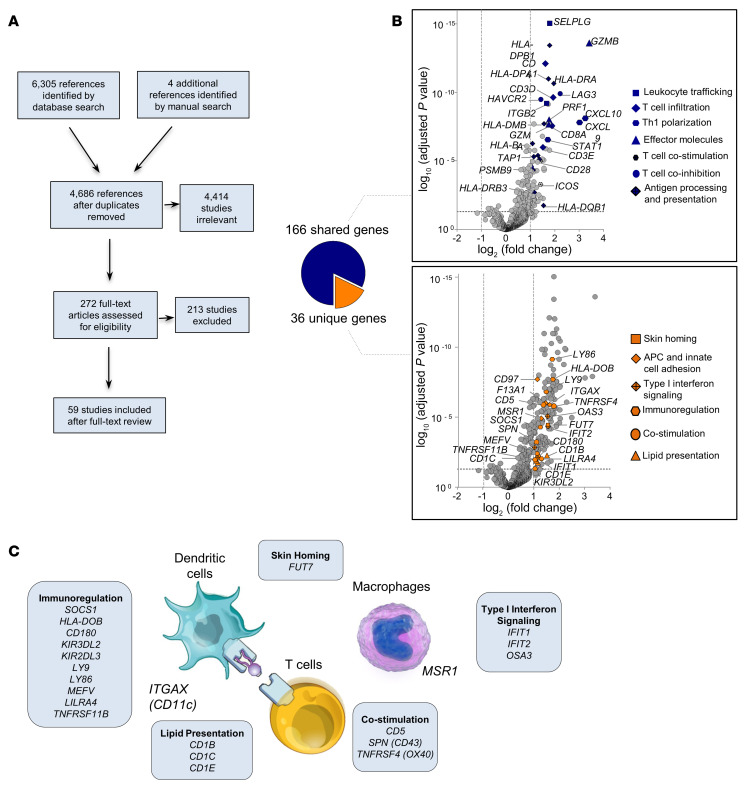
To compare the gene expression signature of face transplant ACR with molecular signatures of acute rejection in human solid organ transplants, we undertook a scoping review of solid organ transplant literature using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (18) (see Methods). Studies measuring mRNA levels in the biopsy specimens across 3 types of human solid organ transplants (kidney, liver, and heart) during established acute rejection and nonrejection were evaluated (Figure 7A and Supplemental Table 11). One hundred sixty-six of 202 genes upregulated in face transplant ACR were shared with genes previously reported to be increased in solid organ transplant biopsies during acute rejection (Figure 7B and Supplemental Table 12). Common elements included leukocyte trafficking, T cell infiltration, Th1 polarization, T cell costimulation, T cell coinhibition, antigen processing and presentation, and effector molecules (Figure 7B). We identified 36 genes that were unique to face transplant rejection, including genes associated with skin homing (FUT7) and APC and innate cell adhesion (MSR1, F13A1, ITGAX, CD97), and genes induced in skin as a result of type I interferon signaling (IFIT1, IFIT2, OAS3), immunoregulation (CD180, HLA-DOB, KIR2DL3, KIR3DL2, LILRA4, LY86, LY9, MEFV, SOCS1, TNFRSF11B), costimulation (CD5, TNFRSF4, SPN), and lipid presentation (CD1B, CD1C, CD1E) (Figure 7, B and C; Supplemental Figure 5; and Supplemental Table 13).
Figure 7. Comparison between human face and solid organ transplant rejection.
(A) Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram of selection of solid organ transplant studies. (B) Volcano plots showing DEGs between grade 3 rejection biopsies (n = 11) and grade 0 nonrejection biopsies (n = 10). DEGs were obtained using normalized gene expression counts as input and the Wald significance test. Genes shared with solid organ transplant rejection are shown in blue, and genes unique to face transplant rejection are shown in orange. A subset of genes is shown; the complete list of shared genes together with associated statistics is shown in Supplemental Table 12, and the complete list of unique genes together with associated statistics is shown in Supplemental Table 13. Each dot represents an individual gene. Horizontal dashed lines represent an adjusted P value cutoff of –log10 (0.05); vertical dashed lines represent log2 fold change of –1 and +1. (C) Genes unique to face transplant rejection. A subset of genes is shown; the complete list of unique genes and their expression are shown in Supplemental Figure 5 and Supplemental Table 13.
GLIPH-predicted CD1b- and CD1c-specific T cell clones are enriched in the skin during episodes of rejection.
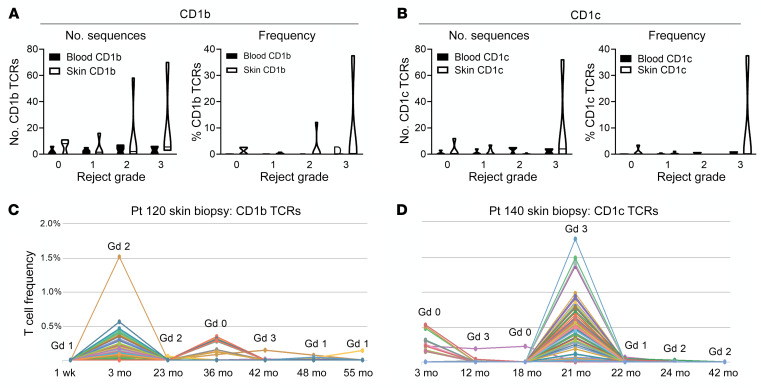
CD1b and CD1c are nonpolymorphic MHC I–like molecules specialized for the presentation of glycolipid antigens derived from self-injury, pathogens, or environmental exposures (19). Mice express CD1d, but CD1a, CD1b, and CD1c are unique to humans. Because we did not have access to living T cells from rejection specimens, we used an indirect approach to evaluate the possible role of CD1b and CD1c in rejection. We carried out high-throughput sequencing of the TCR complementarity-determining region 3 (CDR3), the TCR antigen-specific receptor domain, in skin biopsies from face transplant recipients. High-throughput TCR sequencing measures the number and diversity of T cells and identifies the exact antigen receptor sequences (CDR3) of individual T cells in a biological specimen (20). We then used the grouping of lymphocyte interactions by paratope hotspots (GLIPH) algorithm to identify T cells likely specific for CD1b and CD1c (21). The GLIPH algorithm clusters TCRs with a high probability of recognizing a common antigen based on their conserved motifs and global similarity of CDR3 sequences. GLIPH can reliably group TCRs of common specificity from different donors. We identified 19 CD1b-reactive and 19 CD1c-reactive TCR CDR3 sequences from the scientific literature and ran the GLIPH clustering algorithm on these known sequences together with the CDR3 sequences isolated from face transplant rejection skin biopsy specimens. We identified a total of 285 CDR3 sequences from face transplant skin specimens that clustered with known CD1b-reactive TCRs and 88 sequences that clustered with known CD1c-specific TCRs (Supplemental Table 14). The number and relative frequency of likely CD1b- and CD1c-reactive T cell clones identified by GLIPH increased in rejection skin biopsy specimens (Figure 8, A and B). The frequency and number of CD1b- and CD1c-associated sequences were enriched during rejection episodes in skin but not in blood, suggesting that skin inflammation may drive expansion of these T cells. Interestingly, we observed local expansion of multiple different CD1b- and CD1c-associated T cell clones during rejection episodes (Figure 8, C and D). CD1b- and CD1c-associated T cell clones were expanded in some but not all rejection episodes (Figure 8, C and D), suggesting that CD1-presented glycolipid antigens may drive some episodes of rejection whereas classically presented peptide antigens may drive others.
Figure 8. T cells expressing CD1b- and CD1c-associated TCRs are enriched in the skin but not blood during rejection episodes.
The antigen receptor (CDR3) sequences of T cells infiltrating face transplant rejection specimens were analyzed by high-throughput TCR sequencing. These CDR3 sequences were then clustered with known CD1b- and CD1c-specific sequences using the grouping of lymphocyte interactions by paratope hotspots (GLIPH) algorithm. Two hundred eighty-five CDR3 sequences from face transplant skin specimens that clustered with known CD1b-reactive TCRs and 88 sequences that clustered with known CD1c-specific TCRs were identified (Supplemental Table 14). (A and B) The absolute number (left) and relative frequency of total T cells (right) of CD1b-associated TCRs (A) and CD1c-associated TCRs (B) in blood and skin are shown. Seventeen skin biopsies and 12 skin samples from 3 donors were studied. (C and D) Local enrichment of multiple CD1b-associated (C) and CD1c-associated (D) TCRs occurred in some but not all episodes of rejection. Gd, grade; Pt, patient.
Discussion
Vascularized composite allografts present unparalleled opportunities for restoration as well as unique challenges. Limb and face transplants can restore the ability of patients to eat, speak, and care for themselves. In exchange, recipients commit to a lifetime of immunosuppression and to a certain degree of uncertainty. Rejection leading to graft loss is a potentially fatal complication, and little is known about the biology of VCA rejection in humans. A better understanding of VCA rejection could lead to novel and targeted therapies that could put existing patients on a firmer footing and open up the procedure to broader use.
Face transplants and other skin-containing VCAs are unique from an immunologic perspective. Skin is the primary trigger and the main target of ACR in VCAs (22). Skin is highly immunogenic because it is an immunologically active epithelial barrier tissue designed to sense and respond to danger. It contains a wealth of immune cells, including Langerhans cells, macrophages, dendritic cells, and T cells, the majority of which are long-lived non-recirculating resident memory T cells (5, 23). A 600 cm2 full face transplant transfers approximately 600 million donor-derived memory T cells (24). All of the immunologic components needed to mount a memory response are present in healthy skin. Skin rejection in vascularized composite allotransplantation is, therefore, a study of what happens when 2 mature immune systems come face to face with one another.
Diagnosing rejection histologically can be more complicated in patients with VCAs than in solid organ transplant recipients. The presence of T cells in heart and kidney transplants is always abnormal. However, mild to moderately brisk lymphocytic infiltrates are characteristics of healthy noninflamed skin, and distinguishing rejection from business as usual in the skin is therefore challenging. Histologically, it is only a relative and nonquantified increase in lymphocyte density in the skin that distinguishes nonrejection from grades 1 and 2 rejection by the Banff criteria. Histologic evidence of keratinocyte injury differentiates grade 3 rejection, making it easier to diagnose, but by this point, damage has already occurred. VCA rejection can also be challenging to distinguish from other causes of skin inflammation, including drug eruptions and rosacea. Indeed, a face transplant recipient we reported had biopsies classified as grade 1 and 2 rejections, failed to respond to antirejection treatments, and only improved once it was recognized that inflammation was the result of rosacea; the patient improved with topical metronidazole cream (25). A clearer understanding of early events in VCA rejection is needed to facilitate prompt and accurate diagnosis.
We addressed these challenges by gene expression profiling of prospectively collected skin biopsies from the largest cohort of face transplant recipients at a single center. We found that there were no significant gene expression differences between nonrejection (grade 0) and mild rejection (grade 1) biopsies. Moreover, there were no significant differences in the number of total T cells, proliferative T cells, activated T cells, CD8+ T cells, or T cells expressing granzyme or perforin in grade 0 versus grade 1 rejection as assayed by multiplex immunostaining. Mild rejection is the most commonly reported rejection grade in face transplant recipients, but its clinical impact on long-term graft survival is unclear, and there is no consensus on whether it actually represents rejection requiring treatment or normal local immunologic skin homeostasis (14). Our results suggest that grade 1 rejection does not represent a pathologic state and that the T cells noted in mild rejection biopsies likely reflect immunologic business as usual within the skin. We have discontinued treating mild rejection in face transplants at our center; patients are instead actively observed. It is important to emphasize that this represents a substantial change in clinical practice. Rejection is treated with systemic pulsed steroids and/or increases in maintenance immunosuppressive medications, therapies that put patients at increased risk for infections and cancer.
In grade 2 rejection biopsies, we began to observe features of immune activation, including increased expression of genes associated with leukocyte trafficking, T cell infiltration, costimulation, Th1 polarization, and effector molecules. However, there is no evidence of cell death or tissue injury histologically. Early episodes of face transplant rejection are conceptually similar to cutaneous graft-versus-host disease; both involve an influx of hematopoietic cells into an epithelial barrier tissue colonized with allogeneic T cells and APCs, creating in essence an in vivo mixed lymphocyte reaction. This explosive immunologic combination — allogeneic T cells meeting tissue-resident APCs — is likely why skin allografts are rejected rapidly (26). Given the evidence of T cell and APC activation in grade 2 biopsies, it is surprising that there was no cell death or tissue damage. Clinically, it is also remarkable that recipients of skin-containing face transplants are successfully managed on the same immunosuppressive regimens used for kidney transplants.
The explanation for this behavior may reside in the potent immunoregulatory pathways that build up over time in human skin. Patients receiving immune checkpoint inhibitors for cancer therapy can develop severe skin toxicities (27), demonstrating that checkpoint molecules such as PD-1 and CTLA-4 play critical but normally invisible roles in controlling cutaneous inflammation in response to daily immunologic challenges. Checkpoint molecules are also expressed by tissue-resident memory T cells, not as an indication of exhaustion, but likely as a mechanism to remain poised and functional long-term within peripheral tissues (28, 29). The healthy skin of an adult human contains approximately 20 billion memory T cells that have accumulated over decades of antigen exposure (23, 30, 31). Some of these T cells are immunosuppressive, including FOXP3+ regulatory T cells (Tregs), and others are effector cells programmed to downregulate activation in the presence of checkpoint ligands. We found induction of immunoregulatory pathways in grade 2 rejection, including upregulation of PDCD1 (PD-1), LAG3, BTLA, IL2RA, IDO1, and HLA-DOB. We hypothesize that it is the induction of these diverse immunoregulatory pathways that prevents cytotoxic tissue damage in grade 2 rejection, at a point in time when T cells are clearly responding to allostimulatory APCs. Supporting this, FOXP3+ Tregs have been shown to locally accumulate during rejection in human face and hand transplants (32, 33). These results suggest that therapies that enhance skin-associated immunoregulatory pathways may be useful to combat rejection in VCAs. An antibody that acts as an agonist for the potent inhibitory receptor PD-1 is currently in a phase I trial for treatment of psoriasis (https://clinicaltrials.gov/ct2/show/NCT03337022). Such agonists, if proven effective, could represent a new therapeutic avenue for treating VCA rejection.
There was a marked upregulation of cytotoxicity and antigen presentation genes in grade 3 biopsies that corresponded to the tissue injury observed histologically. Immunoregulatory pathways were further induced, with upregulation of CTLA4, HAVCR2 (TIM-3), CD247, TIGIT, PDCD1LG2, FOXP3, SOCS1, and PTPRC, but were clearly no longer adequate to prevent tissue damage. Induction of genes associated with IFN-γ signaling (STAT1, IRF1) and its downstream effects (TAP1 and numerous HLAs), and genes and signaling pathways that directly or indirectly enhance IFN-γ responses (such as CXCR3 and CCR5 pathways), demonstrated that IFN-γ is the principal cytokine driving face transplant rejection. Multiplex immunostaining confirmed an important role for T cells in mediating tissue injury, with increased numbers of proliferative T cells expressing markers of antigen-specific activation, CD8+ T cells, and T cells expressing perforin and granzyme. Activated NK cells were present and contributed to cytotoxicity, but T cells were the major source of cytotoxic injury in grade 3 rejection. In addition to direct cytotoxic damage, locally produced cytokines may also contribute to tissue injury. Apoptosis of epithelial stem cells is the predominant form of cellular injury in experimental models of acute GVHD (34, 35).
Although face and solid organ transplants share many genes that are overexpressed during acute rejection, 36 genes were uniquely upregulated in face transplant rejection. These genes were unique to rejection and were not upregulated in non–transplant-related inflammatory skin diseases. The unique genes included those associated with skin homing, immunoregulation, and APC and innate cell adhesion, and genes induced in skin as a result of type I interferon signaling, costimulation, and lipid antigen presentation. Ten of the 36 genes unique to face transplant rejection were immunoregulatory, meaning that they are known to participate in the downregulation of inflammation. As an epithelial barrier tissue, the skin is constantly confronted by immunologic challenges, only a fraction of which are dangerous. The skin has developed complex immunoregulatory networks to dampen inappropriate inflammation and maintain homeostasis (36). Our data suggest that these networks are activated in face transplant rejection and could be leveraged further to reduce rejection-associated inflammation. CTLA-4 agonist therapy is a relatively weak but safe immunomodulatory approach that has so far been of limited benefit in vascularized composite allotransplantation (37–39). Antagonizing the level of immunologic activation that occurs in rejection will likely require the stimulation of multiple, highly effective immunoregulatory pathways. For example, SOCS1 negatively regulates the production of inflammatory cytokines such as IFN-γ and stabilizes Treg function (40). SOCS1 mimetics are useful in the treatment of autoimmune and inflammatory skin disorders in animal models (41, 42).
The upregulation of genes encoding CD1 molecules (CD1B, CD1C, and CD1E) is a unique aspect of face transplant rejection. CD1 molecules resemble MHC I structurally and are specialized to present lipid antigens derived from topical exposures, pathogens, or self-lipids generated during tissue injury (19). Unlike other members of the MHC family, CD1 molecules are nonpolymorphic. CD1-specific T cell responses in VCA recipients would not necessarily be driven by antigenic differences between donor and recipient; donor and recipient T cells would be expected to react similarly to antigenic lipids. We analyzed the TCRs from rejection specimens using TCR sequencing and antigenic clustering with known CD1b- and CD1c-reactive TCRs via the GLIPH algorithm. We identified 285 CDR3 sequences from face transplant skin specimens that clustered with known CD1b-reactive TCRs and 88 sequences that clustered with known CD1c-specific TCRs. These sequences were enriched in the skin but not blood during rejection episodes, suggesting a local expansion of these clones in skin. Many different CD1-associated TCRs were upregulated in the skin during some rejection episodes, suggesting a polyclonal response to local lipids. However, CD1-associated sequences were not upregulated in all rejection episodes, demonstrating that some rejection events are entirely driven by classic MHC-based peptide allorecognition. A shortcoming of our study is that we did not have access to living T cells isolated from rejection specimens and we could not directly confirm CD1 reactivity of the TCRs we identified by GLIPH clustering. Future studies of viable T cells stimulated by CD1-expressing APCs or stained with loaded CD1-multimer reagents are needed to definitively confirm CD1 reactivity in rejection specimens.
Our study had additional limitations. The number of samples we were able to study was limited by both the rare nature of this surgical procedure and our stringent inclusion and exclusion criteria, which we employed to avoid confounding factors. We have focused on pretreatment rejection biopsies with the goal of identifying the immunologic events driving rejection. All patients in our cohort were successfully treated with antirejection therapies and returned to nonrejection baseline both clinically and histologically. Additional studies of treated and partially treated rejection episodes have the potential to identify which aspects of rejection are modified by corticosteroids and which require increased immunosuppression. Given the current scarcity of samples, we were not able to carry out these studies, but they should become possible in future years when the number of patients and specimens reaches a critical mass. Our study also focused on cellular but not antibody-mediated rejection; only 1 patient in our cohort had antibody-mediated rejection. Our comparisons of face transplant rejection with published studies of solid organ transplant rejection were necessarily qualitative given the variety of platforms used. Lastly, our findings are associative and require functional validation. We had access only to archived histologic specimens, which limited our ability to use powerful techniques such as single-cell RNA sequencing and functional studies of T cells from rejecting skin samples. Despite these shortcomings, our results represent a meaningful first step in understanding acute rejection in clinical face transplants. Ongoing work in our laboratory focuses on single-cell techniques to comprehensively study APCs and T cells in VCA rejection.
In summary, we find that the unique aspects of face transplant rejection reflect the unique immunologic characteristics of the skin. Our data suggest that grade 1 rejection should not be considered a pathologic state and can be managed with watchful waiting. Grade 2 rejection showed induction of cytotoxic and Th1 responses as well as upregulation of multiple immunoregulatory pathways that likely prevent tissue injury even in the context of allorecognition. Grade 3 rejection was characterized by cytotoxicity and tissue damage but also involves the upregulation of numerous antiinflammatory pathways and evidence for possible lipid antigen presentation. These results demonstrate that transplantation of skin containing VCA is both a challenge and an opportunity. These tissues are heavily populated by donor immune cells but also have complex immunoregulatory networks that could be leveraged to decrease inflammation and combat rejection. Further studies on the details of these immunoregulatory pathways may lead to novel therapies for rejection in patients who have received these life-changing transplants.
Methods
Patient and skin biopsy details
Patients were recruited from among facial allograft recipients seen at the Brigham and Women’s Hospital. Blood samples and 4 mm punch biopsies were obtained from facial allografts. Skin biopsies for rejection monitoring were routinely collected at follow-up clinic appointments. Additional skin biopsies were taken when rejection was clinically suspected at any time after transplant. Skin biopsies were obtained from the allografts in a standardized manner: nonrejection biopsies were taken from esthetically insensitive areas (such as the preauricular area), and rejection biopsies were taken from the most inflamed area within the facial allograft. Because the primary aim was to study acute cellular rejection, we established the following inclusion criteria prospectively for selection of face transplant skin biopsies for our study: (a) absence of circulating donor-specific antibodies at the time of biopsy, (b) absence of infection or treatment for infections at the time of and in the week before biopsy collection, (c) collection of biopsy prior to treatment for rejection, and (d) adequate tissue availability for isolation of RNA of sufficient quality and quantity for use in NanoString analysis. We excluded biopsies collected within 3 weeks of transplantation to prevent confounding by peri- and postoperative proinflammatory events. Because the focus of the study was to study acute cellular rejection, biopsies collected during antibody-mediated rejection (circulating donor-specific antibodies at the time of biopsy), during mixed antibody and cellular rejection, or during periods when a patient had evidence of active infection or was on therapies for infection were excluded. Only samples collected before the initiation of antirejection therapies were used. Thirty-eight face transplant skin biopsies from 7 patients met the inclusion criteria and were included in the study. The biopsies were taken at different time points. Histology grading and gene expression values were obtained for each biopsy independently.
Skin biopsies (4 mm punch biopsies) from nontransplanted patients with rosacea and delayed-type hypersensitivity reaction were obtained as excess material from biopsies obtained for clinical purposes. Normal facial skin was obtained from elective facelift surgeries.
Diagnosis of rejection
Three-micrometer sections were cut from 4 mm skin punch formalin-fixed paraffin-embedded (FFPE) biopsies from face transplants. The sections were stained using H&E. Rejection was graded by a panel of dermatopathologists according to the Banff 2007 classification and agreed upon in dermatopathology departmental case conferences at the Brigham and Women’s Hospital. Histologic analysis was carried out without the knowledge of the molecular findings.
RNA extraction
Six consecutive 10 μm sections were obtained from each FFPE skin biopsy using a microtome. Sections were immediately transferred to sterile 1.5 mL microcentrifuge tubes and stored at room temperature until RNA extraction. Xylene deparaffinization and RNA extraction were performed with the RNeasy FFPE Kit (Qiagen Valencia). Total RNA concentration and purity were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The quality of the RNA was determined using an absorbance ratio of 260/280 nm.
mRNA NanoString gene expression assay
The NanoString nCounter PanCancer Immune Profiling Gene Expression (GX) CodeSet with probes representing 770 genes was used. Samples were processed on the NanoString nCounter Analysis System according to the manufacturer’s protocol (NanoString Technologies). Briefly, 100 ng of total RNA was hybridized to the reporter and capture probes in 30 μL reaction volume for 20 hours at 65°C. The captured targets were then immobilized on the nCounter cartridge on the NanoString PrepStation. The cartridge was scanned at 555 fields of view (FOV) on the nCounter Digital Analyzer. Images were processed into a digital format (RCC files). All samples were analyzed using a single lot of reagents.
Scoping review
Scoping reviews are intended to provide an overview of the literature through structured search strategies and rigorous methods not unlike a systematic review. This review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline (18) adapted for use in a scoping review as appropriate. A search strategy was designed with the assistance of a research support librarian from the Harvard Countway Library of Medicine.
Inclusion and exclusion criteria.
Articles that met the following predetermined criteria were included: (a) peer-reviewed original research article; (b) studies that measured mRNA levels in biopsies of human solid organ transplants (liver, kidney, heart); (c) studies comparing acute rejection with nonrejection specimens. Publications that studied antibody-mediated rejection and chronic rejection were excluded. All articles not in English were excluded.
Search strategy.
PubMed, Embase, Web of Science, and Countway Discovery Medicine, including the EBSCO databases Academic Search Premier and CINAHL Plus, were searched, using Medical Subject Headings (MeSH) terms for PubMed and comparable controlled vocabulary for other databases (Supplemental Table 11).
Data screening and extraction.
References identified through database searches were imported into EndNote (Clarivate Analytics). The complete list of searches run and imported to EndNote is shown in Supplemental Table 11. Duplicates were removed in EndNote, and unique references were then transferred to Covidence systematic review software (Veritas Health Innovation; https://www.covidence.org/; accessed 29 October 2018). Two independent reviewers screened the title and abstract of each record, and conflicts were resolved by discussion and consensus, or by adjudication by a third reviewer. Two hundred seventy-two full-text articles were obtained for full-text review, and assessed against the predetermined inclusion/exclusion criteria. Both reviewers reviewed each full-text article, and conflicts were resolved by discussion and consensus, or by adjudication by the third reviewer. One hundred forty full-text articles met the inclusion/exclusion criteria. Because the purpose of our scoping review was to identify the studies from solid organ transplant literature that investigated the genes of interest from our face transplant study (202 DEGs in grade 3 face transplant ACR compared with grade 0; Supplemental Table 3), 81 of the 140 articles were removed after full-text review as they did not study these genes of interest. Therefore, a total of 59 studies were included after full-text review (a detailed list of the studies is shown in Supplemental Table 12).
Multiplex immunostaining
Five-micrometer sections were cut from 4 mm skin punch FFPE biopsies from face transplants. Antigen retrieval was carried out by incubation in a pressure cooker for 3 minutes at 120°C. Fc receptors were blocked with PerkinElmer Block (ARD1001EA). Primary antibodies were diluted in Dako Antibody Diluent (S3022), and samples were stained overnight. Samples were washed 3 times with 1× TBS/0.1% Tween-20 (1× TBST) and once with dH2O. Secondary fluorescent conjugated antibodies (Invitrogen) were diluted in 1× PBS, and samples were stained for 30 minutes followed by 3 washes with 1× TBST and a final wash in dH2O. Samples were mounted using Invitrogen ProLong Gold Antifade Reagent with DAPI (P36931). Tissue was imaged immediately after mounting on a Mantra Quantitative Pathology Workstation using Mantra Snap 1.0 imaging software and analyzed with inForm 2.4.8 image analysis software (PerkinElmer). Primary and secondary antibodies and the concentrations used are listed in Supplemental Table 15. Cells of interest were quantified in ×200 high-power fields, and a minimum of 3 replicate counts were performed for each sample.
DNA isolation from skin, high-throughput TCR sequencing, and GLIPH analyses.
DNA was isolated from PBMCs and FFPE skin samples using the QIAamp DNA Mini Kit (Qiagen) per the manufacturer’s instructions with overnight tissue digestion for skin samples. Paraffin was first removed from FFPE skin samples by xylene extraction followed by ethanol washes prior to overnight tissue digestion. For each DNA sample, TCRβ CDR3 regions were amplified and sequenced using ImmunoSEQ (Adaptive Biotechnologies) from 100–400 ng of DNA template (43–45). Amino acid sequences of all TCR antigen receptor domains (CDR3) detected in the tissue were obtained. CDR3s obtained from analysis of sequential biopsies from a single patient were grouped together into a single pool of CDR3s. Nineteen sequences published as recognizing CD1b and 19 sequences identified in the literature as recognizing CD1c were identified. Known CD1b sequences were added to the CDR3 pool from each donor, and the GLIPH algorithm was performed on these pools as previously described (21). Sample CDR3 sequences that clustered together with known CD1b sequences were identified as CD1b-associated; a similar process was followed to identify CD1c-associated sequences. The number of CD1-associated sequences and the relative frequencies of all CD1-associated sequences were determined in each skin biopsy sample from each donor. The presence, number, and frequencies of CD1b- and CD1c-associated sequences were then determined in matched blood samples from the same patient using the results of high-throughput sequencing analyses.
Statistics
Quality control, normalization, and filtering of gene expression data.
Quality control metrics and plots were obtained using the NanoStringQCPro package (ref. 46; R package, version 1.10.0). A noise threshold was defined as the median value of expression for the negative controls plus 2 standard deviations. Samples were removed if the median expression of all housekeeping genes in that sample was less than the noise threshold. Three samples did not pass the noise threshold and were therefore excluded, resulting in 35 samples being included in the subsequent analyses (Banff rejection grade 0, n = 10; grade 1, n = 6; grade 2, n = 8; grade 3, n = 11). Raw mRNA counts were normalized with the geometric mean in 2 steps, first using the positive control genes, and second using a subset of housekeeping genes. Housekeeping genes were selected if they had an expression value above the noise threshold and a mean value higher than 200 (a value selected empirically after examination of the average expression of housekeeping genes). After normalization, genes with values lower than the noise threshold in all samples were excluded; 769 genes passed this filter and were included in the analysis. Normalized data were log2-transformed. All the analyses, heatmaps, and PCAs were generated using the R statistical language (version 3.5.1).
Differential expression analysis.
Differentially expressed genes (DEGs) between rejection grades were analyzed with the R package DESeq2-v.1.20.0 (47), using normalized gene expression counts as input and the Wald significance test. Samples were not paired in the analysis. P values were adjusted using the Benjamini-Hochberg false discovery rate (FDR) method to account for multiple testing. A gene was considered differentially expressed when comparison between 2 groups yielded (a) an adjusted P value less than 0.05 and (b) a log2 fold change greater than 1.
Ingenuity pathways analysis.
Data were analyzed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems), with a focus on the top canonical pathways (by P value) and upstream regulators. DEGs (which met both criteria of log2 fold change >1 and adjusted P value <0.05) were used for IPA (detailed lists of the genes are found in Supplemental Tables 3 and 7). The following IPA core analysis settings were used: general settings, reference set = NanoString CodeSet (Supplemental Table 14), relationships to consider = direct and indirect relationships; confidence = experimentally observed only; species = human. Significant enrichment for gene sets in IPA-curated canonical pathways was determined using Fisher’s exact test P values with multiple testing adjustments according to the Benjamini-Hochberg FDR method. The upstream regulator analysis allows identification of potential regulators of genes belonging to a given set. Activation or inhibition status of upstream regulators was determined by the z score, which accounts for both the total number of genes differentially expressed and the direction of activation. A z score ≥ 2 (activated) or ≤ –2 (inhibited) was considered significant.
Gene Ontology analysis.
To assess the main functions of DEGs, overrepresented Gene Ontology (GO) terms were identified using the g:Profiler toolkit (http://biit.cs.ut.ee/gprofiler/). DEGs (which met both criteria of log2 fold change >1 and adjusted P value <0.05) were used as input for the GO analysis (detailed lists of the genes are found in Supplemental Tables 3 and 8). The following g:Profiler settings were used: organism was Homo sapiens; enrichment for GO terms only (biological processes); Fisher’s 1-tailed test as statistical method for enrichment; Benjamini-Hochberg FDR as multiple testing correction; statistical domain size was only annotated genes; no hierarchical filtering was included. Minimum and maximum size of functional category was set to 3 and 500, respectively. Size of query was set to 2. NanoString CodeSet genes (Supplemental Table 14) were used as statistical background.
Pattern analysis.
Patterns of expression were identified using the degPatterns function from DEGreport (ref. 48; R package, version 1.15.4). Differentially expressed genes, with an adjusted P value less than 0.05 using DESeq2 with a likelihood ratio test (LRT) model (48), were classified in pattern clusters based on the correlation of their expression changes between rejection grades.
Functional annotation of DEGs.
We performed a detailed gene-by-gene analysis and functional category assignment based on descriptions given in GeneCards (https://www.genecards.org/) as well as on a literature search (PubMed) using gene symbols and gene names.
Multiplex immunostaining quantification.
For multiplex immunostaining, significance was determined by nested 1-way ANOVA followed by Tukey’s post hoc test with correction for multiple comparisons (Figure 5) or by 2-tailed nested t tests (Figure 6).
Study approval
The protocols of this study were performed in accordance with the Declaration of Helsinki. All patients provided written informed consent to participate in the clinical trial for face transplantation (ClinicalTrials.gov identifier NCT01281267), approved by the human research committee at the Brigham and Women’s Hospital (2008P00055). All patients provided consent for the use of photographs. The research protocol was approved by the Massachusetts General Brigham Institutional Review Board, protocol 2017P001486.
Author contributions
TSW was listed as the first co–first author based on contributions to the manuscript and work on figures. BP, RAC, WJC, and TSW conceived the project and designed the analytic approaches used. TSW, RAC, and WJC drafted, edited, and revised the figures and manuscript. Samples were provided as a result of leadership supplied by BP. LVR provided experimental advice. TSW and WJC carried out the experiments. TSW, WJC, VB, SHS, and RAC analyzed the data. QZ optimized antibodies for immunostaining and provided advice on histologic studies. BDA, RL, and WJC carried out the immunofluorescence staining. JET, SGT, NM, ST, AC, BP, and LVR contributed to the analysis and scientific discussions.
Supplementary Material
Acknowledgments
We thank the patients who made these studies possible, for donating their tissue and for entrusting us with their clinical care. We thank Liyan Guo for his expert assistance with the data analysis and Jacqueline Cellini (Research Support Librarian, Harvard Countway Library of Medicine) for her expert assistance with the scoping review. We thank the Human Skin Disease Resource Center of Brigham and Women’s Hospital and Harvard Medical School, which provided skin biopsies from nontransplanted individuals with rosacea and delayed-type hypersensitivity reaction as well as normal facial skin. The Human Skin Disease Resource Center is supported in part by National Institute of Arthritis and Musculoskeletal and Skin Diseases Resource-based Center Grant 1P30AR069625. This study was supported by funding from US Department of Defense awards to BP (W81XWH-17-1-0278), to LVR (W81XWH-16-1-0647, W81XWH-16-1-0689), and to RAC (W81XWH-18-1-0784, W81XWH-1-810798). TSW is supported by the American Society of Transplantation’s Transplantation and Immunology Research Network Fellowship Research Grant and by the Plastic Surgery Foundation Fellowship from the American Society of Plastic Surgeons. BDA is supported by grants from the Novo Nordisk Foundation (NNF15OC0014092), the Lundbeck Foundation, the Aage Bangs Foundation, and the A.P. Moller Foundation for the Advancement of Medical Science. Biostatistics and bioinformatics support was partially funded by Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH award UL1 RR025758 and financial contributions from participating institutions).
Version 1. 03/05/2021
In-Press Preview
Version 2. 04/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(8):e135166.https://doi.org/10.1172/JCI135166.
Contributor Information
Thet Su Win, Email: thetsuwin828@gmail.com.
William J. Crisler, Email: w.crisler@BWH.HARVARD.EDU.
Beatrice Dyring-Andersen, Email: beatrice@dyring-andersen.dk.
Rachel Lopdrup, Email: rlopdrup@gmail.com.
Jessica E. Teague, Email: jteague@partners.org.
Qian Zhan, Email: qzhan@rics.bwh.harvard.edu.
Victor Barrera, Email: vbarrera@hsph.harvard.edu.
Sotirios Tasigiorgos, Email: s.tasigiorgos@gmail.com.
Naoka Murakami, Email: naokamt@gmail.com.
Anil Chandraker, Email: achandraker@partners.org.
Stefan G. Tullius, Email: stullius@bwh.harvard.edu.
Bohdan Pomahac, Email: BPOMAHAC@PARTNERS.ORG.
Leonardo V. Riella, Email: LRIELLA@bwh.harvard.edu.
References
- 1.Rifkin WJ, et al. Achievements and challenges in facial transplantation. Ann Surg. 2018;268(2):260–270. doi: 10.1097/SLA.0000000000002723. [DOI] [PubMed] [Google Scholar]
- 2.Tasigiorgos S, et al. Face transplantation—current status and future developments. Transpl Int. 2018;31(7):677–688. doi: 10.1111/tri.13130. [DOI] [PubMed] [Google Scholar]
- 3.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 4.Heath WR, Carbone FR. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat Immunol. 2013;14(10):978–985. doi: 10.1038/ni.2680. [DOI] [PubMed] [Google Scholar]
- 5.Clark RA, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176(7):4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 6.Clark RA, Kupper TS. IL-15 and dermal fibroblasts induce proliferation of natural regulatory T cells isolated from human skin. Blood. 2007;109(1):194–202. doi: 10.1182/blood-2006-02-002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaunat O, et al. Immunopathology of rejection: do the rules of solid organ apply to vascularized composite allotransplantation? Curr Opin Organ Transplant. 2015;20(6):596–601. doi: 10.1097/MOT.0000000000000242. [DOI] [PubMed] [Google Scholar]
- 8.Nankivell BJ, Alexander SI. Rejection of the kidney allograft. N Engl J Med. 2010;363(15):1451–1462. doi: 10.1056/NEJMra0902927. [DOI] [PubMed] [Google Scholar]
- 9.Schneeberger S, et al. Achievements and challenges in composite tissue allotransplantation. Transpl Int. 2011;24(8):760–769. doi: 10.1111/j.1432-2277.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 10.Petruzzo P, et al. Clinicopathological findings of chronic rejection in a face grafted patient. Transplantation. 2015;99(12):2644–2650. doi: 10.1097/TP.0000000000000765. [DOI] [PubMed] [Google Scholar]
- 11.Morelon E, et al. Face transplantation: partial graft loss of the first case 10 years later. Am J Transplant. 2017;17(7):1935–1940. doi: 10.1111/ajt.14218. [DOI] [PubMed] [Google Scholar]
- 12.Cendales LC, et al. The Banff 2007 working classification of skin-containing composite tissue allograft pathology. Am J Transplant. 2008;8(7):1396–1400. doi: 10.1111/j.1600-6143.2008.02243.x. [DOI] [PubMed] [Google Scholar]
- 13.Turnbull AK, et al. Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: comparison of gene expression profiling approaches. BMC Bioinformatics. 2020;21(1):30. doi: 10.1186/s12859-020-3365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider M, et al. Vascularized composite allotransplantation: a closer look at the banff working classification. Transpl Int. 2016;29(6):663–671. doi: 10.1111/tri.12750. [DOI] [PubMed] [Google Scholar]
- 15.Benichou G, et al. Natural killer cells in rejection and tolerance of solid organ allografts. Curr Opin Organ Transplant. 2011;16(1):47–53. doi: 10.1097/MOT.0b013e32834254cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medema JP, et al. Cleavage of FLICE (caspase-8) by granzyme B during cytotoxic T lymphocyte-induced apoptosis. Eur J Immunol. 1997;27(12):3492–3498. doi: 10.1002/eji.1830271250. [DOI] [PubMed] [Google Scholar]
- 17.Hébert M-J, Jevnikar AM. The impact of regulated cell death pathways on alloimmune responses and graft injury. Curr Transplantation Rep. 2015;2(3):242–258. doi: 10.1007/s40472-015-0067-4. [DOI] [Google Scholar]
- 18.Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moody DB, Suliman S. CD1: from molecules to diseases. F1000Res. 2017;6:1909. doi: 10.12688/f1000research.12178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etra JW, et al. Mechanisms of rejection in vascular composite allotransplantation. Curr Opin Organ Transplant. 2018;23(1):28–33. doi: 10.1097/MOT.0000000000000490. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe R, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med. 2015;7(279):279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siemionow M, et al. A cadaver study in preparation for facial allograft transplantation in humans: part I. What are alternative sources for total facial defect coverage? Plast Reconstr Surg. 2006;117(3):864–872. doi: 10.1097/01.prs.0000204875.10333.56. [DOI] [PubMed] [Google Scholar]
- 25.Saavedra AP, et al. Transmission of donor-specific skin condition from donor to recipient of facial allograft. Am J Transplant. 2011;11(6):1340. doi: 10.1111/j.1600-6143.2011.03596.x. [DOI] [PubMed] [Google Scholar]
- 26.Dvorak HF, et al. Rejection of first-set skin allografts in man. the microvasculature is the critical target of the immune response. J Exp Med. 1979;150(2):322–337. doi: 10.1084/jem.150.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sibaud V. Dermatologic reactions to immune checkpoint inhibitors: skin toxicities and immunotherapy. Am J Clin Dermatol. 2018;19(3):345–361. doi: 10.1007/s40257-017-0336-3. [DOI] [PubMed] [Google Scholar]
- 28.Djenidi F, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol. 2015;194(7):3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- 29.Petrelli A, et al. PD-1+CD8+ T cells are clonally expanding effectors in human chronic inflammation. J Clin Invest. 2018;128(10):4669–4681. doi: 10.1172/JCI96107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. 2015;7(269):269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark RA. Skin-resident T cells: the ups and downs of on site immunity. J Invest Dermatol. 2010;130(2):362–370. doi: 10.1038/jid.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hautz T, et al. Molecular markers and targeted therapy of skin rejection in composite tissue allotransplantation. Am J Transplant. 2010;10(5):1200–1209. doi: 10.1111/j.1600-6143.2010.03075.x. [DOI] [PubMed] [Google Scholar]
- 33.Borges TJ, et al. Codominant role of interferon-γ- and interleukin-17-producing T cells during rejection in full facial transplant recipients. Am J Transplant. 2016;16(7):2158–2171. doi: 10.1111/ajt.13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilliam AC, et al. Apoptosis is the predominant form of epithelial target cell injury in acute experimental graft-versus-host disease. J Invest Dermatol. 1996;107(3):377–383. doi: 10.1111/1523-1747.ep12363361. [DOI] [PubMed] [Google Scholar]
- 35.Ferrara JL. Cytokine dysregulation as a mechanism of graft versus host disease. Curr Opin Immunol. 1993;5(5):794–799. doi: 10.1016/0952-7915(93)90139-J. [DOI] [PubMed] [Google Scholar]
- 36.Pasparakis M, et al. Mechanisms regulating skin immunity and inflammation. Nat Rev Immunol. 2014;14(5):289–301. doi: 10.1038/nri3646. [DOI] [PubMed] [Google Scholar]
- 37.Grahammer J, et al. Benefits and limitations of belatacept in 4 hand-transplanted patients. Am J Transplant. 2017;17(12):3228–3235. doi: 10.1111/ajt.14440. [DOI] [PubMed] [Google Scholar]
- 38.Krezdorn N, et al. Immunological characteristics of a patient with belatacept-resistant acute rejection after face transplantation. Am J Transplant. 2016;16(11):3305–3307. doi: 10.1111/ajt.13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cendales LC, et al. De novo belatacept in clinical vascularized composite allotransplantation. Am J Transplant. 2018;18(7):1804–1809. doi: 10.1111/ajt.14910. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi R, et al. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-γ and IL-17A production. J Exp Med. 2011;208(10):2055–2067. doi: 10.1084/jem.20110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recio C, et al. Suppressor of cytokine signaling-1 peptidomimetic limits progression of diabetic nephropathy. J Am Soc Nephrol. 2017;28(2):575–585. doi: 10.1681/ASN.2016020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed CM, et al. SOCS1 mimetics and antagonists: a complementary approach to positive and negative regulation of immune function. Front Immunol. 2015;6:183. doi: 10.3389/fimmu.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(issue suppl 1):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefranc MP. IMGT, the international ImMunoGeneTics information system. Cold Spring Harb Protoc. 2011;2011(6):595–603. doi: 10.1101/pdb.top115. [DOI] [PubMed] [Google Scholar]
- 45.Lefranc MP. IMGT, the international ImMunoGeneTics information system, http://imgt.cines.fr. Methods Mol Biol. 2004;248:27–49. doi: 10.1385/1-59259-666-5:27. [DOI] [PubMed] [Google Scholar]
- 46. NanoStringQCPro: Quality metrics and data processing methods for NanoString mRNA gene expression data. R package version 1.22.0. Nickles D, et al.; 2020. https://www.bioconductor.org/packages/release/bioc/html/NanoStringQCPro.html Accessed February 26, 2021.
- 47.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DEGreport: Report of DEG analysis. R package version 1.26.0. Pantano L; 2020. https://www.bioconductor.org/packages/release/bioc/html/DEGreport.html Accessed February 26, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.