Abstract
We provide integrated protein sequence-based predictions via https://bio2byte.be/b2btools/. The aim of our predictions is to identify the biophysical behaviour or features of proteins that are not readily captured by structural biology and/or molecular dynamics approaches. Upload of a FASTA file or text input of a sequence provides integrated predictions from DynaMine backbone and side-chain dynamics, conformational propensities, and derived EFoldMine early folding, DisoMine disorder, and Agmata β-sheet aggregation. These predictions, several of which were previously not available online, capture ‘emergent’ properties of proteins, i.e. the inherent biophysical propensities encoded in their sequence, rather than context-dependent behaviour (e.g. final folded state). In addition, upload of a multiple sequence alignment (MSA) in a variety of formats enables exploration of the biophysical variation observed in homologous proteins. The associated plots indicate the biophysical limits of functionally relevant protein behaviour, with unusual residues flagged by a Gaussian mixture model analysis. The prediction results are available as JSON or CSV files and directly accessible via an API. Online visualisation is available as interactive plots, with brief explanations and tutorial pages included. The server and API employ an email-free token-based system that can be used to anonymously access previously generated results.
Graphical Abstract
Graphical Abstract.
The server provides integrated sequence-based predictions of protein characteristics, including analysis of multiple sequence alignments.
INTRODUCTION
Traditionally, our understanding of how proteins operate and how evolution shapes them is based on the overall protein fold and its amino acid sequence, using multiple sequence alignments (MSAs). The recent impressive performance of AlphaFold (1) demonstrates how well we can now predict the relationship between these two, however a significant part of the proteome shows highly dynamic and/or structurally ambiguous behaviour, which is still very difficult to address. Our single-sequence based DynaMine predictor (2) and its web server (3) attempted to capture the ‘emerging’ protein backbone dynamics property, as determined by local interactions between amino acids. We have since extended the DynaMine range of predictors, based on the same principle, with side-chain dynamics and conformational propensity, and have developed predictors for early folding (4), disorder (5) and aggregation (6) that are dependent on the DynaMine suite predicted biophysical features. We now make all these predictions directly available from a single place in an integrated server, with the side-chain dynamics, conformational propensities, early folding and aggregation predictions not previously directly accessible online.
Within the community, servers already exist that provide a wide variety of predictions for a single protein sequence at a time, notably PredictProtein (7), PSIPRED (8) and SCRATCH (9), whose output includes a variety of predictions such transmembrane regions, solvent accessibility, disulfide bonds, antigenicity, etc.. Other servers predict single features but allow the upload of multiple sequences, for example there are many online predictors of protein disorder, such as Espritz (10), PrDOS (11) and IUPred2A (12), with the CAID competition now providing benchmarking (13). For aggregation predictions, PASTA2.0 (14) and AGGRESCAN3D (15) exist, while for protein flexibility, the PredyFlexy server (16) was developed. In addition, many predictors exist for secondary structure (17), but crucially, these predictions are based on protein structure data from the PDB, whereas the core DynaMine dynamics and conformational propensities predictions are based on the in-solution behaviour of proteins, as observed from their NMR chemical shift values. The DynaMine training data set also includes a wide range of protein behaviour, from fully disordered proteins to well-folded ones, thus enabling it to provide more general predictions on their biophysical behaviour. These predictions then form the basis for the unique EFoldMine early folding predictions we provide, as well as the DisoMine disorder and Agmata aggregation predictions. Conceptually, the information we provide relates to ‘emergent’ properties of proteins, in other words the behavior the protein is capable of, not its final state in a particular context (e.g. when folded).
As a novel feature, to our knowledge not available elsewhere, we now attempt to capture how evolutionary pressure shapes the (predicted) biophysical features of a protein, by calculating multiple sequence alignment (MSA) based statistics. With this approach, we want to highlight conserved biophysical features that cannot be directly observed from a single protein structure, nor from multiple sequences, hence enabling the exploration of protein behaviour and functionality from a new perspective. We also highlight residues that have unusual properties in the 7D ‘biophysical space’ we predict through a Gaussian mixture model that is directly trained on the MSA-based statistics. This functionality is available at https://bio2byte.be/b2btools/, and makes the concept of biophysical features and their conservation available to the scientific community. This website is free and open to all users and there is no login requirement.
MATERIALS AND METHODS
Frontend
The website (https://bio2byte.be/b2btools/) is implemented in Python 3.7 using the Django 3.0 framework. We use the NGINX HTTP server as a gateway and an SQLite 3 database for persistent storage and retrieval of requested data. All modern browsers accept the b2bTools frontend, which combines statically rendered HTML documents with selective layers of interactivity using the jQuery JavaScript library. The website uses ApexCharts as its visualization library, which enables browser-based visualisation of large data sets and provides a flexible API. The requests from the web interface are first sent to NGINX and then sent to RESTful APIs.
The b2bTools RESTful API is based on the Django REST framework 3.11 and provides programmatic access to our predictor tools, for single sequences using a JSON as input, for MSA-based predictions using the multiple sequence alignment file as input (see https://www.bio2byte.be/b2btools/documentation/dynamine/ and links therein). Using only two easy-to-use endpoints (Table 1), the API returns its results in JSON (JavaScript Object Notation) format. To obtain the results, it is necessary to use an alphanumeric hash ID provided by the submission process, thus ensuring greater security and data confidentiality.
Table 1.
REST API endpoints
| Method | Endpoint | Description |
|---|---|---|
| POST | /msatools/api/ | Sends the data in JSON format for processing. |
| GET | /msatools/api/queue/ | Gets the data after processing 1 . |
1It is necessary to use an alphanumeric hash ID provided in the submission to obtain the result.
Job management
The management of computational workflows and batch jobs depends on the Python RQ 1.2 library (Redis Queue), which is supported by the Redis message broker.
Backend
The backend consists of the integrated set of predictors in Python 3.7, available as a standalone package from https://bitbucket.org/bio2byte/bio2byte_server_public/; please see the README.md file therein for a full description. The DynaMine suite of predictors (backbone and sidechain dynamics, secondary structure propensities) forms the core of the predictions, with EFoldMine dependent on these predictions. DisoMine and Agmata are each in turn dependent on EFoldMine. These predictors are available from the singleSeq/ directory in the package. The MSA-based predictions include the DynaMine, EFoldMine and DisoMine predictions, and are available from the multipleSeq/ directory. Additional external dependencies are joblib v0.14.1, numpy v1.19.1, Pillow v7.0.0, pomegranate v0.13.5, scikit-learn v0.21.3, scipy v1.4.1, six v1.14.0, pytorch v1.3.1, torchvision v0.4.2 and matplotlib v3.1.2. General code employed in the server, e.g. a parser for MSA’s, is available in the general/ directory. Ready-to-run python scripts are available in the run/ directory, and additional information on how to run it can be found employing the –h flag while running the script.
The MSA-based predictions can handle most common MSA formats (CLUSTAL, FASTA, STOCKHOLM, BALIBASE, PSI, A3M, BLAST and PHYLIP). This feature was originally developed in the context of the RNAct project (http://rnact.eu/) to compare the biophysical characteristics of different RNA Recognition Motif (RRM) families at the MSA level (available from https://bitbucket.org/bio2byte/seqrrm/), but was adapted and implemented for use on the server. From the MSA, the non-gapped individual sequences are recreated, on which the predictions are computed. The obtained results are then mapped back to the original alignment, so that each position in the MSA is associated with NP-NG prediction values for each prediction type, with NP being the total number of sequences in the MSA, NG being the number of gaps in that column. Per MSA column, 5 statistical values are then calculated; top outlier, first quartile, median, third quartile and bottom outlier. Individual protein predictions that fall within the quartile range are therefore ‘normal’ with respect to the sequences in the MSA, while values outside of this range indicate deviating behaviour for the sequence under study. To quantify this deviating behavior, a further statistical study is performed to assess which residues have biophysical behavior that deviates significantly from the MSA. The biophysical predictions calculated for all the individual sequences in the MSA are fitted on-the-fly using a Gaussian mixture model (GMM) with a single component using the sklearn Python library. This allows reduction of the number of dimensions from the seven biophysical predictions (backbone and sidechain dynamics, helix, sheet, coil and early folding propensities and disorder) to a single scoring value. The more negative the value, the further that residue is from the MSA-based model. This enables determination of residues that are over the 95, 99 and 99.9 percentiles as estimated by the GMM, which are indicated in the visualization.
RESULTS
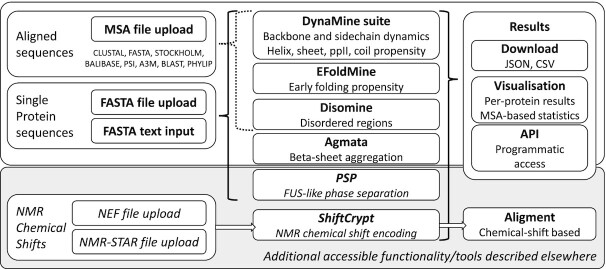
For an overview of the input, predictions and output/visualisation possibilities, see Figure 1. Below we describe in more detail how to use the web server for single sequence predictions, and for predictions based on a multiple sequence alignment (MSA).
Figure 1.
Overview of the joint input for the b2bTools server (left), the predictions that are accessible depending on input type (middle) and the output possibilities (right). The bottom box in grey relates to tools described elsewhere which are also accessible from the joint input page.
Obtaining and displaying predictions
The first step in using the b2bTools web server is generating a unique token or using a previously generated one (top left of page). The token ensures secure web server access; users only have access to their results with the token, and no credentials have to be entered. The token can be re-used to retrieve or delete earlier results when accessing the web server, for example for jobs that take a long time to finish. Without this unique token, earlier results cannot be accessed any more. Note that we provide the option to send the token to the user via e-mail via the ‘Send token to email’ option in the top bar, but this is not obligatory.
-
The second step is to provide the protein sequence(s) for prediction. Examples for each type of protein sequence input are available for direct testing on the left-hand side of the page under ‘Test examples’, which will go directly to step 3. The user can provide their own sequence(s) in the following way:
For single sequences as an input, the user can upload a file containing one or several unaligned sequences in FASTA format (in ‘Choose File’). Alternatively, it is possible to type or paste sequences in FASTA format in the input text box.
For multiple sequence alignments (MSA) as an input, the user can upload a file in a variety of formats, such as CLUSTAL, FASTA, STOCKHOLM, BALIBASE, PSI, A3M, BLAST and PHYLIP. The input text box does not work for MSAs.
At this point, the web page will try to detect the type of file within the frontend browser, and the page will be updated accordingly. When pressing ‘submit’ the protein sequence data will be sent to the server, parsed and validated, and the sequences will be identified, leading to the next page.
-
The user can then decide which predictions to run on their data:
For single sequences, a list of the protein identifiers detected in the FASTA file will be displayed, and on the left-hand side the user can select the predictors to run. Currently, Dynamine, Disomine, EFoldMine, and Agmata can be selected simultaneously to generate integrated results, with PSPer available separately. Brief explanations are provided for each tool under ‘Info’. There is presently a limit of 10 sequences for Agmata, and of 50 sequences for all other predictors, to be revised in the future.
For MSAs, the number of aligned sequences is shown. Click the ‘Run predictions’ button to obtain the DynaMine, EFoldMine and DisoMine predictions, with a brief explanation available under ‘More info’. There is presently a limit of 200 sequences.
The result page should load within a few seconds for uploaded files, although Agmata results are significantly slower, taking up to a couple of minutes.
-
For both single sequence(s) and MSAs, results are shown as interactive plots, with the top plot showing the results for one protein at a time. The protein to display can be selected using the dropdown menu at the top of the page. The plot itself shows the selected tool's predictions along the protein sequence. By default, only the requested results are shown, but additional available predictions can be toggled on by clicking on their name in the legend; in general, predictions can be toggled off or on this way by clicking on their name in the legend underneath the plot. The x-axis range can be changed in two ways: by using the two-way slider above the plot, with the start and end residue indicated, or by entering values manually in the boxes above the slider. When hovering the cursor over the plot, its position is shown, with on the y-axis the current value, and on the x-axis the amino acid one-letter code and sequence position number. The actual predicted values are shown in the legend. Hovering the cursor over a particular label in the legend will highlight the corresponding values in the plot. The menu at the top right of the plot offers a selection of functions, providing from left to right:
Zoom in
Zoom out
Selection zoom: zoom in the selected region of the plot
Panning: drag the view around the plot
Reset zoom: reset the zoom to default view
Download: images as SVG and PNG, data in CSV format
A detailed explanation of the different predictions is available by clicking on ‘Click for explanation’ underneath the plot.
For MSAs, a second plot is available that compares the results of the selected protein (for the top plot) with the distribution of predicted values for all the sequences in the MSA; the black line is the median, the dark grey lines first/third quartile, and the light grey line the outlier range. Only MSA positions that are not gaps in the selected protein are shown, with the predictions for the protein in red. The user can select which prediction values to display from the dropdown menu below the plot. The other options are the same as described for the top plot. A detailed explanation is available by clicking on the ‘Click for explanation’ underneath the plot. In addition, when hovering over the top plot, the results from the GMM are given for the residue corresponding to the x-axis position, with more negative values meaning this residue is unusual in relation to the values observed in the MSA. The displayed values are colour-coded when below the 5% (orange), 1% (red) and 0.1% (dark red, white text) percentile limits calculated from all values for all proteins in the MSA. An overview of the number of residues belonging to these categories, with their percentage in relation to the overall length of the protein, is shown underneath the sequence length information (GMM score analysis), together with the list of relevant residues.
For the predicted results, the user can access the API directly via the ‘API’ button, or they can download the results in JSON or CSV format. For MSA-based predictions, two CSV files are available, one with the per-protein predictions, one with the MSA-based statistics. Both are organised according to the MSA, with gap positions shown as None.
Note that it is also possible to directly access a predictor by using a direct link, for example https://bio2byte.be/disomine/. In this case, step 3 of the above pipeline will be skipped, and the results immediately shown. In addition, it is possible to access all results previously generated with your current token via the ‘Past results’ link in the top bar.
Case studies
Single sequence prediction
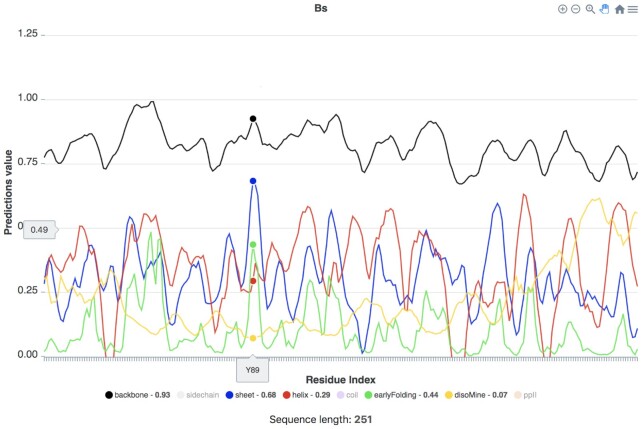
As a case study to highlight the analysis possibilities of the server, we present 10 proteins that form TIM barrel folds (with codes Bs, Ch, Ec, Hu, Lm, Pf, Tb, Tm, Vm, Ye), which were previously compared and their thermostability studied (18). These proteins all adopt TIM barrel folds, but have relatively low shared pairwise sequence identities (37%-55%, with 3 outliers that have higher shared identity, Tb-Lm, Ch-Hu and Ec-Vm). To this dataset we added the sTIM-11 protein, successfully designed de novo to fold as a TIM barrel by using a subunit repeat strategy, and the OctaV1 protein, which was designed to fold as a TIM barrel, but in fact folds differently, even though its secondary structure elements are generally in the intended (TIM barrel) positions (19). An interactive version of these data is available via https://www.bio2byte.be/b2btools/tutorial/singleseq/. By default, the interactive plot displays the predictions (y-axis) in relation to the protein sequence for the selected protein (x-axis). All predictions are in this case turned on, but you can toggle individual predictions on and off by clicking on their name below the plot. When you run a specific predictor, for example DisoMine, only that data will be displayed, but all predictions that were executed to obtain the final one will be available; click on their name below the plot to display them. When you hover over the plot, it will display the amino acid residue that the cursor is currently on (x-axis, Y89 in Figure 2), and the corresponding prediction values are listed next to their name at the bottom of the plot (see Figure 2). The length of the displayed protein sequence is visible underneath the plot (here 253 residues), and a brief explanation of the interpretation for the predictions is available by ‘Click for explanation’.
Figure 2.
Single sequence predictions for the Bs protein as displayed in the server, showing the Dynamine backbone dynamics (black), sheet (blue) and helix (red) propensities, and the early folding (green) and disorder predictions (yellow). The prediction values for the amino acid residue (here Y89) corresponding to the cursor position (large dots) are shown next to their name in the legend at the bottom.
Figure 2 shows the individual DisoMine (and related) predictions, with the ‘sidechain’, ‘coil’, and ‘ppII’ values turned off, for the Bs protein (Uniprot P00943, a structure is available as PDB code 1BTM; note that such structure information is not used in our predictions). This protein has low DisoMine (disorder) values, except for the last 25 C-terminal residues, which do have strong helix propensities. The DynaMine backbone dynamics values are generally above 0.80, in agreement with the fact that this is a well-folded protein (see https://www.ebi.ac.uk/pdbe/pdbe-kb/proteins/P00943 for an overview). Overall, for all the other TIM barrel proteins, there are consistent peaks for beta sheet propensities, which tend to coincide with the early folding peaks (e.g. Y89 in Figure 2), with helix propensities generally of similar maximum height as the beta sheet. Displaying the designed sTIM-11 protein online (data not shown here) will evidence the subunit repeats, as groups of signals are repeated 4 times, with some differences because of the fine-tuning of their final design. This protein is shorter, but the overall pattern of the signals is similar to that of the natural TIM barrel proteins. The OctaV1 protein (data not shown here), despite being designed-based on a TIM barrel structure template (19), shows more differences, with much higher helix propensities in relation to beta sheet and early folding peaks corresponding to the regions with high helical propensity. This indicates that its ‘emergent’ behaviour, or what the OctaV1 sequence is capable of doing based on local amino acid interactions, is quite different from TIM barrel proteins, whilst the designed sTIM-11 remains similar. In other words, the local interactions between amino acids in the OctaV1 sequence seem to determine a different overall behaviour, which might in this case influence the secondary structures that are formed first when folding, and so change the fold it finally obtains. These differences are highlighted in the MSA-based visualisation.
MSA-based prediction
We aligned the 11 sequences of TIM barrel proteins described above based on their structures in the PDB with the PROMALS3D server (20,21), and added to this alignment, based on the location of its secondary structure elements (19), the OctaV1 sequence. We here describe the results for the resulting MSA. An interactive version is available via https://www.bio2byte.be/b2btools/tutorial/msatools/, the MSA used to generate the plots is available from supplementary data. The first plot at the top of the page is the same as described for the single sequence prediction, with the individual sequence recreated without gaps from the MSA information, but now displays the GMM scores in relation to the MSA. The second plot at the bottom shows, for the protein selected at the top of the page, the variation in predicted biophysical parameters within the MSA. This variation is displayed according to simple box plot statistics, with median, first/third quartile, and outlier range of the distributions shown (Figure 3). Columns in the MSA that are ‘gapped’ for the selected protein are not shown here. In other words, what is displayed is how the biophysical prediction for each aligned position varies for all the proteins that are in the MSA, but only for positions that are not gaps in the selected protein. You can select the type of prediction that you want to display in the selection box below the plot and turn each distribution statistic on and off by clicking on its name. The ‘prediction’ field corresponds to the same type of prediction shown in the top plot.
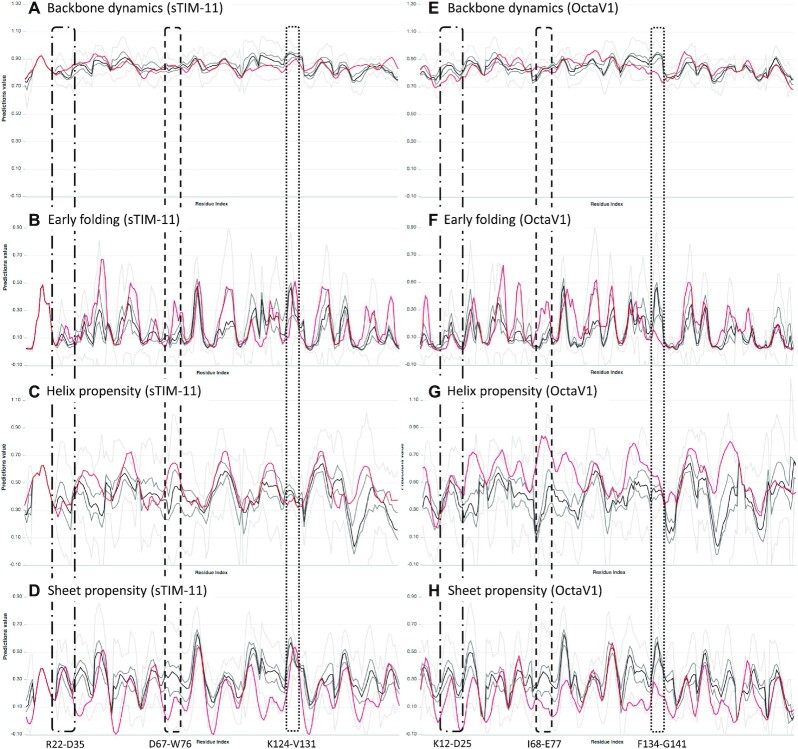
Figure 3.
Multiple sequence alignment statistics for the sTIM-11 (left column) and OctaV1 (right column) proteins for backbone dynamics (A, E), early folding (B, F) and helix (C, G) and sheet propensity (D,H). The regions corresponding to Arg22–Asp35 (sTIM-11) and Lys12–Asp25 (OctaV1) (dash-dot box), Asp67–Trp76 (sTIM-11) and Ile68–Glu77 (OctaV1) (dashed box) and Lys124–Val131 (sTIM-11) and Phe134–Gly141 (OctaV1) (dotted box) are indicated. The red lines show the actual sTIM-11 (left) and OctaV1 (right) predictions, the black line the median, the dark grey lines first/third quartile, and light grey the outlier range for the predicted values for the corresponding position in the MSA. Individual images were created from the ‘Download png’ plot feature.
In Figure 3 we compare the individual predictions (red line) to the MSA statistics (black/grey lines) for backbone dynamics, early folding, helix and sheet propensity for the successfully de novo designed sTIM-11 protein, and for the OctaV1 protein, which was designed using Rosetta (22) based on a TIM barrel structure, but in fact folds differently. For both proteins, the backbone dynamics are very similar to the MSA-based distributions, with the red line mostly falling within its quartile ranges (dark grey lines). The early folding predictions show some immediate differences, with some peaks being higher in both proteins (both proteins, dashed box), many similar (e.g. sTIM-11, dotted box) and one notably absent (OctaV1 dotted box). Note that the dashed and dotted box regions do not correspond exactly to each other due to gaps in the alignment, but they do correspond to the same overall region in the individual sequence. The early folding differences imply that, compared to natural TIM barrel proteins, many regions of both the sTIM-11 and OctaV1 protein will start to fold earlier, but only a few regions will have delayed folding. From the helix and sheet propensities, it is apparent that overall, the helix propensities for sTIM-11 are similar to the MSA-based distributions, whilst OctaV1 has overall higher propensities. Both proteins have generally lower β-sheet propensities, but the pronounced β-sheet propensity peaks are present in sTIM-11, while some notable ones are absent in OctaV1. Relating back to the early folding changes, the dashed region has little β-sheet propensity for both proteins, whilst it has very high helix propensity for OctaV1, but similar propensity for sTIM-11. The dotted region, on the other hand, has similar helix propensity in both proteins, while the β-sheet propensity is similar in sTIM-11, but much reduced in OctaV1 (outside of the quartile range). Also note that the region preceding the dotted box has much higher helix propensity in OctaV1, and much lower β-sheet propensity in both proteins, with similar early folding propensity. These differences are also reflected in the GMM scores for both proteins; for OctaV1, 17.51% of residues belong to the worst 5% of values (for all proteins in the MSA), 5.53% to the worst 1%, and 0.92% to the worst 0.1%, totalling 23.96%. For sTIM-11, only 7.78% belong to the worst 5% category, and 1.11% to the 1% category. Out of the natural TIM barrel proteins, only Bs is more unusual, with 9.56% in the 5% category, 2.79% in the 1% category, with most of these values in the C-terminal end. To illustrate how the GMM picks up regions that are unusual with respect to the MSA-based statistics, consider the K12-D25 region for OctaV1 (Figure 3, dot-dash box), the residues of which are all in the worst 1–5% category. Even though, on sight, the individual predictions seem to be just within the quartile range (except for sheet), the GMM picks up that the combined predictions here have an unusual multi-dimensional distribution, indicating that this region might be relevant for the different folding of OctaV1. Similarly, the L75 and A75 residues in the I68-E77 region are unusual, whereas the F134-G141 region in fact falls within the expected GMM range.
Overall, we therefore expect that this exploratory type of analysis can help to highlight relevant differences of interest in proteins via the inherent biophysical characteristics encoded by protein sequences. For designing sequences, it might help alleviate misfolding by comparing to the biophysical signal of natural sequences, and in addition could help to identify differences between natural proteins of similar structure, for example in relation to changes in folding pathway (4). Neither of these can, to the best of our knowledge, be easily determined from the structure, nor from molecular dynamics simulations. In addition, the analyses described here require only the protein sequences, and a multiple sequence alignment.
DISCUSSION
The integrated set of predictors on this web server are free and open to all, using a token-based system for security without login requirement. Because of the in-solution NMR data on which the core DynaMine predictions are based, as well as the wider set of protein behaviour they incorporate, we provide an integrated set of more general predictions on protein biophysical behaviour. Instead of aiming to predict a final folded state, it is rather the inherent properties present in the protein, due to local interactions of amino acids close to each other in the sequence, that are targeted. Such a type of analysis can provide a new angle to understanding protein behaviour, and is especially important for proteome-scale studies, where the fold or dynamic behaviour of many proteins remains unknown and is likely ambiguous (the ‘dark proteome’) (23).
As a novel feature, to our knowledge not available anywhere, we also attempt to capture how evolutionary pressure shapes the (predicted) biophysical features of a protein, by calculating multiple sequence alignment (MSA) based statistics. With this approach, we want to highlight conserved biophysical features that cannot be directly observed from a single protein structure nor from multiple sequences. This enables the exploration of protein behaviour and functionality from a new perspective, in essence trying to capture the concept of biophysical features and their conservation in a ‘biophysical fingerprint’. This also enables the definition of a 7D ‘biophysical space’ that can be interpreted with methods such as Gaussian mixture models (GMM) to pinpoint which residues in which proteins show unusual behavior compared to all proteins in the MSA. With respect to interpretation of such data, we stress that this is highly dependent on the quality and type of MSA used; a structure-based alignment of similarly folded proteins with highly diverse sequences is likely to highlight features important for that particular protein fold, whereas an alignment of more closely related sequences within a domain of life might rather delineate the biophysical limits of a specific protein function. Overall, this kind of approach might highlight patterns not obvious from protein structures, nor from amino acid conservation in MSAs, and can thus complement computationally intensive approaches to understand protein behaviour, such as molecular dynamics, and enable in silico screenings of, for example, newly designed sequences.
We will continue to expand the website with new capabilities, for example the ability to align proteins based on their biophysical characteristics, similar to what we already provide for the ShiftCrypt NMR chemical shift-based alignments (24,25). In the meantime, we remain very interested in feedback from users to pinpoint case studies, and to provide suggestions for improvements and novel approaches we could include to analyse these data.
DATA AVAILABILITY
The webserver is available at https://bio2byte.be/b2btools/.
The API web service is available at https://bio2byte.be/msatools/api/, with documentation available at https://bio2byte.be/b2btools/dynamine/ (and for other levels of prediction: disomine, efoldmine, msatools).
The backend b2btools is available at https://bitbucket.org/bio2byte/bio2byte_server_public/.
The case study interactive visualisations are available at https://www.bio2byte.be/b2btools/tutorial/singleseq/ and https://www.bio2byte.be/b2btools/tutorial/msatools/.
No accession numbers are available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Francesco Codice and Francesco Tabaro for work on previous web services of the group, which inspired the current implementation, and Kim Van Roey for thorough reading of the manuscript and editorial suggestions.
Notes
Present address: Daniele Raimondi, ESAT-STADIUS, KU Leuven, Leuven 3001, Belgium.
Present address: Gabriele Orlando, Switch laboratory, KU Leuven, Leuven 3000, Belgium.
Contributor Information
Luciano Porto Kagami, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium.
Gabriele Orlando, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium.
Daniele Raimondi, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium.
Francois Ancien, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; 3Bio, Université Libre de Bruxelles, Brussels 1050, Belgium.
Bhawna Dixit, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium.
Jose Gavaldá-García, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium.
Pathmanaban Ramasamy, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium; VIB-UGent Center for Medical Biotechnology, VIB, Ghent 9000, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, 9000, Belgium.
Joel Roca-Martínez, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium.
Konstantina Tzavella, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium.
Wim Vranken, Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium; VIB Structural Biology Research Centre, Brussels, 1050, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement [813239 to J.R.-M. and J.G.-G.]; Research Foundation Flanders (FWO) [G.0328.16N to P.R., G.O., B.D.]; European Regional Development Fund (ERDF); Brussels-Capital Region-Innoviris within the framework of the Operational Programme 2014–2020 through the ERDF-2020 project [ICITY-RDI.BRU to F.A.]; Vrije Universiteit Brussel Research Council under the Interdisciplinary Research Program TumorScope [IRP20 to K.T.]. Funding for open access charge: European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement [813239].
Conflict of interest statement. None declared.
REFERENCES
- 1. AlQuraishi M. A watershed moment for protein structure prediction. Nature. 2020; 577:627–628. [DOI] [PubMed] [Google Scholar]
- 2. Cilia E., Pancsa R., Tompa P., Lenaerts T., Vranken W.F.. From protein sequence to dynamics and disorder with DynaMine. Nat. Commun. 2013; 4:2741. [DOI] [PubMed] [Google Scholar]
- 3. Cilia E., Pancsa R., Tompa P., Lenaerts T., Vranken W.F.. The DynaMine webserver: predicting protein dynamics from sequence. Nucleic. Acids. Res. 2014; 42:W264–W270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raimondi D., Orlando G., Pancsa R., Khan T., Vranken W.F.. Exploring the sequence-based prediction of folding initiation sites in proteins. Sci. Rep. 2017; 7:8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orlando G., Raimondi D., Codicè F., Tabaro F., Vranken W.F.. Prediction of disordered regions in proteins with recurrent neural networks and protein dynamics. 2020; bioRxiv doi:28 May 2020, preprint: not peer reviewed 10.1101/2020.05.25.115253. [DOI] [PubMed]
- 6. Orlando G., Silva A., Macedo-Ribeiro S., Raimondi D., Vranken W.F.. Accurate prediction of protein beta-aggregation with generalized statistical potentials. Bioinformatics. 2020; 36:2076–2081. [DOI] [PubMed] [Google Scholar]
- 7. Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Hönigschmid P., Schafferhans A., Roos M., Bernhofer M.et al.. PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic. Acids. Res. 2014; 42:W337–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchan D.W.A., Jones D.T.. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019; 47:W402–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng J., Randall A.Z., Sweredoski M.J., Baldi P.. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005; 33:W72–W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walsh I., Martin A.J.M., Di Domenico T., Tosatto S.C.E.. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012; 28:503–509. [DOI] [PubMed] [Google Scholar]
- 11. Ishida T., Kinoshita K.. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007; 35:W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mészáros B., Erdos G., Dosztányi Z.. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic. Acids. Res. 2018; 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Necci M., Piovesan D.CAID Predictors, DisProt Curators CAID Predictors, DisProt Curators Tosatto S.C.E.. Critical assessment of protein intrinsic disorder prediction. Nat. Methods. 2021; 18:421–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walsh I., Seno F., Tosatto S.C.E., Trovato A.. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic. Acids. Res. 2014; 42:W301–W307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zambrano R., Jamroz M., Szczasiuk A., Pujols J., Kmiecik S., Ventura S.. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015; 43:W306–W313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Brevern A.G., Bornot A., Craveur P., Etchebest C., Gelly J.-C.. PredyFlexy: flexibility and local structure prediction from sequence. Nucleic Acids Res. 2012; 40:W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y., Gao J., Wang J., Heffernan R., Hanson J., Paliwal K., Zhou Y.. Sixty-five years of the long march in protein secondary structure prediction: the final stretch?. Brief. Bioinformatics. 2018; 19:482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maes D., Zeelen J.P., Thanki N., Beaucamp N., Alvarez M., Thi M.H., Backmann J., Martial J.A., Wyns L., Jaenicke R.et al.. The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures. Proteins. 1999; 37:441–453. [PubMed] [Google Scholar]
- 19. Figueroa M., Sleutel M., Vandevenne M., Parvizi G., Attout S., Jacquin O., Vandenameele J., Fischer A.W., Damblon C., Goormaghtigh E.et al.. The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools. J. Struct. Biol. 2016; 195:19–30. [DOI] [PubMed] [Google Scholar]
- 20. Pei J., Grishin N.V.. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 2014; 1079:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pei J., Tang M., Grishin N.V.. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008; 36:W30–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rohl C.A., Strauss C.E.M., Misura K.M.S., Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004; 383:66–93. [DOI] [PubMed] [Google Scholar]
- 23. Perdigão N., Heinrich J., Stolte C., Sabir K.S., Buckley M.J., Tabor B., Signal B., Gloss B.S., Hammang C.J., Rost B.et al.. Unexpected features of the dark proteome. Proc. Natl Acad. Sci. U.S.A. 2015; 112:15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orlando G., Raimondi D., F Vranken W.. Auto-encoding NMR chemical shifts from their native vector space to a residue-level biophysical index. Nat. Commun. 2019; 10:2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orlando G., Raimondi D., Kagami L.P., Vranken W.F.. ShiftCrypt: a web server to understand and biophysically align proteins through their NMR chemical shift values. Nucleic Acids Res. 2020; 48:W36–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The webserver is available at https://bio2byte.be/b2btools/.
The API web service is available at https://bio2byte.be/msatools/api/, with documentation available at https://bio2byte.be/b2btools/dynamine/ (and for other levels of prediction: disomine, efoldmine, msatools).
The backend b2btools is available at https://bitbucket.org/bio2byte/bio2byte_server_public/.
The case study interactive visualisations are available at https://www.bio2byte.be/b2btools/tutorial/singleseq/ and https://www.bio2byte.be/b2btools/tutorial/msatools/.
No accession numbers are available.