Abstract
Searching and reading relevant literature is a routine practice in biomedical research. However, it is challenging for a user to design optimal search queries using all the keywords related to a given topic. As such, existing search systems such as PubMed often return suboptimal results. Several computational methods have been proposed as an effective alternative to keyword-based query methods for literature recommendation. However, those methods require specialized knowledge in machine learning and natural language processing, which can make them difficult for biologists to utilize. In this paper, we propose LitSuggest, a web server that provides an all-in-one literature recommendation and curation service to help biomedical researchers stay up to date with scientific literature. LitSuggest combines advanced machine learning techniques for suggesting relevant PubMed articles with high accuracy. In addition to innovative text-processing methods, LitSuggest offers multiple advantages over existing tools. First, LitSuggest allows users to curate, organize, and download classification results in a single interface. Second, users can easily fine-tune LitSuggest results by updating the training corpus. Third, results can be readily shared, enabling collaborative analysis and curation of scientific literature. Finally, LitSuggest provides an automated personalized weekly digest of newly published articles for each user's project. LitSuggest is publicly available at https://www.ncbi.nlm.nih.gov/research/litsuggest.
Graphical Abstract
Graphical Abstract.
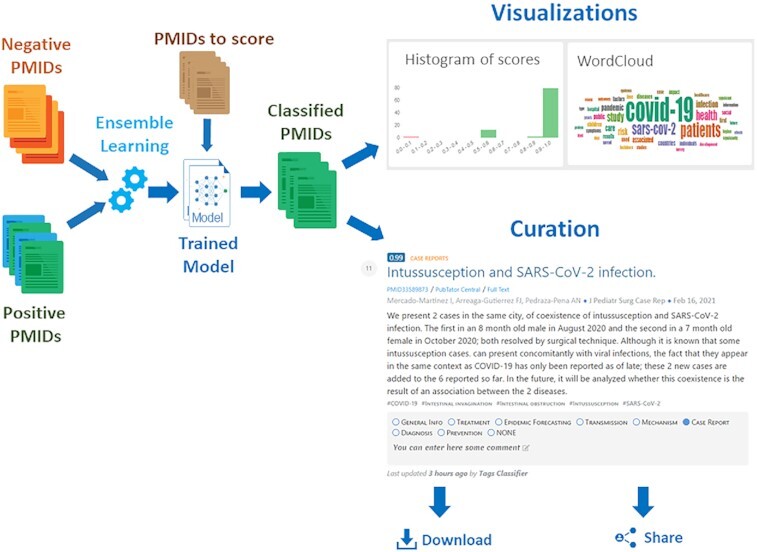
LitSuggest suggests relevant PubMed articles with high accuracy, offers visualization and curation features, in addition to the possibility to download and share classification results.
INTRODUCTION
Searching and reading relevant literature is a routine practice in biomedical research, as new research builds on previous discoveries (1). Indeed, millions of users access daily biomedical resources such as PubMed (2,3), PubMed Central and many others (4,5). With the ever-increasing amount of new literature, it is becoming more important than ever to be able to easily and rapidly find relevant publications in literature repositories such as PubMed (2,3). This is not only essential for individual researchers but as well for curators of biomedical databases such as UniProt (6), GWAS (7), LitCovid (8,9) and ClinVar (10), where finding relevant documents for annotation is the very first step of the curation pipeline (11). However, it is challenging for a user to design optimal search queries using all the keywords related to a given topic. As such, existing query-based search systems such as PubMed and many others often return suboptimal results (8,12): missing some publications while returning some that are irrelevant.
Several computational methods have been proposed as an effective alternative to keyword-based query methods for literature recommendation. Their effectiveness has been demonstrated in multiple publications (12–14), showing that machine learning and natural language processing methods (15,16) allow database curators to retrieve and curate more accurately and efficiently relevant literature, than traditional keyword-based PubMed search methods. For example, Lee et al. (13), used the machine learning (Convolutional Neural Networks) text classification method for a literature classification step for the GWAS Catalog database curation, reducing by two-thirds the curators’ workload of reading newly published papers, without compromising recall. However, those methods require specialized knowledge in machine learning and natural language processing, which can make it difficult for biologists to utilize. In addition, retrieving relevant publications is only the first part of the literature review pipeline, hence managing, curating, annotating, storing, and sharing the list of retrieved publications is often a cumbersome task for researchers. A handful of web applications have been developed to address this issue, such as MedlineRanker (17) and BioReader (18), but there is a strong desire for more user-friendly systems with additional features such as reusing models, curating and sharing classification results.
To assist biomedical researchers with their literature search needs, we propose LitSuggest, a web server that provides an all-in-one literature recommendation and curation service to help biomedical researchers stay up to date with scientific literature. LitSuggest combines advanced machine learning techniques for suggesting relevant PubMed articles with high accuracy. In addition to these innovative text-processing methods, LitSuggest offers a streamlined interface, practical functionality, and several specific advantages over existing tools. First, LitSuggest allows users to easily curate classified publications, organize classification tasks into jobs and projects, and download classification results with related annotations. Second, users can easily fine-tune LitSuggest results by observing the score distribution histogram and updating the training corpus to retrain the model. Third, classification results can be readily shared through a URL, enabling collaborative curation of scientific literature. Finally, users can activate a personalized weekly digest to classify the newly published literature every week automatically. Overall, LitSuggest offers a user-friendly interface with an all-in-one service for the recommendation and curation of biomedical literature.
SYSTEM DESCRIPTION
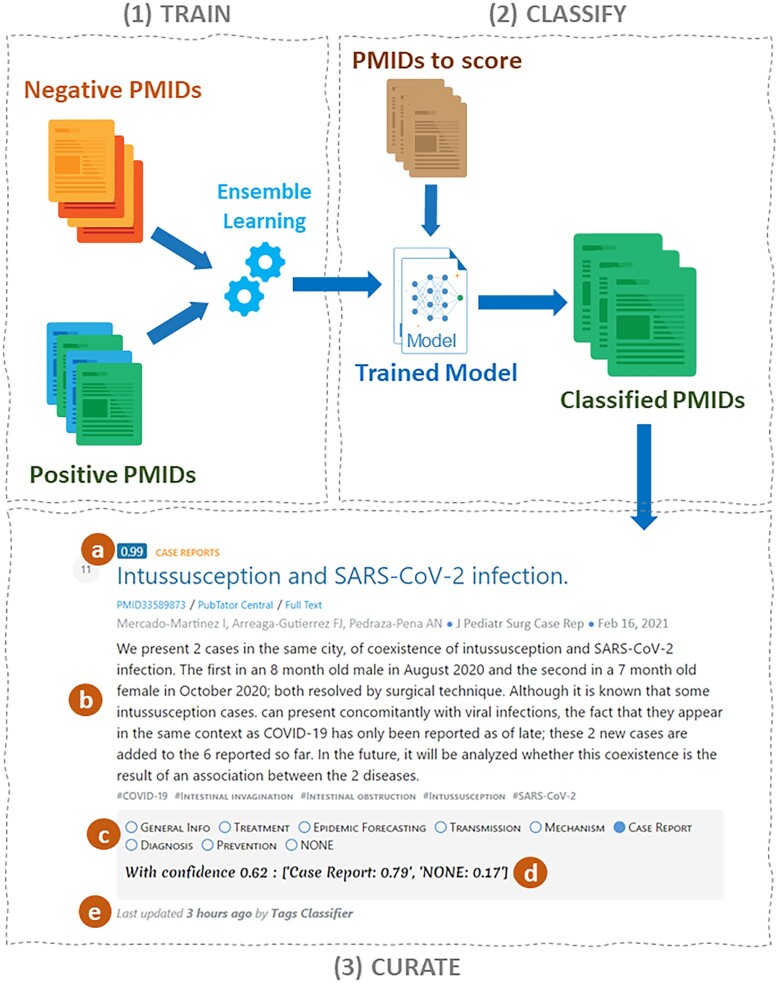
LitSuggest is a machine learning-based document recommendation and management system for biomedical literature. As shown in Figure 1, the workflow of working with LitSuggest can be split into three major steps: training a new model inside a project (Figure 1.1), using that trained model to classify new publications in separate jobs (Figure 1.2), and finally manually curating and annotating those automatically classified publications (Figure 1.3).
Figure 1.
Overview of LitSuggest. LitSuggest trains ensemble learning models based on a set of example articles from users (1). The model is then used to rank and classify new publications (2). Classified publications can then be curated (3) and shared with other users. The curation interface displays the probability score (a) for each publication, publication content (b) such as title, abstract, type, keywords, journal, date, authors, links to external resources, and interface to annotate the publication with custom tags (c), a custom text note (d) and the date and user which made the latest changes (e).
Training interface
The interface represents one topic per project. Users can train a model on a set of publications relevant for that topic and then apply this learned model to classify and rank new articles. All projects created by a user are displayed in the left sidebar of LitSuggest website. For new users, the interface displays a default empty project as well as an example project. The example project has a previously trained model, allowing users to start new classification jobs immediately. Users can add new projects or select an existing project by clicking on its name. Projects can be renamed, and users can add a description for each project.
Users start training a model for a new project by entering a list of ‘positive’ PMIDs (examples of publications they are interested in) as well as an optional list of ‘negative’ PMIDs (examples of publications that are not of interest). If no negative examples are supplied by the user, negative publications will be automatically generated. If the number of negative examples supplied by the user is not enough compared to the number of positive examples, additional negative PMIDs will be generated. There are three convenient ways of supplying positive and negative PMIDs: entering a list of PMIDs separated by spaces, loading PMIDs from a file, or retrieving (a maximum of 10,000) publications from PubMed, based on a query.
To quickly test LitSuggest, clicking on the ‘Try GWAS example’ button allows to easily populate positive or negative fields with example PMIDs. Depending on the size of the training set, training may take between a few seconds to several hours. Models for each project can be easily retrained, after modifying the positive and negative fields, and users can work on multiple projects (and multiple models) at the same time.
Classification interface
After a model has been trained, users can start analyzing new publications by entering a list of PMIDs to be classified and ranked. As with training, there are three ways of supplying positive and negative PMIDs: entering a list of PMIDs separated by spaces, loading PMIDs from a file, or retrieving (a maximum of 10 000) publications from PubMed, based on a query. In addition to the list of PMIDs to classify, it is possible to specify additional filtering options such as to keep only publications mentioning genes, variants, or chemicals, by checking corresponding checkboxes. This filtering is based on data retrieved from the PubTator Central API (19). The project page displays a list of all current and finished jobs with their status, as well as a progress bar showing the progress of each running job.
RESULTS AND CURATION INTERFACE
When an automatic text classification job finishes, it will be marked as completed on the project page. Clicking on it will display classification results on the job page. On the top of each job page, high level visualizations are presented, allowing to assess classification job performance. First, a histogram shows the distribution of scores for all classified publications. The score distributions demonstrate how confidently the models classify the articles (e.g. a score of 0.9 suggests the model is very confident when classifying the article), thereby facilitating user decisions (for example, whether re-training a model is needed).
Second, most discriminative words, enriched in positively and negatively classified publications are calculated using the Z-score and the top enriched words are displayed as a word cloud, so that users can better understand what the models regard as the main characteristics of the positive and negative sets, and easily verify the results.
After reviewing the high-level visual analysis of the classification job, users can start reviewing the publications themselves, classified into three tabs: Positive (positively classified publications, with scores of 0.5 or above), Negative (negatively classified publications), and Filtered (publications ignored by user-set filters).
In each tab, publications are sorted by score (Figure 1.3.a), with the most relevant (highly scored) publications displayed first.
It is possible to assign a curation tag (RELEVANT, IRRELEVANT, or TBD (TO BE DETERMINED)) to each publication (Figure 1.3.c). If the default annotation tags do not suit user needs, it is possible to edit the list of available curation tags on the Project page. The new tags will be available for later classification jobs. In addition to tags, users can add a text note under each publication (Figure 1.3.d).
Multi-user collaboration
Users can easily share any of their projects or classification jobs with other users by sharing the URL. Their collaborators will then be able to see the results of classification jobs and curate relevant publications, but they will not be able to edit the properties of the projects, retrain the model or start new classification jobs within the project. To easily track changes in this multi-user environment, the name of the last user who made a change and the date of the latest change is displayed under the curation interface for each publication (Figure 1.3.e).
Data download
All positively and negatively classified publications can be easily downloaded in TSV format by clicking the ‘Download Publications’ button. In addition to PMIDs and associated classification scores, the file will contain all the tags assigned by the user to each publication and the text notes. LitSuggest can therefore be very useful for building training sets for automated tag classifiers or performing database curation tasks.
Weekly automated digest
When the user checks the appropriate checkbox, LitSuggest automatically scans every week for new literature relevant to the project's model, then adds the result as a new job on the project page.
System implementation
As shown in Figure 2, the LitSuggest platform consists of three main components: Web frontend, Server backend, and processing nodes. Our backend is implemented as a pure JSON API server, based on Django and Django REST framework (https://www.djangoproject.com). Our frontend, consuming JSON data from the backend, is based on the Angular framework (https://angular.io). The data from projects and jobs is stored in a MongoDB database (https://www.mongodb.com/). To train new models and use existing models for classification jobs, we rely on celery workers (https://github.com/celery/celery) for immediate processing and TeamCity (https://www.jetbrains.com/teamcity/) agents for scheduled tasks. New training and classification tasks are sent to the RabbitMQ (https://www.rabbitmq.com/) message broker. The tasks are then processed by celery workers running on two virtual machines (VMs) with four worker instances per VM, allowing up to eight jobs to process simultaneously. This architecture allows LitSuggest to be easily scaled to support additional workload: new VMs with additional celery workers can be added easily as needed.
Figure 2.
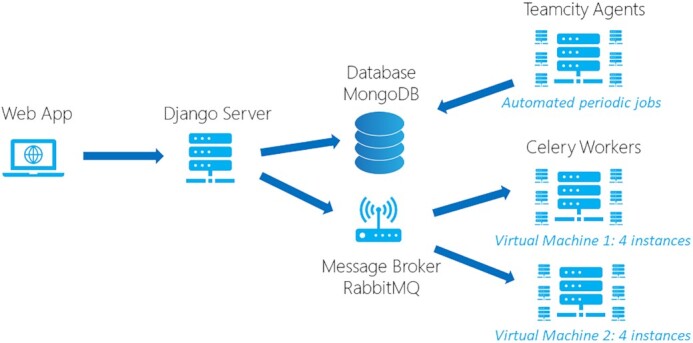
LitSuggest architecture.
Each celery worker uses the ‘stacking ensemble learning method’ to provide the best document classification results, suitable for the variable number of publications and variable nature of training datasets submitted by our users, which might not be suitable for one specific classifier. This method has achieved the best performance in previous biomedical text mining open challenge tasks (20) and is known to show stable results in text classification methods (21,22).
The text contained in the fields Journal, Publication Type, Title, Abstract, Registry Number (identifiers and names of substances mentioned) and Other Term (user submitted keywords) of each publication is concatenated, transformed into bag-of-words representation, then fed to a diverse set of classifiers, widely used in the text-mining community and available through the scikit-learn python library (https://scikit-learn.org/) such as The Ridge classifier, elastic net classifier (23), and others. The output of these classifiers is then used to train a logistic regression classifier, which will generate the final output.
LitSuggest takes several steps to ensure reproducibility of training and classification results. First, all random functions in the classifier code use the same seed. This ensures that even the randomly generated negative PMIDs will always be the same. Second, we add the content of all publications to a cache in our database, to ensure that classification jobs with the same PMIDs and same model will yield the same results even if these publications’ data has been updated in PubMed.
RESULTS
Evaluation
LitSuggest has been evaluated on two different corpora (Table 1). The first corpus is derived from LitCovid (see Use Case 1), and contains publications related to the new coronavirus pandemic, excluding publications mentioning previous coronaviruses, annotated as relevant or irrelevant. The second corpus (see Use Case 2) is centered around liver cancer, epidemiology, and health disparities. The collection of articles was selected from PubMed using ‘liver cancer’ as the query then manually annotated as relevant (containing the intersection of liver cancer, epidemiology and health disparities) or irrelevant.
Table 1.
Evaluation of LitSuggest classifier on two corpora
| Corpus | Training size (positives + negatives) | Test size (positives + negatives) | Precision | Recall | F-1 score | Area under curve |
|---|---|---|---|---|---|---|
| LitCovid | 31 998 + 32 000 | 8000 + 8000 | 0.995 | 0.993 | 0.994 | 0.994 |
| hcc_r11 | 573 + 1099 | 149 + 270 | 0.831 | 0.758 | 0.793 | 0.837 |
The evaluation on the LitCovid dataset is summarized in-depth in (8). This evaluation shows that the model achieved high performance in classifying COVID-19 related articles because many COVID-19 related articles have explicit terms to describe COVID-19 and SARS-COV-2, and the dataset is large enough for machine learning models. Most erroneous cases are articles which do not have abstracts available in PubMed (i.e. only a title is available as the input of models).
For the second corpus (hcc_r11), according to the error analysis of NCI (National Cancer Institute) team, most false positive articles do not discuss health disparities, and false negatives describe health disparities but are misclassified as negatives. The performance is lower than the LitCovid corpus because the size of the hcc corpus is small, and developing a high-performance machine learning algorithm to detect health disparities is a challenge with limited training data, because the health disparity studies encompass many subjects (including gender, age, race, ethnicity, education, income, etc.).
Comparison with other services
In addition to generating high-quality classification results, LitSuggest also offers more useful features when compared to similar tools such as MedlineRanker (17) and BioReader (18), as shown in Table 2. The major difference between LitSuggest and the other tools is that LitSuggest allows to store and reuse both training and classification results, offers powerful curation and collaboration features as well as an automated digest.
Table 2.
Functionality comparison of machine learning-based literature ranking tools
| MedlineRanker (17) | BioReader (18) | LitSuggest | |
|---|---|---|---|
| Support for PubMed query to add publications | Yes | No | Yes |
| Negative dataset | No | Mandatory | Optional |
| Filtering by bio-entities | No | No | Yes (gene, variant, chemical) |
| Automated weekly digest of new literature | No | No | Yes |
| Reusable models | No | No | Yes |
| Possibility to manage multiple models and classifications at the same time | No | No | Yes |
| Stored models and classification results | No | No | Yes |
| Sharable results through URL | No | Partially through Email notification | Yes |
| Histogram of distribution of scores | No | No | Yes |
| Method of showing discriminative words | List | Word cloud | Word cloud |
| Availability of publication abstract and keywords on result page | No | No | Yes |
| Curation Interface | No | No | Yes |
| Multi-user curation | No | No | Yes |
| Downloadable results with curation tags | No | No | Yes |
Compared to BioReader in terms of classification performance (Table 3), LitSuggest shows superior results on the hcc_r11 corpus. 15 PMIDs could not be classified by BioReader. BioReader was not able to classify the LitCovid dataset because of its size. MedlineRanker does not support a negative training set, and so was not evaluated.
Table 3.
Comparison of LitSuggest and BioReader on the hcc_r11 dataset
| Tool | Precision | Recall | F-1 score | AUC |
|---|---|---|---|---|
| LitSuggest | 0.831 | 0.758 | 0.793 | 0.837 |
| BioReader | 0.824 | 0.614 | 0.704 | 0.770 |
USE CASES
LitSuggest classification performance and rich curation features have been used under several real-world circumstances, as shown with two use cases below.
Use case 1: COVID-19 literature triage in LitCovid
The number of COVID-19 related papers has been growing at ∼10,000 articles per month, which accounts for over 7% of articles in PubMed since May 2020 (arXiv:2010.16413). LitCovid (8,9), a literature database keeping track of COVID-19 related papers in PubMed, has been developed for scientists, healthcare professionals, and the general public to stay up to date with the latest COVID-19 literature. It has been publicly available for more than a year, with millions of accesses by users worldwide each month.
LitSuggest has been used as a document triage module of the curation pipeline in LitCovid. Every day, it scans new articles in PubMed, identifies COVID-19 related articles, and adds them to the curation pipeline for further annotations, such as annotating topics and extracting locations mentioned in titles and abstracts. The performance of LitSuggest models on identifying COVID-19 related papers (See evaluation section) was evaluated in detail, and the models achieved an F1-score of 0.99 (the training and testing datasets are also publicly available) (8). We also compared the database coverage using document triage provided by LitSuggest and traditional keyword searches. We found that LitSuggest identified about 30% more COVID-19 related articles than keyword searches, suggesting that machine learning models handle the high variability and ambiguity of terms describing COVID-19 and SARS-COV-2 in the literature (8) more effectively.
In addition, the LitSuggest interface has been used for manual review and curation in LitCovid. The predictions of LitSuggest models on COVID-19 related articles are manually reviewed; the relevant articles are further annotated in the LitSuggest interface. LitSuggest streamlines LitCovid curation and updates it daily.
Use case 2: triage literature at the intersection of liver cancer, epidemiology and health disparities
Liver cancer is one of the most diagnosed cancers and the leading cause of cancer death worldwide, causing about 0.91 million new cases and 0.83 million deaths in 2020 (24). Epidemiologic data show that significant disparities exist by sex, socioeconomic status, and racial and ethnic minorities. There is a large body of epidemiologic literature on liver cancer that describes disparities. This wealth of information can be used to devise strategies to reduce liver cancer disparities. LitSuggest has been used as a literature curation tool in cooperation with the NCI team. This project aims to understand the breadth of literature at the intersection of liver cancer, epidemiology, and health disparities to identify areas of future research inquiry. In the project, the initial collection of articles was selected from PubMed using targeted search strategies and manually annotated as relevant or irrelevant according to established criteria to train an initial machine learning classifier. Then an iterative article classification process was conducted using LitSuggest. For each round, we manually examined the classification results of LitSuggest for the new article collection and added results to retrain the classifier for the next round. To date, we have curated 3132 articles. LitSuggest provides a rich environment to classify relevant articles and review annotations, which speeds up the literature curation process.
CONCLUSION
In summary, LitSuggest combines advanced machine learning techniques to provide a powerful tool for suggesting relevant PubMed articles with a high accuracy. It allows users to curate, organize, download, and share classification results, enabling collaborative curation of scientific literature.
The current version of LitSuggest has some known limitations. First, LitSuggest can currently only process publications available in PubMed. This means that non peer-reviewed publications from bioRxiv or custom PDF files cannot be used for training or classification. Second, LitSuggest currently only operates on abstracts and does not yet support full texts, which might impact the recall if pertinent information is only present in the full text. Classification accuracy is also often limited by the fact that for some publications the abstract is not available, and the classification must be based only on the title and related bibliographic fields. Finally, while recent studies including our own (13,25) have shown promise in improving classification performance with deep learning, such algorithms are not implemented due to the bottleneck of model efficiency in practical use.
In the future, we plan to enhance LitSuggest in several ways. First, we would like to add support for publications from pre-print servers such as bioRxiv. Second, we would like to improve the multi-user experience of LitSuggest by allowing different users to distribute annotation tasks more easily (currently users must agree on a page range that each will review and annotate), create groups of users, etc.
DATA AVAILABILITY
LitSuggest is publicly available at https://www.ncbi.nlm.nih.gov/research/litsuggest/
ACKNOWLEDGEMENTS
The authors would like to thank Alan James Bridge and his team for useful feedback and discussions on LitSuggest. We would also like to thank Travis Hyams and Daniela Seminara at National Cancer Center (NCI) for validating the use-cases of liver cancer health disparities. We would like to thank Robert Leaman for proofreading our manuscript.
Contributor Information
Alexis Allot, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kyubum Lee, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), 8600 Rockville Pike, Bethesda, MD 20894, USA; Department of Biostatistics and Bioinformatics, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Qingyu Chen, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), 8600 Rockville Pike, Bethesda, MD 20894, USA.
Ling Luo, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), 8600 Rockville Pike, Bethesda, MD 20894, USA.
Zhiyong Lu, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), 8600 Rockville Pike, Bethesda, MD 20894, USA.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Khare R., Leaman R., Lu Z.. Accessing biomedical literature in the current information landscape. Methods Mol. Biol. 2014; 1159:11–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fiorini N., Leaman R., Lipman D.J., Lu Z.. How user intelligence is improving PubMed. Nat. Biotechnol. 2018; 36:937–945. [DOI] [PubMed] [Google Scholar]
- 3. Fiorini N., Lipman D.J., Lu Z.. Towards PubMed 2.0. Elife. 2017; 6:e28801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Europe, P.M.C.C. Europe PMC: a full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015; 43:D1042–D1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jensen L.J., Saric J., Bork P.. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006; 7:119–129. [DOI] [PubMed] [Google Scholar]
- 6. UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E.et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Q., Allot A., Lu Z.. LitCovid: an open database of COVID-19 literature. Nucleic Acids Res. 2020; 49:D1534–D1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Q., Allot A., Lu Z.. Keep up with the latest coronavirus research. Nature. 2020; 579:193. [DOI] [PubMed] [Google Scholar]
- 10. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W.et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biocuration I.S.f. Biocuration: distilling data into knowledge. PLoS Biol. 2018; 16:e2002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poux S., Arighi C.N., Magrane M., Bateman A., Wei C.-H., Lu Z., Boutet E., Bye-A-Jee H., Famiglietti M.L., Roechert B.et al.. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics. 2017; 33:3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee K., Famiglietti M.L., McMahon A., Wei C.-H., MacArthur J.A.L., Poux S., Breuza L., Bridge A., Cunningham F., Xenarios I.et al.. Scaling up data curation using deep learning: An application to literature triage in genomic variation resources. PLoS Comput. Biol. 2018; 14:e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu Y.-Y., Clyne M., Wei C.-H., Khoury M.J., Lu Z.. Using deep learning to identify translational research in genomic medicine beyond bench to bedside. Database. 2019; 2019:baz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gobeill J., Caucheteur D., Michel P.A., Mottin L., Pasche E., Ruch P.. SIB Literature Services: RESTful customizable search engines in biomedical literature, enriched with automatically mapped biomedical concepts. Nucleic Acids Res. 2020; 48:W12–W16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lever J., Barbarino J.M., Gong L., Huddart R., Sangkuhl K., Whaley R., Whirl-Carrillo M., Woon M., Klein T.E., Altman R.B.. PGxMine: text mining for curation of PharmGKB. Pac. Symp. Biocomput. 2020; 25:611–622. [PMC free article] [PubMed] [Google Scholar]
- 17. Fontaine J.-F., Barbosa-Silva A., Schaefer M., Huska M.R., Muro E.M., Andrade-Navarro M.A.. MedlineRanker: flexible ranking of biomedical literature. Nucleic Acids Res. 2009; 37:W141–W146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon C., Davidsen K., Hansen C., Seymour E., Barnkob M.B., Olsen L.R.. BioReader: a text mining tool for performing classification of biomedical literature. BMC Bioinformatics. 2019; 19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei C.-H., Allot A., Leaman R., Lu Z.. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 2019; 47:W587–W593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Q., Du J., Kim S., Wilbur W.J., Lu Z.. Deep learning with sentence embeddings pre-trained on biomedical corpora improves the performance of finding similar sentences in electronic medical records. BMC Med. Inform. Decis. Mak. 2020; 20:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia R., Zong C., Li S.. Ensemble of feature sets and classification algorithms for sentiment classification. Inform. Sci. 2011; 181:1138–1152. [Google Scholar]
- 22. Fung G.P.C., Yu J.X., Wang H., Cheung D.W., Liu H.. Sixth International Conference on Data Mining (ICDM'06). 2006; 869–873. [Google Scholar]
- 23. Friedman J., Hastie T., Tibshirani R.. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 24. Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021; 68:394–424. [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y., Chen Q., Yang Z., Lin H., Lu Z.. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci Data. 2019; 6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
LitSuggest is publicly available at https://www.ncbi.nlm.nih.gov/research/litsuggest/