Abstract
Which genes, gene sets or pathways are regulated by certain miRNAs? Which miRNAs regulate a particular target gene or target pathway in a certain physiological context? Answering such common research questions can be time consuming and labor intensive. Especially for researchers without computational experience, the integration of different data sources, selection of the right parameters and concise visualization can be demanding. A comprehensive analysis should be central to present adequate answers to complex biological questions. With miRTargetLink 2.0, we develop an all-in-one solution for human, mouse and rat miRNA networks. Users input in the unidirectional search mode either a single gene, gene set or gene pathway, alternatively a single miRNA, a set of miRNAs or an miRNA pathway. Moreover, genes and miRNAs can jointly be provided to the tool in the bidirectional search mode. For the selected entities, interaction graphs are generated from different data sources and dynamically presented. Connected application programming interfaces (APIs) to the tailored enrichment tools miEAA and GeneTrail facilitate downstream analysis of pathways and context-annotated categories of network nodes. MiRTargetLink 2.0 is freely accessible at https://www.ccb.uni-saarland.de/mirtargetlink2.
Graphical Abstract
Graphical abstract.
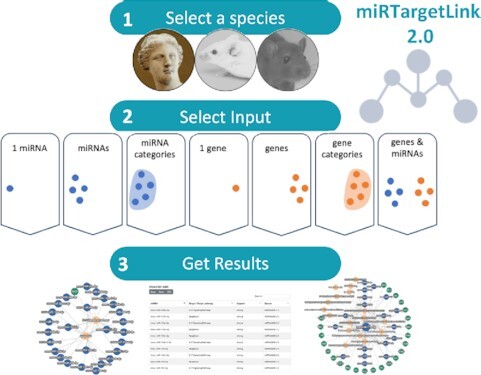
MiRTargetLink 2.0 offers interactive, web-based functionality to dissect networks of miRNAs and their target genes and pathways in three commonly investigated species.
INTRODUCTION
A central question in biomedical and life science research studies is how the expression of genes is modulated in physiological and pathophysiological processes (1). In this context, miRNAs play a central role in orchestrating gene expression. A frequent mechanism is that miRNAs bind to a specific sequence at the 3′ untranslated region (UTR) of a target mRNA to induce translational repression (2). In addition to this common mode of action, miRNA-binding sites in other mRNA regions such as the 5′ untranslated region, coding sequence or promoter regions exist (3). Moreover, RNA-binding proteins have an important role in the regulation of miRNA activity (4). An overview on canonical and noncanonical miRNA targeting (including aspects upstream such as miRNA biogenesis) has been provided by O’Brien et al. (5) and Erkeland (6). Moreover, Bartel provided a comprehensive overview on the current understanding of the defining features for miRNA biogenesis and related genomics (7).
It has become evident that miRNAs target genes in a systematic manner and exhibit a targetome specificity up to the pathway level (8). On the molecular level this effect manifests in different aspects. For example, the 3′ UTR of mRNAs often harbors multiple binding sites of the same miRNA, and the binding to these target sites has a cooperative effect. Likewise, the binding of different miRNAs to the same UTR can have cooperative effects. To facilitate the systematic analysis of miRNAs in the context of target genes or vice versa, i.e., in one-to-many or many-to-many relationships, we implemented miRTargetLink (9). The analysis of putative target genes and target pathways in a systems biology context is an essential step in noncoding RNA studies, including contrasts of experimental groups in disease research. Thus, several research tools with varying scope and functionality, as both stand-alone and web-based tools, have been proposed. Tools4mirs, a popular meta-repository for miRNA analysis methods (10), currently lists 60 target prediction tools and 26 toolboxes for the functional analysis of targets. In the following, we introduce the tools with an application scope closest to miRTargetLink 2 (11,12). For instance, miRTarVis is a tool specifically developed to display co-expression networks of paired miRNA and mRNA data (13). Second, MIENTURNET generates interaction networks by estimating the statistical significance of paired lists of miRNA and mRNA identifiers provided (14). An advantage of this approach is to reduce potentially large input lists to likely core interactions of the putative biological network, while a disadvantage is that weak informative edges might be removed due to statistical instability, potentially introducing a bias to the network. The tool miRViz allows to quickly visualize precomputed networks for multiple species, based on preselected features such as shared seed region identities between related miRNAs from the same or similar families (15). Furthermore, miRNet is a web-based tool supporting statistical analysis and functional interpretation miRNA studies (16,17). Also, it facilitates exploring the results in miRNA–target interaction networks. To analyse miRNA function in a more tissue-specific manner, miTALOS has been developed (18,19). Furthermore, tools that analyse miRNA and gene expression data in an integrated manner are available. One example of such is MMIA (miRNA and mRNA integrated analysis) that processes miRNA and gene expression experiments (20). With a similar application focus, TaLasso (21) and miRTrail were developed (22). Other more specialised web servers such as FFLtool are designed for transcription factor and miRNA feed-forward loop analysis (23).
In general, many methods are based on gene and miRNA expression data to find putative new regulatory edges or integrate known edges from miRNA target gene association databases. To test the significance of putative interactions in a statistical framework, several tools perform miRNA and gene set enrichment analyses to annotate biological function. One research goal in our studies was to provide evidence that miRNAs target genes in a systematic manner. To this end, we released miRPathDB 2.0, indexing thousands of enriched pathways for known miRNAs and miRNA candidates using validated and predicted target genes from the literature (24). Following up on our observations, we published the first comprehensive experimental validation of miRNA target pathway regulation (8).
In 2016, we presented miRTargetLink Human (9), a tool that hierarchically builds miRNA regulatory networks, containing validated and predicted target genes. Here, we present miRTargetLink 2.0, a novel version of our interactive tool for systems biology applications in miRNA research by a dynamic presentation of miRNA target gene and pathway networks. We provide a large set of miRNA gene associations from published repositories [miRTarBase (25), mirDIP (26), miRDB (27) and miRATBase (8)] and extend it by the pathway data from the recent release of miRPathDB 2.0 (24). Besides new analysis-centric features that are described in this manuscript, we want to highlight the new multi-species support, as Mus musculus and Rattus norvegicus are now available for analysis.
MATERIALS AND METHODS
Data selection and processing
The new version of miRTargetLink supports miRNAs and targets for Homo sapiens, M. musculus and R. norvegicus. MiRNA identifiers and annotation records were obtained from the latest release of miRBase (v.22.1) (28), and validated targets were downloaded from miRTarBase (v.8) (25) and miRATBase (8).
As for predicted targets, top 5% predictions (high and very high confidence) from mirDIP (v.4.1) (27) were used for human whereas we used miRDB (v.6) (27) for mouse and rat. miRTargetLink also supports target pathways from miRPathDB 2.0 (24).
Annotations for functional or categorical miRNA sets were obtained from miEAA 2.0 (29) and from GeneTrail 3 (30) for gene sets. MiEAA sets can be used for all three species, but target pathways are only available for human and mouse. Mygene python package (31) was used to translate RefSeq names to gene symbols where required. An overview on the different tools used throughout this work is given in Table 1 along with the type, the task we used the resources for and the respective version.
Table 1.
Overview of in-house and third-party resources included in miRTargetLink 2.0
| Database | Task | Own/third-party | Type | Version |
|---|---|---|---|---|
| miRBase | miRNA annotation | Third-party | Database | 22.1 |
| miRTarBase | miRNA target database | Third-party | Database | 8.0 (2020) |
| mirDIP | miRNA target database | Third-party | Database | 4.1 |
| miRDB | miRNA target database | Third-party | Database | 6.0 |
| miRATBase | miRNA target database | Own | Database | 1.0 |
| miRPathDB | miRNA target pathway database | Own | Database | 2.0 |
| miEAA | miRNA set enrichment analysis | Own | Tool | 2.0 |
| GeneTrail | Gene set enrichment analysis | Own | Tool | 3.0 |
Web server implementation
The web server was implemented using Django v2.2 in a docker environment with a PostgreSQL database, celery for job scheduling and execution and Redis as message broker backend. The frontend was built using common HTML, CSS and Javascript libraries, including the Bootstrap framework (v.4) for the styling, dataTables for the network node and interaction tables, and jQuery and Cytoscape.js (32) to create the interactive network visualizations. Typing-ahead suggestions are generated using the autoComplete JS library.
RESULTS
Data input
We implemented a convenient workflow for miRTargetLink 2 (Figure 1). We observed that most research questions related to miRNAs and genes, respectively, and their interactions can be addressed from a simple yet powerful selection of input types. In the unidirectional mode, the user can decide from six upload options, i.e. whether to select a single miRNA, a single gene, a list of either miRNAs or genes and lastly a predefined miRNA set or a gene set, as for instance, disease-associated miRNAs. In addition, a bidirectional query (paired miRNA gene lists) can be initiated. To prevent unintended results that may occur if the organism is selected automatically (e.g. genes can share the same name in mouse and human), we also ask the user to select the organism. From the input data, a comprehensive network on miRNA targets and target pathways is generated. As background data sets, we integrate four miRNA–target databases (miRTarBase, mirDip, miRDB and miRATBase) complemented with pathway interactions from miRPathDB. Altogether, the miRTargetLink knowledge base hosts ∼553 000 entries from miRTarBase, ∼1 519 000 entries from mirDIP, ∼1 173 000 entries from miRDB, ∼300 targets from miRATBase and ∼13 000 entries from miRPathDB. The detailed distribution of data records per organism and category such as validated or predicted targets, or pathways is presented on the miRTargetLink statistics page.
Figure 1.
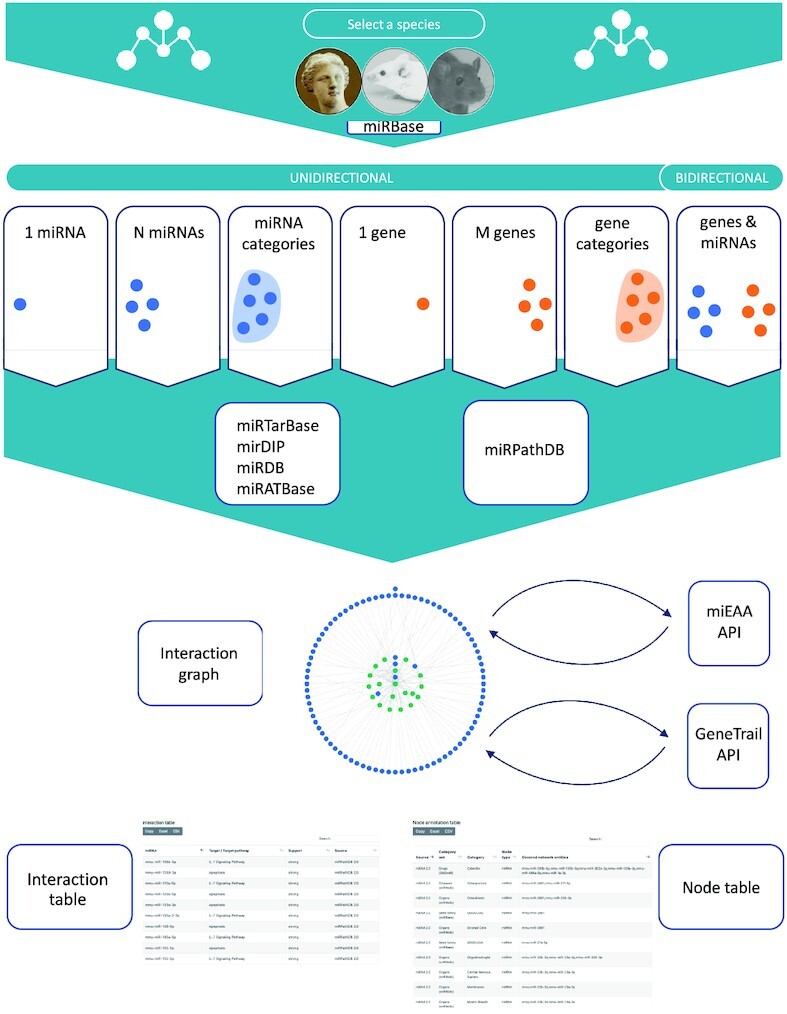
Workflow of miRTargetLink 2.0. The user selects the input species and the data input option. All information is extracted from incorporated databases automatically, and the interaction graph is immediately generated and visualized. APIs to gene and miRNA set enrichment facilitate the interpretation of more complex interaction graphs. Nodes and edges are further annotated in interactive tables.
Representation of results
Based on the input and the information in the knowledge database, the interaction graphs with all edges between miRNAs and targets, and miRNAs and target pathways are generated and visualized. Edges between genes and pathways have been omitted from the graphs, since those would introduce more complexity to the graphs without adding information content for the miRNA-centered application. The network can be shifted, zoomed and node positions and colors can be adjusted. To edit the network, the user can choose from a range of options, e.g. select whether weak/strong evidence or also predicted targets are shown or whether pathways should be added. If pathways are available, then also other miRNAs regulating these pathways can be revealed. Further, the number of shared targets for each miRNA can be increased to highlight the key regulators. Finally, six different layout options for the network are available. The network can be downloaded as JSON file or as image in jpg and png format. Below the network view, interactions are presented as the table where miRNAs, target evidence and sources are given in detail. In addition, available node annotations based on biological categories such as known tissue expression of miRNAs and genes are shown in a separate view. Both tables can be downloaded in either xls or csv format and directly copied to the clipboard.
APIs facilitate gene and miRNA set enrichment analysis
Once the miRNA and target gene network has been generated by miRTargetLink 2, the interpretation of the results becomes important. For small networks and field experts, this task can often be achieved manually. But especially for larger networks with several dozens of genes and miRNAs, it is frequently not obvious where to focus on. Here, the pathway information from miRPathDB supports interpretability, but it focuses on miRNA pathways that are annotated with experimental evidence. To guide researchers and to draw their attention to relevant hits, we integrated miRNA and gene set enrichment through miEAA 2.0 (29) and GeneTrail 3 (30) via external APIs. By selecting the relevant enrichment analysis method, these tools are automatically executed using their standard parameters and the aggregated download of result tables is initiated after completion.
Context dependency
miRNA expression is known to depend on the physiological or pathophysiological contexts. For example, the age of unaffected individuals or patients with different diseases can affect miRNA expression (33). Cholinergic-targeting noncoding RNAs, miRNAs or lncRNAs can also modulate sex-specific- and age-related acetylcholine signals (34). Especially in the context of age-related disorders such as Parkinson’s disease, miRNAs seem to be differentially expressed in specific age windows (1). Moreover, the tissue or different body fluids can confound miRNA expression (35), and regulatory events between miRNAs and target genes seem to depend on the tissue context (36). As the first step to make this information visible, miRTargetLink 2 contains and displays available context information where available. Specifically, we added metadata information from miRTarBase about the experimental setup around which the listed interactions were obtained, e.g. which type of experiment was used along with the source PubMed ID. Moreover, miRTargetLink specifically allows to search for age- and sex-related miRNAs on the unidirectional search. Similarly, users can spawn a network by searching for biological pathways or categories from GeneTrail 3. For instance, users can test systematically for a bias in a current network toward sex- or age-associated miRNAs using the connected miEAA or GeneTrail APIs, respectively. Finally, we added a one-click literature search functionality. Users can initiate a search for a selected miRNA or gene in the context of age, sex or function using PubMed.
Use case 1—mouse miRNA let-7a-5p target network
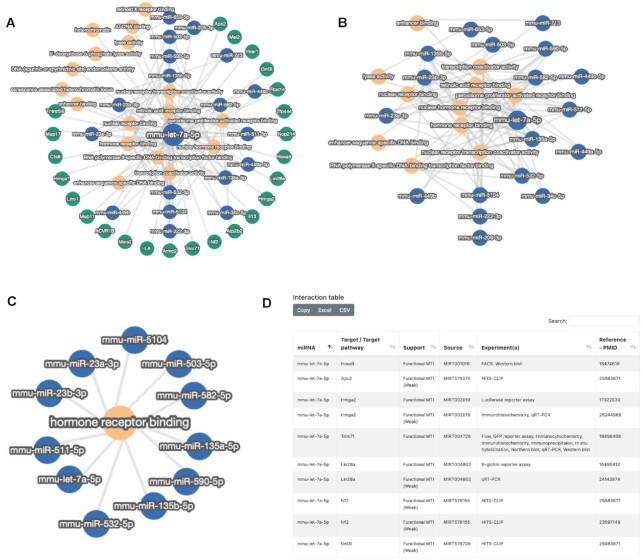
As the first use case, we studied the target gene and target pathway network of mouse miRNA let-7a-5p. This miRNA has previously been described in M. musculus with various functions (37–39). The quick-search functionality highlighted 24 target genes, including IL6 and IL13. Additionally, 17 pathways are targeted by this miRNA (Figure 2A). To examine whether other miRNAs are also targeting these genes, we increased the number of minimal targets. As a result, we got a target network that only contains miRNAs and the aforementioned pathways (Figure 2B). To display all miRNAs in mouse targeting the hormone receptor binding, we applied the third input option presented above, resulting in a condensed set of 11 miRNAs (Figure 2C). The total interaction set for mmu-let-7a-5p contains 129 entries that are displayed below the interaction graph (Figure 2D). The results presented in this use case focus on a single miRNA or a single pathway with an interaction graph of only a few dozens of nodes and edges, which can be well interpreted manually, not yet requiring gene and miRNA set enrichment analysis. To further showcase this application, we next evaluated human aniridia paired miRNA and gene expression data.
Figure 2.
Use case 1—mouse let-7a-5p targetome. (A) For the input option of a single miRNA (mmu-let-7a-5p), the interaction graph is presented. (B) After increasing the number of required target interactions per gene, only the pathways and miRNAs remain. (C) As the second input, the hormone receptor binding was selected, and miRNAs targeting this pathway are shown. (D) Interaction graph for mmu-let-7a-5p as browsable table adjacent to the network representation. The first 10 of 129 entries are displayed.
Use case 2—integrated miRNA/gene analysis in human aniridia
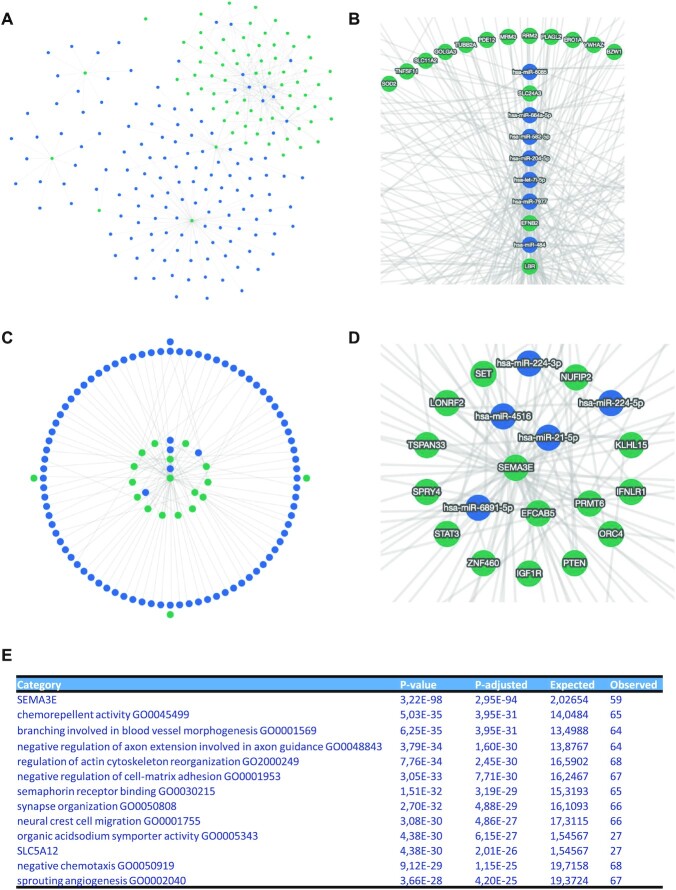
As second use case, we explored miRNA and mRNA expression patterns that were generated from the same case and control samples, facilitating paired analysis of the two RNA classes (40). From this study, we extracted both dysregulated genes and miRNAs. We then performed two analyses, where we computed the network of upregulated genes and downregulated miRNAs and, vice versa, the network of upregulated miRNAs and downregulated genes. This opposite direction of regulation was selected due to the dominant biological role of miRNAs repressing the translation of target mRNAs. In all cases, we limited the input to the top 10 genes and miRNAs. In the first scenario, the interaction graph contained 4609 edges. Even after requiring a minimum shared number of three targets, the graph contains 422 edges (Figure 3A), however, revealing a central component and the genes SLC24A3, EFNB2 and LBR with high node degrees (Figure 3B). The opposite use case still highlighted 2474 entries in the initial interaction graph. Here, 228 edges remained in the collapsed network with a minimal number of three shared targets (Figure 3C). The genes central to the network were SEMA3E, EFCAB5, PRMT6 and several others (Figure 3D). To simplify the analysis of the complex network, we performed miRNA set enrichment analysis. The 10 most significant pathways and categories are provided as results table (Figure 3E). The top hit with an adjusted P-value of 3 × 10−94 was the gene SEMA3E, exactly validating a statistically significant coverage by multiple miRNAs in the interaction graph, more than one would expect for a random enrichment. Second most significant was the Gene Ontology category chemorepellent activity with an adjusted P-value of 4 × 10−31, followed by neuronal pathways, cell adhesion and others. This use case demonstrates how the practical application of miRTargetLink 2 guides researchers to focus on potentially more relevant biological findings.
Figure 3.
Use case 2—aniridia genes and miRNAs. (A) Interactions between downregulated miRNAs (blue nodes) and upregulated genes (green nodes) with a minimum of three shared targets. (B) The enlarged core part of the interaction network shown in (A). (C) The opposite case upregulated miRNAs and downregulated genes. (D) The enlarged central part of the interaction graph shown in (C). (E) Top 10 significant miEAA pathways for the network in panel (C). Following the category name, the raw and adjusted P-value are provided, followed by the expected number of miRNAs by chance and the actual observed number.
DISCUSSION AND CONCLUSION
We present a significant update of our web server miRTargetLink 2 for the integrative analysis of miRNA, target gene and target pathway interaction networks. While the original version was focused on human data, we now offer support for other highly relevant model organisms. Altogether, the integrated knowledge base contains over 3 million entries of regulatory events between miRNAs and genes, and miRNAs and pathways across the three supported species. By adding the layer of validated pathways to the network view and providing quick access to frequently used gene / miRNA set enrichment tools, we lower the boundaries for potential users from life science to generate new insights into driving questions in fundamental biology and biomedicine. Due to the largely increased number of miRNA–target interactions and to prevent major performance issues, we were required to completely reimplement both the front- and backend of the original web server.
One current limitation of miRTargetLink is the restriction of its scope to miRNAs. Other small RNA classes are emerging and should be taken into account. Our tool miRMaster (41) already includes all previously characterized noncoding RNA classes. Among those most similar to miRNAs, tRNA fragments play a remarkable role. For example, tRNA fragments can replace miRNA regulators in diseases, as demonstrated for the cholinergic poststroke immune blockade (42). As such, tRNA fragments will add to a complete picture of how small RNAs regulate genes. Before such information is added to available integrative miRNA tools like miRTargetLink, the development of comprehensive databases containing detailed and experimentally validated regulatory events of tRNAs is mandatory.
In addition to expanding miRTargetLink to other noncoding RNA classes in the future, we will continue to add new features requested by the community. One extension could be to support uploading of expression or fold-change scores together with the identifiers such as to dynamically modify node and edge strength in the inferred network. This way, one could rank and select connected components in the graph according to their importance in a particular research context.
DATA AVAILABILITY
Not applicable.
Contributor Information
Fabian Kern, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Ernesto Aparicio-Puerta, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Yongping Li, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Tobias Fehlmann, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Tim Kehl, Center for Bioinformatics, Saarland Informatics Campus, Saarland University, 66123 Saarbrücken, Germany.
Viktoria Wagner, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Kamalika Ray, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany.
Nicole Ludwig, Center for Human and Molecular Biology, Institute of Human Genetics, Saarland University, 66421 Homburg, Germany.
Hans-Peter Lenhof, Center for Bioinformatics, Saarland Informatics Campus, Saarland University, 66123 Saarbrücken, Germany.
Eckart Meese, Center for Human and Molecular Biology, Institute of Human Genetics, Saarland University, 66421 Homburg, Germany.
Andreas Keller, Chair for Clinical Bioinformatics, Saarland University, 66123 Saarbrücken, Germany; Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford 94304, CA, USA.
FUNDING
Internal funds of Saarland University; Instituto de Salud Carlos III [IFI16/00041, MV19/00058 to E.A.]. Funding for open access charge: Internal funds of Saarland University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kern F., Fehlmann T., Violich I., Alsop E., Hutchins E., Kahraman M., Grammes N.L., Guimarães P., Backes C., Poston K.L.et al.. Deep sequencing of sncRNAs reveals hallmarks and regulatory modules of the transcriptome during Parkinson’s disease progression. Nat. Aging. 2021; 1:309–322. [Google Scholar]
- 2. Huntzinger E., Izaurralde E.. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011; 12:99–110. [DOI] [PubMed] [Google Scholar]
- 3. Xu W., San Lucas A., Wang Z., Liu Y.. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014; 15:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loffreda A., Rigamonti A., Barabino S.M., Lenzken S.C.. RNA-binding proteins in the regulation of miRNA activity: a focus on neuronal functions. Biomolecules. 2015; 5:2363–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Brien J., Hayder H., Zayed Y., Peng C.. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 2018; 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stavast C.J., Erkeland S.J.. The non-canonical aspects of microRNAs: many roads to gene regulation. Cells. 2019; 8,:1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel D.P. Metazoan microRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kern F., Krammes L., Danz K., Diener C., Kehl T., Kuchler O., Fehlmann T., Kahraman M., Rheinheimer S., Aparicio-Puerta E.et al.. Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res. 2021; 49:127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamberg M., Backes C., Fehlmann T., Hart M., Meder B., Meese E., Keller A.. miRTargetLink—miRNAs, genes and interaction networks. Int. J. Mol. Sci. 2016; 17:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lukasik A., Wojcikowski M., Zielenkiewicz P.. Tools4miRs: one place to gather all the tools for miRNA analysis. Bioinformatics. 2016; 32:2722–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G.. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013; 41:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G.. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015; 43:W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung D., Kim B., Freishtat R.J., Giri M., Hoffman E., Seo J.. miRTarVis: an interactive visual analysis tool for microRNA–mRNA expression profile data. BMC Proc. 2015; 9:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Licursi V., Conte F., Fiscon G., Paci P.. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinform. 2019; 20:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giroux P., Bhajun R., Segard S., Picquenot C., Charavay C., Desquilles L., Pinna G., Ginestier C., Denis J., Cherradi N.et al.. miRViz: a novel webserver application to visualize and interpret microRNA datasets. Nucleic Acids Res. 2020; 48:W252–W261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang L., Zhou G., Soufan O., Xia J.. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020; 48:W244–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J.. miRNet: dissecting miRNA–target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016; 44:W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowarsch A., Preusse M., Marr C., Theis F.J.. miTALOS: analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. RNA. 2011; 17:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Preusse M., Theis F.J., Mueller N.S.. miTALOS v2: analyzing tissue specific microRNA function. PLoS One. 2016; 11:e0151771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nam S., Li M., Choi K., Balch C., Kim S., Nephew K.P.. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009; 37:W356–W362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muniategui A., Nogales-Cadenas R., Vazquez M., Aranguren X.L., Agirre X., Luttun A., Prosper F., Pascual-Montano A., Rubio A.. Quantification of miRNA–mRNA interactions. PLoS One. 2012; 7:e30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laczny C., Leidinger P., Haas J., Ludwig N., Backes C., Gerasch A., Kaufmann M., Vogel B., Katus H.A., Meder B.et al.. miRTrail–a comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC Bioinform. 2012; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie G.Y., Xia M., Miao Y.R., Luo M., Zhang Q., Guo A.Y.. FFLtool: a web server for transcription factor and miRNA feed forward loop analysis in human. Bioinformatics. 2020; 36:2605–2607. [DOI] [PubMed] [Google Scholar]
- 24. Kehl T., Kern F., Backes C., Fehlmann T., Stöckel D., Meese E., Lenhof H.-P., Keller A.. miRPathDB 2.0: a novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2019; 48:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H.-Y., Lin Y.-C.-D., Li J., Huang K.-Y., Shrestha S., Hong H.-C., Tang Y., Chen Y.-G., Jin C.-N., Yu Y.et al.. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019; 48:D148–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tokar T., Pastrello C., Rossos A.E.M., Abovsky M., Hauschild A.-C., Tsay M., Lu R., Jurisica I.. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2017; 46:D360–D370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y., Wang X.. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020; 48:D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kern F., Fehlmann T., Solomon J., Schwed L., Grammes N., Backes C., Van Keuren-Jensen K., Craig D.W., Meese E., Keller A.. miEAA 2.0: integrating multi-species microRNA enrichment analysis and workflow management systems. Nucleic Acids Res. 2020; 48:W521–W528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gerstner N., Kehl T., Lenhof K., Muller A., Mayer C., Eckhart L., Grammes N.L., Diener C., Hart M., Hahn O.et al.. GeneTrail 3: advanced high-throughput enrichment analysis. Nucleic Acids Res. 2020; 48:W515–W520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xin J., Mark A., Afrasiabi C., Tsueng G., Juchler M., Gopal N., Stupp G.S., Putman T.E., Ainscough B.J., Griffith O.L.et al.. High-performance web services for querying gene and variant annotation. Genome Biol. 2016; 17:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fehlmann T., Lehallier B., Schaum N., Hahn O., Kahraman M., Li Y., Grammes N., Geffers L., Backes C., Balling R.et al.. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun. 2020; 11:5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Madrer N., Soreq H.. Cholino-ncRNAs modulate sex-specific- and age-related acetylcholine signals. FEBS Lett. 2020; 594:2185–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stahler C., Meese E.et al.. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016; 44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmieri V., Backes C., Ludwig N., Fehlmann T., Kern F., Meese E., Keller A.. IMOTA: an interactive multi-omics tissue atlas for the analysis of human miRNA-target interactions. Nucleic Acids Res. 2018; 46:D770–D775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Z., Wang H., Zhong J., Yang J., Darwazeh R., Tian X., Huang Z., Jiang L., Cheng C., Wu Y.et al.. Significant changes in circular RNA in the mouse cerebral cortex around an injury site after traumatic brain injury. Exp. Neurol. 2019; 313:37–48. [DOI] [PubMed] [Google Scholar]
- 38. Martyniuk C.J., Martinez R., Kostyniuk D.J., Mennigen J.A., Zubcevic J.. Genetic ablation of bone marrow beta-adrenergic receptors in mice modulates miRNA-transcriptome networks of neuroinflammation in the paraventricular nucleus. Physiol. Genomics. 2020; 52:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seifer B.J., Su D., Taylor H.S.. Circulating miRNAs in murine experimental endometriosis. Reprod. Sci. 2017; 24:376–381. [DOI] [PubMed] [Google Scholar]
- 40. Latta L., Ludwig N., Krammes L., Stachon T., Fries F.N., Mukwaya A., Szentmary N., Seitz B., Wowra B., Kahraman M.et al.. Abnormal neovascular and proliferative conjunctival phenotype in limbal stem cell deficiency is associated with altered microRNA and gene expression modulated by PAX6 mutational status in congenital aniridia. Ocul. Surf. 2021; 19:115–127. [DOI] [PubMed] [Google Scholar]
- 41. Fehlmann T., Backes C., Kahraman M., Haas J., Ludwig N., Posch A.E., Wurstle M.L., Hubenthal M., Franke A., Meder B.et al.. Web-based NGS data analysis using miRMaster: a large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017; 45:8731–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winek K., Lobentanzer S., Nadorp B., Dubnov S., Dames C., Jagdmann S., Moshitzky G., Hotter B., Meisel C., Greenberg D.S.et al.. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc. Natl. Acad. Sci. USA. 2020; 117:32606–32616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.