Abstract
The COVID-19 pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Similar to other coronaviruses, its particles are composed of four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. S, E, and M proteins are glycosylated, and the N protein is phosphorylated. The S protein is involved in the interaction with the host receptor human angiotensin-converting enzyme 2 (hACE2), which is also heavily glycosylated. Recent studies have revealed several other potential host receptors or factors that can increase or modulate the SARS-CoV-2 infection. Interestingly, most of these molecules bear carbohydrate residues. While glycans acquired by the viruses through the hijacking of the host machinery help the viruses in their infectivity, they also play roles in immune evasion or modulation. Glycans play complex roles in viral pathobiology, both on their own and in association with carrier biomolecules, such as proteins or glycosaminoglycans (GAGs). Understanding these roles in detail can help in developing suitable strategies for prevention and therapy of COVID-19. In this review, we sought to emphasize the interplay of SARS-CoV-2 glycosylated proteins and their host receptors in viral attachment, entry, replication, and infection. Moreover, the implications for future therapeutic interventions targeting these glycosylated biomolecules are also discussed in detail.
Graphical abstract
Keywords: SARS-CoV-2 glycosylation, Spike protein glycosylation, Envelope protein glycosylation, Membrane protein glycosylation, Glycosaminoglycan SARS-CoV-2, Human ACE2 SARS-CoV-2
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can induce fever, severe respiratory illness, and various multi-organ disease manifestations [1]. The SARS-CoV-2 virus predominantly attacks lung cells, which can lead to pneumonia and acute respiratory distress syndrome [2].
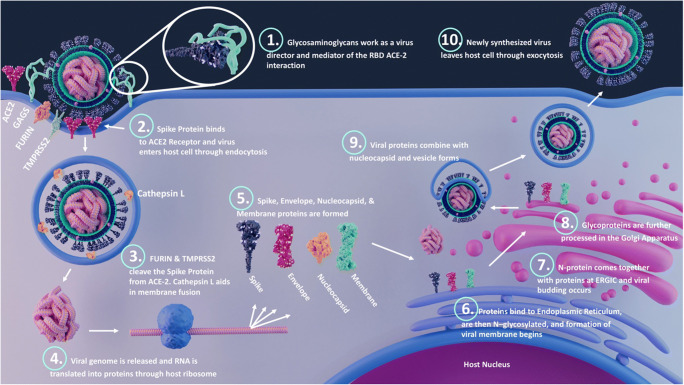
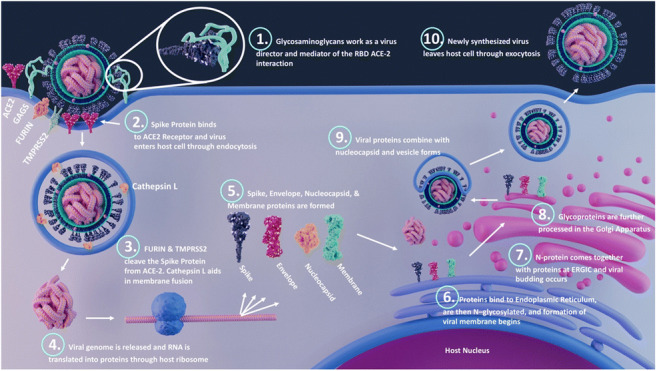
SARS-CoV-2 viral particles, which range from 60 to 140 nm size, contain single positive-stranded ribonucleic acid (RNA) with a length of 26 to 32 kb. Sequencing of the virus revealed six major open-reading frames and several accessory genes encoding spike (S) protein, 3-chymotrypsin-like cysteine protease (also termed main protease), papain-like protease, and RNA-dependent RNA polymerase [3, 4]. Structurally, SARS-CoV-2 virus contains a S protein on its surface, which, upon binding with host receptors, becomes enzymatically activated via host proteases, leading to viral fusion and endocytosis [5]. Inside the host cell, the viral, RNA-dependent RNA polymerase transcribes the viral genome, which is then translated by host ribosomes to biosynthesize viral proteins. Further, new virions are formed by budding into the lumen of the endoplasmic reticulum-Golgi intermediate compartments (ERGIC), and finally, they are primed for exocytosis [6] (Fig. 1).
Fig. 1.
Different processes during SARS-CoV-2 infection. Viral binding: the receptor-binding domain (RBD) of spike (S) protein interacts with the host cell surface receptors such as ACE2, GAGs, and other potential receptors; fusion: host proteases such as TMPRSS2, cathepsins, and furin cleave the S1 and S2 subunits, and the S2 subunit mediates the viral fusion; and entry: the virus enter the host cell by endocytosis or membrane fusion. Once inside the host cell, the RNA uses the host machinery to translate the viral proteins. The post-translational modifications happen on structural proteins by hijacking the host system and viral budding occurs at endoplasmic reticulum-Golgi intermediate compartments (ERGIC). Finally, the viral assembly occurs, and the virus is released by exocytosis
Recent structural analysis of the SARS-CoV-2 S protein by cryo-EM shows that it is extensively glycosylated, similar to SARS-CoV-1 S protein [7–9]. Moreover, the site-specific glycosylation analysis of S protein by our group and other groups through mass spectrometry revealed both N- and O-glycosylation [10–16]. Based on recent reports, each trimeric spike presents up to 66 N-linked glycosylation sites and several O-linked glycosylation sites [12, 16]. Site-specific glycosylation on virus-derived, wild-type non-stabilized and recombinant stabilized spike glycoproteins was compared in a latest study [16].
Glycans on the viral surface proteins are involved in the process of viral binding to host cells for entry, viral fusion, shielding of specific epitopes, and in the folding, stability, and protection of viral proteins [6, 17–20]. In general, the glycans in the vicinity of receptor-binding regions can negatively impact viral binding. Thus, the comparatively slim glycan shield of coronaviruses, in contrast to other viruses, may be advantageous for more efficient receptor binding [17]. An elevation of glycan shield densities and oligomannose abundance was observed in certain types of viruses, such as HIV-1, which can evade the immune system response very effectively [6]. The glycosylation on the surface proteins of viruses can hinder antibodies from binding by shielding the surface antigens with glycan envelopes. Glycans can undergo large internal motions, leading to challenges in their accurate description by any single three-dimensional (3D) shape [21, 22]. Recently, MD simulations have been conducted on glycoproteins to accurately predict the 3D shapes and glycan motions, similar to characterization of oligosaccharide conformations and dynamics by NMR [22].
The structural and functional roles of glycosylation on viral pathogenesis are enumerated in several studies. In Hendra virus, a bat-borne virus causing a highly fatal infection in horses and humans, fusion (F) protein contains five N-linked glycosylation sites, and the glycans at N414 are critical for fusion protein folding and transport [19]. A study on the Nipah virus showed that the removal of N-glycans on the fusion protein resulted in significant increase in viral fusogenicity but improved sensitivity to antibody neutralization [20]. Glycans decorating the viral envelope proteins account for half of the molecular weight of these carrier glycoproteins [23]. The glycosylation on the viral surface, where host glycans decorate the viral surface proteins, facilitates immune evasion by blocking humoral and cellular innate immune systems [24, 25]. Moreover, since the virus hijacks the host cellular machinery for its replication and protein glycosylation, the viral surface glycans are composed of host glycans that are recognized as self by the immune system and thus suppress the anti-carbohydrate immune response [26]. However, the immune system responds to glycosylated pathogens in several ways, such as either increasing or decreasing the expression of certain endogenous lectins during infections and thereby fighting against the pathogens through lectin-mediated defense mechanisms [27]. Differential hemagglutinin N-glycosylation affects T cell activation and cytokine production and can thus challenge the development of vaccines [28]. These types of immune evasion and shielding of the receptor-binding site with glycans are shown by viral glycoproteins of HIV-1 Env, influenza hemagglutinin, and Lassa virus envelope glycoprotein complex (LASV GPC) [29]. All this demonstrates how glycosylation helps the virus evade host innate and adaptive immune responses.
In general, during the viral evolution, protein sequences in viruses undergo mutations (antigenic drift), which can lead to loss of species specificity in the virus [30]. This can also lead to modulation of viral infectivity and surface protein antigenicity [6, 17]. These mutations can alter the glycosylation of the protein by generation of new or removal of existing glycosylation sites as reported in the case of influenza viruses [31, 32]. These changes in glycosylation can lead to new virus strains with increased ability to evade the host immune response, and this can attenuate vaccine efficacy [28, 31]. While these observations are mostly on viruses such as influenza, mutation studies on SARS-CoV-2 S protein show that some glycosylation sites are crucial for viral infectivity [33]. The most common mutation reported on the S protein is D614G mutation, although a total of 9654 mutations have been detected on 400 distinct sites on S protein [34].
In this review, we discuss the functional and structural roles glycans play in SARS-CoV-2 pathobiology and how these understandings can lead to future therapeutic interventions to tackle COVID-19.
Roles played by the structural proteins (S, M, E, and N proteins) in viral assembly
There are four major structural proteins of SARS-CoV-2, including the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. E and M proteins regulate intracellular trafficking of the S protein, as well as its intracellular processing [35]. When inducing the retention of the S protein inside cells, the E and M proteins provide a mechanism that allows targeting close to the virion assembly site, as well as limits processing to a fusion-active conformation and cell surface expansion, preventing syncytia formation. E or M protein co-expression with S protein alters the N-glycans of S, independent of their effect on S retention (Fig. 1). The E protein induces the retention of S protein by slowing down the cell secretory pathway independently of the retrieval motif harbored by the S protein cytoplasmic tail. All four structural proteins are required for optimal production of SARS-CoV-2 virus-like particles (VLPs). S expression alone does not induce secretions of VLPs; however, when this expression was combined with any other structural protein (E, M, or N), there was an increased formation on VLPs. Co-expression of all four structural proteins in combination induces VLP secretion more powerfully than other combinations of the four proteins [35]. Combined studies on different coronavirus strains suggest that this virus family utilizes their M proteins to evade the host innate immune system [36]. The M protein is important for the coronavirus budding process. During virus particle assembly, the M protein interacts with the N protein, E protein, S protein, and itself [37]. The M protein cooperates with the S protein during cell attachment and entry in alpha-coronaviruses [38]. A study comparing SARS-CoV-2 E and M proteins in humans with that found in bats and pangolins shows that the E protein is identical in these species. Structural similarities of human SARS-CoV-2 M and E proteins to their counterparts in bat and pangolin isolates, as well as differences specific to SARS-CoV-2 proteins could explain the cross-species transmission and properties of the virus [39].
SARS-CoV-2 spike protein glycosylation
The S protein of SARS-CoV-2 is a heavily glycosylated homotrimer protein with two subunits, S1 and S2, linked through transmembrane protease, serine 2 (TMPRSS2), and furin cleavage sites [7]. Subunit S1 facilitates attachment to host cell receptors through a receptor-binding domain (RBD), and subunit S2 is involved in fusion of viral and human cellular membranes [5, 7]. The glycosylation of the S protein could mediate S protein folding and modulates conformational dynamics of S protein, priming by host proteases, and immune evasions through glycan shielding [7, 40–42]. One of the earlier studies on coronaviruses showed that inhibition of N-glycosylation by treatment with tunicamycin led to spikeless virions because of improper protein folding [7, 42]. Two mutations in the spike of bat coronavirus HKU4 and consequent introduction of a new N-glycan site mediated entry of Middle East respiratory syndrome (MERS) coronavirus into human cells by allowing it to be primed by human proteases [41]. It was demonstrated that the coronaviruses employ conformational masking and glycan shielding of S trimer to evade immune recognition [7, 43].
Site-specific N-glycosylation of S protein
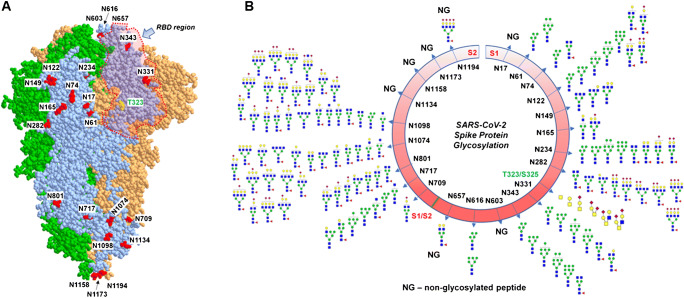
The S protein of SARS-CoV-2 expressed in human HEK-293 cells is extensively glycosylated on 22 N-glycan sites and a number of O-glycosylation sites (Figs. 2 and 3) [10, 11]. Site-specific analyses of N-linked glycosylation on S proteins expressed in the human cell line HEK-293 revealed extensive heterogeneity by showing high mannose-type glycans to highly processed complex-type glycans with sialylation and fucosylation [10, 11]. Site N234, which is adjacent to the RBD of SARS-CoV-2 S protein, displays high mannose-type glycans [6, 10]. Complex-type N-glycans with bi- and triantennary glycans and high mannose-type glycans were identified on sites N165, N331, and N343 [10, 11]. While sites N331 and N343 in the RBD region showed predominantly high mannose glycans when S protein is expressed in individual subunits S1 and S2, the same sites showed complex-type glycans (with 98% fucosylation) on trimeric form of S protein expressed on HEK293 cells [6, 10]. A recent quantitative N-glycan analysis on S protein subunit S1 isolated from SARS-CoV-2-infected Calu-3 cells via immunoaffinity purification showed high prevalence of complex-type N-glycans (79%) and 21% high mannose and/or hybrid structures [16]. The same study compared the different glycans on vaccine candidates and recombinant S protein with the wild-type virus S protein with an aim to help the vaccine design strategies and thereby enable high-quality immune response through correct immunogen presentation. The authors demonstrated that distinctive cellular secretion pathways result in variation in protein glycosylation.
Fig. 2.
The glycosylation of SARS-CoV-2 spike (S) protein. A 3D model of SARS-CoV-2 spike protein trimer showing the RBD region and glycosylation sites (only labelled on one monomer). B The site-specific N- and O-glycosylation of S protein [10, 11]
Fig. 3.
Distribution of N- (A) and O- (B) glycosylation on specific sites of SARS-CoV-2 spike (S) protein expressed in HEK293 cells [10]
An NMR study on the RBD domain of SARS-CoV-2 S protein allowed identification of the chemical nature and structural details, such as glycosidic linkages, and sulfation of the RBD glycans. They observed fucoses and GalNAc at the RBD domain and several unexpected glycan motifs such as 4-O-sulfated LacDiNAc, α2,6-sialylated LacDiNAc, LewisX (LeX), and fucosylated terminal GalNAcβ1-4GlcNAc (LacdiNAcFuc) along with terminal LacNAc, LacDiNAc, α2,3-linked sialyl (3′SLacNAc), and α2,6-linked sialyl (6′SLacNAc) fragments [44]. A recent report on the characterization of S protein glycosylation by a newly developed liquid chromatography-mass spectrometry methodology showed the presence of LacdiNAc structural motifs on all occupied N-glycopeptides and polyLacNAc structures on six glycopeptides [12].
Roles of S protein glycosylation in viral binding, fusion, entry, and immunogenicity
Specific types of glycans were observed at each site of the S protein, and the type of glycosylation in the RBD region, such as sites N331 and N343, are critical for viral infectivity [33]. By blocking N-glycan biosynthesis at the high mannose stage through both genetic manipulation and use of the small-molecule kifunensine, only minor changes were noted in spike-ACE2 binding. However, S protein N-glycosylation is important for viral entry to human cell models as the viruses lacking N-glycans enter the host cell less efficiently [45]. The glycans at sites N165 and N234 have roles on RBD conformational plasticity, as their presence stabilizes the RBD “up” conformation, permitting efficient binding to human angiotensin-converting enzyme 2 (hACE2) receptor. Deletion of these glycan residues through N165A and N234A mutations significantly reduced binding of S protein to ACE2 as a result of a conformational shift of the RBD toward the “down” state, hampering accessibility to ACE2 [40]. Coronaviruses have less high mannose-type N-glycans on their surface proteins than other viruses such as HIV-1 [6]. The structural mapping of glycans of MERS-CoV S proteins revealed that glycans contribute to the formation of a cluster of high-density oligomannose-type glycans at specific regions of the S protein [9]. N-Glycosylation sites N331 and N343, which are in the RBD region, and N165 and N234, which are adjacent to it, are suggested to be critical for immune recognition [29]. In addition, the N234Q mutant was significantly resistant to neutralizing antibodies and the N165Q mutant became more sensitive to antibody neutralization [46].
O-linked glycosylation on S protein
The presence of O-glycosylation at T323 and the plausible glycosylation at S325 have previously been reported by our group, and this was confirmed by several later studies (Fig. 3) [10, 14, 47]. More recently, O-glycosylation has been detected at residues T678, S686, and T1160 of SARS-CoV-2 S glycoprotein [47]. O-linked glycans such as Tn, core 1, mono and di-sialyl core 1, and sialylated core are reported on the S protein [10, 14]. The O-glycans located in the hinge region of RBD (T323 and S325) and those adjacent to the furin cleavage site (S686) have been suggested to play critical roles in viral binding and the membrane fusion, respectively [10, 47].
Interaction of SARS-CoV-2 with the host receptors
Recent extensive research on SARS-CoV-2 revealed multiple sources of viral entry and enhanced our understanding how the host system helps the viruses throughout the process of infection (Table 1). Walls et al. demonstrated that ACE2 is acting as the functional receptor for SARS-CoV-2 S-mediated entry into cells [7]. They showed that both SARS-CoV-1 and SARS-CoV-2 bind to hACE2 with comparable affinity. Both of these viruses depend on the hACE2 receptor for binding and the host membrane serine protease TMPRSS2 and cathepsin for S protein cleavage for subsequent activation [5, 29]. Interestingly, the presence of a furin cleavage site at the S1/S2 boundary of SARS-CoV-2, in contrast to SARS-CoV-1, which does not have such a cleavage site, is implicated as cause of increased infectivity of SARS-CoV-2. Abrogation of the furin cleavage motif reduced S-mediated SARS-CoV-2 entry into VeroE6 or BHK cells [7].
Table 1.
Potential cell surface human receptors or factors for SARS-CoV-2 infection
| Receptor/factor | Interaction | Effect | Publications |
|---|---|---|---|
| hACE2 | Cell surface receptor of SARS-CoV-2 | Viral binding | Hoffmann et al. [5] |
| Tyrosine-protein kinase receptor UFO (AXL) | Cell surface receptor of SARS-CoV-2 | Viral binding | Wang et al. [48] |
| Neuropilin-1 (NRP1) | Binding with the furin-cleaved S1 fragment of the S protein | Promoting viral fusion | Daly et al. [49] |
| TMPRSS2 | Cleavage of S1 and S2 subunit of S protein | Viral fusion | Hoffmann et al. [5] |
| C-type lectins—DC-SIGN, L-SIGN, MR, and MGL | Cell surface receptor of SARS-CoV-2 | Viral binding | Gao et al. [47] |
| GAGs | Cell surface receptor of SARS-CoV-2 | Viral binding | Kim et al. [50] |
Glycosylation of SARS-CoV-2 host receptor human angiotensin-converting enzyme 2
hACE2, which is a type I transmembrane protein, comprises an extracellular, a transmembrane, and a cytosolic domain with a total of 805 amino acids [51]. The transmembrane region of hACE2 can be cleaved into a soluble form of hACE2 (sACE2)—lacking the transmembrane and cytosolic domains—that is enzymatically active having a catalytic site and a zinc-binding motif [52]. This secreted hACE2 is involved in the renin-angiotensin system (RAS) [53].
hACE2 acts as a receptor for human coronaviruses SARS-CoV-1 and SARS-CoV-2, as well as human coronavirus NL63/HCoV-NL63 [5, 54, 55] (Figs. 1 and 4). Multiple studies have shown efficient infection of SARS-CoV-2 on cell lines and mouse models expressing hACE2 [7, 56].
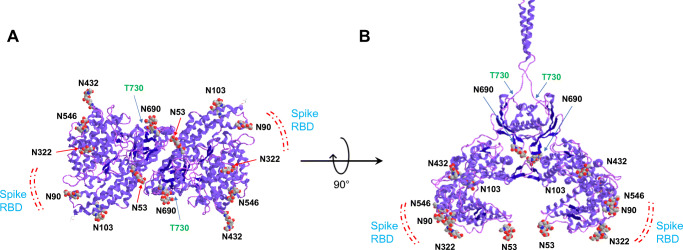
Fig. 4.
3D model of human ACE2 showing the S protein binding region and the distribution of N- and O-glycosylation sites (A) and (B) are showing different orientations
The N-glycosylation site at N90 of hACE2 plays an important part in the interaction of the coronavirus with the ACE2 receptor (Fig. 4). An in silico study predicted the role N90 in the viral interaction with ACE2, and removal of the N-glycosylation motif by in silico mutation of N90 and T92 was associated with stronger interaction with SARS-CoV-2 virus, suggesting that glycosylation at N90 may impose steric hindrance for the RBD binding [57]. Another study also highlighted that all substitutions of N90 and T92 other than S92 (which retains N-glycosylation motif) enhances the RBD binding, and this enhancement may depend on the type of glycans on ACE2, which changes with expression cell types [58]. Devaux et al. showed in silico that species with ACE2 sequence containing K31, Y41, N90, and K353 are likely to be susceptible to SARS-CoV-2 infection [59]. The importance of several hinge regions and N-glycosylations including N90 of ACE2 is suggested based on the crystal structure analysis of ACE2 [59]. This study by Devaux et al. contradicts other studies on N90-mediated ACE2 binding mentioned above and thus demands more detailed experimental evidence on the roles of ACE2 glycosylation in viral binding. The glycan at N322 enabled tight interaction of ACE2 with the RBD, an opposite effect to that of N90 [29].
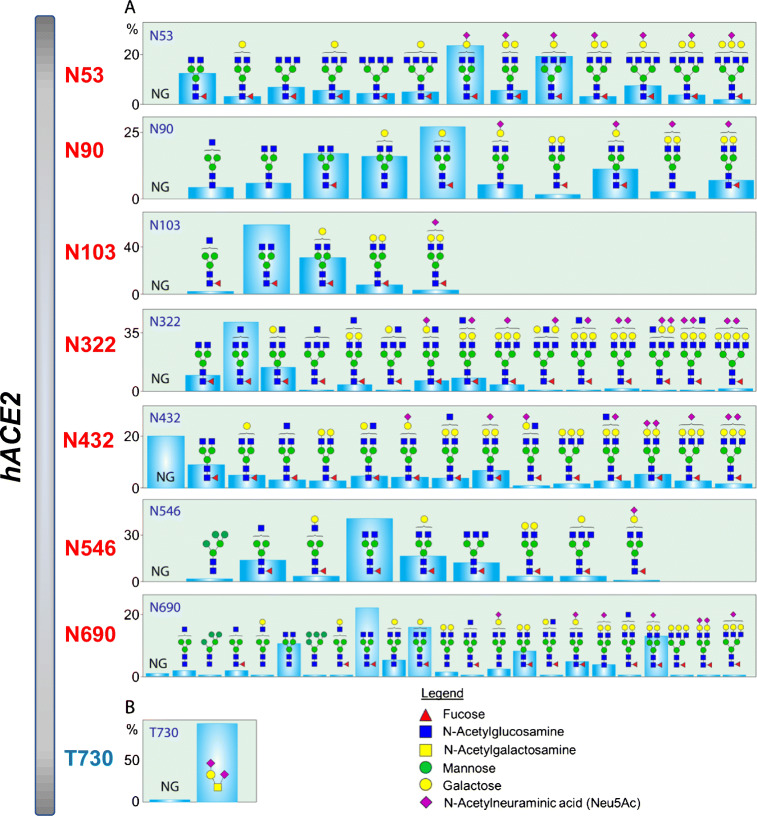
Quantitative site-specific N-linked and O-linked glycosylation of hACE2 has recently been reported by our group and others through glycomic and glycoproteomic approaches [60, 61]. Glycosylation at all seven potential N-glycosylation sites on hACE2 and one O-glycosylation site was described in detail (Figs. 4 and 5). Moreover, evidences for the presence of both core and antennal fucosylation, bisecting GlcNAc, and prevalence of 2,3-linked sialic acid were demonstrated in our study [60]. However, a recent study found that the effect of hACE2 sialic acids on the viral interaction is smaller than anticipated based on previous crystal structure and molecular modeling studies [45].
Fig. 5.
A, B Distribution of N- and O-glycosylation on specific sites of human ACE2 expressed in HEK293 cells [60]
SARS-CoV-2 and glycosaminoglycan interactions
Glycosaminoglycans (GAGs) are linear polysaccharides involved in a variety of biological processes, including wound healing, anticoagulation, cell signaling, and pathogenesis [62–64]. GAGs are covalently bound to a core protein, making up proteoglycans (PGs). PGs are found inside cells, on the surface of cells, and in the extracellular matrix [50]. The four main groups of GAGs are heparin/heparan sulfate (Hp/HS), chondroitin sulfate/dermatan sulfate (CS/DS), keratin sulfate (KS), and hyaluronic acid (HA) [65]. GAG chains can be modified with acetylation and sulfation. The uronic acid residues combined with sulfation modifications result in a net negative charge [65]. Hp is an FDA-approved anticoagulant, which exerts its effect by binding to antithrombin III [66, 67]. The WHO recommends the use of Hp in COVID-19 patients to reduce incidence of venous thromboembolism [50]. GAG-binding proteins contain amino acid sequences, Cardin-Weintraub motifs, which correspond to ‘XBBXBX’ and ‘XBBBXXBX’ where X is a hydropathic residue and B is a basic residue [50]. The basic residue is responsible for interacting with the sulfate groups present on the GAG chain.
SARS-CoV-1 and numerous other pathogens are known to utilize host cell surface GAGs during host cell entry. Kim et al. reported GAG-binding and GAG-binding-like motifs at sites 1–3 (453–459 (YRLFRKS), 681–686 (PRRARS), and 810–816 (SKPSKRS), respectively) of the SARS-CoV-2 S glycoprotein [50]. Human lung cells predominantly contain HS and CS GAGs, and the mast cells are rich in Hp. In a surface plasmon resonance (SPR) direct binding assay, both monomeric and trimeric SARS-CoV-2 S proteins (KD = 40 pM and 73 pM, respectively) bind more tightly to immobilized Hp than SARS-CoV-1 and MERS-CoV S protein (500 nM and 1 nM, respectively) [50]. Specific degree and position of sulfation are imperative for binding, with N-, 2-O, and 6-O sulfation required for binding to SARS-CoV-2. Hp, which has 6-O, 2-O, 3-O, and N-sulfation, and trisulfated (TriS) HS, which has 6-O, 2-O, and N-sulfation, both have therapeutic potential as competitive inhibitors against SARS-CoV-2 infection. When the receptor-binding domain is in open conformation, HS interacts with the GAG-binding motif at the S1/S2 site 2 (681–686 (PRRARS)) and at site 1 (453–459 (YRLFRKS)) [50]. A model displaying how GAGs influence SARS-CoV-2 host cell entry is shown in Fig. 6. A follow-up study to these findings investigated if the high binding affinity of Hp to the S protein of SARS-CoV-2 translates to a potent antiviral activity [68]. In vitro antiviral properties of Hp, HS, and CS were tested, as well as fucoidan, a sulfated polysaccharide composed of monomers. When testing the binding affinity of these polysaccharides to the SARS-CoV-2 S protein, two varieties of fucoidans, trisulfated (TriS) Hp and unfractionated USP-Hp, were able to compete with cell surface Hp for S protein binding. Alternatively, other GAGs, such as HS, CS, and KS, showed no competitive binding. When testing efficacy, a fucoidan-like, branched polysaccharide was substantially more potent than remdesivir (EC50 = 83 nM and 770 nm, respectively), which is an approved therapeutic for severe COVID-19 infections. Hp and TriS Hp, which differ only in one sulfation (Hp has 3-O-sulfation whereas TriS Hp does not), had lower activity than fucoidan-like compounds (2.1 μM and 5 μM, respectively). The higher activity of the fucoidan-like samples could be due to multivalent interactions between these polysaccharides and the virus. These results suggest that certain polysaccharide structures can be used as decoys to prevent SARS-CoV-2 S protein binding to the HS co-receptor in host tissues [68].
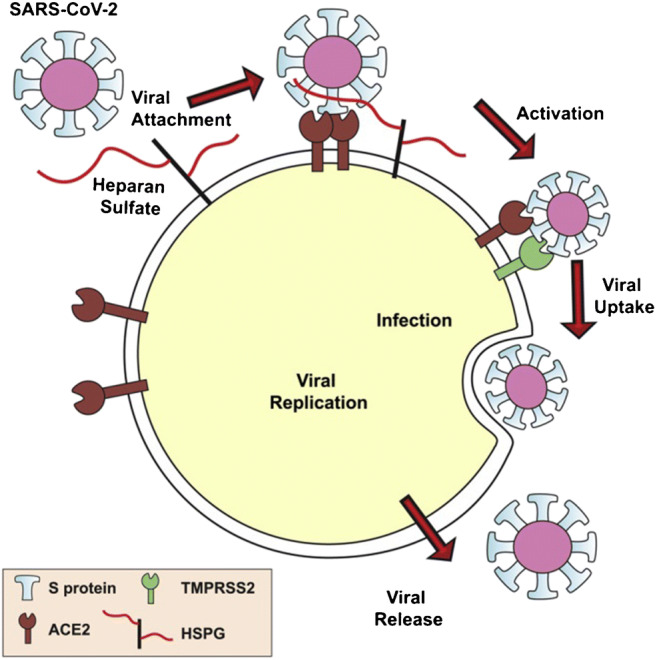
Fig. 6.

Kim et al. proposed model of SARS-CoV-2 host cell entry. A Virion binds to heparan sulfate. B Cell surface protease digests S protein, initiating viral-host cell membrane fusion via conformational change by host cell receptor binding to heparan sulfate and ACE2. C Virion enters host cell and experiences further proteolytic processing. Reprinted with permission from Elsevier [50]
Clausen et al. investigated the SARS-CoV-2 S protein interactions with cellular HS and ACE2 through its RBD [69]. The RBD of the SARS-CoV-2 S protein was found to bind to Hp/HS, likely through a docking site composed of positively charged amino acid residues. This is a separate docking site than that involved in ACE2 binding. SARS-CoV-2 S protein binds cell surface HS in a cooperative manner to ACE2 receptors. The binding of Hp/HS to S trimers enhanced the binding to ACE2. This suggests that cell surface HS works as a virus collector and mediator of the RBD-ACE2 interaction, resulting in more efficient viral infection. HS structures vary across tissue and cell types, and gender and age, possibly shedding some light on the different susceptibility of virus infection by different patient populations. Cell surface HS removed using a mixture of heparin lyases I, II, and III (HSase) in multiple cell types before SARS-CoV-2 infection prevented infection of cells. However, SARS-CoV-1 infection was not blocked by the removal of cell surface HS, which confirms the tighter binding of SARS-CoV-2 S protein to Hp than SARS-CoV-1. Hp binding to the SARS-CoV-2 S protein increased interactions with ACE2 [69]. A mechanism for SARS-CoV-2 infection utilizing host cell HS is shown in Fig. 7. Further work by Tandon et al. built upon these studies and utilized a lentiviral pseudotyping system to screen potential viral entry inhibitors [70]. When testing a variety of GAG structures, it was determined that 6-O sulfation is not necessary for inhibitors, as 6-O-desulfated Hp/HS showed no change in their ability to inhibit infection. Researchers also found that HS bound tightly with the pseudotyped lentivirus, making it a possible candidate as an adhesion co-receptor. These works outline the possibility of utilizing HS mimetics, degrading lyases and metabolic inhibitors of HS biosynthesis for therapeutic components against COVID-19 [69, 70].
Fig. 7.
Clausen et al. proposed mechanism of SARS-CoV-2 viral attachment facilitated by host cell heparan sulfate. Reprinted with permission from Elsevier [69]
Aside from Hp/HS, hyaluronic acid (HA) has also been found to influence SARS-CoV-2 infection. Specifically, genes encoding enzymes involved in upregulation of HA and GAG metabolism in bronchoalveolar cells infected by SARS-CoV-2 establish that inhibition of these GAG’s synthesis could contribute toward management of severe COVID-19 cases [71]. T CD4+ lymphocytes, neutrophils, and macrophages were also found infiltrating the lungs of COVID-19 patients. Increased amounts of macrophages have been identified in the lungs of deceased COVID-19 patients and are likely responsible for the inflammatory process [72]. Blood mononuclear cells were also tested and displayed a proliferative state. Control studies also displayed a dramatic reduction of NK and T lymphocytes and an increase in monocytes, which supports previous findings of changes in myeloid, NK, and B cells in COVID-19 patients [73]. These data show multiple molecular events that are likely involved in SARS-CoV-2 infection and the pulmonary complications known to occur with COVID-19 [71].
Other host receptors for SARS-CoV-2 S protein
Several recent studies have shown that many neutralizing human antibodies that bind to SARS-CoV-2 S do not bind the RBD, which suggests the possibility of other important host receptors and/or co-receptors that bind to different domain(s) of SARS-CoV-2 S protein and promote the entry of virus into host cells [46, 48, 74].
In a recent study, Wang et al. demonstrated that the tyrosine-protein kinase receptor UFO (AXL) specifically interacts with SARS-CoV-2 S protein, and overexpression of AXL promoted viral entry as efficiently as ACE2 overexpression. Significant reduction of pulmonary cell infection by SARS-CoV-2 was observed by downregulating AXL, but not ACE2. Moreover, they showed that soluble human recombinant AXL could block SARS-CoV-2 infection in cells expressing high levels of AXL, whereas soluble ACE2 did not show such an effect [48]. In another study, the roles played by neuropilin-1 (NRP-1) in increasing SARS-CoV-2 infectivity by binding with the furin-cleaved S1 fragment of the S protein were shown, as well as how blocking such interaction with a small-molecule inhibitor or monoclonal antibodies reduces the viral infection in cell culture [49].
It has been reported that the S protein of SARS-CoV-2 potentially binds to several innate immune receptors such as C-type lectin receptors (CLRs) [47]. CLRs bind to specific glycans via a Ca2+-dependent interaction [75]. Several CLRs such as DC-SIGN/CD209, L-SIGN/CD209L/CLEC4M, mannose receptor/MR/MRC1/CD206, MGL/CLEC10A/CD301, and Dectin-2/CLEC6A, which act as first line of defense against invading pathogens, are highly expressed in the human immune system, including monocytes, dendritic cells, and macrophages [76, 77]. Gringhuis et al. reported that CLRs like DC-SIGN can modulate Toll-like receptor–induced activation and thus direct host immune responses against pathogens in a glycan-specific manner [78]. A recent study showed that multiple CLRs including DC-SIGN, L-SIGN, MR (C-type lectin domains 4–7), and MGL can bind strongly to the recombinant full-length S produced in human embryonic kidney HEK293 cells [47].
Effects of glycan termini in viral binding
Viruses often target sialylated glycans and cell adhesion molecules to gain entry into the host cell, and the redundancy in such receptor preference indicates evolutionary conservation in the viral targeting to take advantage of host cellular function [79, 80]. Glycans that are terminated by sialic acids are expressed on cell surfaces and act as ligands for intrinsic or extrinsic sialic acid–specific lectins [79]. Most pathogens express sialic acid–specific lectins on their cell surfaces, which facilitates sialic acid–mediated host cell attachment and immune evasion [81]. Several RNA viruses and DNA viruses gain access to the host cells through sialylated glycans. Interestingly, host cell receptors evolve to combat rapidly emerging pathogens without affecting critical endogenous functions. Viruses express sialidases cleaving the sialic acids that enable virus binding to the cell in the first place, thereby affecting their release from infected host cells. Even though sialic acids may mediate virus binding and infection of cells, they can bind to virions as decoy receptors and thus prevent their access to host epithelial cells [82].
There are several strong indications that sialylated glycans can play important roles in COVID-19 infection [83]. In silico studies have shown evidence that the sialic acid termini on receptors can act as potential entry receptors for the SARS-CoV-2 [84]. Glycans are suggested to play crucial roles on specific sites on the receptor-binding domain in viral binding with hACE2 [5, 7, 85] (Fig. 4). A bioreporter based assay showed that deglycosylation of RBD glycans resulted in lack of interaction of RBD with hACE2 [86]. Several viruses, including coronavirus, bind more avidly to host glycoproteins such as hACE2 featuring α-2,3-linked sialylated glycans [60, 79, 87, 88]. In a study on avian coronavirus ligand interaction, six out of 10 N-glycosylation site mutants lost binding to host chicken trachea tissue, and an ELISA showed specific loss of interaction with ligand α-2,3-linked sialic acid [89]. A study demonstrated that human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acid (9-O-Ac-Sia) and identified the specific site through which this binding occurs [90].
Recent developments in the understanding of structural variants of glycans in the human system could guide in deducing the intricate interactions among different endogenous and exogenous sialic acid–specific ligands and host receptors. Moreover, such knowledge can help in understanding how pathogenic viruses fight the host system by modulating immune tolerance.
Post-translational modifications of M, E, and N proteins
M protein glycosylation
Unlike other components of the SARS-CoV-2 virus, the membrane protein (M protein) and envelope protein (E protein) of the SARS-CoV-2 virus have not been extensively studied and characterized. The M protein is a 222-amino acid glycosylated structural protein containing three N-terminal membrane-spanning domains that are essential for viral particle assembly [36]. Additionally, the M protein is the most abundant envelope protein of SARS-CoV-2 [91]. The M protein of SARS-CoV-2 resembles the M protein of bat and pangolin SARS-CoV-1. In silico analysis showed the SARS-CoV-2 M protein and bat and pangolin SARS-CoV-1 M protein resemble SWEET (sugars will eventually be exported transporter). SWEETs and semiSWEETs are unique sugar transporters with homologs found in all kingdoms of life [92]. SWEETs and semiSWEETs catalyze diffusion of sugars driven by their concentration gradients [93]. SWEETs of eukaryotes have 6–7 transmembrane helices; however, the M protein only has 3 transmembrane helices. This difference means the M proteins of SARS-CoV-2 more closely resemble semiSWEET, which possess only 3 transmembrane helices. SWEETs and semiSWEETs are predominantly found in organisms requiring a high efflux of sugars [93]. Enveloped viruses typically use a two-step procedure to infect and release genetic material into the cell. First, they bind to surface receptors of the target cell membrane; then, they fuse the viral and cell membranes. It is currently unknown how the M proteins are fused to the host cell membrane; however, if they do fuse, it is possible they function as a sugar transporter [91]. The presence of a sugar transporter may influence sucrose entry into the endosome, lysosome, and/or autophagosome, aiding in virus release into cells. The presence of this semiSWEET glucose transporter may be an efficient mechanism to induce rapid viral proliferation and immune evasion [91]. Additionally, in silico experiments were used to identify eight novel N-glycosylation sites of the M protein: N5, N21, N41, N43, N117, N121, N203, and N216. Six of these eight sites were common to both human SARS-CoV-2 and SARS-CoV-1. The main difference worth noting is the mutation in SARS-CoV-2 resulting in the addition of one amino acid. Therefore, though there are six sites in common, the location of these sites differs by 1 amino acid [94].
E protein glycosylation
The E protein is the smallest of the four major structural proteins and has the lowest copy number of the membrane proteins found in the lipid envelope of mature virus particles. The E protein has a short outer amino acid terminal domain, a single helix, and a long inner carboxy-terminal domain [91]. For other coronaviruses, the E protein has been shown to be critical for pathogenesis [95]. A comparison of the amino acid sequences of E proteins across six human coronaviruses is shown in Fig. 8, which displays small changes between the SARS-CoV-2 and the SARS-CoV-1 E proteins. The E protein is possibly mono-glycosylated at site N66, which could serve as a C-terminal translocation reporter. Based on the sequence, N48 could also be glycosylated; however, due to this site’s close proximity to the membrane if the hydrophobic region is recognized as transmembrane by the translocon, it is likely not glycosylated. Both sites, N48 and N66, are located C-terminally in the transmembrane segment [95]. A mutant with a highly efficient glycosylation acceptor site at the N-terminus was designed to test N-terminal translocation. When E protein constructs were translated in vitro in the presence of microsomes, the protein was significantly glycosylated when the N-terminal designed glycosylation site was present. However, when the glycosylation acceptor site was absent, E protein molecules were minimally glycosylated [95].
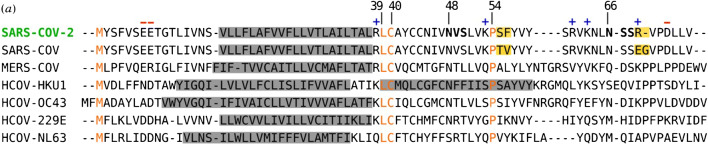
Fig. 8.
Alignment of E protein amino acid sequences comparing SARS-CoV-2 to six other human coronaviruses. Gray boxes highlight predicted transmembrane segments. SARS-CoV-2 native predicted glycosylation acceptor sites are shown bolded, with + or – symbols depicting charge. Orange highlighted residues are conserved; yellow highlighted boxes display differences between SARS-CoV-2 and SARS-CoV-1. Reprinted with permission from Royal Society Publishing [95]
Post-translational modification on N-protein
Unlike the other structural proteins of SARS-CoV-2, the N protein is located in the nucleocapsid, and does not go through the secretory pathway [96]. The N protein facilitates entering the host cell, binding to viral RNA genome, and forms the ribonucleoprotein core [97]. The N protein is able to form high-order oligomers in the absence of RNA. N protein is secreted in the presence of S protein, but independently of E and M protein expression. This indicates the N protein may help virion budding when co-expressed with S protein [35]. The N protein is capable of forming or regulating biomolecular condensates in vivo by interaction with RNA and other key host cell proteins [98]. This activity could then be harnessed to regulate viral life cycle and host cell response to viral infection. Cascarina et al. proposed that the N protein could harness the ability to form or join biomolecular condensates to dysregulate stress granules, enhance viral replication or translation of viral proteins, and package the viral RNA genome into new virions. Targeting host cell kinases or membraneless organelles could modulate N protein regulation and could be a viable solution for combating existing SARS-CoV-2 infections due to the vital role the N protein plays in multiple stages of the viral lifecycle [98]. N proteins of other coronaviruses have similar crystal structures and sequence homology to that of SARS-CoV-2 and are heavily phosphorylated [99]. As the N protein does not go through the secretory pathway, it is not expected to be glycosylated. The N protein of SARS-CoV-2 expressed on HEK293 is phosphorylated at S176 and not glycosylated unless it is forced through the secretory pathway by the addition of a leader sequence during expression in HEK293. The latter form of N protein is also phosphorylated at a different site (T393) [100]. Intriguingly, a study based on MS on the COVID-19 patient samples showed that N protein is detected in patient saliva after deglycosylation [101]. However, another study demonstrated that the recombinant N proteins which are expressed in the mammalian system with leader sequences for protein secretion can lead to a glycosylated form of N protein. Such artificially glycosylated N protein needs deglycosylation for ELISA-based detection of COVID-19 patient anti N protein antibodies. This also makes it clear that the viral N protein present on COVID-19 patients are unglycosylated, and thus the anti N protein antibodies can only recognize the unglycosylated form of N protein [99, 100].
Conclusions
Since the beginning of the SARS-CoV-2 pandemic, the virus and its host infection mechanism have been the focus of many research articles. The four structural proteins of the SARS-CoV-2 virus, spike (S), membrane (M), envelope (E), and nucleocapsid (N), have been extensively analyzed to determine their function within SARS-CoV-2 infection and severity of the resultant COVID-19 disease. Specifically, the glycans located on these proteins as well as at the cell surface of hosts have been shown to facilitate viral entry. Understanding the glycosylation pattern and the role this plays in viral attachment and infection can lead to therapeutic possibilities.
Recently, several vaccine candidates based on either RBD domain or full-length S proteins were approved and are being administered worldwide [29, 102]. The S protein is heavily glycosylated with both N- and O-glycans and directly interacts with hACE2 to facilitate infection. Several therapeutic strategies involving glycans or based on carbohydrate molecules were found effective in addressing SARS-CoV-2 infection. Employing vaccine candidates that could elicit effective immune responses through appropriate glycan display and viral immunogen with glycans that are unique to viruses are recommended for therapeutic purposes [29]. Preventing or reducing the host N-glycan biosynthesis and thereby preventing the viral glycosylation are other approaches being explored [103]. Molecules such as chitosan which can interact with viral surface glycans, particularly the S2 subunit of the S protein, were shown to reduce SARS-CoV-2 infection [104]. Several aminoglycoside antibiotics and polysaccharides (acarbose) displayed therapeutic effectivity against SARS-CoV-2 by binding with viruses, by preventing viral protein translation, or by improving host immune defense [29]. hACE2 acts as a receptor for human coronaviruses, and N-glycosylation of hACE2 is imperative for infection. The study of glycosylation on hACE2 and other known receptors of SARS-CoV-2 could help in understanding how these receptors present their glycan epitopes to the viruses. Such knowledge will help in development of therapeutics which can act as viral decoys in the case of infections. In addition to hACE2, it has been shown that cell surface GAGs also contribute to host cell binding to SARS-CoV-2. Specifically, cell surface heparan sulfate (HS) facilitates SARS-CoV-2 attachment to human host cells. Researchers have studied the possibility of using GAG and GAG-like structures for competitive binding with the cell surface HS. Treatment with heparin (Hp)/HS as well as with fucoidan-like structures results in a lower percentage of SARS-CoV-2 viral attachment to host cells compared to no treatment [68, 69].
In silico experiments have suggested glycosylation of the E and M proteins, but extensive glycosylation mapping is still needed to confirm these assignments and to help determine their importance. The M protein structure is shown to resemble a sugar transporter, which may influence sucrose entry into the endosome, lysosome, and/or autophagosome, aiding in virus release into cells. This sugar transporter may be an efficient mechanism to induce rapid viral proliferation and immune evasion [91]. Recent studies showed that the N protein of SARS-CoV-2 is not glycosylated but phosphorylated. The N protein can form or join biomolecular condensates to dysregulate stress granules, enhance viral replication or translation of viral proteins, and package the viral RNA genome into new virions [96–98]. The ability to modulate N protein regulation could be a viable solution for treating existing SARS-CoV-2 infections, due to the vital role the N protein plays in multiple stages of the viral lifecycle. Understanding the glycosylation and other post translational modifications of these proteins and receptors can help determine viable options to prevent SARS-CoV-2 infection and to treat COVID-19.
Herein, we have reviewed current studies on SARS-CoV-2 and the processes by which the viral proteins influence infection. By understanding these processes, therapeutic methods can be developed to help combat SARS-CoV-2 infection and to treat COVID-19.
Author contribution
Asif Shajahan: conceptualization, writing—original draft, review and editing.
Lauren E. Pepi: writing—original draft, review and editing.
Daniel Rouhani: writing—figure development.
Christian Heiss: writing—review and editing.
Parastoo Azadi: conceptualization, writing—review and editing.
Funding
This work received funding from the US National Institutes of Health (R24GM137782, S10OD018530) and US Department of Energy, Office of Science, Basic Energy Sciences (under Award DE-SC0015662 to DOE—Center for Plant and Microbial Complex Carbohydrates at the Complex Carbohydrate Research Center, USA).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asif Shajahan, Email: asifklm@uga.edu.
Lauren E. Pepi, Email: lauren.pepi@uga.edu
Daniel S. Rouhani, Email: daniel.rouhani@uga.edu
Christian Heiss, Email: cheiss@ccrc.uga.edu.
Parastoo Azadi, Email: azadi@ccrc.uga.edu.
References
- 1.Ge HP, Wang XF, Yuan XN, Xiao G, Wang CZ, Deng TC, et al. The epidemiology and clinical information about COVID-19. Eur J Clin Microbiol. 2020;39(6):1011–1019. doi: 10.1007/s10096-020-03874-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. 2020;213(2):54–56. doi: 10.5694/mja2.50674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe Y, Berndsen ZT, Raghwani J, Seabright GE, Allen JD, Pybus OG, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183(6):1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D, Wang NS, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Y, Cao DF, Zhang YF, Ma J, Qi JX, Wang QH, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8. [DOI] [PMC free article] [PubMed]
- 10.Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanda M, Morrison L, Goldman R. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal Chem. 2021;93(4):2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhao W, Mao Y, Chen Y, Wang S, Zhong Y, et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol Cell Proteomics. 2020;100058. [DOI] [PMC free article] [PubMed]
- 14.Antonopoulos A, Broome S, Sharov V, Ziegenfuss C, Easton RL, Panico M, et al. Site-specific characterization of SARS-CoV-2 spike glycoprotein receptor-binding domain. Glycobiology. 2021;31(3):181–187. doi: 10.1093/glycob/cwaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou DP, Tian XX, Qi RB, Peng C, Zhang W. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: implications for vaccination and antibody therapeutics. Glycobiology. 2021;31(1):69–80. doi: 10.1093/glycob/cwaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brun J, Vasiljevic S, Gangadharan B, Hensen MV, Chandran A, Hill ML, et al. Assessing antigen structural integrity through glycosylation analysis of the SARS-CoV-2 viral spike. ACS Central Science. 2021;7(4):586–593. doi: 10.1021/acscentsci.1c00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe Y, Bowden TA, Wilson IA, Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Bba-Gen Subjects. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter JR, Pager CT, Fowler SD, Dutch RE. Role of N-linked glycosylation of the Hendra virus fusion protein. J Virol. 2005;79(12):7922–7925. doi: 10.1128/JVI.79.12.7922-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80(10):4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homans SW, Dwek RA, Rademacher TW. Solution conformations of N-linked oligosaccharides. Biochemistry-Us. 1987;26(21):6571–6578. doi: 10.1021/bi00395a001. [DOI] [PubMed] [Google Scholar]
- 22.Grant OC, Montgomery D, Ito K, Woods RJ. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep-Uk. 2020;10(1). [DOI] [PMC free article] [PubMed]
- 23.Depetris RS, Julien JP, Khayat R, Lee JH, Pejchal R, Katpally U, et al. Partial enzymatic deglycosylation preserves the structure of cleaved recombinant HIV-1 envelope glycoprotein trimers. J Biol Chem. 2012;287(29):24239–24254. doi: 10.1074/jbc.M112.371898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses-Basel. 2014;6(3):1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helle F, Duverlie G, Dubuisson J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses-Basel. 2011;3(10):1909–1932. doi: 10.3390/v3101909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crispin M, Doores KJ. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casals C, Campanero-Rhodes MA, Garcia-Fojeda B, Solis D. The role of collectins and galectins in lung innate immune defense. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed]
- 28.Hutter J, Rodig JV, Hoper D, Seeberger PH, Reichl U, Rapp E, et al. Toward animal cell culture-based influenza vaccine design: viral hemagglutinin N-glycosylation markedly impacts immunogenicity. J Immunol. 2013;190(1):220–230. doi: 10.4049/jimmunol.1201060. [DOI] [PubMed] [Google Scholar]
- 29.Lardone RD, Garay YC, Parodi P, Fuente S, Angeloni G, Bravo EO, et al. How glycobiology can help us treat and beat the COVID-19 pandemic. J Biol Chem. 2021;100375. [DOI] [PMC free article] [PubMed]
- 30.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 31.Altman MO, Angel M, Kosik I, Trovao NS, Zost SJ, Gibbs JS, et al. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. Mbio. 2019;10(2). [DOI] [PMC free article] [PubMed]
- 32.Zost SJ, Parkhouse K, Gumina ME, Kim K, Perez SD, Wilson PC, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. P Natl Acad Sci USA. 2017;114(47):12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021;89(5):569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset F-L, et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J Biol Chem. 2021;296:100111. doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y-Z, Wang S-Y, Zheng Z-Q, Yi H, Li W-W, Xu Z-S, et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18(3):613–620. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsaadi EA, Jones IM. Membrane binding proteins of coronaviruses. Futur Virol. 2019;14(4):275. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naskalska A, Dabrowska A, Szczepanski A, Milewska A, Jasik KP, Pyrc K. Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. J Virol. 2019;93(19). [DOI] [PMC free article] [PubMed]
- 39.Bianchi M, Benvenuto D, Giovanetti M, Angeletti S, Ciccozzi M, Pascarella S. SARS-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed Res Int. 2020;2020. [DOI] [PMC free article] [PubMed]
- 40.Casalino L, Gaieb Z, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. Acs Central Science. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Liu C, Du L, Jiang S, Shi Z, Baric RS, et al. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(17):9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossen JW, de Beer R, Godeke GJ, Raamsman MJ, Horzinek MC, Vennema H, et al. The viral spike protein is not involved in the polarized sorting of coronaviruses in epithelial cells. J Virol. 1998;72(1):497–503. doi: 10.1128/JVI.72.1.497-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls AC, Tortorici MA, Frenz B, Snijder J, Li WT, Rey FA, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenza MP, Oyenarte I, Diercks T, Quintana JI, Gimeno A, Coelho H, et al. Structural characterization of N-linked glycans in the receptor binding domain of the SARS-CoV-2 spike protein and their interactions with human lectins. Angew Chem Int Ed Eng. 2020;59(52):23763–23771. doi: 10.1002/anie.202011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q, Hughes TA, Kelkar A, Yu X, Cheng K, Park S, et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife. 2020;9. [DOI] [PMC free article] [PubMed]
- 46.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao C, Zeng J, Jia N, Stavenhagen K, Matsumoto Y, Zhang H, et al. SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv. 2020. 10.1101/2020.07.29.227462.
- 48.Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Anton-Plagaro C, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SY, Jin W, Sood A, Montgomery DW, Grant OC, Fuster MM, et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020;181:104873. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang F, Yang J, Zhang Y, Dong M, Wang S, Zhang Q, et al. Angiotensin-converting enzyme 2 and angiotensin 1-7: novel therapeutic targets. Nat Rev Cardiol. 2014;11(7):413–426. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275(43):33238–43. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 53.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87(5):E1–E9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 54.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. P Natl Acad Sci USA. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 57.Suryamohan K, Diwanji D, Stawiski EW, Gupta R, Miersch S, Liu J, et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun Biol. 2021;4(1):475. doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, et al. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devaux CA, Pinault L, Osman IO, Raoult D. Can ACE2 receptor polymorphism predict species susceptibility to SARS-CoV-2? Front Public Health. 2021;8. [DOI] [PMC free article] [PubMed]
- 60.Shajahan A, Archer-Hartmann S, Supekar NT, Gleinich AS, Heiss C, Azadi P. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin-converting enzyme 2. Glycobiology. 2020. [DOI] [PMC free article] [PubMed]
- 61.Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor, bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 62.Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al. Essentials of glycobiology. 3rd ed: Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015. [PubMed]
- 63.Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat Prod Rep. 2002;19(3):312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 64.Clegg DO, Reda DJ, Harris CL, Klein MA, O’Dell JR, Hooper MM, et al. Glucosamine, chondroitin sulfate, and the two in combination for painful knee osteoarthritis. The New England Jounral of Medicine. 2006;354(8):795–808. doi: 10.1056/NEJMoa052771. [DOI] [PubMed] [Google Scholar]
- 65.Pepi LE, Sanderson P, Stickney M, Amster IJ. Developments in mass spectrometry for glycosaminoglycan analysis: a review. Mol Cell Proteomics. 2021;20:100025. doi: 10.1074/mcp.R120.002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y, Lin L, Agyekum I, Zhang X, Ange KS, Yu Y, et al. Structural analysis of heparin-derived 3-O-sulfated tetrasaccharides: antithrombin binding site variants. J Pharm Sci. 2017;106(4):973–981. doi: 10.1016/j.xphs.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y, Singh A, Li L, Linhardt RJ, Xu Y, Liu J, et al. Investigating changes in the gas-phase conformation of antithrombin III upon binding of Arixtra using traveling wave ion mobility spectrometry (TWIMS) Analyst. 2015;140(20):6980–6989. doi: 10.1039/C5AN00908A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwon PS, Oh H, Kwon S-J, Jin W, Zhang F, Fraser K, et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discovery. 2020;6(1):1–4. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–57.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tandon R, Sharp JS, Zhang F, Pomin VH, Ashpole NM, Mitra D, et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2021;95(3). [DOI] [PMC free article] [PubMed]
- 71.Andonegui-Elguera S, Taniguchi-Ponciano K, Gonzalez-Bonilla CR, Torres J, Mayani H, Herrera LA, et al. Molecular alterations prompted by SARS-CoV-2 infection: induction of hyaluronan, glycosaminoglycan and mucopolysaccharide metabolism. Arch Med Res. 2020;51(7):645–653. doi: 10.1016/j.arcmed.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. MedRxiv. 2020.
- 73.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discovery. 2020;6(1):31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drouin M, Saenz J, Chiffoleau E. C-type lectin-like receptors: head or tail in cell death immunity. Front Immunol. 2020;11. [DOI] [PMC free article] [PubMed]
- 76.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9(6):593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 77.Routhu NK, Lehoux SD, Rouse EA, Bidokhti MRM, Giron LB, Anzurez A, et al. Glycosylation of Zika virus is important in host-virus interaction and pathogenic potential. Int J Mol Sci. 2019;20(20). [DOI] [PMC free article] [PubMed]
- 78.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TBH. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10(10):1081–1U58. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 79.Kuchipudi SV, Nelli RK, Gontu A, Satyakumar R, Nair MS, Subbiah M. Sialic acid receptors: the key to solving the enigma of zoonotic virus spillover. Viruses-Basel. 2021;13(2). [DOI] [PMC free article] [PubMed]
- 80.Maginnis MS. Virus-receptor interactions: the key to cellular invasion. J Mol Biol. 2018;430(17):2590–2611. doi: 10.1016/j.jmb.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nadano D, Iwasaki M, Endo S, Kitajima K, Inoue S, Inoue Y. A naturally-occurring deaminated neuraminic acid, 3-deoxy-D-glycero-D-galacto-nonulosonic acid (Kdn) - its unique occurrence at the nonreducing ends of oligosialyl chains in polysialoglycoprotein of rainbow-trout eggs. J Biol Chem. 1986;261(25):1550–1557. doi: 10.1016/S0021-9258(18)67278-3. [DOI] [PubMed] [Google Scholar]
- 82.Wasik BR, Barnard KN, Parrish CR. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 2016;24(12):991–1001. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wielgat P, Rogowski K, Godlewska K, Car H. Coronaviruses: is sialic acid a gate to the eye of cytokine storm? From the entry to the effects. Cells. 2020;9(9). [DOI] [PMC free article] [PubMed]
- 84.Morniroli D, Gianni ML, Consales A, Pietrasanta C, Mosca F. Human sialome and coronavirus disease-2019 (COVID-19) pandemic: an understated correlation? Front Immunol. 2020;11:1480. doi: 10.3389/fimmu.2020.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26(6):481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azad T, Singaravelu R, Taha Z, Jamieson TR, Boulton S, Crupi MJF, et al. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 S for viral entry. Mol Ther. 2021. [DOI] [PMC free article] [PubMed]
- 87.Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W, et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114(40):E8508–E8E17. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hao W, Ma B, Li Z, Wang X, Gao X, Li Y, et al. Binding of the SARS-CoV-2 spike protein to glycans. Sci Bull (Beijing). 2021. [DOI] [PMC free article] [PubMed]
- 89.Parsons LM, Bouwman KM, Azurmendi H, de Vries RP, Cipollo JF, Verheije MH. Glycosylation of the viral attachment protein of avian coronavirus is essential for host cell and receptor binding. J Biol Chem. 2019;294(19):7797–7809. doi: 10.1074/jbc.RA119.007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hulswit RJG, Lang Y, Bakkers MJG, Li W, Li Z, Schouten A, et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A. 2019;116(7):2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas S. The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter semiSWEET. Pathogens and Immunity. 2020;5(1):342. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng L, Frommer WB. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci. 2015;40(8):480–486. doi: 10.1016/j.tibs.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 93.Jia B, Zhu XF, Pu ZJ, Duan YX, Hao LJ, Zhang J, et al. Integrative view of the diversity and evolution of SWEET and SemiSWEET sugar transporters. Front Plant Sci. 2017;8(2178). [DOI] [PMC free article] [PubMed]
- 94.Dawood AA. Glycosylation, ligand binding sites and antigenic variations between membrane glycoprotein of COVID-19 and related coronaviruses. Vacunas. 2021;22(1):1–9. doi: 10.1016/j.vacun.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duart G, García-Murria MJ, Grau B, Acosta-Cáceres JM, Martínez-Gil L, Mingarro I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10(9):200209. doi: 10.1098/rsob.200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahman MS, Islam MR, Alam ASMRU, Islam I, Hoque MN, Akter S, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. 2021;93(4):2177–2195. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- 97.Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cascarina SM, Ross ED. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020;34(8):9832–9842. doi: 10.1096/fj.202001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rump A, Risti R, Kristal M-L, Reut J, Syritski V, Lookene A, et al. Dual ELISA using SARS-CoV-2 nucleocapsid protein produced in E. coli and CHO cells reveals epitope masking by N-glycosylation. Biochem Biophys Res Commun. 2021;534:457–460. doi: 10.1016/j.bbrc.2020.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Supekar NT, Shajahan A, Gleinich AS, Rouhani D, Heiss C, Azadi P. SARS-CoV-2 Nucleocapsid protein is decorated with multiple N- and O-glycans. bioRxiv. 2020; 2020.08.26.269043.
- 101.Nikolaev EN, Indeykina MI, Brzhozovskiy AG, Bugrova AE, Kononikhin AS, Starodubtseva NL, et al. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J Proteome Res. 2020;19(11):4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
- 102.Irwin A. What it will take to vaccinate the world against COVID-19. Nature. 2021;592(7853):176–178. doi: 10.1038/d41586-021-00727-3. [DOI] [PubMed] [Google Scholar]
- 103.Williams SJ, Goddard-Borger ED. Alpha-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem Soc Trans. 2020;48(3):1287–1295. doi: 10.1042/BST20200505. [DOI] [PubMed] [Google Scholar]
- 104.Milewska A, Chi Y, Szczepanski A, Barreto-Duran E, Dabrowska A, Botwina P, et al. HTCC as a polymeric inhibitor of SARS-CoV-2 and MERS-CoV. J Virol. 2021;95(4). [DOI] [PMC free article] [PubMed]