ABSTRACT
Regulation of flagellum biosynthesis is a hierarchical process that is tightly controlled to allow for efficient tuning of flagellar expression. Flagellum-mediated motility directs Salmonella enterica serovar Typhimurium toward the epithelial surface to enhance gut colonization, but flagella are potent activators of innate immune signaling, so fine-tuning flagellar expression is necessary for immune avoidance. In this work, we evaluate the role of the LysR transcriptional regulator YeiE in regulating flagellum-mediated motility. We show that yeiE is necessary and sufficient for swimming motility. A ΔyeiE mutant is defective for gut colonization in both the calf ligated ileal loop model and the murine colitis model due to its lack of motility. Expression of flagellar class 2 and 3 but not class 1 genes is reduced in the ΔyeiE mutant. We linked the motility dysregulation of the ΔyeiE mutant to repression of the anti-FlhD4C2 factor STM1697. Together, our results indicate that YeiE promotes virulence by enhancing cell motility, thereby providing a new regulatory control point for flagellar expression in Salmonella Typhimurium.
KEYWORDS: Salmonella, flagellar gene regulation, gastrointestinal infection, host-pathogen interactions, transcriptional regulation
INTRODUCTION
Flagellum-mediated motility is a key virulence determinant facilitating gut colonization by many organisms, including nontyphoidal salmonellae (1, 2). Flagellum-mediated motility allows movement through the intestinal mucus layer toward the epithelium, allowing Salmonella to scan epithelial cells for permissive entry sites (1, 3). Upon contact with the epithelium, Salmonella invades intestinal cells, induces a neutrophilic inflammatory response, and replicates intracellularly, processes that require secretion of effector proteins through the two type 3 secretion systems (4). During early intracellular growth, Salmonella enterica serovar Typhimurium downregulates the expression of flagella to avoid immune activation induced by sensing intracellular flagellin (5). However, flagella are expressed during late intracellular infection in preparation for escape from the intracellular environment (6, 7). The dynamic expression of flagellar genes requires tight control to facilitate gut colonization while avoiding inappropriate stimulation of the innate immune response.
Flagellar biosynthesis is regulated by a transcriptional hierarchy comprised of genes in three classes. Class 1 genes encode the heterodimeric master transcriptional regulator FlhD4C2, required for expression of class 2 and 3 genes that encode the flagellar apparatus, motor force-generating elements, chemotaxis proteins, and numerous regulatory proteins (8–15). Global transcriptional regulators integrate environmental signals at the single class 1 promoter to control expression of flhDC (16). Small RNAs and the RNA binding protein CsrA exert posttranscriptional control of flhDC (17, 18). Further control of flagellar biosynthesis is mediated by regulation of FlhD4C2 function. Two proteins, each containing EAL-like domains, YdiV and STM1697 (STM14_2047), inhibit motility using different mechanisms to regulate FlhD4C2 function (19–22). YdiV binds FlhD to prevent FlhD4C2 from binding DNA and targets it for proteolysis (21, 23), whereas STM1697 restricts FlhD4C2 from recruiting σ70 RNA polymerase to promoters of class 2 flagellar genes (20). Together, the extensive transcriptional, posttranscriptional, and functional control of the flagellar master regulator enables tight control over the initiation of flagellar biosynthesis.
In prior work, we found a motility defect for a Salmonella Typhimurium mutant in the putative LysR-type transcriptional regulator (LTTR) yeiE (STM2201/STM14_2717) (24). LTTRs comprise the largest family of transcriptional regulators among prokaryotes and can regulate local or global gene expression in response to small-molecule ligands (25, 26). There are at least 44 annotated LTTRs encoded in the Salmonella Typhimurium genome (27–29). Although YeiE is poorly characterized in S. Typhimurium, it is highly conserved across related organisms. In Escherichia coli, there are no genes with clear roles that regulate flagellar biosynthesis in the YeiE regulon and no motility defect identified for a ΔyeiE mutant (30, 31). However, YeiE homologs improve the in vivo fitness of other bacterial pathogens, including Cronobacter sakazakii (gpESA_01081), Pseudomonas aeruginosa (PA3398/finR), and Vibrio cholerae (VC2324/tehR) (32–34). These data indicate an important role for YeiE in the pathogenesis of numerous organisms.
Despite extensive study of the regulation of flagellar biogenesis, the role of YeiE in flagellar regulation has not previously been described. The purpose of this study was to determine the role of yeiE in the regulation of S. Typhimurium flagellum-mediated motility and gastrointestinal colonization. We hypothesized that yeiE impacts flagellar gene expression and that dysregulated motility results in reduced gut colonization in animal models. Our work demonstrates the critical role of yeiE in the complex pathway regulating flagellum-mediated motility in Salmonella Typhimurium.
RESULTS
yeiE is required for swimming motility.
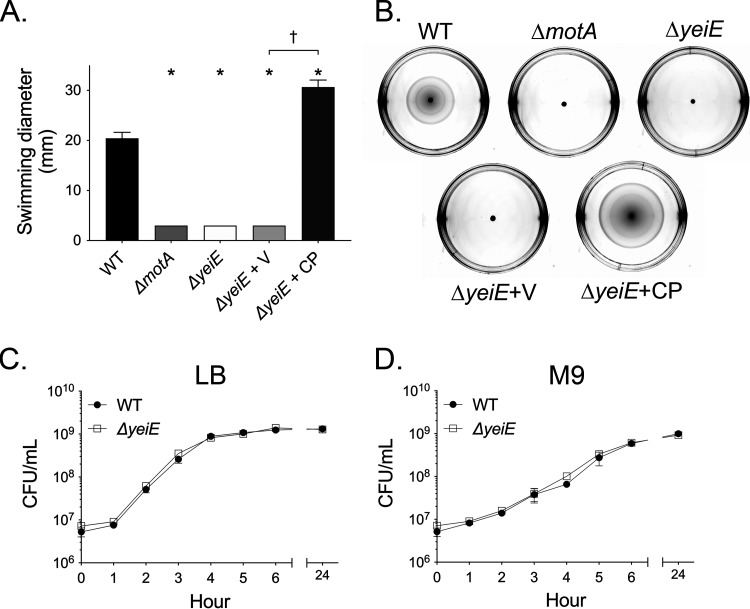
Flagellum-mediated motility is a potent agonist of the neutrophil respiratory burst (35). In a screen of Salmonella Typhimurium mutants, we found that a ΔyeiE mutant elicited a decreased neutrophil respiratory burst due to its lack of motility (24). To definitively link the observed motility defect to the disruption of yeiE, we assessed the swimming motility of the complemented ΔyeiE mutant. Complementation in trans restored the swimming motility of the ΔyeiE mutant to levels greater than that of the wild-type (WT) organism (Fig. 1A and B). The ΔyeiE mutant grew normally in both rich and minimal media (Fig. 1C and D), ruling out any potential effects of abnormal growth on swimming motility. These data demonstrate that yeiE plays an important role in S. Typhimurium swimming motility.
FIG 1.
yeiE is required for swimming motility. (A) Normalized overnight cultures of the WT (HA420) and the ΔmotA mutant (JE1202), ΔyeiE mutant (JE973), ΔyeiE mutant with an empty vector (ΔyeiE + V; JE1511), and the ΔyeiE mutant with a complementing plasmid (ΔyeiE + CP; JE1513) were spotted onto swimming plates. Diameters of cell spread were measured 4 h postinoculation. Each assay was performed in 3 replicates on 3 independent occasions. Bars represent means ± standard errors of the means (SEM). Statistical significance was determined by Student's t test (P of <0.05). * indicates a significant difference between the WT and mutant, and † indicates a significant difference between the indicated mutants. (B) Representative photographs of swimming plates 5 h postinoculation from one experiment. Growth curves of the WT (HA420) and the ΔyeiE mutant (JE973) in rich (C) and minimal (D) media.
yeiE is required for gastrointestinal colonization.
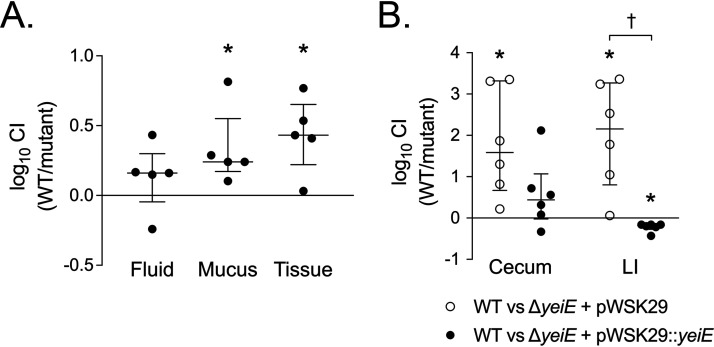
Since flagellum-mediated motility is required for efficient colonization of the gut, we hypothesized that the ΔyeiE mutant would have a colonization defect in animal models of enteric salmonellosis. We used the calf ligated ileal loop model of infection because calves are natural hosts for Salmonella, infections occur in the presence of an intact microbiota, and pathology is consistent with that of enteric salmonellosis in humans (36). In competitive infections with the virulent WT organism, we found that the ΔyeiE mutant was defective for colonization of the intestinal mucus layer and tissue but not for survival within luminal fluid (Fig. 2A). The lack of a defect in the fluid compartment suggests no effect of the ΔyeiE mutation on fitness in the inflamed gut, and the failure of the ΔyeiE mutant to penetrate the mucus layer and colonize intestinal tissue is consistent with a defect in flagellum-mediated motility.
FIG 2.
yeiE is required for efficient gut colonization. (A) Ligated ileal loops from five 3- to 6-week-old calves were inoculated with ∼109 CFU of an equivalent mixture of the WT (HA420) and ΔyeiE mutant (JE973). Loops were harvested 12 h after inoculation, and luminal fluid, mucus, and intestinal tissue were processed for enumeration of CFU. (B) Six C57BL/6J mice were treated with streptomycin prior to infection with ∼108 CFU of an equivalent mixture of the WT and ΔyeiE mutant bearing the empty plasmid (JE1511) or the complemented mutant (JE1513) by gavage. Mice were euthanized 72 h after infection and the cecum and large intestine (LI) processed for enumeration of CFU. The competitive index (CI) was determined by dividing the ratio of WT cells to mutant cells in each tissue compartment by the ratio in the inoculum. Each data point represents a single animal, with median and interquartile ranges indicated. Statistical significance was determined by Student's t test (P < 0.05). * indicates a significant difference in competitive indexes, and † indicates a significant difference between groups.
Next, we used the murine colitis model to investigate the mechanism of the ΔyeiE mutant gut colonization defect. In the murine colitis model, antibiotic treatment disrupts the gut microbiota prior to Salmonella infection, allowing development of a neutrophilic typhlitis (37). As with our findings from the calf model, the ΔyeiE mutant had a colonization defect in the murine cecum and large intestine (Fig. 2B). The colonization defect was reversed by complementation in trans in both the large intestine and the cecum (Fig. 2B), definitively linking yeiE to the observed gut colonization defect.
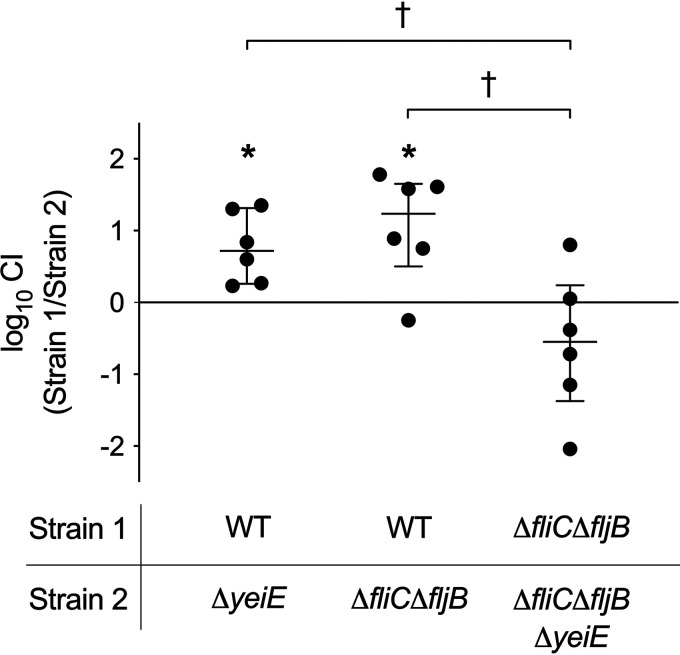
We hypothesized that the motility defect of the ΔyeiE mutant was the likely cause of the observed gut colonization defect. To investigate the role flagellum-mediated motility played in the ΔyeiE gut colonization defect, we compared the competitive index (CI) of the aflagellated ΔfliC ΔfljB mutant with that of the ΔyeiE mutant. We found that the ΔyeiE mutant and the ΔfliC ΔfljB mutant had similar colonization defects in competition with the WT organism (Fig. 3). To establish whether abnormal motility was the reason for the fitness defect of the ΔyeiE mutant, we performed a competitive infection experiment with the ΔfliC ΔfljB mutant and a ΔfliC ΔfljB ΔyeiE mutant. We found no significant alteration in fitness of the ΔfliC ΔfljB ΔyeiE mutant from that of the ΔfliC ΔfljB mutant (Fig. 3). Together, these data demonstrate that the ΔyeiE mutant poorly colonizes the mammalian intestine, likely as a result of defective flagellum-mediated motility.
FIG 3.
The ΔyeiE mutant gut colonization defect is linked to defective flagellar motility. The CI between the indicated strains in cecum from the murine colitis model was determined as described in the legend for Fig. 2. Statistical significance was determined by Student's t test (P < 0.05). * indicates a significant difference in competitive indexes, and † indicates a significant difference between groups.
Role of YeiE in flagellar regulation.
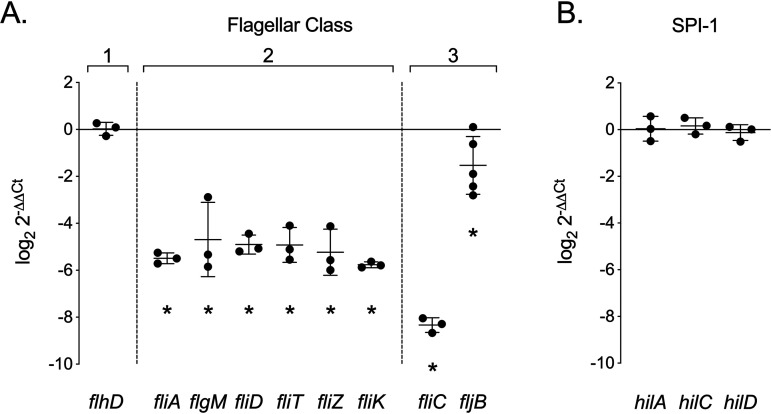
Although the E. coli YeiE regulon does not include genes responsible for flagellar biogenesis (30), our data demonstrate a role for yeiE in S. Typhimurium flagellum-mediated motility. We hypothesized that YeiE influences the expression of flagellar genes. To investigate the level at which YeiE affects the transcriptional hierarchy of flagellar biogenesis, we measured the mRNA expression of a subset of genes from each of the three flagellar regulatory classes in the WT and ΔyeiE mutant. Expression of the class 1 gene flhD was not affected by deletion of yeiE (Fig. 4A). In contrast, expression of genes from both class 2 and class 3 was significantly downregulated in the ΔyeiE mutant (Fig. 4A).
FIG 4.
Flagellar class 2 and 3 genes are downregulated in the ΔyeiE mutant. Relative expression of the indicated genes from the ΔyeiE mutant compared with that of the WT in late exponential growth in rich medium. Each data point represents the mean from triplicate samples from one biological replicate; means ± standard deviations (SD) are indicated. Statistical significance was determined by Student's t test (P < 0.05). * indicates a significant difference in relative levels of expression of the given gene.
There is regulatory cross talk between the expression of flagellar genes and those borne in Salmonella pathogenicity island 1 (SPI-1), encoding type 3 secretion system 1 and effector proteins needed for the invasion of epithelial cells (38–40). To rule out an effect of yeiE on SPI-1 gene expression, we measured the mRNA expression of three SPI-1 regulators in the WT and ΔyeiE mutant. We observed no difference in expression of hilA, hilC, or hilD in the ΔyeiE mutant (Fig. 4B), suggesting that yeiE does not impact SPI-1 gene expression. These data are consistent with our prior work demonstrating no role for yeiE in the activation of a promoter for a type 3 secretion system 1 apparatus protein (24). These findings suggest that YeiE promotes the expression of flagellar class 2 genes through an SPI-1-independent process.
Characterization of the ΔyeiE suppressor mutant.
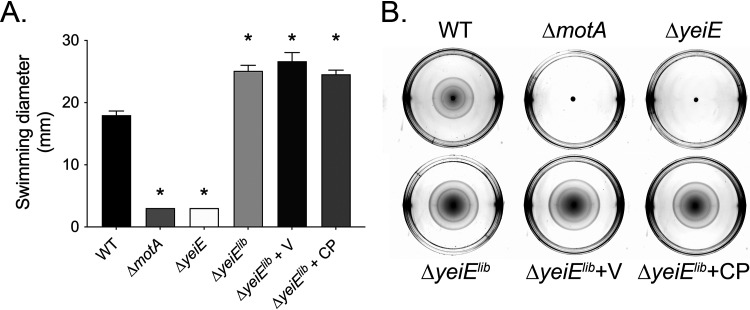
In prior work evaluating the role of S. Typhimurium genes in the stimulation of the neutrophil respiratory burst, we found that the effects of the ΔyeiE mutation in a strain obtained from the kanamycin-resistant Salmonella Typhimurium ATCC 14028s single-gene-deletion library (ΔyeiElib strain) differed from those of the ΔyeiE mutation in a clean genetic background (24, 41). To establish whether the differential effects were due to a motility difference between the ΔyeiE mutants, we measured the motility of the two ΔyeiE mutants on semisolid agar. The ΔyeiE mutant in the clean genetic background was amotile, whereas the ΔyeiElib mutant was hypermotile compared to the WT organism (Fig. 5). Furthermore, the complemented ΔyeiElib mutant remained hypermotile (Fig. 5), suggesting that the hypermotility is not due to disruption of yeiE in the ΔyeiElib mutant. Therefore, we hypothesized that a suppressor mutation was the likely cause of the observed hypermotility of the ΔyeiElib mutant.
FIG 5.
The ΔyeiElib mutant is hypermotile. Normalized overnight cultures of the WT (HA420), ΔmotA mutant (JE1202), ΔyeiE mutant (JE973), ΔyeiElib mutant (JE1681), ΔyeiElib mutant with the empty vector (ΔyeiElib + V; JE2014), and complemented ΔyeiElib mutant (ΔyeiElib + CP; JE2016) were spotted onto swimming plates. Statistical significance was determined by Student's t test (P < 0.05). * indicates a significant difference between the WT and mutant. (B) Representative photographs of swimming plates from one experiment.
We performed whole-genome sequencing of the amotile ΔyeiE mutant and the hypermotile ΔyeiElib suppressor mutant to establish the identity of the suppressor mutation. In a comparison with the published ATCC 14028s genome sequence (42), we identified three single-nucleotide variants (SNVs) in the amotile ΔyeiE mutant and three SNVs in the ΔyeiElib suppressor mutant that were different between the two genomes (see Tables S1 and S2 in the supplemental material). In the amotile mutant, we found an SNV in gyrA (260T>C) which confers nalidixic acid resistance and is consistent with the phenotypic nalidixic acid resistance in the amotile ΔyeiE mutant (Table 1). We also identified an SNV in eaeH on the 94-kb virulence plasmid and a single-nucleotide deletion in cyoB leading to a frameshift resulting in a predicted truncation of the protein from 664 amino acids to 278 amino acids. CyoB encodes cytochrome bo ubiquinol oxidase subunit I, which helps generate the proton-motive force (43). Since the proton-motive force drives flagellum-mediated motility, the predicted CyoB truncation may play a contributing role in its decreased motility (44). However, complementation in trans reverses the ΔyeiE mutant motility defect (Fig. 1), suggesting that CyoB truncation is not the likely cause of the motility defect.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| HA420 | ATCC 14028.s (spontaneous Nalr) | 59 |
| JE973 | HA420 ΔyeiE::kan (Nalr Kanr) | 24 |
| JE1202 | HA420 ΔmotA::kan (Nalr Kanr) | 35 |
| JE1511 | JE973 carrying pWSK29 (Nalr Kanr Ampr) | This study |
| JE1513 | JE973 carrying pWSK29::yeiE (Nalr Kanr Ampr) | This study |
| JE1681 | 14028.s ΔyeiE::kan (Kanr) | 41 |
| JE1389 | HA420 ΔyeiE::frt (Nalr) | This study |
| JE1699 | HA420 ΔSTM1697::cm (Nalr Cmr) | This study |
| JE1907 | JE973 ΔSTM1697::cm (Nalr Kanr Cmr) | This study |
| JE1915 | HA420 ΔfliC::kan ΔfljB::cm (Nalr Kanr Cmr) | This study |
| JE1919 | HA420 ΔfliC::frt ΔfljB::frt (Nalr) | This study |
| JE1921 | HA420 ΔfliC::frt ΔfljB::frt ΔyeiE::kan (Nalr Kanr) | This study |
| JE2014 | JE1681 carrying pWSK29 (Kanr Ampr) | This study |
| JE2016 | JE1681 carrying pWSK29::yeiE (Kanr Ampr) | This study |
| Plasmids | ||
| pCP20 | flp recombinase, Ampr | 51 |
| pWSK29 | Cloning vector, Ampr | 53 |
| pWSK29::yeiE | pWSK29::yeiE Ampr | This study |
Genome sequencing variants. Variants are differences between the ΔyeiE mutants and the ATCC 14028s genome (GenBank accession no. CP001363, chromosome; accession no. CP001362, plasmid). Bold variants are those with reads in both strands (forward/reverse balance of >0.05). Variants in red are those that were identified in both the amotile and suppressor ΔyeiE mutants. Download Table S1, XLSX file, 0.01 MB (12.8KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of single-nucleotide variants in the ΔyeiE and ΔyeiElib mutants identified by whole-genome sequencing. The impact of SNV on the target amino acid sequence of 6 genes was established. *, variants were supported by reads in both strands with over 50% frequency. Download Table S2, XLSX file, 0.01 MB (10.4KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Three SNVs were identified in the ΔyeiElib suppressor mutant (Tables S1 and S2). An SNV identified in hrpA (248G>T) resulted in a threonine-to-asparagine amino acid sequence change that is unlikely to alter protein function due to replacement of one polar amino acid for another. Two other SNVs in the ΔyeiElib mutant were in hupA and STM1697. Published work demonstrated that deletion of hupA causes an ∼10% motility reduction compared with the motility of its wild-type parent (45). The STM1697 mutation (565G>A) resulted in a stop codon after amino acid 188, causing early termination of the protein. Deletion of STM1697 caused hypermotility (19). Since the ΔyeiElib mutant is hypermotile, the SNV in STM1697 was considered the most likely cause of the observed phenotype.
The association between YeiE and STM1697.
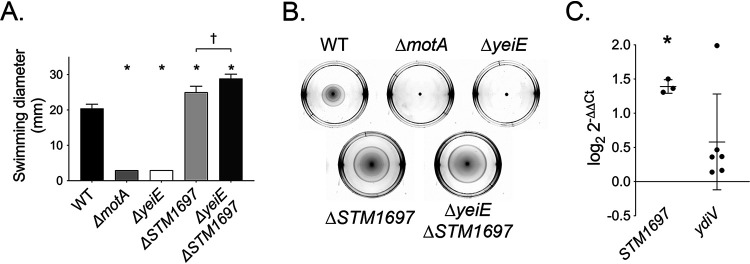
STM1697 inhibits cell motility by preventing RNA polymerase recruitment to FlhD4C2, leading to repressed flagellar class 2 and 3 gene expression (20). Similarly, flagellar class 2 and 3 genes are downregulated by deletion of yeiE without an effect on class 1 gene expression (Fig. 4). Therefore, we hypothesized that there was an interaction between STM1697 and yeiE. We found that the ΔSTM1697 mutant is hypermotile on semisolid agar (Fig. 6A and B), consistent with published reports (19). Deletion of STM1697 exhibited a dominant effect on the ΔyeiE mutant, resulting in the hypermotility of the ΔyeiE ΔSTM1697 mutant (Fig. 6A and B). These data are similar to our observations of hypermotility for the ΔyeiElib suppressor mutant and suggest that YeiE may repress the expression of the STM1697 FlhD4C2 repressor.
FIG 6.
YeiE interacts with STM1697 to alter cell motility. (A) Normalized overnight cultures of the WT (HA420), ΔmotA mutant (JE1202), ΔyeiE mutant (JE973), ΔSTM1697 mutant (JE1699), and ΔyeiE ΔSTM1697 mutant (JE1907) were spotted onto swimming plates. Data analysis was as described in the legend of Fig. 1. (B) Representative photographs of swimming plates from one experiment. (C) Gene expression from the ΔyeiE mutant compared with that of the WT as described in the legend of Fig. 4. Each data point represents the mean from triplicate samples from one biological replicate; means ± SD are indicated.
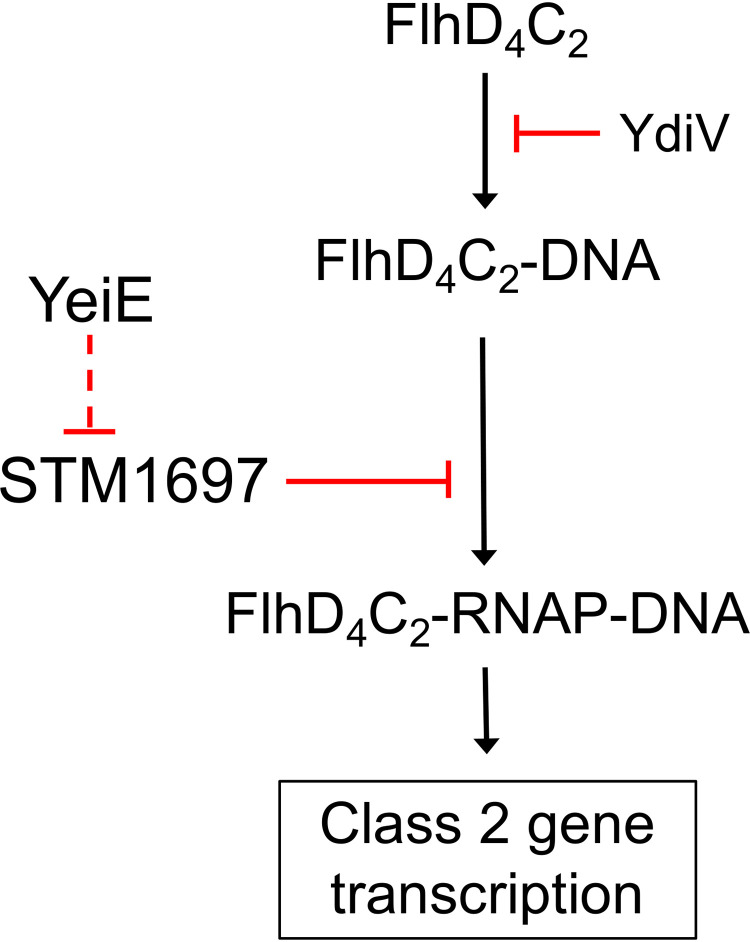
To test whether YeiE influences the expression of STM1697, we determined the relative expression of STM1697 in the WT and the ΔyeiE mutant. We found that the relative expression of STM1697 is upregulated in the ΔyeiE mutant (Fig. 6C). Since YdiV is a related EAL domain-containing protein which also inhibits FlhD4C2 function, we measured the effects of deletion of yeiE on the expression of ydiV. We found no significant alteration from ydiV expression in the ΔyeiE mutant under the conditions tested (Fig. 6C). These data demonstrate that YeiE regulates motility by repressing the expression of the FlhD4C2 functional repressor STM1697 (Fig. 7).
FIG 7.
Proposed mechanism for YeiE regulation of cell motility. YeiE acts as a repressor of STM1697 to promote flagellum-mediated motility. The dotted line indicates an unknown mechanism of repression. RNAP, RNA polymerase.
DISCUSSION
The putative LysR-type transcriptional regulator YeiE is critical for Salmonella Typhimurium flagellum-mediated motility and gut colonization. We demonstrate that flagellar class 2 gene expression is reduced in a ΔyeiE mutant. Furthermore, we link the positive effect of YeiE on cell motility with STM1697, an anti-FlhD4C2 factor, and propose that YeiE is a repressor of STM1697 (Fig. 7).
E. coli and S. Typhimurium share a core genome, with approximately 70% of their genetic material conserved between the two species (29). S. Typhimurium YeiE (STM2201/STM14_2717) has 89% amino acid identity and 92% similarity with its E. coli homolog, suggesting high functional similarity (46). YeiE is both an activator and a repressor of transcription, binds DNA upstream and downstream of the promoter, and can occupy the same sequence as RNA polymerase (30). YeiE has more than 100 predicted binding sites in the E. coli MG1655 genome, with target genes enriched in functional groups including energy production, amino acid and inorganic ion transport and metabolism, and iron transport (30). The E. coli yeiE regulon also includes numerous genes with roles in the regulation of transcription and translation and genes with unknown or poorly characterized functions (30). There were no genes with a direct relationship to flagellar biogenesis in the E. coli YeiE regulon and no motility defects in a ΔyeiE mutant (30, 31). Unlike in E. coli, the S. Typhimurium ΔyeiE mutant has a severe motility defect that we linked to dysregulation of the FlhD4C2 inhibitor STM1697. STM1697 restricts bacterial motility and adds a layer of flagellar regulation that promotes Salmonella intracellular survival and evasion of the host immune system (20). There is no ortholog of STM1697 in the E. coli MG1655 genome (19) or in the genomes of several thousand other E. coli and Shigella strains (data not shown), explaining the different phenotypes observed for ΔyeiE mutants of S. Typhimurium and E. coli. There are multiple different mechanisms by which YeiE may influence STM1697 expression. YeiE may directly repress STM1697 expression by binding to its promoter to inhibit transcription. YeiE may indirectly influence STM1697 expression by regulating the expression of an activator or inhibitor of STM1697 transcription. Further work is needed to establish the full YeiE regulon of S. Typhimurium and to establish how YeiE represses STM1697 expression in S. Typhimurium.
We observed a significant gut colonization defect for the S. Typhimurium ΔyeiE mutant that we linked to defective motility. Since YeiE is likely to have numerous regulatory targets within the S. Typhimurium genome, it is possible that YeiE also regulates other genes that influence S. Typhimurium interactions with the host. For example, yeiE responds to DNA damage in E. coli, suggesting that it may be activated during exposure to oxidative and nitrosative stresses encountered by S. Typhimurium during infection (47). Furthermore, iron import genes are included in the E. coli yeiE regulon, suggesting a potential role for yeiE in iron homeostasis during S. Typhimurium host colonization (30). Although there are numerous potential mechanisms by which yeiE may influence pathogenesis, elimination of the flagellar filament restored virulence to the ΔyeiE mutant, suggesting that the regulation of flagellum-mediated motility is the primary YeiE target facilitating S. Typhimurium colonization during acute enterocolitis. To our knowledge, this is the first demonstration of a role for YeiE in S. Typhimurium pathogenesis.
The related organisms Yersinia enterocolitica and Cronobacter sakazaki each encode homologs of both yeiE and STM1697. The Cronobacter sakazakii yeiE homolog (gpESA_01081) shares 84% amino acid identity with S. Typhimurium YeiE and is required for enterocyte invasion, biofilm formation, neutrophil recruitment, and virulence in neonatal rats (32, 48). The Yersinia enterocolitica yeiE homolog (rscR) shares 75% identity with S. Typhimurium YeiE and limits systemic dissemination of the organism (48, 49). The effects of the C. sakazaki and Y. enterocolitica yeiE homologs (gpESA_01081 and rscR, respectively) on motility have not yet been characterized. We hypothesize that the role of the yeiE homologs in virulence is due, in part, to regulation of the STM1697 homolog in each of these organisms.
Evaluation of the ΔyeiElib suppressor mutant led to the identification of STM1697 as a target for YeiE motility regulation. STM1697 is an anti-FlhD4C2 factor that represses cell motility by binding FlhD, thereby preventing RNA polymerase recruitment to the FlhD4C2-DNA complex (20). The C terminus of STM1697 is critical for its function, as truncation of the 235-amino-acid protein at residue 192 results in a nonfunctional protein and hypermotility (19). Although the crystal structure of the STM1697-FlhD complex suggests that Tyr180 is the residue closest to the C terminus required for stable STM1697-FlhD interaction (20), our data demonstrate an SNV in STM1697 causing a stop codon after amino acid 188, and deletion of STM1697 resulted in the same hypermotile phenotype, confirming prior reports that the C terminus is critical for STM1697 function (see Table S2 in the supplemental material). These data, in combination with prior work (19), suggest that one or more residues located even closer to the C terminus may be required for STM1697 stability or function in vivo.
Salmonella uses flagellum-mediated motility to interact with host cells, driving successful gut colonization; however, it is critical for Salmonella to repress flagellin expression when residing within host cells to evade immune detection (2, 4, 7, 50). Our data suggest that YeiE functions as a regulatory control point to promote flagellar motility by inhibiting the anti-FlhD4C2 factor STM1697. The promotility effect of STM1697 inhibition by YeiE likely facilitates initial gut colonization, whereas STM1697-mediated repression of flagellin aids immune evasion once Salmonella is located intracellularly (20). The regulatory functions of LysR regulators are influenced by small-molecule coinducers that sense environmental conditions (25, 26). Further molecular characterization of YeiE and the identification of its coinducer will help to delineate how this LysR regulator exerts control over cell motility during infection.
We definitively link yeiE with S. Typhimurium motility and demonstrate its requirement for gut colonization using two animal models of enterocolitis. We propose that YeiE serves as a control point for flagellar regulation through inhibition of the anti-FlhD4C2 factor STM1697, although additional work is needed to establish the mechanism by which YeiE influences STM1697 expression. Tight control of flagellar biogenesis is critical to facilitate pathogenesis. Whereas the flagellum-based motility state is needed for gut colonization, elimination of flagellins is critical to evade immune detection. Our work demonstrates that YeiE serves as a key regulatory control point for flagellar biogenesis to facilitate S. Typhimurium enteropathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains are derivatives of Salmonella enterica serotype Typhimurium ATCC 14028s (Table 1). Mutations were moved into a clean genetic background by P22 transduction, and antibiotic cassettes were removed by flp-mediated recombination as previously described (51, 52). Bacteria were grown on Luria-Bertani (LB)-Miller agar or in LB broth at 37°C with agitation (225 rpm) unless otherwise noted. Medium was supplemented with the following antibiotics, as appropriate: nalidixic acid (50 mg/liter), chloramphenicol (20 mg/liter), kanamycin (50 mg/liter), and carbenicillin (100 mg/liter).
Complementing plasmid construction.
Genomic DNA was isolated from the wild-type organism using the GenElute bacterial genomic DNA kit (Sigma-Aldrich). A 1.5-kb PCR product of yeiE, including its native promoter, was generated from genomic DNA by PCR using Q5 polymerase (New England Biolabs), with an annealing temperature of 72°C and an extension time of 40s for 30 cycles. Restriction sites for endonucleases were incorporated into the following primers to facilitate cloning: STM2201BamH1Fwd, 5′-GTCGGATCCTGCCTGTCCAGACCAAAGA-3′, and STM2201Kpn1Rev, 5′-GTCGGTACCTGCGCGGTTATAAGAGACCT-3′. The expected size of the PCR product was confirmed by agarose gel electrophoresis. The PCR product was digested with restriction endonucleases BamHI and KpnI (New England Biolabs) and purified with the QIAquick PCR purification kit (Qiagen). The insert was cloned into the pWSK29 vector, sequentially digested with BamHI and KpnI (53). Ligation was performed overnight at 14°C with T4 DNA ligase (New England Biolabs). The resulting construct was transformed into DH5α Escherichia coli by heat shock. Transformants were obtained by selection on LB agar with carbenicillin and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml). Plasmids were isolated (QIAprep miniprep kit; Qiagen), and the correct insert size was confirmed by agarose gel electrophoresis of digested plasmids. The insert sequence was confirmed by Sanger sequencing (Eton Bioscience). The complementing plasmid (CP) was transformed into restriction-deficient modification-positive S. Typhimurium LB5000 by electroporation, and transformants were isolated by selection on LB agar with carbenicillin (54). The complementing plasmid and empty vector were isolated and then transformed into the indicated mutants by electroporation. The resulting strains were purified by streaking them twice for single colonies on LB with carbenicillin and stored in glycerol stocks at −80°C.
Growth curves.
Overnight cultures were diluted 1:100 into 50 ml of LB or M9 minimal medium and grown at 37°C with agitation (225 rpm) for 24 h. M9 minimal medium (48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 0.1 mM CaCl2, and 2 mM MgSO4) was supplemented with 0.2% (wt/vol) dextrose as a carbon source (55). Samples were taken hourly for 6 h and once at 24 h, serially diluted, and plated for enumeration of CFU per milliliter.
Bacterial motility assays.
Swimming assays were performed on semisolid agar as previously described (56). Swimming motility was assayed on plates containing 0.3% Difco Bacto agar (LB-Miller base, 25 g/liter). Overnight cultures were grown at 37°C with agitation, and the cell concentration was normalized by optical density. Bacterial strains were spotted (3 μl) onto swimming plates and incubated at 37°C. The widest diameter of each colony was measured in two intersecting planes after 4 h of incubation. Images of swimming plates were obtained on ChemiDoc MP (Bio-Rad) after 5 h of incubation. Each assay was performed in triplicate on three independent occasions.
Calf infections.
Calf infections were approved by the North Carolina State University Institutional Animal Care and Use Committee (protocol 15-047-B). Calves were obtained from the university teaching herd and transferred to individual AALAC-approved housing within 8 h following birth. Calves were administered a commercial colostrum replacer (AgriLabs Colostrx CR) by an esophageal feeder upon arrival and 2 h following initial administration. Calves were fed milk replacer twice daily and provided hay and water ad libitum. At 1 day of age, adequate passive transfer of immunity was estimated by measuring serum total solids with a refractometer. Fecal cultures were performed at least twice weekly using selective media to ensure that calves were not shedding Salmonella prior to experimental infection (57).
In preparation for the ligated ileal loop surgery, bacteria were grown overnight at 37°C with agitation in LB broth. Overnight cultures were subcultured 1:100 into LB broth and incubated for 3 to 4 h at 37°C with agitation. Bacteria were washed twice in sterile LB broth, and the mutant and WT were mixed in a 1:1 ratio based on optical density (600 nm). Loops were inoculated with approximately 109 CFU of the mixture in 3 ml LB broth.
Ligated ileal loop surgery was performed on 5 bull calves (3 Jersey and 2 Holstein) aged 3 to 6 weeks, as previously described (57). At 12 h postinfection, the luminal contents, mucus overlying the epithelium, and epithelial tissue were harvested, collected in phosphate-buffered saline (PBS), homogenized, serially diluted, and plated to determine numbers of CFU. The competitive index (CI) between the WT and mutant was determined as the ratio of WT to mutant bacteria after infection normalized to the ratio in the inoculum.
Mouse infections.
Mouse infections were approved by the University of Wisconsin—Madison Institutional Animal Care and Use Committee (protocol no. V006255). The acute murine colitis model was used as previously described, with 10- to 12-week-old female C57BL/6J mice obtained from Jackson Laboratories (strain 000664) (37). Mice were administered 20 mg streptomycin in sterile water by oral gavage 24 h prior to infection. Overnight bacterial cultures were washed in PBS, and mice were infected with approximately 108 CFU of a 1:1 mixture of the two competing strains by oral gavage. Mice were euthanized 72 h postinfection. Organs were harvested, homogenized, serially diluted, and plated for enumeration of CFU. The competitive index was determined as for calf infections.
Genome sequencing and analysis.
Genomic DNA from the amotile (JE973) and hypermotile (JE1681) ΔyeiE mutants was isolated (GenElute bacterial genomic DNA kit; Sigma-Aldrich) and submitted to the North Carolina State University Genomic Sciences Laboratory. Genomic DNA (gDNA) quality was analyzed using TapeStation (Agilent), and library preparation and whole-genome shotgun sequencing was performed using a MiSeq platform (PE300; Illumina). Raw sequencing data were provided as demultiplexed .fastq files. Variants were found using the CLC Genomics Workbench (Qiagen) resequencing analysis module. Reads were first mapped to the published ATCC 14028s genome (42) (NCBI accession no. NC_016856.1), followed by local realignment. Variants were then detected using the fixed-ploidy variant detection tool. Variants that were supported by reads in both strands with more than 50% frequency were further investigated.
Gene expression analyses.
Overnight cultures of the WT and ΔyeiE mutant were diluted 1:100 into LB broth and grown at 37°C with agitation for 3.5 h. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA quantity and integrity were determined using a Qubit 4 fluorometer (Invitrogen), and samples with an RNA IQ value of >7.5 (Qubit RNA IQ assay; Invitrogen) were considered of good quality and were used for downstream applications. Removal of contaminating DNA was performed using TURBO DNase (Invitrogen) according to the manufacturer’s instructions.
Reverse transcription (RT) of RNA to cDNA was performed by random-hexamer-dependent amplification using TaqMan reverse transcription reagents according to the manufacturer’s instructions (Invitrogen). Real-time PCR was performed using probe-based 5′ nuclease assays (IDT PrimeTime quantitative PCR [qPCR]). Primer and probe sets (see Table S3 in the supplemental material) were designed using the IDT PrimerQuest tool (https://www.idtdna.com/SciTools) and incorporated 6-carboxyfluorescein (FAM)/ZENTM (IDT trademarked dark quencher)/Iowa black fluorescent quencher (IBFQ) doubly quenched probes (Integrated DNA Technologies). Cycling parameters were an initial 95°C polymerase activation step for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, performed on a StepOnePlus real-time PCR system (Applied Biosystems). Amplification efficiency was determined to be 90 to 100% for all primer/probe sets (Table S4). All qPCR products were confirmed to have a single product of the expected size by agarose gel electrophoresis. All real-time PCR assays were performed in three technical repeats using cDNA from at least 3 biological replicates. The change in threshold cycle (ΔCt) for each gene of interest was determined for each strain, using rpoD as the reference gene for normalization (58). The relative expression of each gene of interest was determined by comparing the ΔCt of the mutant to that of the WT using the comparative threshold (ΔΔCt) method. All quantitative RT-PCR data are available in Table S4.
Primer and probe sequences for qRT-PCR. Download Table S3, XLSX file, 0.01 MB (10.6KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
qRT-PCR primer efficiencies and raw Ct data. Download Table S4, XLSX file, 0.06 MB (66.5KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data analysis.
Statistical significance was determined by a two-tailed Student t test, with significance set at a P of <0.05. Analyses were performed using GraphPad Prism version 8.0.
Data availability.
The raw DNA sequences were deposited in GenBank (BioProject accession no. PRJNA704982).
ACKNOWLEDGMENTS
We thank M. Mitsu Suyemoto, Craig Radi, Steffen Porwollik, and WeiPing Chu for assistance with experiments associated with this work.
This work was supported by startup funds from the University of Wisconsin—Madison (to J.R.E.). J.R.E. was supported, in part, by USDA-NIFA grant 2018-67017-27632 and NIH grant K08AI108794. T.L.W. was supported by grant 5T32OD01113. M.M. was supported, in part, by U.S. Department of Agriculture, National Institute of Food and Agriculture grants 2015-67017-23360 and 2017-67015-26085, NIFA Hatch grant CA-d-PLS-2327-H, NIFA-BARD award 2017-67017-26180, and NIH grant AI139557.
Footnotes
Citation Westerman TL, McClelland M, Elfenbein JR. 2021. YeiE regulates motility and gut colonization in Salmonella enterica serotype Typhimurium. mBio 12:e03680-20. https://doi.org/10.1128/mBio.03680-20.
Contributor Information
J. R. Elfenbein, Email: jelfenbein@wisc.edu.
Cagla Tukel, Temple University.
N. Luisa Hiller, Carnegie Mellon University
REFERENCES
- 1.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt CK, Ikeda JS, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, O'Brien AD. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect Immun 69:5619–5625. doi: 10.1128/iai.69.9.5619-5625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dibb-Fuller MP, Allen-Vercoe E, Thorns CJ, Woodward MJ. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023–1031. doi: 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Bäumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect Immun 71:1–12. doi: 10.1128/iai.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 6.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology (Reading) 156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutsukake K, Ohya Y, Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol 172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagihara S, Iyoda S, Ohnishi K, Iino T, Kutsukake K. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet Syst 74:105–111. doi: 10.1266/ggs.74.105. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene FliA encodes an alternative sigma factor specific for flagellar operons in Salmonella-Typhimurium. Mol Gen Genet 221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 12.Kutsukake K, Ikebe T, Yamamoto S. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet Syst 74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- 13.Yokoseki T, Kutsukake K, Ohnishi K, Lino T. 1995. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]
- 14.Ohnishi K, Kutsukake K, Suzuki H, Lino T. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol Microbiol 6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Matsumura P. 1995. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene 164:81–84. doi: 10.1016/0378-1119(95)00480-t. [DOI] [PubMed] [Google Scholar]
- 16.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. doi: 10.1128/JB.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, Anwar N, Chuah ML, Lunsdorf H, Frank R, Rhen M, Liang ZX, Lindqvist Y, Romling U. 2013. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella typhimurium. Mol Microbiol 90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Yue Y, Yuan Z, Zhang F, Li P, Song N, Lin W, Liu Y, Yang Y, Li Z, Gu L. 2017. Salmonella STM1697 coordinates flagella biogenesis and virulence by restricting flagellar master protein FlhD4C2 from recruiting RNA polymerase. Nucleic Acids Res 45:9976–9989. doi: 10.1093/nar/gkx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang H-W, Gu L. 2012. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res 40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD(4)C(2) factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol 83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westerman TL, Sheats MK, Elfenbein JR. 5 April 2021. Sulfate import in Salmonella Typhimurium impacts bacterial aggregation and the respiratory burst in human neutrophils. Infect Immun doi: 10.1128/IAI.00701-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol 47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 26.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (Reading) 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 27.Pareja E, Pareja-Tobes P, Manrique M, Pareja-Tobes E, Bonal J, Tobes R. 2006. ExtraTrain: a database of extragenic regions and transcriptional information in prokaryotic organisms. BMC Microbiol 6:29. doi: 10.1186/1471-2180-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahiri A, Das P, Chakravortty D. 2009. Salmonella Typhimurium: insight into the multi-faceted role of the LysR-type transcriptional regulators in Salmonella. Int J Biochem Cell Biol 41:2129–2133. doi: 10.1016/j.biocel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 29.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y, Yurkovich JT, Seo SW, Kabimoldayev I, Dräger A, Chen K, Sastry AV, Fang X, Mih N, Yang L, Eichner J, Cho B-K, Kim D, Palsson BO. 2018. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res 46:10682–10696. doi: 10.1093/nar/gky752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol 189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi Y, Kim KP, Kim K, Choi J, Shin H, Kang DH, Ryu S. 2012. Possible roles of LysR-type transcriptional regulator (LTTR) homolog as a global regulator in Cronobacter sakazakii ATCC 29544. Int J Med Microbiol 302:270–275. doi: 10.1016/j.ijmm.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Boonma S, Romsang A, Duang-Nkern J, Atichartpongkul S, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S. 2017. The FinR-regulated essential gene fprA, encoding ferredoxin NADP+ reductase: roles in superoxide-mediated stress protection and virulence of Pseudomonas aeruginosa. PLoS One 12:e0172071. doi: 10.1371/journal.pone.0172071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pei B, Wang Y, Katzianer DS, Wang H, Wu H, Zhong Z, Zhu J. 2013. Role of a TehA homolog in Vibrio cholerae C6706 antibiotic resistance and intestinal colonization. Can J Microbiol 59:136–139. doi: 10.1139/cjm-2012-0673. [DOI] [PubMed] [Google Scholar]
- 35.Westerman TL, Bogomolnaya L, Andrews-Polymenis HL, Sheats MK, Elfenbein JR. 2018. The Salmonella type-3 secretion system-1 and flagellar motility influence the neutrophil respiratory burst. PLoS One 13:e0203698. doi: 10.1371/journal.pone.0203698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos RL, Zhang S, Tsolis RM, Bäumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet Pathol 39:200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- 37.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chubiz JEC, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol 192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol 185:5096–5108. doi: 10.1128/jb.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer HM, Kühne C, Deditius JA, Hughes KT, Erhardt M. 2014. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol 196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang H-J, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura H, Saiki K, Mogi T, Anraku Y. 1997. Assignment and functional roles of the cyoABCDE gene products required for the Escherichia coli bo-type quinol oxidase. J Biochem 122:415–421. doi: 10.1093/oxfordjournals.jbchem.a021769. [DOI] [PubMed] [Google Scholar]
- 44.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A 74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangan MW, Lucchini S, T OC, Fitzgerald S, Hinton JCD, Dorman CJ. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology (Reading) 157:1075–1087. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 46.Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 47.Hong J, Ahn JM, Kim BC, Gu MB. 2009. Construction of a functional network for common DNA damage responses in Escherichia coli. Genomics 93:514–524. doi: 10.1016/j.ygeno.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson KM, Young GM, Miller VL. 2001. Identification of a locus involved in systemic dissemination of Yersinia enterocolitica. Infect Immun 69:6201–6208. doi: 10.1128/IAI.69.10.6201-6208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 7:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sternberg NL, Maurer R. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol 204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 53.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 54.Bullas LR, Ryu JI. 1983. Salmonella typhimurium LT2 strains which are r– m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol 156:471–474. doi: 10.1128/JB.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 56.Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, Andrews-Polymenis HL. 2014. Identification of novel factors involved in modulating motility of Salmonella enterica serotype Typhimurium. PLoS One 9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elfenbein JR, Endicott-Yazdani T, Porwollik S, Bogomolnaya LM, Cheng P, Guo J, Zheng Y, Yang HJ, Talamantes M, Shields C, Maple A, Ragoza Y, Deatley K, Tatsch T, Cui P, Andrews KD, McClelland M, Lawhon SD, Andrews-Polymenis H. 2013. Novel determinants of intestinal colonization of Salmonella enterica serotype Typhimurium identified in bovine enteric infection. Infect Immun 81:4311–4320. doi: 10.1128/IAI.00874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. 2008. ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol Microbiol 70:1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome sequencing variants. Variants are differences between the ΔyeiE mutants and the ATCC 14028s genome (GenBank accession no. CP001363, chromosome; accession no. CP001362, plasmid). Bold variants are those with reads in both strands (forward/reverse balance of >0.05). Variants in red are those that were identified in both the amotile and suppressor ΔyeiE mutants. Download Table S1, XLSX file, 0.01 MB (12.8KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of single-nucleotide variants in the ΔyeiE and ΔyeiElib mutants identified by whole-genome sequencing. The impact of SNV on the target amino acid sequence of 6 genes was established. *, variants were supported by reads in both strands with over 50% frequency. Download Table S2, XLSX file, 0.01 MB (10.4KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primer and probe sequences for qRT-PCR. Download Table S3, XLSX file, 0.01 MB (10.6KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
qRT-PCR primer efficiencies and raw Ct data. Download Table S4, XLSX file, 0.06 MB (66.5KB, xlsx) .
Copyright © 2021 Westerman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The raw DNA sequences were deposited in GenBank (BioProject accession no. PRJNA704982).